Abstract
When Pseudomonas aeruginosa is grown with organosulfur compounds as sulfur sources, it synthesizes a set of proteins whose synthesis is repressed in the presence of sulfate, cysteine, or thiocyanate (so-called sulfate starvation-induced proteins). The gene encoding one of these proteins, PA13, was isolated from a cosmid library of P. aeruginosa PAO1 and sequenced. It encoded a 381-amino-acid protein that was related to several reduced flavin mononucleotide (FMNH2)-dependent monooxygenases, and it was the second in an operon of three genes, which we have named msuEDC. The MsuD protein catalyzed the desulfonation of alkanesulfonates, requiring oxygen and FMNH2 for the reaction, and showed highest activity with methanesulfonate. MsuE was an NADH-dependent flavin mononucleotide (FMN) reductase, which provided reduced FMN for the MsuD enzyme. Expression of the msu operon was analyzed with a transcriptional msuD::xylE fusion and was found to be repressed in the presence of sulfate, sulfite, sulfide, or cysteine and derepressed during growth with methionine or alkanesulfonates. Growth with methanesulfonate required an intact cysB gene, and the msu operon is therefore part of the cys regulon, since sulfite utilization was found to be CysB independent in this species. Measurements of msuD::xylE expression in cysN and cysI genetic backgrounds showed that sulfate, sulfite, and sulfide or cysteine play independent roles in negatively regulating msu expression, and sulfonate utilization therefore appears to be tightly regulated.
Sulfonates are chemically stable compounds which are common xenobiotics released into the environment (22) but which are also natural products that contribute significantly to the global biogeochemical sulfur cycle. They constitute a large proportion of the sulfur found in aerobic soils and together with sulfate esters make up >95% of the sulfur content of soil environments (2). Sulfonates have also been found to accumulate to levels of 20 to 40% of the total organic sulfur in near-surface marine sediments (46). Methanesulfonic acid is the main biogenic sulfur component in the atmosphere, being generated by photooxidation of dimethyl sulfide released by marine ecosystems and subsequently returned to terrestrial environments in precipitation. Bacteria which can utilize methanesulfonate and other naturally occurring alkanesulfonates, such as cysteate, taurine, or isethionate, as sources of sulfur for growth can readily be isolated from soil environments (25, 40), even under nonselective conditions, and several strains that can use these compounds as carbon sources have also been reported (16, 43, 44).
With the exception of taurine (2-aminoethanesulfonate), the detailed enzymology of desulfonation of alkanesulfonates within the sulfur cycle has not yet been reported. In Escherichia coli, taurine is desulfonated to aminoacetaldehyde by the α-ketoglutarate-dependent taurine dioxygenase encoded by the tauD gene (10). Taurine sulfur is released by taurine dioxygenase as sulfite (10). Sulfite also appears to be the direct desulfurization product of cysteate and isethionate in E. coli, since utilization of these compounds by E. coli required an intact sulfite reductase enzyme (45). Synthesis of TauD is repressed in the presence of sulfate or cysteine in the growth medium and is dependent on two very similar LysR-type transcriptional activators, CysB and Cbl (48).
For Pseudomonas aeruginosa, physiological data suggest that release of alkanesulfonate sulfur is also catalyzed by an oxygenase (21), although the enzyme responsible for the reaction has not yet been identified or characterized. As in E. coli, desulfonation activity in whole cells is repressed in the presence of cysteine or sulfate in the growth medium. In pseudomonads thiocyanate also leads to repression of desulfonation (in contrast, E. coli is unable to grow with thiocyanate as a sulfur source [24]). This pattern of regulation corresponds with that observed for a set of 10 proteins in P. aeruginosa whose synthesis is controlled by the sulfur supply to the cell, being synthesized only in the absence of sulfate, cysteine, or thiocyanate (17). These proteins, called sulfate starvation-induced proteins (SSI proteins), have been found in several gram-positive and gram-negative species (24) and have been studied in detail in E. coli, where they constitute a subset of the cysteine regulon products (35, 48, 49).
In this study we have carried out a more detailed investigation of the PA13 protein, one of the SSI proteins of P. aeruginosa. The gene encoding this protein was identified, and the putative operon in which it is located was sequenced. Overexpression studies showed that the encoded proteins are involved in methanesulfonate metabolism and constitute a novel reduced flavin mononucleotide (FMNH2)-dependent desulfonating sulfonatase system.
MATERIALS AND METHODS
Chemicals, bacterial strains, and growth conditions.
All chemicals used as sulfur sources were of the highest quality available and were obtained from Fluka (Buchs, Switzerland). Oligonucleotides were obtained from Microsynth (Balgach, Switzerland). Bacterial strains, plasmids, and phages used in this study are listed in Table 1. All P. aeruginosa strains were grown at 37°C either in Luria-Bertani medium (1) or in MMAA medium, which was a succinate-salts medium supplemented with all of the proteinogenic amino acids except methionine and cysteine, as previously described (3). Sulfur sources were added as described in Results, to a final concentration of 250 to 500 μM. Antibiotics were added at the following concentrations (micrograms per milliliter): ampicillin, 100 for E. coli; tetracycline, 25 for E. coli and 125 for P. aeruginosa; gentamicin, 15 for E. coli and 200 for P. aeruginosa; streptomycin and carbenicillin, 500 for P. aeruginosa; and kanamycin, 25 for E. coli. Sulfur-limited solid media were prepared by addition of 0.6% SeaPlaque agarose (FMC BioProducts). 5-Bromo-4-chloro-3-indolylgalactoside (X-Gal) (80 μg/ml) was added when necessary. Growth in liquid cultures was monitored as the optical density at 650 nm (OD650), and cultures used for enzyme assays were harvested in the mid-exponential phase (OD650 = 0.5 to 0.7).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant features | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| S17-1 | hsdR thi pro recA; RP4 integrated into the chromosome (kan::Tn7 ter::Mu) | 41 |
| BL21(DE3) | F−ompT [lon] hsdSB (rB− mB−; B strain); λDE3 lysogen with T7 RNA polymerase | Novagen |
| P. aeruginosa | ||
| PAO1 | Prototroph | 14 |
| PAO-CB | cysB::Gm | This study |
| CHA-CB | cysB::Gm | 6 |
| AX18 | cysI::miniTn5Tc | 17 |
| AC309 | cysN::miniTn5Tc | 17 |
| SLF2 | msuD::xylE, xylE antiparallel to msuD | This study |
| SLF3 | msuD::xylE, xylE parallel to msuD | This study |
| SLF4 | cysI::miniTn5Tc msuD::xylE | This study |
| SLF5 | cysN::miniTn5Tc msuD::xylE | This study |
| Plasmids | ||
| pBluescript II KS | Cloning vector, Ampr | Stratagene |
| pET24b | Expression vector, Kmr | Novagen |
| pIA3 | pUC18Mob cysB::Gm | 6 |
| pEX100T | ColE1 AmprsacB oriT | 39 |
| pUCGM1 | ColE1 Ampr Ωaac1 | 38 |
| pX1918GT | ColE1 AmprxylE Gmr | 39 |
| pME3087 | ColE1 TcroriT | 29 |
| pME4006 | pLAFR3 cosmid carrying the msu locus | This study |
| pME4088 | pME3087 carrying an 8-kb BamHI/HindIII fragment with msuD::xylE | This study |
| pME4095 | pBluescript KS carrying a 7-kb EcoRV fragment with msuEDC | This study |
| pME4503 | pET24b with msuEDC genes | This study |
| pME4529 | pET24b with the msuC gene | This study |
| pME4531 | pET24b with the msuE gene | This study |
| pME4532 | pET24b with msuED genes | This study |
| Oligonucleotides | ||
| 960377 | 5′-CCGAGACGGTCGGCGAAA-3′ | This study |
| 960873 | 5′-CAGGAAACATGGCTTGGGAAAC-3′ | This study |
| M13 | 5′-GTAAAACGACGGCCAGT-3′ | This study |
| SlfA-N | 5′-CCACAGGGAAATCATATGACCAGCC-3′a | This study |
| SlfC-N | 5′-GAAACCCCATATGAATGCGAAG-3′ | This study |
| orfM500 | 5′-CAASRGCCGYATCCGCC-3′ | This study |
| orfM970rev | 5′-TCTTCYTRRTGCGGATAGCC-3′ | This study |
| PA13F1 | 5′-CAAAGTCGTCGCCGTCTCC-3′ | This study |
| PA13R1 | 5′-AAGTCCACCGCCTCGCC-3′ | This study |
Nucleotides in boldface were changed from the template sequence to introduce restriction sites.
