Abstract
Abstract
Phenotypic plasticity describes the ability of cancer cells to undergo dynamic, nongenetic cell state changes that amplify cancer heterogeneity to promote metastasis and therapy evasion. Thus, cancer cells occupy a continuous spectrum of phenotypic states connected by trajectories defining dynamic transitions upon a cancer cell state landscape. With technologies proliferating to systematically record molecular mechanisms at single-cell resolution, we illuminate manifold learning techniques as emerging computational tools to effectively model cell state dynamics in a way that mimics our understanding of the cell state landscape. We anticipate that “state-gating” therapies targeting phenotypic plasticity will limit cancer heterogeneity, metastasis, and therapy resistance.
Significance:
Nongenetic mechanisms underlying phenotypic plasticity have emerged as significant drivers of tumor heterogeneity, metastasis, and therapy resistance. Herein, we discuss new experimental and computational techniques to define phenotypic plasticity as a scaffold to guide accelerated progress in uncovering new vulnerabilities for therapeutic exploitation.
INTRODUCTION: PHENOTYPIC PLASTICITY AS A SOURCE OF HETEROGENEITY
Any given tumor is made up of a unique collection of cancer cells that exist within a spectrum of diverse genotypic and phenotypic cell states. Understanding the scale and functional implications of such heterogeneity is a significant scientific challenge because this diversity is clinically associated with aggressive disease, resistance to conventional chemotherapies, and poor overall survival (1–6). Yet this implies that targeting specific cell populations within a tumor, or preventing the emergence of particularly aggressive subpopulations, could lead to enhanced therapies and improved survival outcomes for patients. To reach this goal, strategies to identify and therapeutically target key intratumoral cell populations are vital.
It is long established that genetic diversity is a critical mediator of intratumoral heterogeneity (5). Studies in glioma demonstrate that intratumoral heterogeneity may consist of anywhere between 0 and 8,000 coding mutations (7) that provide tumors with crucial survival advantages like permitting survival in evolving microenvironments or in response to therapeutic insults (5, 7–10). In some instances, genetic heterogeneity is already present within the primary tumor, such that chemotherapy favors the emergence of otherwise minor or dormant clones (11). In other cases, disease progression is driven by the therapy-induced emergence of clones with de novo mutations that are not present in the primary tumor (12). Given the spontaneous, random, and stochastic nature of such mutations, designing therapies and clinical trials to prevent their initial emergence or selection remains challenging (5).
Importantly, there is now substantial evidence that nongenetic mechanisms can also significantly increase intratumoral heterogeneity and disease progression (4, 13, 14). In this setting, distinct cancer cell states are created by a process termed “phenotypic plasticity,” in which epigenetic, transcriptional, and/or translational mechanisms—rather than mutations—drive phenotypic cell state switching in a manner akin to normal development, wound healing, and organ regeneration (4, 13, 15–17). In fact, phenotypic plasticity is now recognized as a hallmark of cancer (18).
In contrast to genetically driven heterogeneity, phenotypic plasticity is dynamic, reversible, and responsive to regulation (19–24). Implicit here is the notion that phenotypic plasticity reflects molecular program switching under the control of specific pathways that may be therapeutically targetable. Understanding the molecular mechanisms regulating phenotypic plasticity thus constitutes the first step toward establishing what we term “state-gating strategies” for cancer therapy—controlling entry into or exit from specific phenotypic cell states. Given the failure of cytotoxic therapies to treat advanced-stage disease, mastering phenotypic plasticity to prevent cells from entering into highly aggressive or resistant cell states is a promising strategy to curtail both metastasis and therapy escape. Such therapeutic advances may prove clinically transformative (25, 26) but demand a new systematic understanding of cancer cell states, their plastic dynamics, and molecular regulation.
LINKING SINGLE-CELL TECHNOLOGIES WITH MANIFOLD LEARNING TO RESOLVE THE CANCER CELL LANDSCAPE
In this review, we consider the biology of nongenetic heterogeneity from the perspective of a cancer cell state landscape, whereupon cancer cells occupy a continuous spectrum of phenotypic states connected by trajectories defining dynamic cell state transitions, herein termed “plasticity.” We illustrate how single-cell “omics” measurement technologies now synergize with manifold learning–based analytical methods to illuminate the topology of this cancer cell state landscape. This reflects the recent proliferation of single-cell technologies that are providing unprecedented capacity to systematically characterize molecular mechanisms governing phenotypic plasticity at the cellular scale (27–30). Leveraging this revolutionary data, manifold learning techniques are also emerging to effectively model cell state dynamics in a way that mimics our understanding of the cell state landscape, being both globally complex (nonlinear) and locally continuous. Building on this tripartite synthesis of (i) the cancer cell state landscape paradigm, (ii) single-cell omics measurement methods, and (iii) manifold learning analysis tools, we further consider how this systematic framework will inform biological advances leading to novel therapeutics. Indeed, we believe this synthesis will promote not only new individual therapies but ultimately a new class of therapies that strategically target cancer cell plasticity itself to restrict cancer cell heterogeneity and adaptability.
CELLULAR PLASTICITY AS A DRIVER OF CANCER PROGRESSION
Although there is mounting evidence that phenotypic plasticity drives disease progression in multiple cancer types (refs. 14, 16, 20, 27–30; for a recent review on this topic, see ref. 4), it remains a complex process to dissect. The temporal dynamics governing cell state changes can encompass trajectories spanning months to years, in line with disease progression or therapeutic treatment and response. Yet plasticity may also result from rapid changes in transcription, protein expression, and even localization within days, hours, or minutes. Cancer cell plasticity is also modulated by spatially resolved cues spanning organ, tissue, cellular, and subcellular scales. Thus, resolving both temporal and spatial dimensions is critical to understanding cancer cell plasticity and its role in both tumor progression and therapy resistance.
Much of our understanding of the molecular programs governing cancer cell plasticity derives from early observations revealing the co-option of developmental cell plasticity programs by cancer cells, such as transitions between epithelial and mesenchymal cell states. This includes epithelial-to-mesenchymal transition (EMT; refs. 31, 32) and the reverse process, mesenchymal-to-epithelial transition (MET; refs. 33, 34). In this paradigm, cancer cells defined as epithelial-like are considered less tumorigenic and generally lacking in cancer stem cell characteristics. Upon undergoing EMT to acquire a full mesenchymal or even hybrid epithelial/mesenchymal state, they are thought to gain stem cell characteristics that permit promotion of metastasis and chemotherapy resistance (35–37). These early studies have also revealed important information about the significance of ongoing cell plasticity dynamics. For example, EMT at the primary tumor site increases metastatic potential; yet among metastatic cells, the ability to also undergo the reverse MET process, even if only partially, better enables secondary tissue colonization and metastasis growth (38–43). Thus, metastatic efficiency is greatest when cancer cells retain the plasticity to use epithelial or mesenchymal attributes best suited at distinct stages of the metastatic cascade.
The epigenetic landscape of a cancer cell is an important determinant of its intrinsic propensity to undergo EMT or MET. For example, a bivalent chromatin configuration at the promoter of the EMT transcription factor ZEB1 predisposed basal breast cancer cells lacking stemness features to spontaneous ZEB1 activation and subsequent entrance into a cancer stem cell state. In contrast, the equivalent cells isolated from luminal breast cancer maintain repressed chromatin at the ZEB1 promoter and do not readily change cell state (19). Bivalent chromatin configuration also underlies phenotypic plasticity within cancer stem cell populations themselves, providing cancer stem cells with additional adaptability (44). Epigenetic modulation plays an important role not only in a cell's predisposition to invoke phenotypic plasticity but in the execution of the process. The transcription factors that execute EMT—for example, SNAI1—bind to the promoter of the CDH1 gene encoding E-cadherin, a gatekeeper of the epithelial cell state. This binding recruits components of the polycomb repressor complex and histone deacetylases to modify chromatin and mediate the silencing of CDH1 expression (45, 46). Accordingly, chromatin remodelers, writers, and readers provide important regulatory control over phenotypic plasticity and are the subject of therapeutic investigation (47, 48).
