Abstract
In the yeast Saccharomyces cerevisiae lipid particles harbor two acyltransferases, Gat1p and Slc1p, which catalyze subsequent steps of acylation required for the formation of phosphatidic acid. Both enzymes are also components of the endoplasmic reticulum, but this compartment contains additional acyltransferase(s) involved in the biosynthesis of phosphatidic acid (K. Athenstaedt and G. Daum, J. Bacteriol. 179:7611–7616, 1997). Using the gat1 mutant strain TTA1, we show here that Gat1p present in both subcellular fractions accepts glycerol-3-phosphate and dihydroxyacetone phosphate as a substrate. Similarly, the additional acyltransferase(s) present in the endoplasmic reticulum can acylate both precursors. In contrast, yeast mitochondria harbor an enzyme(s) that significantly prefers dihydroxyacetone phosphate as a substrate for acylation, suggesting that at least one additional independent acyltransferase is present in this organelle. Surprisingly, enzymatic activity of 1-acyldihydroxyacetone phosphate reductase, which is required for the conversion of 1-acyldihydroxyacetone phosphate to 1-acylglycerol-3-phosphate (lysophosphatidic acid), is detectable only in lipid particles and the endoplasmic reticulum and not in mitochondria. In vivo labeling of wild-type cells with [2-3H, U-14C]glycerol revealed that both glycerol-3-phosphate and dihydroxyacetone phosphate can be incorporated as a backbone of glycerolipids. In the gat1 mutant and the 1-acylglycerol-3-phosphate acyltransferase slc1 mutant, the dihydroxyacetone phosphate pathway of phosphatidic acid biosynthesis is slightly preferred as compared to the wild type. Thus, mutations of the major acyltransferases Gat1p and Slc1p lead to an increased contribution of mitochondrial acyltransferase(s) to glycerolipid synthesis due to their substrate preference for dihydroxyacetone phosphate.
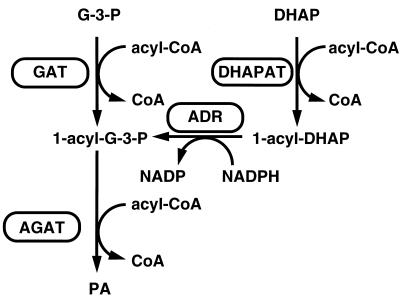
Phosphatidic acid (PA) is a key intermediate in the formation of glycerophospholipids and triacylglycerols. Two precursors, namely, glycerol-3-phosphate (G-3-P) and dihydroxyacetone phosphate (DHAP), can serve as substrates for acylation reactions; the first step of acylation leads to the formation of 1-acylglycerol-3-phosphate (lysophosphatidic acid; LPA) and 1-acyl-DHAP, respectively. Prior to the second step of acylation, 1-acyl-DHAP is reduced in an NADPH-dependent reaction yielding LPA. In the last step of this reaction sequence, LPA formed from either G-3-P or DHAP is converted to PA as summarized in Fig. 1.
FIG. 1.
Pathways of phosphatidic acid (PA) biosynthesis in the yeast. Metabolites: G-3-P, glycerol-3-phosphate; DHAP, dihydroxyacetone phosphate; 1-acyl-G-3-P, 1-acylglycerol-3-phosphate; 1-acyl-DHAP, 1-acyldihydroxyacetone phosphate; PA, phosphatidic acid. Enzymes: GAT, glycerol-3-phosphate acyltransferase; DHAPAT, dihydroxyacetone phosphate acyltransferase; ADR, 1-acyldihydroxyacetone phosphate reductase; AGAT, 1-acylglycerol-3-phosphate acyltransferase; CoA, coenzyme A.
In mammalian cells, G-3-P acyltransferase (GAT), DHAP acyltransferase (DHAPAT) and 1-acylglycerol-3-phosphate acyltransferase (AGAT) activities are distributed among mitochondria, the microsomal fraction, and peroxisomes (11, 13, 27). Whereas in mitochondria only G-3-P and in peroxisomes only DHAP serve as substrates for the acylation reaction, both precursors can be utilized for acylation reactions in the microsomal fraction (3). In plant cells, GAT activity was localized to mitochondria, microsomes, and plastids (20). In contrast to animal cells, plant cells and bacteria lack the DHAP acylation pathway (8, 27).
