SUMMARY
Peripheral nervous system (PNS) injuries initiate transcriptional changes in glial cells and sensory neurons that promote axonal regeneration. While the factors that initiate the transcriptional changes in glial cells are well characterized, the full range of stimuli that initiate the response of sensory neurons remain elusive. Here, using a genetic model of glial cell ablation, we find that glial cell loss results in transient PNS demyelination without overt axonal loss. By profiling sensory ganglia at single-cell resolution, we show that glial cell loss induces a transcriptional injury response preferentially in proprioceptive and Aβ RA-LTMR neurons. The transcriptional response of sensory neurons to mechanical injury has been assumed to be a cell-autonomous response. By identifying a similar response in non-injured, demyelinated neurons, our study suggests that this represents a non-cell-autonomous transcriptional response of sensory neurons to glial cell loss and demyelination.
In brief
Using a genetic mouse model of glial cell ablation and demyelination, Elbaz et al. found that demyelinated sensory neurons initiate an injury-induced transcriptional program suggestive of active participation in the remyelination process. Among the sensory neurons, this transcriptional response occurred predominantly in large myelinated proprioceptive and Aβ RA-LTMR neurons.
Graphical abstract
INTRODUCTION
Peripheral nervous system (PNS) myelin facilitates rapid nerve conduction velocities and provides trophic support to axons. During development, neural crest cells comigrate with the growing axons and differentiate to satellite glial cell (Hall and Landis, 1992) and Schwann cell precursor cells that later differentiate to myelinating or non-myelinating Schwann cells (Jessen et al., 2015). Satellite glial cells are PNS-specific glial cells that wrap around neuronal cell bodies in sensory, sympathetic, and parasympathetic ganglia, where they control neuronal homeostasis and the microenvironment (Hanani and Spray, 2020). Schwann cells support the long-term survival of peripheral axons and the formation and organization of nodes of Ranvier, while the axons provide trophic and mitogenic factors to the Schwann cells (Corfas et al., 2004). Non-myelinating Schwann cells engulf low-caliber (<1 μm) axons. High levels of neuregulin 1 type III expressed on the surface of high-caliber (>1 μm) axons triggers the Schwann cell myelination program, resulting in a one-to-one ratio between myelinating Schwann cells and axons (Michailov et al., 2004; Taveggia et al., 2005). Mutations in Schwann cell-specific genes, such as Connexin-32 (Cx32) and Myelin Protein Zero (Mpz), that affect normal Schwan cell function, result in demyelination and axonal degeneration (Berger et al., 2006). In addition, impaired Schwann cell function and demyelination accompanied by axon damage is a debilitating result of chemotherapy treatment (Imai et al., 2017) and of both type 1 and type 2 diabetes (Goncalves et al., 2017). Strikingly, despite the clinical importance, the effect of PNS demyelination on sensory neuron gene expression is largely unknown.
PNS injury initiates transcriptional changes in Schwann cells, satellite glial cells, and sensory neurons that promote regeneration. Schwann cells that lose their axon-glia interaction, upon injury, dedifferentiate and transform into repair Schwann cells that aid axonal regeneration. This transcriptional reprograming is controlled mainly by the early transcription factor c-Jun (Arthur-Farraj et al., 2012), the transcription factor STAT3 (Benito et al., 2017), by chromatin modifications (Ma and Svaren, 2018), and by Merlin (Mindos et al., 2017). Satellite glial cells respond to injury by changing the expression of transcripts related to fatty acid synthesis, which is mediated by the peroxisome proliferator-activated receptor (PPARα) pathway (Avraham et al., 2020). In parallel, sensory neurons respond to mechanical axonal injury by transcriptional reprograming that suppresses sensory neuron cell identity and promotes neuronal regeneration. In sensory neurons, this reprograming depends on the early transcription factor ATF3 (Renthal et al., 2020). Upon completion of neuronal regeneration and reinnervation, the original neuronal and glial cell identity is reestablished, and Schwann cell axon-glia interactions are restored (Jessen and Mirsky, 2016, 2019).
While signals from the injured axon to its counterpart Schwann cell contribute to the neuronal injury response (Catenaccio et al., 2017; Vaquie et al., 2019), the precise signals that initiate the transcriptional reprograming in sensory neurons upon injury are poorly understood. Previous studies suggest that this transcriptional reprograming is a cell-autonomous property of sensory neurons, activated solely by mechanical injury of the neurons (Cheng et al., 2021; Cho et al., 2013). In these studies, injury-induced transcriptional reprograming in sensory neurons was documented in spinal nerve transection, sciatic nerve transection, and sciatic nerve crush, and was not observed in nonmechanical models that promote chronic pain, such as models of inflammatory and chemotherapy-induced pain, where only a few genes were activated in sensory neurons (Renthal et al., 2020). These findings raise the possibility that disruption of axon-glia interactions may drive the transcriptional reprogramming observed after mechanical axonal injury.
To study whether peripheral glial cells have a specific role in modulating the transcriptional state of sensory neurons, we characterized a genetic mouse model of Schwann cell and satellite glial cell ablation. We find that glial cell loss results in a transient PNS demyelination without overt axonal loss. Using a combination of bulk and single-nucleus RNA sequencing (snRNA-seq), we find that this glial cell loss and demyelination results in a transcriptional response in myelinated neurons that is very similar to the transcriptional reprograming observed in sensory neurons after mechanical axonal injury, suggesting that glial cell loss and demyelination contribute to the transcriptional reprograming in sensory neurons.
RESULTS
Genetic ablation of PNS glial cells results in neuropathy and demyelination of the PNS without overt axonal loss
For our studies, we focused on the well-characterized PLP-CreERT;ROSA26-eGFP-DTA (DTA) model, in which the expression of the diphtheria toxin A subunit (DT-A) is prevented by upstream lox-stop-lox (LSL) cassette, and only upon tamoxifen-induced PLP-CreERT-mediated recombination is the stop sequence removed and the expression of DTA released, resulting in cell loss. CNS demyelination in DTA mice has been characterized in detail (Traka, 2019; Traka et al., 2010, 2016). Nevertheless, Pip is also expressed in Schwann cells (Doerflinger et al., 2003) and satellite glial cells (Avraham et al., 2020), suggesting possible loss of Schwann cells, PNS demyelination, and loss of satellite glial cells in tamoxifen-treated animals.
To explore the cellular specificity of recombination driven by the PLP-CreERT line, we used the reporter line ROSA26-Stop-EYFP, in which an LSL cassette followed by an enhanced yellow fluorescent protein (EYFP) gene was inserted into the ROSA26 locus (Srinivas et al., 2001). Using the PLP-CreERT-ROSA26-Stop-EYFP line, we found that tamoxifen treatment resulted in EYFP expression in non-myelinating Schwann cells and myelinating Schwann cells in the sciatic nerve (Figures 1A–1D and S1). In the DRGs, we found EYFP expression in satellite glial cells but not in sensory neurons (Figures 1A–1D and S1), as previously described (Kim et al., 2016).
Figure 1. Glial cell ablation results in demyelination without axonal loss.
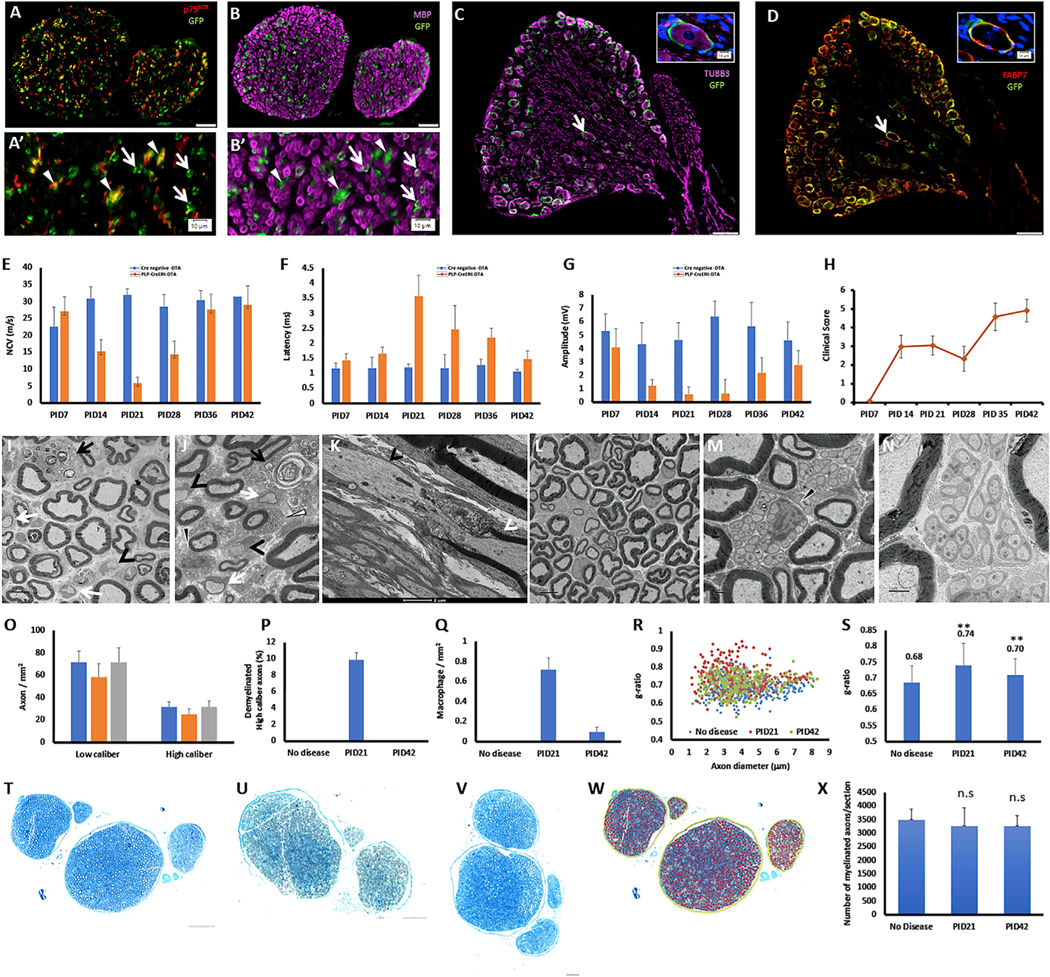
Eight-week-old PLP-CreERT;ROSA26-EYFP mice were injected with tamoxifen, and the sciatic nerve and the L3-L5 DRGs were dissected.
