Abstract
Recent epidemiological studies have shown that inflammatory bowel disease is associated with periodontal disease. The oral–gut microbiota axis is a potential mechanism intersecting the two diseases. Porphyromonas gingivalis is currently considered a keystone oral pathogen involved in periodontal disease pathogenesis and disease progression. Recent studies have shown that oral ingestion of P. gingivalis leads to intestinal inflammation. However, the molecular underpinnings of P. gingivalis-mediated gut inflammation have remained elusive. In this study, we show that the oral administration of P. gingivalis indeed leads to ileal inflammation and alteration in gut microbiota with significant reduction in bacterial alpha diversity despite the absence of P. gingivalis in the lower gastrointestinal tract. Utilizing an antibiotic-conditioned mouse model, cecal microbiota transfer experiments were performed to demonstrate that P. gingivalis-induced dysbiotic gut microbiota is sufficient to reproduce gut pathology. Furthermore, we observed a significant expansion in small intestinal lamina propria IL9+ CD4+ T cells, which was negatively correlated with both bacterial and fungal alpha diversity, signifying that P. gingivalis-mediated intestinal inflammation may be due to the subsequent loss of gut microbial diversity. Finally, we detected changes in gene expression related to gut epithelial barrier function, showing the potential downstream effect of intestinal IL9+CD4+ T-cell induction. This study for the first time showed the mechanism behind P. gingivalis-mediated intestinal inflammation where P. gingivalis indirectly induces intestinal IL9+ CD4+ T cells and inflammation by altering the gut microbiota. Understanding the mechanism of P. gingivalis-mediated intestinal inflammation may lead to the development of novel therapeutic approaches to alleviate the morbidity from inflammatory bowel disease patients with periodontal disease.
Keywords: inflammatory bowel disease, microbiome, periodontitis, Porphyromonas gingivalis
1 |. BACKGROUND
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract that is driven by abnormal host immune response toward dysbiotic gut microbiota (Ni et al., 2017). Periodontal disease is similarly mediated by host immune response toward the plaque biofilm and affects the periodontal tissue, resulting in progressive loss of tooth-supporting tissues (Lamont et al., 2018). Recent epidemiological studies have shown that IBD is associated with periodontal diseases (Lira-Junior & Figueredo, 2016; She et al., 2020). As one of the most prevalent chronic diseases in the United States, periodontitis affects more than half of American adults aged over 30 years with prevalence increasing with age (Eke et al., 2015). Worldwide, IBD patients have higher prevalence, severity, and extent of periodontal diseases compared to healthy populations (Brito et al., 2008; Habashneh et al., 2012; Vavricka et al., 2013; Yu et al., 2018). Therefore, understanding the molecular pathological mechanisms behind the association between the two diseases may lead to discovery of important therapeutic targets for the management of IBD patients with chronic periodontitis.
Oral microbiota is a potential mechanism linking the two diseases. As the average human adult ingests approximately 1012 oral bacteria everyday through saliva, changes in oral microbiota have the potential to affect the gut microbiota (Kitamoto et al., 2020; Sohn et al., 2020). Porphyromonas gingivalis is a one of the keystone species that proliferates in subgingival periodontal microenvironment surrounding teeth known to play a major critical role in the pathogenesis of periodontal disease (How et al., 2016). Previous studies have shown that the oral administration of P. gingivalis in mice alters the gut microbiota and induces intestinal and systemic inflammation through impairing the gut barrier function (Arimatsu et al., 2014; Kato et al., 2018; Nakajima et al., 2015). However, these studies do not establish mechanistic basis for P. gingivalis-mediated intestinal inflammation. P. gingivalis do not colonize the lower intestinal tract, which suggests that there exists an alternate mechanism for the observed intestinal pathology (Geva-Zatorsky et al., 2017). We hypothesize that P. gingivalis induces intestinal inflammation by disrupting the gut microbiota. We utilized antibiotic-conditioned mouse model raised and maintained in positive pressure cages to perform microbiota transplantation studies to determine the underlying mechanism behind P. gingivalis-mediated intestinal inflammation.
2 |. METHODS
2.1 |. Animals
All animal experiments were performed in accordance with relevant guidelines and regulations. The study protocol was approved by Institutional Animal Care and Use Committee (IACUC) of the University at Buffalo and all methods were carried out in compliance with the ARRIVE guidelines. Wild-type BALB/cJ mice (Jackson Laboratory; Cat#000651) were given 1 mg/ml of Kanamycin sulfate (IBI Scientific; Cat# 25389–94-0) via autoclaved drinking water for 5 days to suppress host microbiota. After 2 days of washout period, approximately 1 × 109 CFU of P. gingivalis was administered via oral gavage using stainless steel feeding tube (see below for P. gingivalis cultivation method). Interleukin 10 (Il10)-deficient BALB/cJ mice (Jackson Laboratory; Cat#004333) were given antibiotic cocktail of 1 mg/ml each of ampicillin (Teknova; Cat#50–841-073), neomycin sulfate (Fagron; Cat#804599), metronidazole (Spectrum Chemical; Cat#M1284), and 0.5 mg/ml of Vancomycin hydrochloride (Fagron; Cat#804147) via drinking water for 4 weeks (refilled every 96 h) to ablate host microbiota (See below for cecal microbiota transplantation method). All animals were single housed and raised in sealed positive pressure cages (Allentown).
2.2 |. gingivalis cultivation
P. gingivalis strain ATCC 33277 was cultivated anaerobically (85% N2, 10% H2, 5% CO2) at 37°C in tryptic soy broth (BD; Cat#211825) medium supplemented with 2% (w/v) yeast extract, haemin (5 μg/ml), and vitamin K (5 μg/ml) for 2 days, removed, and washed with PBS solution once.
2.3 |. gingivalis detection by PCR
P. gingivalis-specific primers F-CTTGACTTCAGTGGCGGCAG and R-AGGGAAGACGGTTTTCACCA were used for detection of P. gingivalis in cecal samples. PCR was performed using Phire Green Hot Start II DNA Polymerase (Thermo Scientific; Cat# F124L) according to manufacturer’s instruction.
2.4 |. Histology
Distal ileal tissues were formalin-fixed, parafilm embedded, horizontally sectioned into 5 μm sections, and stained with hematoxylin and eosin (H&E). Samples were blinded and scored based on inflammation severity, inflammation extent, epithelial changes, and mucosal architecture by two individuals (Erben et al., 2014). Statistical significance was calculated using Student’s t-test.
2.5 |. Microbiome sequencing and analysis
DNA from cecal samples were extracted using DNeasy PowerSoil Kit (Qiagen; Cat#12888). 16S rRNA gene and internal transcribed spacer (ITS) metagenomic sequencing library was prepared as according to Illumina’s metagenomic sequencing protocol. Briefly, V3–V4 and ITS1 hypervariable region was first amplified and Illumina adaptor overhang sequences were appended using 2x KAPA HiFi HotStart Ready Mix (KapaBiosystems; Cat#KK2602) and T100 Thermal Cycler (Biorad). Library was cleaned using the Ampure XP Bead kit, quantified using Quant-iT™ PicoGreen™ dsDNA Reagent (Invitrogen; Cat#P7581), and pooled at equal concentration. Library quality control was done using Fragment Analyzer (Advanced Analytical Technologies). 2 × 300-cycle paired end sequencing was performed using Illumina Miseq System (Illumina).
16S rRNA gene and ITS sequencing data were analyzed using QIIME2 2020.2 (Bolyen et al., 2019). Raw sequence data were demultiplexed and filtered using demux plugin and denoised using DADA2 plugin (Callahan et al., 2016). Multiple sequence alignment was performed using mafft plugin (Katoh et al., 2002). Measurements of alpha diversity and beta diversity were calculated using diversity plugin. PERMANOVA test was used to determine differences in beta diversity. Taxonomic classification was performed using classify-sklearn against genome taxonomy database (GTDB) r86 reference sequence for 16S rRNA gene data and international society for human and animal mycology barcoding (ISHAM) database for ITS data (Bokulich et al., 2018; Irinyi et al., 2015). Differential abundance testing was done with ANCOM (Mandal et al., 2015).