In the yeast Saccharomyces cerevisiae the highest specific activity of GAT was detected in lipid particles (4, 29). This subcellular compartment consists of a hydrophobic core formed from neutral lipids (mainly triacylglycerols and steryl esters) which is surrounded by a phospholipid monolayer with only a small amount of proteins embedded in it (17). Recent experiments conducted at our laboratory (1) demonstrated that lipid particles contain only one GAT activity catalyzed by the putative Gat1p and one AGAT activity that was attributed to the product of SLC1. The second subcellular site of G-3-P acylation in yeast is the endoplasmic reticulum; mitochondria and other organelles did not show significant acylation activities (1, 29). GAT activity of the endoplasmic reticulum is mainly catalyzed by Gat1p, which is thus located in two distinct compartments. Like GAT, AGAT activity was detected not only in lipid particles but also in the endoplasmic reticulum (microsomes obtained at 30,000 × g [30,000 × g microsomes]). The microsomal AGAT activity was partially attributed to Slc1p, but LPA acylation is also catalyzed by additional acyltransferase(s) (1). These results were obtained by using acylation mutants (21, 27) in combination with techniques of yeast subcellular fractionation (see reference 28). Earlier work by Tillman and Bell (26) and Racenis et al. (22) with total yeast membrane fractions had not been able to distinguish clearly between different GAT and AGAT activities.
In animal cells, the DHAP pathway is obligatory for ether lipid biosynthesis (10). In the yeast, DHAP was shown to be incorporated into PA (12), which serves as a precursor for glycerophospholipid and triacylglycerol formation. The possible occurrence of ether lipids in yeast has been a long-standing matter of dispute. Recently, it was shown by analysis of isolated yeast organelle membranes that alkylether lipids may indeed be present in the yeast S. cerevisiae (24). DHAPAT activity was detected in Saccharomyces carlsbergensis (12) and S. cerevisiae (23). Tillman and Bell (26) suggested that acylation of the precursor G-3-P and that of the precursor DHAP are catalyzed by the same enzyme. In contrast, Racenis et al. (22) favored the view that GAT and DHAPAT activities are attributed to different enzymes.
In this work we tested GAT and DHAPAT activities in different subcellular fractions of the yeast by making use of the availability of gat1 and slc1 mutants and the elaborate methods of subcellular fractionation applied in our laboratory. We addressed the question of whether there is only one enzyme in yeast acylating both precursors, G-3-P and DHAP. In addition, we tested the utilization of G-3-P and DHAP as precursors of glycerolipids in vivo in wild-type strains and gat1, slc1, and gat1slc1 mutant strains. Furthermore, the subcellular localization of 1-acyl-DHAP reductase was studied to obtain a more detailed view of the enzymatic steps involved in the biosynthesis of PA in yeast.
MATERIALS AND METHODS
Strains and culture conditions.
The wild-type yeast strains SJ21R (MATa, ura3-52, leu2-3,112, ade1) and DBY746 (MATα, his3-Δ1, leu2-3, leu2-112, ura3-52, trpl-289), the yeast mutants YMN5 (MATa, ura3-52, leu2-3,112, ade1, slc1Δ2::LEU2) (kindly provided by M. Nagiec and R. Dickson) and TTA1 (MATα, his3-Δ1, leu2-3, leu2-112, ura3-52, trpl-289) (kindly provided by R. Bell), and the gat1slc1 double mutant (1) were used throughout this study.
Cells were grown aerobically in 2-liter Erlenmeyer flasks to the late logarithmic phase at 30°C in YPD medium, pH 5.5, containing 1% yeast extract (Oxoid), 2% peptone (Oxoid), and 2% glucose (Merck). Five hundred milliliters of culture medium was inoculated with 0.5 ml of a preculture grown aerobically for 48 h in YPD medium.
Labeling of yeast cells with [2-3H, U-14C]glycerol.
Yeast cells were grown aerobically in 500 ml of minimal medium (25) containing 2% ethanol–0.2% glucose supplemented with the appropriate amino acids. Cells were inoculated with a preculture to an optical density at 600 nm of 0.1, cultivated to an optical density at 600 nm of 4 to 5, harvested, and resuspended in 500 ml of the same fresh medium containing 10 μCi of [2-3H]glycerol (specific activity, 200 mCi/mmol) and 10 μCi of [U-14C]glycerol (specific activity, 180 mCi/mmol). Labeling was continued for 12 h at 30°C with vigorous shaking.
Isolation and characterization of subcellular fractions.
Lipid particles at high purity were obtained by the method of Leber et al. (17) from cells grown to the late logarithmic phase. Microsomal fractions used in this study were prepared as described by Zinser et al. (29), and mitochondria were isolated by the method of Daum et al. (5). Relative enrichment of markers and cross-contamination were similar to those described by Zinser and Daum (28).