Enzyme assays.
Arylsulfatase was assayed by using 4-nitrocatecholsulfate as the substrate as described previously (3). Catechol-2,3-dioxygenase was quantified by measuring the A375 in a whole-cell continuous assay (19). NADH-dependent flavin mononucleotide (FMN) reductase activity was measured as the disappearance of NADH at 340 nm, in a reaction mixture (1 ml) consisting of 100 μM FMN, 100 μM NADH, and 50 mM Tris-HCl, pH 7.5. The reaction was started by addition of a suitable amount of enzyme, and the mixture was incubated at 25°C. No electron acceptor other than oxygen was provided, and so initial, linear reduction rates were determined, before oxygen became rate limiting. Sulfonate desulfonation was assayed in a reaction mixture (0.5 ml) consisting of 5 mM substrate, 100 μM FMN, 5 mM NADH, and 50 mM Tris-HCl (pH 7.5), and the reaction was started by addition of cell extract (ca 0.5 mg of protein). Sulfite release was quantified at 430 nm after a suitable time by diluting samples into stop buffer [125 μg of 5,5′-dithio-bis(2-nitrobenzoate) per ml, 62 mM EDTA]. Desulfonation assay mixtures were shaken at 37°C (180 rpm). α-Ketoglutarate-dependent taurine dioxygenase activity was measured as previously described (10). Oxygen consumption was measured with a Clark oxygen electrode (Rank Bros, Bottisham, United Kingdom). Formaldehyde was determined by chemical derivatization, and formaldehyde dehydrogenase was measured in an NAD-dependent assay (31). Protein was measured by the method of Bradford (4), with bovine serum albumin as the standard.
DNA manipulations.
For plasmid isolation, restriction enzyme digestion, and transformation, published procedures were used (1). Where required, DNA fragments were isolated from agarose gels with GeneClean (Bio101, La Jolla, Calif.) or Qiaquick (Qiagen, Basel, Switzerland) spin columns. PCR was carried out in a Trio Block (BioMetra, Göttingen, Germany). Standard reaction mixtures consisted of 50 pmol of primers, 200 nmol of deoxynucleoside triphosphates, 0.2 U of Taq DNA polymerase (Fermentas, Vilnius, Lithuania), and 1 to 100 ng of template in a final volume of 50 μl. Dimethyl sulfoxide (10%, vol/vol) was routinely added, and for cloning purposes the Taq polymerase was supplemented with 1 U of Vent DNA polymerase (New England Biolabs, Bad Schwalbach, Germany). When required, PCR-derived fragments were sequenced to confirm that no point mutations had occurred. Southern analysis was carried out by using digoxigenin-labelled probes which had been labelled either by random-primed labelling (1) or by using PCR of a suitable DNA fragment to incorporate digoxigenin-11-dUTP. Hybridization on nylon membranes (Hybond; Amersham) was carried out at 68°C in 500 mM sodium phosphate (pH 7.2)–7% sodium dodecyl sulfate (SDS) for 24 to 48 h, and the membranes were washed at 65°C with 50 mM sodium phosphate (pH 7.2)–1% SDS twice for 30 min each.
Isolation of the msuEDC genes and construction of insertion and deletion mutants.
The cosmid pME4006, which contained the msuEDC operon, was identified in a gene bank of P. aeruginosa PAO1 constructed in the pLAFR3 cosmid vector (50) by PCR with primers PA13F1 and PA13R1. A 7.5-kb EcoRV fragment of pME4006 was subcloned into pBluescript, yielding plasmid pME4095. The xylE reporter gene was inserted into the NcoI site in the msuD gene as a 2.2-kb SmaI fragment from plasmid pX1918GT. Allele replacement was performed by subcloning into the ColE1 suicide vector pME3087 (29) to yield plasmid pME4088, followed by conjugative transfer into P. aeruginosa PAO1 by plate mating. Recombinants were sought by selection for a Gmr Tcs phenotype. Correct allele replacement in strain SLF3 was confirmed by PCR and Southern analysis. An identical strategy was used to construct strain SLF2, which had the xylE-Gmr cassette inserted in the reverse orientation.
Overexpression of MsuE, MsuD, and MsuC.
For overexpression and partial characterization of the MsuE, MsuD, and MsuC proteins, pET-24b-based plasmids were used. To make pME4503, the 5′ end of the msuE gene was amplified by PCR with primers SlfA-N and 960377. The resulting fragment was digested with NdeI and DrdI. This fragment was then ligated with NdeI- and XhoI-digested pET-24b and the 2,940-bp DrdI/XhoI fragment of pME4095 containing the rest of the msuE gene and the msuD and msuC genes, yielding plasmid pME4503.
The MsuC protein was also expressed alone, using plasmid pME4525. The 5′ portion of the msuC gene was amplified by PCR with primers SlfC-N and 960873, to yield a 1.3-kb DNA fragment which was digested with NdeI and BamHI and ligated into plasmid pET-24b. The resulting construct was recleaved with KpnI and XhoI, and the KpnI/XhoI fragment of pME4095 was introduced, yielding plasmid pME4525.
For expression studies, both plasmids were transformed into E. coli BL21(DE3), which was cultivated at 30°C. At an OD650 of 0.7, expression of T7 RNA polymerase was induced with isopropylthiogalactoside (50 μM), and cultivation was continued for 2.5 h. The cells were harvested and washed with 50 mM Tris-HCl (pH 7.5)–10% (vol/vol) glycerol, and DNase I (50 μg/ml) and RNase (10 μg/ml) were added. The cells were disrupted by three passes through a chilled French pressure cell (135 MPa), and cell debris was removed by centrifugation (20,000 × g, 30 min, 4°C). Cell extracts of P. aeruginosa were also made with the French pressure cell, using cells harvested in the mid-exponential phase.
Sequence analysis.
Nucleotide and protein sequences were analyzed with the University of Wisconsin Genetics Computer Group package version 9.1 and compared to the GenEMBL database and the SWISS-PROT database, respectively.
Nucleotide sequence accession number.
The nucleotide sequence of the msuEDC operon has been deposited in the GenBank database under accession no. AF026067.
RESULTS
Cloning and sequence analysis of the msuEDC genes.