Notably, there are now several examples of phenotypic plasticity programs that drive cancer progression outside of those associated with developmental differentiation. Metabolic plasticity enables cancer cells to alter their utilization of metabolic programs to sustain growth and survival. These changes may occur in response to hyperproliferative cell states, changes in microenvironmental nutrient availability, or the requirement for de novo rate-limiting macromolecule synthesis (49). The capacity to change metabolic dependencies not only favors survival but may also favor metastatic outgrowth in distinct organs—for example, elevated fatty acid synthesis (50) or de novo serine synthesis (51) is enhanced in breast cancer brain metastases compared with primary tumors, whereas the pyruvate carboxylase–dependent resupply of metabolites is enhanced in breast cancer lung metastasis compared with primary tumors (52, 53). Beyond the impact of those changes on the cell's metabolism, epigenome regulators are also highly sensitive to changes in intracellular metabolism. Indeed, a cancer cell's epigenetic and transcriptional landscape can be altered, for example, by iron endocytosis (54), proline metabolism (55), or the metabolite 2-hydroxyglutarate (56).
Entrance and exit from slow-cycling, quiescent, dormant, or even senescent cell states are phenotypic plasticity programs that are particularly consequential for tumor relapse or recurrence (57–59). Cells that escape these latent states may reenter the cell cycle via activation of proteins controlling cycling (e.g., cyclin-dependent kinases; ref. 59), through upregulation of stemness markers (60), or by changing extracellular matrix composition in the dormant niche (61). Returning to the importance of metabolic plasticity in cell state determination, reactivation of cell cycling among therapy-resistant persister cells has been linked to the upregulation of fatty acid metabolism (62). Interestingly, survival in a dormant cell state has been linked to the upregulation of immune genes that provide a “cloak” to cancer cells to avoid immune detection. Similarly, downregulation of that immune program enables dormant cells to reenter the cell cycle (63).
Immune evasion is not only a critical component of tumor dormancy, it is also fundamental to robust tumor growth. Indeed, this biology underpins the breakthrough success of immunotherapy (64). Like most cancer therapeutics, however, intrinsic or acquired resistance to immunotherapy ensues, with evidence that phenotypic plasticity plays a significant role (65). Alongside the ability of EMT to drive cancer cells into more aggressive cell states, it also promotes immune evasion by decreasing levels of MHC-I and promoting high levels of PD-L1 on cancer cells (66, 67). These changes are in line with the direct impact of EMT-inducing cytokines, for example, TGFb, that drive EMT in cancer cells while suppressing innate and adaptive immune responses in the tumor microenvironment (67, 68). In fact, clear differences in the immune landscape have been noted in epithelial versus mesenchymal tumors, including a change from CD8+ T cells and M1 antitumor macrophages to M2 protumor macrophages, increased CD8+ T-cell exhaustion, and increased regulatory T-cell recruitment (67). These studies suggest that therapies preventing EMT, or conversely, enhancing MET, may be effective in sensitizing tumors to immunotherapy.
Perhaps the most alarming manifestation of phenotypic plasticity is in facilitating therapy evasion and the emergence of therapy-resistant disease (69, 70). A prime example of this is evidence demonstrating that a partial EMT is associated with entrance into a chemotherapy-resistant state (71). Similarly, radiotherapy can promote phenotypic plasticity in the stroma of distant metastatic organs, establishing a positive feedback loop with metastatic cancer cells, which in turn undergo a phenotypic transition that enhances their capacity to grow and evade therapy (72). Moreover, multiple studies have now proven that chemotherapies themselves—spanning anthracyclines, topoisomerase inhibitors, platinum-based therapies, and taxanes—all induce components of the EMT program and thereby chemotherapy resistance (73). Similarly, the evidence of partial MET at metastatic sites also leads to chemotherapy-resistant cell states (74). These studies show that chemotherapies affect multiple cell states and stages of metastasis to actively induce therapy-resistant phenotypes, as opposed to only creating resistance through a gradual selection of preexisting resistant clones. This active induction of resistant cancer cell states through nongenetic plasticity programs suggests a targetable vulnerability, such that the elucidation and modulation of these chemotherapy-induced plasticity programs will permit novel therapeutic strategies (25).
Together, these studies highlight the complex interplay of plasticity programs spanning developmental, metabolic, immune evasion, epigenetic, and drug resistance systems that cooperate to optimize cancer cell growth and survival (75–77). Beyond these insights, we are only on the cusp of understanding how underlying genetic mutations contribute additional dimensions to the control of phenotypic plasticity (78–81).
TEMPORAL AND SPATIAL REGULATION OF PHENOTYPIC PLASTICITY
Given the sensitivity of phenotypic plasticity programs to environmental cues, it is interesting to consider how short or transient exposure can lead to long-term, stable phenotypic changes. For example, if a cancer cell is exposed to an EMT-inducing signal at the primary tumor site, how is the phenotype maintained once the cell transits into the circulation or to a distant metastatic site? Recent insights into miRNA/EMT transcription factor regulatory loops, for example, the miR200–ZEB1 feedback, demonstrate how negative feedback loops create a sensitive yet noise-resistant bistable response to short-term cytokine exposure (82). In this example, a 5-minute exposure to the EMT-inducing cytokine TGFb reinforces ZEB1 signaling to stabilize an EMT that persists for days and is capable of increasing metastatic potential. The bistability revealed here is emblematic of a hysteretic process in which differences in the histories of individual cells dictate divergent responses to equivalent stimuli. Such hysteresis raises further questions as to whether particular state transitions are (i) bidirectional, in which cells in state A can transition to state B and cells in state B can transition to state A; (ii) reversible, in which a single cell in state A can transition to state B and then back to state A; and (iii) asymmetrical, in which cells transition from state A to state B via a different trajectory versus cells transitioning from state B to state A, as in the hysteresis example above and depicted in the epithelial-to-hybrid epithelial/mesenchymal state and hybrid epithelial/mesenchymal-to-epithelial example in Fig. 1A. Ultimately, understanding how these plasticity pathways are regulated, together with defining permissive epigenetic states, is vital for a systematic understanding of the temporal dynamics giving rise to cancer cell heterogeneity and to the adaptive capacity of individual cancer cells.
