Abstract
Regulation of gene expression in the domain Archaea, and specifically hyperthermophiles, has been poorly investigated so far. Biochemical experiments and genome sequencing have shown that, despite the prokaryotic cell and genome organization, basal transcriptional elements of members of the domain Archaea (i.e., TATA box-like sequences, RNA polymerase, and transcription factors TBP, TFIIB, and TFIIS) are of the eukaryotic type. However, open reading frames potentially coding for bacterium-type transcription regulation factors have been recognized in different archaeal strains. This finding raises the question of how bacterial and eukaryotic elements interact in regulating gene expression in Archaea. We have identified a gene coding for a bacterium-type transcription factor in the hyperthermophilic archaeon Sulfolobus solfataricus. The protein, named Lrs14, contains a potential helix-turn-helix motif and is related to the Lrp-AsnC family of regulators of gene expression in the class Bacteria. We show that Lrs14, expressed in Escherichia coli, is a highly thermostable DNA-binding protein. Bandshift and DNase I footprint analyses show that Lrs14 specifically binds to multiple sequences in its own promoter and that the region of binding overlaps the TATA box, suggesting that, like the E. coli Lrp, Lrs14 is autoregulated. We also show that the lrs14 transcript is accumulated in the late growth stages of S. solfataricus.
In the last few years, it has been clearly shown that the basal transcription apparatus of the domain Archaea is of a eukaryotic type. Indeed archaeal promoter sequences and core proteins (RNA polymerase, transcription factors TBP, TFIIB, and TFIIS) are structurally and functionally related to their eukaryotic counterparts (recently reviewed in references 27 and 30).
In particular, a TATA box-like element is typically found about 25 bp upstream of transcription start sites; this sequence is sufficient, in the presence of purified TBP, TFIIB, and RNA polymerase, for correct in vitro transcription initiation in cell-free transcription systems.
In the current view, the transcription initiation complex in Archaea is a simple, ancient version of the eukaryal one and is composed of only absolutely essential factors—the starting point for the evolution of the actual, complex eukaryotic transcription machinery. However, several lines of evidence suggest that this might be an oversimplification. Regulation of gene expression in Archaea, and specifically hyperthermophiles, has been poorly investigated; in the few examples reported so far, no eukaryote-type regulation factor (such as enhancer or silencer binding proteins) has been described, and no clear evidence of DNA sequences (besides the TATA box) involved in transcription regulation has been provided.
Instead, from the emerging sequences of archaeal genomes, a large number of open reading frames (ORFs) potentially coding for bacterium-type transcription regulation factors has been identified (3, 17, 29). This finding suggests that bacterium- and eukaryote-type elements cooperate in the regulation of gene expression in Archaea. Given the chimeric nature of the archaeal transcription apparatus, neither the bacterial nor the eukaryotic model can be simply applied to the regulation of gene expression in these organisms. Regulatory circuits need to be elucidated to explain the interplay among eukaryote- and bacterium-type elements.
Putative homologs of the leucine-responsive regulation protein of Escherichia coli (Lrp) have been found in Archaea. Lrp is the prototype, and most extensively studied member, of the Lrp-AsnC family of regulators of gene expression in both Gram-positive and Gram-negative bacteria (reviewed in references 4 and 23). Lrp is a homodimer containing two identical subunits of 18 kDa, and it acts either as an activator or as a repressor on a number of different genes and operons. Besides its role as a specific transcriptional regulator, Lrp also acts as a chromosomal organizer, inducing conformational changes in DNA and promoting the formation of higher-order DNA-protein complexes (32).
ORFs potentially coding for Lrp-AsnC homologs are widely distributed in the archaeal domain, since they have been found in the following members of two distant subdomains, Crenarchaeota and Euryarchaeota: Sulfolobus acidocaldarius (reported as S. solfataricus in reference 6 and corrected in reference 5); Sulfolobus solfataricus (10), Pyrococcus furiosus (9, 20), Methanococcus jannaschii (3), and Archaeoglobus fulgidus (17). Interestingly, they are present in multiple copies within the same genome. No functional studies of such proteins have been published so far.
We have identified in the genome of the hyperthermophilic archaeon S. solfataricus a gene coding for a bacterium-type transcription factor we named Lrs14, which is distantly related to the Lrp-AsnC family. Lrs14, expressed in E. coli, is a highly thermostable DNA-binding protein which binds to specific sequences in its own promoter. Characterization of the Lrs14 DNA-binding activity in vitro and analysis of the lrs14 gene transcription in vivo are presented in this report.
MATERIALS AND METHODS
DNA manipulations.
