Abstract
Corona virus disease 2019 (COVID-19) testing relies on traditional screening methods, which require a lot of manpower and material resources. Recently, to effectively reduce the damage caused by radiation and enhance effectiveness, deep learning of classifying COVID-19 negative and positive using the mixed dataset by CT and X-rays images have achieved remarkable research results. However, the details presented on CT and X-ray images have pathological diversity and similarity features, thus increasing the difficulty for physicians to judge specific cases. On this basis, this paper proposes a novel coronavirus pneumonia classification model using the mixed dataset by CT and X-rays images. To solve the problem of feature similarity between lung diseases and COVID-19, the extracted features are enhanced by an adaptive region enhancement algorithm. Besides, the depth network based on the residual blocks and the dense blocks is trained and tested. On the one hand, the residual blocks effectively improve the accuracy of the model and the non-linear COVID-19 features are obtained by cross-layer link. On the other hand, the dense blocks effectively improve the robustness of the model by connecting local and abstract information. On mixed X-ray and CT datasets, the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), area under curve (AUC), and accuracy can all reach 0.99. On the basis of respecting patient privacy and ethics, the proposed algorithm using the mixed dataset from real cases can effectively assist doctors in performing the accurate COVID-19 negative and positive classification to determine the infection status of patients.
Keywords: Adaptive region enhancement, COVID-19, Deep learning, Dense block, Mixed dataset
1. Introduction
The director-general of the World Health Organization announces [1] that the 2019 infectious coronavirus is named corona virus disease 2019 (COVID-19) on February 11, 2020. As of August 31, Beijing time, China, 219 million people worldwide have novel coronavirus pneumonia and 4 million 480 thousand deaths have been reported [2]. Medical imaging [3] has become an indispensable part of disease diagnosis and treatment. The latest research shows that with the development of medical imaging [4], [5], computed tomography (CT) [6], [7], [8], [9], and X-ray images [10], [11], [12] can effectively assist doctors to judge the COVID-19 infection status. Here, the CT images are the sensitive positive COVID-19 diagnostic method, which has good timeliness. Conversely, X-ray images with fast imaging speed and low radiation damage have become more and more important in the process of the COVID-19 diagnosis [13], [14].
Recently, because deep learning [15] can be used to assist doctors in medical image processing, significant research results have been achieved in the classification of negative and positive COVID-19 [16]. At present, the existing deep learning models [17] for negative and positive COVID-19 classification include Alexnet [18], Vgg [22], Resnet [26], and Densenet [27]. The traditional deep learning model mainly includes the following issues:
-
•
As the network deepens, the gain in model performance is not visible, and major network degradation problems may develop as a result of the overzealous pursuit of network depth and the expression capabilities of the non-linear model.
-
•
The feature maps in the convolution kernel with different scales are coupled to improve the feature dimension of the single layer, resulting in a considerable amount of duplicated information. The computational complexity has increased exponentially as the number of network levels has grown.
-
•
To increase the model's fitting speed, a large-scale kernel is utilized to retrieve the lesion's local feature. Meanwhile, the feature maps cannot be separated precisely during the convolution phase, resulting in a loss of coherence in the retrieved feature.
-
•
Due to the means of the sudden drop in the feature dimension, information loss may occur in some traditional networks, which reduces the accuracy of the model.
Here, Maghdid et al. [19] combine the Alexnet model with transfer learning, which can effectively classify negative and positive COVID-19 on mixed X-ray and CT images. Then, Turkoglu et al. [20] can effectively improve the classification accuracy through mitigation feature selection algorithms and support vector machine (SVM). However, this structure contains more layers and increases memory consumption. To solve this problem, Loey et al. [21] combine the Generic Advantageous Network (GAN) with the Alexnet model. The above networks [19], [20], [21] improve the Alexnet model through preprocessing and transfer learning, which use dropout randomly inactivated neurons in the full connection layer to prevent the over-fitting phenomenon. However, due to the non-differentiable points in the Alexnet model, some neurons are not activated after updating the parameters. On this basis, the Vgg model replaces the receptive domain with the same size by reusing the convolution kernel with the same size. Sitaula et al. [23] propose a novel Vgg model using the attention module, which can effectively fine-tune the parameters in the classification process by capturing the spatial relationship. Shibly et al. [24] can effectively detect COVID-19 patients from the chest X-ray images by fast regions with a revolutionary neural networks framework. Lee et al. [25] use the deep convolution neural network to optimize the parameters of the Vgg network and fine-tune each backbone network. However, because most of the training parameters come from the first full connection layer, the storage cost of the Vgg network training may be large.
Resnet model realizes feature connection across layers through bottleneck convolution, which can directly transfer the weight without any intermediate transformation to the next layer. To reduce the impact of weight noise on the classification accuracy, Zhou et al. [28] input the regrouped images into the residual blocks for feature extraction and then input the extracted features into the support vector machine for recognition. Sakib et al. [29] classify negative and positive COVID-19 by using Gan and general data enhancement methods. Hira et al. [30] effectively obtain the low-level features of COVID-19 by using the pre-trained network weights and fine-tuning the whole model at an appropriate learning rate. The Resnet model obtains more features based on the residual blocks to deepen the network and avoids network degradation through the cross-layer link. However, because the convolution kernel of the Resnet model can not be divided by the corresponding upper feature map, the problem of feature loss will occur.
To solve this problem, the Densenet model based on the dense blocks connects the non-linear extracted information by comprehensively using the shallow features. Tabrizchi et al. [32] and Zhang YuDong et al. [33] improve the Densenet model and transfer learning to improve the classification accuracy of COVID-19. Chowdhury et al. [34] use the pre-trained deep learning model to automatically detect COVID-19 from chest X-ray images. It can overcome the problem of feature loss by dense blocks and fine-tuning the pre-trained network. The Densenet model connects the lesion features to obtain a decision function with good generalization performance. However, a large amount of redundant information will appear in the connection process, which can change feature dimensions through bottleneck convolution.
Thus, a novel coronavirus pneumonia classification model based on the mixed dataset by X-ray and CT images is proposed in this paper. The adaptive region enhancement algorithm then enriches the COVID-19 features. By using a cross-layer link, the residual blocks can yield non-linear COVID-19 properties. Besides, the robustness can be improved by the dense blocks by connecting abstract and local properties. The mixed COVID-19 dataset can be classified using the proposed COVID-RDNet model. Its main contributions are as follows:
-
•
The proposed COVID-RDNet model may successfully tackle the problem of network degradation in conventional deep learning by obtaining a network with the ability to represent local and global features.
-
•
Based on a mixed dataset of public X-ray and CT images, the proposed model can identify negative and positive COVID-19.
-
•
The proposed method can effectively assist doctors in the diagnosis and treatment of COVID-19 patients.
The paper is organized as follows. In Section II, the structure and research significance of the proposed model are introduced. Section III presents the adaptive region enhancement algorithm and the proposed COVID-RDNet model. Section IV analyzes the experimental results. The proposed model is compared with state-of-the-art classification algorithms. Finally, the conclusion is given in Section V.
2. Methodology and theory
2.1. Related work
2.1.1. The Resnet model
To effectively solve the network degradation in traditional deep learning, the Resnet model [26] obtains local and global feature expression by adjusting the number of channels and stacking layers in the residual block. The Resnet model [27], [28], [29], [30] connects an across the layer to the next layer directly without any change of intermediate weight matrix. Besides, it can make the connection and propagation of local and global features of lesions smoother. Because the local feature of COVID-19 can be directly transmitted to the corresponding abstract information without any intermediate weight matrix transformation, the Resnet model has strong depth adaptive ability. The corresponding structure is shown in Fig. 1 .
Fig. 1.
The structure of the Resnet model.
By connecting the extracted abstract features with the corresponding upper features maps, the gradient vanishing problem can be effectively overcome. For the layer of the Resnet model, its partial derivative is as follows:
| (1) |
When the -th layer and the corresponding contain coefficient, the -th layer can be expressed as:
| (2) |
However, the Resnet model uses convolution kernels with odd sizes for feature extraction. When the kernel size set by the model cannot be completely divided by the step size, it is very easy to increase the redundancy information of feature maps and result in the uneven overlap. Especially for the COVID-19 images, due to the small gray difference between the COVID-19 region and the normal region, this overlapping redundancy can be enlarged. Therefore, the proposed algorithm uses the residual blocks to extract COVID-19 local information, which can not only effectively obtain the non-linear features of lesions, but also overcome the problem of feature loss caused by a convolution operation.