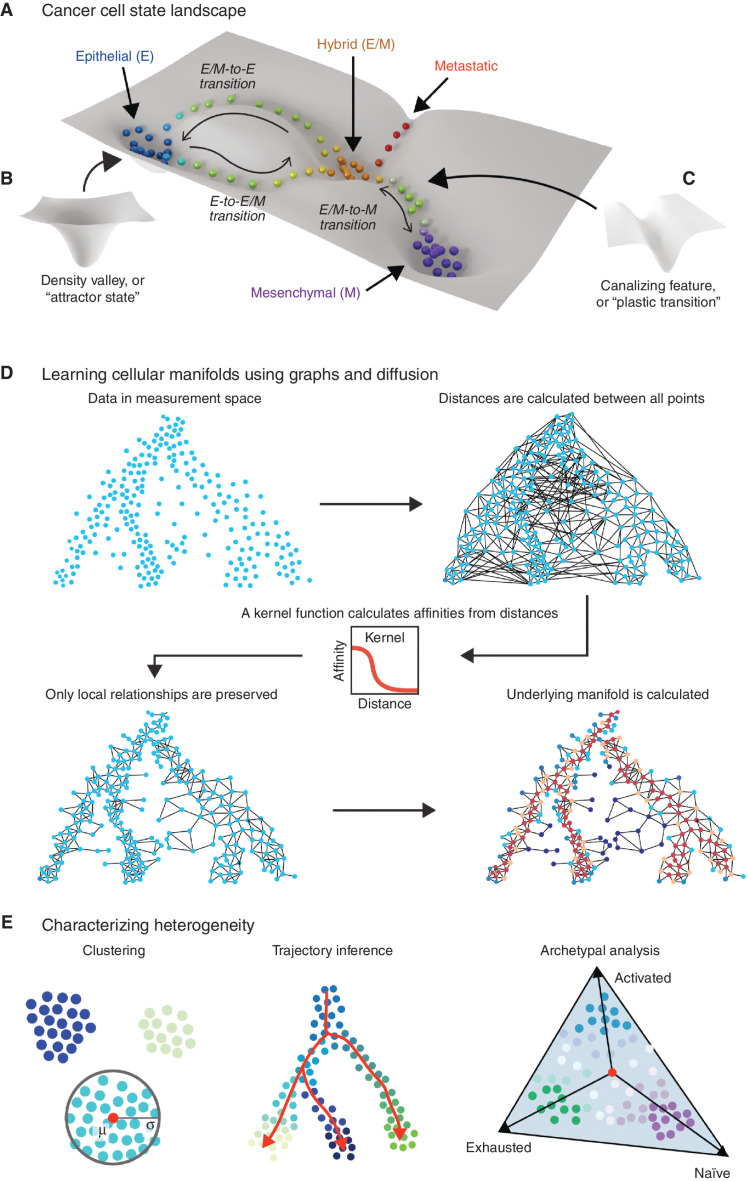
Figure 1.
Estimating the topology of the cancer cell state landscape via manifold modeling. Cancer cell populations reside in a high-dimensional state space that can be conceived as a landscape (A) in which highly populated “attractor states” constitute valleys (B) and the trajectories of “plastic transitions” between these states follow canalizing features such as channels (C). The height or depth of topologies on this landscape reflect the relative favorability of the corresponding cell state in thermodynamic or informational terms. D, The topology of cell state landscapes can be modeled using graph representations that approximate nonlinear but locally continuous “cellular manifolds.” Learning a graph from high-dimensional data such as single-cell RNA sequencing involves calculating global distances and then connecting adjacent neighborhoods of cells using a kernel function. Methods like diffusion, modeling random walks on the connected graph, can be used to estimate recurring trajectories within the data, reflecting plastic cell state transitions. E, Given the estimation of a manifold representing the cell state landscape, tasks like clustering (left), trajectory inference (center), and archetypal analysis (right) of phenotype composition can be performed to extend biological inferences. E, epithelial; E/M, epithelial/mesenchymal; M, mesenchymal.
Of course, phenotypic plasticity does not arise purely through cell-autonomous decision-making or stochasticity. Rather, it emerges as part of the complex, spatially distributed milieu comprising each evolving tumor site (83). Just as the recent history of a given cell may be encoded in hysteretic signaling motifs of the kinds discussed above, or in gene expression or epigenetic patterns, each cell's history is also manifest in its local spatial context. This context incorporates factors such as cell–cell interactions (homotypic; with other cancer cells and/or heterotypic; with fibroblasts, immune cells, vasculature, etc.); cell–extracellular matrix (ECM) interactions; extracellular soluble factors (autocrine/paracrine signaling ligands or cytokines); and chemical properties such as pH and the levels of O2, CO2, or reactive oxygen species. Importantly, the reciprocal relationship between each cell and its spatial context not only reflects the cell's history but also shapes its future, guiding decisions between processes such as proliferation, programmed death, motility, or particular cell state transitions. As a consequence, a robust understanding of cancer cell state dynamics—and how these give rise to population heterogeneity—cannot be disentangled from a parallel understanding of intratumoral spatial organization, since in the evolution of tumors, temporal and spatial determinants are fundamentally interwoven (84). This basic concept underpins established yet more specific questions about the existence and nature of so-called cancer stem cell niches, the composition and properties of tumor invasive fronts, and the interfaces between tumors and macrostructures such as vasculature or organ fascia. The mapping of phenotypic cell states via single-cell spatial transcriptomics (85, 86) and proteomic analyses (87) has enormous potential to resolve these questions.
RESOLVING CANCER CELL STATES AND PLASTICITY TRAJECTORIES: MAPPING THE CANCER CELL STATE LANDSCAPE
Given the potential complexity and interplay of phenotypic plasticity programs, how do we begin to unravel, resolve, and ultimately understand how those programs contribute to the creation and evolution of a heterogeneous tumor? We can begin by thinking of cancer cells as occupying a cancer cell state “landscape” (Fig. 1A–C). On this landscape, cancer cells reside within a phenotypic continuum. We hypothesize that there may be multiple stable states connected by transitional trajectories permitting traversal of the cell state continuum and that some states and transitions will be favored at different sites or stages of cancer progression. Accordingly, we expect a final understanding of cancer cell states to reveal a more complex and multifaceted landscape—much like the landscape underpinning cellular differentiation envisaged by Conrad Waddington, as revisited by Sui Huang and colleagues (88), and like that proposed as an explanation of nongenetic cancer driver dynamics (89).
In the paradigm of the cancer cell state landscape, high-stability cell states constitute “valleys,” or “attractor states,” into whose orbit adjacent cells tend to fall (Fig. 1B). Conversely, unstable or unfavorable cell states constitute “mountain ranges” that cells cannot easily traverse. These valleys and mountain ranges, thus, partition the cellular landscape into accessible and inaccessible state spaces that collectively give rise to cancer cell heterogeneity. Plastic phenotypic transitions occur between cell attractor states via “canalising” features, for example, channels linking valleys (Fig. 1C). Crucially, it is becoming clear that specific molecular mechanisms underpin the plastic state transitions that constitute these canalizing features, making them prime targets for therapeutic intervention to control tumor heterogeneity.
A significant but often overlooked aspect of cellular dynamics within this landscape paradigm is that the landscape topology “shapes probabilistic outcomes” but is not wholly deterministic. Specifically, although the dynamics of an individual cell are heavily influenced by the local landscape topology (e.g., uphill or downhill gradients), the stochastic nature of cellular information processing nonetheless permits some movements counter to these gradients, meaning that cells can occasionally climb “uphill.” This accounts for the ability of cells to traverse landscapes that have multiple local minima without becoming permanently trapped in these. This “stochastic jittering” provides a degree of flexibility that maintains cell heterogeneity, while nonetheless being guided by the evolved topology or “logic” of the cell state landscape. This mirrors the classic concept of the “explore versus exploit” trade-off considered in evolutionary and complexity theories, in which exploration corresponds to stochastic jittering and exploitation corresponds to a strict following of topological cues (90).
Intriguingly, the level of stochastic jittering—essentially, noise in cellular information processing—is evidently a meta-property that is subject to evolutionary tuning. At the genomic level, mechanisms exist to rapidly raise this noise level in response to stress—for example, stress-induced chromosomal instability increases mutation rates, thereby promoting greater heterogeneity and thus improved population survivability in the presence of stressors (91). Understanding how this meta-property of “noise levels” may be embodied and tuned in the context of nongenetic cellular plasticity could provide an alternative lens through which to interpret and potentially modulate cell state landscapes and the cancer cell dynamics emerging upon them. For example, distinct epigenetic states, or epigenetic priming, such as, a bivalent chromatin configuration at critical gene mediators of phenotypic plasticity, increase the likelihood of normal and cancer cells changing states (19, 92).
Overall, defining tumor-specific cancer cell state landscapes is critical to understanding the diversity of cell states that exist, the transitional trajectories that link significant states, and how these topologies contribute to disease progression and site-specific metastasis. Using such knowledge, we may define the molecular programs controlling plasticity, thereby informing the design of novel therapeutics to block deleterious trajectories or activate beneficial ones. By thus effectively remodeling cancer landscape topologies, we envision a new treatment strategy to prevent cancer progression and therapy resistance. Yet to enact this vision, sophisticated new combinations of experimental and computational tools must now elucidate the specific molecular programs driving plastic transitions that bridge the continuum of cancer cell states.