All enzymatic DNA reactions were performed according to standard techniques. DNA fragments and oligonucleotides were end labelled either with [γ-32P]ATP and T4 polynucleotide kinase or with the Klenow enzyme and the appropriate 32P-labelled deoxynucleotide. DNA amplifications were obtained by standard techniques using the Pfu DNA polymerase (Stratagene); amplified fragments were always checked by DNA sequencing. For nucleic acid hybridization, DNA probes were labelled with the Random Primer kit from Boehringer; RNA probes were obtained with T7 RNA polymerase as reported previously (26).
Cloning of the lrs14 gene.
The ORF coding for Lrs14 was identified during the sequencing of the S. solfataricus (P2 strain) genome; the gene lrs14 was amplified from S. solfataricus MT4 DNA by using the oligonucleotides N-FULL (5′CGGGATCCAATGCAAGTAGAGAATATAAG) and C-FULL (5′CTGTCGACTTACTTTTCTTTCAATTCTTG), which match the 5′- and the 3′-terminal ends of the coding sequence, respectively, with the addition of a BamHI site tail at the 5′ end and a SalI site tail at the 3′ end. The BamHI-SalI fragment was subcloned in vector pGEM3 (Promega), producing plasmid pGEM-Hom; the insert of this plasmid was checked by DNA sequencing.
Computer analysis.
The deduced protein sequence of Lrs14 was compared with those in the Swiss Prot, PIR, and GenBank-EMBL libraries by using standard software (programs FASTA, BLAST, and Gapped-BLAST). The programs MultALIN (7) and CLUSTALW were used to obtain sequence alignments. The program HTH, available at the NPSA server (22a), has been used to predict the helix-turn-helix motif (8).
Northern analysis.
S. solfataricus cultures (500 ml) were grown at different optical densities at 600 nm (OD600s) as indicated; RNA was extracted and analyzed as previously described (26). The amount of RNA loaded was normalized by the fluorescence of rRNAs in ethidium bromide-stained gels and by staining the filters with methylene blue. The same filters were hybridized sequentially with the 390-bp BamHI-SalI DNA fragment from pGEM-Hom, which contains the whole lrs14 coding sequence, labelled by random primer, and with a riboprobe prepared from the 185-bp HindIII-BglII fragment internal to the lacS gene, as described previously (26).
Purification of Lrs14.
The BamHI-SalI fragment from pGEM-Hom was subcloned in the BamHI-XhoI sites of pRSET B (Invitrogen), producing plasmid pRSET-390. This plasmid (after resequencing of the insert) was transformed into E. coli JM109 (DE3). Protein purification through Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen) was performed with modifications of the manufacturer’s protocol. Briefly, a 1-liter culture was grown at an OD600 of 0.9 in Luria-Bertani medium supplemented with 50 μg of ampicillin per ml, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and grown overnight. Cells, together with a 10-ml uninduced sample, were centrifuged for 20 min at 4,000 rpm in a Sorvall GSA rotor, resuspended in 10 ml of lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole), and broken with a French press. The lysate was clarified by centrifugation at 10,000 rpm in a Sorvall S534 rotor for 10 min. Three milliliters of 50% Ni-NTA matrix was added to the supernatant (12 ml), and the mixture was incubated for 1 h at 4°C with gentle shaking and packed onto a column. The column was washed with 30 ml of washing buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole), and eluted with elution buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl) containing stepwise increasing amounts of imidazole (50, 150, and 300 mM); fractions were collected and checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions 11 to 13, which eluted at 150 mM imidazole, contained a single 14-kDa protein band which was absent in other fractions (data not shown); the fractions were pooled, and protein content was determined by the Bio-Rad method (2). The total amount of pure Lrs14 recovered was 1.5 mg.
Bandshift assays.
Standard reaction mixtures (10 μl) contained 1× binding buffer (20 mM Tris-HCl [pH 7.5], 10% glycerol, 50 mM KCl, 0.1 mM dithiothreitol), purified Lrs14, cold competitor DNA as indicated, and about 2 × 103 to 5 × 103 cpm of the appropriate end-labelled DNA. After incubations, samples were immediately loaded on native 5% or 7% polyacrylamide gels in 0.5× Tris-borate buffer and run at room temperature. The gels were dried, and the autoradiograms were exposed at −80°C. The probes UP and DOWN were obtained by PCR amplification with the following pairs of primers: 480Hinc (5′AAATCACGTTAACTT) and 537Nde (5′TTGCATATGAATATAA) and N-FULL (5′CGGGATCCAATGCAAGTAGAGAATATAAG) and 595Acc (5′GTGCGTCTACTAATC).