2.1.2. The Densenet model
With the increasing number of deep network layers, batch normalization can alleviate the problems of gradient vanishing. However, because the model calculation increases with the increase of network depth, the training efficiency is low and there is still the problem of network degradation. The Resnet model superimposes the output of identity mapping and non-linear transformation by numerical addition, which destroys the information flow in the network to a certain extent. To solve the above problems, the Densenet model [31] proposed in 2017 deepens the number of network layers to improve performance, as shown in Fig. 2 . It alleviates the network degradation problem extent by l reusing local features and adding dimension. The Densenet model [32], [33], [34] can effectively enhance the combination of COVID-19 information by connecting local and abstract features. Besides, the network can extract more features from relatively fewer data.
Fig. 2.
The structure of Densenet model.
Different from the Resnet model, the Densenet model limits the realization of convolution operation by combining all relevant outputs before layer (from layer to layer ):
| (3) |
In the Densenet model, the dense blocks contain two convolution operations and the kernel size is 1*1@128 and 1*1@32. Through 1*1 convolution operation, it can effectively realize the fusion of local features, which can enhance the classification accuracy of the model. In each dense block, assuming that the feature dimension of each non-linear transform is, the layer is. Because the local features extracted from each layer are connected with the previous ones, the training weights are learned through the current global information, which ensures the amount of information updated by the global state. Therefore, compared with the traditional depth model, the Densenet model can adopt fewer feature maps as the output of the network layer through the extreme utilization of extracted features.
However, because the Densenet model reduces the size of the current feature map to the next stage by average pooling, the feature dimension is half of the size of the previously extracted one. With the increase in model depth, the dimension of the small-size feature map is larger and the model adopts 1*1@128 and 1*1@32. The sudden drop of feature dimension and the sudden rise of connection dimension, it may lead to the lack of robustness of the COVID-19 features. Therefore, to enhance the ability of feature extraction, the proposed algorithm obtains COVID-19 local information through the dense blocks, in which the minimum amount of data is used through continuous bottleneck convolution operation.
2.2. The proposed network
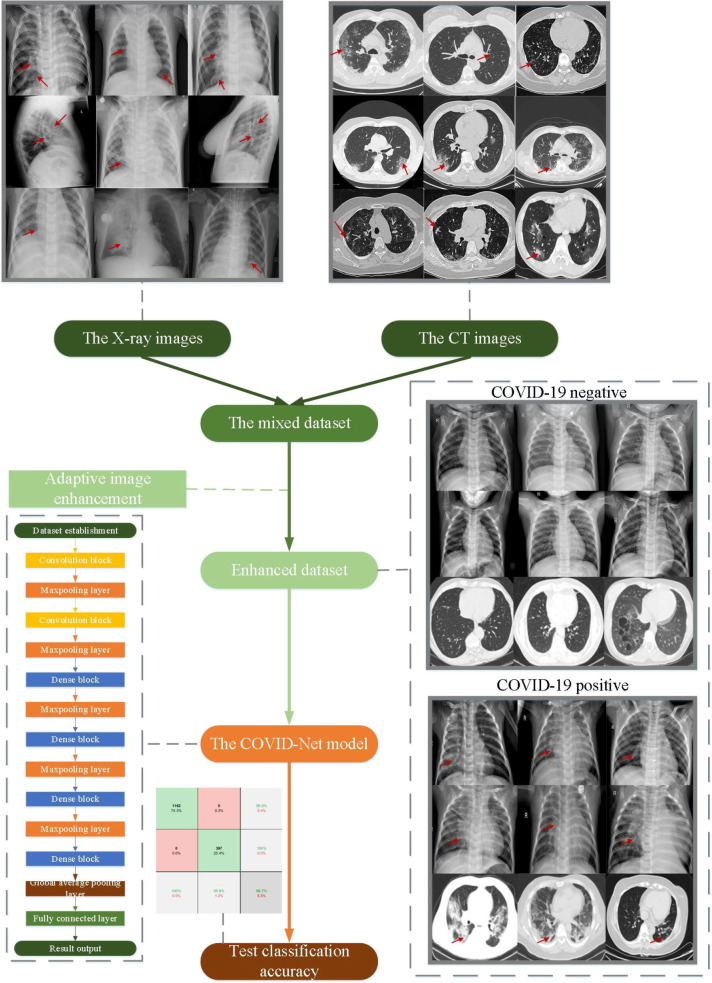
This paper proposes a COVID-19 classification model COVID-RDNet based on the mixed X-ray and CT images. The experimental process contains three steps: acquisition of the mixed dataset, enhancement of the adaptive COVID-19 feature, and establishment of the proposed COVID-RDNet model, as shown in Fig. 3 . Firstly, a mixed COVID-19 dataset with the X-ray images and the CT images is built through multiple image sources, which can ensure the authenticity and effectiveness of the proposed model in detecting novel coronavirus pneumonia cases. Then, the region containing the COVID-19 feature is enhanced and separated from the background by an adaptive threshold algorithm. Next, the COVID-RDNet model proposed in this paper is constructed. By combining the advantages of residual blocks and dense blocks, the non-linear connection between local features and global ones is realized.
Fig. 3.
The flow chart of the proposed algorithm.
2.2.1. Acquisition of the mixed dataset
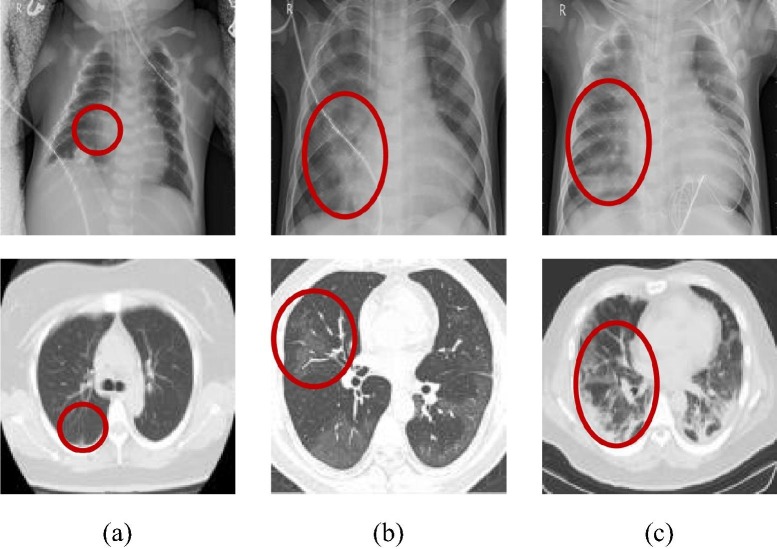
The imaging features of X-ray and CT images will change with the severity and duration of infection [35], [36], as shown in Fig. 4 . In general, the lesions of early COVID-19 patients mostly occur at the bottom of the lung and below the pleura, mainly in the peripheral zone of the lung. It may present as a single or double lung multiple patchy ground-glass shadows, sometimes accompanied by a slight bronchial inflation sign, as shown in Fig. 4(a) [37], [38]. With the further progress of the lesion, consolidation changes may occur. The scope and types of inflammatory lesions increase, which can begin to spread to multiple lung lobes. At this time, the patient's images show a mixture of ground glass shadow and solid change shadow. Besides, some patients will also have mild cord shadow and a small amount of pleural effusion, as shown in Fig. 4(b) [39], [40]. Severe novel coronavirus pneumonia patients have larger lesion regions and diffuse interstitial changes will appear. The symptoms of the white lung will appear at the later stage, often accompanied by a solid change shadow, which will always endanger the patient's life and health, as shown in Fig. 4(c) [41].
Fig. 4.
Imaging features of COVID-19 in different periods. (The first row is the X-ray images and the second one is the CT images. The red region indicates the position of the COVID-19 signs.) (a) early stage; (b) middle stage; (c) later stage.