2.6 |. RNAseq
RNA was extracted from ileal tissues using Qiagen RNeasy Mini Kit (Qiagen; Cat#74104) and treated with DNase (Qiagen; Cat#79254) according to manufacturer’s protocol. RNA libraries were generated using TruSeq Stranded RNA Library Preparation (Illumina) and sequenced with 2 × 75 bp paired-end reads in an Nextseq 500 (Illumina). Sequence reads were aligned using HISAT2 (2.2.0) and GRCm38 reference sequence. Differential expression analysis was performed with DESeq2 Bioconductor package in R (3.6.3) Canonical pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen) (Love et al., 2014).
2.7 |. Cecal microbiota transplantation
Approximately 50 mg of cecal contents from each mouse were pooled in 1 ml of liquid dental transport medium (Anaerobe Systems; Cat#AS-916) and vortexed. Pooled samples were filtered using 100 μm pore cell strainer (Falcon; Cat#08–771) and the plunger end of a sterile 5 ml syringe to remove any debris. Two hundred microliters of filtered pooled cecal contents were administered into Il10-deficient BALB/c mice via oral gavage using stainless steel feeding tube.
2.8 |. Isolation of lamina propria lymphocyte and flow cytometry
Small intestinal lamina propria lymphocytes were isolated using lamina propria dissociation kit (Miltenyi Biotec; Cat#130–097-410) and gentleMACS™ Octo Dissociator with Heaters (Miltenyi Biotec; Cat#130–096-427) according to manufacturer’s instruction. The dissociated tissue was resuspended in 5 ml of 40% Percoll (GE Healthcare; Cat#17–0891-01) and underlaid with 5 ml of 80% Percoll in 15 ml Falcon tube. Percoll gradient separation was performed by centrifuging at 600 RCF for 20 min at room temperature with break off. The lymphocytes at interphase were collected and washed with 10 ml 1× PBS once and resuspended in RPMI with 10% FBS. Cell Activation Cocktail (with Brefeldin A) (Biolegend; Cat#423303) was used to stimulate cells according to manufacturer’s instruction. LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen; Cat#L34955) was used to distinguish live and dead cells. Rat Anti-Mouse CD16/CD32 (BD; Cat#553141, Clone: 2.4G2) was used block nonspecific staining. Extracellular staining was performed with anti-CD3 (Miltenyi Biotec; Cat#130–102-943, Clone: REA641), anti-CD4 (Invitrogen; Cat#25–0041-82, Clone: GK1.5), and anti-CD8 (Miltenyi Biotec; Cat#130–120-806, REA601). After cells were permeabilized using Foxp3/Transcription Factor Staining Buffer Set (Invitrogen; Cat#00–5523-00), cells were stained with anti-IL17 (Miltenyi Biotec; Cat#130–102-262, Clone: TC11–18H10), anti-IL9 (Miltenyi Biotec; Cat#130–102-442, Clone: RM9A4), anti-IL4 (Miltenyi Biotec; Cat#130–102-435, Clone: BVD4–1D11), and anti-IFN-y (Miltenyi Biotec; Cat#130–102-388, Clone: AN.18.17.24).
2.9 |. Focused transcriptomic analyses
Ileal gene expression associated with tight junction pathway was measured using nCounter® Custom CodeSets (NanoString Technologies) (Supplementary Table 1). RNA counts were normalized to a group of housekeeping genes, and differentially expressed genes were calculated using Student’s t-test.
3 |. RESULTS
Oral administration of P. gingivalis induces ileal inflammation and alters gene expression profile associated with T-cell signaling pathway. To initially confirm whether oral administration of P. gingivalis leads to intestinal inflammation, 109 CFU of P. gingivalis was administered to wild-type Balb/c mice via oral gavage every other day for 14 days (Figure 1a). Histopathological analysis revealed significant inflammation in terminal ileum (Figure 1b). To further elucidate the mechanism behind P. gingivalis-induced intestinal inflammation, metatranscriptomic analysis was performed on ileal tissue. Differential gene expression analysis revealed 4 significantly upregulated and 30 significantly downregulated genes (Figure 1c). Notably, pathway analysis showed upregulation of T-cell signaling-related pathways as well as downregulation of liver X receptor (LXR)/retinoid X receptor (RXR) activation pathway (Figure 1d).
FIGURE 1.
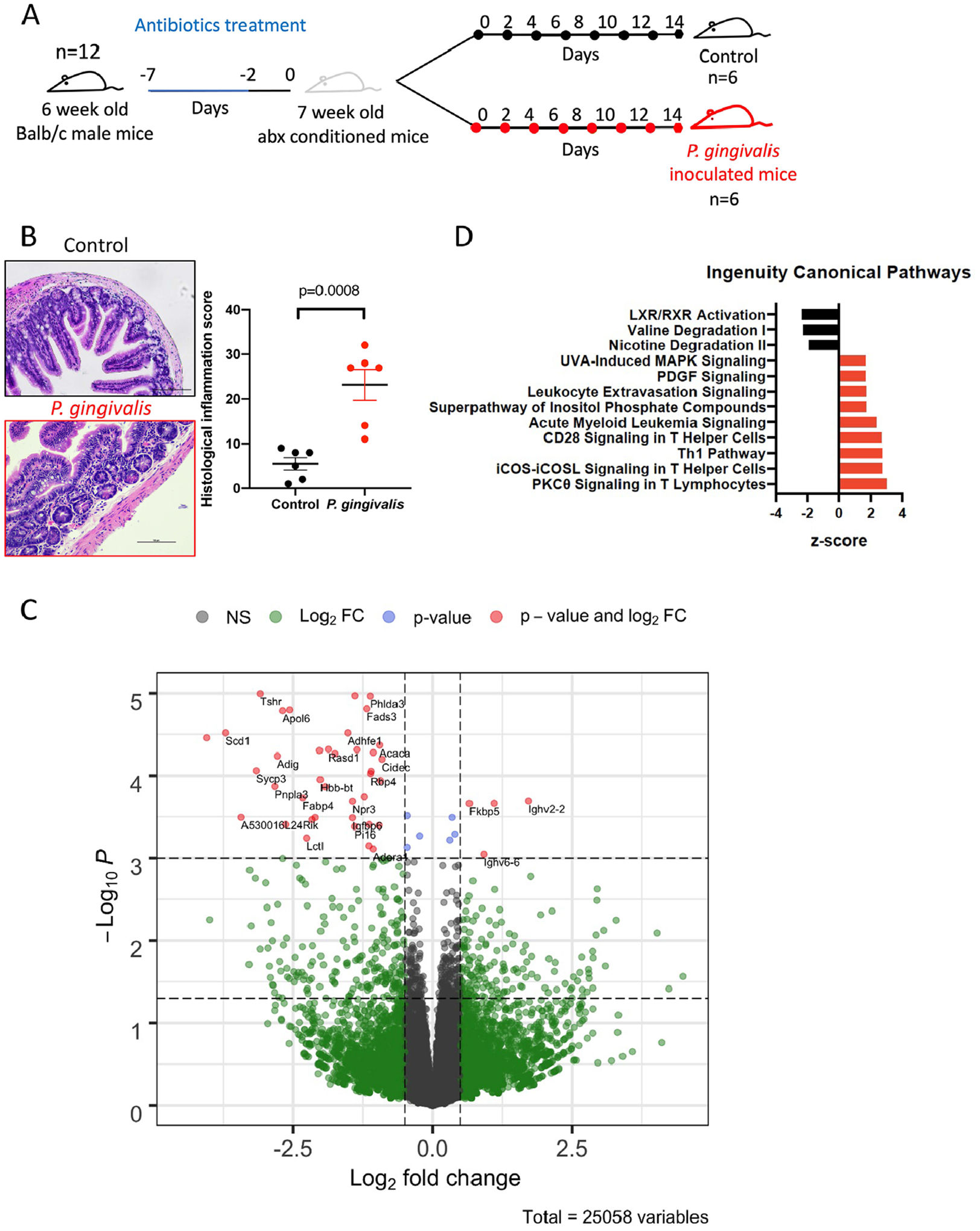
Oral administration of P. gingivalis induces intestinal inflammation. (a) Study design: 6-week-old Balb/c mice were given Kanamycin via autoclaved drinking water (1 mg/ml) for 5 days before the oral administration of either PBS control or P. gingivalis every consecutive day for 14 days. (b) Distal ileal samples were processed for H&E stain and scored for histological inflammation score. (c) Volcano plot showing the differentially expressed ileal genes in mice given P. gingivalis. Y-axis represents −log10 p value. Two horizontal dotted lines represent p = 0.05 and q = 0.05. X-axis represents Log2 fold-change. Two vertical dotted lines represent Log2 fold-change cutoff of −0.25 and 0.25. Dots labeled and colored in red represent genes with q < 0.05 and Log2 fold-change greater than 0.25 or less than −0.25. (d) Differentially expressed ileal gene expression data (p < 0.05) were used to perform pathway analysis using Ingenuity Pathway Analysis
Oral administration of P. gingivalis alters the gut microbiota and decreases bacterial alpha diversity but P. gingivalis does not colonize the intestinal tract. Next, we characterized the changes in the gut microbial community with oral administration of P. gingivalis by performing 16S rRNA gene and ITS sequencing on the mouse cecal samples. Results indicated that there was significant reduction in gut bacterial but not fungal alpha diversity in mice that received P. gingivalis compared to control mice (Figure 2a). Furthermore, we observed significant difference in bacterial beta diversity (Figure 2b). Administration of P. gingivalis led to increased Acholeplasmataceae in mice cecum (Figure 2c) (Supplementary Table 2). There were no significant differences in abundance of fungal taxa (Supplementary Table 3). P. gingivalis was not detected in the cecum of mice that received P. gingivalis via oral gavage (Figure 2d), indicating that P. gingivalis indirectly induces intestinal inflammation by disrupting the gut microbiota.