Amplified fragments were end labelled with [γ-32P]ATP and T4 polynucleotide kinase. The oligonucleotides cLRS and TATA (see Fig. 3) were purchased from PRIMM (Milan, Italy) and were annealed and end labelled as previously reported (12).
FIG. 3.
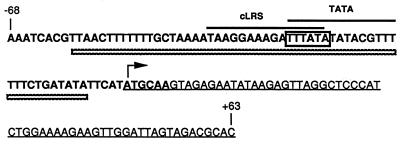
DNA sequence of the region spanning nt −68 to +63 of lrs14 relative to ATG, which is indicated by an arrow. The putative TATA-like sequence is boxed. The probes used in bandshift analysis are shown as follows: UP, boldface; DOWN, underlined. cLRS and TATA are the oligonucleotides used in the experiment described in the legend to Fig. 6. The whole fragment shown, labelled at the 5′ end, was used in the DNase I footprinting experiment; the region protected from DNase I cleavage is indicated by dashed box underlines.
DNase I footprinting.
A fragment spanning nucleotides −68 to +63 relative to the ATG was amplified by using the oligonucleotides 480Hinc and 595Acc (shown above). The primers were alternatively end labelled. The Lrs14 protein was preincubated for 1 h at 75°C; the probe was added, and binding reactions (5′ at room temperature) were performed as described above. DNase I digestion and electrophoresis were performed as reported previously (32).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBL, EMBL, and GenBank nucleotide sequence databases under accession no. AF098294.
RESULTS
Identification and transcriptional analysis of lrs14.
During the sequencing of the S. solfataricus genome, we have identified an ORF, which we named lrs14, potentially coding for a 123-amino-acid protein (molecular mass of 14 kDa). A database search showed that the predicted Lrs14 protein shares significant sequence similarity with three archaeal hypothetical proteins, and low similarity with bacterial transcription regulation factors of the Lrp-AsnC family (Fig. 1). In particular the protein most similar to Lrs14 is an A. fulgidus ORF (11) which, in turn, gives a significant hit (E value of <e-4 in a BLAST search) with established Lrp homologs. Interestingly, the xylose repressor of Bacillus subtilis (19) also appears to be related somehow to Sulfolobus Lrs14 and E. coli Lrp.
FIG. 1.
Alignment of Lrs14 with some of the best matches from a BLAST search. Shown are results for three sequences of hypothetical archaeal proteins (MJ Orf, MJO82 in the M. jannaschii database [3]; Hs Orf, Halobacterium sp. strain NRC-1 [15]; Af Orf, A. fulgidus [11]) E. coli Lrp (Ec Lrp [36]), and the B. subtilis xylose repressor (Bs XylR; 200 C-terminal amino acids are not shown [19]). The helix-turn-helix motif of E. coli Lrp (25) and that of Lrs14, predicted by the program HTH (8), are underlined; asterisks indicate C termini. Amino acid residues that are identical or similar in at least three sequences are boxed.
Figure 1 shows that conservation is higher in a region corresponding to the helix-turn-helix motif of E. coli Lrp, which is responsible for its DNA-binding activity (25). A potential helix-turn-helix motif in the corresponding region of Lrs14 (amino acids 45 to 66) has been predicted by the program HTH (8).
We have analyzed the transcription of the lrs14 gene by Northern blotting (Fig. 2A). Total RNA was extracted from S. solfataricus cells at different growth stages and was probed with a DNA fragment corresponding to the lrs14 coding sequence. The probe hybridizes to a 0.4-kb-long transcript, accounting for a monocistronic transcription of the gene. This finding suggests that the lrs14 promoter is adjacent to its coding sequence; indeed the sequence TTTATA, which matches exactly the consensus for the archaeal TATA box, is located 31 bp upstream of the presumptive first ATG (Fig. 3), a distance rather common in archaeal genes (for a recent review, see reference 31). This result also suggests that eventual transcription regulatory sequences, if present, should be located within 100 bp upstream of the coding sequence (see below).
FIG. 2.
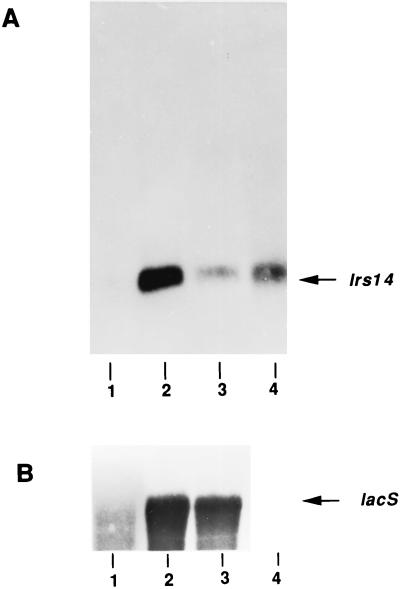
(A) Northern blot showing the lrs14 transcript at different growth stages. S. solfataricus RNAs were extracted from cultures grown at different OD600s: lane 1, 0.2; lane 2, 0.4; lane 3, 0.6; lane 4, 0.8. The same amount of RNA (2 μg) was loaded in each lane; the filter was hybridized with the 390-bp BamHI-SalI DNA fragment from pGEM-Hom, which contained the whole lrs14 coding sequence. (B) The same filter was hybridized with a riboprobe prepared from the HindIII-BglII fragment of the lacS gene (26).