(A) In the sciatic nerve, the expression of EYFP was detected by anti-GFP antibody in non-myelinating Schwann cells, marked by p75NTR. (A′) Higher magnification; white triangles mark EYFP-positive cells that are positive for p75NTR, and white arrows mark EYFP-positive cells that are p75NTR negative (Scale bar, 50 μm, A’- scale bar, 10 μm).
(B) In the sciatic nerve, the expression of EYFP was detected also in myelinating Schwann cells, marked by MBP. (B′) Higher magnification; white triangles mark EYFP-positive cells that are negative for MBP, and white arrows mark EYFP-positive cells that are MBP positive (Scale bar, 50 μm, B’- scale bar, 10 μm).
(C) In the DRG, the expression of EYFP was not detected in sensory neurons, marked by TUBB3. White arrow marks the cell presented in higher magnification in the insertion (Scale bar, 100 μm, scale bar in the insertion 10 μm).
(D) In the DRG, the expression of EYFP was detected in satellite glial cells, marked by the satellite glial cell marker FABP7 (Avraham et al., 2020). White arrow marks the cell presented in higher magnification in the insertion (Scale bar, 100 μm, scale bar in the insertion 10 μm).
(E-G) Eight-week-old PLP-CreERT;ROSA26-eGFP-DTA mice and Cre-negative ROSA26-eGFP-DTA mice (used as control) were injected with tamoxifen and the sciatic nerve were subjected to electrophysiological examination at days 7–42 post tamoxifen injection (PID). (E) Nerve conduction velocity. (F) Distal latency. (G) Distal amplitude Values represent mean ± SEM.
(H) Clinical scores of the demyelinated mice. Clinical scores were based on a scoring system developed for phenotypic characterization of demyelination in this line (Traka, 2019; Traka et al., 2010, 2016). Values represent mean ± SEM.
(I-K) EM of the sciatic nerve at PID21: demyelinated large-caliber axons (black arrowheads), remyelinated large-caliber axons (white arrows), phagocytic macrophages (black arrows), apoptotic myelinating Schwann cells (J, white triangle), and Remak bundles that are not fully engulfed by basal lamina (J, black triangle). (K) Segmented demyelination: on the same axon, the left internode was absent (black arrowhead), while the right internode was present (white arrowhead) (Scale bars: panel I= 5 μm, panel K= 2 μm).
(L-N) At PID42, the large-caliber axons were remyelinated. (M and N) Remak bundle at PID42. Black trianle in M-Remak bundle that is not fully engulfed by basal lamina at PID42 (Scale bars: panel L= 5 μm, panel M- 1 μm, panel K= 2 μm).
(O) No axonal loss was observed, values represent mean ± SEM.
(P) About 10% of the high-caliber axons (diameter > 1 μm) were demyelinated at PID21, values represent mean ± SEM.
(Q) Quantification of phagocytic macrophages in the sciatic nerve, values represent mean ± SEM.
(R) Reduced myelin thickness at PID21 (red) and PID42 (green) compared with control (blue). For (A)-(F) PLP-CreERT;ROSA26-EYFP mice; n = 3.
(S) Myelin thickness, quantified results, values represent mean ± SEM, (**p < 0.01; unpaired Student’s t test).
(T-V) Semi-thin blue sections were harvested from the sciatic nerve at 21 and 42 days post tamoxifen injection (PID) and from control (no disease). (T) No disease. (U) PID21. (V) PID42 (scale bars in T-V, 100 μm).
(W and X) The total number of myelinated fibers was quantified from the semi-thin blue sections by the software HALO. (W) An example of the fibers that were counted (red). (X) Quantified results. (A-D) n = 3 PLP-CreERT;ROSA26-EYFP mice. (E-H) PLP-CreERT;ROSA26-eGFP-DTA mice; n = 14–8. For the Cre-negative ROSA26-eGFP-DTA mice; n = 9–5. (I-N) n = 3for no disease, n = 4for PID21, and n = 3for PID42. (T-W) n = 3for no disease, n = 3forPID21, and n = 3for PID42.
About 80% of the myelinated axons of the sciatic nerve are sensory axons (Schmalbruch, 1986). Based on this, we focused on demyelination of the sciatic nerve and on the transcriptional response of the sensory neurons in the lumbar DRGs (L3-L5) that project through the sciatic nerve (Rigaud et al., 2008). Our phenotypic characterization of the PLP-CreERT;ROSA26-eGFP-DTAline(herein DTA mice) revealed that, in DTAmice,the conduction velocity and the distal latency in the sciatic nerve is substantially reduced 21 days post tamoxifen administration (PID21), and full functional recovery is achieved at PID42 (Figures 1E and 1F). This was accompanied by reduction in the distal amplitude (Figure 1G), likely due to loss of motor unit function, caused by the loss of the terminal Schwann cells at the neuromuscular junctions, which was reported previously in a similar PLP-CreERT-driven diphtheria toxin mouse model (Hastings et al., 2020). Interestingly, the clinical score of these mice peaks at PID21, before the onset of the reported CNS pathology, and reduces at PID28, in parallel to the functional recovery in the PNS (Figure 1H). While complete functional recovery of the PNS is achieved at PID42 (Figure 1E), the clinical score dramatically increases at this time point (Figure 1H), due to widespread CNS demyelination.
Based on these electrophysiological results, we focused on the PID21 and PID42 time points, and assessed the morphology of the sciatic nerve by electron microscopy (EM). We found that, at PID21, the sciatic nerve is characterized by appearance of demyelinated and remyelinated large-caliber axons, aberrant morphology of Remak bundles, infiltration of phagocytic macrophages, and reduced myelin thickness (increased g-ratio) (Figures 1I–1S). In addition, we observed segmented demyelination in which, on the same axon, one internode was absent, due to loss of the myelinating Schwann cell, and the adjacent internode was still present (Figure 1K). This segmented demyelination suggests that, even for the axons that seem myelinated in cross section, the axons are likely demyelinated at one or more points along the long axon, which results in the observed severe drop in nerve conduction. This demyelination did not change the axon diameter distribution or the size of NEFH-positive sensory neurons’ cell somas (Figure S1). The DRGs at PID21 were characterized by a reduction in the expression of the satellite glial cell marker fatty acid binding protein 7 (FABP7) (Avraham et al., 2020) (Figure S1). At PID42, all large-caliber axons in the sciatic nerves were well myelinated (Figure 1L), but an aberrant morphology of Remak bundles that are not fully engulfed by basal lamina was still observed (Figure 1M and 2N). We did not detect a reduction in axonal density by EM (Figure 1O), nor did we detect a reduction in the total number of myelinated fibers in our samples (Figure 1T–2X), suggesting the lack of axonal loss. We also did not observe Bungner bands, which characterize loss of myelinated axons, or collagen pockets, which characterize loss of nonmyelinated fibers.
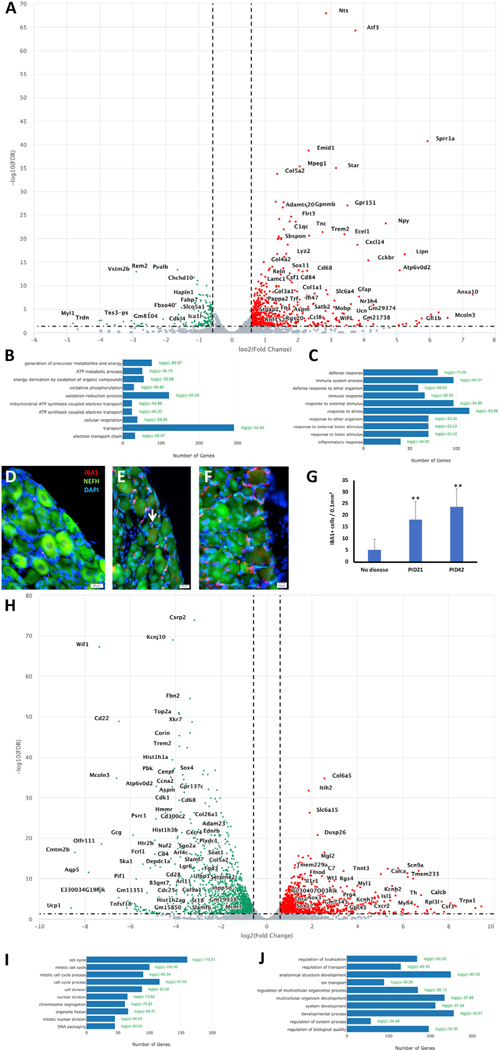
Figure 2. Glial cell loss induces the expression of regeneration-associated genes in DRG neurons.
Eight-week-old PLP-CreERT;ROSA26-eGFP-DTA mice and Cre-negative ROSA26-eGFP-DTA mice (used as control) were injected with tamoxifen, and L3-L5 DRGs were dissected for bulk RNA-seq at PID21.
(A) Volcano plot of the differentially expressed genes (DEGs) in the DRG at PID21. Green, downregulated genes; red, upregulated genes.
(B) Biological processes analysis of downregulated genes.
(C) Biological processes analysis of upregulated genes.
(D-G) Verification by immunohistochemistry (IHC). (D) Very few IBAI-positive cells in the control (no disease). (E and F) Infiltration of IBAI-positive cells into the DRG at PID21 (E), and PID42
(F).White arrow marks IBAI-positive cell (Scale bar in D and E = 20 μm, in F= 10 μm).(G) Quantified results, values represent mean ± SEM, (**p < 0.01; unpaired Student’s t test).
(H) Volcano plot of the DEGs in the sciatic nerve at PID21. Green, downregulated genes; red, upregulated genes.
(I) Biological processes analysis of downregulated genes.
(J) Biological processes analysis of upregulated genes. n = 2 biologically independent experiments (FDR ≤ 0.05, fold change ≥2). (D-G) n = 3 for no disease, n = 3 for PID21, and n = 3 for PID42.
