Abstract
The proliferation of synthetic data in artificial intelligence for medicine and healthcare raises concerns about the vulnerabilities of the software and the challenges of current policy.
As artificial intelligence (AI) for applications in medicine and healthcare undergoes increased regulatory analysis and clinical adoption, the data used to train the algorithms are undergoing increasing scrutiny. Scrutiny of the training data is central to understanding algorithmic biases and pitfalls. These can arise from datasets with sample-selection biases — for example, from a hospital that admits patients with certain socioeconomic backgrounds, or medical images acquired with one particular type of equipment or camera model. Algorithms trained with biases in sample selection typically fail when deployed in settings sufficiently different from those in which the trained data were acquired1. Biases can also arise owing to class imbalances — as is typical of data associated with rare diseases — which degrade the performance of trained AI models for diagnosis and prognosis. And AI-driven diagnostic-assistance tools relying on historical data would not typically detect new phenotypes, such as those of patients with stroke or cancer presenting with symptoms of COVID-19 (coronavirus disease 2019)2. Because the utility of AI algorithms for healthcare applications hinges on the exhaustive curation of medical data with ground-truth labels, the algorithms are as effective or as robust as the data they are supplied with.
Therefore, large datasets that are diverse and representative (of the heterogeneity of phenotypes, in the gender, ethnicity and geography of the individuals or patients, and in the healthcare systems, workflows and equipment used) are necessary to develop and refine best practices in evidence-based medicine involving AI (ref.3). To overcome the paucity of annotated medical data in real-world settings, synthetic data are being increasingly used. Synthetic data can be created from perturbations via accurate forward models (that is, models that simulate outcomes given specific inputs), physical simulations or AI-driven generative models. As with the development of computer-vision algorithms for self-driving cars to emulate scenarios such as road accidents and harsh driving environments for which collecting data can be challenging4, in medicine and healthcare accurate synthetic data can be used to increase diversity in datasets and to increase the robustness and adaptability of AI models. However, synthetic data can also be used maliciously, as exemplified by fake impersonation videos (also known as deepfakes), which can propagate misinformation and fool facial-recognition software5.
The United States Federal Drug and Administration (FDA) has put forward an approval pathway for AI-based software as a medical device (AI-SaMD). Over 70 algorithms have been approved as of today, with uses ranging from the detection of atrial fibrillation to the clinical grading of pathology slides6,7. By incorporating synthetic data emulating the phenotypes of underrepresented conditions and individuals, AI algorithms can make better medical decisions in a wider range of real-world environments. In fact, the use of synthetic data has attracted mainstream attention as a potential path forward for greater reproducibility in research and for implementing differential privacy for protected health information8,9 (PHI). With the increasing digitization of health data and the market size of AI for healthcare expected to reach $45 billion by 2026 (ref.8), the role of synthetic data in the health-information economy needs to be precisely delineated in order to develop fault-tolerant and patient-facing health systems10. How do synthetic data fit within existing regulatory frameworks for modifying AI algorithms in healthcare? To what ends can synthetic data be used to protect or exploit patient privacy, and to improve medical decision-making? In this Comment, we examine the proliferation of synthetic data in medical AI and discuss the associated vulnerabilities and necessary policy challenges.
Fidelity tests
Beyond improved image classification and natural language processing, one promise of AI involves learning algorithms, also known as deep generative models, that can emulate how data are generated in the real world12. Generative adversarial networks (GANs) are a type of generative model that learn probability distributions of how high-dimensional data are likely to be distributed. GANs consist of two neural networks — a generator and a discriminator — that compete in a minimax game (that is, a game of minimizing the maximum possible loss) to fool each other. For instance, in a GAN being trained to produce paintings in the style of Claude Monet, the generator would be a neural network that aims to produce a Monet counterfeit that fools a critic discriminator network attempting to distinguish real Monet paintings from counterfeits. As the game progresses, the generator learns from the poor counterfeits caught by the discriminator, progressively creating more realistic counterfeits. GANs have shown promise in a variety of applications, ranging from synthesizing paintings of modern landscapes in the style of Claude Monet to generating realistic images of skin lesions13 (Fig. 1, top), pathology slides14, colon mucosa15, chest X-rays16–18 (Fig. 1, top), and from a range of imaging modalities19–22.
Fig. 1 |. Synthetic medical data in action.