FIGURE 2.
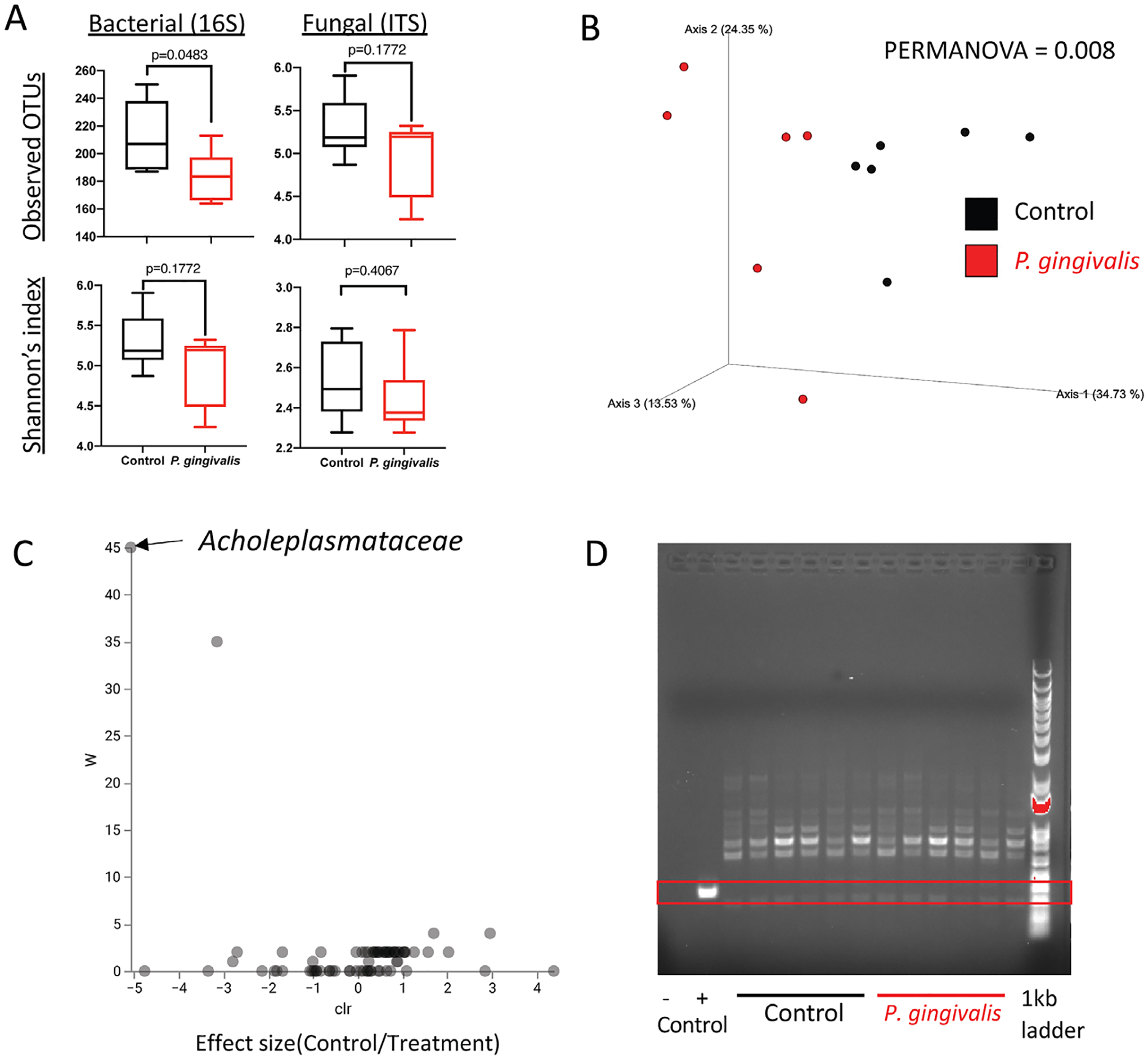
The effect of oral administration of P. gingivalis in the composition of gut microbiota. (a) Measurements of alpha diversity. (b) Beta diversity represented by Principal coordinates analysis (PCoA) using unweighted UNIFrac distance. (c) Volcano plot representing differentially abundant microbial taxa determined by ANCOM analysis. W statistic, strength of ANCOM test statistics, is on the y-axis, and the effect size, F-score, is on the x-axis. Statistically significant bacterial taxa are labeled. (d) PCR amplification of P. gingivalis-specific probe shown in gel electrophoresis
3.1 |. P. gingivalis-induced dysbiotic gut microbiota is sufficient to induce ileal inflammation in genetically susceptible mice
To investigate whether P. gingivalis induces intestinal inflammation by altering the gut microbiota, we performed a cecal microbiota transplantation study using antibiotic-conditioned Il10-deficient mice raised in positive pressure cages (Figure 3a). Histology analysis of the ileal tissue showed more ileal inflammation in mice that received P. gingivalis-altered cecal microbiota compared to control cecal transfer mice (Figure 3b). 16S rRNA gene and ITS sequencing analysis of the cecal samples confirmed the successful transfer of cecal microbiota from the previous experiment. There was a significant reduction in bacterial but not fungal alpha diversity and alteration in bacterial beta diversity in P. gingivalis-altered cecal microbiota recipient mice (Figure 4a, b). Increase in Acholeplasmataceae was observed in the control cecal microbiota recipient as previously seen (Figure 4c) (Supplementary Table 4). No fungal taxa were significantly altered (Supplementary Table 5).
FIGURE 3.