When it is cultivated in the absence of sulfate, cysteine, or thiocyanate, P. aeruginosa synthesizes a set of additional SSI proteins (17). The reported N-terminal amino acid sequences of these SSI proteins yielded little information about their function in the cell, and it was speculated that they were involved in desulfurization of organosulfur compounds, similarly to the SSI proteins of E. coli (10, 49). One of the reported P. aeruginosa SSI proteins, PA13, was further investigated. With the reported N-terminal sequence as a probe, a DNA fragment encoding protein PA13 was identified in a P. aeruginosa cosmid library (50), carried on cosmid pME4006. Southern analysis showed that the PA13 N-terminal region was encoded on a 7.2-kb EcoRV fragment of pME4006. This fragment was therefore subcloned into pBluescript to give plasmid pME4095, and the nucleotide sequence of a 5,000-bp region centered on PA13 was determined. Analysis of the sequence revealed the presence of three open reading frames, arranged in a putative operon structure, which have been designated msuE, msuD, and msuC, for methanesulfonate sulfur utilization (Fig. 1). All three msu reading frames were preceded by acceptable ribosome binding sites, and their codon usage corresponded to that reported previously for P. aeruginosa (51). The overall G+C content of the coding region was 70.3%, which is within the range reported for this species (51).
FIG. 1.
Genetic organization of the msu locus of P. aeruginosa. The position of the msuD::xylE fusion is shown, as are selected restriction sites: (B, BamHI; D, DrdI; E, EcoRI; K, KpnI; N, EcoNI; Nc, NcoI; S, SphI; Sa, SauI; V, EcoRV; and X, XhoI). The DNA fragments in several plasmids described in the text are shown at the bottom.
The msuE gene encoded a protein of 186 amino acids with a predicted molecular mass of 20.0 kDa. Searches of the database yielded no clear indication of the function of this protein, although it was found to be related to the E. coli Ssi4 protein (33% identity) (35) and to the E. coli YieF protein (23% identity), both of which are of unknown function. The encoded protein contained no cysteine or methionine residues, a feature common to proteins expressed under sulfate limitation conditions (3).
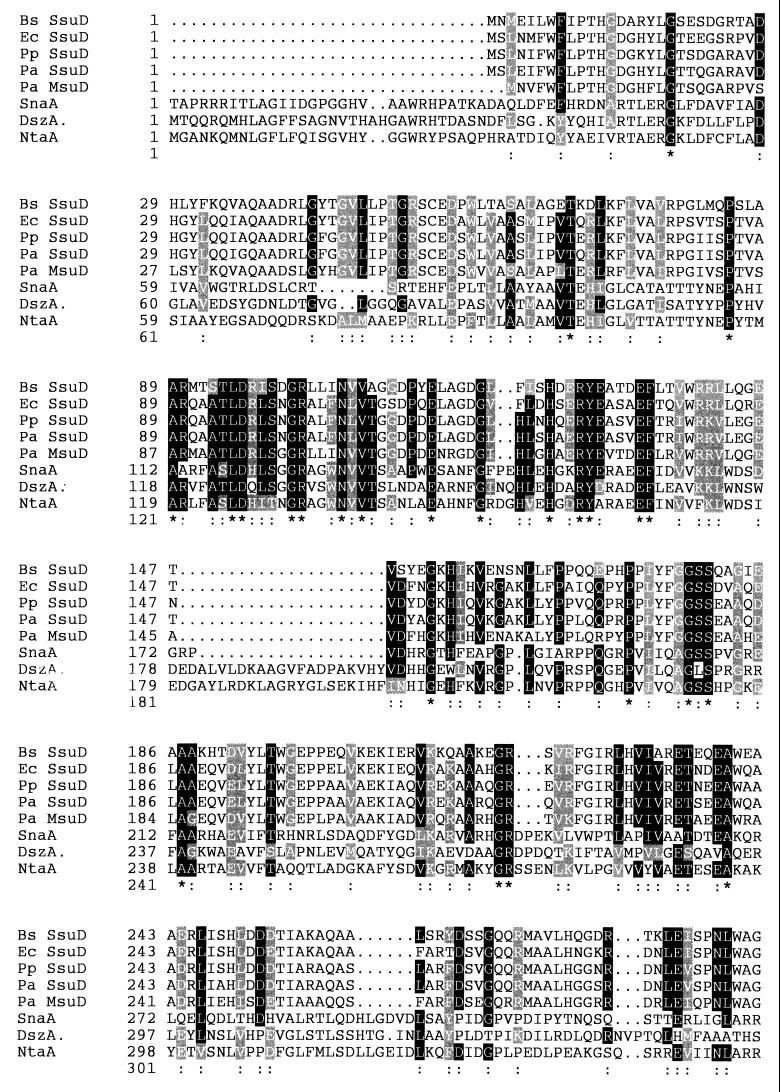
The second open reading frame in the msu gene cluster, msuD, encoded a protein with a predicted molecular mass of 41.6 kDa (381 amino acids) and an isoelectric point of 6.6. These values were consistent with the reported values for SSI protein PA13 (molecular mass, 44.5 kDa; pI, 6.0) (17), and the N-terminal peptide sequence determined for PA13 (17) was identical to that deduced from the msuD gene sequence. MsuD was found to be closely related to two other proteins under study in our laboratory, the Ssi6 protein of E. coli (35) (accession no. P80645) and OrfM of Bacillus subtilis (36, 47), and it displayed 67 and 64% amino acid identity to these two proteins, respectively. These two proteins have recently been renamed SsuD (47), for sulfonate sulfur utilization, and a copy of the ssuD gene also appears to be present in P. aeruginosa (Fig. 2). MsuD appeared from sequence comparison to be a member of the family of FMNH2-dependent oxygenases, since it was also 22 to 30% identical to DszA (dibenzothiophene [DBT] dioxide monooxygenase) (11), NtaA (nitrilotriacetate monooxygenase) (26), and SnaA (pristinamycin synthase, A subunit) (42) (Fig. 2). The first two of these enzymes catalyze oxygenation reactions adjacent to sulfinate and carboxylate groups, respectively, whereas SnaA is involved in oxygenation of the proline ring in the final biosynthetic step of the antibiotic pristinamycin IIA. All three of these proteins are 50 kDa in size and therefore are slightly larger than MsuD.
FIG. 2.
Sequence comparison of FMNH2-dependent monooxygenases. Sequence alignment was done with the program CLUSTALW. Amino acid identities (*) and similarities (:) are shown. The proteins are B. subtilis (Bs) SsuD (OrfM) (accession no. L16808), E. coli (Ec) SsuD (Ssi6) (accession no. P80645), P. putida (Pp) SsuD (accession no. AF075709), P. aeruginosa (Pa) SsuD (preliminary results from the Pseudomonas sequencing project [34]), P. aeruginosa MsuD (accession no. AF026067), Streptomyces pristinaespiralis pristinamycin synthase A (SnaA) (accession no. P54991), R. erythropolis DBT oxide monooxygenase (DszA [SoxA]) (accession no. P54995), and Chelatobacter heintzii nitrilotriacetate monooxygenase (NtaA) (accession no. P54989).