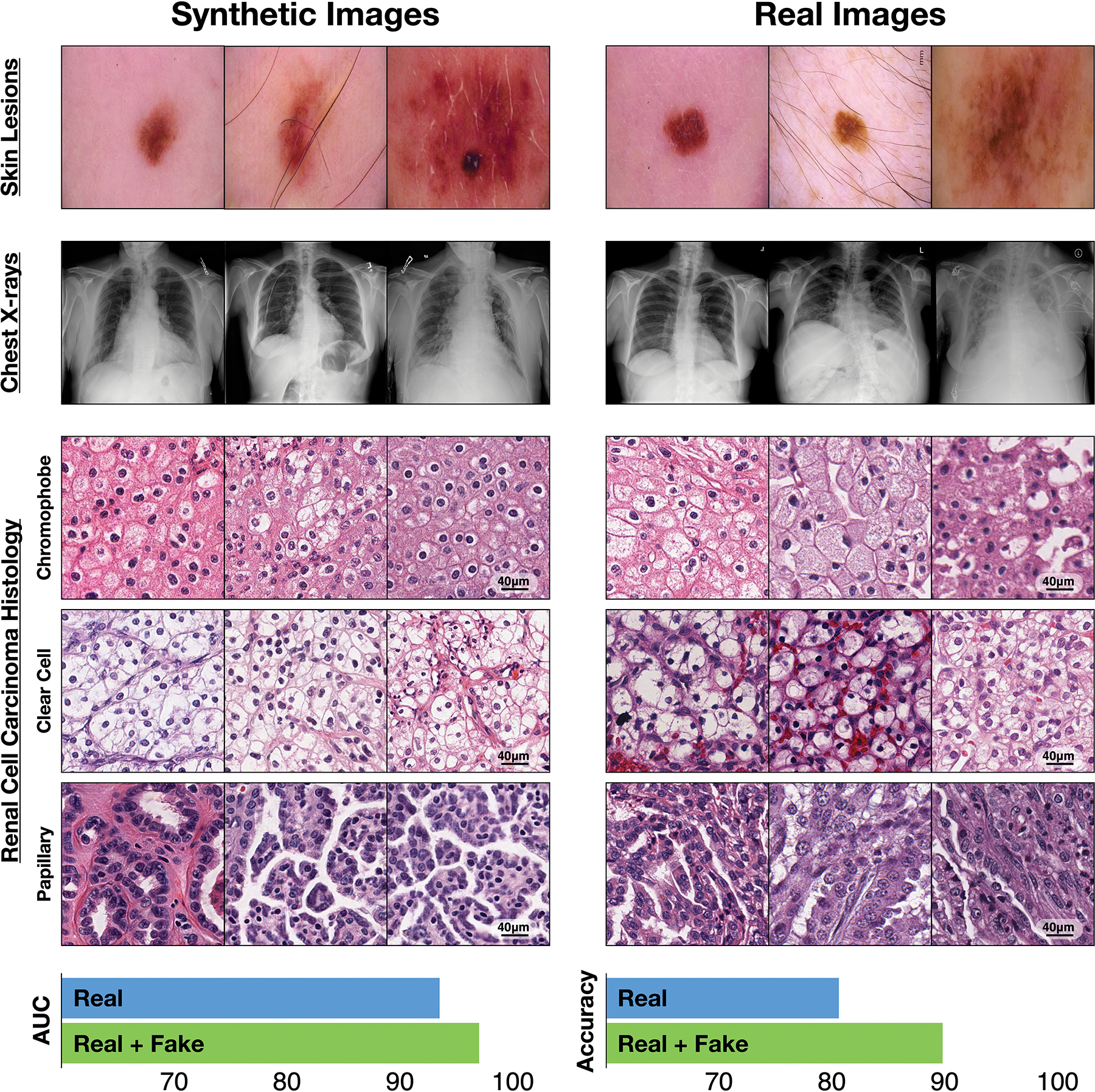
Top, synthetic and real images of skin lesions and of frontal chest X-rays. Middle, Synthetic and real histology images of three subtypes of renal cell carcinoma. Bottom, Areas under the receiver operating characteristic curve (AUC) for the classification performance of an independent dataset of the histology images by a deep-learning model trained with 10,000 real images of each subtype and by the same model trained with the real-image dataset augmented by 10,000 synthetic images of each subtype. Methodology and videos are available as Supplementary Information.