Protein was quantitated by the method of Lowry et al. (19) by using bovine serum albumin as a standard. Proteins were precipitated from the aqueous phase with trichloroacetic acid (final concentration, 10%). The protein pellet was solubilized in 0.1% sodium dodecyl sulfate–0.1% NaOH. Prior to protein analysis of the lipid particle fraction samples were delipidated. Nonpolar lipids were extracted with 2 volumes of diethyl ether, the organic phase was withdrawn, residual diethyl ether was removed under a stream of nitrogen, and proteins were precipitated from the extracted aqueous phase as described above.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out by the method of Laemmli (16). Samples were dissociated at 37°C. Western blot analysis was performed as described by Haid and Suissa (9), and immunoreactive proteins were detected by enzyme-linked immunosorbent assay with rabbit antisera as the first antibody and goat anti-rabbit immunoglobulin G linked to peroxidase or phosphatase as the second antibody.
Enzyme analysis.
Enzymatic activities of GAT and AGAT were measured as described by Schlossman and Bell (23). Total yeast homogenate (200 μg of protein), lipid particles (20 μg of protein), or microsomes or mitochondria (50 μg of protein) were used as the enzyme sources. The assay mixture contained 75 nmol of [14C]G-3-P (0.4 μCi), 8 nmol of oleoyl-coenzyme A, 15 μg of dithiothreitol, and 0.2 mg of bovine serum albumin in 0.2 ml of 4 mM NaF–2 mM MgCl2–37.5 mM Tris-Cl− (pH 7.5). After 3 and 6 min of incubation at 30°C lipids were extracted with 3 ml of chloroform-methanol (1:2 [vol/vol]) in the presence of 0.7 ml of 1% perchloric acid. The organic phase was washed three times, with 2 ml of 1% perchloric acid each time, and radioactivity was measured by liquid scintillation counting (1500 Tricarb, Beckman). The lipid extract was applied to high-performance thin-layer chromatography (HPTLC) plates (silica gel 60 plates; Merck, Darmstadt, Germany), and chromatograms were developed in an ascending manner by using the solvent system chloroform/methanol/water/acetic acid (65:35:3.8:0.2; per volume). After chromatographic separation the radioactively labeled lipids formed during the assay were detected by TLC scanning with a Tracemaster 20 Automatic TLC-Linear Analyzer (Berthold). In addition, lipids were visualized on HPTLC plates by staining with iodine vapor, bands were scraped off, and radioactivity was measured by liquid scintillation counting by using LSC Safety (Baker, Deventer, The Netherlands) plus 5% water as a scintillation cocktail.
Enzyme activity of DHAPAT was measured as described by Bates and Saggerson (2). Total yeast homogenate (2 to 3 mg of protein), lipid particles (40 to 80 μg of protein), or microsomes or mitochondria (0.3 to 1.0 mg of protein) were used as the enzyme source. Samples were incubated at 30°C in a final volume of 1 ml of a solution containing 120 mM KCl, 50 mM Tris-Cl− (pH 7.5), 4 mM MgCl2, 8 mM NaF, 4 mg of bovine serum albumin, 65 nmol of oleoyl-CoA, 500 nmol of [U-14C]fructose-1,6-bisphosphate (0.4 μCi), 0.44 U of aldolase, and 28 U of triose phosphate isomerase. A 600-μl portion of the reaction mixture was preincubated for 16 min at 30°C prior to the addition of 400 μl of samples containing each enzyme source. When measurement of 1-acyl-DHAP reductase (ADR) activity was included, 80 mM NADPH was added to the reaction mixture. The assay was performed for 10 min at 30°C and was terminated by the addition of 3 ml of chloroform-methanol (1:2 [vol/vol]) and 0.7 ml of 1% perchloric acid. Lipid extracts were washed as described above for the GAT assay. 1-Acyl-DHAP, LPA, and PA were separated by TLC (on HPTLC plates) by using the solvent system chloroform/methanol/acetic acid/5% sodium metabisulfate (100:40:12:4; per volume).
Lipid analysis.
Lipids of whole yeast cells were extracted by the procedure of Folch et al. (6) after disruption of cells with glass beads. Individual phospholipids were separated by two-dimensional TLC on Silica gel 60 plates (Merck) by using chloroform–methanol–25% NH3 (65:35:5; per volume) as the first developing solvent and chloroform/acetone/methanol/acetic acid/water (50:20:10:10:5; per volume) as the second developing solvent. Phospholipids were visualized on TLC plates by staining with iodine vapor, scraped off, and extracted from the silica gel three times with 2 ml of chloroform-methanol (1:4 [vol/vol]) each time. The organic phase was removed under a stream of nitrogen, and radioactivity was measured by liquid scintillation counting by using LSC Safety (Baker) plus 5% water as a scintillation cocktail.