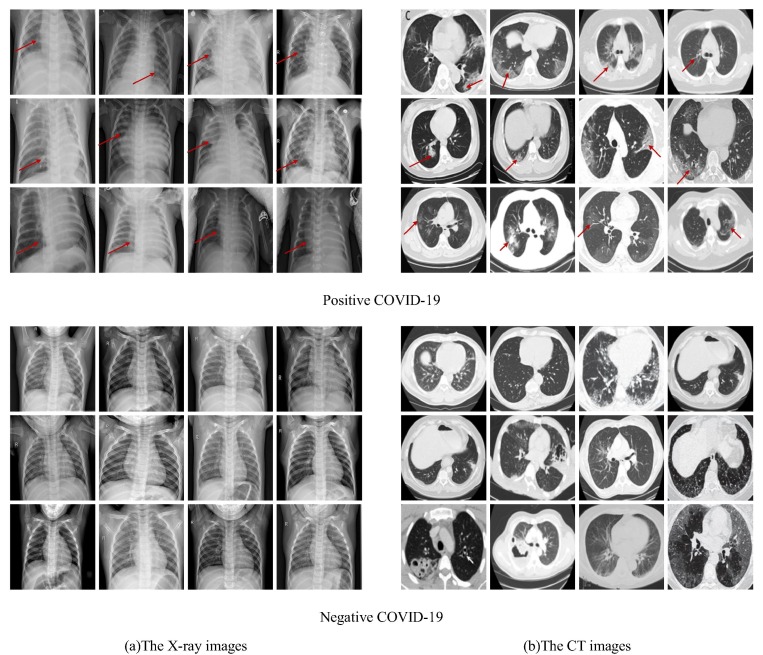
To effectively improve the detection of the novel coronavirus pneumonia, the mixed dataset is established by X-ray and CT images. Here, the X-ray images [42] are from the public dataset provided by Guangzhou Women's and children's medical center [43], as shown in Fig. 5 (a). The images are from the routine clinical examination of children aged 1 to 5 years. They are screened by removing all low-quality or unreadable X-ray images for quality control. To ensure the authenticity and effectiveness of the results, all the negative and positive are classified by two experts. The evaluation set is also checked by a third expert. There are 8447 images in the X-ray image dataset, which is composed of 1583 negative images and 6864 positive images. It is divided into three parts for training, verification, and testing. The CT images are from an open-source COVID-CT dataset [44], as shown in Fig. 5(b). Some researchers from the University of California at San Diego select images of clinical symptoms by artificial screening. The CT images of 333 cases with the positive detection of COVID-19 and 397 cases with the positive detection are provided.
Fig. 5.
The negative and positive COVID-19 of the X-ray images and the CT images (Red arrows indicate the location of the COVID-19).
The dataset used in this paper is a mixed public X-ray and CT images. The established database contains 5216 images, which consist of 1341 non-novel coronavirus pneumonia and 3875 novel coronavirus pneumonia. Here, 70 % is used for training, 20 % for testing, and 10 % for validation, as shown in Table 1 . To facilitate the training of the proposed COVID-RDNet model, the image size in the dataset is set to 224*224.
Table 1.
The dataset structure distribution.
| Training(70 %) | Testing(20 %) | Validation(10 %) | Total(100 %) | |
|---|---|---|---|---|
| Negative COVID-19 | 939 | 268 | 134 | 1341 |
| Positive COVID-19 | 2713 | 775 | 387 | 3875 |
| Total | 3652 | 1043 | 521 | 5216 |
Due to the complex and diverse features of COVID-19, this paper uniformly preprocessed CT and X-ray images through matrix normalization. It can not only increase the ratio of the brightest to the darkest location while maintaining the original gray-scale distribution features of the COVID-19 images. Meanwhile, it can also effectively overcome the problem of too slow running speed caused by the same increase and subtraction of the connected weights, so that the operation efficiency of the algorithm can be accelerated when the mean of all signals is infinitely close to zero.
2.2.2. The adaptive enhancement algorithm of the COVID-19 region
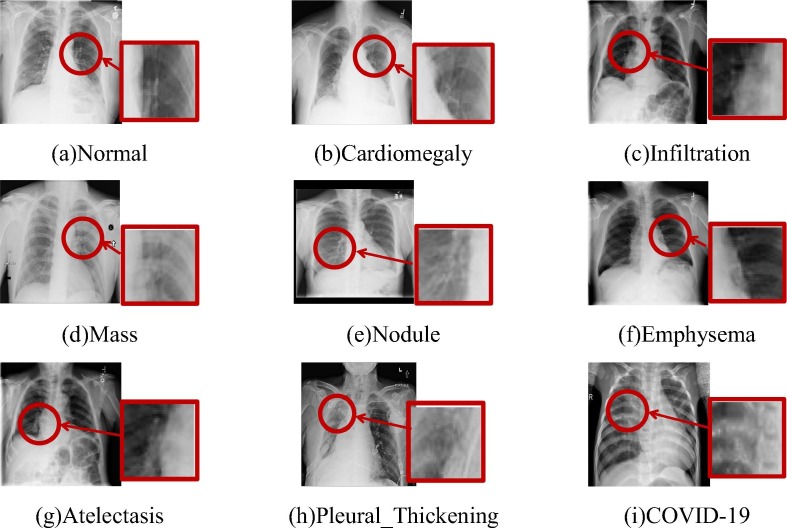
Fig. 6 shows the image features of different lung diseases taking the X-ray images as an example [45]. From this figure, one can see that the novel coronavirus has very similar image features to other diseases. Besides, there is no clear boundary between the lesion region and normal lung tissue, where it is in a gradual transition state. To improve the negative and positive COVID-19 classification accuracy, the proposed algorithm enhances the contrast between the COVID-19 region and the normal region by an adaptive enhancement algorithm [46], [47], [48], [49].
Fig. 6.
Imaging features of different lung diseases (The red region indicates the position of the lesion signs).
To effectively enhance the COVID-19 region, the proposed algorithm firstly divides the image into the COVID-19 region and normal region through threshold. Here, the obtained region is represented by:
| (4) |
| Algorithm 1 Adaptive threshold algorithm |
|---|
| Input: Image, is used to represent the coordinates of the pixel. and represent the number of pixels in the COVID-19 region and the normal region, respectively. and represent the average gray levels of the two regions, respectively. |
| Output: Threshold |
| Initialization: =0 |
| 1 while <=255 |
| 2 if |
| 3 =; |
| 4 bleak; |
| 5 else = + 1; |
| 6 end |
To obtain the threshold, the weighted average value of the adjacent region around each pixel is calculated by the adaptive threshold algorithm. It can better process the images with large light and dark differences, which can retain more lesion features. The basic idea is to calculate the average value of each pixel by traversing the images under the current cyclic variable. When the variance between the COVID-19 region and the normal region is the largest, the current value is the threshold. The calculation process is shown in Algorithm 1. Here, and represent the number of pixels in the COVID-19 region and the normal region, respectively. Similarly, and represent the average gray levels of the corresponding regions.
Then, the COVID-19 images are enhanced by enlarging the feature difference between the COVID-19 region and the normal region [50], [51], [52]. The proposed algorithm segments the COVID-19 region into. Here, represent the pixel value, and are the minimum and the maximum pixel value of the image, respectively. For, the -th histogram is mapped to the new dynamic range:
| (5) |
| (6) |
Here,
| (7) |
where and represent the maximum and the minimum pixel value in each histogram, respectively. is the corresponding number of pixels and represents the gray level. For, the corresponding equalized histogram is obtained:
| (8) |
where represents the number of current pixels to. Thus, the final output is obtained:
| (9) |
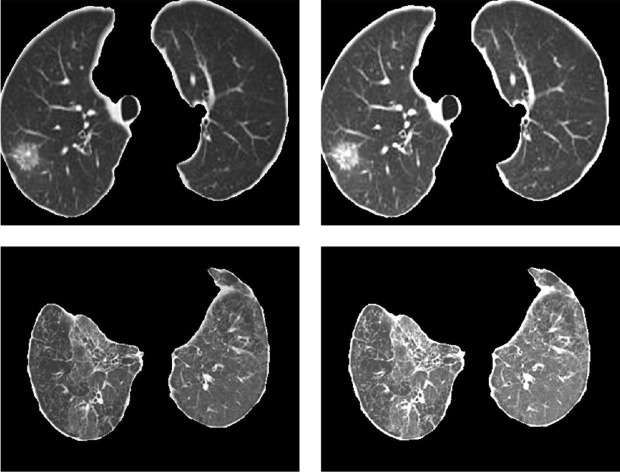
where and represent the average brightness of the COVID-19 region before and after equalization, respectively. The corresponding enhancement results are shown in Fig. 7 .