The protein encoded by the last gene in the msu operon, msuC (bp 2902 to 4119), had a predicted molecular mass of 44.8 kDa (405 amino acid residues) and an isoelectric point of 8.9. The msuC start codon was GTG, rather than the more usual ATG. The MsuC protein was closely related (42% identity) to another sulfur-regulated enzyme, the DBT monooxygenase of Rhodococcus erythropolis IGTS8, which catalyzes the initial oxidation of DBT to DBT dioxide in the DBT desulfurization pathway (11). However, it presumably has another function in P. aeruginosa, since this species is unable to utilize DBT as a source of sulfur (17). MsuC was also related to a number of eukaryotic acyl coenzyme A dehydrogenases (24 to 25% identity over nearly the full length of the protein), but the significance of this is still unclear.
Upstream of the msu genes, part of an additional open reading frame, designated orf1, was detected, which is transcribed in the same direction as the msu genes. The protein encoded by orf1 showed 45 to 50% identity to transcriptional regulators of the nifA family. Since orf1 is separated from msuE by over 600 bp, it is unlikely to be cotranscribed with the msu operon, and it is not yet known whether it is involved in expression of the msu genes.
Chromosomal location of the msuEDC genes.
The msuEDC genes were localized on the chromosome of P. aeruginosa PAO by hybridization of a digoxigenin-labelled SphI/KpnI fragment (msuED) (Fig. 1) to genomic DNA which had been digested with SpeI or DpnI. The probe hybridized strongly to chromosomal SpeI fragment J and DpnI fragment B (37), corresponding to a map position of 46.5 to 49.5 min on the chromosomal map (15). To date, the only genes in this region that have been characterized are related to pyoverdine synthesis and transport. A second, weak signal was also detected on SpeI fragment C, corresponding to a map position of 32 to 37 min. The presence of this second signal suggests that a second copy of closely related genes exists on the chromosome of P. aeruginosa, and this second copy may well correspond to the ssuD gene mentioned above.
Analysis of mutants with mutations in the msuEDC region.
In order to investigate the function of the msu genes, we constructed strain SLF3, in which the msuD gene was interrupted by a cassette carrying a promoterless xylE reporter gene and a gentamicin resistance gene. The gentamicin resistance gene in this cassette is flanked by transcriptional termination signals, so a polar effect on expression of msuC was also expected. Since SSI protein PA13 was not synthesized at all in the presence of inorganic sulfate, we anticipated that strain SLF3 would be deficient in utilization of some alternative sulfur source. Unexpectedly, the mutant was able to grow with any of over 20 sulfur sources tested, including sulfate, thiocyanate, sulfonates, sulfate esters, sulfamates, sulfur amino acids, and glucosinolates. However, the fact that the msu genes hybridized to the chromosome at two sites (see above) suggested a gene duplication, and so the lack of an observable phenotype for this mutant was perhaps not surprising.
To confirm the above-described result, we attempted to delete the entire gene cluster by replacing all three msu genes with a gentamicin resistance cassette and to construct an in-frame deletion in the msuE gene. Suicide plasmids carrying these constructs were transferred into P. aeruginosa by conjugation, and the cointegrates were readily obtained. However, extensive attempts to resolve these yielded only the wild-type strain, and the deletion mutants could not be isolated. Whereas it is possible that MsuE is required for cell growth, this seems unlikely in view of the low expression of the operon seen during growth with sulfate (see below).
Overexpression of the MsuEDC proteins and characterization of enzyme activities.
To examine the enzyme activities encoded by the msuEDC genes, the entire operon was inserted into the expression vector pET-24b, placing the msu genes under the control of a T7 promoter. After overexpression of the operon, two strongly overexpressed proteins corresponding to MsuE and MsuD were observed, together with a weakly expressed band corresponding to MsuC (Fig. 3). The weak expression of msuC observed with plasmid pME4503 (Fig. 3, lane 2) was probably due to the GTG start codon of msuC being poorly recognized in E. coli, since expression of msuC alone under T7 control gave high levels of the corresponding protein (Fig. 3, lane 3).
FIG. 3.
Overexpression of the Msu proteins. The Msu proteins were overexpressed in E. coli BL21(DE3) containing various pET24b-derived constructs, as described in Materials and Methods. Total cell extracts were separated by SDS-polyacrylamide gel electrophoresis (12% gel), and the proteins were visualized with Coomassie blue. The positions of the MsuE, MsuD, and MsuC proteins are indicated. Lanes: M, molecular weight marker; 1, BL21(pET24b); 2, BL21(pME4503); 3, BL21(pME4529). Approximately 10 μg of protein was loaded per lane.
When FMN and NADH were added to cell extracts containing the overexpressed MsuE, MsuD, and MsuC proteins, immediate reduction of the FMN was observed, accompanied by rapid oxygen consumption. This suggested that one of the Msu proteins encoded a flavin reductase activity. Deletion analysis demonstrated that MsuE was the protein responsible for FMN reduction (Table 2). Flavin reduction by MsuE was dependent on the presence of NADH, and no activity was observed when NADPH was substituted. Both FMN and flavin adenine dinucleotide (FAD) were reduced by the protein, but the rate was highest for FMN (Table 3); no activity was seen with lumiflavin or riboflavin. MsuE therefore seems to be very similar enzymatically to the cB components of the nitrilotriacetic acid and EDTA monooxygenases (26, 52), although no sequence data are yet available for these enzymes or for the corresponding genes.
TABLE 2.
Flavin reduction and desulfonation by MsuE, MsuD, and MsuCa
| Plasmid(s) | Encoded protein(s) | Flavin reduction | Desulfonation (%)b |
|---|---|---|---|
| pET24b | − | 0 | |
| pME4503 | MsuE, MsuD, MsuC | + | 100 |
| pME4529 | MsuC | − | 0 |
| pME4531 | MsuE | + | 0 |
| pME4532 | MsuE, MsuD | + | 95 |
| pME4529, pME4531 | MsuE, MsuC | + | 0 |
| pME4532, pME4529 | MsuE, MsuD, MsuC (excess) | + | 150 |
Cell extracts containing the overexpressed proteins were prepared as described in Materials and Methods. Flavin reductase activity in cell extracts was monitored as FMN disappearance and by oxygen uptake; reduction of the 100 μM FMN present in the assay was complete within seconds of extract addition. Desulfonation was measured as described in Materials and Methods.
Desulfonation results are normalized to the activity value obtained for the extract from E. coli BL21(pME4503) (100% = 35 ± 2.5 nmol/min/mg of protein). The data represent averages from three independent experiments.
TABLE 3.
Substrate spectrum of NADH-dependent flavin reductase
| Nucleotide | Flavin reductase activity (μmol/min/mg of protein)a with:
|
|||
|---|---|---|---|---|
| FMN | FAD | Riboflavin | Lumiflavin | |
| NADH | 1.85 | 0.83 | 0 | 0 |
| NADPH | 0.07 | 0.03 | 0 | 0 |
Flavin reductase activity in extracts of overexpressed MsuE (plasmid pME4531) was measured as described in Materials and Methods. No activity was seen with a similar extract prepared with cells containing pET24b. The values given were obtained after correction for the reduction rates of NADH and NADPH in the absence of flavin, which were 0.20 and 0.013 μmol/min/mg, respectively.
