Abstract
Expression of the degradative tryptophanase (tna) operon of Escherichia coli is regulated by catabolite repression and tryptophan-induced transcription antitermination. In cultures growing in the absence of added tryptophan, transcription of the structural genes of the tna operon is limited by Rho-dependent transcription termination in the leader region of the operon. Tryptophan induction prevents this Rho-dependent termination, and requires in-frame translation of a 24-residue leader peptide coding region, tnaC, that contains a single, crucial, Trp codon. Studies with a lacZ reporter construct lacking the spacer region between tnaC and the first major structural gene, tnaA, suggested that tryptophan induction might involve cis action by the TnaC leader peptide on the ribosome translating the tnaC coding region. The leader peptide was hypothesized to inhibit ribosome release at the tnaC stop codon, thereby blocking Rho’s access to the transcript. Regulatory studies with deletion constructs of the tna operon of Proteus vulgaris supported this interpretation. In the present study the putative role of the tnaC stop codon in tna operon regulation in E. coli was examined further by replacing the natural tnaC stop codon, UGA, with UAG or UAA in a tnaC-stop codon-tnaA′-′lacZ reporter construct. Basal level expression was reduced to 20 and 50% when the UGA stop codon was replaced by UAG or UAA, respectively, consistent with the finding that in E. coli translation terminates more efficiently at UAG and UAA than at UGA. Tryptophan induction was observed in strains with any of the stop codons. However, when UAG or UAA replaced UGA, the induced level of expression was also reduced to 15 and 50% of that obtained with UGA as the tnaC stop codon, respectively. Introduction of a mutant allele encoding a temperature-sensitive release factor 1, prfA1, increased basal level expression 60-fold when the tnaC stop codon was UAG and 3-fold when this stop codon was UAA; basal level expression was reduced by 50% in the construct with the natural stop codon, UGA. In strains with any of the three stop codons and the prfA1 mutation, the induced levels of tna operon expression were virtually identical. The effects of tnaC stop codon identity on expression were also examined in the absence of Rho action, using tnaC-stop codon-′lacZ constructs that lack the tnaC-tnaA spacer region. Expression was low in the absence of tnaC stop codon suppression. In most cases, tryptophan addition resulted in about 50% inhibition of expression when UGA was replaced by UAG or UAA and the appropriate suppressor was present. Introduction of the prfA1 mutant allele increased expression of the suppressed construct with the UAG stop codon; tryptophan addition also resulted in ca. 50% inhibition. These findings provide additional evidence implicating the behavior of the ribosome translating tnaC in the regulation of tna operon expression.
Tryptophanase is a multifunctional enzyme that degrades l-tryptophan to indole, pyruvate, and ammonia (27) by a β-elimination reaction (34). Bacteria with tryptophanase activity can utilize tryptophan as a source of carbon, nitrogen, and energy (23). In addition, since the β-elimination reaction is reversible, tryptophanase can synthesize l-tryptophan from indole and l-serine, l-cysteine, or pyruvate and ammonia (34, 53).
Transcription of the tna operon of E. coli has been shown to be regulated by catabolite repression of transcription initiation (1, 39, 40) and by tryptophan-induced transcription antitermination (46). The tryptophanase (tna) operon has been cloned and sequenced from Escherichia coli (10), Proteus vulgaris (24), Enterobacter aerogenes (26), Symbiobacterium thermophilum (22), and the pathogen Haemophilus influenzae type b (30). The tna operon of E. coli contains two major structural genes, the promoter proximal gene, tnaA, encoding tryptophanase, and a distal gene, tnaB, encoding a low-affinity, high-capacity, tryptophan permease (10, 12). tnaA is preceded by a transcribed regulatory leader region containing a short open reading frame, tnaC, specifying a 24-residue leader peptide. Between tnaC and tnaA there is a ca. 200-bp spacer region containing several transcription pause sites. When cells grow in the absence of inducer, tryptophan, transcription is subject to Rho-dependent transcription termination (45) at these pause sites. In the presence of inducer, termination at these sites is prevented and expression is elevated 15- to 100-fold (46). Induction requires translation of tnaC, which encodes a peptide with a single Trp residue at position 12. Tryptophan induction is not observed when the tnaC start codon is replaced by a stop codon, or when the Trp codon at position 12 is replaced by codons specifying other amino acids (13, 17, 47). Additional residues in the TnaC peptide are also essential for induction, particularly several residues near the Trp residue (15).
Several classes of tna operon constitutive mutants have been isolated; these show elevated expression of the operon in the absence of added tryptophan (46). Many constitutive mutants have single base pair changes in a 9-bp sequence at the distal end of tnaC (46) which has homology to boxA of bacteriophage λ (14). boxA is critical for proper antitermination, not termination, in phage λ (9) and other systems (21, 50). Deletion of a putative Rho utilization (rut) site located immediately following the tnaC UGA stop codon also results in constitutive expression of the operon (17). Because the boxA-like sequence in the tna operon does not behave like a typical boxA, it is conceivable that recognition of both the boxA-like sequence and the presumed rut site are required for Rho-dependent transcription termination in this operon. A third class of constitutive mutants contains +1 or −1 frameshift mutations in tnaC that allow translation to proceed well beyond the presumed rut site that follows the tnaC stop codon (17). Continued translation presumably interferes with Rho action. However, it is unlikely that tryptophan induction involves frameshifting, because stop codons introduced in the +1 and −1 reading frames between Trp codon 12 and the tnaC stop codon do not affect either basal level expression or tryptophan induction (15). Similarly, introducing stop codons in the +1 or −1 reading frame immediately following tnaC does not interfere with the effects of induction (25).
The precise mechanism by which added tryptophan induces tna operon expression is not known. Previous studies indicated that no unique regulatory factor other than TnaC is needed for induction (15, 16). Specifically, a plasmid containing a foreign promoter, the E. coli tna leader regulatory region, and a lacZ translational fusion to the first 20 codons of tnaA responded to tryptophan induction when introduced into two bacterial species that do not produce tryptophanase (15). Thus, if additional factors are required for induction, they are likely to be components common to many bacterial species, including those that lack a tna operon (15). All tests of TnaC trans-activation have given negative results (46).