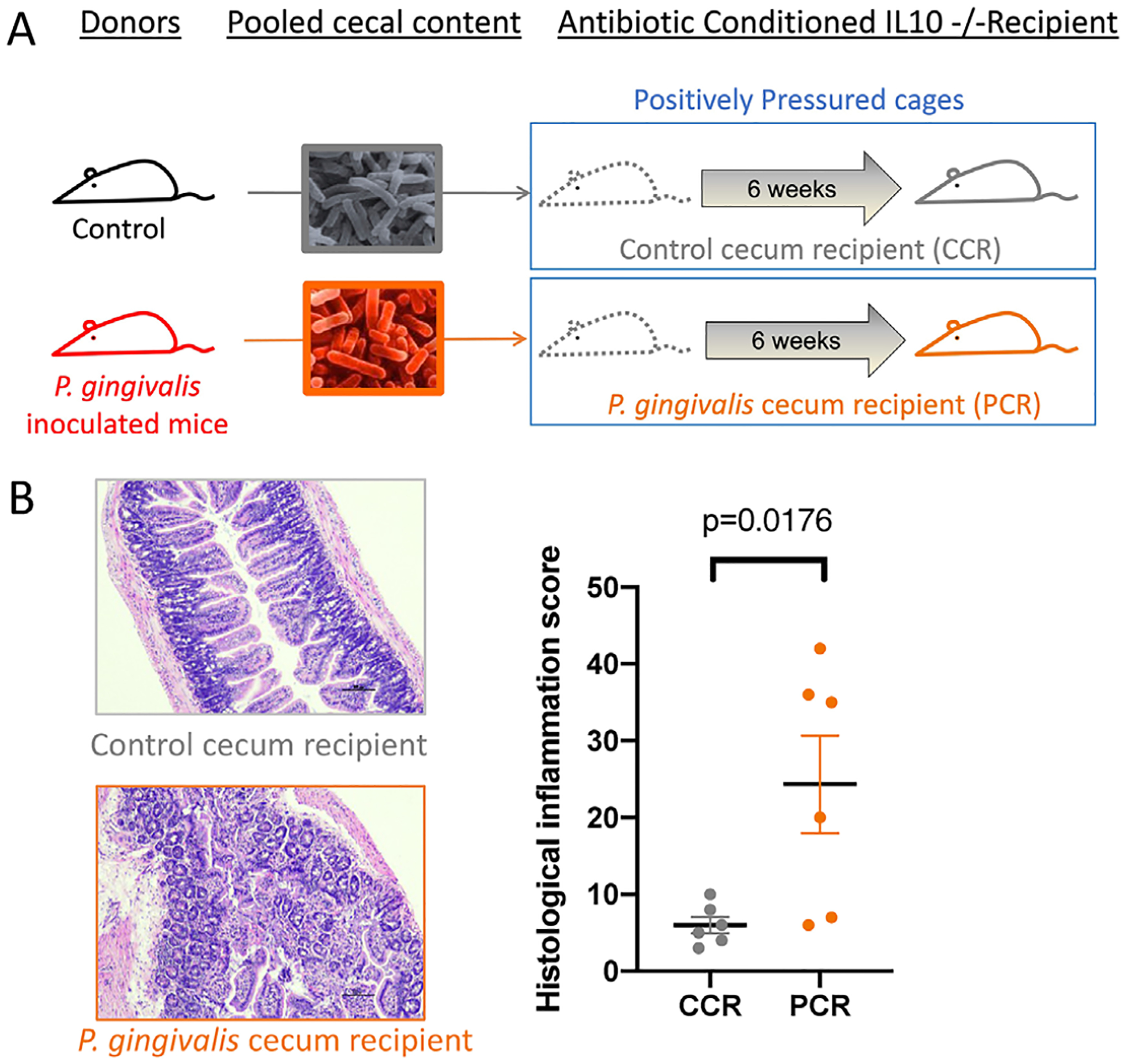
P. gingivalis-mediated dysbiotic gut microbiota induces intestinal inflammation. (a) Study design: 4-week-old Il10-deficient Balb/c mice were given antibiotic cocktail via autoclaved drinking water for 4 weeks before the oral administration of pooled-cecal contents from PBS or P. gingivalis treated mice. Mice were maintained in positively pressured cages for 6 weeks during the intestinal disease development experimental period. (b) Distal ileal samples were processed for H&E stain and scored for histological inflammation score
FIGURE 4.
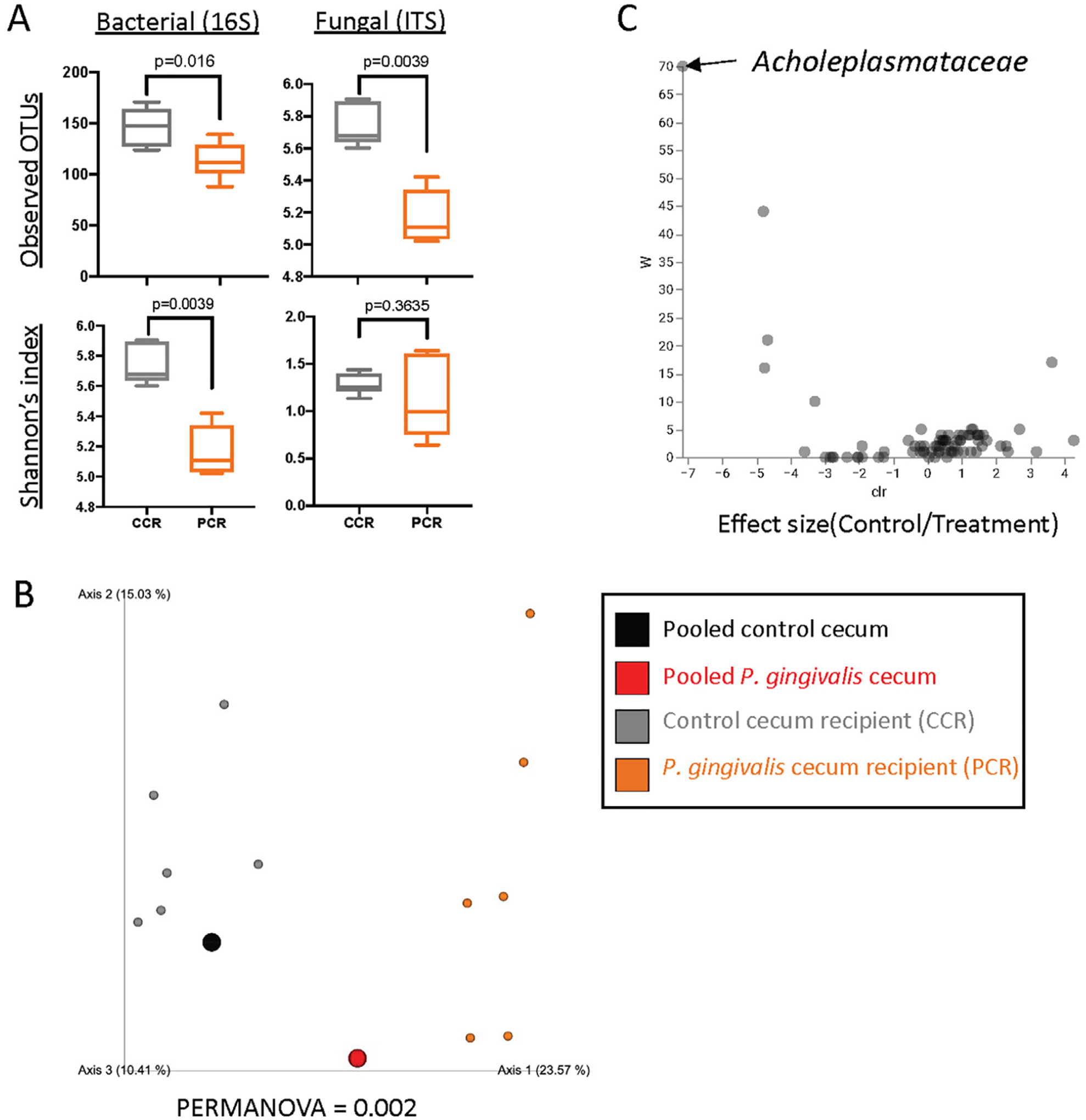
The gut microbiota composition of P. gingivalis-altered gut microbiota recipient mice. (a) Measurements of alpha diversity. (b) Beta diversity represented by Principal coordinates analysis (PCoA) using unweighted UNIFrac distance. (c) Volcano plot representing differentially abundant microbial taxa determined by ANCOM analysis. W statistic, strength of ANCOM test statistics, is on the y-axis, and the effect size, F-score, is on the x-axis. Statistically significant bacterial taxa are labeled
3.2 |. gingivalis-altered gut microbiota induces IL9+ CD4 T cells and disrupts the gut barrier function
To elucidate the cellular mechanisms mediating P. gingivalis-induced intestinal inflammation, flow cytometry analysis was performed to characterize the small intestinal T-cell population from the lamina propria. There was significant increase in the percentage of IL9+ CD4+ T cells in P. gingivalis-altered cecal microbiota recipient (Figure 5a). However, there was no change in IL17+, IFN-y+, or IL4+ CD4 T cells (Supplementary Figure 1). Intriguingly, the percentage of IL9+ CD4+ T cells was negatively correlated with both bacterial and fungal measurements of alpha diversity (Figure 5b). To further investigate the underlying mechanism, differential gene expression analysis was performed using custom NanoString panel consisting of genes that have been shown to play a role in intestinal barrier function. Notably, Cldn4, Cldn2, and Tjp2 mRNAs were significantly altered (Figure 5c).