COMPUTATIONAL FRAMEWORK FOR DEFINING CONTINUUM CELL STATE SPACES: MAPPING LANDSCAPE TOPOLOGY
Recent advances in measurement technologies have facilitated the measurement of gene expression, DNA accessibility, protein content, or genomic mutations across tens of thousands of single cells and now even allow for the measurement of multiple modalities in the same cell. This is a revolutionary change from the previous half century of research where techniques such as flow cytometry recorded only one or two dozen features per cell and involved iterative gating of cells into high or low expression on a per-marker basis. The recent explosion in the number of features provided by cutting-edge single-cell techniques now requires a new framework for data analysis.
One emerging approach is manifold learning (Fig. 1D), which models the cellular landscape as a “manifold” to elucidate the states that cells can occupy. The manifold assumption holds that, despite the measurement space of cells being very high dimensional [∼20,000 gene dimensions in the case of single-cell RNA sequencing (scRNA-seq)], the data actually lie in an intrinsically lower dimensional space due to informational redundancy, that is, dependency between the genes. Thus, by discovering the real intrinsic (often nonlinear) axes of variation, we can understand the true shape of the data. This in turn can be used for denoising data (i.e., restoring data to its manifold-intrinsic dimensions), clustering data (discretizing the lower dimensional representations; Fig. 1E, left), and analyzing trajectories (by associating the intrinsic axes with gene expression, etc.).
The term “manifold” comes from Riemannian geometry and describes a space that is smooth, differentiable, and locally Euclidean (93). For example, measured features (e.g., gene counts in an scRNA-seq space) define a space with tens of thousands of dimensions in which relationships between genes, as well as between genes and phenotypes (94–96), may be nonlinear. We understand, however, that cells progress smoothly between states rather than jumping discretely as an electron might change orbitals, implying that the landscape of cellular states consists of locally smooth patches of cells. Each individual patch can be thought of as analogous to Waddington's landscape, in terms of gene configuration, wherein valleys represent the viable cell states. Intrinsic dimensionality in the gene space is further reduced by informational redundancy resulting from gene interactions and coregulation of gene modules. This suggests the cell phenotypic manifold has lower dimensionality than that implied by the combinatorial diversity of genes or proteins comprising each cell, a prediction validated, for instance, by the quantification of discernible phenotypic diversity produced by large-scale chemical compound screening (97, 98). These constraints on the gene space mean that the cellular landscape is in many cases suitable to be computed from scRNA-seq data using manifold learning techniques. Manifold learning may be limited in settings with very small or disconnected data. For example, bulk sequencing, though high dimensional, typically contains only limited samples (<20). In such cases, the cellular state space or landscape cannot be mapped, and more traditional statistical testing may be applied to understand differential expression signatures. Alternatively, if the data contain selectively sampled points from disparate clusters or curated markers designed to decide between only known cell types, then a supervised classification approach may be preferable.
A particularly elegant framework available within the manifold theory is data diffusion (ref. 99; Fig. 1D). The goal of diffusion geometry is to identify the major trajectories and density centers of a manifold. Diffusion geometry can be defined by the creation of a Markovian diffusion operator, which involves the computation of distances between data points, converted to affinities via a kernel function (such as a Gaussian function), with the subsequent normalization of the affinities. Eigenvectors of the diffusion operator, also called diffusion components (or diffusion maps), reveal major pathways or trajectories within the data. If the original kernel is anisotropic—that is, adapts to the effect of data density—the diffusion operator then shows diffusion, similar to Brownian motion in ambient space, and delineates the geometry of the cellular landscape. On the other hand, if the kernel is isotropic, the Markovian diffusion operator shows a combination of Brownian motion and drift toward density centers. Both modalities can be useful for mapping the cellular landscape in cancer. The diffusion operator applied to an initial distribution of cells can show the pathways that lead to favorable or “low energy” attractor states where many cells gather. Thus, energy changes in the landscape can be revealed to a large extent simply by looking at data density, as this approximates potential energy at that landscape location. Overall, diffusion-based manifold learning has been used successfully for many types of cellular analysis, including trajectory and clustering pseudotime analysis (refs. 100–106; Fig. 1E, center), data visualization (107), data denoising (108), differentiation potency analysis (109), and comparative analysis of single-cell data types (110), and joint embedding from multimodal data (111). Thus, diffusion operators are used to analyze both steady state and transitional behaviors in these systems.
Another promising approach for learning cellular manifolds is deep learning. Self-supervised deep learning methods such as autoencoders re-create their (high-dimensional) input as (high-dimensional) output following encoding to a low-dimensional “latent space” embedding. Methods such as SAUCIE (112), sCVI (113), VASC (114), and a recent two-stage variational autoencoder architecture (115) developed for single-cell phenotypic imaging data constrain (typically smoothing) the structure of the latent space to produce state space manifold representations suitable for biological inference.
Archetypal analysis (116) constitutes a third analytical approach (Fig. 1E, right) wherein a convex hull is fitted to the data such that corners of the convex hull represent extrema and other data points are convex mixtures of the extrema (116, 117). In contrast to clustering, which assumes cells occupy distinct and disconnected spaces, the archetypal analysis describes a spectrum of cell states. Recently, archetypal analysis has been advanced with neural networks AAnet and DeepAA (118), providing scalable and nonlinear extensions of the classic archetypal analysis framework. Taken together, the manifold model provides an expansive and growing framework for single-cell data analysis.
MODELING TEMPORAL DYNAMICS OF PHENOTYPIC PLASTICITY: TRAVERSING THE LANDSCAPE
With methodologic advances in data acquisition and manifold learning in place to map the “static” topology of the cancer cell state landscape, including through the integration of data from several measurement modalities (Fig. 2A) (119), it is now vital to understand how cells dynamically traverse this landscape. At the molecular level, understanding the mechanisms that control, initiate, or inhibit phenotypic plasticity constitutes an opportunity to gain therapeutic control over those cancer cell dynamics and thereby curb disease progression.
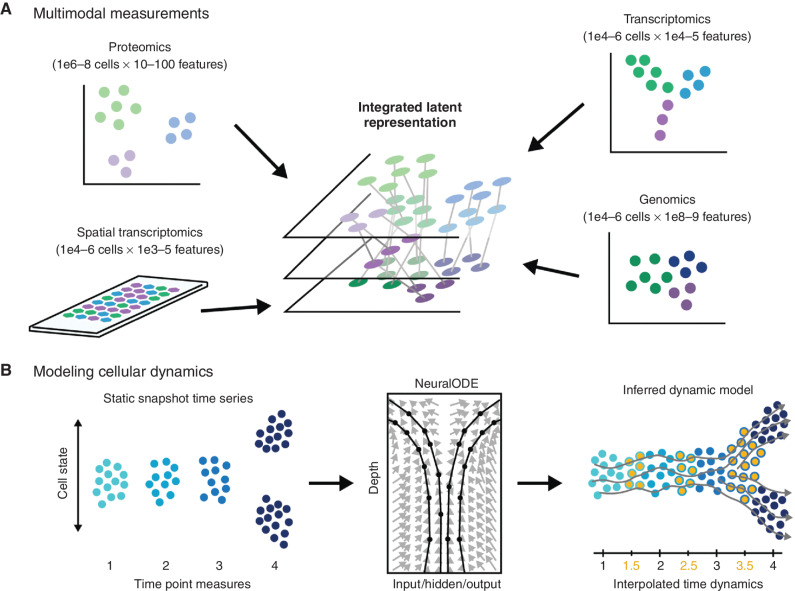
Figure 2.