For the analysis of neutral lipids, extracts were applied to Silica gel 60 plates, and chromatograms were developed in an ascending manner by using the solvent system light petroleum/diethyl ether/acetic acid (70:30:2; per volume). Neutral lipids were visualized on TLC plates by staining with iodine vapor and extracted three times with 2 ml of chloroform-methanol (2:1 [vol/vol]) each time, and radioactivity was measured by liquid scintillation counting as described above.
RESULTS
One aim of this study was to investigate GAT and DHAPAT activities in different organelles of the yeast S. cerevisiae. The strain TTA1, which contains a point mutation in the GAT1 gene (27), and the slc1 deletion strain YMN5 (21), which is defective in AGAT (1), were used to address this question.
Table 1 shows the enrichment of GAT and DHAPAT activities in various yeast subcellular fractions. For these experiments lipid particles, microsomes, and mitochondria were prepared from the same culture although by using the different protocols appropriate for the isolation of each organelle. This strategy was chosen to avoid the influence of variable growth and spheroplasting conditions on enzyme activities. In the wild-type strain DBY746, the highest enrichment of both GAT and DHAPAT activities was detected in lipid particles, although the enrichment of GAT was approximately three- to fourfold higher than that of DHAPAT in this compartment. In microsomal fractions of DBY746, GAT activity was three- to fivefold more highly enriched than DHAPAT activity. In contrast, mitochondria of the wild-type strain were only slightly enriched in GAT activity, whereas the enrichment factor for DHAPAT was approximately 2.5 to 3. Thus, mitochondria appear to be the only compartment with a preference for DHAP over G-3-P as an acylation substrate. Other subcellular fractions, such as vacuoles or 100,000 × g microsomes, were practically devoid of acyltransferase activities with both substrates.
TABLE 1.
GAT and DHAPAT activities in subcellular fractions of wild-type strain and the gat1 mutant strain TTA1
| Fraction | DBY746 (wild type)
|
TTA1 (gat1)
|
||
|---|---|---|---|---|
| Relative specific activity ofa:
|
Relative specific activity ofa:
|
|||
| GAT | DHAPAT | GAT | DHAPAT | |
| Homogenate | 1 (0.68 ± 0.20) | 1 (0.15 ± 0.04) | 1 (0.05 ± 0.01) | 1 (0.03 ± 0.01) |
| Lipid particles | 31.0 | 9.3 | Trace | NDb |
| Microsomes (30,000 × g) | 5.8 | 2.4 | 4.0 | 1.3 |
| Microsomes (40,000 × g) | 5.0 | 0.9 | 1.2 | 0.4 |
| Mitochondria | 1.5 | 2.6 | 0.9 | 1.0 |
Mean values from three independent experiments are shown; the mean deviation was ±15%. Specific activities (nanomoles per milligram per min) are shown in parentheses.
ND, not detectable.
Lipid particles of the gat1 mutant TTA1 contain neither significant activity of GAT nor that of DHAPAT (Table 1). A negligible amount of PA which is formed from G-3-P as precursor can be attributed to catalysis of Slc1p. This result was in agreement with previous experiments (1) which had demonstrated that a gat1slc1 double mutant did not show any activity of G-3-P acylation in lipid particles. Microsomes of TTA1, which lack Gat1p but contain at least one other acylating system (1), showed acyltransferase activity with both substrates, G-3-P and DHAP (Table 1). The ratios of the enrichment factors of GAT:DHAPAT in 30,000 × g and 40,000 × g microsomes were similar to those of the corresponding wild-type strain, DBY746. In mitochondria of TTA1 a distinct preference for DHAP acylation was not observed. It has to be mentioned, however, that the specific activities of GAT and DHAPAT are very low in this organelle of the mutant strain. Similar to those of the wild-type strain, mitochondria of the slc1 deletion strain YMN5 exhibit a twofold higher enrichment of DHAPAT activity as compared to GAT activity.