FIGURE 5.
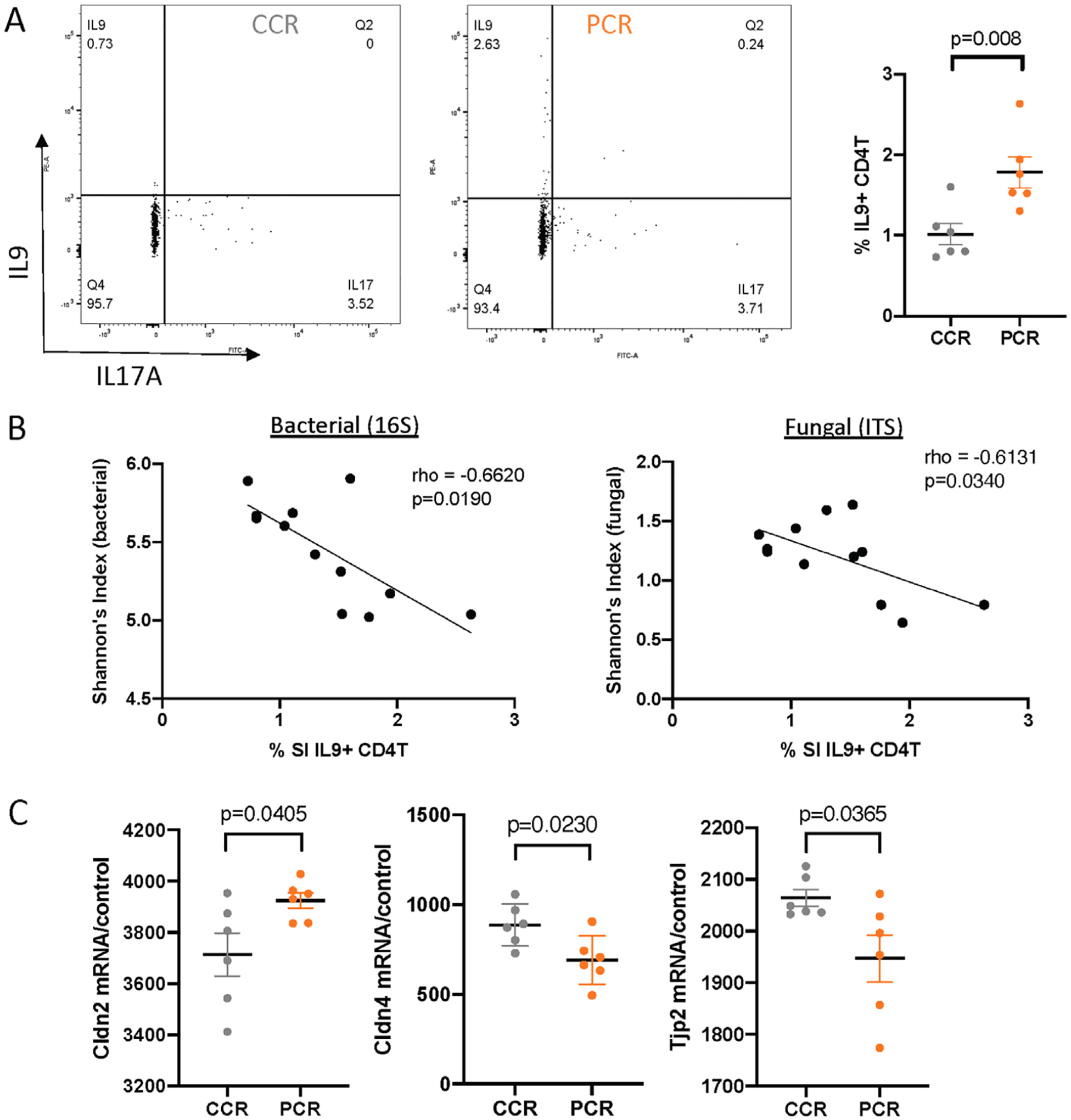
P. gingivalis-altered gut microbiota induces IL9+ CD4+ T cells and alters gut barrier function. (a) % IL9+ CD4+ T cells in the lamina propria of small intestine measured by flow cytometry analysis. (b) Spearman’s correlation analysis between % IL9+ CD4+ T cells and measurements of bacterial and fungal alpha diversity. (c) Gene expression levels of selected genes associated with tight junction pathway measured by NanoString analysis
4 |. DISCUSSION
A number of studies have attempted to investigate the oral–gut microbiota axis as the potential link that connects periodontal disease and inflammatory bowel disease (IBD; reviewed in Sohn et al. [2020]). In particular, the role of the oral pathogen, P. gingivalis, in initiating intestinal inflammation has been previously addressed. The Yamazaki group has shown that the oral administration P. gingivalis alters the gut microbiota and disrupts the gut barrier function to cause inflammation and alter host metabolism (Arimatsu et al., 2014; Kato et al., 2018; Nakajima et al., 2015). Subsequent studies by others have reproduced similar results where P. gingivalis is capable of disrupting the gut barrier function, leading to exacerbation of a variety of systemic diseases, including rheumatoid arthritis (Flak et al., 2019; Sato et al., 2017) and Parkinson’s disease (Feng et al., 2020). However, only few studies have attempted to address how P. gingivalis induces gut barrier dysfunction despite several data sets, including one from this study, showing that P. gingivalis is not capable of colonizing the lower gastrointestinal tract in mice (Geva-Zatorsky et al., 2017). Previous studies have shown that lipopolysaccharide (LPS), found in the outer membrane of Gram-negative bacteria, can disrupt the gut barrier function and induce intestinal inflammation (Guo et al., 2015; Guo et al., 2013; Im et al., 2012). Therefore, P. gingivalis-mediated intestinal inflammation may be due to the effect of ingesting P. gingivalis-derived LPS. However, a recent study shows that P. gingivalis-derived LPS paradoxically ameliorates DSS-induced colitis suggesting that the P. gingivalis-derived LPS is not responsible for the observed intestinal inflammation (Seo et al., 2019). Gut dysbiosis has also been implicated in intestinal inflammation (Buttó & Haller, 2016). IBD disease severity is significantly correlated with low species richness (Gevers et al., 2014). Furthermore, gnotobiotic animal studies show evidence for the causal role of gut dysbiosis in intestinal inflammation (Schaubeck et al., 2016). Therefore, we hypothesized that P. gingivalis induces intestinal inflammation by disrupting the gut microbial community.
To test the hypothesis, we first showed that oral administration of P. gingivalis indeed alters the gut microbiota and induces intestinal inflammation. Only one bacteria of the family, Acholeplasmataceae, which is a commensal facultative anaerobe, was identified to be increased in control mice by ANCOM analysis of the 16S rRNA gene sequencing data (Stephens et al., 1983). There is limited information on the role of Acholeplasmataceae in IBD, however one study investigating the anti-inflammatory effect of Lactobacillus mucosae in neuropsychiatric disorder, indicated that an increase in intestinal Acholeplasmataceae and amelioration of Escherichia coli K1-induced colitis with administration of L. mucosae (Kim et al., 2020). Overall, there was significant reduction in bacterial species richness and difference in unweighted UNIFrac distance as seen by others (Nakajima et al., 2015). We also characterized, for the first time, the changes in the fungal community by performing ITS sequencing as previous studies have indicated the potential role of altered fungal communities in both IBD and periodontal disease (Peters et al., 2017; Sokol et al., 2017). However, there were no significantly differentially abundant fungal taxa as well as differences in alpha diversity.