RNA samples in Fig. 2 were prepared from cultures grown at OD600s of 0.2, 0.4, 0.6, and 0.8, respectively. Despite the small difference in OD, these conditions reflect very different growth stages, since S. solfataricus has a very slow growth rate (doubling time of about 7.5 h in rich medium) and grows at low density; in the experiment shown in Fig. 2, the maximal OD600 was 0.8, which was therefore assumed to be stationary phase. Steady-state lrs14 RNA shows a growth phase-regulated pattern: it is undetectable in the early growth stages (Fig. 2A, lane 1), is highly induced during the exponential phase (lane 2), and declines in the late logarithmic phase (lane 3), but is maintained at relatively high levels in the stationary phase (lane 4). This accumulation in late stages of growth is specific for the lrs14 transcript, as shown by control hybridization of the same filter with a probe from the lacS gene, which is not expressed at an OD600 of 0.8 (Fig. 2B) (26). Moreover, a transcription pattern similar to that of lacS is shown by the Sso7d gene of S. solfataricus, coding for a nonspecific DNA-binding protein (22). To our knowledge, lrs14 is the first Sulfolobus gene whose transcript accumulates in late stages of growth described so far. Interestingly, the E. coli Lrp transcription is also sustained during the stationary phase (21).
Lrs14 is a DNA-binding protein which binds to its own promoter.
In order to characterize the Lrs14 protein, we have expressed it in E. coli by using vector pRSETB (see Materials and Methods). In this system, the protein is obtained as a fusion with a histidine tag at the N terminus, and can be purified by affinity chromatography on an Ni-NTA-agarose column (14). Using this single-step purification, we have obtained Lrs14 >95% pure, as verified by SDS-PAGE (not shown).
The Lrs14 protein purified from E. coli shows the ability to bind total S. solfataricus DNA in nitrocellulose binding assays (not shown). In order to characterize its DNA-binding activity, we needed to identify its target sequences. The E. coli Lrp protein binds to specific sequences in the promoters of many different genes, including its own (34); we reasoned that Lrs14 might have the same autoregulatory function.
We therefore performed bandshift analysis with a probe spanning 68 bp immediately upstream of the first ATG of the lrs14 gene (probe UP [Fig. 3]). As a control, we used a fragment of similar length located immediately downstream of the ATG (probe DOWN [Fig. 3]). The purified Lrs14 protein binds to the probe UP (Fig. 4, lanes 2 to 4), forming a single band at a low protein concentration and two bands at increasing protein concentrations (also described below). Both bands are specific, since they do not form with the probe DOWN (lane 9), and they are effectively competed by an excess of cold UP (lanes 5 and 6), but not DOWN (lanes 7 and 8), fragments.
FIG. 4.
Binding of purified Lrs14 expressed in E. coli to the UP and DOWN probes, as analyzed by bandshift assay. Lanes 1 to 8, probe UP; lane 1, naked probe; lane 2, 0.1 μg of purified Lrs14; lane 3, 0.6 μg of purified Lrs14; lane 4, 1.2 μg of purified Lrs14; lanes 5 to 8, 0.6 μg of purified Lrs14 preincubated with cold competitors; lane 5, 10× excess of UP; lane 6, 100× excess of UP; lane 7, 10× excess of DOWN; lane 8, 100× excess of DOWN; lanes 9 and 10, probe DOWN; lane 9, DOWN with 1.2 μg of purified Lrs14; lane 10, naked probe. Prebinding and binding reactions were performed for 10 min at 25°C and run on 5% polyacrylamide gels.
Lrs14 is a highly thermostable DNA-binding protein.