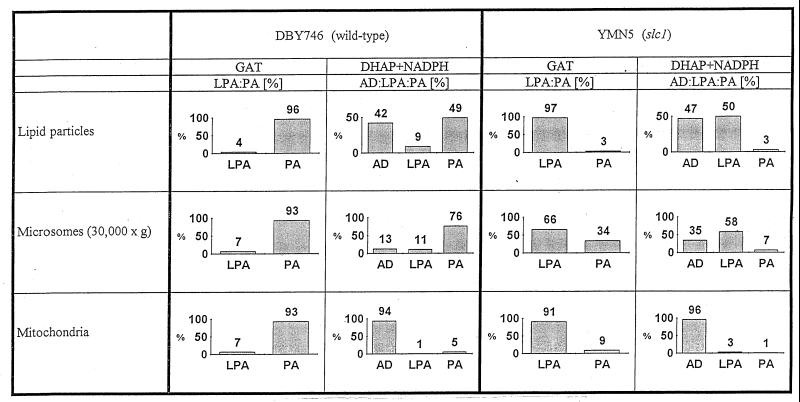
When G-3-P was used as a substrate for acylation, all enzyme sources from the wild-type strain used for in vitro assays formed the intermediate LPA only at low concentrations (Fig. 2); in all cases the main product was PA. Acylation of DHAP led to the formation of 1-acyl-DHAP; when NADPH was present in the assay mixture, LPA and PA were formed in the assay with lipid particles or 30,000 × g microsomes of wild-type cells as the enzyme source (Fig. 2). In the absence of NADPH, however, 1-acyl-DHAP was not further converted to LPA and PA. In contrast, mitochondria were not able to catalyze the reduction reaction in the presence of NADPH, and 1-acyl-DHAP accumulated in this organelle in vitro. Minor amounts of PA formed in this assay can be attributed to cross-contamination of the mitochondrial fraction with microsomes. These experiments demonstrated that only lipid particles and the endoplasmic reticulum (30,000 × g microsomes) harbor ADR activity. The dual localization of this enzyme is reminiscent to that of other proteins present in lipid particles and the endoplasmic reticulum, such as Gat1p and Slc1p (1), Erg6p (17), and Erg1p (18).
FIG. 2.
Products of GAT and DHAPAT in subcellular fractions of the wild-type strain DBY746 and the slc1 deletion strain YMN5. LPA, lysophosphatidic acid (1-acylglycerol-3-phosphate); AD, 1-acyldihydroxyacetone phosphate; PA, phosphatidic acid.
Lipid particles of the slc1 deletion strain YMN5 are unable to further acylate LPA to PA due to the defect in AGAT (1). As a consequence, the reaction sequence stopped after the first step of acylation and LPA accumulated when G-3-P was used as a precursor (Fig. 2). When DHAP was used as a substrate and NADPH was added to the assay mixture both intermediates, 1-acyl-DHAP and LPA, appeared, confirming the presence of ADR activity in lipid particles. Microsomes of the slc1 deletion strain, which harbor at least a second AGAT activity, were still able to form PA from both precursors, G-3-P and DHAP. Similar to the findings for the assay with lipid particles, 1-acyl-DHAP and LPA appeared as the main products in the DHAP assay with 30,000 × g microsomes of the mutant as an enzyme source. When G-3-P was used as the substrate mitochondria of YMN5 mainly formed LPA and only a small amount of PA. DHAP acylation with mitochondria of YMN5 as the enzyme source resulted mainly in formation of 1-acyl-DHAP, even in the presence of NADPH. The small amounts of PA and LPA which were formed from DHAP can be attributed to contamination of mitochondria with microsomes. These results confirmed data obtained with the wild-type strain.
To address the question of whether the DHAP pathway is relevant also in vivo cells were labeled with [2-3H, U-14C]glycerol for 12 h and the 3H/14C ratio in major glycerolipids was measured. If G-3-P is used for glycerolipid formation the [2-3H] labeling of glycerol should be retained; if glycerolipids are synthesized via the DHAP pathway the [2-3H] labeling of glycerol should be lost during conversion of glycerol to DHAP. As a consequence, a lower 3H/14C ratio is indicative of an increased contribution of the DHAP pathway to glycerolipid biosynthesis. Control experiments demonstrated that only the glycerol backbone of glycerolipids, not the fatty acids, was labeled in this in vivo assay. In Table 2, the 3H/14C ratios of glycerolipids formed during such experiments conducted by using the wild-type and mutant strains are shown. In the gat1 mutant, TTA1, 3H/14C values for all glycerolipids tested were lower than in the corresponding wild-type strain. This result suggests that lack of the major GAT, Gat1p, in TTA1 led to a moderate but significant preference for use of DHAP as a substrate for glycerolipid formation. Similarly, deletion of the SLC1 gene encoding the major AGAT resulted in a decrease of the 3H/14C ratio as compared to the corresponding wild-type strain, SJ21R (Table 2). For the gat1slc1 double mutant the 3H/14C ratios of all glycerolipids are intermediate in value between those of TTA1 and YMN5, reflecting the different wild-type backgrounds of the gat1 and slc1 mutations.
TABLE 2.