The expression pattern observed for the msuD gene was very similar to that previously described in in vivo studies of alkanesulfonate metabolism in P. aeruginosa (21), and we therefore suspected that the msuEDC gene products might play a role in sulfonate utilization. Incubation of the overexpressed Msu proteins with a variety of alkanesulfonates led to release of sulfite. This reaction required FMN, NADH, and oxygen (vigorous shaking) and was not seen in extracts of strain BL21(pET24b). Desulfonation was also observed in the absence of the MsuC protein (plasmid pME4532) and was not seen when extracts containing MsuE and MsuC were mixed, demonstrating that MsuD is the active desulfonating oxygenase in the complex. However, when extract containing MsuC was mixed with extract containing MsuE and MsuD, the desulfonation rate was about 1.5-fold greater than that with MsuE and MsuD alone, suggesting that MsuC is involved but not essential in the desulfonation process. Of the substrates tested, only alkanesulfonates were desulfonated by MsuEDC (Table 4). The best substrate for the enzyme was methanesulfonate, and alkanesulfonates with charged side chains (cysteate, sulfoacetate, and taurine) were not desulfonated by the enzyme. The sequence similarity between MsuC and DBT monooxygenase (30) suggested that the Msu proteins might be involved in metabolism of dimethyl sulfide via dimethyl sulfoxide, as has previously been reported for Rhodococcus sp. strain SY1 (33). However, neither of these compounds was desulfonated by MsuEDC, and P. aeruginosa was unable to grow with dialkyl or diaryl sulfides as a sulfur source, even at levels which were demonstrated to be nontoxic to the cell (100 μM).
TABLE 4.
Substrate spectrum of desulfonation by the MsuED enzymes
| Substrate | Desulfonation rate (%)a |
|---|---|
| Methanesulfonate | 100 |
| PIPESb | 31 ± 2 |
| Ethanesulfonate | 27 ± 3 |
| Isethionate | 19 ± 2 |
| MOPSc | 17 ± 2 |
| Pentanesulfonate | 13 ± 2 |
| HEPES | 9 ± 0.7 |
| Sulfoacetate | 1.8 ± 0.7 |
| Taurine | 1.5 ± 0.7 |
| Allylsulfonate | 1.3 ± 0.9 |
| Cysteate | 0 |
| Sulfate | 0 |
| Benzenesulfonate | 0 |
| Dimethyl sulfoxide | 0 |
| Methionine | 0 |
Desulfonation was measured with cell extracts of E. coli BL21(pME4503) as described in Materials and Methods. The results are the averages and standard deviations of three determinations and are normalized to the values obtained with methanesulfonate as the substrate (100% = 35 ± 2.5 nmol/min/mg of protein).
PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid).
MOPS, morpholinepropanesulfonic acid.
Desulfonation of methanesulfonate was also tested with extracts of P. aeruginosa PAO1 which had been grown in minimal medium with pentanesulfonate or sulfate as a sulfur source (500 μM). Using the same assay as described above, we found a methanesulfonate-desulfonating specific activity of 2.7 nmol/min/mg of protein in extracts of cells grown with pentanesulfonate. However, since the msuD mutant described above was able to grow with methanesulfonate, it is clear that there is more than one methanesulfonate sulfonatase activity in cells grown under these conditions, and so the observed desulfonation activity in the wild-type strain is probably due to both MsuD and other desulfonating enzymes. As expected, no methanesulfonate-desulfonating activity was seen with sulfate-grown cells. Extracts of pentanesulfonate-grown cells also showed significant α-ketoglutarate-dependent taurine dioxygenase activity (6 nmol/min/mg of protein), but no α-ketoglutarate-dependent methanesulfonate desulfonation was found.
Transcriptional regulation of msuEDC.
The msuD::xylE transcriptional fusion in strain SLF3 was used to investigate the pattern of expression of the msuD gene during growth with several different sulfur sources (Table 5). Catechol oxygenase (XylE) specific activity was measured in whole cells harvested in the mid-exponential growth phase and compared with the specific activity of the well-characterized, sulfate-regulated enzyme arylsulfatase (3). The specific activities of both enzymes were much higher during growth with organosulfur sources such as sulfonates or methionine than with sulfate as the sole sulfur source. After growth with a mixture of pentanesulfonate and sulfate, enzyme activity was low, confirming that regulation was mediated by repression by sulfate or a related metabolite (see below). This corresponds well with the qualitative behavior previously reported for protein PA13 on two-dimensional electropherograms (17). In the presence of cysteine, expression of msuD and atsA was not completely repressed, perhaps due to partial oxidation to cystine, which acts as a derepressing growth substrate in this species (17). When the xylE-Gmr cassette was inserted in the reverse orientation (strain SLF2), no catechol oxygenase activity was observed (data not shown).
TABLE 5.
Expression of msuD::xylE and atsA in strain SLF3a
| Sulfur source for growth (250 μM) | Catechol-2,3-dioxygenase activity (μmol/min/mg of protein) | Arylsulfatase activity (nmol/min/mg of protein) |
|---|---|---|
| Sulfate | 0.6 ± 0.2 | 0.29 ± 0.04 |
| Cysteine | 47 ± 4 | 1.72 ± 0.03 |
| Methionine | 473 ± 5 | 17.7 ± 1.0 |
| Pentanesulfonate | 118 ± 12 | 14.6 ± 0.9 |
| Pentanesulfonate + sulfate | 2.0 ± 1.4 | 0.27 ± 0.04 |
Cells were harvested in mid-exponential phase, and catechol oxygenase and arylsulfatase activities in cell extracts were measured as described in Materials and Methods. The results represent means and standard deviations from four measurements.
Effect of cys mutations.
Several genetic systems involved in sulfur metabolism are regulated by metabolites involved in the cysteine biosynthetic pathway. Thus, cysteine biosynthesis in E. coli is positively regulated by O-acetylserine and is negatively controlled by cysteine, sulfide, and thiosulfate (27). In P. aeruginosa, repression of arylsulfatase synthesis under sulfur-replete conditions has been shown to be under the control of at least two effectors, probably sulfite and sulfide (17). To investigate the dependence of msuD expression on cysteine biosynthetic intermediates, the msuD::xylE allele was transduced into cysI and cysN genetic backgrounds (strains AX18 and AC309, respectively) to give strains SLF4 and SLF5. The cells were grown under derepressing conditions (methionine or pentanesulfonate [100 μM] as a sulfur source), and the negative effect of sulfate, sulfite, or sulfide (500 μM) on arylsulfatase and catechol oxygenase synthesis was tested (Table 6). msuD expression was negatively regulated in response to all three sulfur-containing compounds tested, in both the cysI and cysN backgrounds, in contrast to the case for arylsulfatase, expression of which was not directly affected by sulfate. It therefore appears that sulfate, sulfite, and sulfide or cysteine play independent roles in the negative regulation of the msu genes and that multiple negative regulatory inputs are involved.
TABLE 6.
Expression of msuD::xylE and atsA in cysI and cysN genetic backgroundsa
| Sulfur sourceb | Strain SLF3
|
Strain SLF4 (cysI)
|
Strain SLF5 (cysN)
|
|||
|---|---|---|---|---|---|---|
| C23O (%) | AtsA (%) | C23O (%) | AtsA (%) | C23O (%) | AtsA (%) | |
| Pn/Met | 100 ± 5 | 100 ± 6 | 100 ± 7 | 100 ± 2 | 100 ± 8 | 100 ± 1 |
| Pn/Met + sulfate | 1.3 ± 0.1 | 11 ± 2 | 1.5 ± 1 | 9.2 ± 2 | 1.8 ± 0.4 | 99 ± 1 |
| Pn/Met + sulfite | 3.3 ± 1 | 9 ± 0.7 | 2.0 ± 0.6 | 5.9 ± 0.2 | 5.5 ± 2 | 4.7 ± 0.1 |
| Pn/Met + sulfide | 2.0 ± 2 | 10 ± 0.7 | 4.5 ± 0.4 | 3.3 ± 1 | 7.9 ± 2 | 1.5 ± 0.3 |
Cells were harvested in mid-exponential phase, and catechol oxygenase (C23O) and arylsulfatase (AtsA) activities in cell extracts were measured as described in Materials and Methods. The results were normalized to 100% for the value obtained after growth with pentanesulfonate or methionine (Pn/Met) (Table 5) and are means and standard deviations from four separate experiments.