Fig. 7.
Results of adaptive local COVID-19 region enhancement. (Each row corresponds to a group of images. The first and second columns represent the images before and after enhancement, respectively).
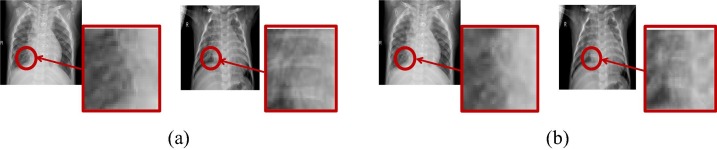
The proposed adaptive enhancement algorithm converts the COVID-19 region into an output with the same number of pixels at each gray level, which can effectively improve the dynamic range of pixel gray value by balancing the gray level distribution in the COVID-19 region. Besides, it can make the difference between the COVID-19 region and the normal region larger, as shown in Fig. 8 .
Fig. 8.
Results of adaptive enhancement algorithm (Red regions indicate the lesion before and after the enhancement). (a) Before enhancement; (b) After enhancement.
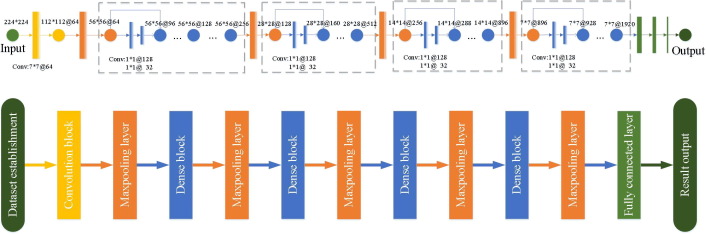
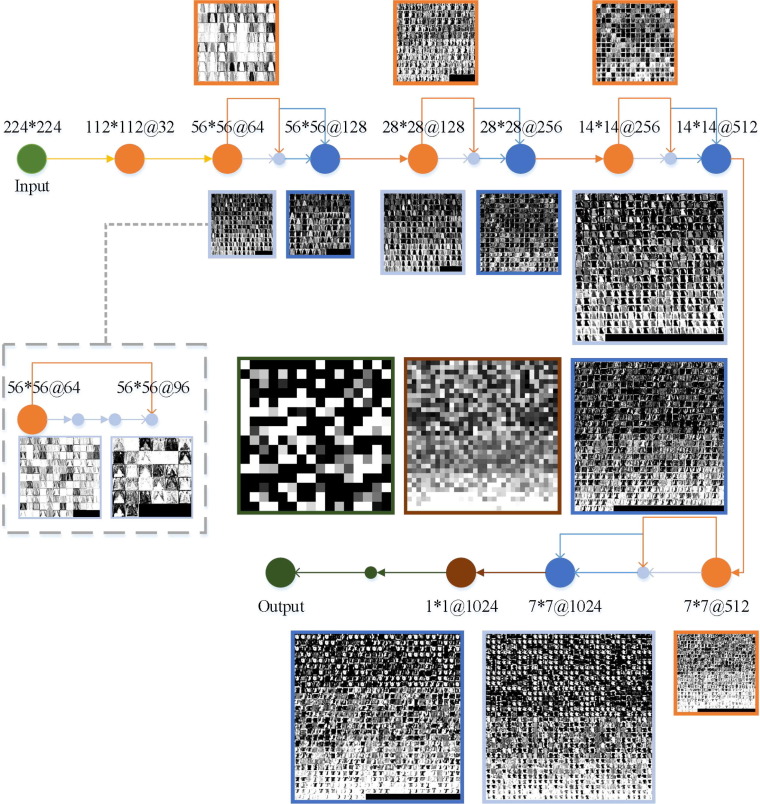
2.2.3. Establishment of the proposed COVID-RDNet model
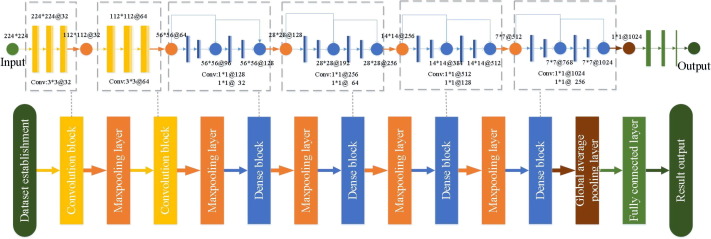
To overcome the problems in traditional deep learning, the proposed COVID-RDNet model is shown in Fig. 9 and its innovations are as follows:
-
•
Model construction: this paper constructs a depth COVID-19 classification model COVID-RDNet. The proposed model uses residual blocks to adjust the dimension of the feature maps, which can effectively broaden the depth of local and global feature extraction. To obtain a network with strong generalization ability, the proposed model connects the non-linear transformation extracted by the upper layer with the extracted features of the lower layer. It can overcome the network degradation problem caused by overfitting and improves the robustness of the algorithm.
-
•
Model performance: To enhance the non-linear extended ability of the proposed model, this paper uses continuous 3*3 expansion convolution to achieve the depth extraction operation. It can obtain more abstract local features through the superposition of convolution layers. Meanwhile, to effectively utilize the local features with relatively low shallow complexity, the feature reuse in the proposed model is realized by continuously using 1*1 bottleneck convolution, which greatly reduces the calculation parameters and improves the generalization performance.
-
•
Data training: the proposed network can effectively train and test a mixed dataset containing both public X-ray and CT images, which can meet the actual needs of novel coronavirus detection in terms of diagnosis cost and determination speed.
Fig. 9.
The structure of the proposed COVID-RDNet model.
In the proposed algorithm, each convolution block contains three 3*3 expansion convolutions. After each convolution layer, a batch normalization layer and a correction linear unit layer are added to correct the weight of single-layer input. Thus, it alleviates the decline of model accuracy caused by the extraction of lesion features in the convolution process. To speed up the training and reduce the sensitivity to network initialization, the proposed model uses a batch normalization layer between the convolution layer and non-linear features:
| (10) |
| (11) |
| (12) |
| (13) |
where represents the size of the batch. and represent the average and variance of batch data, respectively. and are training parameters, initially, and. Through batch normalization, the original dense data distribution is more uniform and the non-linear transformation function falls into the region sensitive to input, which can avoid the problem of gradient disappearance. Then, the threshold operation is performed on each element through the non-linear activation layer, which effectively improves the accuracy of the COVID-19 negative and positive classification network.
The proposed COVID-RDNet model has the following advantages:
First, the proposed model obtains the local lesion feature by continuously reusing 3*3 expansion convolution of the same size, which effectively enhances the generalization ability of the proposed model. The novel coronavirus pneumonia feature reuse is achieved through the bottleneck convolution of 1*1 in four dense blocks and the computation amount of the training process is reduced to a great extent by means of dimension reduction in the proposed COVID-RDNet model.
Second, because the difference between the COVID-19 region and the normal region is not very significant, the traditional Resnet model adjusts the depth and width by adding cross-layer values. However, in the convolution process, the input of the upper layer cannot divide the convolution kernel, which results in the loss of the feature details. The proposed model overcomes the loss of details by cross-layer links.
Next, because it is used every time 1*1@128 and 1*1@32, the convolution kernel for feature extraction leads to the sudden drop and rise of the convolution layer scale with large dimensions of the subsequent feature maps, which are not conducive to the stability of the model. The proposed model uses the cross-layer link to gradually improve the extracted feature dimension, realizes the non-linear connection between local features and global features, and overcomes the problem of network degradation caused by the increase of model depth.
Finally, the 1*1 bottleneck convolution operation is used to map multiple feature components into two categories of labels. Thus, the mixed dataset can be classified, which makes the COVID-19 detection of the proposed COVID-RDNet model more accurate. Besides, the initial epochs, learning rate, and corresponding decay factors of the proposed model are set as 30,, and 0.1, respectively. The gradient moment estimation is calculated by the adaptive moment estimation optimizer to stabilize the parameters in a dynamic range. The final output is obtained by .