Advancements in computer graphics and in information theory have driven progress in generative modelling from the generation of 28 × 28 (pixels per side) black-and-white images of handwritten digits to simulating life-like human faces with 1,024 × 1,024 high-fidelity images23. To show the capabilities of the AI-driven generation of synthetic medical data, we produced images of three histological subtypes of renal cell carcinoma (chromophobe, clear cell, and papillary carcinoma) by training a GAN with 10,000 real images of each subtype, and then compared the performance of the model with another model trained using both real and synthetic data (Fig. 1, middle and bottom panels). The synthetic images generated by the GAN finely mimic the characteristic thin-walled ‘chicken wire’ vasculature of the clear-cell carcinoma subtype and the unique features of the other two subtypes. The GAN also improves the accuracy of classification.
By closely mimicking real-world observational data, synthetic data could transform interoperability standards in the sharing of health data, and contribute to improving reproducibility24. For example, in lieu of revealing actual patient data, synthetic datasets that accurately capture the original distribution of the data would substantially lessen patient-privacy concerns and could be freely shared. Unfortunately, current generative models are not ready for the off-the-shelf generation of synthetic data, and may even create vulnerabilities (which could lead to patient re-identification) if adopted carelessly across healthcare ecosystems. For example, if a clinician working with developmental disorders and using a generative model to capture phenotype diversity in adolescents with de novo mutations makes the weights of the trained GAN model publicly available, the GAN could be used by a third party to synthesize real faces of the adolescents, thus leaking PHI. This is an example of information leakage (and in particular of a membership interference attack25,26), one of many failure modes of generative models, wherein samples from the training dataset can be recovered from the probability distribution, owing to overfitting. Although information leakage can be mitigated with sophisticated modelling techniques such as differential privacy, the adaptation to clinical scenarios would require expertise in machine learning as well as medical-domain knowledge27–30. As best practices for generative models continue to be developed, better privacy guarantees should be put forward to minimize the possibility of a PHI leak31.
Challenges in implementation and adoption
The generation of synthetic data has garnered significant attention in medicine and healthcare13,14,17,32–34 because it can improve existing AI algorithms via data augmentation. For instance, among renal cell carcinomas, the chromophobe subtype is rare; it accounts for merely 5% of all cases, and has a global incidence of one case per two million patients with renal cancer. By providing synthetic histology images of renal cell carcinoma as additional training input to a convolutional neural network, the detection accuracy of the subtype can be improved (Fig. 1, bottom).
However, the wider roles of synthetic data in AI systems in healthcare remain unclear. Unlike traditional medical devices, the function of AI-SaMDs may need to be adaptive to data streams that evolve over time, as is the case for health data from smartphone sensors35,36. Researchers may be tempted to use synthetic data as a stopgap for the fine-tuning of algorithms; however, policymakers may find it troubling that there are not always clinical-quality measures and evaluation metrics for synthetic data. In a proposed FDA regulatory framework for software modifications in adaptive AI-SaMDs, guidance for updating algorithms would mandate reference standards and quality assurance of any new data sources6. However, when generating synthetic data for rare or novel disease conditions, there may not even be sufficient samples to establish clinical reference standards. As with other data-driven deep-learning algorithms, generative models are constrained by the size and quality of the training dataset used to model the data distribution, and models trained with biased datasets would still be biased toward overrepresented conditions. How can we assess whether synthetic data are emulating the correct phenotype and are free from artifacts that would bias the deployed AI-SaMDs? Current quantitative metrics for the evaluation of generative models use probability likelihood and divergence scores that are not easy to interpret by clinicians and that do not reflect specific failure modes in the generation of synthetic data37. This complicates the adoption of synthetic data for AI-SaMDs.
Synthetic data could be evaluated via visual Turing tests; in fact, human-eye perceptual evaluation metrics have been proposed for evaluating generative models on real and synthetic images3,7. These metrics can be adapted to assessing synthesized radiology and pathology images by expert radiologists and pathologists, yet they may be prone to large intra-observer variability. Another practical barrier to visual Turing Tests relates to intractable data-curation protocols; for example, assessing thousands or millions of synthetic images would be as tedious as collecting and labelling actual real images. These problems would be further exacerbated with synthetic data that cannot be as readily assessed, such as electrocardiograms, voice measurements, longitudinal disease trajectories, and entire electronic medical records39–43 (EMRs).