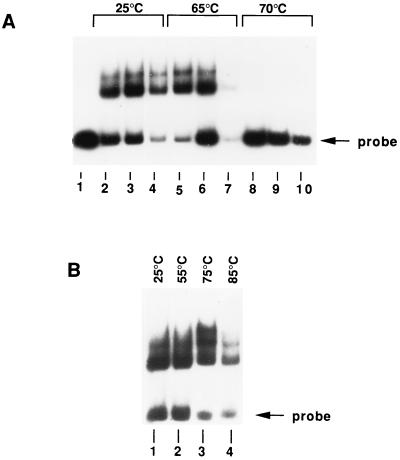
S. solfataricus is a hyperthermophilic microorganism, with optimum growth temperature above 80°C; to date, only nonspecific DNA-binding activities from such organisms have been characterized (1, 12). The DNA-binding activity of the S. solfataricus Sso7d protein is not affected by temperatures up to 70°C (12). In order to test the effect of temperature on the binding of Lrs14 to DNA, we performed binding experiments at different temperatures (Fig. 5A). Lrs14 binds with the same efficiency to the UP probe at 25°C and 65°C; in contrast, no DNA-protein complex is formed if the reaction is carried out at 70°C. This result might be due to protein inactivation or to local denaturation of the DNA probe, which is A/T rich (Fig. 3). To test the thermostability of Lrs14, we incubated the protein in the absence of DNA at different temperatures; subsequently we added the probe and carried out the binding reaction at room temperature for 5 min. The DNA-binding activity of Lrs14 is completely stable to 1 h of incubation at 85°C (Fig. 5b). Interestingly, we noticed that incubation at 75°C results in a more efficient binding, and a third, slower DNA-protein complex appears (also described below). This observation suggests that the binding activity of Lrs14 is activated by preincubation at 75°C and can be explained assuming that high temperatures induce subtle conformational changes in the protein. Experiments are in progress to determine the effect of high temperature on the structural stability and flexibility of Lrs14.
FIG. 5.
Lrs14 is a thermostable DNA-binding protein. (A) Binding of Lrs14 to the probe UP at different temperatures. Lane 1, naked probe; lanes 2 to 10, 0.8 μg of Lrs14. Lanes 2 to 4 were incubated for 2, 5, and 10 min, respectively, at room temperature. Lanes 5 to 7 were incubated for 2, 5, and 10 min, respectively, at 65°C. Lanes 8 to 10 were incubated for 2, 5, and 10 min, respectively, at 70°C. Loss of material occurred in lane 7; repetion showed the same result as in lane 6. (B) Lrs14 (0.8 μg) was preincubated at the indicated temperatures for 1 h. Lane 1, room temperature; lane 2, 55°C; lane 3, 75°C; lane 4, 85°C. The assay for DNA binding with the UP probe was performed at room temperature for 5 min.
Lrs14 binds to multiple sites in its promoter.
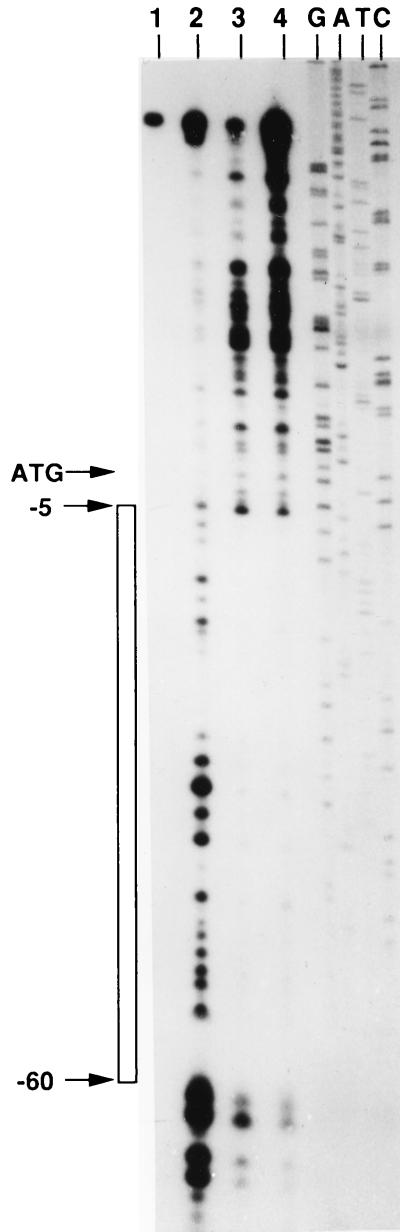
DNase I footprinting was performed in order to further define the region of Lrs14 binding. Lrs14 protects a continuous region of about 60 bp, spanning from −5 to −60 relative to the ATG (Fig. 6). This extended protection suggests that Lrs14 might bind to multiple sites in the region.
FIG. 6.
DNase I protection of the lrs14 promoter by Lrs14. A 130-bp fragment spanning nt −68 to +63 of the lrs14 gene was labelled at the distal (−68) end and incubated with no protein (lanes 1 and 2) or with 2 μg (lane 3) or 4 μg (lane 4) of Lrs14, preincubated for 1 h at 75°C. After binding, 10 ng of DNase I was added to lanes 2 to 4; in lane 1, no DNase was added. GATC indicates a sequence ladder used as a molecular weight marker. The open bar indicates the region protected by Lrs14. The position of ATG is indicated.