3. Experiment and analysis
3.1. Dataset and evaluation indexes
To verify the effectiveness of the proposed model, two open datasets are used to train and test. Here, the X-ray images are from the public dataset provided by Guangzhou Women's and children's medical center. There are normal regions in the X-ray images, while the images of patients with COVID-19 usually show focal consolidation. Besides, the bilateral lung regions with severe viral pneumonia show diffuse interstitial. The CT images are from the open-source COVID-CT dataset provided by some researchers at the University of California, San Diego. The chest CT images of the novel coronavirus pneumonia usually show patchy or segmental ground glass density. Most of the lesions are gridded in shape and a few cases are associated with a small amount of pleural effusion or pericardial effusion. Because the image sizes in the two datasets are different, for the sake of fairness, this paper uniformly sets the size of all X-ray and CT images to 224*224. For fairness, this experiment employs Intel(R)Core(TM) i5-7200UCPU@2.50 GHz, 4 GB of memory, Windows 10 Professional, and 64-bit operating systems. Besides, to evaluate the performance of the proposed COVID-RDNet model to classify negative and positive COVID-19 [53], [54], sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy [55], [56] are used as follows:
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
, , , and . and represent the classification results of the proposed model and ground truth, respectively ( represents normal people and COVID-19, respectively). The relevant code of the proposed algorithm is available on the web: https://pan.baidu.com/s/1bgwT-4k_5bmmBNYIaBKkww.
3.2. The results of the proposed model
3.2.1. The feature maps extracted by the residual block
The proposed algorithm contains two residual blocks, which may successfully extract different lesion features by extending the depth and width, as shown in Fig. 10 . The larger the weight of the retrieved negative and positive COVID-19 features, the lighter the hue in the feature maps. The residual blocks can significantly reduce feature error and the likelihood of network degradation by connecting feature maps from different convolution layers. As a result of gradually developing the network model, residual blocks can acquire more and more negative and positive COVID-19 properties, effectively improving non-linear expression ability.
Fig. 10.
The COVID-19 feature maps extracted from the residual blocks. (Columns 1–3 represent the convolution results of layers 1–3 of the residual blocksfrom left to right). (a) The 1st residual block; (b) The 2nd residual block.
3.2.2. The extraction feature by the dense block
Due to the particularity of medical image sampling, the amount of medical images is limited and public datasets are difficult to match the deep learning model's feature extraction requirements. The proposed model connects the non-linear information gathered from each layer with dense blocks to effectively handle this challenge. Thus, the proposed model does not depend on the lesion features extracted from the last layer in a small dataset. Meanwhile, the local features with low shallow complexity can be comprehensively used, which can better obtain the feature maps with strong generalization performance and then improve the classification accuracy. Besides, to alleviate the feature loss caused by the sudden drop of the feature dimension, the proposed algorithm uses the way of slowly rising the dimension to ensure abstract information. The dimension is reduced by a 1*1 convolution kernel simultaneously, which effectively reduces calculation. Fig. 11 shows the results of COVID-19 feature maps extracted from different dense blocks (blue points). Through the reuse and connection of feature maps, the coupling of the obtained lesion information is reduced.
Fig. 11.
The COVID-19 feature maps extracted from the dense blocks.
3.2.3. The classification results of the proposed model
The proposed network can effectively combine local information with abstract features through the residual blocks and dense blocks, which can ensure the classification accuracy of negative and positive COVID-19. Meanwhile, the COVID-19 features are reused through continuous bottleneck convolution, which greatly reduces the calculation in network training. Fig. 12 shows the COVID-19 visual features extracted at each layer of the residual blocks and the dense blocks, respectively. As can be seen from Fig. 12, the proposed network obtains color, edge, and other information in the residual blocks of layers 1–2. From the third layer of the dense blocks, the proposed model starts to obtain advanced texture features. With the increase of network depth, it can obtain useful features to distinguish between negative and positive COVID-19.
Fig. 12.
The COVID-19 visualized features extracted from the proposed model: (a) The residual block (Columns 1–3 represent the results of layers 1–2 of the residual blocks from left to right.); (b) The dense block.
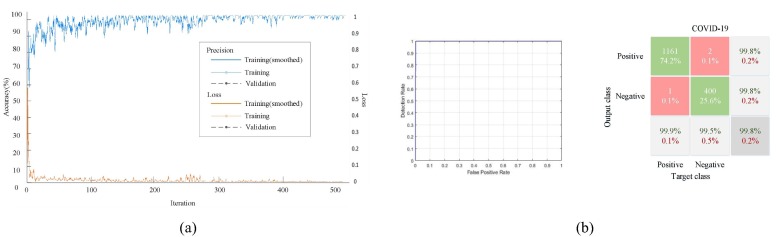
The proposed COVID-RDNet model can classify negative and positive COVID-19, the average value of the accuracy, AUC, sensitivity, specificity, PPV, and NPV can reach 0.99, 0.9998, 0.999, 0.995, 0.998, and 0.998, respectively, as shown in Fig. 13 . Here, Fig. 13(a) shows the loss rate and accuracy of the proposed model. It can be seen that the accuracy and loss rate with the number of iterations increasing can improve the optimized classification weight, which has a good convergence speed. Fig. 13(b) shows the testing results. The ROC curve on the left can effectively express the influence of different learning rates on the generalization performance. In general, the closer the ROC curve is to the upper left corner, the less the total number of false positives and negatives. The confusion matrix on the right can directly get the data under different parameters and effectively assess the COVID-19 classification.
Fig. 13.
The results of the proposed COVID-RDNet model: (a) The accuracy and loss rate; (b) The testing results. (The left is the ROC curve and the right is the confusion matrix).
3.3. Analysis and comparison
For fairness, all experiments are repeated 10 times in the same environment. Besides, the average values are taken while removing the maximum and minimum ones, thus effectively measuring the effectiveness of the proposed algorithm.
3.3.1. Analysis of the mixed datasets
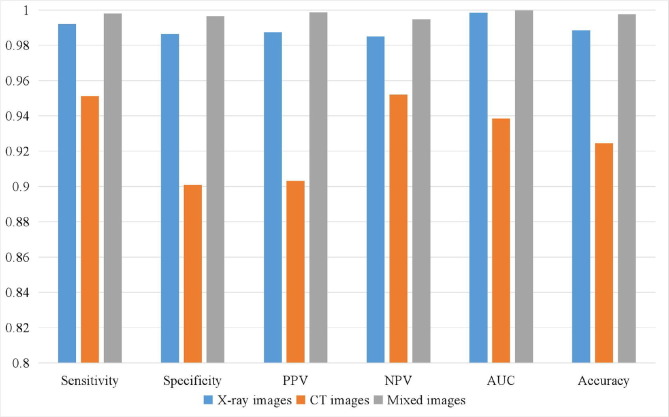
The proposed COVID-RDNet model can classify the negative and positive COVID-19 of individual X-ray images, CT images, and mixed datasets, as shown in Fig. 14 . It can be seen that the proposed COVID-RDNet model has the best classification performance in sensitivity, specificity, PPV, NPV, area under curve (AUC), and accuracy on the mixed X-ray and CT datasets. Besides, the classification performance of the proposed model using the X-ray images is slightly better than that of the CT images and the corresponding AUC results can reach 0.99 and 0.94, respectively. Therefore, the proposed algorithm can operate on the individual or the mixed dataset, which has good robustness and can obtain an assisted depth network that meets the actual medical diagnosis of doctors.
Fig. 14.
The performance comparison of the proposed model under different datasets.