Training generative models with multi-institutional datasets that capture a larger diversity of clinical phenotypes and outcomes can improve model generalization and reduce biases. And larger training datasets naturally lead to more robust algorithms. However, sharing data and models between institutions is complex, owing to the regulated nature of PHI and to privacy concerns regarding model-information leakage. Synthetic data can certainly facilitate reproducibility and transparency, and minimize biases, yet data-driven generative models can be trapped in a catch-22 dilemma: the data-paucity problem that generative modelling aims to solve is unfortunately constrained by the inherent stagnant interoperability of EMRs, and this prevents large and diverse datasets from being curated in the first place.
Privacy and security
Deepfakes are an increasingly pervasive form of AI-synthesized media (images, audio and video); in fact, GANs such as those used by the deepfake software faceswap allow users to impersonate any individual through appearance and voice, and have been convincing enough to defraud a UK-based energy firm out of $243,000 (ref.44). Regulation around the creation and distribution of deepfakes has engaged policymakers, digital-forensics experts and technology companies, and recent legislation in the United States has prohibited the distribution of malicious synthetic media in order to protect political candidates5. However, in healthcare the proliferation of deepfakes is a blind spot; current measures to preserve patient privacy, authentication and security are insufficient. For instance, algorithms for the generation of deepfakes can also be used to potentially impersonate patients and to exploit PHI, to falsely bill health insurers relying on imaging data for the approval of insurance claims45, and to manipulate images sent from the hospital to an insurance provider so as to trigger a request for reimbursement for a more expensive procedure.
Yet algorithms for the generation of deepfakes can also be used to anonymize patient data. A GAN architecture similar to that used by the software faceswap can be used to de-identify faces in live videos46, and similar methods could be used for the de-identification of EMRs, medical images and other PHI47. In healthcare settings, and in particular in clinical research, it may be necessary to video record patient interactions in order to detect phenotypes for early disease prognosis (for example, saccadic eye movements in autism spectrum disorders, and speech defects in mild cognitive impairment and in Alzheimer’s disease). As clinical practice is increasingly adopting telemedicine for remote health monitoring48, software that may leak PHI needs to be regulated with mandatory security measures, such as synthetic-data-driven differential-privacy systems27.
Paths forward
What constitutes authenticity, and how would the lack of authenticity shape our perception of reality? The science-fiction American write Philip K. Dick posited similar questions throughout his literary career and in particular in his 1972 essay ‘How to build a universe that doesn’t fall apart two days later’, where he commented on the dangerous ‘blur’ replacing reality with synthetic-like constructs49. As if he were describing the dilemmas of today’s technology, Dick wrote ”What is real? Because unceasingly we are bombarded with pseudo-realities manufactured by very sophisticated people using very sophisticated electronic mechanisms. I do not distrust their motives; I distrust their power. They have a lot of it. And it is an astonishing power: that of creating whole universes, universes of the mind.” In healthcare, this power lies in the creation of realistic data that can influence the perception of clinicians and healthcare policymakers regarding what is clinical ground truth, and that affect the deployment of AI algorithms used to make decisions influencing human lives50. The advancements made are maturing so rapidly that we should carefully understand what control we cede if we allow for ‘spurious imitations’ to gain a foothold in healthcare decision-making. For instance, since the start of the COVID-19 pandemic, there has been an explosion of interest around the development of synthetic data, with use cases such as the training AI algorithms17,51, epidemiological modelling and digital contact tracing52–54, and data sharing between hospitals55. Because synthetic data will undoubtedly be soon used to solve pressing problems in healthcare, it is urgent to develop and refine regulatory frameworks involving synthetic data and the monitoring of its impact in society.
Algorithms grounded on real data.
To make synthetic data more compliant with existing clinical regulations, algorithms for the generation of synthetic data should be developed with accurate forward models of existing data collections15. Generative models are one of many data-generation techniques that have pushed AI ‘over the precipice’ into product deployment across industries (most prominently, in digital advertising and in autonomous vehicles; companies developing self-driving vehicles can simulate tens of millions of driven miles every day56). Indeed, synthetic data have already been widely adopted to develop algorithms that can make better decisions than most humans in unseen narrow scenarios32, 57–60.