A hypothesis consistent with all the observations made to date is that in the presence of inducer, the nascent TnaC peptide acts in cis on the ribosome translating tnaC and inhibits its release at the tnaC stop codon. The inhibited ribosome would interfere with Rho’s binding to the tna transcript, thereby preventing transcription termination. If ribosome release or stalling were essential in the regulation of tna operon expression, then peptide chain release factors and the ribosome release factor could be influential in setting the basal or induced level of expression of the operon. In E. coli, two codon-specific peptide chain release factors (RF1 and RF2) direct termination of protein synthesis; RF1 recognizes UAG and UAA stop codons (54), whereas RF2 acts at the UGA and UAA stop codons (6). Unlike other prokaryotic RF2s, E. coli RF2 terminates translation weakly at UGA and UAA stop codons (33, 48). The third release factor, RF3, enhances the activity of RF1 and RF2 and lacks nonsense codon specificity (18, 31).
In previous studies performed to examine models invoking ribosome release or stalling, we designed and tested a construct that would not be subject to Rho regulation. This construct, tnaC-UGA-′lacZ, lacks the tnaC-tnaA spacer region within which Rho-dependent termination occurs (28). Its expression is dependent on translation of the UGA codon and synthesis of a TnaC-′LacZ fusion protein. Using this construct and various nonsense suppressors, we showed that addition of inducer inhibited β-galactosidase (β-Gal) formation and that this inhibition was dependent upon the presence of Trp residue 12 in the TnaC portion of the fusion protein (28). We found that inactivation of the structural gene for RF3 (55), increased β-Gal production 30-fold, and this increase was also reduced by the presence of tryptophan (28). In a parallel study with deletion derivatives of the tna operon of P. vulgaris, it was observed that a deletion that places the Shine-Dalgarno region for tnaA (in tnaA′-′lacZ fusion) near the tnaC stop codon results in tryptophan inhibition of tnaA′-′lacZ expression (25).
In this report, we extend our examination of the role of the tnaC stop codon by replacing the natural UGA stop codon with UAG and UAA. We show that the presence of UAG or UAA leads to a reduction of both basal and induced expression with tnaC-stop codon-tnaA′-′lacZ constructs containing the intact tnaC-tnaA spacer region. We also show that a temperature-sensitive mutation altering RF1 increases basal level expression in strains with UAG or UAA as the tnaC stop codon, but not in a strain with UGA. The induced levels of expression in these same strains are indistinguishable. We also examine the role of the three stop codons in a tnaC-stop codon-′lacZ fusion construct and show that tryptophan addition inhibits reading past the stop codon; however, this inhibition is not as pronounced when UAG or UAA replaces UGA. An RF1 mutation increased expression of the UAG construct in the presence and absence of tryptophan. These findings support the hypothesis that in the presence of tryptophan, TnaC peptide inhibition of ribosome release at the tnaC stop codon may be the crucial event that regulates Rho-dependent termination in the tna operon leader region.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and the plasmids used in this study are listed in Table 1. Strains VK700, VK800, and VK900 and their derivatives are all single lysogens carrying λRS45 (44) with various inserts. To prepare these strains, the tnaC-UA(G/A)-′lacZ and the tnaC-UAG-tnaA′-′lacZ fusions from pRS552 (Table 1) were independently crossed into phage λRS45 (44) and the recombinant phage genome was inserted into the chromosome of CY15076 (Table 1). prfA1(Ts) (37, 38) or the serU suppressors [su UAG or su UG(A/G)] (2, 28) were introduced into VK100 (28), VK700, or VK800 by P1 transduction. Mutant trpA9761 is an amber mutant of trpA in E. coli. The trpA9761 allele was introduced into appropriate strains by transduction. Plasmids were introduced into various strains by transformation (41), selecting for the appropriate antibiotic resistance marker.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| CY15076 | W3110 tnaA2 ΔlacU169 | This study |
| PDG1158 | SVS1100 (λtnap tnaC-UAA-tnaA′-′lacZ) | 17 |
| PDG1184 | SVS1100 (λtnap tnaC-UUA-tnaA′-′lacZ) | 17 |
| SVS1100 | W3110 bglR55Δ (lac-argF)U169 | 46 |
| SVS1144 | SVS1100 (λtnap tnaA′-′lacZ) | 46 |
| VK100 | SVS1100 (λtnap tnaC-UGA-′lacZ) | 28 |
| VK100/SerUUGA | VK100 serU [Ser UG(A/G)] suppressor | 28 |
| VK700 | CY15076 (λtnap tnaC-UAG-′lacZ) | This study |
| VK700/SerUUAG | VK700 serU (Ser UAG suppressor) | This study |
| VK701 | VK700 prfA1(Ts) | This study |
| VK800 | CY15076 (λtnap tnaC-UAG-tnaA′-′lacZ) | This study |
| VK801 | VK800 prfA1(Ts) | This study |
| VK805 | VK801 trpA9761 | This study |
| VK900 | CY15076 (λtnap tnaC-UAA-′lacZ) | This study |
| VK1100 | CY15076 (λtnap tnaC-UAG UUUGAC-tnaA′-′lacZ) | This study |
| VK1101 | VK1100 prfA1(Ts) | This study |
| VK1145 | SVS1144 prfA1(Ts) | This study |
| VK1159 | PDG1158 prfA1(Ts) | This study |
| VK1185 | PDG1184 prfA1(Ts) | This study |
| VK1200 | CY15076 (λtnap tnaC-UAG UUGACC-tnaA′-′lacZ) | This study |
| VK1201 | VK1200 prfA1(Ts) | This study |
| Plasmids | ||
| pGFIBI | hisT su (UAG suppressor) | 35 |
| pMY228 | trpT su7 (UAG suppressor) | 36 |
| pRS552 | pBR322 derivative, lac-based vector | 44 |
| pSWC115 | trpT su7 (UAA suppressor) | 36 |
Media and enzyme assay.