P. gingivalis affects the oral and gut microbiota by dissimilar mechanisms. In the oral cavity, P. gingivalis produces gingipain, which inhibits leukocyte killing of periodontal pathogens, thereby contributing to the increased microbial diversity of periodontal pockets (Hajishengallis, 2015). In fact, increased microbial diversity in the oral cavity is positively correlated with the severity of periodontal disease (Genco et al., 2019). This increased diversity seen in periodontal disease is distinct from infections seen from other sites of the body, where the progression of infection leads to reduction in diversity due to increasing dominance of the specific pathogen (Van Dyke et al., 2020). The results from this study and others have shown that ingestion of P. gingivalis leads to decreased gut bacterial diversity (Nakajima et al., 2015). The decreased gut bacterial diversity has been shown by multiple studies to be associated with various gastrointestinal pathologies such as IBD and colorectal cancer (Cheng et al., 2020; Gong et al., 2016). Further studies exploring the mechanism in which P. gingivalis reduces the gut bacterial diversity is needed to understand the role P. gingivalis has in gastrointestinal disease pathogenesis.
Next, we further expanded the phenotypic description of P. gingivalis-induced intestinal inflammation by characterizing the metatranscriptomic profile of the ileal tissue. Among the differentially regulated genes, Mrap, melanocortin receptor accessory protein, which inhibits the activation of nuclear transcription factor NF-κB and thus exert marked anti-inflammatory effects, was notably downregulated (Catania et al., 2010). The critical role of NF-κB activation in the development of IBD is well studied and Mrap has been proposed as a therapeutic target for IBD management (Lee et al., 2007; Podolsky, 2002). The top upregulated pathways were PKC-theta signaling in T lymphocyte, iCOS-iCOSL signaling in T helper cells, Th1 pathway, and CD28 signaling in T helper cells pathways, which highlighted the critical role of T cells in the observed intestinal pathology. Cellular immunity mediated by T lymphocyte is critical in maintaining mucosal immunity and dysregulation of T cells and their secreted mediators have shown to be play an essential role in IBD pathogenesis (Chen & Sundrud, 2016). In fact, the most effective therapies for IBD are the biologics, which target critical mediators of key T-cell immunological pathways (Paramsothy et al., 2018). The top downregulated pathway was liver X receptor/retinoid X receptor (LXR/RXR) activation, which is part of nuclear receptor network involved in gastrointestinal mucus secretion and expression of tight junction proteins autophagy (Klepsch et al., 2019). Both LXR and RXR induce anti-inflammatory effects and their deletion have been attributed to worsening intestinal inflammation or IBD phenotype in animal models and human IBD (Andersen et al., 2011; Jakobsson et al., 2014; Knackstedt et al., 2014).
Next, we utilized antibiotic-conditioned Il10-deficient mice raised and maintained in positively pressure cages to perform a cecal microbiota transplantation study to test whether the dysbiotic gut microbial community from P. gingivalis ingestion plays a causal role in the intestinal inflammation. Metagenomics sequencing data showed a successful transference of donor cecal microbiota to the recipient. Histological results clearly exhibited a transference of phenotype despite the absence of P. gingivalis in the cecal inoculum, suggesting that P. gingivalis induces intestinal inflammation by altering the gut microbiota. We further elucidated the mechanism whereby P. gingivalis mediates intestinal inflammation by performing flow cytometry analyses on the lamina propria lymphocytes isolated from the small intestine and shows an increase in the percentage of IL9+ CD4+ T cells. Recent translocation of microbes and microbial products to cause intestinal inflammation (Gerlach et al., 2014; Vyas & Goswami, 2018). To test whether intestinal barrier integrity is altered, we performed a focus transcriptomic analysis on custom panel consisting of genes involved in tight junction pathways and observed significant alteration in gene expression profile. Notably, Cldn4 and Cldn2 and Tjp2 were differentially expressed with same expression pattern as seen in human IBD patients (Gurram et al., 2016; Zhu et al., 2019). Studies have shown that host–microbiota interaction is necessary for expansion of IL9-producing T cells (Almeida et al., 2020). Our results not only highlight the crucial role of gut bacterial communities in IL9-producing T-cell expansion but also the fungal communities by showing the negative correlation between both bacterial and fungal species richness to the percent IL9+ CD4+ T cells. Anti-IL9 is currently being considered for therapeutic approach for the management of IBD (Zhang et al., 2018). Our results indicate that intestinal IL9-producing T cells could be modulated by manipulating the gut microbiota, possibly through altering the oral microbiota by treating periodontal disease.
5 |. CONCLUSION
Overall, this study fills a critical gap in knowledge by providing a mechanistic connection between periodontal diseases and IBD pathology. We have elucidated the mechanistic underpinnings of P. gingivalis-mediated intestinal inflammation. Specifically, we demonstrated that P. gingivalis induces ileal inflammation and IL9+ CD4+ lamina propria T cells and reduces gut epithelial barrier function through induction of gut dysbiotic microbial community (Figure 6).
FIGURE 6.
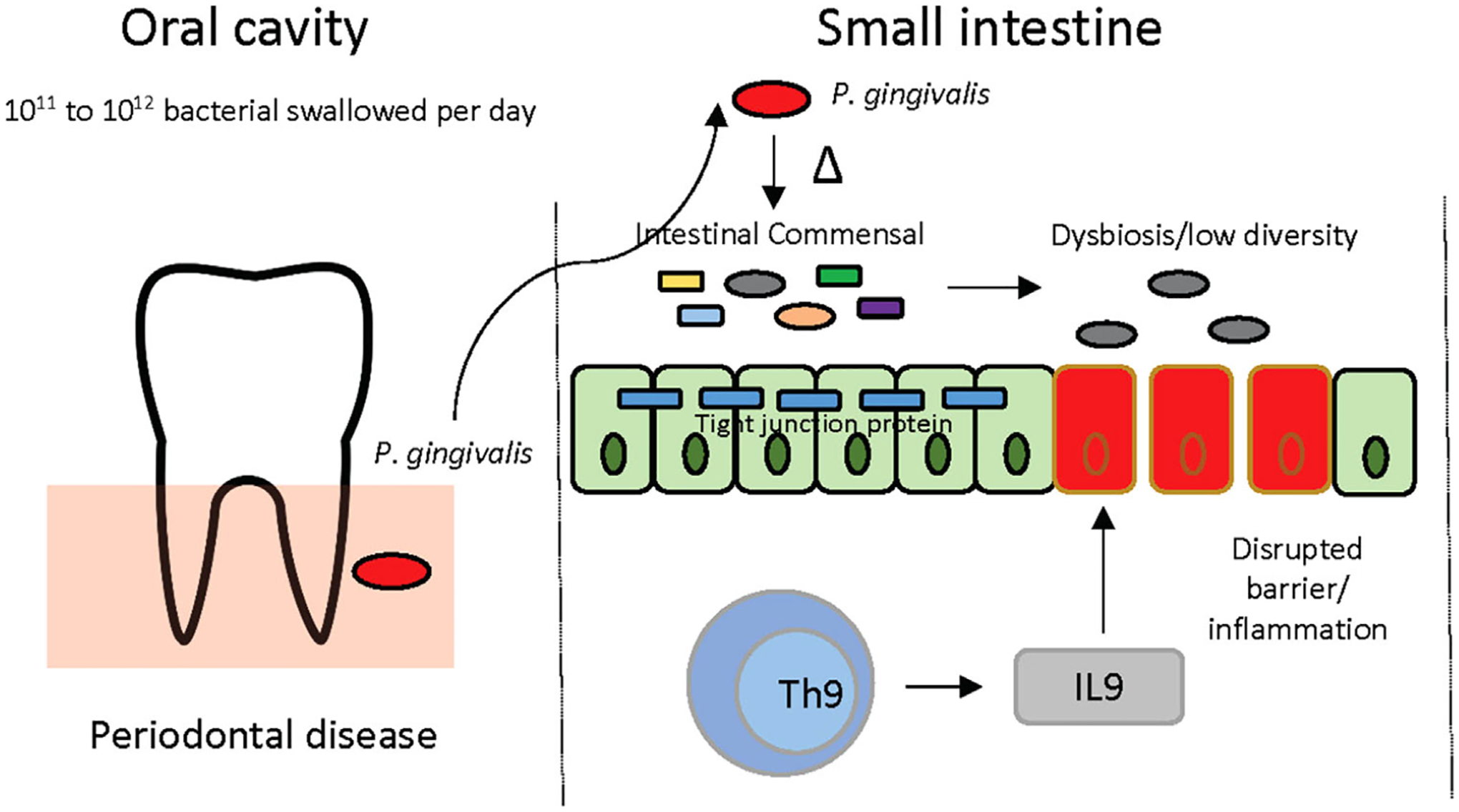
Schematic view of the proposed mechanism behind P. gingivalis-induced intestinal inflammation. Ingestion of P. gingivalis through saliva swallowing alters the gut microbiota and lowers the diversity, which causes induction of IL9+ CD4+ T cells in the small intestine, disrupts the gut barrier function, and causes ileal inflammation
P. gingivalis is one of many periodontal pathogens known to contribute to the pathogenesis of periodontal diseases. Thus, it is likely that other microorganisms exist that can migrate to the lower intestinal tract and induce dysbiosis to cause intestinal inflammation. Additional studies are needed to understand if other bacteria contribute to the oral–gut connection responsible for periodontal disease-associated IBD.