Loss of glial cells in the PNS induces expression of regeneration-associate genes in lumbar DRG neurons
Based on the morphological changes in both myelinated fibers and Remak bundles in the sciatic nerve detected in the PLPDTA mouse, we hypothesized that the Schwann cell loss might result in changes in gene expression in the sensory neurons in the lumber DRGs that project through the sciatic nerve. Based on the electrophysiology and EM studies, we decided to focus our transcriptomic studies on PID21 and PID42, and harvested the sciatic nerve and the lumbar DRGs (L3-L5) for bulk RNA sequencing (RNA-seq). In the DRGs, we found that, at peak disease (PID21), expression of the known key injury-induced regeneration-associated genes (RAGs) Atf3, Sox11, and Sprr1a is induced (Figure 2A). In addition, transcripts involved in inflammation (such as Trem2, Stab1, and Tgfb1) were also induced (Figures 2A, 2C, and S2). We hypothesized that these changes might reflect infiltration of macrophages into the DRGs. To explore this possibility by immunohistochemistry, we used the macrophage marker IBA1. We found macrophage infiltration into the DRGs at peak disease (PID21) and in the recovery phase (PID42) (Figures 2D–2G), as suggest by the bulk RNA-seq of the DRGs at PID21 (Figure 2) and PID42 (Figure S2). Transcripts involved in oxidative phosphorylation and ATP production were downregulated in the DRGs (Figure 2B), suggesting that the cells in the DRGs suffer from mitochondrial damage. Nevertheless, this may also reflect death of DRG resident glial cells. In the sciatic nerve, transcripts involved in cell mitosis and cell division were downregulated at the peak of demyelination (Figures 2H and 2I), and transcripts involved in developmental processes were upregulated (Figure 2J). This probably reflects the effect of concurrent demyelination and remyelination observed at the peak of demyelination in the sciatic nerves (Figure 1). The induction of RAGs Atf3, Sox11, and Sprr1a was specific for the DRG and was not observed in bulk RNA-seq of the sciatic nerve, suggesting that the induction of RAGs is specific to the cells in the DRGs. To directly assess whether the induction of RAGs is specific to sensory neurons, we analyzed the transcriptome of DRG neurons directly by snRNA-seq.
Glial cells loss results in an injured transcriptional state in DRG neurons
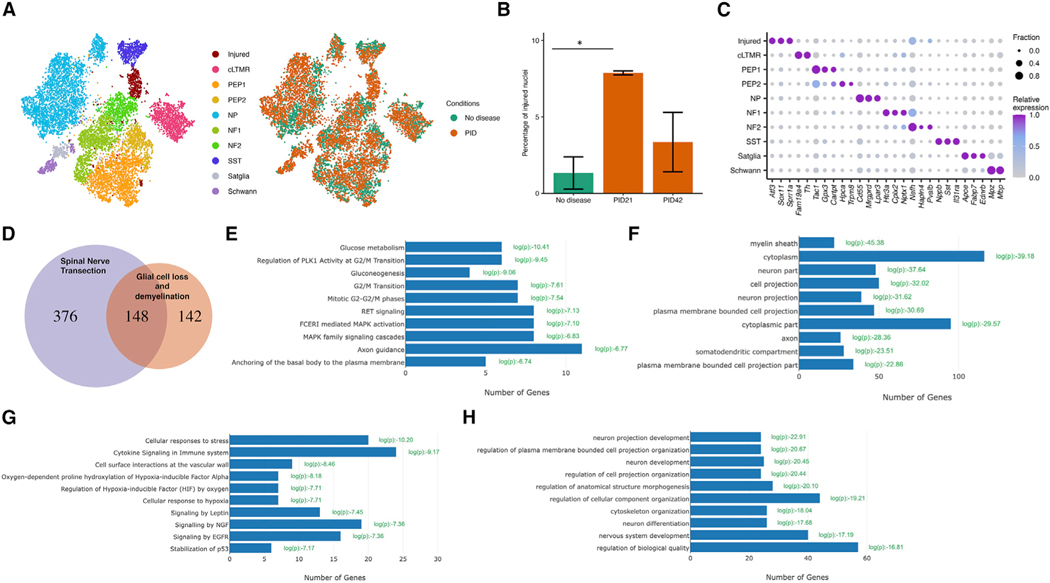
To characterize the transcriptional changes in sensory neurons in response to glial cell loss and demyelination at the single-cell level, we sequenced DRG nuclei from naive mice (no disease), from mice at the peak of demyelination (PID21), and from mice following PNS remyelination (PID42). For these studies, we harvested the cell bodies of the sensory neurons that project through the sciatic nerve (L3-L5 DRGs). Using a method that enriches for neuronal nuclei (Yang et al., 2021), we profiled 14,065 nuclei that passed quality control. The number of cells analyzed in each condition is provided in Table S2. To classify cell types, DRG nuclei from naive and demyelinated DRGs were initially clustered together based on their gene expression patterns. Dimensionality reduction (t-distributed stochastic neighbor embedding [tSNE]) revealed distinct clusters of cells, with neuronal clusters expressing Rbfox3, and non-neuronal clusters expressing Sparc (Figure S4). We identified nine naive cell types, including two clusters of glial cells (725 nuclei) and six different subtypes of sensory neurons (12,726 nuclei). We observed six naive DRG neuron subtypes: Fam19a4+/Th+ C-fiber LTMRs (cLTMR1), Tac1+/Gpx3+ peptidergic nociceptors (PEP1), Tac1+/Hpca+ peptidergic nociceptors (PEP2), Mrgprd+ non-peptidergic nociceptors (NP), Nefh+ A fibers including Aβ low-threshold mechanoreceptors (Aβ-LTMRs) (NF1) and Pvalb+ proprioceptors (NF2), and Sst+ pruriceptors (SST). We also observed non-neuronal cell types, including Apoe+ satellite glia and Mpz+ Schwann cells (Figure 3A; Table S1). Moreover, we identified an additional cluster of nuclei (614 nuclei, ~4% total) that are derived mostly from mice at the peak of PNS demyelination (86.8%). Nuclei in this cluster express high levels of the known peripheral nerve injury-induced genes Atf3, Sox11, and Sprr1a, such that they were defined as “injured” (Renthal et al., 2020) (Figure 3A). In line with previous studies (Nguyen et al., 2019; Renthal et al., 2020), we found that 1.3% of the nuclei at “no disease” exhibited an injured transcriptional state, which may represent routinely occurring fight wound injuries in group-housed mice. The percentage of the injured nuclei increased to 7.9% upon demyelination and was reduced to 3.4% upon remyelination (Figure 3B).
Figure 3. Induction of an injured transcriptional state in DRG neurons after glial cell loss.
(A) Eight-week-oldPLP-CreERT;ROSA26-eGFP-DTA mice and Cre-negative ROSA26-eGFP-DTA mice (used as control) were injected with tamoxifen, and L3-L5 DRGs were dissected for snRNA-seq studies at PID21 and PID42. tSNE plot of 14,065 nuclei from Cre-negative ROSA26-eGFP-DTA mice (no disease) and PLP-CreERT;ROSA26-eGFP-DTA mice at PID21 and PID42. Nine naive cell types, including glial cells and different subtypes of sensory neurons, and one cluster of injured nuclei were identified. Nuclei are colored by cell type (left) or condition (right).
(B) Bar plot showing average percentage of nuclei that are in the injured cluster across duplicates in each condition (n = 2). Error bars denote standard deviations. Two-way ANOVA was conducted to compare the percentages across conditions. F(2, 3) = 13.74, p = 0.03. PID21 shows significant increase in injured nuclei compared with no disease control (Bonferroni post hoc, p = 0.03).
(C) Dot plot displaying the relative gene expression of marker genes (columns) in the injured cluster and each naive cell type (rows). The fraction of nuclei expressing a marker gene is calculated as the number of nuclei in each cell type that express a gene (>0 counts) divided by the total number of nuclei in the respective cell type. Relative expression in each cell type is calculated as mean expression of a gene relative to the highest mean expression of that gene across all cell types.
(D) Overlap between the common neuronal injury-induced genes in SpNT (see STAR Methods) and marker genes in the injured cluster (log2FC > 0.25, p < 0.05, comparing nuclei from the injured cluster with all other nuclei) (hypergeometric test, p = 2e-102).
(E) Reactome pathway analysis of the genes induced only by demyelination.
(F) Cellular component analysis of the genes induced only by demyelination.
(G) Reactome pathway analysis of the genes induced only by SpNT.
(H) Reactome pathway analysis of the genes induced by both SpNT and demyelination.
The DRG cell types identified here express distinctive patterns of ion channels, G protein-coupled receptors (GPCRs), neuropeptides, and transcription factors that are consistent with previous studies (Figure 3C) (Renthal et al., 2020; Sharma et al., 2020; Usoskin et al., 2015; Zeisel et al., 2018; Zheng et al., 2019). Comparison between the neuronal injury-induced genes in spinal nerve transection (Renthal et al., 2020) and the marker genes in the injured cluster observed here upon demyelination revealed that the two injured transcriptional states are very similar (Figure 3D). Ontological enrichment analyses of the transcripts induced only by demyelination and not by mechanical injury revealed that demyelination induced in neurons the expression of transcripts related to glucose metabolism, such as malate dehydrogenase 1 (Mdh1) (Figure 3E), perhaps reflecting loss of metabolic support from the lost glial cells. Surprisingly, cellular component analysis revealed that demyelination induced in injured NF2 neurons the expression of transcripts related to cellular component myelin sheath, such as spectrin alpha (Sptanl) and carbonic anhydrase 2 (Car2) (Figure 3F). Network analysis of the transcripts induced specifically by demyelination revealed that the myelin sheath component transcripts compose the center of the network of the transcripts induced specifically by demyelination (Figure S3). Ontological enrichment analyses of the transcripts induced only by mechanical injury revealed enrichment for categories related to cell stress and hypoxia, such as hypoxia inducible factor 1 subunit alpha (Hifla) (Figure 3G), without enrichment for a specific cellular component (Figure S3). The transcripts that were induced by both demyelination and mechanical injury were enriched for categories related to neuron projections and development, such as adenylate cyclase activating polypeptide 1 (Adcyapl) (Figure 3I), with enrichment for cellular components related to neuron and neuron projection (Figure S3). The lists of the differentially expressed transcripts induced by glial cell loss, by spinal nerve transection, or by both is provided in Table S3. Gene set motif enrichment analysis revealed that the genes induced specifically by demyelination were enriched for motifs of the transcription factors Atf3 and Klf6, similar to the genes induced only by spinal nerve transection (SpNT), which were enriched for motifs of the transcription factors Atf3, Klf6, Jun, and Sox11 (Figure S3).