Vogel and Bonner minimal medium (49) was used throughout. For β-Gal assays (32), cultures were generally grown with shaking at 30 or 37°C in minimal medium plus 0.2% glycerol–0.05% acid-hydrolyzed casein, with or without l-tryptophan (100 μg/ml). When appropriate, media were supplemented with kanamycin (30 μg/ml), tetracycline (15 μg/ml), or ampicillin (100 μg/ml). β-Gal assays were performed as described by Miller (32); β-Gal activity is reported in Miller units (32). For tryptophan synthetase (TSase) α and β2 assays, cultures were grown overnight at 37°C in the same minimal medium supplemented with 0.2% glycerol, 0.05% acid-hydrolyzed casein, and indole (4 μg/ml). Cells were collected by centrifugation, washed, and frozen. Each frozen pellet was resuspended in 0.1 M Tris buffer, pH 7.8, and disrupted by sonication. The sonication extracts were centrifuged and the cell supernatants were assayed for TSase activity in the indole plus serine to tryptophan reaction (8) in the presence and absence of excess wild-type TSase α or β2. For a definition of the unit of specific activity, and for other assay conditions, see the work of Creighton and Yanofsky (8).
Site-directed mutagenesis.
Two PCR-based methods (28) were used to introduce point mutations in the tnaC region. In the standard PCR approach, the VK1 primer, 5′-CGG AAT TCA GCT TCT GTA TTG GTA AG-3′, has a sequence of nucleotides identical to the region upstream of tnaC and contains an EcoRI site at its 5′ end. The mutagenic TNAC-UAG primer, 5′-GGG ATC CCC GGG AAT CTA AGG GCG GTG ATC-3′, and TNAC-UAA primer, 5′-TCC CCC GGG AAT TTA AGG GCG GTG-3′, contain a BamHI site and a SmaI site at their respective 5′ ends. Primer pairs VK1 and TNAC-UAG or VK1 and TNAC-UAA were used with Taq polymerase (Boehringer Mannheim Co., Indianapolis, Ind.) to amplify the 353- and 347-bp PCR products, respectively. These products were individually cloned into the pCRII vector (Invitrogen Co., San Diego, Calif.), and sequences were confirmed by dideoxy sequencing (42). The 353-bp insert in the pCRII vector was digested with EcoRI and BamHI, purified by using the GENECLEAN II kit (BIO 101 Inc., La Jolla, Calif.), and subcloned into the EcoRI-, BamHI- cleaved pRS552 (44) to give the tnaC-UAG-′lacZ construct. To prepare the tnaC-UAA-′lacZ construct, the 347-bp insert in the pCRII vector was first cloned into the EcoRI-, and SmaI- cleaved sites of pBluescript (41). The resulting recombinant was digested with EcoRI and BamHI, and the insert was subcloned into the EcoRI-, BamHI-cut sites of pRS552 (44).
The megaprimer PCR method (43) was used to make the tnaC-UAG-tnaA′-′lacZ construct. First, the LACZ-RT primer, 5′-GCG ATT AAG TTG GGT AAC GCC AGG-3′, and the mutagenic TNAC-UAG-TNA primer, 5′-CAC CGC CCT TAG TTT GCC CTT CTG-3′, were used with Taq polymerase to amplify a 305-bp product. This product was then purified by using a PCR purification kit (QIAGEN Inc., Chatsworth, Calif.) and combined with the VK1 primer, 5′-CGG AAT TCA GCT TCT GTA TTG GTA AG-3′, to amplify the final 675-bp PCR product. This product was also cloned into the pCRII vector and sequenced (42). The resulting 675-bp insert was cleaved with EcoRI and BamHI, purified by using the GENECLEAN II kit, and subcloned into the EcoRI-, BamHI-cleaved pRS552 vector (44). This approach was also used to introduce the primer sequences 5′-CGC CCT TAG TTT GAC CTT CTG TAG CCA TCA-3′ and 5′-CAC CGC CCT TAG TTG ACC CTT CTG TAG CCA TCA-3′ (with mutational changes underlined) into the tnaC-UAG-tnaA′-′lacZ construct.
RESULTS
Basal and induced expression from tnaC-stop codon-tnaA′-′lacZ constructs in which UAG or UAA replaces UGA as the tnaC stop codon.
To determine whether the identity of the tnaC stop codon influences basal or induced expression of the tna operon, constructs containing each of the three stop codons were prepared and integrated into the E. coli chromosome, and the β-Gal activities of the resulting strains were determined. The natural tnaC stop codon of E. coli is UGA (Fig. 1) (10, 46). With constructs containing the UAG and UAA stop codons, basal expression of the operon was 20 and 50%, respectively, of that observed with the UGA stop codon (Table 2). These findings are consistent with evidence demonstrating that in E. coli, RF1 terminates translation more efficiently than RF2 (7, 48). Following growth with tryptophan as inducer, the same pattern was observed; induced expression was only 15 or 60%, respectively, of that obtained with the tnaC-UGA stop codon construct (Table 2). These findings establish that both basal and induced expression of the tna operon are influenced by the identity of the tnaC stop codon.
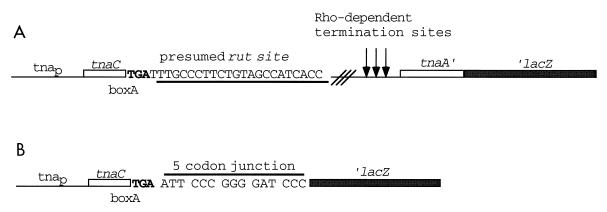
FIG. 1.
