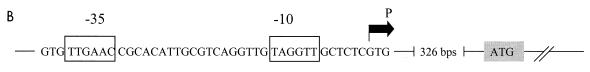Abstract
Examination of nolA revealed that NolA can be uniquely translated from three ATG start codons. Translation from the first ATG (ATG1) predicts a protein (NolA1) having an N-terminal, helix-turn-helix DNA-binding motif similar to the DNA-binding domains of the MerR-type regulatory proteins. Translation from ATG2 and ATG3 would give the N-terminally truncated proteins NolA2 and NolA3, respectively, lacking the DNA-binding domain. Consistent with this, immunoblot analyses of Bradyrhizobium japonicum extracts with a polyclonal antiserum to NolA revealed three distinct polypeptides whose molecular weights were consistent with translation of nolA from the three ATG initiation sites. Site-directed mutagenesis was used to produce derivatives of nolA in which ATG start sites were sequentially deleted. Immunoblots revealed a corresponding absence of the polypeptide whose ATG start site was removed. Translational fusions of the nolA mutants to a promoterless lacZ yielded functional fusion proteins in both Escherichia coli and B. japonicum. Expression of NolA is inducible upon addition of extracts from 5-day-old etiolated soybean seedlings but is not inducible by genistein, a known inducer of the B. japonicum nod genes. The expression of both NolA2 and NolA3 requires the presence of NolA1. NolA1 or NolA3 is required for the genotype-specific nodulation of soybean genotype PI 377578.
The understanding of gene expression was initially guided by the one-gene–one-enzyme hypothesis (24). Since then, it has become apparent that multiple proteins can be derived from one gene. This is well documented in eukaryotic and viral systems. However, very few examples of this phenomenon in prokaryotes have been reported. In a few cases, one gene has been shown to encode two proteins. Examples of these include tipA, infB, clpB, clpA, and fbcH (23, 33, 37, 44, 52). To our knowledge, there have been only two reports (for celA and PPI3316) describing cases in which three proteins are encoded by one gene (3, 34). Here, we describe the characterization of the Bradyrhizobium japonicum nolA gene, which possesses the rare capacity to encode three distinct functional proteins.
nolA (16, 40) is one of three regulatory genes essential for the establishment of a nitrogen-fixing symbiosis between B. japonicum and its host plants. The other regulatory genes include nodD1, which encodes a LysR-type regulator, NodD1 (5, 19, 54), and nodVW, which encode a two-component regulatory system, NodVW (18, 28, 43). These regulatory proteins control the expression of the bacterial nodulation genes (nod, nol, and noe) in response to host plant signals such as flavonoids. The products of the nodulation genes are involved in the synthesis of lipochitooligosaccharide signals, which, when applied to the plant roots, are able to initiate many of the early nodulation events elicited by the bacterial symbiont (reviewed in reference 11).
nolA was first identified by Sadowsky et al. (40) as a genotype-specific nodulation gene since it was able to extend the host range of B. japonicum serogroup 123 strains to certain soybean genotypes (e.g., PI 377578) that normally restrict nodulation by these strains. The importance of nolA in the nodulation process is also supported by recent data (16), which demonstrated that B. japonicum mutants with nolA deleted are grossly defective in nodulation and nitrogen fixation on cowpea. However, the absence of nolA in these strains did not affect the nodulation of soybean plants. Microscopic examination of cowpea nodules infected with the nolA mutant showed that the bacteroids had an atypical morphology. These results indicate that nolA plays a significant role not only in the early stages of infection but also during the later stages of bacteroid development and maintenance within the host cell. A nolA homolog has been identified in Bradyrhizobium (Arachis) sp. strain NC 92 (17). Similar to B. japonicum, mutations to nolA resulted in a reduced ability of this bacterium to nodulate its plant host, the peanut.
Analysis of the nolA gene predicts a protein product that shares an N-terminal helix-turn-helix DNA-binding motif, similar to that of the conserved DNA-binding domains of the MerR family of regulatory proteins (40, 50). Members of this regulatory family initiate the transcription of genes they regulate upon binding of an inducer molecule (22, 23, 36). Interestingly, the inducer molecules (e.g., mercury and superoxide) are generally toxic to the bacterial cell. Binding of the MerR regulators occurs between the −35 and −10 consensus sequences of the target promoters. These promoters have a unique feature in that the −35 and −10 consensus sequences are separated by 19 bp of DNA rather than the usual 16 or 17 bp. An inverted repeat is contained within this 19 bp and is thought to be the site of protein binding (1, 22, 23, 36).
Several MerR-type regulatory proteins autoregulate their own expression. A notable example is TipA, which positively regulates tipA expression in Streptomyces lividans in response to the toxic protein thiostrepton. Interestingly, TipA exists in two forms, TipAL and TipAS. TipAL, which contains the DNA-binding motif, is thought to be a transcriptional regulator, while TipAS, which contains the same carboxyl terminus as TipAL, is believed to be important for thiostrepton binding. Transcription of tipA is initiated at a single site, and the formation of TipAL or TipAS appears to be regulated posttranscriptionally. Recently, we have shown that NolA is also positively autoregulated (16). In this paper, we detail studies to further characterize the regulation and expression of the nolA gene. Notably, we report the presence of three molecular forms of NolA (i.e., NolA1, NolA2, and NolA3) that are derived from the nolA gene. The expression of these proteins appears to be regulated at both the transcriptional and posttranscriptional levels.