Glial cell loss induces transcriptional reprograming preferentially in proprioceptive and Aβ rapidly adapting low-threshold-mechanoreceptor neurons
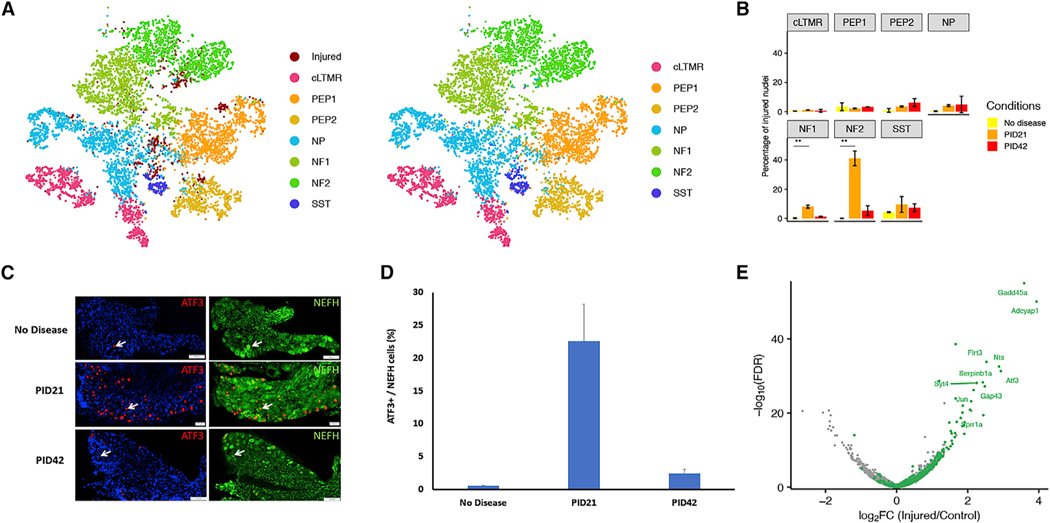
The sensory neurons in the DRGs represent a heterogeneous population of neurons that vary in their degree of myelination. We therefore hypothesized that, unlike mechanical injury, demyelination may have a specific impact preferentially on heavily myelinated sensory neurons (A fibers) and little to no effect on non-myelinated sensory neurons (C fibers). To test this hypothesis, we investigated whether the gene expression program induced by demyelination differs between distinct DRG neuronal subtypes. To classify the cell types of nuclei in the injured cluster whose cell-type-specific marker genes are downregulated, we both co-clustered the injured nuclei identified in our study to those previously classified after SpNT (Renthal et al., 2020) (Figure 4A) and anchored the injured nuclei to the naive nuclei in our study whose cell identity has been identified previously. Cell type assignments from the two approaches had 85.7% concordance (Figure S3). Consistent with prior observations (Nguyen et al., 2019; Renthal et al., 2020), a small fraction of the NF1 and NF2 cells (0.18% and 0%, respectively) display an injured transcriptional state in non-diseased mice, but 8.1% of the cells in the NF1 cluster and 40.1% of those in the NF2 cluster exhibited the injured state at the peak of demyelination (PID21). The percentage of injured NF1 and NF2 nuclei declines to 1.3% and 5.3% at the remyelination (PID42), respectively (Figure 4B). These data suggest that the transcriptional response to demyelination among DRG neurons is preferentially induced in the neurons that compose clusters NF1 and NF2, which are the heavily myelinated A fibers including Nefh+ Aβ LTMRs and Pvalb+ proprioceptors. To validate these results by immunohistochemistry, we used the marker neurofilament heavy polypeptide (NEFH) that marks Aβ-LTMRs and proprioceptors neurons in the DRG (Renthal et al., 2020; Usoskin et al., 2015). In line with the snRNA-seq results, the expression of ATF3 is strongly induced at the time of peak disease (PID21), specifically in large-diameter A fibers marked by NEFH (Figures 4B and 4C). Similar induction was observed also for the protein encoded by the injury-induced transcript Nts (Table S3). To confirm the temporal expression of the injury-induced transcripts observed by snRNA-seq, we analyzed bulk-RNA-seq derived from DRGs at PID42. This independently confirmed the upregulation of injury-induced transcripts at peak disease compared with naive, and their normalization upon recovery (Figure S2). Analysis of the differentially expressed genes in the NF2 cluster upon demyelination revealed that the transcriptional changes, which are characterized by reduced expression of transcripts that are important for neuronal cell identity and enhanced expression of injury-induced transcripts, are very similar to transcriptional changes that occur in sensory neurons upon mechanical transection or crush injury (Renthal et al., 2020) (Figure 4E).
Figure 4. Glial cell loss induced transcriptional reprograming selectively in proprioceptive and A² rapidly adapting low-threshold-mechanoreceptor Neurons.
(A) Classification of injured DRG nuclei after demyelination by co-clustering with injured nuclei after SpNT. Nuclei after demyelination are in dark red, and nuclei after SpNT are colored by their respective cell types (left). Nuclei after demyelination were annotated by projecting the SpNT neuronal subtype classification onto the cluster. All nuclei are colored by their respective cell types (right).
(B) Bar plot showing average percentage of nuclei that are in the injured cluster for each subtype in each condition. Error bar shows the standard deviation across biological replicates. ANOVA was conducted to compare the average percentage of injured nuclei across conditions in each subtype. NF1: F(2, 3) = 87.68, p = 0.002. NF2: F(2, 3) = 81.53, p = 0.002. PID21 shows an increase in percentage of injured nuclei compared with no disease in both NF1 and NF2 (Bonferroni post hoc, p < 0.01).
(C and D) Verification of the results by IHC. (C) The expression of ATF3 is induced at peak disease (PID21) specifically in Aβ-LTMR and proprioceptor neurons, marked by NEFH. (D) Quantitative analysis, values represent mean ± SEM.
(E) Volcano plot displays DEGs between injured NF2 nuclei from PID21 and nuclei from no disease. Common injury-induced genes in SpNT are colored green. Top 10 most differentially expressed genes that are also common injury-induced genes in SpNT are labeled. (C–D) n = 3 for no disease, n = 3 for PID21, and n = 3 for PID42.
DISCUSSION
Mechanical injury of peripheral nerve axons leads to Wallerian degeneration followed by a regrowth of axons toward their targets and their reinnervation. This process is tightly regulated by a cascade of transcriptional events that result in the conversion of mature sensory neurons into actively growing cells (Abe and Cavalli, 2008; Chandran et al., 2016). In this study, we used a genetic model of glial cell ablation to study the effect of PNS demyelination and satellite glial cell loss on the transcriptome of sensory neurons, without the confounding effect of neuronal/axonal injury. We found that ablation of peripheral glial cells results in a transient PNS demyelination, in which about 10% of large-caliber (>1 μm) axons are demyelinated, as observed in similar models of induced Schwann cell loss (Gerber et al., 2019). We observed that this glial cell loss and peripheral demyelination results in transcriptional changes in a distinct set of sensory neurons, including the induction of RAGs and the downregulation of genes that define transcriptional identity. We found that this transcriptional change is transient and reversible, as the transcriptional states of injured neuronal nuclei return to their naive states within weeks (Figures 4 and S2), coinciding with remyelination (Figure 1).
Previous studies revealed a core transcriptional reprograming in sensory neurons in response to various peripheral nerve mechanical injuries (Renthal et al., 2020). We found a very similar transcriptional response in sensory neurons upon glial cell loss and demyelination (Figures 3 and 4). The similar transcriptional changes in both conditions suggests that this pattern of reprograming may represent a non-cell-autonomous transcriptional response of sensory neurons to glial cell loss and demyelination rather than simply being a reaction to axonal damage. A similar phenomenon was recently observed following optic nerve injury, where the response of retinal ganglion cells to optic nerve injury turns out to be non-cell autonomous, dictated by amacrine cells (Sergeeva et al., 2021; Zhang et al., 2019). Nevertheless, while mechanical axonal injury produces transcriptional changes in effectively all the injured sensory neurons, our studies here reveal a neuronal subtype vulnerability to PNS demyelination of Aβ rapidly adapting low-threshold mechanoreceptors (RA-LTMR) and proprioceptor neurons, which have the highest axon caliber and thickest myelin, suggesting that the transcriptional reprograming in these neurons can be evoked by demyelination and satellite glial cell loss without direct physical axonal injury. Interestingly, despite the marked aberrant phenotype in nonmyelinating Schwann cells following Schwann cell ablation (Figure 1), we did not observe substantial changes in the transcriptome of C-fiber neurons (Figure 4). A similar neuronal specificity is present in Charcot-Marie-Tooth disease type 1A (CMT1A) patients suffering from PNS demyelination. In these patients, despite the presence of abnormal non-myelinating Schwann cells, no effect on the unmyelinated axons was noted (Koike et al., 2007).
Detailed transcriptional studies revealed that demyelination and remyelination are characterized by vast transcriptional changes in myelinating glia. Nevertheless, the transcriptional changes in the specific neurons that are demyelinated or remyelinated are less well characterized. Our cellular component analysis revealed that glial cell loss and demyelination induces in injured Aβ RA-LTMR and proprioceptor neurons the expression of transcripts related to the myelin sheath, such as spectrin alpha (Sptan1) and carbonic anhydrase 2 (Car2) (Figure 3F). Previous studies found that Sptan1 has a cell-autonomous role in large-diameter myelinated sensory neurons, where it mediates the assembly of nodes of Ranvier (Huang et al., 2017). Car2 is expressed at early developmental stages by neurons and glia (Cammer and Tansey, 1987), and Car2 is one of the proteins that comprise the PNS myelin proteome (Siems et al., 2020). In addition, although not one of the 10 most significantly enriched categories, the cellular component node of Ranvier (Gene Ontology [GO]: 0033268) was also enriched in the transcripts induced specifically by glial cell loss and demyelination (FDR 0.0083) (data not shown). The induced expression of transcripts related to the myelin sheath and node of Ranvier in neurons specifically upon glial cell loss and demyelination may suggest that demyelinated neurons are actively participating in the remyelination process by activating the expression of transcripts that mediate myelin sheath and node of Ranvier formation.