Modeling temporal dynamics. A, Single-cell populations can be characterized via different omics modalities capturing genomic, transcriptomic, or proteomic information. Resulting data sets may vary significantly in the number of observations and the number of features, and different sets of relationships may exist between the same set of cells depending on which set of features is being examined. Data integration algorithms must be used to merge data sets for joint analysis of multiple data domains. B, In the context of single-cell time-series analyses comprising discrete time point data sets (left), dynamical models based on optimal transport or neural ordinary differential equations (NeuralODE; center) have been used to improve our understanding of biological dynamics by interpolating intervening time point data (orange points, right) to allow inference of dynamic trajectory models (gray lines).
Biomedical technologies usually negotiate a trade-off of the ability to measure many cellular components (genes, proteins, etc.), with the ability to allow the cell to continue to carry out its function. Thus, it is very difficult to devise technologies that can follow a cell unperturbed in time with a high-dimensional measurement. However, high-dimensional data can be collected as static snapshot measurements, and newly emerging computational techniques, particularly involving deep learning, can allow us to learn continuous dynamics and causal effects even using discrete time-series data (Fig. 2B).
Among these approaches, pseudotime trajectory (101, 102, 109) methods were first applied to learn biological dynamics, in which data from a single time point are used to infer a “pseudotime” dimension along which cells transition. Effectively, if the cellular data were collected under a transitional or differentiation condition, the largest axis of variation could be associated with the developmental process. Since the term “pseudotime” was coined in 2014, over 50 methods have been described for trajectory inference. Yet a major drawback of these techniques is that they learn a simple ordering of cells without modeling the dynamics that underlie the probability that a cell will follow one path or another. A newer data-augmented technique for learning cellular transitions is RNA velocity (120, 121). RNA velocity learns transcriptional dynamics based on the ratio between spliced and unspliced transcripts that are naturally found within scRNA-seq data, where the ratio of spliced to unspliced RNA is used to determine if the cell is increasing or decreasing the expression of each gene, that is, learning a velocity vector for each cell. However, this is a very localized snapshot of a global process.
To elucidate underlying dynamic processes across the cellular state space, some groups have leveraged advances in computational optimal transport. Optimal transport is a mathematical technique that quantifies the minimum cost mapping between two distributions. For example, Waddington optimal transport (122) is a method that has been used to describe cellular dynamics during induced pluripotent stem cell reprogramming. However, Waddington optimal transport restricts these dynamics to linear shifts in the ambient space.
To learn nonlinear dynamics on an underlying cellular manifold, we recently described TrajectoryNet, which combines RNA velocity (capturing local transitions) with the global transport ability of optimal transport (123). TrajectoryNet uses a newly devised class of neural network called a NeuralODE (ordinary differential equation). NeuralODEs (Fig. 2B, center) learn a time-varying derivative of a complex dynamic system parameterized via a neural network by setting network weights using an ODE solver instead of standard back-propagation and gradient descent. The resulting network can predict the future state of a measured cell based on the underlying geometry of the data set and predicted intracellular dynamics based on RNA velocity. This framework can enforce directionality through time and along a manifold representation of the data.
We anticipate that emerging models of cellular dynamics will increasingly leverage stochasticity to more closely model cellular processes. Currently, optimal transport models used in TrajectoryNet and Waddington optimal transport are deterministic, meaning that two cells with identical transcriptional profiles will produce identical future state prediction. However, it is possible that these cells have different posttranscriptional regulatory regimes and will therefore diverge in their specific fates. Stochastic differential equation models (124) might be more suited to capture such divergent potentials among similar cells.
THERAPEUTICS TO TARGET CELL PLASTICITY: STRATEGICALLY REMODELING THE CELL STATE LANDSCAPE
Although we do not yet fully understand the intricacies of the tumor ecosystem's dynamism, we posit that cancer progression, metastasis, and therapy resistance all depend heavily on the emergence of landscape topologies permitting adaptive phenotypic cell state dynamics (Fig. 3A). It is vital to understand that, even as individual cells move dynamically upon these landscapes, the landscape topologies are themselves mutable, being sensitive to microenvironmental changes, metastatic relocation, or therapeutic challenges (125). In the clinical context, the inherently adaptive nature of the tumor ecosystem, thus, actively enables therapeutic evasion.
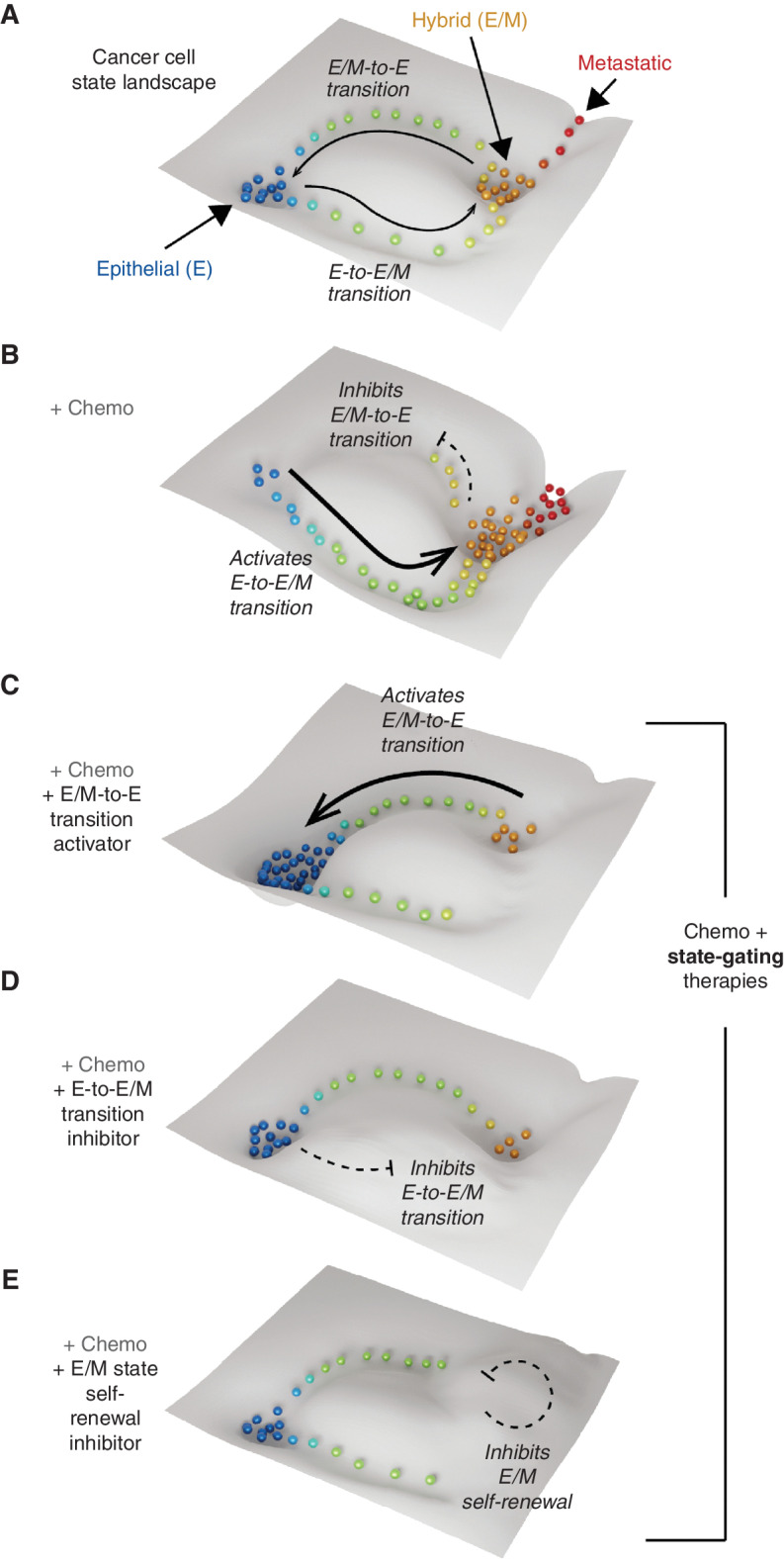
Figure 3.