3H/14C ratios in different glycerolipids after labeling of wild-type and acyltransferase mutant strains with [2-3H, U-14C]glycerol
| Glycerolipid |
3H/14C ratio after labeling ofa:
|
||||
|---|---|---|---|---|---|
| DBY746 (wild type) | TTA1 (gat1) | SJ21R (wild type) | YMN5 (slc1) | gat1slc1 | |
| Phosphatidic acid | 3.82 | 2.99 | 2.47 | 2.22 | 2.96 |
| Phosphatidylcholine | 3.47 | 3.02 | 2.80 | 2.09 | 2.79 |
| Phosphatidylethanolamine | 3.67 | 3.02 | 2.77 | 1.99 | 2.66 |
| Phosphatidylinositol | 3.42 | 3.02 | 2.64 | 2.12 | 2.86 |
| Phosphatidylserine | 3.72 | 3.35 | 2.60 | 2.10 | 3.10 |
| Cardiolipin | 3.16 | 2.50 | 2.13 | 1.73 | 2.45 |
| Diacylglycerols | 3.57 | 2.96 | 3.24 | 2.28 | 2.48 |
| Triacylglycerols | 3.58 | 3.10 | 3.07 | 2.24 | 3.55 |
Double labeling experiments with [2-3H, U-14C]glycerol were carried out as described in the Materials and Methods section. Decrease of the 3H/14C ratio is indicative of increased contribution of the DHAP pathway to glycerolipid formation. Experiments were carried out in triplicate; the mean deviation was ±5%.
DISCUSSION
In the yeast S. cerevisiae, PA, a key intermediate in lipid metabolism, can be formed in vitro from the precursors G-3-P and DHAP. Contradictory evidence has been presented about the number of enzymes involved in the first step of acylation forming LPA or 1-acyl-DHAP and the substrate specificity of the enzyme(s). Tillman and Bell (26) concluded from their experiments that one enzyme can acylate both G-3-P and DHAP. Racenis et al. (22), on the other hand, argued that although temperature optima and kinetics of inactivation by heat were the same for GAT and DHAPAT activities some differences exist, such as the inhibitory effect of N-ethylmaleimide and pH optima. Tillman and Bell (26) and Racenis et al. (22) used crude yeast membrane preparations as enzyme sources for their in vitro assays. Thus, their results reflected an “overall” situation of acylation activities in yeast. More recent studies of G-3-P acylation with highly purified subcellular fractions and the use of the two acyltransferase mutant strains TTA1 and YMN5 revealed that more than one acyltransferase system contributes to PA formation in yeast (1). One acylation machinery is dually localized to lipid particles and the endoplasmic reticulum; at least one other acylation system was detected only in the endoplasmic reticulum.
Results presented in this study demonstrate that the major GAT of the yeast, the hypothetical Gat1p, which is located in lipid particles and the endoplasmic reticulum, can acylate both G-3-P and DHAP. Lipid particles of the wild-type strain can acylate both precursors, whereas those of the gat1 mutant strain TTA1 showed neither GAT nor DHAPAT activity. Since lipid particles do not contain a second GAT, both enzymatic activities can be attributed to the same protein. These results are in line with the finding of Tillman and Bell (26) that one acylating enzyme, namely Gat1p, has GAT activity as well as DHAPAT activity.
The results are more complicated for GAT and DHAPAT activities of other subcellular compartments. It has been shown during previous work in our laboratory (1) that Gat1p is not exclusively localized to lipid particles but indeed contributes most prominently to GAT activity of the endoplasmic reticulum (30,000 × g microsomes). Using the gat1 mutant strain TTA1 it was shown, however, that at least another GAT must be present in the microsomal fraction. Although the GAT:DHAPAT activity ratios are similar in 30,000 × g microsomes of wild-type and TTA1 strains (Table 1), the possibility that microsomal acyltransferase activity is derived from more than one enzyme cannot be excluded. In contrast to other organelles tested, mitochondria exhibit a low ratio of GAT activity to DHAPAT activity that suggests the presence of a distinct mitochondrial DHAPAT. This activity cannot be attributed to contamination with other subcellular fractions.