The transcriptional control of neuronal responses to glial cell loss and demyelination, described here, warrants further investigation. Previous studies found that transcription factors Sox11, Atf3, Jun, Bhlhe41, Klf6, Stat2, Klf7, Smad1, and Ets2 are induced by mechanical injury, and gene set motif enrichment analysis revealed that their binding motifs are enriched in the genes induced after mechanical injury (Renthal et al., 2020). Similar to SpNT, glial cell loss and demyelination induced the expression of the transcription factors Klf6, Atf3, Smad1, Jun Zfp367, and Sox11 (see Table S3). Nevertheless, glial cell loss and demyelination also induced the expression of the transcription factors Arnt2, Tsc22d1, and Pbx1, which were not induced by SpNT. Gene set motif enrichment analysis revealed that the genes induced by glial cell loss and demyelination are enriched for motifs of Atf3 and Klf6, but not for motifs of Jun and Sox11, that were enriched in the transcripts induced only by SpNT (Figure S3). Further investigation is required to determine the relative role of these transcription factors in axonal regeneration after SpNT and the response to glial cell loss and demyelination.
Peripheral axonal injury is a clear driver of the transcriptional reprogramming in injured sensory neurons (Cheng et al., 2021; Cho et al., 2013). The results here show that, for neurons with large, myelinated axons, these transcriptional changes can be induced by demyelination and satellite glial cell loss without axonal injury. Further studies are required to determine the mechanism(s) by which Schwann cells and satellite glial cells dictate the transcriptional state of sensory neurons. Such mechanisms may include an injury signal from Schwann cells to axons provoked by demyelination, such as the release of the toxic metabolite acylcarnitines from Schwann cells (Viader et al., 2013) with demyelination-induced mitochondrial dysfunction (Della-Flora Nunes et al., 2021), or signals from myelinating Schwann cells to axons that continuously prevent injury-induced transcription reprograming. Another mechanism by which Schwann cells affect axons is the metabolic support from the myelinating cells to axons (Beirowski et al., 2014; Fünfschilling et al., 2012; Lee et al., 2012). In line with the different effects of demyelination on specific sensory neurons described here, recent studies revealed that this metabolic support is differentially important for specific neurons (Jha et al., 2020). The ontological enrichment of categories related to glucose metabolism in the transcripts induced specifically by demyelination (Figure 3E) further supports this idea. In addition, satellite glial cell ablation may result in loss of metabolic support or other neuronal survival factors such as neurotrophic factors. The axon survival factor nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) catalyzes the synthesis of nicotinamide adenine dinucleotide (NAD+) from nicotinamide mononucleotide (NMN) (Gilley and Coleman, 2010). Axonal injury induces the loss of the axonal NMNAT2, resulting in an impaired NMN to NAD+ ratio, which in turn activates the constitutively expressed protein Sterile alpha and Toll/interleukin-1 receptor motif-containing 1 (SARM1) (Figley et al., 2021), resulting in axonal destruction (Coleman and Hoke, 2020; Figley and DiAntonio, 2020). Interestingly, maintenance of the mature, myelinating state of Schwann cells also depends on NAD+ homeostasis (Sasaki et al., 2018). Nevertheless, a direct link between Schwann cell and axonal factors of survival or destruction has yet to be established.
In peripheral nerve injury, the interaction between Schwann cells and axons proximal to the injury site presumably persist. This implies that other cell-autonomous mechanisms in the axons, unrelated to axon-glia interactions, may trigger injury-induced transcriptional changes. This possibility is supported by the ontological enrichment of categories related to stress and hypoxia in the transcripts induced specifically by mechanical injury (Figure 3G), and by the observation that enrichment for the myelin sheath compartment was found only in glial cell loss and demyelination, and not in mechanical injury. This possibility is further supported by previous studies that suggested that mechanical-injury-induced transcriptional reprograming is a cell-autonomous property of sensory neurons, activated by injury-induced calcium influx into the axoplasm and nuclear export of histone deacetylase 5 (HDAC5) (Cho et al., 2013) and by DNA breaks (Cheng et al., 2021).
The concept that glial cell loss and demyelination are the cause for the observed transcriptional changes is supported by previous ethidium bromide-induced demyelination studies (Hollis et al., 2015) that demonstrated similar transcriptional changes in sensory neurons. The ethidium bromide that was used to induce demyelination in these studies, however, is not cell type specific. Moreover, the neuronal subtype of the injured sensory neurons was not resolved in this study (Hollis et al., 2015). Interestingly, ethidium bromide-induced demyelination did not activate macrophages at the demyelinated sciatic nerve site (Hollis et al., 2015), whereas the glial cell loss described here resulted in the infiltration of macrophages to the demyelinated nerve (Figure 1). In the DRG, we observed macrophage infiltration and immune cell activation at both the peak of disease and during recovery (Figures 2 and S2), when the expression of the injury-induced transcripts is downregulated and neuronal cell identity is restored. These data suggest that the presence of infiltrating macrophages in the DRG does not have a direct impact on neuronal cell identity. Nevertheless, macrophages have been shown to be important for sensory neuron regeneration (Kwon et al., 2013). It is possible that the macrophages that we observed in the sciatic nerve (Figure 1) and in the DRG (Figure 2) provide metabolic support to the demyelinated axons (Jha et al., 2021).
We found that glial cell loss and demyelination does not cause overt widespread axonal loss (Figures 1 and S1), which is consistent with a previous study that used the lysolecithin model to induce PNS demyelination (Wallace et al., 2003). Although the authors of the lysolecithin model found that PNS demyelination mainly affects A fibers, they did not observe induction of ATF3 in the DRG upon PNS demyelination. This might be the result of the inherent differences between the focal demyelination caused by lysolecithin and the widespread demyelination caused by genetic ablation of Schwann cells and may also be attributed to the difference in lumber DRGs analyzed. Using the same PLP-CreERT line used here (Doerflinger et al., 2003), a previous study demonstrated that terminal Schwann cells at the neuromuscular junctions are ablated in a similar diphtheria toxin mouse model (Hastings et al., 2020). Consistent with our results, terminal Schwann cell ablation does not cause motor axon degeneration (Hastings et al., 2020). The reduced compound muscle action potential that we observed at the peak of the disease in our model is likely due to a similar loss of terminal Schwann cells and motor unit function.
Our study has implications for the ongoing effort to identify therapeutic targets to mitigate PNS neuropathies caused by demyelination, such as CMT1A. Currently, the only available therapy for CMT1A is the experimental drug PXT3003, which is a combination of baclofen, naltrexone, and sorbitol (Attarian et al., 2021). Surprisingly, although CMT1A is caused by demyelination, the beneficial effects of PXT3003 are mainly achieved by targeting axons (Prukop et al., 2020). While the current efforts to identify therapies for PNS demyelination are focused on Schwann cells, aiming to enhance PNS myelination (Fledrich et al., 2018; Zhao et al., 2018), our study suggests that sensory neurons might also be a therapeutic target in demyelinating diseases and that small molecules that target the injury response of sensory neurons may be beneficial for PNS demyelination.
Limitations of the study
Our data suggest that the PLP-CreERT line that was used here (Doerflinger et al., 2003) mediates recombination in oligodendrocytes (Traka et al., 2010), Schwann cells, and satellite glial cells (Figure 1), such that the generation of a mouse model would be required to separate the loss of satellite glial cells from the loss of Schwann cells, and to separate CNS versus PNS demyelination. In addition, since we were interested here mainly in the transcriptional response of neurons, we used for our snRNA-seq studies a protocol that enriches for neuronal nuclei (Yang et al., 2021). Additional sequencing would be required to directly study the satellite glial response in this model. Lastly, we are unable to functionally characterize the DRG transcriptional response observed here, because the DTA model used is dependent on Cre-mediated recombination, such that the evaluation of conditional alleles of differentially expressed genes is not currently possible.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Brian Popko (brian.popko@northwestern.edu).
Materials availability
The PLP-CreERt-ROSA26-eGFP-DTA mouse model used in this study is available from Brian Popko (brian.popko@northwestern.edu) upon request.
Data and code availability
Raw and processed data have been deposited at the Gene Expression Omnibus (GEO) repository and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources table. This paper also analyzes our previously published data (Renthal et al., 2020). This existing data is publicly available, and the accession number is listed in the Key Resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit anti-ATF3 | Sigma-Aldrich | Cat#HPA001562; RRID:AB_1078233 |
| Chicken anti-Neurofilament, H | Millipore | Cat#AB5539; RRID:AB_11212161 |
| Rabbit anti Neurotensin | ImmunoStar | Cat# 20072; RRID:AB_572254 |
| Rabbit Anti-Iba1 | Wako Pure Chemical | Cat # 019–19741; RRID:AB_839504 |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA-seq data | This paper | GEO: GSE205461 |
| snRNAseq data | This paper | GEO: GSE205561 |
| snRNAseq data of DRG following traumatic | Renthal et al. (2020) | GEO: GSE154659 |
| injury | ||
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: ROSA26-eGFP-DTA line. | The Jackson Laboratory | Cat#JAX:032087; RRID:IMSR_JAX:032087 |
| Mouse: PLP-CreERt line | The Jackson Laboratory | Cat# JAX: 005975; RRID:IMSR_JAX:005975 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | National Institutes of Health | RRID: SCR_003070 |
| Adobe Illustrator CC | Adobe Systems | RRID: SCR_010279 |
| Indrops mapping | (Klein et al., 2015) | https://github.com/indrops/indrops |
| R for statistical computing- 3.5.0 | R Core Team (2013) | https://www.r-project.org/ |
| Seurat - 2.3.4 | Satija et al., 2015 | https://satijalab.org/seurat/ |
| SCENIC −1.1.1–9 | Aibar et al. (2017) | https://aertslab.org/#scenic |
| edgeR - 3.24.3 | Robinson et al. (2010) | http://bioinf.wehi.edu.au/edgeR/ |
| ggplot2 – 3.2.0 | Wickham, 2016 | https://ggplot2.tidyverse.org/ |
| gplots - 3.0.1.1 | Warnes et al., 2009 | https://github.com/talgalili/gplots |
| topGO | Alexa and Rahnenfuhrer (2016) | https://bioconductor.org/packages/release/bioc/html/topGO.html |
| R for statistical computing- 3.5.0 | R Core Team (2013) | https://www.r-project.org/ |
| HALO | Indica Labs Inc. | RRID:SCR_018350 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animals used in this study were housed under pathogen-free conditions at controlled temperatures and relative humidity with a 12/12 h light/dark cycle and free access to pelleted food and water. All animal experiments were conducted in accordance with ARRIVE guidelines and in complete compliance with the guidelines of Northwestern University Institutional Animal Care and Use Committee (IACUC). All mice lines were on the C57BL/6 background, and both female and male mice were used. Six to eight weeks old mice were used for tamoxifen injections.