State-gating strategies to control cancer cell plasticity by remodeling the cancer landscape. A, A subspace of the cancer cell state landscape containing epithelial (E) and hybrid epithelial/mesenchymal (E/M) attractor states linked by plastic transitions. B, Chemotherapy (Chemo) remodels the landscape favoring the transition from the E to the E/M state while inhibiting the reverse process. This increases the population of E/M cells, which promote metastasis and therapy resistance. C–E, Potential antiplasticity state-gating strategies: activating E/M-to-E transition (C), inhibiting E-to-E/M transition (D), and inhibiting E/M self-renewal (E). These will have dual actions, preventing the amplification of E/M cells by chemotherapy while favoring the E state that is sensitive to chemotherapy.
Yet recognition of the mutability of cancer cell state landscapes illuminates a new therapeutic strategy: intentional remodeling of the landscape topology to “corral” cancer cells into states that are more therapeutically vulnerable. By designing therapies that erect barriers to undesirable cell states or shape new channels toward preferable states, we envisage a new class of state-gating therapeutics designed to control the cancer cell state equilibrium. Moreover, we foresee potential for powerful synergies when combining such state-gating therapies with existing anticancer therapies.
For example, as detailed above, it is becoming apparent that some chemotherapies fail because they actively promote plastic transitions to resistant cancer cell states by creating or enhancing transitional trajectories (i.e., channels; Fig. 3B). Given this, multiple approaches can be conceived to enhance and sustain the effectiveness of chemotherapies by activating transitions from the resistant to chemosusceptible states (Fig. 3C), inhibiting transition to the resistant state (Fig. 3D), or preventing self-renewal of the resistant state (Fig. 3E). To design such state-gating strategies, however, now demands that we understand the molecular programs enabling specific trajectories of cell plasticity.
Significantly, state-gating strategies differ fundamentally from traditional targeted therapies. They are designed to target topological “channels” to modulate state plasticity rather than targeting specific topological “wells” to attack particular cancer cell states. Considering this, we may conceive of a future scenario where state-gating approaches are used to shift cells toward, or corral cells within, cell-attractor states that are vulnerable to chemotherapy or other targeted treatments.
CLINICAL MANIFESTATIONS OF PHENOTYPIC HETEROGENEITY AND PLASTICITY
Developing clinical assays to define and quantify distinct tumor cell phenotypic states, and their evolution in response to specific treatments, remains a significant clinical challenge. With clinical pathology tests often limited to IHC stains of a small number of proteins (generally in the range of one to three), it is not yet generally possible to define phenotypic cell states in a clinically sufficient manner or to use that information to inform therapeutic strategy. Despite these limitations, several studies have demonstrated how the simultaneous analysis of even a limited set of markers can successfully define a range of phenotypic cell states in human carcinomas and their association with more aggressive disease, poorer clinical outcome, and resistance to therapy (73, 126). For example, b-catenin, E-cadherin, and Vimentin define the evolution of EMT cell states in untreated and treated human carcinomas (including breast, prostate, non–small cell lung, esophageal carcinomas). CD104 and CD44 can define differing tumorigenicities of cancer cell subpopulations in breast cancer models (35), whereas CD51, CD61, and CD106 define different tumorigenic and metastatic potential in models of squamous cell carcinoma (21). Although such data hint at the clinical potential of defining phenotypic cell state spaces, we are only beginning to understand the diversity of cell phenotypes that exist within tumors, the levels of plasticity coupling these states, and how therapies change the phenotypic landscape. As the field continues to develop technologies for systematic single-cell measurement of this complex biology, we envisage that manifold learning methods will also continue to develop in parallel to enable the full exploitation of these profoundly transformative data. These methods hold great promise to delineate both intrinsic and extrinsic factors underlying phenotypic plasticity, enabling the next generation of diagnostic and therapeutic capabilities.
FUTURE DIRECTIONS
The cancer cell state landscape paradigm, matched to the analytical framework of manifold learning, provides a coherent prism through which to explore the adaptive phenotypic plasticity of cancer cell populations. The complementarity detailed herein between single-cell measurement modalities and computational approaches for manifold learning is vital to advance our understanding of the integrated signaling, genetic, and epigenetic regulatory networks that control cancer cell state plasticity and thus the cancer cell state landscape. In turn, such understanding is critical for the design of novel state-gating therapeutics whose capacity to constrain specific trajectories of plasticity could be used to lock cancer cells into vulnerable states that are targetable with existing chemotherapies and targeted therapies. We believe this dual strategy of combining state-gating and state-targeting therapies will substantially undermine one of cancer's most problematic capabilities—its adaptability—thereby transformatively improving cancer patient outcomes.
Acknowledgments
D.B. Burkhardt was supported, in part, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH (F31HD097958). J.G. Lock received funding from the National Health and Medical Research Council of Australia (GNT1184009, GNT1181230, and GNT2012848), the Australian Research Council (DP220104036), and the Ramaciotti Foundation Biomedical Research Award. C.L. Chaffer and B.P. San Juan were supported by the National Health and Medical Research Council of Australia (GNT1088122 and GNT1181230), National Breast Cancer Foundation research grant IIRS-19-092, and philanthropic support from the Nelune Foundation Rebecca Wilson Fellowship. C.L. Chaffer was supported by a Cancer Institute New South Wales Fellowship (CDF181243). S. Krishnaswamy is supported by Chan-Zuckerberg Initiative grants 182702 and CZF2019-002440, National Science Foundation career grant 2047856, Sloan Fellowship FG-2021-15883, and NIH grants R01GM135929 and R01GM130847.