Schematic representations of the basic tna operon-′lacZ fusions employed in this study. (A) tnaC-UGA-tnaA′-′lacZ construct in strain SVS1144. The underlined rut site is part of the natural tnaC-tnaA spacer region, which also contains Rho-dependent termination sites (45). (B) tnaC-UGA-′lacZ construct in strain VK100. tnaC, its UGA stop codon, and a five-codon junction are in phase with lacZ lacking its first eight codons. The UGA stop codon (shown in boldface type) in both constructs was also changed to UAG or UAA.
TABLE 2.
Basal and induced expression in strains with constructs containing one of three stop codons (at 37°C)
| Strain | Relevant features | Avg β-Gal activity (Miller units) ± SDb
|
||
|---|---|---|---|---|
| −Trp | +Trp | +Trp/ −Trpc | ||
| SVS1144 | tnaC-UGA-tnaA′-′lacZ | 518 ± 20 | 14,019 ± 1,159 | 27 |
| VK800 | tnaC-UAG-tnaA′-′lacZ | 99 ± 12 | 1,919 ± 252 | 19 |
| VK800/ pMY228a | VK800 with su UAG (Trp) | 367 ± 33 | 1,199 ± 82 | 3 |
| PDG1158 | tnaC-UAA-tnaA′-′lacZ | 223 ± 17 | 8,344 ± 940 | 37 |
| PDG1184 | tnaC stop to UUAd | 239 ± 9 | 2,076 ± 97 | 9 |
pMY228, a plasmid carrying a Trp-inserting tRNATrp UAG suppressor
Cultures were grown at 37°C in minimal medium (49) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+Trp) or without (−Trp) 100 μg of l-Trp per ml. For β-Gal assay conditions, see Materials and Methods. Values are for at least five cultures of each type.
Ratio of β-Gal activities with and without Trp.
Adds five sense codons to tnaC to a UAG.
In strain PDG1184, the tnaC stop codon was replaced by a leucine codon, UUA (17). This change allows translation to proceed five codons beyond the last sense codon of tnaC, to a UAG termination codon (17). Such a construct exhibits expression levels much like those of the UAG construct VK800; however, basal level expression is two-fold higher (Table 2).
Effects of the presence of a mutant RF1 on tna operon expression.
Mutant allele prfA1 encodes a temperature-sensitive RF1. This allele was introduced into appropriate strains, and basal and induced expression were measured (Table 3). Since strains with prfA1 are temperature sensitive for growth, they were grown at the permissive temperature, 30°C (38). The prfA1 allele increased basal expression of the constructs with the UAG and UAA stop codons 60- and 3-fold, respectively; it reduced both basal and induced expression of the construct with the wild-type tnaC UGA stop codon by 50%. Induced expression levels of strains with the prfA1 allele and any of the three tnaC stop codons were virtually identical (Table 3). The prfA1 allele increased basal level expression of the construct in strain PDG1184 sevenfold, but there was little induced expression (Table 3).
TABLE 3.
Effects of a prfA1 mutation on basal and induced expression in strains with one of three stop codons (at 30°C)
| Strain | Relevant features | Avg β-Gal activity (Miller units) ± SDa
|
||
|---|---|---|---|---|
| −Trp | +Trp | +Trp/ −Trpb | ||
| SVS1144 | tnaC-UGA-tnaA′-′lacZ | 729 ± 24 | 16,244 ± 729 | 22 |
| VK1145 | SVS1144 prfA1 | 421 ± 52 | 6,833 ± 875 | 16 |
| VK800 | tnaC-UAG-tnaA′-′lacZ | 49 ± 3 | 1,102 ± 104 | 22 |
| VK801 | VK800 prfA1 | 3,093 ± 295 | 7,096 ± 548 | 2.3 |
| PDG1158 | tnaC-UAA-tnaA′-′lacZ | 163 ± 3 | 6,554 ± 357 | 40 |
| VK1159 | PDG1158 prfA1 | 459 ± 21 | 7,634 ± 861 | 17 |
| PDG1184 | tnaC stop to UUAc | 72 ± 4 | 626 ± 80 | 9 |
| VK1185 | PDG1184 prfA1 | 513 ± 23 | 881 ± 45 | 1.7 |
Cultures were grown at 30°C in minimal medium (49) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+Trp) or without (−Trp) 100 μg of l-Trp per ml. For β-Gal assay conditions, see Materials and Methods. Values are for at least five cultures of each type.
Ratio of β-Gal activities with and without Trp.
Adds five codons to tnaC to a UAG.
Test for suppression of the trpA9761 amber mutation in VK801 derivatives.
The high basal level expression observed in strain VK801 (tnaC-UAG-tnaA′-′lacZ/prfA1) (Table 3) could be due to specific suppression of the tnaC UAG stop codon by a resident UAG suppressor. Suppression would allow translation to continue four codons downstream from the tnaC stop codon. Previous findings argue against this possibility. First, suppression of the tnaC stop codon should at best approach expression in strain PDG1184, in which a sense codon replaces the tnaC stop codon. As shown in Table 2, strain PDG1184 does not exhibit high basal expression in the absence of tryptophan. Second, suppression of the tnaC UAG stop codon in strain VK800 (tnaC-UAG-tnaA′-′lacZ) gives basal and induced expression levels comparable to those of strain PDG1184 (Table 2).
Two tests were performed to rule out significant amber suppression in strain VK801 containing prfA1. An amber mutation that is known to respond to all tested amber and ochre suppressors, trpA9761, was introduced into VK801, yielding strain VK805 (Tables 1 and 4). Isolates of VK805 were observed to be tryptophan-dependent, indicating that the prfA1 alteration does not allow natural suppression of the trpA9761 amber codon by insertion of an amino acid that restores TSase α activity. Although trpA9761 is known to be suppressed to prototrophy by known amber and ochre suppressors, it is conceivable that the prfA1 mutation allows insertion of an amino acid that does not restore TSase α activity. The trpA system provides an excellent test of this possibility since virtually all trpA missense mutants produce an enzymatically inactive protein that nevertheless can complex with TSase β2 and activate this subunit 30-fold in the indole plus serine to tryptophan reaction. Two isolates of strain VK805 were examined, and no significant TSase α protein was observed, demonstrating the absence of significant amber suppression (Table 4). Other data also indicate that the increased tna operon expression associated with the prfA1 allele is not due to significant amber suppression (see Table 7).