Supplementary Material
ACKNOWLEDGMENTS
Authors would like to thank Dr. Robert J. Genco who initially led this project but sadly passed away before its completion. Flow cytometry data was acquired at the Optical Imaging and Analysis Facility, School of Dental Medicine, State University of New York at Buffalo. Sequencing was performed at University at Buffalo Genomics and Bioinformatics Core at New York State Center of Excellence in Bioinformatics and Life Sciences.
FUNDING
We gratefully acknowledge support of the grant from Sunstar, Inc. (to J.S.) and the National Institutes of Health Grant R01DE02825803 (K.L.K.), R01DE02825803-S1 (to K.L.K.), R01DE029497 (to A.S.), and K18 DE029526 (to K.L.K.).
Funding information
the National Institutes of Health, Grant/Award Numbers: R01DE02825803, R01DE02825803-S1, R01DE029497, K18 DE029526
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE167088, reference number PRJNA701907.
REFERENCES
- Almeida RR, Vieira R. D.e S., Castoldi A, Terra FF, Melo ACL, Canesso MCC, Lemos L, Cipelli M, Rana N, Hiyane MI, Pearce EL, Martins FDS, Faria A. M. C. D.e, & Câmara NOS (2020). Host dysbiosis negatively impacts IL-9-producing T-cell differentiation and antitumour immunity. British Journal of Cancer, 123(4), 534–541. 10.1038/s41416-020-0915-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen V (2011). Polymorphisms in NF-κB, PXR, LXR, PPARγ and risk of inflammatory bowel disease. World Journal of Gastroenterology, 17(2), 197–206. 10.3748/wjg.v17.i2.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, Gotoh K, Motooka D, Nakamura S, Iida T, & Yamazaki K (2014). Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Scientific Reports, 4(1), 4828. 10.1038/srep04828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, & Gregory Caporaso J (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome, 6(1), 90. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J,…Caporaso JG (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito F, de Barros FC, Zaltman C, Carvalho AT, Carneiro AJ, Fischer RG, Gustafsson A, & Figueredo CM (2008). Prevalence of periodontitis and DMFT index in patients with Crohn’s disease and ulcerative colitis. Journal of Clinical Periodontology, 35(6), 555–560. 10.1111/j.1600-051X.2008.01231.x [DOI] [PubMed] [Google Scholar]
- Buttó LF, & Haller D (2016). Dysbiosis in intestinal inflammation: Cause or consequence. International Journal of Medical Microbiology, 306(5), 302–309. 10.1016/j.ijmm.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson A. J.o A., & Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania A, Lonati C, Sordi A, Carlin A, Leonardi P, & Gatti S (2010). The melanocortin system in control of inflammation. ScientificWorldJournal [Electronic Resource], 10, 1840–1853. 10.1100/tsw.2010.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, & Sundrud MS (2016). Cytokine networks and T-cell subsets in inflammatory bowel diseases. Inflammatory Bowel Diseases, 22(5), 1157–1167. 10.1097/MIB.0000000000000714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ling Z, & Li L (2020). The intestinal microbiota and colorectal cancer. Frontiers in Immunology, 11(3100). 10.3389/fimmu.2020.615056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, & Genco RJ (2015). Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. Journal of Periodontology, 86(5), 611–622. 10.1902/jop.2015.140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben U, Loddenkemper C, Doerfel K, Spieckermann S, Haller D, Heimesaat MM, Zeitz M, Siegmund B, & Kühl AA (2014). A guide to histomorphological evaluation of intestinal inflammation in mouse models. International Journal of Clinical and Experimental Pathology, 7(8), 4557–4576. [PMC free article] [PubMed] [Google Scholar]
- Feng Y-K, Wu Q-L, Peng Y-W, Liang F-Y, You H-J, Feng Y-W, Li G.e, Li X-J, Liu S-H, Li Y-C, Zhang Y, & Pei Z (2020). Oral P. gingivalis impairs gut permeability and mediates immune responses associated with neurodegeneration in LRRK2 R1441G mice. Journal of Neuroinflammation, 17(1), 347. 10.1186/s12974-020-02027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flak MB, Colas RA, Muñoz-Atienza E, Curtis MA, Dalli J, & Pitzalis C(2019). Inflammatory arthritis disrupts gut resolution mechanisms, promoting barrier breakdown by Porphyromonas gingivalis. JCI Insight, 4(13). 10.1172/jci.insight.125191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genco RJ, Lamonte MJ, Mcskimming DI, Buck MJ, Li L, Hovey KM, Andrews CA, Sun Y, Tsompana M, Zheng W, Banack HR, Murugaiyan V, & Wactawski-Wende J (2019). The subgingival microbiome relationship to periodontal disease in older women. Journal of Dental Research, 98(9), 975–984. 10.1177/0022034519860449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach K, Hwang Y, Nikolaev A, Atreya R, Dornhoff H, Steiner S, Lehr H-A, Wirtz S, Vieth M, Waisman A, Rosenbauer F, Mckenzie ANJ, Weigmann B, & Neurath MF (2014). TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nature Immunology, 15(7), 676–686. 10.1038/ni.2920 [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, & Kasper DL (2017). Mining the human gut microbiota for immunomodulatory organisms. Cell, 168(5), 928–943.e11. 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J,…Xavier RJ (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host & Microbe, 15(3), 382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gong X, Wang L, Yu X, & Dong Q (2016). Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterology Research and Practice, 2016, 1. 10.1155/2016/6951091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Al-Sadi R, Said HM, & Ma TY (2013). Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. American Journal of Pathology, 182(2), 375–387. 10.1016/j.ajpath.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Nighot M, Al-Sadi R, Alhmoud T, Nighot P, & Ma TY (2015). Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and MyD88. Journal of Immunology, 195(10), 4999–5010. 10.4049/jimmunol.1402598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurram B, Salzman NH, Kaldunski ML, Jia S, Li BUK, Stephens M, Sood MR, & Hessner MJ (2016). Plasma-induced signatures reveal an extracellular milieu possessing an immunoregulatory bias in treatment-naive paediatric inflammatory bowel disease. Clinical and Experimental Immunology, 184(1), 36–49. 10.1111/cei.12753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashneh RA, Khader YS, Alhumouz MK, Jadallah K, & Ajlouni Y (2012). The association between inflammatory bowel disease and periodontitis among Jordanians: A case-control study. Journal of Periodontal Research, 47(3), 293–298. 10.1111/j.1600-0765.2011.01431.x [DOI] [PubMed] [Google Scholar]
- Hajishengallis G (2015). Periodontitis: From microbial immune subversion to systemic inflammation. Nature Reviews Immunology, 15(1), 30–44. 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- How KY, Song KP, & Chan KG (2016). Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Frontiers in Microbiology, 7, 53. 10.3389/fmicb.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E, Riegler FM, Pothoulakis C, & Rhee SH (2012). Elevated lipopolysaccharide in the colon evokes intestinal inflammation, aggravated in immune modulator-impaired mice. American Journal of Physiology Gastrointestinal and Liver Physiology, 303(4), G490–G497. 10.1152/ajpgi.00120.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand A-C, Giraldo A, Da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, De Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P,…Meyer W (2015). International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycology, 53(4), 313–337. 10.1093/mmy/myv008 [DOI] [PubMed] [Google Scholar]