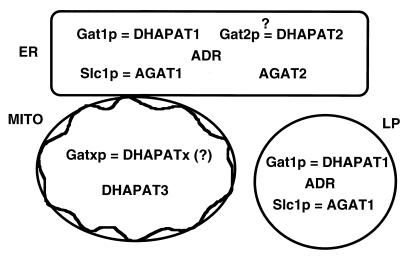
Cell biological and biochemical experiments presented in this paper led us to propose a model of the subcellular distribution of acyltransferase reactions involved in the formation of PA in yeast (Fig. 3). One GAT, namely Gat1p, which can use G-3-P as well as DHAP as a substrate, is located in lipid particles. Gat1p is also present in the endoplasmic reticulum, which in addition contains at least another GAT, a hypothetical Gat2p. This enzyme may also have DHAPAT activity. Alternatively, besides Gat1p additional distinct enzymes for G-3-P and DHAP acylation may be present in the endoplasmic reticulum. The third site of acyltransferase activity is mitochondria, but this compartment appears to harbor a distinct DHAPAT.
FIG. 3.
Scheme of subcellular distribution of enzymes involved in PA biosynthesis in yeast. ER, endoplasmic reticulum; MITO, mitochondria; LP, lipid particles.
Double labeling experiments with [2-3H, U-14C]glycerol demonstrated that both GAT and DHAPAT reactions are relevant in vivo. The fact that the 3H/14C ratios in all glycerolipid species of the gat1 mutant strain TTA1 were lower than in the corresponding wild-type strain indicates that in the absence of Gat1p the overall contribution of the DHAP pathway to glycerolipid biosynthesis is increased. This observation is in good agreement with the cell biological model of PA formation shown in Fig. 3: when Gat1p, which is the only GAT and DHAPAT in lipid particles and the major GAT and DHAPAT in the endoplasmic reticulum, is inactivated, the ratio of cellular GAT activity to DHAPAT activity is shifted towards the DHAP pathway due to the contribution of the mitochondrial DHAPAT.
Similar to the result for TTA1, which lacks Gat1p, deletion of SLC1 results in a shift of the acylation pathway towards utilization of DHAP. This result may serve as another indication that Slc1p can at least in part replace Gat1p, as was suggested earlier (1). A gat1slc1 double mutant exhibited intermediate 3H/14C ratios in all glycerolipids as compared to those for TTA1 and YMN5. Thus, residual acyltransferases present in the gat1slc1 mutant compensate for the lack of Gat1p and Slc1p by catalyzing formation of PA at a level sufficient for balanced cellular growth. This result also indicates that both the G-3-P pathway and the DHAP pathway are active in the double mutant and that the latter is preferred over the former, as contrasted with the situation for the wild-type.
Another important finding revealed during this investigation was the subcellular localization of ADR activity. ADR catalyzes the NADPH-dependent reduction of 1-acyl-DHAP to LPA. Among the subcellular fractions which harbor DHAPAT activity only lipid particles and 30,000 × g microsomes showed ADR activity; mitochondria lack this reductase activity.
An interesting result obtained through in vivo labeling experiments (Results section and Table 2) was the relatively low 3H/14C ratio in cardiolipin of all strains tested compared to the other glycerolipid species. The decreased 3H/14C ratio indicates a greater contribution of the DHAP pathway than the G-3-P pathway to cardiolipin formation. Cardiolipin synthesis occurs in mitochondria, the organelles which have a preference for DHAP acylation over G-3-P acylation. A greater contribution of the DHAP pathway to the synthesis of cardiolipin could be due to the utilization of PA mainly formed from mitochondrion-derived 1-acyl-DHAP. For further metabolic conversion of 1-acyl-DHAP formed by DHAPAT of mitochondria, however, the acylation intermediate has to be transferred to a site of ADR activity where it is reduced to LPA prior to the second step of acylation. 1-Acyl-DHAP is reasonably soluble in an aqueous environment and might thus migrate rather easily from one compartment to another. In respect to the greater contribution of the DHAP pathway to cardiolipin synthesis, however, an alternative possibility of 1-acyl-DHAP migration may be envisaged. 1-Acyl-DHAP formed in mitochondria could be translocated to the “closest” site of ADR activity, which is the mitochondrion-associated endoplasmic reticulum, the so-called MAM fraction (7). At this site, reduction of 1-acyl-DHAP and further acylation could occur. CDP-diacylglycerol formed in the subsequent metabolic step in the MAM subfraction of the endoplasmic reticulum or in mitochondria (14, 15, 29) might serve as a substrate for cardiolipin synthesis in mitochondria. As a result of this (hypothetical) sequence of translocation and conversion steps DHAP might be preferentially incorporated into cardiolipin. Since PA formed in the MAM fraction of the endoplasmic reticulum from G-3-P can also be used for CDP-diacylglycerol formation and incorporation into cardiolipin, part of the glycerol moieties of cardiolipin is also derived from G-3-P.
The complexity of PA formation in yeast requires further characterization of enzymes involved in the first step of acylation of G-3-P and/or DHAP and in the subsequent steps involved in this process. Thus, future efforts will be focused on the identification of the enzymes involved in PA biosynthesis, their characterization at the molecular level, and the interplay of organelles during initial steps of glycerolipid biosynthesis.