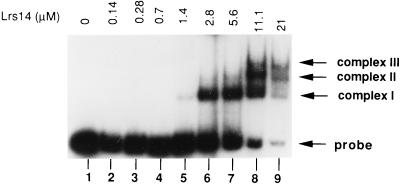
To test this hypothesis, we performed a bandshift experiment with increasing concentrations of Lrs14 (Fig. 7). At a relatively low protein concentration, only one complex (I) was formed, whereas two slower complexes (II and III) appeared at increasing protein concentrations; this result suggests that Lrs14 binds to multiple sites in the upstream region of its gene. A similar behavior is shown by the E. coli Lrp protein, which binds to several (up to six) distinct sites in upstream regions of its target genes, producing extended footprints (28, 33, 34, 35).
FIG. 7.
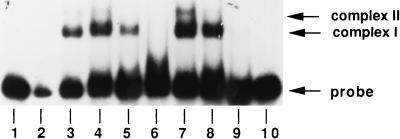
Binding of different amounts of Lrs14 to the UP probe. The protein was preincubated for 1 h at 75°C; binding was for 5 min at room temperature. Lane 1, naked probe; lane 2, 0.02 μg of Lrs14; lane 3, 0.04 μg of Lrs14; lane 4, 0.1 μg of Lrs14; lane 5, 0.2 μg of Lrs14; lane 6, 0.4 μg of Lrs14; lane 7, 0.8 μg of Lrs14; lane 8, 1.6 μg of Lrs14. In lane 9, 3 μg of Lrs14 was used in the reaction, but only one-fifth of the sample was loaded in order to resolve the three complexes.
The affinity of Lrs14 for its promoter, as calculated from data in Fig. 7, is quite low for a sequence-specific DNA-binding protein (apparent Kd of about 5 μM). The Kd is 2 or 3 orders of magnitude higher than that of the E. coli Lrp (28, 33, 35), but is comparable to that measured for the TATA binding protein of the hyperthermophile Pyrococcus woesei (24). Different possibilities could explain how this low affinity is coupled with sequence-specific recognition in vivo; for instance, the intracellular concentration of Lrs14 might be very high, or the affinity might be increased by physical and chemical parameters (ionic strength, pH, temperature, and so on) or by specific mechanisms (posttranslational modifications, interaction with other proteins). Moreover, because Lrp binding affinities for different sites greatly vary (28, 33, 35), we cannot rule out the possibility that Lrs14 higher-affinity sites exist (showing, for instance, palindromic sequences or bending).
Binding of E. coli Lrp to some, but not all, sites within a promoter is cooperative (33). By comparing the relative affinity with which Lrs14 binds to the three different sites, we could not find any cooperative effect (data not shown).
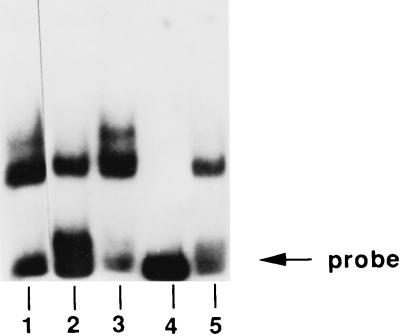
Bacterial Lrp proteins bind to A/T-rich sequences whose degenerate consensus is (c/t)AG(a/t/c)A(a/t)ATT(a/t)T(a/c/t)CT(a/g) (28). The A/T-rich upstream region of lrs14 contains sequences related to this consensus (Fig. 3). We constructed an oligonucleotide containing one of these sites (cLRS) and a control oligonucleotide overlapping the TATA box (TATA [Fig. 3]), and we used them as competitors in bandshift experiments (Fig. 8). Binding of Lrs14 to the fragment UP was competed by the cLRS (lanes 4 and 5), but not TATA (lanes 2 and 3), oligonucleotide, suggesting that Lrs14 recognizes sequences similar to those in the Lrp consensus. We noticed that complete competition is obtained when a large excess of cLRS is used, suggesting that additional sequences are required for the binding to be more effective.
FIG. 8.
Lrs14 binds to Lrp consensus sequences. Binding of Lrs14 (0.8 μg) to the UP probe in the presence of different competitor oligonucleotides (Fig. 3). Lane 1, no competitors; lane 2, oligonucleotide TATA (1 μg); lane 3, oligonucleotide TATA (0.1 μg); lane 4, oligonucleotide cLRS (1 μg); lane 5, oligonucleotide cLRS (0.1 μg). Prebinding and binding reactions were performed at room temperature for 10 min.