Plp-CreERt;ROSA26-eGFP-DTA mice and Cre negative ROSA26-eGFP-DTA mice (used as control) were generated by crossing the ROSA26-eGFP-DT-A mice (Cat#JAX:032087; RRID:IMSR_JAX:032087) with the PLP-CreERt mice (Doerflinger et al., 2003) (Cat# JAX: 005975; RRID:IMSR_JAX:005975). Plp-CreERt;ROSA26-EYFP mice and Cre negative ROSA26-EYFP mice (used as control) were generated by crossing the ROSA26-EYP mice (Srinivas et al., 2001) (Cat# JAX_006148,RRID:IMSR_JAX:006148) with the PLP-CreERt mice (Doerflinger et al., 2003) (Cat# JAX: 005975; RRID:IMSR_JAX:005975).
METHOD DETAILS
Dissection procedures
Tamoxifen treated mice were euthanized by CO2 asphyxiation and decapitation. Lumbar L3-L5 ganglia were collected at various 21 days and 42 days post injection. Ganglia from 3–5 mice per sample were immediately frozen on dry ice, then pooled for subsequent snRNA-seq profiling or histology. There were 2–3 biological replicates of each pooled condition.
Tamoxifen injections
0.8 mg/day 4-hydroxytamoxifen (H-6278, Sigma) was injected for 3 consecutive days for females, and for 4 consecutive days for males, based on protocol developed for the Plp-CreERt;ROSA26-eGFP-DTA mice (Traka, 2019). 1 mg/day 4-hydroxytamoxifen (H-6278, Sigma) was injected for 5 consecutive days for Plp-CreERt;ROSA26-EYFP mice (Traka, 2019).
Single-nuclei isolation from mouse DRG
Single-nuclei suspensions of lumbar DRGs from treated mice were collected using a recently described protocol (Yang et al., 2021). This method increases the isolation of neuronal nuclei compared to commonly used nuclear extraction protocols (Mo et al., 2015; Renthal et al., 2018). Briefly, DRGs were removed from dry ice and placed into homogenization buffer (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM tricine-KOH, pH 7.8, 1 mM DTT, 5 μg/mL actinomycin). After a brief incubation on ice, the samples were briefly homogenized using a Tissue-Tearor and transferred to a Dounce homogenizer for an additional ten strokes with a tight pestle in a total volume of 5 mL homogenization buffer. After ten strokes with a tight pestle, a 5% IGEPAL (Sigma) solution was added to a final concentration of 0.32% and five additional strokes with the tight pestle were formed. The tissue homogenate was then passed through a 20-mm filter, and diluted 1:1 with OptiPrep (Sigma) and layered onto an OptiPrep gradient. After ultracentrifugation, nuclei were collected between the 30–40% OptiPrep layers. This layer contains DRG nuclei as well as some membrane fragments likely from Schwann cells that have the same density as nuclei. We diluted this layer in 30% OptiPrep to a final concentration of 80,000 – 90,000 nuclei /mL for loading into the inDrops microfluidic device. All buffers and gradient solutions for nuclei extraction contained 0.01U/ul RNase inhibitor (Promega) and 0.04% BSA.
Single-nucleus RNA sequencing (inDrops)
Single-nuclei suspensions were encapsulated into droplets and the RNA in each droplet was reverse transcribed using a unique oligonucleotide barcode for each nucleus as described previously (Renthal et al., 2020). Nuclei encapsulation was performed in a blinded fashion and the order of sample processing was randomized. The total number of droplets collected per sample varied based on available reagents and line integrity. After encapsulation, each sample was divided into pools of approximately 3,000 droplets and library preparation was performed on each pool of droplets. Libraries were sequenced on an Illumina Nextseq 500 to a depth of 500 million reads per ~30,000 droplets collected, resulting in at least 5 reads per UMI on average per sample. Sequencing data was processed and mapped to the mouse genome using the pipeline described in https://github.com/indrops/indrops (Klein et al., 2015). Counts tables from each library were then combined and processed as described below. The metrics for all sequencing samples is presented in Figure S4.
Initial quality control, clustering and visualization of snRNA-seq
To be included for analysis, nuclei were required to contain counts of greater than 500 unique genes, fewer than 15,000 total UMI, and fewer than 5% of the counts deriving from mitochondrial genes. We used the Seurat package (version 2.3.4) in R (R Core Team, 2013) to perform clustering of these nuclei as previously described (Satija et al., 2015). Raw counts were scaled to 10,000 transcripts per nucleus to control the sequencing depth between nuclei. Counts were centered and scaled for each gene. The effects of total UMI and percent of mitochondrial genes in each nucleus were regressed out using a linear model in the Scaledata() function. Highly variable genes were identified using the MeanVarPlot() with default settings. The top 20 principal components were retrieved with the RunPCA() function using default parameters. Nuclei clustering was performed using FindClusters() based on the top 20 principal components, with resolution at 1 for the initial clustering of all nuclei and the sub-clustering of non-neuronal nuclei except where otherwise specified. For dimension reduction and visualization, tSNE coordinates were calculated in the PCA space by using the implemented function runTSNE() in Seurat.
Doublet identification and classification of DRG subtypes
Marker genes for each cluster were identified by running FindAllMarkers() in Seurat comparing nuclei in one cluster to all other nuclei. Doublet or low-quality clusters were identified as clusters which either are significantly enriched for at least two mitochondrial genes (Log2FC >0.5, FDR < 0.05), or have no significantly enriched cluster marker genes (FDR < 0.05, log2FC > 1) other than Rgs11. Nuclei in those clusters were excluded from the dataset. The remaining nuclei were clustered again, and marker gene analysis was run as described above. Significant enrichment (FDR <0.05, log2FC >0.5) of known subtype marker genes (peptidergic nociceptors (PEP) = Tac1, non-peptidergic nociceptors (NP) = Cd55, pruriceptors (SST) = Sst, cLTMR = Fam19a4, A-LTMR (NF) = Nefh, Satglia = Apoe, Schwann cells = Mpz) within a cluster of nuclei compared to all other nuclei was used to assign the subtype to each of the naive cluster. In our samples Cadps2+ Aδ-LTMRs nuclei were rare and were not classified as a separate cluster.
Identification of injured clusters and subtype classification of of injured nuclei
Clusters of cells that are in a transcriptionally injured state were identified as the clusters that are significantly enriched of Atf3 (FDR <0.05, log2FC> 1) compared to all other clusters. To identify the subtypes for injured nuclei, we took two different approaches. First, All injured nuclei from the demyelination model were co-clustered with injured nuclei from spinal nerve transection (SpNT) as described previously (Renthal et al., 2020), and clusters were identified. Having classified SpNT nuclei from the previous study, we were able to project those neuronal subtypes onto the injured state nuclei from the demyelination model. For each cluster, the subtype pf the most abundant previously-classified injured SpNT was projected onto all unclassified nuclei in that cluster (95 ± 3% of previously-classified injured SpNT nuclei in each cluster share the same subtype label). Cell type labels classified using this approach were used for all analysis unless otherwise specified. As an independent approach, we also anchored all injured nuclei from the demyelination model to all other nuclei that are in a transcriptionally uninjured state from the same model. FindTransferAnchors(reduction = “cca”) in Seurat was then used to identify anchors between injured and mouse data. TransferData() was used to transfer uninjured subtype labels to each nucleus in the injured state.
Differential expression analysis
To identify genes that are differentially expressed in injured NF2 nuclei, differential expression analysis was done with edgeR (version 3.24.3) as we recently described (Renthal et al., 2020). Briefly, edgeR uses the raw counts as input, and genes detected in fewer than 5% of nuclei selected for each comparison were excluded from analysis. Counts within each nucleus were normalized by the trimmed mean of M-values (TMM) method to adjust for total RNA differences between nuclei. Dispersion was estimated by fitting a quasi-likelihood negative binomial generalized log-linear model (glmQLFit) with the conditions being analyzed. The QL F-test was used to determine statistical significance between differentially expressed genes in the experimental and control group. The experimental group was identified as 166 injured NF2 nuclei from PID21 samples. The control group was selected by randomly sampling the same number of NF2 nuclei from no disease samples. Sampling of control group and differential expression analysis was repeated ten times and median Log2FC and FDR for each gene was reported.
Gene set motif enrichment analysis
To identify motifs that are significantly enriched in a gene set, motif enrichment analysis was run using the SEA (“Simple Enrichment Analysis”) algorithm (Bailey and Grant, 2021), in the MEME Suite of sequence analysis tools. Motif analysis was performed for 400 bp upstream and 100 bp downstream of the transcription start site of each gene. Enrichment for the transcription factors that are induced by glial cell loss and demyelination (Jun, Sox11, Atf3, Smad1, Klf6, Pbx1 and Arnt2) was determined. Motif enrichment analysis for Tsc22d1 and Zfp367 was not performed due to lack of motif data. Enrichment was determent against control sequences, composed of the same sequences shuffled by the algorithm. The relative enrichment ratio of the motif in the primary vs. control sequences, defined as Ratio = ((TP+1)/(NPOS+1)) / ((FP+1)/(NNEG+1)), where NPOS is the number of primary sequences in the input, and NNEG is the number of control sequences in the input, TP is the number of control sequences matching the motif, and FP is the number of control sequences matching the motif.
Toluidine blue stained section and electron microscopy (EM) analysis
Morphometric analysis was performed essentially as we described previously (Elbaz et al., 2016; Traka et al., 2013; Xu et al., 2020). In brief, mice were perfused with saline, followed by perfusion with a solution of 2.5% glutaraldehyde and 4% paraformaldehyde in a 0.1 M sodium cacodylate buffer. Sciatic nerves were harvested and post-fixed in the same fixative at 4°C for 2 weeks and sciatic nerve samples were embedded in epoxy resin. Semithin section were stain with toluidine blue and observed on a slide scanner microscope. The total number of myelinated fibers was quantified from the toluidine blue sections using the image analysis software HALO (Indica Labs Inc). Ultrathin sections were double stained with uranyl acetate and lead citrate and observed on a FEI Tecnai F30 scanning transmission electron microscope. G-ratios were calculated from the EM images, as described in (Auer, 1994).