In computer-aided diagnostics, forward models can be used to create photorealistic environments for training AI algorithms when data collection is unfeasible. Instead of using data-driven approaches such as generative modelling, algorithms that explicitly model physical properties (for example, light scattering in tissue) can be used to generate biologically accurate synthetic data20,61,62. For complicated medical procedures such as colonoscopies, virtual environments akin to those used to develop self-driving cars could be used to train AI-based capsule endoscopes to navigate the gastrointestinal tract63. Unlike synthetic data from generative models, simulation-based synthetic data from forward models are created from existing clinical reference standards, medical prior knowledge and physical laws. This strategy may have regulatory advantages, especially regarding the adoption of AI software that can be regularly modified.
For the real-world deployment of active-learning systems, synthetic data may be used in regulatory ‘stress tests’ before AI algorithms can be used by physicians and patients. For example, during the deployment of an algorithm for the automated screening of diabetic retinopathy in clinical centres across Thailand64, the algorithm failed to analyse some eye scans owing to variable lighting conditions, camera angles and deficient image quality. Such issues of ‘domain shift’ (that is, of unmatched training-data and test-data distributions) in healthcare applications may be addressed by adopting best practices developed by the autonomous-vehicle industry. For instance, for self-driving cars, virtual environments that generate synthetic data for data augmentation and that simulate non-expert behaviours that human drivers cannot feasibly create are a substitute for scarce data from real-world collisions and other potentially harmful scenarios65–67. In the case of diabetic-retinopathy screening, a solution would be to simulate challenging scenarios, such as variable lighting and camera distortions during model training, to make the model robust against changes in lighting, image acquisition and patient pose. Such environments would also benefit other settings and systems, such as computer-assisted surgical procedures: AI algorithms could be trained to learn from incorrect surgery techniques without putting patients at risk.
Evaluation metrics and human-in-the-loop tests.
In addition to creating regulatory standards for synthetic-data quality, regulations and evaluation metrics should also be developed for models that assess not only realism, but also failure modes such as information leakage. Although no consensus for a universal quantitative metric has been reached, recent discussions have pointed toward rethinking the evaluation of generative models as if facing a bias–variance trade-off — that is, models biased towards emulating only one label would fail to capture the multimodal nature of probability distributions, and models with high variance would generate data outside of the distributions68–70. This analogy gives rise to two qualities for scoring synthetic datasets: fidelity for assessing realism of synthetic samples, and diversity for capturing the variability of real data. The privacy issues in synthetic data can also define authenticity, a measurement of the number of copies of real data made by the model. In experimentation with synthetic EMR data in the context of COVID-19, these three metrics were used to understand the fidelity–diversity and privacy–utility trade-offs in ranking generative models70. It was seen that prioritizing diversity and privacy-preserving performance decreased fidelity and downstream classification tasks using synthetic data.
In grounding synthetic data with biological priors, the generation of synthetic data can also be used as a tool for scientific discovery. This is exemplified by AlphaFold (developed by the company DeepMind), an algorithm that uses generative models to predict the 3D structure of proteins71. Although the adoption of AI-generated protein structures as therapeutic candidates may be improbable in the short term, sequence-based synthetic data can be experimentally validated in animal models (out of the millions of potential protein candidates that can be generated using GANs, only a handful of samples need to pass fidelity tests for physical experimentation72). Such an ‘auditing’ process for synthetic data has led to a relay-race of biotechnology start-ups and pharmaceutical companies collaborating together to use AI for drug discovery73. In 2018, the biotech start-up In silico Medicine showed the use of GANs to generate small-molecule inhibitors for a protein target and their in vitro and in vivo validation in only 46 days74.
Regulations for the use of synthetic data in medicine and healthcare need to be developed and specifically adapted for different use cases. And although there may always exist unknown unknowns during algorithm deployment, experimentation and human-in-the-loop evaluation can be used to iteratively refine AI-SaMDs so that they become more fault-tolerant.