TABLE 4.
TSase α and β2 activities of trpA prfA1(Ts) strainsa
| Strain | Characteristics | Sp act ofb:
|
TSase α/ TSase β2c | |
|---|---|---|---|---|
| TSase α | TSase β | |||
| VK800 | tnaC-UAG-tnaA′-′lacZ | 2.8 | 3.3 | 0.85 |
| VK801 | VK800 prfA1 | 3.4 | 4.9 | 0.69 |
| VK805d | VK801 trpA9761 | 0.17e | 15 | 0.01 |
| VK805d | VK801 trpA9761 | 0.07e | 13 | 0.01 |
Cultures were grown overnight in minimal medium (49) supplemented with 0.2% glycerol, 0.05% acid-hydrolyzed casein, and 4 μg indole per ml. For TSase assay conditions, see Materials and Methods.
Indole plus serine to tryptophan reaction.
Ratio of specific activities of TSase α and TSase β2.
Two different VK805 isolates were used.
Corrected for the inherent activity of TSase β2 in the indole to tryptophan reaction.
TABLE 7.
Effects of a prfA1 mutation on tryptophan inhibition of translation of the tnaC-UAG-′lacZ construct (at 30°C)
| Strain | Relevant features | Avg β-Gal activity (Miller units) ± SDb
|
+Trp/ −Trpc | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| VK700 | tnaC-UAG-′lacZ | 0.05 | <0.01 | |
| VK700/pGFIBIa | VK700 with su UAG (His) | 274 ± 18 | 173 ± 22 | 0.6 |
| VK701 | VK700 prfA1 | 4 ± 0.8 | 1.5 ± 0.4 | 0.4 |
| VK701/pGFIBIa | VK701 with su UAG (His) | 715 ± 42 | 355 ± 59 | 0.5 |
pGFIBI, plasmid carrying a His-inserting tRNAHis UAG suppressor.
Cultures were grown at 30°C in minimal medium (49) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+Trp) or without (−Trp) 100 μg of l-Trp per ml. For β-Gal assay conditions, see Materials and Methods.
Ratio of activities with and without Trp.
Effects of introducing UGA stop codons in the −1 or +1 potential reading frames immediately following the tnaC UAG stop codon.
Point mutations were introduced in the spacer region of the tnaC-UAG-tnaA′-′lacZ construct to test whether there is ribosomal frameshifting beyond the tnaC UAG stop codon in the prfA1 mutant background (Table 5). The resulting constructs tnaC-UAG UUUGAC and tnaC-UAG UUGACC, introduce out-of-frame UGA stop codons (shown in boldface type). In a prfA1 mutant background, any −1 or +1 frameshift would allow translation to terminate at a UGA codon immediately following the normal UAG stop codon and would presumably result in β-Gal production similar to that obtained in strain PDG1184 (Table 3). In Table 5, the presence of the prfA1 allele resulted in a 5- to 10-fold increase in the basal level of β-Gal activity in strains with the constructs tnaC-UAG UUUGAC (strain VK1101) and tnaC-UAG UUGACC (strain VK1201), respectively; a similar increase (18-fold) in the basal level of β-Gal expression was observed with the control strain VK801 (Table 5). These findings indicate that ribosomal frameshifting beyond the tnaC UAG stop codon is not responsible for the increase in the basal level expression in a prfA1 mutant background. Note that the changes introduced following the tnaC stop codon do influence the absolute levels of basal and induced expression.
TABLE 5.
Introducing the UGA stop codon in the −1 or +1 reading frame immediately following tnaC-UAG does not appreciably alter the response to prfA1
| Strain | Relevant featuresa | Avg β-Gal activity (Miller units) ± SDb
|
+Trp/ −Trpc | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| VK800 | tnaC-UAG-tnaA′-′lacZ | 52 ± 4 | 1,218 ± 164 | 23 |
| VK801 | VK800 prfA1 | 953 ± 83 | 3,090 ± 434 | 3.2 |
| VK1100 | tnaC-UAG UUUGCC to tnaC-UAG UUUGAC | 182 ± 15 | 1,270 ± 101 | 7 |
| VK1101 | VK1100 prfA1 | 953 ± 118 | 2,074 ± 300 | 2.2 |
| VK1200 | tnaC-UAG UUUGCC to tnaC-UAG UUGACC | 27 ± 5 | 857 ± 94 | 32 |
| VK1201 | VK1200 prfA1 | 261 ± 18 | 841 ± 81 | 3.2 |
Underlined changes introduce stop codons in the −1 or +1 reading frame following tnaC-UAG. The out-of-frame UGA stop codon is in boldface type.
Cultures were grown at 30°C in minimal medium (49) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+Trp) or without (−Trp) 100 μg of l-Trp per ml. For β-Gal assay conditions, see Materials and Methods. Values are for at least five cultures of each type.
Ratio of activities with and without Trp.
Tryptophan inhibition of translational readthrough beyond the tnaC stop codon in constructs lacking the tnaC-tnaA spacer region.
In a previous study (28) we designed and tested a construct that we believed would permit analysis of the effects of inducer in the absence of Rho-dependent termination. This construct, tnaC-UGA-′lacZ, lacks the tnaC-tnaA spacer region. It contains the tna promoter through the tnaC UGA stop codon, has an added in-phase five-codon junction, and is followed by lacZ minus its first eight codons (5) (Fig. 1). Since this construct lacks the tnaC-tnaA spacer region, Rho action should be eliminated. Our previous findings supported this expectation (28).