- Jakobsson T, Vedin L-L, Hassan T, Venteclef N, Greco D, D’amato M, Treuter E, Gustafsson J-Å, & Steffensen KR (2014). The oxysterol receptor LXRβ protects against DSS- and TNBS-induced colitis in mice. Mucosal Immunology, 7(6), 1416–1428. 10.1038/mi.2014.31 [DOI] [PubMed] [Google Scholar]
- Kato T, Yamazaki K, Nakajima M, Date Y, Kikuchi J, Hase K, Ohno H, & Yamazaki K (2018). Oral administration of porphyromonas gingivalis alters the gut microbiome and serum metabolome. mSphere, 3(5), e00460–18. 10.1128/mSphere.00460-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14), 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-K, Lee K-E, Lee S-A, Jang H-M, & Kim D-H (2020). Interplay between human gut bacteria Escherichia coli and Lactobacillus mucosae in the occurrence of neuropsychiatric disorders in mice. Frontiers in Immunology, 11(273), 10.3389/fimmu.2020.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG, Hayashi A, Imai J, Sugihara K, Miyoshi M, Brazil JC, Kuffa P, Hill BD, Rizvi SM, Wen F, Bishu S, Inohara N, Eaton KA, Nusrat A, Lei YL, Giannobile WV, & Kamada N (2020). The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell, 182(2), 447–462.e14. 10.1016/j.cell.2020.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepsch V, Moschen AR, Tilg H, Baier G, & Hermann-Kleiter N (2019). Nuclear receptors regulate intestinal inflammation in the context of IBD. Frontiers in immunology, 10, 1070. 10.3389/fimmu.2019.01070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knackstedt R, Shaoli S, Moseley V, & Wargovich M (2014). The importance of the retinoid X receptor alpha in modulating inflammatory signaling in acute murine colitis. Digestive Diseases and Sciences, 59(4), 753–759. 10.1007/s10620-013-2902-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, & Hajishengallis G (2018). The oral microbiota: Dynamic communities and host interactions. Nature Reviews Microbiology, 16(12), 745–759. 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim JS, Kim JM, Kim N, Jung HC, & Song IS (2007). Simvastatin inhibits NF-kappaB signaling in intestinal epithelial cells and ameliorates acute murine colitis. International Immunopharmacology, 7(2), 241–248. 10.1016/j.intimp.2006.10.013 [DOI] [PubMed] [Google Scholar]
- Lira-Junior R, & Figueredo CM (2016). Periodontal and inflammatory bowel diseases: Is there evidence of complex pathogenic interactions? World Journal of Gastroenterology, 22(35), 7963–7972. 10.3748/wjg.v22.i35.7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, & Peddada SD (2015). Analysis of composition of microbiomes: A novel method for studying microbial composition. Microbial Ecology in Health and Disease, 26, 27663. 10.3402/mehd.v26.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, Ohno H, & Yamazaki K (2015). Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of Enterobacteria to the liver. Plos One, 10(7), e0134234. 10.1371/journal.pone.0134234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Wu GD, Albenberg L, & Tomov VT (2017). Gut microbiota and IBD: Causation or correlation? Nature Reviews Gastroenterology & Hepatology, 14(10), 573–584. 10.1038/nrgastro.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramsothy S, Rosenstein AK, Mehandru S, & Colombel J-F (2018). The current state of the art for biological therapies and new small molecules in inflammatory bowel disease. Mucosal Immunology, 11(6), 1558–1570. 10.1038/s41385-018-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, Wu J, Hayes RB, & Ahn J (2017). The oral fungal mycobiome: Characteristics and relation to periodontitis in a pilot study. Bmc Microbiology, 17(1), 157. 10.1186/s12866-017-1064-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolsky DK (2002). Inflammatory bowel disease. New England Journal of Medicine, 347(6), 417–429. 10.1056/NEJMra020831 [DOI] [PubMed] [Google Scholar]
- Sato K, Takahashi N, Kato T, Matsuda Y, Yokoji M, Yamada M, Nakajima T, Kondo N, Endo N, Yamamoto R, Noiri Y, Ohno H, & Yamazaki K (2017). Aggravation of collagen-induced arthritis by orally administered Porphyromonas gingivalis through modulation of the gut microbiota and gut immune system. Scientific Reports, 7(1), 6955. 10.1038/s41598-017-07196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, Basic M, Dupont A, Hornef M, von Bergen M, Bleich A, & Haller D (2016). Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut, 65(2), 225. 10.1136/gutjnl-2015-309333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y, Oh S-J, Ahn J-S, Shin YY, Yang JW, & Kim H-S (2019). Implication of Porphyromonas gingivalis in colitis and homeostasis of intestinal epithelium. Laboratory Animal Research, 35(1), 26. 10.1186/s42826-019-0029-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Y-Y, Kong X-B, Ge Y-P, Liu Z-Y, Chen J-Y, Jiang J-W, Jiang H-B, & Fang S-L (2020). Periodontitis and inflammatory bowel disease: A meta-analysis. BMC Oral Health, 20(1), 67. 10.1186/s12903-020-1053-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Sun Y, Genco RJ, & Kirkwood KL (2020). The periodontal microenvironment: A potential reservoir for intestinal pathobionts in Crohé s Disease. Current Oral Health Reports, 7, 37–44. 10.1007/s40496-020-00251-9 [DOI] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, & Beaugerie L (2017). Fungal microbiota dysbiosis in IBD. Gut, 66(6), 1039. 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens EB, Aulakh GS, Rose DL, Tully JG, & Barile MF (1983). Intraspecies genetic relatedness among strains of Acholeplasma laidlawii and of Acholeplasma axanthum by nucleic acid hybridization. Microbiology (Reading, England), 129(6), 1929–1934. 10.1099/00221287-129-6-1929 [DOI] [PubMed] [Google Scholar]
- Van Dyke TE, Bartold PM, & Reynolds EC (2020). The nexus between periodontal inflammation and dysbiosis. Frontiers in Immunology, 11(511), 10.3389/fimmu.2020.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavricka SR, Manser CN, Hediger S, Vögelin M, Scharl M, Biedermann L, Rogler S, Seibold F, Sanderink R, Attin T, Schoepfer A, Fried M, Rogler G, & Frei P (2013). Periodontitis and gingivitis in inflammatory bowel disease: A case-control study. Inflammatory Bowel Diseases, 19(13), 2768–2777. 10.1097/01.MIB.0000438356.84263.3b [DOI] [PubMed] [Google Scholar]
- Vyas SP, & Goswami R (2018). A decade of Th9 cells: Role of Th9 cells in inflammatory bowel disease. Frontiers in Immunology, 9, 1139. 10.3389/fimmu.2018.01139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-C, Chen T-P, & Chang Y-C (2018). Inflammatory bowel disease as a risk factor for periodontitis under Taiwanese National Health Insurance Research database. Journal of Dental Sciences, 13(3), 242–247. 10.1016/j.jds.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li L, Huang S, Feng T, Zhou G, & Chen M (2018). IDDF2018-ABS-0193 Cytokine IL9 mediates the pathogenesis of Crohn’s disease through the MIR21-CLDN8 pathway. Gut, 67(Suppl 2), A11. 10.1136/gutjnl-2018-IDDFabstracts.25 [DOI] [PubMed] [Google Scholar]
- Zhu L, Han J, Li L.i, Wang Y, Li Y, & Zhang S (2019). Claudin family participates in the pathogenesis of inflammatory bowel diseases and colitis-associated colorectal cancer. Frontiers in immunology, 10, 1441. 10.3389/fimmu.2019.01441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE167088, reference number PRJNA701907.