Total RNA-seq
RNA-seq was performed essentially as we described previously (Aaker et al., 2016; Elbaz et al., 2018; Xu et al., 2020). In brief, RNA samples were prepared and isolated from sciatic nerve or DRG using RNA isolation kit following the manufacture’s protocol (Bio-Rad, Cat# 732–6820). RNA quality was confirmed by 2100 Bioanalyzer using a model 6000 Nano kit (Agilent technologies, Cat# 5067–1511). Samples with RNA integrity number >8 were used. Libraries were prepared and sequenced at the University of Chicago Genomics Core facility using the Illumina HiSeq 4000. Reads were mapped using both STAR v2.6.1a and Kallisto v.0.44.0 using bowtie 2 aligner (Bray et al., 2016; Dobin et al., 2013). Mapped reads were further analyzed with the Bioconductor suite v3.7 (Huber et al., 2015) by the University of Illinois at Chicago Bioinformatics Core facility. Q-values were determined as false discovery rate adjusted p values using the method previously described (Benjamini and Yekutieli, 2005).
Electrophysiology
Electrophysiology was performed with a Nicolet Viking Quest Laptop System (VikQuest Port 4ch-7) from Nicolet Biomedical (Madison, WI), as previously described (Elbaz et al., 2016). Briefly, recording needle electrodes were placed subcutaneously in the footpad. Supramaximal stimulation of sciatic nerves was performed with a 0.1–0.2 ms pulse, stimulating distally at the ankles and proximally at the sciatic notch with needle electrodes. Latencies, conduction velocities and amplitudes of compound muscle action potentials were measured. Results from stimulation of bilateral sciatic nerves were averaged for each animal, with n representing the number of animals in each group.
Immunohistochemistry
L3-L5 DRGs were harvested from treated mice perfused with cold PBS followed by cold 4% PFA. The perfused tissues were fixed in 4% PFA for 1hrat 4°C and cryoprotected with 30% sucrose in PBS overnight. DRGs were sectioned into 12 μm sections, which were blocked and permeabilized with 1% Triton X-100 in blocking buffer for 1hr at 25°C. Sections were incubated with rabbit polyclonal antibody against ATF3 ([1:1000]; Santa Cruz Biotech; Cat#sc-188; RRID:AB_2258513) and chicken polyclonal antibody against NF200 ([1:2000]; Millipore; AB5539; RRID:AB_11212161) at 4°C overnight and then incubated with Alexa Fluor 568 goat antibody against rabbit IgG and Alexa Fluor 488 goat antibody against chicken IgG for 1hr at 25°C. Images were acquired using a Slide Scanner microscope.
Data visualization
Plots were generated using R version 3.5.0 with ggplot2 package (version 3.2.0) (Wickham, 2016). Heatmaps were generated using gplots package (version 3.0.1.1) (Warnes et al., 2009). Figures were made using Adobe Illustrator (Adobe Systems; RRID: SCR_010279).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses including the number of animals or cells (n) and p values for each experiment are noted in the figure legends. For the snRNAseq experiments, statistics were performed using R version 3.5.0. Hypergeometric tests were used to test the significance of overlap between two gene sets. It was conducted by calling the phyper() function in R version 3.5.0. For the immunohistochemistry and electron microscopy data, statistics were performed using two-tailed unpaired Student’s t test. A p value of less-than 0.05 was considered significant.
Supplementary Material
Highlights.
Peripheral glial cell ablation results in transient demyelination
PNS demyelination initiates a transcriptional injury response in sensory neurons
The injury response occurs predominantly in proprioceptive and Aβ RA-LTMR neurons
Traumatic injury and glial cell loss induce similar neuronal transcriptional responses
ACKNOWLEDGMENTS
This research was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (C.J.W. and B.P.). The authors would like to thank the team of the AMRF Functional Genomics Common Research Resource at the Department of Psychiatry and Neurology, University of California Los Angeles, for their expert help with the sequencing of the Drop-seq libraries. The authors would like to thank Dr. Erik P. Pioro for the helpful discussion regarding the electrophysiological studies. B.P. is supported by NIH R01NS067550, R01NS109372, R01NS099334, 1RF1AG072080, and by the National Multiple Sclerosis Society (RG-1807-32005). B.E. is supported by NIH R01NS067550 and by DOD W81XWH2210386. C.J.W. is supported by NIH R35NS105076. W.R. is supported by the Burroughs Wellcome Fund, NIH K08NS101064, R01NS119476, NIDA DP1DA054343, Teva Pharmaceuticals, and BWH Women’s Brain Initiative and Neurotechnology Studio
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111130.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aaker JD, Elbaz B, Wu Y, Looney TJ, Zhang L, Lahn BT, and Popko B (2016). Transcriptional fingerprint of hypomyelination in zfp191null and shiverer (mbpshi) mice. ASN Neuro 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N, and Cavalli V (2008). Nerve injury signaling. Curr. Opin. Neurobiol 18, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, and Rahnenfuhrer J (2016). topGO: Enrichment analysis for Gene Ontology. R package version 2.28. 0. Cranio. [Google Scholar]
- Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, Woodhoo A, Jenkins B, Rahman M, Turmaine M, et al. (2012). c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 75, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attarian S, Young P, Brannagan TH, Adams D, Van Damme P, Thomas FP, Casanovas C, Tard C, Walter MC, Pereon Y, et al. (2021). Adouble-blind, placebo-controlled, randomized trial of PXT3003 for the treatment of Charcot-Marie-Tooth type 1A. Orphanet J. Rare Dis 16, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer RN (1994). Automated nerve fibre size and myelin sheath measurement using microcomputer-based digital image analysis: theory, method and results. J. Neurosci. Methods 51, 229–238. [DOI] [PubMed] [Google Scholar]
- Avraham O, Deng P-Y, Jones S, Kuruvilla R, Semenkovich CF, Klyachko VA, and Cavalli V (2020). Satellite glial cells promote regenerative growth in sensory neurons. Nat. Commun 11, 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, and Grant CE (2021). SEA: Simple Enrichment Analysis of Motifs. Preprint at BioRxiv. 10.1101/2021.08.23.457422. [DOI] [Google Scholar]
- Beirowski B, Babetto E, Golden JP, Chen Y-J, Yang K, Gross RW, Patti GJ, and Milbrandt J (2014). Metabolic regulator LKB1 is crucial for Schwann cell-mediated axon maintenance. Nat. Neurosci 17, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito C, Davis CM, Gomez-Sanchez JA, Turmaine M, Meijer D, Poli V, Mirsky R, and Jessen KR (2017). STAT3 controls the long-term survival and phenotype of repair schwann cells during nerve regeneration. J. Neurosci 37, 4255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Yekutieli D (2005). False discovery rate-adjusted multiple confidence intervals for selected parameters. J. Am. Stat. Assoc 700, 71–81. [Google Scholar]
- Berger P, Niemann A, and Suter U (2006). Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease). Glia 54, 243–257. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Cammer W, and Tansey FA (1987). Immunocytochemical localization of carbonic anhydrase in myelinated fibers in peripheral nerves of rat and mouse. J. Histochem. Cytochem 35, 865–870. [DOI] [PubMed] [Google Scholar]
- Catenaccio A, Llavero Hurtado M, Diaz P, Lamont DJ, Wishart TM, and Court FA (2017). Molecular analysis of axonal-intrinsic and glial-associated co-regulation of axon degeneration. Cell Death Dis. 8, e3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, et al. (2016).Asystems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y-C, Snavely A, Barrett LB, Zhang X, Herman C, Frost DJ, Riva P, Tochitsky I, Kawaguchi R, Singh B, et al. (2021). Topoisomerase I inhibition and peripheral nerve injury induce DNA breaks and ATF3-associated axon regeneration in sensory neurons. Cell Rep. 36, 109666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Sloutsky R, Naegle KM, and Cavalli V (2013). Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell 155, 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, and Höke A (2020). Programmed axon degeneration: from mouse to mechanism to medicine. Nat. Rev. Neurosci 21, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfas G, Velardez MO, Ko C-P, Ratner N, and Peles E (2004). Mechanisms and roles of axon-Schwann cell interactions. J. Neurosci 24, 92509260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Flora Nunes G, Wilson ER, Hurley E, He B, O’Malley BW, Poitelon Y,Wrabetz L, and Feltri ML (2021). Activation ofmTORC1 and c-Jun by Prohibitin1 loss in Schwann cells may link mitochondrial dysfunction to demyelination. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, and Popko B (2003). Inducible site-specific recombination in myelinating cells. Genesis 35, 63–72. [DOI] [PubMed] [Google Scholar]
- Elbaz B, Traka M, Kunjamma RB, Dukala D, Brosius Lutz A, Anton ES, Barres BA, Soliven B, and Popko B (2016). Adenomatous polyposis coli regulates radial axonal sorting and myelination in the PNS. Development 143,2356–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz B, Aaker JD, Isaac S, Kolarzyk A, Brugarolas P, Eden A, and Popko B (2018). Phosphorylation state of ZFP24 controls oligodendrocyte differentiation. Cell Rep. 23, 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, and DiAntonio A (2020). The SARM1 axon degeneration pathway: control of the NAD+ metabolome regulates axon survival in health and disease. Curr. Opin. Neurobiol 63, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, Gu W, Nanson JD, Shi Y, Sasaki Y, Cunnea K, Malde AK, Jia X, Luo Z, Saikot FK, et al. (2021). SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration. Neuron 109,1118–1136.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledrich R, Abdelaal T, Rasch L, Bansal V, Schütza V, Brügger B, Lüchtenborg C, Prukop T, Stenzel J, Rahman RU, et al. (2018). Targeting myelin lipid metabolism as a potential therapeutic strategy in a model of CMT1A neuropathy. Nat. Commun 9, 3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fünfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, Brinkmann BG, Kassmann CM, Tzvetanova ID, Möbius W, et al. (2012). Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber D, Ghidinelli M, Tinelli E, Somandin C, Gerber J, Pereira JA, Ommer A, Figlia G, Miehe M, Nageli LG, et al. (2019). Schwann cells, but not oligodendrocytes, depend strictly on dynamin 2 function. Elife 8, e42404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, and Coleman MP (2010). Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves NP, Vægter CB, Andersen H, Østergaard L, Calcutt NA, and Jensen TS (2017). Schwann cell interactions with axons and microvessels in diabetic neuropathy. Nat. Rev. Neurol 13, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AK, and Landis SC (1992). Division and migration of satellite glia in the embryonic rat superior cervical ganglion. J. Neurocytol 21, 635–647. [DOI] [PubMed] [Google Scholar]