In strain VK100 containing this construct (tnaC-UGA-′lacZ) with or without UGA suppressors, the presence of tryptophan led to an 80% decrease in β-Gal activity (28). To measure expression in strains with derivatives of this construct in which the UGA stop codon was replaced by UAG or UAA, strains VK700 (tnaC-UAG-′lacZ) and VK900 (tnaC-UAA-′lacZ) were prepared (Table 6). When grown in minimal medium with or without tryptophan, strains VK100 (tnaC-UGA-′lacZ), VK700 (tnaC-UAG-′lacZ), and VK900 (tnaC-UAA-′lacZ) had very low β-Gal levels, indicating that these strains lack active nonsense suppressors (Table 6). Introducing appropriate suppressors into these strains did lead to an increase in expression, in the presence or absence of tryptophan. In contrast to our results with the tnaC-UGA-′lacZ construct (Table 6), the presence of the UAG or UAA stop codon coupled with the appropriate suppressor allowed only modest tryptophan inhibition of expression (30 to 60% inhibition with UAG or UAA versus 80% inhibition with UGA).
TABLE 6.
Tryptophan inhibition of translation beyond the tnaC stop codon in constructs lacking the tnaC-tnaA spacer region
| Strain | Relevant features | Avg β-Gal activity (Miller units) ± SDf
|
+Trp/ −Trpg | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| SVS1144 | tnaC-UGA-tnaA′-′lacZ | 465 ± 42 | 15,728 ± 1,353 | 34 |
| VK100 | tnaC-UGA-′lacZ | 3 | 0.4 | 0.1 |
| VK100/SerUa | VK100 with su UGA (Ser) | 85 ± 6 | 18 ± 2 | 0.2 |
| VK700 | tnaC-UAG-′lacZ | 0.1 | 0.03 | 0.3 |
| VK700/pMY228b | VK700 with su UAG (Trp) | 430 ± 42 | 490 ± 39 | 1 |
| VK700/SerUc | VK700 with su UAG (Ser) | 190 ± 21 | 130 ± 9 | 0.7 |
| VK700/pGFIBId | VK700 with su UAG (His) | 709 ± 63 | 475 ± 48 | 0.6 |
| VK900 | tnaC-UAA-′lacZ | 5 ± 0.5 | 2 ± 0.4 | 0.4 |
| VK900/pSWC115e | VK900 with su UAA (Trp) | 9 ± 1 | 4 ± 0.4 | 0.4 |
SerU, Ser-inserting tRNASer UGA suppressor.
pMY228, plasmid carrying a Trp-inserting tRNATrp UAG suppressor.
SerU, Ser-inserting tRNASer UAG suppressor.
pGFIBI, plasmid carrying a His-inserting tRNAHis UAG suppressor.
pSWC115, plasmid carrying a Trp-inserting tRNATrp UAA suppressor.
Cultures were grown at 37°C, in minimal medium (49) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+Trp) or without (−Trp) 100 μg of l-Trp per ml. For β-Gal assay conditions, see Materials and Methods.
Ratio of activities with and without Trp.
To examine the effects of the temperature-sensitive RF1 on translation beyond the tnaC UAG stop codon, prfA1 was introduced into strain VK700 (tnaC-UAG-′lacZ), yielding strain VK701 (tnaC-UAG-′lacZ/prfA1) (Table 7). Note that all the strains in Table 7 were grown at 30°C. VK701 showed very low levels of expression, with or without inducer. This result supports the finding that strain VK801 (Table 3) lacks an effective UAG suppressor. Addition of a plasmid containing a UAG-reading tRNAHis suppressor (35) into control strain VK700 increased basal expression appreciably and allowed 40% inhibition by the inducer (Table 7). Introduction of this suppressor into strain VK701 (containing prfA1) increased expression almost threefold compared to VK700 bearing a suppressor and allowed 50% inhibition of expression by tryptophan.
DISCUSSION
Studies with the tna operon of E. coli have identified features of the tna leader region that are necessary for this operon’s regulation by transcriptional attenuation. In the absence of an inducer, transcription is terminated by the action of Rho factor, at one of several transcription pause sites located in the leader region preceding tnaA, the first major structural gene of the operon (45). Mutational changes that allow high-level constitutive expression of the operon in the absence of inducer have identified cell components and sequences that are necessary for transcription termination. These mutations alter Rho factor, change bases in critical regulatory sequences in the tna leader region (boxA, the rut site), or allow translation from the tnaC coding sequence to proceed in other reading frames, beyond the presumed rut site in the tna transcript. These observations and other experimental findings suggest that when cells grow without inducer, Rho factor generally binds to the leader segment of the tna transcript, moves 3′ on the transcript until it interacts with a paused polymerase molecule in the leader region, and then directs the polymerase to terminate transcription.
In the presence of added tryptophan, Rho-dependent termination in the leader region of the tna operon is prevented. Induction requires translation of a 24-residue peptide coding region, tnaC, located near the 5′ end of the transcript (17). Several residues of TnaC are essential for induction, suggesting that the peptide as such participates in the induction process. Some tryptophan analogs which do not appear to be incorporated into protein also function as inducers; thus, an inducer may act without being incorporated into the TnaC leader peptide (15, 16). Stop codons introduced in the +1 or −1 reading frames within tnaC, or following tnaC, do not affect either basal or induced expression, establishing that tryptophan induction does not involve frameshifting (15, 16, 25). Introducing an in-phase stop codon within the distal segment of tnaC does prevent induction (15).
How added tryptophan is used as the signal that leads to inhibition of Rho-dependent termination is a basic unanswered question. One possibility is that the TnaC peptide is modified in some manner when produced in cells growing with excess tryptophan and that the altered peptide blocks Rho action. Alternatively, in the presence of added tryptophan some cell component could interact with the TnaC peptide, altering its properties so that it blocks Rho action. Two previous studies (28, 55) and this report have focused on how the peptide may act in preventing termination. In prior analyses with a suppressed tnaC-stop codon-′lacZ fusion lacking the tnaC-tnaA spacer region, the addition of tryptophan led to inhibition of translation beyond the tnaC stop codon (28). Tryptophan addition also reversed the increased expression resulting from inactivation of RF3 (28). In related studies with the tna operon of P. vulgaris, it was shown that added tryptophan inhibited expression of a tnaA′-′lacZ translational fusion in which the tnaA ribosome binding site was placed close to the tnaC stop codon (25). These effects of inducer were not observed when the critical tryptophan codon in the leader peptide coding region was replaced by some other codon (28). The most straightforward interpretation of these observations is that in the presence of tryptophan, the TnaC leader peptide acts in cis on the translating ribosome, preventing its release at the tnaC stop codon. The stalled ribosome would block Rho’s access to the transcript, thereby inhibiting termination.