Supplementary Material
Acknowledgements
This work was supported in part by internal funds from BWH Pathology, a Google Cloud Research Grant, the Nvidia GPU Grant Program, and NIGMS R35GM138216 (F.M.). R.J.C. was supported by an NSF Graduate Fellowship. The content is solely the responsibility of the authors and does not reflect the official views of the National Science Foundation or the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Pencina MJ, Goldstein BA & D’Agostino RB The New England Journal of Medicine 382, 1583 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Oxley TJ et al. New England Journal of Medicine 0, e60 (0). [Google Scholar]
- 3.Trister AD JAMA oncology 5, 1429–1430 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Wang X et al. IEEE Transactions on Intelligent Transportation Systems 19, 910–920 (2017). [Google Scholar]
- 5.Chesney B & Citron D California Law Review 107, 1753 (2019). [Google Scholar]
- 6.Machine Learning (AI/ML)-based Software as a Medical Device (SaMD), FDA, https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf (2019).
- 7.Benjamens S, Dhunnoo P & Mesko B NPJ digital medicine 3, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abowd JM & Vilhuber L In International Conference on Privacy in Statistical Databases, 239–246 (Springer, 2008). [Google Scholar]
- 9.Beaulieu-Jones BK et al. Circulation: Cardiovascular Quality and Outcomes 12, e005122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markets & Markets. $45.2 billion worldwide artificial intelligence in healthcare industry to 2026 Research and Markets; (2020). [Google Scholar]
- 11.LeCun Y, Bengio Y & Hinton G Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Goodfellow I et al. In Advances in Neural Information Processing Systems, 2672–2680 (2014). [Google Scholar]
- 13.Ghorbani A, Natarajan V, Coz D & Liu Y In Proceedings of the Machine Learning for Health NeurIPS Workshop, 155–170 (2020). [Google Scholar]
- 14.Mahmood F et al. IEEE Transactions on Medical Imaging (2019). [Google Scholar]
- 15.Mahmood F, Chen R & Durr NJ IEEE Transactions on Medical Imaging 37, 2572–2581 (2018). 9 [DOI] [PubMed] [Google Scholar]
- 16.Teixeira B et al. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 9059–9067 (2018). [Google Scholar]
- 17.Waheed A et al. IEEE Access 8, 91916–91923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, Tang Y, Zhu Y, Xiao J & Summers RM Medical Image Analysis 67, 101839 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Costa P et al. IEEE Transactions on Medical Imaging 37, 781–791 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Frangi AF, Tsaftaris SA & Prince JL IEEE Transactions on Medical Imaging 37, 673–679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie D et al. IEEE Transactions on Biomedical Engineering 65, 2720–2730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou T, Fu H, Chen G, Shen J & Shao L IEEE Transactions on Medical Imaging 39, 2772–2781 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Karras T, Aila T, Laine S & Lehtinen J In International Conference on Learning Representations (2018). [Google Scholar]
- 24.El Emam K & Hoptroff R Cutter Executive Update 19 (2019). [Google Scholar]
- 25.Chen D, Yu N, Zhang Y & Fritz M In Proceedings of the 2020 ACM SIGSAC Conference on Computer and Communications Security, 343–362 (2020). [Google Scholar]
- 26.Cheng V, Suriyakumar VM, Dullerud N, Joshi S & Ghassemi M In Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, 149–160 (2021). [Google Scholar]
- 27.Xu C et al. IEEE Transactions on Information Forensics and Security 14, 2358–2371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torkzadehmahani R, Kairouz P & Paten B In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, 0–0 (2019). [Google Scholar]
- 29.Chang Q et al. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 13856–13866 (2020). [Google Scholar]
- 30.Yale A et al. Neurocomputing 416, 244–255 (2020). [Google Scholar]
- 31.Jordon J, Yoon J & Van Der Schaar M In International Conference on Learning Representations (2018). [Google Scholar]
- 32.Movshovitz-Attias Y, Kanade T & Sheikh Y In European Conference on Computer Vision, 202–217 (Springer, 2016). [Google Scholar]
- 33.Wan C & Jones DT Nature Machine Intelligence 2, 540–550 (2020). [Google Scholar]
- 34.Bolanos LA et al. Nature Methods 1–4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R et al. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 2145–2155 (2019). [Google Scholar]
- 36.Shapiro A et al. Patterns 2, 100188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salimans T et al. In Advances in Neural Information Processing Systems (2016).