Strains SLF3 and SLF5 were grown with pentanesulfonate as a sulfur source (100 μM), and strain SLF4 was grown with methionine (100 μM). (SLF4 is unable to utilize the sulfite derived from pentanesulfonate, whereas SLF5 cannot grow with methionine [17].) Sulfate, sulfite, or sulfide (500 μM) was added as appropriate.
To test the cysB dependence of methanesulfonate utilization, a cysB mutant of P. aeruginosa PAO1 was created by introduction of the cysB::Gm allele, described by Delic-Attree et al. (6), to give strain PAO-CB. Strain PAO-CB did not grow with sulfate as a sulfur source but grew normally with cysteine, methionine, or homocysteine, as expected. No growth was seen with sulfate esters or with sulfonates (methanesulfonate, pentanesulfonate, isethionate, or taurine) or with dimethyl sulfoxide. Unexpectedly, however, strain PAO-CB grew when sulfide or sulfite was supplied as a sulfur source. Identical results were obtained with a previously reported cysB mutant of a clinical isolate of P. aeruginosa (strain CHA-CB), suggesting that cysteine biosynthesis is regulated differently in P. aeruginosa than in enteric bacteria.
Distribution of the msu genes.
Analysis of the gene sequence of msuD revealed that it was closely related to the ssuD genes of B. subtilis and E. coli. A closely related gene is also present in Pseudomonas putida (accession no. AF075709), and the desulfonation function that these genes encode therefore seems to be widespread. To test this, a pair of oligonucleotides was designed from the regions of the msuD sequence which were most similar to the homologous sequences from E. coli and B. subtilis, and these primers were used to carry out PCR with chromosomal DNAs from a variety of bacterial species (Fig. 4). A band corresponding to the msuD gene was observed in a range of species, including enteric bacteria, a variety of pseudomonads, and a Bacillus species. Indeed, in a number of species, two bands were observed, consistent with the observation above that the msuD gene maps to two positions on the P. aeruginosa genome (in Fig. 4 the single band obtained with P. aeruginosa was shown to be two superimposed bands by digestion of the PCR product with NcoI or PvuI, both of which cut within the msuD gene). The specificity of the PCR was confirmed by Southern hybridization, which demonstrated that all of the bands shown hybridized with the msuD gene (not shown). No PCR signal was observed for anaerobic bacteria such as Clostridium or Methanobacterium or for rhizobial species such as Bradyrhizobium or Rhizobium. This is not surprising, since the gene products encode an oxygenative sulfonatase and these species live in environments containing low or no oxygen. However, two of the species tested (Salmonella typhimurium and Stenotrophomonas maltophilia) cannot grow with alkanesulfonates as a sulfur source (Fig. 4), although PCR analysis showed a band corresponding to the msuD gene. In S. typhimurium the gene may have become cryptic during evolution, whereas S. maltophilia is by nature a cysteine/methionine auxotroph.
FIG. 4.
Distribution of the msuD gene. PCR was done with genomic DNA of the indicated species, using primers orfM500 and orfM970rev, and the PCR products were separated on a 2% agarose gel. Growth of the various species in succinate-minimal medium with taurine or methane- or pentanesulfonate (Me/Pn sulfonate) is indicated. a, S. maltophilia will grow only with cysteine or methionine as a sulfur source.
DISCUSSION
Alkanesulfonates are ubiquitous in nature, but very few enzyme systems for their metabolism have yet been characterized. In this report, we have shown that the msuD and msuE genes encode an FMNH2-dependent sulfonatase with specificity for methanesulfonate and the NADH-dependent FMN reductase required to supply reduced FMN to the sulfonatase, respectively. A third gene, msuC, is involved in the desulfonation reaction but is not an essential component. Methanesulfonate is a natural oxidation product of dimethyl sulfide; it is the main biogenic organic sulfur compound in the atmosphere and is present in significant quantities in rainwater. The regulatory pattern exhibited by the msu genes (Tables 4 and 5) indicates that they have evolved to allow bacteria to use this compound as a source of sulfur under conditions where more favored sulfur sources (sulfate, sulfide, or cysteine) are not available.
The expected product of methanesulfonate desulfonation is formaldehyde (20) (cf. previous work with other alkanesulfonates [44] and with taurine [10]). Although this compound was not observed in the present study, it may have been metabolized too quickly for determination, since control experiments showed that formaldehyde disappeared rapidly when added to the E. coli cell extracts used. In vivo, P. aeruginosa does not release an aldehyde as the desulfonation product of alkanesulfonates but rather releases its oxidation product, the corresponding carboxylic acid (21). It therefore seemed possible that MsuC might be involved in oxidation of formaldehyde produced in the desulfonation reaction. This reaction could involve either a direct dehydrogenation or an FMNH2-dependent oxygenation reaction (MsuC shows a high level of similarity to DszC, which catalyzes the FMNH2-dependent monooxygenolytic oxidation of DBT). However, overexpressed MsuC protein did not show formaldehyde dehydrogenase activity, nor was any formaldehyde oxidation observed when reduced FMN was provided by addition of overexpressed MsuE protein. The role of MsuC is therefore still unclear and is being further investigated in our laboratory.
The biochemistry of methanesulfonate metabolism has previously been studied with a methylotrophic soil isolate, Methylosulfonomonas methylovora M2 (16, 20), and with two marine methylotrophs, strain TR3 and Marinosulfonomonas methylotropha PSCH4 (16, 43). Those studies differed from the present one in that the strains investigated were isolated by enrichment for their ability to utilize methanesulfonate as a source of carbon and energy, whereas the P. aeruginosa strain used in this study is a standard laboratory strain and can utilize methanesulfonate only as a sulfur source. The key enzyme responsible for methanesulfonate metabolism in methylotrophs is an NADH-dependent monooxygenase (MSAMO) which requires FAD and Fe2+ and cleaves the substrate into formaldehyde and sulfite. The MSAMO enzyme system consists of three components, one of which has been characterized as a [2Fe-2S] ferredoxin (13). No further information is yet available on the remaining components, and a molecular-level comparison with the Msu system is therefore not yet possible. The reconstituted MSAMO enzyme is highly specific for short-chain (C1 to C3) sulfonates as substrates and therefore displays a specificity similar to that of the MsuED system (Table 4). The narrow substrate specificity of MsuED contrasts with the broad substrate tolerance that has been reported for other enzymes that are involved in the sulfur cycle, e.g., the desulfurization of aromatic sulfonates as sulfur source by P. putida S-313 (23, 53) or the arylsulfatase of P. aeruginosa (3, 9). In this sense the MsuED methanesulfonic acid desulfurization system appears to be unusual, and it could be argued that the msu genes may have been recruited from the carbon cycle to exploit methanesulfonic acid as a sulfur source. However, given the similarity between the msu genes and the more widely distributed ssu genes (see below), this seems unlikely. Indeed, since E. coli K-12 possesses the ssu genes yet does not grow with methanesulfonate (our unpublished results), it is more probable that msuD represents a mutant form of ssuD which has adapted its substrate specificity to allow utilization of the common, sulfur-containing methanesulfonate.