Authors’ Disclosures
D.B. Burkhardt reports personal fees from Cellarity during the conduct of the study, as well as personal fees from Cellarity outside the submitted work. B.P. San Juan reports grants from St Vincents Clinic Foundation and Tour de Cure outside the submitted work; a patent for methods of treating cancer (PCT/AU2020/051146) pending; and is about to commence a phase I/II clinical trial (4CAST) based on preclinical work (https://www.medrxiv.org/content/10.1101/2022.03.21.22269988v1) that is proof of principle of some of the ideas and therapeutic strategies defined in the manuscript. J.G. Lock reports grants from the National Health and Medical Research Council, the Australian Research Council, and the Ramaciotti Foundation during the conduct of the study. C.L. Chaffer reports grants from the National Health and Medical Research Council of Australia, the Cancer Institute of New South Wales, and the National Breast Cancer Foundation and other support from the Nelune Foundation during the conduct of the study; a patent for methods of treating cancer (PCT/AU2020/051146) pending; and is a managing director of and shareholder in Kembi Therapeutics Pty Ltd, which owns certain patents including WO/2012/064943 and WO/2019/113301. No disclosures were reported by the other author.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275–91. [DOI] [PubMed] [Google Scholar]
- 3. Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 2012;4:127ps10. [DOI] [PubMed] [Google Scholar]
- 4. Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20:743–56. [DOI] [PubMed] [Google Scholar]
- 5. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 6. Greaves M. Evolutionary determinants of cancer. Cancer Discov 2015;5:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343:189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23–8. [DOI] [PubMed] [Google Scholar]
- 9. Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer 2012;12:487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013;339:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010;464:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science 2017;357:eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 17. Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31. [DOI] [PubMed] [Google Scholar]
- 19. Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013;154:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A 2011;108:7950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463–8. [DOI] [PubMed] [Google Scholar]
- 22. Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013;152:25–38. [DOI] [PubMed] [Google Scholar]
- 23. Morel A-P, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, et al. A stemness-related ZEB1–MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med 2017;23:568–78. [DOI] [PubMed] [Google Scholar]
- 24. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Esposito M, Ganesan S, Kang Y. Emerging strategies for treating metastasis. Nat Cancer 2021;2:258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discovery 2020;19:39–56. [DOI] [PubMed] [Google Scholar]
- 27. LaFave LM, Kartha VK, Ma S, Meli K, Del Priore I, Lareau C, et al. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 2020;38:212–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016;539:309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simeonov KP, Byrns CN, Clark ML, Norgard RJ, Martin B, Stanger BZ, et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell 2021;39:1150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marjanovic ND, Hofree M, Chan JE, Canner D, Wu K, Trakala M, et al. Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 2020;38:229–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21–45. [DOI] [PubMed] [Google Scholar]
- 32. Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer 2021;21:325–38. [DOI] [PubMed] [Google Scholar]
- 33. Pei D, Shu X, Gassama-Diagne A, Thiery JP. Mesenchymal-epithelial transition in development and reprogramming. Nat Cell Biol 2019;21:44–53. [DOI] [PubMed] [Google Scholar]
- 34. Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs 2007;185:7–19. [DOI] [PubMed] [Google Scholar]
- 35. Bierie B, Pierce SE, Kroeger C, Stover DG, Pattabiraman DR, Thiru P, et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Nat Acad Sci U S A 2017;114:E2337–E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jolly MK, Somarelli JA, Sheth M, Biddle A, Tripathi SC, Armstrong AJ, et al. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol Ther 2019;194:161–84. [DOI] [PubMed] [Google Scholar]
- 37. Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A 2019;116:7353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Castano Z, San Juan BP, Spiegel A, Pant A, DeCristo MJ, Laszewski T, et al. IL-1beta inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol 2018;20:1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 2016;351:aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 2006;131:830–40. [DOI] [PubMed] [Google Scholar]
- 41. Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat Rev Cancer 2019;19:716–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmidt JM, Panzilius E, Bartsch HS, Irmler M, Beckers J, Kari V, et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep 2015;10:131–9. [DOI] [PubMed] [Google Scholar]
- 43. Ocaña OH, Córcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012;22:709–24. [DOI] [PubMed] [Google Scholar]
- 44. Maruyama R, Choudhury S, Kowalczyk A, Bessarabova M, Beresford-Smith B, Conway T, et al. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet 2011;7:e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008;28:4772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004;24:306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serresi M, Kertalli S, Li L, Schmitt MJ, Dramaretska Y, Wierikx J, et al. Functional antagonism of chromatin modulators regulates epithelial-mesenchymal transition. Sci Adv 2021;7:eabd7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013;19:1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer 2021;21:669–80. [DOI] [PubMed] [Google Scholar]
- 50. Ferraro GB, Ali A, Luengo A, Kodack DP, Deik A, Abbott KL, et al. Fatty acid synthesis is required for breast cancer brain metastasis. Nat Cancer 2021;2:414–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ngo B, Kim E, Osorio-Vasquez V, Doll S, Bustraan S, Liang RJ, et al. Limited environmental serine and glycine confer brain metastasis sensitivity to PHGDH inhibition. Cancer Discov 2020;10:1352–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Elia I, Rossi M, Stegen S, Broekaert D, Doglioni G, van Gorsel M, et al. Breast cancer cells rely on environmental pyruvate to shape the metastatic niche. Nature 2019;568:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christen S, Lorendeau D, Schmieder R, Broekaert D, Metzger K, Veys K, et al. Breast cancer-derived lung metastases show increased pyruvate carboxylase-dependent anaplerosis. Cell Rep 2016;17:837–48. [DOI] [PubMed] [Google Scholar]
- 54. Müller S, Sindikubwabo F, Cañeque T, Lafon A, Versini A, Lombard B, et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat Chem 2020;12:929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. D'Aniello C, Patriarca EJ, Phang JM, Minchiotti G. Proline metabolism in tumor growth and metastatic progression. Front Oncol 2020;10:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kusi M, Zand M, Lin LL, Chen M, Lopez A, Lin CL, et al. 2-Hydroxyglutarate destabilizes chromatin regulatory landscape and lineage fidelity to promote cellular heterogeneity. Cell Rep 2022;38:110220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010;141:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roberson RS, Kussick SJ, Vallieres E, Chen S-YJ, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005;65:2795–803. [DOI] [PubMed] [Google Scholar]
- 59. Saleh T, Tyutyunyk-Massey L, Murray GF, Alotaibi MR, Kawale AS, Elsayed Z, et al. Tumor cell escape from therapy-induced senescence. Biochem Pharmacol 2019;162:202–12. [DOI] [PubMed] [Google Scholar]
- 60. Milanovic M, Fan DNY, Belenki D, Dabritz JHM, Zhao Z, Yu Y, et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018;553:96–100. [DOI] [PubMed] [Google Scholar]
- 61. Di Martino JS, Nobre AR, Mondal C, Taha I, Farias EF, Fertig EJ, et al. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat Cancer 2022;3:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oren Y, Tsabar M, Cuoco MS, Amir-Zilberstein L, Cabanos HF, Hütter J-C, et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 2021;596:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khoo WH, Ledergor G, Weiner A, Roden DL, Terry RL, McDonald MM, et al. A niche-dependent myeloid transcriptome signature defines dormant myeloma cells. Blood 2019;134:30–43. [DOI] [PubMed] [Google Scholar]
- 64. Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity 2020;52:55–81. [DOI] [PubMed] [Google Scholar]
- 65. Li J, Stanger BZ. How tumor cell dedifferentiation drives immune evasion and resistance to immunotherapy. Cancer Res 2020;80:4037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 2017;6:e1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res 2017;77:3982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tauriello DVF, Sancho E, Batlle E. Overcoming TGFβ-mediated immune evasion in cancer. Nat Rev Cancer 2022;22:25–44. [DOI] [PubMed] [Google Scholar]
- 69. Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, et al. Toward minimal residual disease-directed therapy in melanoma. Cell 2018;174:843–55. [DOI] [PubMed] [Google Scholar]