- 38.Zhou S et al. In International Conference on Learning Representations (2019). [Google Scholar]
- 39.Choi E et al. In Machine Learning for Healthcare, 286–305 (PMLR, 2017). [Google Scholar]
- 40.Chen J, Chun D, Patel M, Chiang E & James J BMC Medical Informatics and Decision Making 19, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ive J et al. npj Digital Medicine 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tucker A, Wang Z, Rotalinti Y & Myles P npj Digital Medicine 3, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Yan C, Lasko TA, Sun J & Malin BA Journal of the American Medical Informatics Association 28, 596–604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stupp C Fraudsters used ai to mimic ceo’s voice in unusual cybercrime case. Wall Street Journal (2018). [Google Scholar]
- 45.Finlayson SG et al. Science 363, 1287–1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gafni O, Wolf L & Taigman Y In Proceedings of the IEEE/CVF International Conference on Computer Vision, 9378–9387 (2019). [Google Scholar]
- 47.Zhu B, Fang H, Sui Y & Li L In Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society, 414–420 (2020). [Google Scholar]
- 48.Wosik J et al. Journal of the American Medical Informatics Association (2020). [Google Scholar]
- 49.Dick PK How to build a universe that doesn’t fall apart two days later. I Hope I Shall Arrive Soon. Doubleday; (1978). [Google Scholar]
- 50.Tzachor A, Whittlestone J, Sundaram L et al. Nature Machine Intelligence 2, 365–366 (2020). [Google Scholar]
- 51.Jiang Y, Chen H, Loew M & Ko H IEEE Journal of Biomedical and Health Informatics (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Chen J & Marathe M ACM Transactions on Spatial Algorithms and Systems 6, 1–39 (2020). [Google Scholar]
- 53.Bao H, Zhou X, Zhang Y, Li Y & Xie Y In Proceedings of the 28th International Conference on Advances in Geographic Information Systems, 273–282 (2020). [Google Scholar]
- 54.Bengio Y et al. In International Conference on Learning Representations (2020). [Google Scholar]
- 55.El Emam K, Mosquera L, Jonker E & Sood H JAMIA open 4, ooab012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waymo. Off road, but not offline: How simulation helps advance our waymo driver. Waymo; (2020). [Google Scholar]
- 57.Wu J, Yildirim I, Lim JJ, Freeman B & Tenenbaum J In Advances in Neural Information Processing Systems, 127–135 (2015). [Google Scholar]
- 58.Varol G et al. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 109–117 (2017). [Google Scholar]
- 59.Shrivastava A et al. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2107–2116 (2017). [Google Scholar]
- 60.Sankaranarayanan S, Balaji Y, Jain A, Nam Lim S & Chellappa R In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 3752–3761 (2018). [Google Scholar]
- 61.Prakosa A et al. IEEE Transactions on Medical Imaging 32, 99–109 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Mahmood F, Chen R, Sudarsky S, Yu D & Durr NJ Physics in Medicine & Biology 63, 185012 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Incetan K et al. Medical Image Analysis (2021). [DOI] [PubMed] [Google Scholar]
- 64.Beede E et al. In Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems, 1–12 (2020). [Google Scholar]
- 65.Johnson-Roberson M et al. In Proceedings in the IEEE International Conference on Robotics and Automation, 746–753 (2017). [Google Scholar]
- 66.Qiu W & Yuille A In European Conference on Computer Vision, 909–916 (Springer, 2016). [Google Scholar]
- 67.Ramanagopal MS, Anderson C, Vasudevan R & Johnson-Roberson M IEEE Robotics and Automation Letters 3, 3860–3867 (2018). [Google Scholar]
- 68.Naeem MF, Oh SJ, Uh Y, Choi Y & Yoo J In International Conference on Machine Learning, 7176–7185 (PMLR, 2020). [Google Scholar]
- 69.Sajjadi MS, Bachem O, Lucic M, Bousquet O & Gelly S In Advances in Neural Information Processing Systems (2018).
- 70.Alaa AM, van Breugel B, Saveliev E & van der Schaar M In International Conference on Machine Learning (2021). [Google Scholar]
- 71.Senior AW et al. Nature 1–5 (2020). [Google Scholar]
- 72.Ogden PJ, Kelsic ED, Sinai S & Church GM Science 366, 1139–1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheridan C Novartis, sarepta join dyno’s enterprise to boldly go to new gene therapy frontier. BioWorld (2020). [Google Scholar]
- 74.Zhavoronkov A et al. Nature Biotechnology 37, 1038–1040 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Rotemberg V et al. Scientific data 8, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X et al. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2097–2106 (2017). [Google Scholar]
- 77.Lu MY et al. Nature Biomedical Engineering (2021). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