MATERIALS AND METHODS
Bacterial culture media and growth conditions.
For routine growth and nucleic acid extraction, B. japonicum strains were grown at 30°C in modified RDY (48). For conjugations or for obtaining cell lysates for Western blot analysis, B. japonicum was grown in HM salt medium (10) supplemented with 0.1% arabinose. B. japonicum was grown in minimal medium (7) for β-galactosidase activity assays. Escherichia coli strains were cultured in Luria-Bertani or M9 medium (41) at 37°C. Antibiotics were used at the following concentrations: for E. coli, ampicillin, 200 μg/ml; tetracycline, 25 μg/ml; streptomycin, 100 μg/ml; spectinomycin, 30 μg/ml; for B. japonicum, tetracycline, streptomycin, and spectinomycin, 100 μg/ml; chloramphenicol, 30 μg/ml.
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. Previously, we reported the construction of a plasmid, pBGAlac1, which encodes a C-terminal nolA-lacZ fusion (16). In the present work, modifications of pBGAlac1 were constructed in which the putative ATG start codons at nucleotides +1, +142, and +228 of nolA were modified. The bases are numbered such that +1 is the first base in the nolA coding or nodD2 (see below) coding region. The nolA constructs were made as follows. To mutate the nolA gene, pBGAlac1 was digested with BamHI and the resultant 1.5-kb fragment containing nolA was cloned into the BamHI site of the pAlter-1 vector (Promega, Madison, Wis.). Mutagenesis reactions were then carried out as specified by the manufacturer. The primers used for these reactions were 5′-GAAATTGAACAACGTTAACAGAGCTACACC-3′ for ATG1 mutagenesis, 5′-GGTCACCGGGCATATGATAGAGAAAGCGG-3′ for ATG2 mutagenesis, and 5′-GATCCGTAAAGCTCTCGAGGGGACG-3′ for ATG3 mutagenesis. In these primers, the putative ATG start codons were replaced with sequences encoding either valine, alanine, or leucine (i.e., GTT, GCA, or CTC), respectively. In addition, the replacement of the ATG codons with either GTT, GCA, or CTC resulted in the insertion of an HpaI, NdeI, or XhoI restriction site in these mutagenic primers, respectively. Clones arising from the mutagenesis reactions were screened for the presence of these restriction sites. Putative clones were then sequenced, and the following plasmids were found to contain the desired combination of mutations: pAlt1 (ATG1 mutation), pAlt2 (ATG2), pAlt3 (ATG3), pAlt12 (ATG1 and ATG2), pAlt13 (ATG1 and ATG3), pAlt23 (ATG2 and ATG3), and pAlt123 (ATG1, ATG2, and ATG3). For nomenclature purposes, subsequent plasmids generated were also subjected to the same numbering system wherein the type of ATG mutation harbored by the plasmid is denoted by the numbers following the plasmid type. To generate the mutant nolA-lacZ fusions, the pAltnolA plasmids were digested with BamHI and the nolA-containing fragments were inserted into the BamHI site of pNM480 (32). DNA sequencing was used to confirm an in-frame fusion between nolA and the lacZ gene. To conjugate the plasmids into B. japonicum, the resultant plasmids were digested with EcoRI and ligated into the EcoRI site of pRK290 (12). The resultant plasmid was transformed into E. coli S17-1 (46) and mobilized by biparental mating (5) into B. japonicum USDA110 or the B. japonicum nolA mutant BjB3 (16).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| B. japonicum | ||
| USDA110 | Wild type | USDA,b Beltsville, Md. |
| BjB3a | nolA, Spr, Smr | 16 |
| USDA438 | B. japonicum serogroup 123 | 40 |
| BJL123a | nolA mutant ATG1, ATG2, ATG3 | This study |
| BJL83a | nolA; ATG1, ATG2; Spr, Smr | This study |
| BJL81a | nolA; ATG2, ATG3; Spr, Smr | This study |
| BJL82a | nolA; ATG1, ATG3; Spr, Smr | This study |
| BJL823a | nolA; ATG1; Spr, Smr | This study |
| BJL813a | nolA; ATG2; Spr, Smr | This study |
| BJL812a | nolA; ATG3; Spr, Smr | This study |
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) (F′, traD36 proAB lacIqZΔM15) | Promega |
| S17-1 | RPR 2-Tc::Mu-Km::Tn7 pro hsdR recA | 46 |
| Plasmids | ||
| pAlter | Tcr | Promega |
| pBG103 | Ampr | 16 |
| pTE3 | Tcr | 13 |
| pTE3A | Tcr, pTE3::nolA | This study |