3.3.2. Analysis of the learning rate
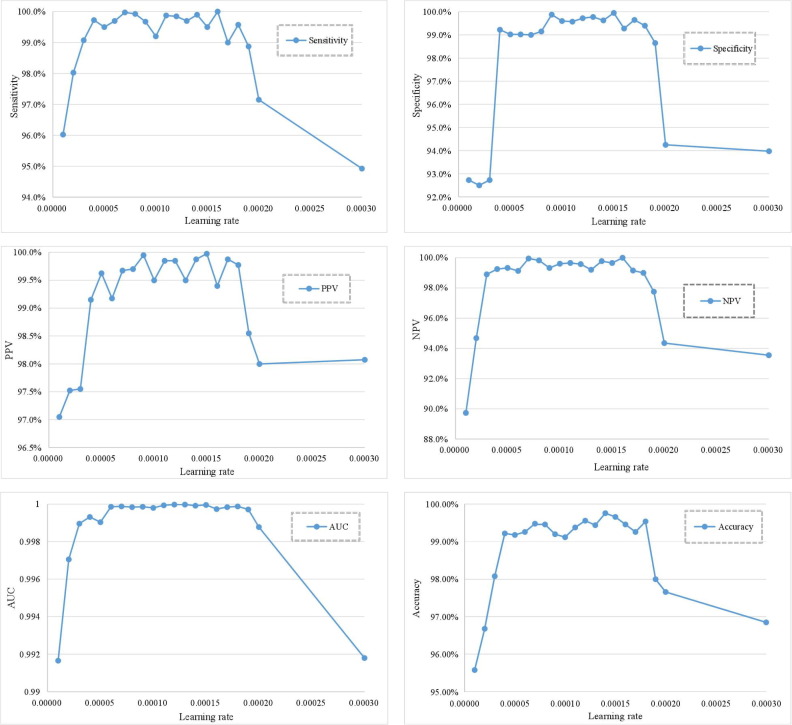
In general, the size of the learning rate setting will directly affect the final classification accuracy of the proposed model. If the learning rate is too large, it will lead to serious network oscillation and the model can not get the global optimal value. On the contrary, if the learning rate is too small, it will easily lead to the slow decline of the loss rate, which will not only reduce the classification accuracy, but also obtain the local optimal weight. Therefore, the proposed model obtains the COVID-19 local features through the residual blocks and connects it with the abstract information of lesions through the dense blocks, which can effectively reduce the impact of learning rate on model training. Fig. 15 shows sensitivity, specificity, PPV, NPV, AUC, and accuracy of the proposed COVID-RDNet model under different learning rates. It can be seen that when the learning rate is in the range of 0.00004 to 0.00018, the accuracy of the proposed model can be maintained at 0.99 and the AUC can reach 0.9998. The average results of sensitivity, specificity, PPV, and NPV are also relatively stable. While maintaining a high classification accuracy, this model also has strong robustness and fault tolerance. When the learning rate is greater than 0.00018 or less than 0.00004, the performance of the proposed network will decrease. Therefore, this paper selects the value in the range of 0.00004 to 0.00018 as the initial learning rate. After five rounds of model training, the learning rate [57], [58], [59] decreases to 1/10 of the initial value.
Fig. 15.
The performance of the proposed model under different learning rates. (From the upper left corner to the lower right corner are sensitivity, specificity, PPV, NPV, AUC, and accuracy, respectively).
3.3.3. Performance comparison of residual blocks and dense blocks in different models
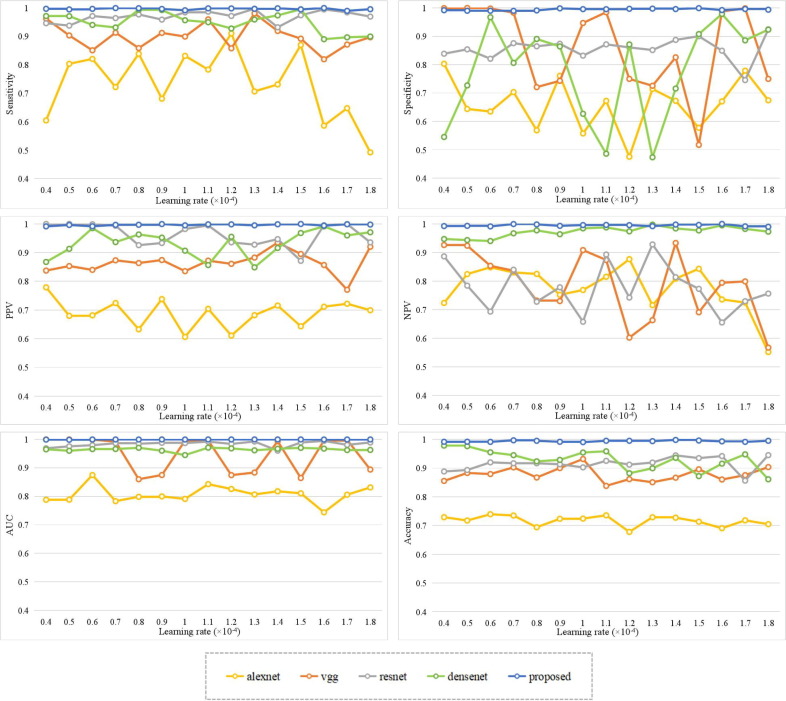
To further verify the performance of the proposed algorithm, this paper measures the network by comparing the sensitivity, specificity, PPV, NPV, AUV, and accuracy of the mixed X-ray and CT images by different models, as shown in Fig. 16 . There are no residual blocks or dense blocks in the Alexnet [19] and the Vgg [22], [23] models. The Resnet network contains residual blocks and the values of adjacent feature maps are added through the cross-layer link, which effectively enhances the COVID-19 features and has high sensitivity and PPV. However, the classification accuracy of the Resnet [26] network is related to the initial learning rate and does not have good stability, especially in the specificity and the NPV. The Densenet model contains dense blocks, which effectively enhance the stability of the network by connecting local and global information. It performs smoothly in sensitivity, PPV, and NPV. However, the Densenet [31] network reduces the feature dimension in a sudden way, which results in the loss of COVID-19 features. The Resnet and Densenet models contain residual blocks and dense blocks, respectively, which improve the performance compared with the Alexnet and the Vgg models. To overcome the above problems, the proposed COVID-RDNet model effectively improves the accuracy and accuracy through the linkage reuse of the residual blocks and the dense blocks, which can realize the efficient classification of the negative and positive COVID-19.
Fig. 16.
The performance of different models. (From the upper left corner to the lower right corner are sensitivity, specificity, PPV, NPV, AUC, and accuracy, respectively).
3.3.4. Comparison with the results of the state-of-the-art algorithms
This paper compares the results of the proposed model with the state-of-the-art networks under the same dataset, as shown in Table 2 . Here, Maghdid et al. [19] enhance the classification accuracy by combining the Alexnet model with the CNN model of transfer learning. Besides, Turkoglu et al. [20] and Loey et al. [21] enhance the ability to extract COVID-19 features by improving the Alexnet model. However, the step size in the above models is larger than the pooling size, which may result in redundancy. To solve this problem, the Vgg model replaces the large convolution kernel in the Alexnet model by reusing a small kernel with equal size, which can effectively enhance computational efficiency. Sitaula et al. [23] obtain spatial features of lesions by adding an attention mechanism to the Vgg model. On this basis, Shibly et al. [24] and Lee et al. [25] enhance the efficiency of the Vgg network by integrating a fast CNN network. However, most of the parameters come from the first fully connected layer, which may result in the need for large storage space. To effectively improve the utilization of COVID-19 features, the Resnet model can save space costs through cross-layer links. Hira et al. [30] improved the classification accuracy through transfer learning, but the stability of model training is slightly insufficient. On the contrary, Sakib et al. [29] have good stability, but at the expense of accuracy. Based on refs. [29], [30], Zhou et al. [28] input the regrouped images into the residual blocks for lesion feature extraction, which can effectively improve both robustness and accuracy. However, due to the improper size of the convolution kernel in the Resnet model, the problem of feature loss occurs. To overcome the problem, Tabrizchi et al. [32] and Zhang YuDong et al. [33] connect abstract features with local features by combining the Densenet model and transfer learning. Chowdhury et al. [34] improve the classification accuracy of the Densenet model through image enhancement, but too many layers may lead to redundancy. The proposed model can effectively solve the problem of feature loss through residual blocks and dense blocks. The accuracy of negative and positive COVID-19 classification can reach 0.99.
Table 2.