DISCUSSION
We have identified a new gene of S. solfataricus (lrs14) coding for a protein related to the Lrp-AsnC family of transcriptional regulators. The predicted Lrs14 protein contains a potential helix-turn-helix motif typical of this class of DNA-binding proteins. We have expressed the Lrs14 protein in E. coli fused to a histidine tag at the N terminus and purified it by affinity chromatography. We have shown that Lrs14 is a highly thermostable DNA-binding protein, which specifically binds to multiple sites in its own promoter and which, in DNase I footprinting experiments, protects a large region of about 60 bp immediately upstream of the ATG. At least one of the Lrs14 binding sites is related to A/T-rich binding sequences of bacterial Lrp proteins. Finally, we have shown that the lrs14 gene is transcribed in a monocistronic RNA which accumulates in the midexponential and late growth stages.
Despite the low sequence similarity, most of these features are reminiscent of the E. coli Lrp protein and suggest that the function and regulation of the two proteins might be similar. Lrp regulates more than 40 genes and operons in E. coli; target genes are involved in cellular processes as diverse as amino acid biosynthesis, transport, and degradation, fimbrial synthesis, tRNA synthesis, maltose transport, outer membrane structure, osmoregulation, and others (4, 23). Unfortunately, due to the lack of molecular tools for the construction of targeted mutants in S. solfataricus, the role of Lrs14 in cell physiology is currently only a matter of speculation. Apparently multiple Lrp-AsnC homologs are present in archaeal strains (3, 17, 29), and, in particular, an S. solfataricus Lrp-related ORF distinct from Lrs14 is present in the database (10); this fact suggests either that the functions of these factors in Archaea are redundant or that each factor is specialized to accomplish a specific set of functions.
E. coli Lrp activity is modulated by its effector ligand, leucine. Leucine is required for binding of Lrp to some operators, whereas it inhibits or has no effect on binding to other sites. The opposite effects of leucine on binding of Lrp to different operator sites can be reproduced in vitro (35). Similarly, the homologous AsnC protein is regulated by asparagine (18). Neither leucine nor asparagine affects binding of Lrs14 to its promoter under the conditions tested (unpublished results).
Based on its global role and the fact that its levels vary inversely with the growth rate of cells, it has been suggested that Lrp is involved in adaptation to changes in the nutritional environment (21). The lrs14 transcript is also accumulated during late growth stages, suggesting a similar regulation for the Lrs14 protein.
Lrp is negatively autoregulated (34); although we do not have such direct functional evidence, the fact that the region of the footprint of Lrs14 overlaps the TATA box of the gene suggests that it may act as a repressor of its own transcription, reducing the affinity of the transcription machinery to the promoter or reducing its ability to initiate transcription.
The presence of Lrp-AsnC-like transcription regulators in Archaea is puzzling from an evolutionary point of view. Most likely, these factors have evolved before the divergence of Bacteria and Archaea, and it is tempting to speculate that they are among the oldest transcription regulators. Such ancient factors, still conserved in two domains of life, apparently have been lost (or dramatically changed) in eukaryotes.
According to current models of regulation of gene expression, in both prokaryotes and eukaryotes, a key role is played by protein-protein interactions between activators (and, in some cases, repressors) and components of the basal transcription machinery (TFIID, carboxy-terminal domain of the RNA polymerase, sigma factors) or other factors (coactivators, corepressors, adapters) (13, 16).
The molecular mechanism of transcriptional activation or repression by Lrp and AsnC proteins has not been elucidated, and, to date, no interactions with components of the basal transcription machinery or other factors has been reported. Identification of the Lrs14 target sites, as well as analysis of possible interactions of Lrs14 with other proteins involved in the transcription initiation process, will be addressed in future experiments. Such studies will help clarify mechanisms of archaeal gene expression and, possibly, those of the Lrp-AsnC class of factors.
ACKNOWLEDGMENTS
We thank Maria Riflettivo who participated in some experiments during work on her thesis, Marco Moracci for helpful discussion, and Giovanni Imperato and Ottavio Piedimonte for technical assistance.
This work was partially supported by EU project “Biotechnology of Extremophiles” contract no. BIO2-CT93-0274 and by MURST project “Biomolecole per la salute umana.”
Footnotes
NRCC publication 42281.
REFERENCES
- 1.Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. Solution structure and DNA-binding properties of a small thermostable protein from the archaeon Sulfolobus solfataricus. Nat Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlier, D. Personal communication.