ACKNOWLEDGMENTS
We thank R. Dickson and M. Nagiec, and we thank R. Bell for providing yeast strains.
This work was financially supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (project 11491 to G.D. and project 12260 to F.P., and SFB Biomembranes F 706).
REFERENCES
- 1.Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates E J, Saggerson E D. A study of the glycerol phosphate acyltransferase and dihydroxyacetone phosphate acyltransferase activities in rat liver mitochondrial and microsomal fractions. Biochem J. 1979;182:751–762. doi: 10.1042/bj1820751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell R M, Coleman R A. Enzymes of triacylglycerol formation in mammals. In: Boyer P D, editor. The enzymes. New York, N.Y: Academic Press; 1983. pp. 87–111. [Google Scholar]
- 4.Christiansen K. Triacylglycerol synthesis in lipid particles from baker’s yeast (Saccharomyces cerevisiae) Biochim Biophys Acta. 1978;530:78–90. doi: 10.1016/0005-2760(78)90128-5. [DOI] [PubMed] [Google Scholar]
- 5.Daum G, Boehni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 6.Folch J, Lees M, Sloane-Stanley G H. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 7.Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- 8.Gurr M I. The biosynthesis of triacylglycerols. In: Stumpf P K, editor. The biochemistry of plants. 4. Lipids: structure and function. New York, N.Y: Academic Press, Inc.; 1980. pp. 205–248. [Google Scholar]
- 9.Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 10.Hajra A K. Glycerolipid biosynthesis in peroxisomes (microbodies) Prog Lipid Res. 1995;34:343–364. doi: 10.1016/0163-7827(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 11.Hajra A K. Dihydroxyacetone phosphate acyltransferase. Biochim Biophys Acta. 1997;1348:27–34. doi: 10.1016/s0005-2760(97)00120-3. [DOI] [PubMed] [Google Scholar]
- 12.Johnston J M, Paltauf F. Lipid metabolism in inositol deficient yeast, Saccharomyces carlsbergensis. II. Incorporation of labeled precursors into lipids by whole cells and activities of some enzymes involved in lipid formation. Biochim Biophys Acta. 1970;218:431–440. [PubMed] [Google Scholar]
- 13.Jones C L, Hajra A K. Properties of guinea pig liver peroxisomal dihydroxyacetone phosphate acyltransferase. J Biol Chem. 1980;255:8289–8295. [PubMed] [Google Scholar]
- 14.Kelley M J, Carman G M. Purification and characterization of CDP-diacylglycerol synthase from Saccharomyces cerevisiae. J Biol Chem. 1987;262:14563–14570. [PubMed] [Google Scholar]
- 15.Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- 18.Leber R, Landl K, Zinser E, Ahorn H, Spök A, Kohlwein S D, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Morlock K R, McLaughlin J J, Lin Y-P, Carman G M. Phosphatidate phosphatase from Saccharomyces cerevisiae. Isolation of 45- and 104-kDa forms of the enzyme that are differentially regulated by inositol. J Biol Chem. 1991;266:3586–3593. [PubMed] [Google Scholar]
- 21.Nagiec M M, Wells G B, Lester R L, Dickson R C. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- 22.Racenis P V, Lai J L, Das A K, Mullick P C, Hajra A K, Greenberg M L. The acyl dihydroxyacetone phosphate pathway enzymes for glycerolipid biosynthesis are present in the yeast Saccharomyces cerevisiae. J Bacteriol. 1992;174:5702–5710. doi: 10.1128/jb.174.17.5702-5710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlossman D M, Bell R M. Glycerolipid biosynthesis in Saccharomyces cerevisiae: sn-glycerol-3-phosphate and dihydroxyacetone phosphate acyltransferase activities. J Bacteriol. 1978;133:1368–1376. doi: 10.1128/jb.133.3.1368-1376.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneiter, R. Personal communication.
- 25.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 26.Tillman T S, Bell R M. Mutants of Saccharomyces cerevisiae defective in sn-glycerol-3-phosphate acyltransferase. J Biol Chem. 1986;261:9144–9149. [PubMed] [Google Scholar]
- 27.Van den Bosch H. Phosphoglyceride metabolism. Annu Rev Biochem. 1974;43:243–277. doi: 10.1146/annurev.bi.43.070174.001331. [DOI] [PubMed] [Google Scholar]
- 28.Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- 29.Zinser E, Sperka-Gottlieb C D M, Fasch E-V, Kohlwein S D, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eucaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]