Sulfonate utilization seems to be a more important part of sulfur metabolism for bacteria than has previously been recognized. Homologues of the msu genes are present in the majority of strains selected for testing from our strain collection (Fig. 4), and several of these bacteria even contain several different desulfonation systems, with partially redundant substrate specificities. In E. coli, desulfonation of alkanesulfonates is catalyzed by both TauD and SsuD (10), and a desulfonation-negative phenotype was seen only in a double mutant (unpublished data). In P. aeruginosa, our mapping studies revealed the presence of an msuD homologue, and this has been confirmed by preliminary data from the Pseudomonas genome sequencing project, which has demonstrated the presence in this species of an ssuD gene very similar to those of B. subtilis and E. coli (Fig. 2). A comparison of the SsuD and MsuD proteins with other FMNH2-dependent monooxygenases (Fig. 2) shows a conserved domain (residues 89 to 135 of MsuD) which may form part of the active site of the enzyme. However, this domain does not contain any known protein motifs (as determined with PROSITE), and further clarification of the enzyme mechanism will require detailed biochemical characterization of these proteins.
In enteric bacteria, sulfate assimilation is controlled by the CysB protein, a LysR-type transcriptional activator which is required for expression of the genes of the cys regulon. Recent work has shown that the cys regulon includes not only the genes for sulfate activation and reduction but also those for uptake and desulfonation of taurine (tauABCD) (49). Expression of the latter genes, however, is dependent not only on CysB but also on the Cbl protein, a second regulator of the LysR family that is closely related to CysB (18, 48). The cysB gene has recently also been identified in P. aeruginosa, and cysB mutants of this species were found to be auxotrophic for cysteine, as expected. Our results show that CysB is also required for utilization of several organosulfur sources, including sulfonates and sulfate esters, and a cysB mutant of P. aeruginosa (strain PAO-CB) could not grow with sulfate but retained the ability to grow with sulfite or sulfide as a sulfur source. This suggests that the genes encoding sulfite reductase and O-succinylhomoserine sulfhydrylase are not part of the cys regulon in this species. Growth with methionine as a sulfur source (via the reverse transsulfuration pathway [12]) was not affected by the cysB mutation, and it is likely that this pathway is separately regulated. CysB is also important in regulating alginate biosynthesis in this species (6), since it acts as an activator in expression of the algD gene, but the physiological significance of this is not yet fully understood. Sequencing of the P. aeruginosa genome is still in progress, but from preliminary analysis there appears to be only one copy of cysB, and there is no evidence for an additional cbl gene.
Recent work has shown that alkanesulfonates not only can be desulfonated by aerobic microorganisms but also can be utilized by a variety of anaerobic bacteria, either as a source of sulfur (5, 7) or to satisfy energy requirements for growth as electron donors or electron sinks (8, 28, 32). The presence of duplicate sulfonate-assimilatory pathways in aerobic bacteria, and of a variety of desulfonation pathways in anaerobes, serves to underline the importance of natural sulfonates in the bacterial life cycle.
ACKNOWLEDGMENTS
We are grateful to Thomas Leisinger for his support and to Paul Vermeij and Jan van der Ploeg for helpful discussions. Our thanks are also due to Franz Narberhaus for providing genomic DNA samples from a number of species, to Laura Serino for the P. aeruginosa gene bank, to Christian Hulen for sharing unpublished results with us, and to Ina Delic-Attree and H. Schweizer for several constructs.
This work was supported in part by Swiss National Science Foundation grant no. 31-41873.94.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. [Google Scholar]
- 2.Autry A R, Fitzgerald J W. Sulfonate S—a major form of forest soil organic sulfur. Biol Fertil Soils. 1990;10:50–56. [Google Scholar]
- 3.Beil S, Kehrli H, James P, Staudenmann W, Cook A M, Leisinger T, Kertesz M A. Purification and characterization of the arylsulfatase, synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA) Eur J Biochem. 1995;229:385–394. doi: 10.1111/j.1432-1033.1995.0385k.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chien C C, Leadbetter E R, Godchaux W., III Sulfonate-sulfur can be assimilated for fermentative growth. FEMS Microbiol Lett. 1995;129:189–193. [Google Scholar]
- 6.Delic-Attree I, Toussaint B, Garin J, Vignais P M. Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol Microbiol. 1997;24:1275–1284. doi: 10.1046/j.1365-2958.1997.4121799.x. [DOI] [PubMed] [Google Scholar]
- 7.Denger K, Cook A M. Assimilation of sulfur from alkyl and arylsulfonates by Clostridium spp. Arch Microbiol. 1997;167:177–181. [PubMed] [Google Scholar]
- 8.Denger K, Laue H, Cook A M. Anaerobic taurine oxidation: a novel reaction by a nitrate reducing Alcaligenes sp. Microbiology. 1997;6:1919–1924. doi: 10.1099/00221287-143-6-1919. [DOI] [PubMed] [Google Scholar]
- 9.Dodgson K S, White G F, Fitzgerald J W. Sulfatases of microbial origin. Boca Raton, Fla: CRC Press; 1982. [Google Scholar]
- 10.Eichhorn E, van der Ploeg J R, Kertesz M A, Leisinger T. Characterization of α-ketoglutarate dependent taurine dioxygenase from Escherichia coli. J Biol Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- 11.Gray K A, Pogrebinsky O S, Mrachko G T, Xi L, Monticello D J, Squires C H. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat Biotechnol. 1996;14:1705–1709. doi: 10.1038/nbt1296-1705. [DOI] [PubMed] [Google Scholar]
- 12.Günther E, Petruschka L, Herrmann H. Reverse transsulfuration pathway in Pseudomonas aeruginosa. Z Allg Mikrobiol. 1979;19:439–442. doi: 10.1002/jobm.3630190610. [DOI] [PubMed] [Google Scholar]
- 13.Higgins T P, Demarco P, Murrell J C. Purification and molecular characterization of the electron transfer protein of methanesulfonic acid monooxygenase. J Bacteriol. 1997;179:1974–1979. doi: 10.1128/jb.179.6.1974-1979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway B. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 15.Holloway B W, Römling U, Tümmler B. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology. 1994;140:2907–2929. doi: 10.1099/13500872-140-11-2907. [DOI] [PubMed] [Google Scholar]
- 16.Holmes A J, Kelly D P, Baker S C, Thompson A S, De Marco P, Kenna E M, Murrell J C. Methylosulfonomonas methylovora gen. nov., sp. nov., and Marinosulfonomonas methylotropha gen. nov., sp. nov.: novel methylotrophs able to grow on methanesulfonic acid. Arch Microbiol. 1997;167:46–53. doi: 10.1007/s002030050415. [DOI] [PubMed] [Google Scholar]
- 17.Hummerjohann J, Kuttel E, Quadroni M, Ragaller J, Leisinger T, Kertesz M A. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology. 1998;5:1375–1386. doi: 10.1099/00221287-144-5-1375. [DOI] [PubMed] [Google Scholar]