- 6.Charlier D, Roovers M, Thia-Toong T L, Durbecq V, Glansdorff N. Cloning and identification of the Sulfolobus solfataricus Lrp gene encoding an archaeal homologue of the eubacterial leucine-responsive global transcription regulator Lrp. Gene. 1997;201:63–68. doi: 10.1016/s0378-1119(97)00428-9. [DOI] [PubMed] [Google Scholar]
- 7.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggen R I, Geerling A C, Wadkotter K, Antranikian G, de Vos W M. The glutamate-dehydrogenase-encoding gene of the hyperthermophilic archaeon Pyrococcus furiosus: sequence, transcription and analysis of the deduced amino acid sequence. Gene. 1993;132:143–148. doi: 10.1016/0378-1119(93)90527-a. [DOI] [PubMed] [Google Scholar]
- 10.GenBank. Accession no. e283948.
- 11.GenBank. Accession no. AE001064.
- 12.Guagliardi A, Napoli A, Rossi M, Ciaramella M. Annealing of complementary DNA strands above the melting point of the duplex promoted by an archaeal protein. J Mol Biol. 1997;267:841–848. doi: 10.1006/jmbi.1996.0873. [DOI] [PubMed] [Google Scholar]
- 13.Hochschild A, Dove S L. Protein-protein contacts that activate or repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 14.Janknecht R, de Martynoff G, Lou J, Hispkind R A, Nordheim A, Stunnenberg H G. Rapid and efficient purification of native histidine-tagged protein expressed in recombinant vaccinia virus. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones J G, Hackett N R, Halladay J T, Scothorn D J, Yang C F, Ng W L, DasSarma S. Analysis of insertion mutants reveals two new genes in the pNRC100 gas vesicle gene cluster of Halobacterium halobium. Nucleic Acids Res. 1989;17:7785–7793. doi: 10.1093/nar/17.19.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 17.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 18.Kölling R, Lother H. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreuzer P, Gärtner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989;171:3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyrpides N C, Ouzounis C A. The eubacterial transcriptional activator Lrp is present in the archaeon Pyrococcus furiosus. Trends Biochem Sci. 1995;20:140–141. doi: 10.1016/s0968-0004(00)88988-4. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf J R, Wu J, Calvo J M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napoli, A., and M. Ciaramella. Unpublished results.
- 22a.NPSA. 21 December 1998, revision date. [Online.] HTH program. http://pbil.ibcp.fr/NPSA/npsa_hth.html. [19 January 1999, last date accessed.]
- 23.Newman E B, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 24.O’Brien R, DeDecker B, Fleming K G, Sigler P B, Ladbury J E. The effects of salt on the TATA binding protein-DNA interaction from a hyperthermophilic archaeon. J Mol Biol. 1998;279:117–125. doi: 10.1006/jmbi.1998.1743. [DOI] [PubMed] [Google Scholar]
- 25.Platko J V, Calvo J M. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J Bacteriol. 1993;175:1110–1117. doi: 10.1128/jb.175.4.1110-1117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prisco A, Moracci M, Rossi M, Ciaramella M. A gene encoding a putative membrane protein homologous to the major facilitator superfamily of transporters maps upstream of the β-glycosidase gene in the archaeon Sulfolobus solfataricus. J Bacteriol. 1995;177:1614–1619. doi: 10.1128/jb.177.6.1614-1619.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeve J N, Sandman K, Daniels C J. Archaeal histones, nucleosomes and transcription initiation. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 28.Rhee K Y, Parekh B S, Hatfield G W. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli. J Biol Chem. 1996;271:26499–26507. doi: 10.1074/jbc.271.43.26499. [DOI] [PubMed] [Google Scholar]
- 29.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 31.Van der Oost J, Ciaramella M, Moracci M, Pisani F M, Rossi M, de Vos W M. Molecular biology of hyperthermophilic Archaea. Adv Biochem Eng Biotechnol. 1998;61:87–115. doi: 10.1007/BFb0102290. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Calvo J M. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993;12:2495–2501. doi: 10.1002/j.1460-2075.1993.tb05904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Calvo J M. Lrp, a global regulatory protein of Escherichia coli, binds co-operatively to multiple sites and activates transcription of ilvIH. J Mol Biol. 1993;229:306–318. doi: 10.1006/jmbi.1993.1036. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Wu J, Friedberg D, Platko J, Calvo J M. Regulation of the Escherichia coli lrp gene. J Bacteriol. 1994;176:1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiese D E, Ernsting B R, Blumenthal R M, Matthews R G. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J Mol Biol. 1997;270:152–168. doi: 10.1006/jmbi.1997.1057. [DOI] [PubMed] [Google Scholar]
- 36.Willins D A, Ryan C W, Platko J V, Calvo J M. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]