- Hanani M, and Spray DC (2020). Emerging importance of satellite glia in nervous system function and dysfunction. Nat. Rev. Neurosci 21, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings RL, Mikesh M, Lee YI, and Thompson WJ (2020). Morphological remodeling during recovery of the neuromuscular junction from terminal Schwann cell ablation in adult mice. Sci. Rep 10, 11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis ER, Ishiko N, Tolentino K, Doherty E, Rodriguez MJ, Calcutt NA, and Zou Y (2015). A novel and robust conditioning lesion induced by ethidium bromide. Exp. Neurol 265, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY-M, Zhang C, Zollinger DR, Leterrier C, and Rasband MN (2017). An αll spectrin-based cytoskeleton protects large-diameter myelinated axons from degeneration. J. Neurosci 37, 11323–11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Koyanagi M, Azimi Z, Nakazato Y, Matsumoto M, Ogihara T, Yonezawa A, Omura T, Nakagawa S, Wakatsuki S, et al. (2017). Taxanes and platinum derivatives impair Schwann cells via distinct mechanisms. Sci. Rep 7, 5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, and Mirsky R (2016). The repair Schwann cell and its function in regenerating nerves. J. Physiol. (Lond.) 594, 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, and Mirsky R (2019). The success and failure of the schwann cell response to nerve injury. Front. Cell. Neurosci 13, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, and Lloyd AC (2015). Schwann cells: development and role in nerve repair. Cold Spring Harb. Perspect. Biol 7, a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Lee Y, Russell KA, Yang F, Dastgheyb RM, Deme P, Ament XH, Chen W, Liu Y, Guan Y, et al. (2020). Monocarboxylate transporter 1 in Schwann cells contributes to maintenance of sensory nerve myelination during aging. Glia 68, 161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Passero JV, Rawat A, Ament XH, Yang F, Vidensky S, Collins SL, Horton MR, Hoke A, Rutter GA, et al. (2021). Macrophage monocarboxylate transporter 1 promotes peripheral nerve regeneration after injury in mice. J. Clin. Invest 10.1172/JCI141964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Anderson M, Park K, Zheng Q, Agarwal A, Gong C, Saijilafu, Young L, He S, LaVinka PC, et al. (2016). Coupled activation of primary sensory neurons contributes to chronic pain. Neuron 91, 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H, Iijima M, Mori K, Yamamoto M, Hattori N, Katsuno M, Tanaka F, Watanabe H, Doyu M, Yoshikawa H, et al. (2007). Nonmyelinating Schwann cell involvement with well-preserved unmyelinated axons in Charcot-Marie-Tooth disease type 1A. J. Neuropathol. Exp. Neurol 66, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kwon MJ, Kim J, Shin H, Jeong SR, Kang YM, Choi JY, Hwang DH, and Kim BG (2013). Contribution of macrophages to enhanced regenerative capacity of dorsal root ganglia sensory neurons by conditioning injury. J. Neurosci 33, 15095–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang P-W, et al. (2012). Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KH, and Svaren J (2018). Epigenetic control of schwann cells. Neuroscientist 24, 627–638. [DOI] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, and Nave K-A (2004). Axonal neuregulin-1 regulates myelin sheath thickness. Science 304, 700–703. [DOI] [PubMed] [Google Scholar]
- Mindos T, Dun X-P, North K, Doddrell RDS, Schulz A, Edwards P, Russell J, Gray B, Roberts SL, Shivane A, et al. (2017). Merlin controls the repair capacity of Schwann cells after injury by regulating Hippo/YAP activity. J. Cell Biol 216, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Le Pichon CE, and Ryba N (2019). Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prukop T, Wernick S, Boussicault L, Ewers D, Jäger K, Adam J, Winter L, Quintes S, Linhoff L, Barrantes-Freer A, et al. (2020). Synergistic PXT3003 therapy uncouples neuromuscular function from dysmyelination in male Charcot-Marie-Tooth disease type 1A (CMT1A) rats. J. Neurosci. Res 98, 1933–1952. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. 201. [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Renthal W, Boxer LD, Hrvatin S, Li E, Silberfeld A, Nagy MA, Griffith EC, Vierbuchen T, and Greenberg ME (2018). Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat. Neurosci 21, 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Tochitsky I, Yang L, Cheng Y-C, Li E, Kawaguchi R, Geschwind DH, and Woolf CJ (2020). Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron 108, 128–144.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud M, Gemes G, Barabas M-E, Chernoff DI, Abram SE, Stucky CL, and Hogan QH (2008). Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain 136, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Hackett AR, Kim S, Strickland A, and Milbrandt J (2018). Dysregulation of NAD+ metabolism induces a schwann cell dedifferentiation program. J. Neurosci 38, 6546–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalbruch H (1986). Fiber composition of the rat sciatic nerve. Anat. Rec 215, 71–81. [DOI] [PubMed] [Google Scholar]
- Sergeeva EG, Rosenberg PA, and Benowitz LI (2021). Non-cell-autonomous regulation of optic nerve regeneration by amacrine cells. Front. Cell. Neurosci 15, 666798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, and Ginty DD (2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siems SB, Jahn O, Eichel MA, Kannaiyan N, Wu LMN, Sherman DL, Kusch K, Hesse D, Jung RB, Fledrich R, et al. (2020). Proteome profile of peripheral myelin in healthy mice and in a neuropathy model. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, et al. (2005). Neuregulin-1 type III determines the ensheathment fate of axons. Neuron 47, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M (2019). The DTA mouse model for oligodendrocyte ablation and CNS demyelination. Methods Mol. Biol 1936, 295–310. [DOI] [PubMed] [Google Scholar]
- Traka M, Arasi K, Avila RL, Podojil JR, Christakos A, Miller SD, Soliven B, and Popko B (2010). A genetic mouse model of adult-onset, pervasive central nervous system demyelination with robust remyelination. Brain 133, 3017–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Millen KJ, Collins D, Elbaz B, Kidd GJ, Gomez CM, and Popko B (2013). WDR81 is necessary for purkinje and photoreceptor cell survival. J. Neurosci 33, 6834–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Podojil JR, McCarthy DP, Miller SD, and Popko B (2016). Oligodendrocyte death results in immune-mediated CNS demyelination. Nat. Neurosci 19, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci 18, 145–153. [DOI] [PubMed] [Google Scholar]
- Vaquié A, Sauvain A, Duman M, Nocera G, Egger B, Meyenhofer F, Falquet L, Bartesaghi L, Chrast R, Lamy CM, et al. (2019). Injured axons instruct schwann cells to build constricting actin spheres to accelerate axonal disintegration. Cell Rep. 27, 3152–3166.e7. [DOI] [PubMed] [Google Scholar]
- Viader A, Sasaki Y, Kim S, Strickland A, Workman CS, Yang K, Gross RW, and Milbrandt J (2013). Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron 77, 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VCJ, Cottrell DF, Brophy PJ, and Fleetwood-Walker SM (2003). Focal lysolecithin-induced demyelination of peripheral afferents results in neuropathic pain behavior that is attenuated by cannabinoids. J. Neurosci 23, 3221–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. (2009). gplots: Various R Programming Tools for Plotting Data. R Package Version 2 [Google Scholar]
- Wickham H (2016). ggplot2 - Elegant Graphics for Data Analysis (SpringerVerlag; ). [Google Scholar]
- Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng Y-L, Elbaz B, Fei Q, Jones JS, Li YI, Zhuang X, et al. (2020). m6A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron 105, 293–309.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Tochitsky I, Woolf CJ, and Renthal W (2021). Isolation of nuclei from mouse dorsal root ganglia for single-nucleus Genomics. Bio. Protoc 11, e4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Williams PR, Jacobi A, Wang C, Goel A, Hirano AA, Brecha NC, Kerschensteiner D, and He Z (2019). Elevating growth factor responsiveness and axon regeneration by modulating presynaptic inputs. Neuron 103, 39–51.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HT, Damle S, Ikeda-Lee K, Kuntz S, Li J, Mohan A, Kim A, Hung G, Scheideler MA, Scherer SS, et al. (2018). PMP22 antisense oligonucleotides reverse Charcot-Marie-Tooth disease type 1A features in rodent models. J. Clin. Invest 128, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, and Ginty DD (2019). Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron 103, 598–616.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data have been deposited at the Gene Expression Omnibus (GEO) repository and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources table. This paper also analyzes our previously published data (Renthal et al., 2020). This existing data is publicly available, and the accession number is listed in the Key Resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit anti-ATF3 | Sigma-Aldrich | Cat#HPA001562; RRID:AB_1078233 |
| Chicken anti-Neurofilament, H | Millipore | Cat#AB5539; RRID:AB_11212161 |
| Rabbit anti Neurotensin | ImmunoStar | Cat# 20072; RRID:AB_572254 |
| Rabbit Anti-Iba1 | Wako Pure Chemical | Cat # 019–19741; RRID:AB_839504 |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA-seq data | This paper | GEO: GSE205461 |
| snRNAseq data | This paper | GEO: GSE205561 |
| snRNAseq data of DRG following traumatic | Renthal et al. (2020) | GEO: GSE154659 |
| injury | ||
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: ROSA26-eGFP-DTA line. | The Jackson Laboratory | Cat#JAX:032087; RRID:IMSR_JAX:032087 |
| Mouse: PLP-CreERt line | The Jackson Laboratory | Cat# JAX: 005975; RRID:IMSR_JAX:005975 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | National Institutes of Health | RRID: SCR_003070 |
| Adobe Illustrator CC | Adobe Systems | RRID: SCR_010279 |
| Indrops mapping | (Klein et al., 2015) | https://github.com/indrops/indrops |
| R for statistical computing- 3.5.0 | R Core Team (2013) | https://www.r-project.org/ |
| Seurat - 2.3.4 | Satija et al., 2015 | https://satijalab.org/seurat/ |
| SCENIC −1.1.1–9 | Aibar et al. (2017) | https://aertslab.org/#scenic |
| edgeR - 3.24.3 | Robinson et al. (2010) | http://bioinf.wehi.edu.au/edgeR/ |
| ggplot2 – 3.2.0 | Wickham, 2016 | https://ggplot2.tidyverse.org/ |
| gplots - 3.0.1.1 | Warnes et al., 2009 | https://github.com/talgalili/gplots |
| topGO | Alexa and Rahnenfuhrer (2016) | https://bioconductor.org/packages/release/bioc/html/topGO.html |
| R for statistical computing- 3.5.0 | R Core Team (2013) | https://www.r-project.org/ |
| HALO | Indica Labs Inc. | RRID:SCR_018350 |