In the present study we examined the effect of changing the natural tnaC UGA stop codon of E. coli to the other stop codons, UAG and UAA. We also analyzed the effects of introducing a temperature-sensitive mutant allele, prfA1, that alters RF1. With a lacZ reporter construct that allows Rho-dependent termination, changing the natural UGA stop codon to UAG lowered basal and induced expression of the operon by ca. 80%. Changing the UAA stop codon to UGA reduced basal and induced expression by 50%. The reduced expression observed with the UAG stop codon relative to the UGA stop codon could be explained most simply if RF1 dissociates the translating ribosome from the tnaC UAG stop codon more rapidly than does RF2 at the UGA stop codon. More rapid release could allow more rapid binding of Rho factor. This interpretation is consistent with the conclusion that in E. coli RF1 terminates translation more efficiently than RF2 (33, 48). Support for this interpretation is also provided by the observation that a mutant allele specifying a temperature-sensitive RF1, when examined at the permissive temperature, led to a 60-fold increase in basal level expression when the tnaC stop codon was UAG (Table 3). In fact, expression was increased only twofold further by the addition of tryptophan.
It is also conceivable that in the presence of the mutant RF1, the nucleotide sequence including the UAG stop codon permits efficient translational frameshifting and reading into the presumed rut site. However, introduction of a UGA stop codon in the −1 or +1 reading frame immediately following the tnaC UAG stop codon did not eliminate the increased basal level expression of the tnaC-UAG construct in a prfA1 mutant background. Furthermore, our findings with an in-phase tnaC-UAG-5 codon junction-′lacZ construct (Table 6) that requires suppression of the UAG stop codon for β-galactosidase production also argue against this possibility. Thus, it appears that the rate of ribosome release at the tnaC stop codon (and not ribosomal frameshifting beyond the tnaC UAG stop codon) may be the key event that determines whether Rho factor will terminate transcription in the leader region of the operon.
In an early study with the tnaC-UGA-5 codon junction-′lacZ construct, we observed that under a variety of conditions allowing in-phase reading of the tnaC UGA stop codon, addition of tryptophan to the culture medium led to ca. 80% inhibition of β-Gal production (28). In this study, the UGA stop codon was changed to UAG or UAA, and the resulting constructs were tested in the presence of appropriate suppressors. We found that in all but one strain (containing a Trp-inserting UAG suppressor) tryptophan addition had a 30 to 60% inhibitory effect on β-Gal production (Table 6). Introduction of the prfA1 alteration resulted in a threefold increase in translation of the UAG stop codon (Table 7). In this case, the altered RF1 clearly allows increased in-phase translation. Tryptophan addition to this strain led to 50% inhibition of β-Gal production (Table 7). These findings demonstrate that stop codon identity does influence the ability of tryptophan to inhibit ribosome movement beyond the suppressed tnaC stop codon.
Inhibition of ribosome function by a leader peptide has been documented in other systems. In prokaryotes, the five-residue leader peptide, MVKTD, encoded by the cat leader sequence (19, 29), and the eight-residue peptide, MSTSKNAD, encoded by the cmlA leader sequence, act in cis to block movement of the translating ribosome (29) by inhibiting its peptidyltransferase activity (20, 29). In both cases, blocking the translating ribosome allows downstream translation of a structural gene conferring drug resistance. In eukaryotes, the 24-residue upstream open reading frame (uORF) encoded upstream of the arg-2 gene of Neurospora crassa (51, 52) and the 22-residue uORF2 encoded by the gp48 gene of cytomegalovirus (3, 4, 11) have been reported to act in cis to block release of their translating ribosomes. In these cases, stalling of the translating ribosome at the respective uORF termination codon appears to inhibit translation of a downstream ORF. Additional analyses focused on the TnaC peptide of the tna operon should reveal how TnaC acts to increase this operon’s expression.
ACKNOWLEDGMENTS
prfA1(Ts) was kindly supplied by S. M. Rydén. serU [su UG(A/G)] and serU (su UAG) were gifts from E. Murgola and J. R. Beckwith, respectively. We are indebted to Virginia Horn for her technical assistance with E. coli strain production. We are also grateful to Peter Margolis, Kristin Black, and M. C. Yee for their critical reading of this manuscript.
These studies were supported by grant GM09738 to C.Y. from the United States Public Health Service. K.V.K. is a Postdoctoral Fellow supported by grant GM09738.
REFERENCES
- 1.Botsford J L, DeMoss R D. Catabolite repression of tryptophanase in Escherichia coli. J Bacteriol. 1971;105:303–312. doi: 10.1128/jb.105.1.303-312.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner S, Beckwith J R. Ochre mutants, a new class of suppressible nonsense mutants. J Mol Biol. 1965;13:629–637. [Google Scholar]
- 3.Cao J, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caskey C T, Forrester W C, Tate W, Ward C D. Cloning of the Escherichia coli release factor 2 gene. J Bacteriol. 1984;158:365–368. doi: 10.1128/jb.158.1.365-368.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craigen W J, Cook R G, Tate W P, Caskey T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creighton T E, Yanofsky C. Chorismate to tryptophan (Escherichia coli)-anthranilate synthetase, PR transferase, PRA isomerase, InGP synthetase, tryptophan synthetase. Methods Enzymol. 1970;17:365–380. [Google Scholar]
- 9.Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA poymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992;174:6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeley M C, Yanofsky C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degnin C R, Schleiss M R, Cao J, Geballe A P. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp480) transcript. J Virol. 1993;67:5514–5522. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards R M, Yudkin M D. Location of the gene for the low-affinity tryptophan-specific permease of Escherichia coli. Biochem J. 1982;204:617–619. doi: 10.1042/bj2040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eshoo, M., and C. Yanofsky. Unpublished results.