- 18.Iwanicka Nowicka R, Hryniewicz M M. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene. 1995;166:11–17. doi: 10.1016/0378-1119(95)00606-8. [DOI] [PubMed] [Google Scholar]
- 19.Karkhoff-Schweizer R R, Schweizer H P. Utilization of a mini-Dlac transposable element to create an alpha-complementation and regulated expression system for cloning in Pseudomonas aeruginosa. Gene. 1994;140:7–15. doi: 10.1016/0378-1119(94)90724-2. [DOI] [PubMed] [Google Scholar]
- 20.Kelly D P, Baker S C, Trickett J, Davey M, Murrell J C. Methanesulphonate utilization by a novel methylotrophic bacterium involves an unusual monooxygenase. Microbiology. 1994;140:1419–1426. [Google Scholar]
- 21.Kertesz M A. Desulfonation of aliphatic sulfonates by Pseudomonas aeruginosa PAO. FEMS Microbiol Lett. 1996;137:221–225. doi: 10.1111/j.1574-6968.1996.tb08109.x. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz M A, Cook A M, Leisinger T. Microbial metabolism of sulfur- and phosphorus-containing xenobiotics. FEMS Microbiol Rev. 1994;15:195–215. doi: 10.1111/j.1574-6976.1994.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 23.Kertesz M A, Kölbener P, Stockinger H, Beil S, Cook A M. Desulfonation of linear alkylbenzenesulfonate surfactants and related compounds by bacteria. Appl Environ Microbiol. 1994;60:2296–2303. doi: 10.1128/aem.60.7.2296-2303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kertesz M A, Leisinger T, Cook A M. Proteins induced by sulfate limitation in Escherichia coli, Pseudomonas putida, or Staphylococcus aureus. J Bacteriol. 1993;175:1187–1190. doi: 10.1128/jb.175.4.1187-1190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King J E, Quinn J P. The utilization of organosulphonates by soil and freshwater bacteria. Lett Appl Microbiol. 1997;24:474–478. [Google Scholar]
- 26.Knobel H R, Egli T, van der Meer J R. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J Bacteriol. 1996;178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 514–527. [Google Scholar]
- 28.Laue H, Denger K, Cook A M. Fermentation of cysteate by a sulfate-reducing bacterium. Arch Microbiol. 1997;168:210–214. doi: 10.1007/s002030050502. [DOI] [PubMed] [Google Scholar]
- 29.Laville J. Régulation du metabolisme secondaire et génétique de la cyanogénèse de Pseudomonas fluorescens CHA0, une souche antagoniste de champignons phytopathogénes. Dissertation. Zurich, Switzerland: Swiss Federal Institute of Technology; 1993. p. 10251. [Google Scholar]
- 30.Lei B, Tu S-C. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J Bacteriol. 1996;178:5699–5705. doi: 10.1128/jb.178.19.5699-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leisinger T, Kohler-Staub D. Dichloromethane dehalogenase from Hyphomicrobium DM2. Methods Enzymol. 1990;188:355–361. [Google Scholar]
- 32.Lie T J, Pitta T, Leadbetter E R, Godchaux W., III Sulfonates: novel electron acceptors in anaerobic respiration. Arch Microbiol. 1996;66:204–211. doi: 10.1007/s002030050376. [DOI] [PubMed] [Google Scholar]
- 33.Omori T, Saiki Y, Kasuga K, Kodama T. Desulfurization of alkyl and aromatic sulfides and sulfonates by dibenzothiophene-desulfurizing Rhodococcus sp. strain SY1. Biosci Biotechnol Biochem. 1995;59:1195–1198. [Google Scholar]
- 34.Pseudomonas Genome Project. 15 December 1998, posting date. [Online.] Internet web site http://www.pseudomonas.com/. [15 January 1999, last date accessed.]
- 35.Quadroni M, Staudenmann W, Kertesz M, James P. Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis. Eur J Biochem. 1996;239:773–781. doi: 10.1111/j.1432-1033.1996.0773u.x. [DOI] [PubMed] [Google Scholar]
- 36.Quirk P G, Guffanti A A, Clejan S, Cheng J, Krulwich T A. Isolation of Tn917 insertional mutants of Bacillus subtilis that are resistant to the protonophore carbonyl cyanide m-chlorophenylhydrazone. Biochim Biophys Acta. 1994;1186:27–34. doi: 10.1016/0005-2728(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 37.Römling U, Duchêne M, Essar D W, Galloway D, Guideo-Rontani C, Hill D, Lazdunski D, Miller R V, Schleifer K H, Smith D W, Toschka H Y, Tümmler B. Localization of alg, opr, phn, pho, 4.5S RNA, 6S RNA, tox, trp, and xcp genes, rrn operons, and the chromosomal origin on the physical genome map of Pseudomonas aeruginosa PAO. J Bacteriol. 1992;174:327–330. doi: 10.1128/jb.174.1.327-330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer H P. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 39.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 40.Seitz A P, Leadbetter E R, Godchaux W., III Utilization of sulfonates as sole sulfur source by soil bacteria including Comamonas acidovorans. Arch Microbiol. 1993;159:440–444. [Google Scholar]
- 41.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 42.Thibaut D, Ratet N, Bisch D, Faucher D, Debussche L, Blanche F. Purification of the two-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin II-B during the last step of pristinamycin II-A biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson A S, Owens N J P, Murrell J C. Isolation and characterization of methanesulfonic acid-degrading bacteria from the marine environment. Appl Environ Microbiol. 1995;61:2388–2393. doi: 10.1128/aem.61.6.2388-2393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thysse G J E, Wanders T H. Degradation of n-alkane-1-sulfonates by Pseudomonas. Antonie Leeuwenhoek. 1972;38:53–63. doi: 10.1007/BF02328077. [DOI] [PubMed] [Google Scholar]
- 45.Uria-Nickelsen M R, Leadbetter E R, Godchaux W., III Sulfonate-sulfur utilization involves a portion of the assimilatory sulfate reduction pathway in Escherichia coli. FEMS Microbiol Lett. 1994;123:43–48. doi: 10.1111/j.1574-6968.1994.tb07199.x. [DOI] [PubMed] [Google Scholar]
- 46.Vairavamurthy M A, Zhou W, Eglinton T, Manowitz B. Sulfonates: a new class of organic sulfur compounds in marine sediments. Geochim Cosmochim Acta. 1994;58:4681–4687. [Google Scholar]
- 47.van der Ploeg J R, Cummings N J, Leisinger T, Connerton I F. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology. 1998;144:2555–2561. doi: 10.1099/00221287-144-9-2555. [DOI] [PubMed] [Google Scholar]
- 48.van der Ploeg J R, Iwanicka-Nowicka R, Kertesz M A, Leisinger T, Hryniewicz M M. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J Bacteriol. 1997;179:7671–7678. doi: 10.1128/jb.179.24.7671-7678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Ploeg J R, Weiss M A, Saller E, Nashimoto H, Saito N, Kertesz M A, Leisinger T. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol. 1996;178:5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West S E, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9325. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witschel M, Nagel S, Egli T. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J Bacteriol. 1997;179:6937–6943. doi: 10.1128/jb.179.22.6937-6943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zürrer D, Cook A M, Leisinger T. Microbial desulfonation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl Environ Microbiol. 1987;53:1459–1463. doi: 10.1128/aem.53.7.1459-1463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]