The proposed model results are compared with the state-of-the-art network.
| Method | Reference | Evaluating indexes | ||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Accuracy | ||
| Alexnet | Alexnet model [18] | 0.86 | 0.92 | 0.91 | 0.94 | 0.91 |
| Maghdid et al. [19] | 0.92 | 0.96 | 0.95 | 0.97 | 0.95 | |
| Turkoglu et al. [20] | 0.92 | 0.97 | 0.97 | 0.91 | 0.94 | |
| Loey et al. [21] | 0.92 | 0.96 | 0.93 | 0.93 | 0.93 | |
| Vgg | Vgg16 model [22] | 0.92 | 0.94 | 0.91 | 0.97 | 0.93 |
| Vgg19 model [22] | 0.96 | 0.96 | 0.95 | 0.97 | 0.96 | |
| Sitaula et et al. [23] | 0.96 | 0.96 | 0.94 | 0.97 | 0.96 | |
| Shibly et al. [24] | 0.97 | 0.95 | 0.93 | 0.98 | 0.96 | |
| Lee et al. [25] | 0.87 | 0.89 | 0.89 | 0.87 | 0.89 | |
| Resnet | Resnet18 model [26] | 0.98 | 0.94 | 0.93 | 0.98 | 0.96 |
| Resnet50 model [26] | 0.97 | 0.98 | 0.98 | 0.98 | 0.97 | |
| Resnet101 model [26] | 0.97 | 0.94 | 0.98 | 0.99 | 0.97 | |
| Zhou et al. [28] | 0.88 | 0.97 | 0.936 | 0.93 | 0.93 | |
| Sakib et al. [29] | 0.89 | 0.91 | 0.78 | 0.98 | 0.90 | |
| Hira et al. [30] | 0.96 | 0.94 | 0.97 | 0.87 | 0.90 | |
| Densenet | Densenet121 model [31] | 0.97 | 0.97 | 0.97 | 0.97 | 0.97 |
| Densenet201 model [31] | 0.98 | 0.91 | 0.95 | 0.98 | 0.97 | |
| Tabrizchi et al. [32] | 0.87 | 0.95 | 0.97 | 0.91 | 0.95 | |
| Mangal et al. [33] | 0.98 | 0.87 | 0.74 | 0.99 | 0.92 | |
| Chowdhury et al. [34] | 0.99 | 0.97 | 0.97 | 0.98 | 0.98 | |
| Proposed | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | |
4. Conclusion
In this paper, a deep learning network is proposed to effectively improve the classification accuracy of negative and positive COVID-19. Firstly, to ensure the authenticity and effectiveness of COVID-19 detection, a mixed X-ray and CT dataset are established through the published COVID-19 images. Then, the COVID-19 region features are effectively enhanced by the adaptive region enhancement algorithm. The non-linear COVID-19 features are obtained through the cross-layer link of the residual blocks. The local and abstract information is connected through the dense blocks. Compared with the state-of-the-art COVID-19 classification models, the proposed COVID-RDNet model has good negative and positive COVID-19 classification performance through sensitivity, specificity, PPV, NPV, AUC, and accuracy on the mixed X-ray and CT dataset. When the learning rate is in the range of 0.00004 to 0.00018, the accuracy of the proposed model can be maintained at 0.99 and the AUC can reach 0.9998.
Based on respect for patient privacy and ethics, the proposed model obtains high accurate classification results by manipulating the mixed dataset containing real cases, which can effectively assist doctors to determine the infection status of COVID-19 patients and has good application prospects. However, the proposed algorithm can not guarantee the computational complexity while improving the classification accuracy, which is the next research focus. Meanwhile, this paper also aims to overcome the problem that the number of medical annotation images is insufficient and does not meet the conditions of deep learning through unsupervised or self-supervised models, which can further meet the actual medical needs.
Ethical approval
This paper avoid distorting the research results and refuse to do anything that harms the trust of journals, the professionalism of scientific authors and the ultimate scientific efforts. Ensuring that the rules of good scientific practice are followed is conducive to maintaining the integrity of research and its statements.
Funding details
This work was supported by Natural Science Foundation of Liaoning Province under Grant 2021-MS-272 and Educational Committee project of Liaoning Province under Grant LJKQZ2021088.
Informed consent
All the authors of this paper, Lingling Fang and Xin Wang, agreed to submit the “COVID-RDNet: A novel coronavirus pneumonia classification model using the mixed dataset by CT and X-rays images” to the “Biocybernetics and Biomedical Engineering” journal.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bialek S., Bowen V., Chow N., Curns A., Gierke R., Hall A., et al. Geographic Differences in COVID-19 Cases, Deaths, and Incidence — United States, February 12–April 7, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(15):465–471. doi: 10.15585/mmwr.mm6915e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y., Dong Y., Wang L., et al. Characteristics and prognostic factors of disease severity in patients with COVID-19: The Beijing experience. J Autoimmun. Apr. 2020;112 doi: 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suri J S, Puvvula A, Biswas M, et al., “COVID-19 pathways for brain and heart injury in comorbidity patients: A role of medical imaging and artificial intelligence-based COVID severity classification: A review,” Comput Biol Med, pp. 103960, Aug. 2020. [DOI] [PMC free article] [PubMed]
- 4.Farhat H., Sakr G.E., Kilany R. Deep learning applications in pulmonary medical imaging: recent updates and insights on COVID-19. Mach Vis Appl. Sept. 2020;31(6):1–422020. doi: 10.1007/s00138-020-01101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedaghat A., Gity M., Radpour A., Karimi M.A., Haghighatkhah H.R., Keshavarz E., et al. COVID-19 protection guidelines in outpatient medical imaging centers. Acad Radiol. 2020;27(6):904. doi: 10.1016/j.acra.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K., Fang Y., Li W., et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur Radiol. Mar. 2020;30(8):4407–4416. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko H., Chung H., Kang W.S., Kim K.W., Shin Y., Kang S.J., et al. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J Med Intern Res. Jan. 2020;22(6) doi: 10.2196/19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y., Chen H., Loew M., Ko H. Covid-19 ct image synthesis with a conditional generative adversarial network. IEEE J Biomed Health Inf. 2021;25(2):441–452. doi: 10.1109/JBHI.2020.3042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abd Elaziz M., Ewees A.A., Yousri D., et al. An improved Marine Predators algorithm with fuzzy entropy for multi-level thresholding: Real world example of COVID-19 CT image segmentation. IEEE Access. Jul. 2020;8:125306–125330. doi: 10.1109/ACCESS.2020.3007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. Jan. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intellig. Feb. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos, Solit Fract. Jul. 2020;140 doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Civit-Masot J., Luna-Perejón F., Domínguez Morales M., Civit A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci. Jul. 2020;10(13):4640. [Google Scholar]
- 14.Nishio M., Noguchi S., Matsuo H., et al. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: combination of data augmentation methods. Sci Rep. Jan. 2020;10(1):1–6. doi: 10.1038/s41598-020-74539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim D.A., Zebari D.A., Mohammed H.J., et al. Effective hybrid deep learning model for COVID-19 patterns identification using CT images. Exp Syst. 2022:e13010. doi: 10.1111/exsy.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shamim S., Awan M.J., Mohd Zain A., Naseem U., Mohammed M.A., Garcia-Zapirain B., et al. Automatic COVID-19 Lung Infection Segmentation through Modified Unet Model. J Healthcare Eng. 2022;2022:1–13. doi: 10.1155/2022/6566982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Abdulkareem K.H., Mostafa S.A., Al-Qudsy Z.N., Mohammed M.A., Al-Waisy A.S., Kadry S., et al. Automated System for Identifying COVID-19 Infections in Computed Tomography Images Using Deep Learning Models. J Healthcare Eng. 2022;2022:1–13. doi: 10.1155/2022/5329014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Krizhevsky A, Sutskever I, Hinton G E, “Imagenet classification with deep convolutional neural networks,” Advances in neural information processing systems, pp. 1097-1105, Jan. 2012.
- 19.Maghdid H.S., Asaad A.T., Ghafoor K.Z., et al. “Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms”, Multimodal Image Exploitation and Learning 2021. Int Soc Optics Photon. Apr. 2021;11734:117340E. [Google Scholar]
- 20.Turkoglu M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl Intellig. Jan. 2021;51(3):1213–1226. doi: 10.1007/s10489-020-01888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loey M., Smarandache F.M., Khalifa N.E. M Khalifa N E, “Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning”. Symmetry. Apr. 2020;12(4):651. [Google Scholar]
- 22.Sengupta A., Ye Y., Wang R., Liu C., Roy K. Going Deeper in Spiking Neural Networks: Vgg and Residual Architectures. Front Neurosci. Jan. 2019;13 doi: 10.3389/fnins.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sitaula C., Hossain M.B. Attention-based Vgg-16 model for COVID-19 chest X-ray image classification. Applied Intelligence. Jan. 2021;51(5):2850–2863. doi: 10.1007/s10489-020-02055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibly K.H., Dey S.K., Islam M.T.U., et al. COVID faster R-CNN: A novel framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-Ray images. Inf Med Unlocked. Aug. 2020;20 doi: 10.1016/j.imu.2020.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K.-S., Kim J.Y., Jeon E.-T., Choi W.S., Kim N.H., Lee K.Y. Evaluation of scalability and degree of fine-tuning of deep convolutional neural networks for COVID-19 screening on chest X-ray images using explainable deep-learning algorithm. J Personal Med. Nov. 2020;10(4):213. doi: 10.3390/jpm10040213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He K, Zhang X, Ren S, et al., “Deep residual learning for image recognition,” Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 770-778; Jun. 2016
- 27.He K, Zhang X, Ren S, et al., “Identity mappings in deep residual networks,” European conference on computer vision. Springer, Cham, pp. 630-645; Oct. 2016.