- 14.Friedman D I. Regulation of phage gene expression by termination and antitermination of transcription. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 263–319. [Google Scholar]
- 15.Gish K, Yanofsky C. Evidence suggesting cis action by the tnaC leader peptide in regulating transcription attenuation in the tryptophanase operon of Escherichia coli. J Bacteriol. 1995;177:7245–7254. doi: 10.1128/jb.177.24.7245-7254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gish K, Yanofsky C. Inhibition of expression of the tryptophanase operon in Escherichia coli by extrachromosomal copies of the tna leader region. J Bacteriol. 1993;175:3380–3387. doi: 10.1128/jb.175.11.3380-3387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gollnick P, Yanofsky C. tRNATrp translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J Bacteriol. 1990;172:3100–3107. doi: 10.1128/jb.172.6.3100-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham R. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Harrod R, Rogers E J, Lovett P S. Anti-peptidyl transferase leader peptides of attenuation regulated chloramphenicol resistance genes. Proc Natl Acad Sci USA. 1994;91:5612–5616. doi: 10.1073/pnas.91.12.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Z, Rogers E J, Lovett P S. Peptidyltransferase inhibition by the nascent leader peptide of an inducible cat gene. J Bacteriol. 1993;173:5309–5313. doi: 10.1128/jb.175.17.5309-5313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinrich T, Condon C, Pfeiffer T, Hartmann R K. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J Bacteriol. 1995;177:3793–3800. doi: 10.1128/jb.177.13.3793-3800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirahara T, Suzuki S, Horinouchi S, Beppu T. Cloning, nucleotide sequences, and overexpression in Escherichia coli of tandem copies of a tryptophanase gene in an obligately symbiotic thermophile, Symbiobacterium thermophilum. Appl Environ Microbiol. 1992;58:2633–2642. doi: 10.1128/aem.58.8.2633-2642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins F G, Cole S W. A contribution to the chemistry of proteid. Part II. The constitution of tryptophanase, and the action of bacteria upon it. J Physiol (London) 1903;29:451–466. doi: 10.1113/jphysiol.1903.sp000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamath A V, Yanofsky C. Characterization of the tryptophanase operon of Proteus vulgaris. J Biol Chem. 1992;267:19978–19985. [PubMed] [Google Scholar]
- 25.Kamath A V, Yanofsky C. Roles of the tnaC-tnaA spacer region and Rho factor in regulating expression of the tryptophanase operon of Proteus vulgaris. J Bacteriol. 1997;179:1780–1786. doi: 10.1128/jb.179.5.1780-1786.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawasaki K, Yokota A, Oita S, Kobayashi C, Yoshikawa S, Kawamoto S, Takao S, Tomita F. Cloning and characterization of a tryptophanase gene from Enterobacter aerogenes SM-18. J Gen Microbiol. 1993;139:3275–3281. doi: 10.1099/00221287-139-12-3275. [DOI] [PubMed] [Google Scholar]
- 27.Kazarinoff M N, Snell E E. Essential arginine residues in tryptophanase from Escherichia coli. J Biol Chem. 1977;252:7598–7602. [PubMed] [Google Scholar]
- 28.Konan K V, Yanofsky C. Regulation of the Escherichia coli tna operon: nascent leader peptide control at the tnaC stop codon. J Bacteriol. 1997;179:1774–1779. doi: 10.1128/jb.179.5.1774-1779.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin K, Morlin G, Smith A, Nordike A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 33.Nakamura Y, Ito K. How protein reads the stop codon and terminates translation. Genes Cells. 1998;3:265–278. doi: 10.1046/j.1365-2443.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 34.Newton W A, Snell E E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci USA. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Normanly J, Masson J-M, Kleina L G, Abelson J. Contruction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci USA. 1986;83:6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raftery L A, Egan J B, Cline S W, Yarus M. Defined set of cloned termination suppressors: in vivo activity of isogenic UAG, UAA, and UGA suppressor tRNAs. J Bacteriol. 1984;158:849–859. doi: 10.1128/jb.158.3.849-859.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roesser J R, Yanofsky C. Ribosome release modulates basal level expression of the trp operon of Escherichia coli. J Biol Chem. 1988;263:14251–14255. [PubMed] [Google Scholar]
- 38.Ryden S M, Isaksson L A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 39.Saier M H J. Regulatory interactions involving the proteins of the phosphotransferase system in enteric bacteria. J Cell Biochem. 1993;51:62–68. doi: 10.1002/jcb.240510112. [DOI] [PubMed] [Google Scholar]
- 40.Saier M H J, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 44.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 45.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart V, Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1985;164:731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart V, Yanofsky C. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1986;167:383–386. doi: 10.1128/jb.167.1.383-386.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 49.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 50.Vogel U, Jensen K F. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J Biol Chem. 1995;270:18335–18340. doi: 10.1074/jbc.270.31.18335. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Sachs M S. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J Biol Chem. 1997;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translation attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Snell E E. Reversibility of the tryptophanase reaction: synthesis of tryptophan from indole, pyruvate and ammonia. Proc Natl Acad Sci USA. 1972;69:1086–1090. doi: 10.1073/pnas.69.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss R B, Murphy J P, Gallant J A. Genetic screen for cloned release factor genes. J Bacteriol. 1984;158:362–364. doi: 10.1128/jb.158.1.362-364.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanofsky C, Horn V, Nakamura Y. Loss or overproduction of polypeptide release factor 3 influences expression of the tryptophanase operon of Escherichia coli. J Bacteriol. 1996;178:3755–3762. doi: 10.1128/jb.178.13.3755-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]