- 70. Su Y, Wei W, Robert L, Xue M, Tsoi J, Garcia-Diaz A, et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A 2017;114:13679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nolan E, Bridgeman VL, Ombrato L, Karoutas A, Rabas N, Sewneth CAN, et al. Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization. Nat Cancer 2022;3:173–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Redfern AD, Spalding LJ, Thompson EW. The Kraken wakes: induced EMT as a driver of tumour aggression and poor outcome. Clin Exp Metastasis 2018;35:285–308. [DOI] [PubMed] [Google Scholar]
- 74. Ma B, Wells A, Clark AM. The pan-therapeutic resistance of disseminated tumor cells: role of phenotypic plasticity and the metastatic microenvironment. Semin Cancer Biol 2020;60:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goldman A, Khiste S, Freinkman E, Dhawan A, Majumder B, Mondal J, et al. Targeting tumor phenotypic plasticity and metabolic remodeling in adaptive cross-drug tolerance. Sci Signal 2019;12:eaas8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer 2019;19:405–14. [DOI] [PubMed] [Google Scholar]
- 77. Ahmed N, Escalona R, Leung D, Chan E, Kannourakis G. Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin Cancer Biol 2018;53:265–81. [DOI] [PubMed] [Google Scholar]
- 78. Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015;525:119–23. [DOI] [PubMed] [Google Scholar]
- 79. Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 2015;525:114–8. [DOI] [PubMed] [Google Scholar]
- 80. Lacroix M, Riscal R, Arena G, Linares LK, Le Cam L. Metabolic functions of the tumor suppressor p53: implications in normal physiology, metabolic disorders, and cancer. Mol Metab 2020;33:2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Blandino G, Valenti F, Sacconi A, Di Agostino S. Wild type- and mutant p53 proteins in mitochondrial dysfunction: emerging insights in cancer disease. Semin Cell Dev Biol 2020;98:105–17. [DOI] [PubMed] [Google Scholar]
- 82. Celià-Terrassa T, Bastian C, Liu DD, Ell B, Aiello NM, Wei Y, et al. Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat Commun 2018;9:5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Abdelmoula WM, Balluff B, Englert S, Dijkstra J, Reinders MJ, Walch A, et al. Data-driven identification of prognostic tumor subpopulations using spatially mapped t-SNE of mass spectrometry imaging data. Proc Natl Acad Sci U S A 2016;113:12244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res 2012;72:4875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82. [DOI] [PubMed] [Google Scholar]
- 86. Vickovic S, Eraslan G, Salmen F, Klughammer J, Stenbeck L, Schapiro D, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods 2019;16:987–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, et al. The single-cell pathology landscape of breast cancer. Nature 2020;578:615–20. [DOI] [PubMed] [Google Scholar]
- 88. Huang S. The molecular and mathematical basis of Waddington's epigenetic landscape: a framework for post-Darwinian biology? Bioessays 2012;34:149–57. [DOI] [PubMed] [Google Scholar]
- 89. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90. [DOI] [PubMed] [Google Scholar]
- 90. Hills TT, Todd PM, Lazer D, Redish AD, Couzin ID, Cognitive Search Research G. Exploration versus exploitation in space, mind, and society. Trends Cogn Sci 2015;19:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol 2007;42:399–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eckersley-Maslin MA, Parry A, Blotenburg M, Krueger C, Ito Y, Franklin VNR, et al. Epigenetic priming by Dppa2 and 4 in pluripotency facilitates multi-lineage commitment. Nat Struct Mol Biol 2020;27:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Moon KR, Stanley JS, Burkhardt D, van Dijk D, Wolf G, Krishnaswamy S. Manifold learning-based methods for analyzing single-cell RNA-sequencing data. Curr Opin Syst Biol 2018;7:36–46. [Google Scholar]
- 94. Farkash-Amar S, Zimmer A, Eden E, Cohen A, Geva-Zatorsky N, Cohen L, et al. Noise genetics: inferring protein function by correlating phenotype with protein levels and localization in individual human cells. PLos Genet 2014;10:e1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kiss A, Gong X, Kowalewski JM, Shafqat-Abbasi H, Strömblad S, Lock JG. Non-monotonic cellular responses to heterogeneity in talin protein expression-level. Integr Biol 2015;7:1171–85. [DOI] [PubMed] [Google Scholar]
- 96. Kowalewski JM, Shafqat-Abbasi H, Jafari-Mamaghani M, Endrias Ganebo B, Gong X, Strömblad S, et al. Disentangling membrane dynamics and cell migration; differential influences of F-actin and cell-matrix adhesions. PLoS One 2015;10:e0135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bryce NS, Failes TW, Stehn JR, Baker K, Zahler S, Arzhaeva Y, et al. High-content imaging of unbiased chemical perturbations reveals that the phenotypic plasticity of the actin cytoskeleton is constrained. Cell Syst 2019;9:496–507. [DOI] [PubMed] [Google Scholar]
- 98. Bryce NS, Hardeman EC, Gunning PW, Lock JG. Chemical biology approaches targeting the actin cytoskeleton through phenotypic screening. Curr Opin Chem Biol 2019;51:40–7. [DOI] [PubMed] [Google Scholar]
- 99. Coifman RR, Lafon S. Diffusion maps. Appl Comput Harmon Anal 2006;21:5–30. [Google Scholar]
- 100. Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 2016;13:845–8. [DOI] [PubMed] [Google Scholar]
- 102. Bendall S C, Davis K L, Amir el- AD, Tadmor M D, Simonds E F, Chen T J, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 2014;157:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kuchroo M, Huang J, Wong P, Grenier JC, Shung D, Tong A, et al. Multiscale PHATE identifies multimodal signatures of COVID-19. Nat Biotechnol 2022;40:681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Brugnone N, Gonopolskiy A, Moyle MW, Kuchroo M, van Dijk D, Moon KR, et al. Coarse graining of data via inhomogeneous diffusion condensation. Proc IEEE Int Conf Big Data 2019;2019:2624–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech: Theory Exp 2008;2008:P10008. [Google Scholar]
- 106. Ng AY, Jordan MI, Weiss Y. On spectral clustering: analysis and an algorithm. Adv Neural Inf Process Syst 2002;14:849–56. [Google Scholar]
- 107. Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol 2019;37:1482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, et al. Recovering gene interactions from single-cell data using data diffusion. Cell 2018;174:716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 2016;34:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Burkhardt DB, Stanley JS, Perdigoto AL, Gigante SA, Herold KC, Wolf G, et al. Quantifying the effect of experimental perturbations in single-cell RNA-sequencing data using graph signal processing. bioRxiv 2019:532846. [Google Scholar]
- 111. Kuchroo M, Godavarthi A, Tong A, Wolf G, Krishnaswamy S. Multimodal data visualization and denoising with integrated diffusion. IEEE Int Workshop Mach Learn Signal Process 2021;2021: 10.1109/mlsp52302.2021.9596214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Amodio M, van Dijk D, Srinivasan K, Chen WS, Mohsen H, Moon KR, et al. Exploring single-cell data with deep multitasking neural networks. Nat Methods 2019;16:1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods 2018;15:1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang D, Gu J. VASC: dimension reduction and visualization of single-cell RNA-seq data by deep variational autoencoder. Genomics Proteomics Bioinformatics 2018;16:320–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wei W, Haidinger S, Lock J, Meijering E. Deep representation learning for image-based cell profiling. In: Lian C, Cao X, Rekik I, Xu X, Yan P, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2021. p. 487–97. [Google Scholar]
- 116. Korem Y, Szekely P, Hart Y, Sheftel H, Hausser J, Mayo A, et al. Geometry of the gene expression space of individual cells. PLoS Comput Biol 2015;11:e1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Adler M, Korem Kohanim Y, Tendler A, Mayo A, Alon U. Continuum of gene-expression profiles provides spatial division of labor within a differentiated cell type. Cell Syst 2019;8:43–52. [DOI] [PubMed] [Google Scholar]
- 118. van Dijk D, Burkhardt DB, Amodio M, Tong A, Wolf G, Krishnaswamy S. Finding archetypal spaces using neural networks. In: Proceedings of the 2019 IEEE International Conference on Big Data (Big Data); 2019 Dec 9–12; Los Angeles, CA. New York: IEEE; 2019. p. 2634–43. [Google Scholar]
- 119. Argelaguet R, Cuomo ASE, Stegle O, Marioni JC. Computational principles and challenges in single-cell data integration. Nat Biotechnol 2021;39:1202–15. [DOI] [PubMed] [Google Scholar]
- 120. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408–14. [DOI] [PubMed] [Google Scholar]
- 122. Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 2019;176:928–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tong A, Huang J, Wolf G, Dijk DV, Krishnaswamy S. TrajectoryNet: a dynamic optimal transport network for modeling cellular dynamics. In: Hal D III, Aarti S, editors. Proceedings of the 37th International Conference on Machine Learning; 2020 Jul 13–18; Virtual. Volume 119. Proceedings of Machine Learning Research: PMLR; 2020. p. 9526–36. [PMC free article] [PubMed] [Google Scholar]
- 124. Li X, Wong TKL, Chen RTQ, Duvenaud D. Scalable gradients for stochastic differential equations. In: Silvia C, Roberto C, editors. Proceedings of the 23rd International Conference on Artificial Intelligence and Statistics; 2020 Aug 26–28; Virtual. Volume108. Proceedings of Machine Learning Research: PMLR; 2020. p. 3870–82. [Google Scholar]
- 125. Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21:309–22. [DOI] [PubMed] [Google Scholar]
- 126. Navas T, Kinders RJ, Lawrence SM, Ferry-Galow KV, Borgel S, Hollingshead MG, et al. Clinical evolution of epithelial–mesenchymal transition in human carcinomas. Cancer Res 2020;80:304–18. [DOI] [PMC free article] [PubMed] [Google Scholar]