- 28.Zhou C., Song J., Zhou S., Zhang Z., Xing J. COVID-19 Detection based on Image Regrouping and Resnet-SVM using Chest X-ray Images. IEEE Access, Jan. 2021;9:81902–81912. doi: 10.1109/ACCESS.2021.3086229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakib S., Tazrin T., Fouda M.M., et al. DL-CRC: deep learning-based chest radiograph classification for COVID-19 detection: a novel approach. IEEE Access. Sept. 2020;8:171575–171589. doi: 10.1109/ACCESS.2020.3025010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hira S, Bai A, Hira S, “An automatic approach based on CNN architecture to detect Covid-19 disease from chest X-ray images,” Appl Intellig, vol. 51, no. 5, pp. 2864-2889, May. 2021. [DOI] [PMC free article] [PubMed]
- 31.Huang G, Liu Z, Van Der Maaten L, et al., “Densely connected convolutional networks,” Proceedings of the IEEE conference on computer vision and pattern recognition. Pp. 4700-4708, Aug. 2017.
- 32.Tabrizchi H, Mosavi A, Vamossy Z, et al., “Densely Connected Convolutional Networks (Densenet) for Diagnosing Coronavirus Disease (COVID-19) from Chest X-ray Imaging,” 2021 IEEE International Symposium on Medical Measurements and Applications (MeMeA). IEEE, pp. 1-5, Sept. 2021.
- 33.Y.-D. Zhang S.C. Satapathy X. Zhang S.-H. Wang COVID-19 Diagnosis via DenseNet and Optimization of Transfer Learning Setting. [DOI] [PMC free article] [PubMed]
- 34.Chowdhury M.E.H., Rahman T., Khandakar A., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. Jan. 2020;8:132665–132676. [Google Scholar]
- 35.Allioui H., Mohammed M.A., Benameur N., Al-Khateeb B., Abdulkareem K.H., Garcia-Zapirain B., et al. A multi-agent deep reinforcement learning approach for enhancement of COVID-19 CT image segmentation. J Personal Med. 2022;12(2):309. doi: 10.3390/jpm12020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S.-H., Govindaraj V.V., Górriz J.M., Zhang X., Zhang Y.-D. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Informat Fusion. 2021;67:208–229. doi: 10.1016/j.inffus.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z., Zhao W., Xie X., et al. Severity assessment of COVID-19 using CT image features and laboratory indices. Phys Med Biol. Jan. 2021;66(3) doi: 10.1088/1361-6560/abbf9e. [DOI] [PubMed] [Google Scholar]
- 38.Hasoon J.N., Fadel A.H., Hameed R.S., Mostafa S.A., Khalaf B.A., Mohammed M.A., et al. COVID-19 anomaly detection and classification method based on supervised machine learning of chest X-ray images. Results Phys. 2021;31 doi: 10.1016/j.rinp.2021.105045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuncer T., Dogan S., Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image. Chemomet Intellig Laborat Syst. Jan. 2020;203 doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen S., Saha S., Chatterjee S., Mirjalili S., Sarkar R. A bi-stage feature selection approach for COVID-19 prediction using chest CT images. Appl Intell. 2021;51(12):8985–9000. doi: 10.1007/s10489-021-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam N.-A.-A., Ahsan M., Based M.A., Haider J., Kowalski M. COVID-19 detection from chest X-ray images using feature fusion and deep learning. Sensors. Feb. 2021;21(4):1480. doi: 10.3390/s21041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kermany D., Zhang K., Goldbaum M. Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data. 2018;2(2) [Google Scholar]
- 43.Bhatt R., Yadav S., Sarvaiya J.N. Springer; Singapore: 2020. Convolutional neural network based chest X-ray image classification for pneumonia diagnosis//International Conference on Emerging Technology Trends in Electronics Communication and Networking; pp. 254–266. [Google Scholar]
- 44.Yang X, He X, Zhao J, et al. COVID-CT-Dataset: A CT Scan Dataset about COVID-19. 2020. doi: 10.48550/arXiv.2003.13865.
- 45.Demir F., Sengur A., Bajaj V. Convolutional neural networks based efficient approach for classification of lung diseases. Health Informat Sci Syst. Dec. 2020;8(1):1–8. doi: 10.1007/s13755-019-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dash L., Chatterji B.N. Adaptive contrast enhancement and de-enhancement. Pattern Recogn. Jan. 1991;24(4):289–302. [Google Scholar]
- 47.Zhou Z., Sang N., Hu X. Global brightness and local contrast adaptive enhancement for low illumination color image. Optik. Jan. 2014;125(6):1795–1799. [Google Scholar]
- 48.Mohamed A.W., Suganthan P.N. Real-parameter unconstrained optimization based on enhanced fitness-adaptive differential evolution algorithm with novel mutation. Soft Comput. Jan. 2018;22(10):3215–3235. [Google Scholar]
- 49.Ahmed J., Salam Z. An enhanced adaptive P&O MPPT for fast and efficient tracking under varying environmental conditions. IEEE Trans Sustain Energy. Jun. 2018;9(3):1487–1496. [Google Scholar]
- 50.Meng Z., Pan J.S., Kong L. Parameters with adaptive learning mechanism (PALM) for the enhancement of differential evolution. Knowl-Based Syst. Jan. 2018;141:92–112. [Google Scholar]
- 51.Issa M., Hassanien A.E., Oliva D., Helmi A., Ziedan I., Alzohairy A. ASCA-PSO: Adaptive sine cosine optimization algorithm integrated with particle swarm for pairwise local sequence alignment. Expert Syst Appl. 2018;99:56–70. [Google Scholar]
- 52.Shi Z., Feng Y., Zhao M., Zhang E., He L. Normalised gamma transformation-based contrast-limited adaptive histogram equalisation with colour correction for sand–dust image enhancement. IET Image Proc. 2020;14(4):747–756. [Google Scholar]
- 53.Yu Z., Zheng J., Ma L., et al. The predictive accuracy of the black hole sign and the spot sign for hematoma expansion in patients with spontaneous intracerebral hemorrhage. Neurol Sci. Sept. 2017;38(9):1591–1597. doi: 10.1007/s10072-017-3006-6. [DOI] [PubMed] [Google Scholar]
- 54.Hajibandeh S., Hajibandeh S., Deering R., et al. Accuracy of routinely collected comorbidity data in patients undergoing colectomy: a retrospective study. Int J Colorectal Dis. Aug. 2017;32(9):1341–1344. doi: 10.1007/s00384-017-2830-8. [DOI] [PubMed] [Google Scholar]
- 55.Yao R.J.R., Andrade J.G., Deyell M.W., et al. Sensitivity, specificity, negative and positive predictive values of identifying atrial fibrillation using administrative data: a systematic review and meta-analysis. Clin Epidemiol. Jan. 2019;11:753. doi: 10.2147/CLEP.S206267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delate T., Jones A.E., Clark N.P., Witt D.M. Assessment of the coding accuracy of warfarin-related bleeding events. Thrombos Res. 2017;159:86–90. doi: 10.1016/j.thromres.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Nayak S.R., Nayak D.R., Sinha U., et al. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomed Signal Process Control. Feb. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duffey R.B., Zio E. Analysing recovery from pandemics by Learning Theory: the case of CoVid-19. IEEE Access. Jun. 2020;8:110789–110795. doi: 10.1109/ACCESS.2020.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toğaçar M, Ergen B, Cömert Z, “COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches,” Comput Biol Med, vol. 121, p. 103805, May. 2020. [DOI] [PMC free article] [PubMed]