| pTE3A12 | Tcr, pTE3::nolA; NolA3 expressed from trp promoter of pTE3 | This study |
| pTE3A13 | Tcr, pTE3::nolA; NolA2 expressed from trp promoter of pTE3 | This study |
| pTE3A23 | Tcr, pTE3::nolA; NolA1 expressed | This study |
| pTE3A123 | Tcr, pTE3::nolA; nolA mutation in ATG1, ATG2, ATG3 | This study |
| pRK290 | Tcr, RP4, Mob+ | 12 |
| pNM480 | Apr, promoterless lacZ | 32 |
| pNMAlac1 | Apr, nolA-lacZ, fusion | 16 |
| pNMAlac12 | Apr, nolA-lacZ; fusion expressed from ATG3 | This study |
| pNMAlac13 | Apr, nolA-lacZ; fusion expressed from ATG2 | This study |
| pNMAlac23 | Apr, nolA-lacZ; fusion expressed from ATG1 | This study |
| pHP45Ω | Spr-Smr cassette | 39 |
| pUC19 | Apr | Stratagene |
| pSUP202 | RP4 mob, Tcr, Apr, Cmr | 46 |
| pJLDA | 4.0-kb EcoRI-PstI fragment with Ω insertion between nodD2 and nolA cloned into pSUP202, Smr, Spr, Tcr | This study |
| pJLDA12 | pJLDA, nolA mutation in ATG1 and ATG2 | This study |
| pJLDA13 | pJLDA, nolA mutation in ATG1 and ATG3 | This study |
| pJLDA23 | pJLDA, nolA mutation in ATG2 and ATG3 | This study |
| pJLDA1 | pJLDA, nolA mutation in ATG1 | This study |
| pJLDA2 | pJLDA, nolA mutation in ATG2 | This study |
| pJLDA3 | pJLDA, nolA mutation in ATG3 | This study |
| pJLDA123 | pJLDA, nolA mutation in ATG1, ATG2, and ATG3 | This study |
Mutant strains of B. japonicum are derived from USDA 110.
USDA, U.S. Department of Agriculture.
Plasmids pTE3A12, pTE3A13, and pTE3A23 were generated to express exclusively NolA3, NolA2, and NolA1, respectively, from the trp promoter of the broad-host-range vector pTE3 (13). To obtain these constructs, plasmid pBG23 harboring nolA was digested with SmaI-SalI and the resultant 1.2-kb nolA fragment was ligated into pUC129 digested with SalI-EcoRV, creating plasmid pJLAS. Replacement of the wild-type 1-kb nolA SalI-StyI fragments of pJLAS with 1-kb mutant fragments derived from pCBnolA12, pCBnolA13, and pCBnolA23 (see below), digested with the same restriction enzymes, resulted in the construction of pJLAS12, pJLAS13, and pJLAS23, respectively. These plasmids were subsequently digested with NsiI and PstI, and the nolA fragments were cloned into the PstI site of pTE3, creating pTE3A, pTE3A12, pTE3A13, pTE3A23, and pTE3A123. These plasmids were then conjugated into the nolA mutant strain BjB3 (16) or the wild-type strain USDA438 (serogroup 123 [40]) as described above. Plasmid pCBnolA was obtained by digesting plasmid pBG103 harboring nolA with ClaI-BglII and ligating the 1.7-kb nolA fragment into the ClaI-BamHI site of pUC129. pCBnolA plasmids harboring ATG mutations in the nolA gene were generated by cloning mutant BamHI nolA fragments derived from the pAlt plasmids into pCBnolA digested with BamHI.
In previous work, we described the construction of two B. japonicum nolA mutants by interposon mutagenesis (16). In the present study, additional B. japonicum nolA mutants containing specific mutations to the putative ATG start codons of the nolA gene were constructed. These strains were constructed as follows. A 2-kb fragment ClaI-StyI fragment from pBG103 containing nolA and the 3′ end of nodD2 was released by digestion of pBG103. This fragment was blunt ended with Klenow DNA polymerase and inserted into the HincII-SmaI site of pUC19 to generate plasmid pJLDA. Derivatives of pJLDA containing mutations to ATG1, ATG2, or ATG3 were obtained by replacing the BamHI wild-type fragment of pJLDA with the corresponding BamHI fragment of the pAlt plasmids harboring mutations to the nolA gene. The pJLDA plasmids were digested at the EagI site located in the intergenic region between nodD2 and nolA. The 5′ overhang sites were blunt ended with Klenow DNA polymerase, and the 2-kb SmaI fragment of pHP45Ω (39) containing an Smr-Spr cassette was ligated into this site. Digestion of this plasmid with EcoRI and PstI released the nolA fragment, which was then cloned into the EcoRI-PstI site of the suicide vector pSUP202 (46). These suicide plasmids were transformed into E. coli S17-1 and conjugated into B. japonicum USDA 110. Transconjugants were selected based on Spr-Smr resistance and Tcs, the latter being an indication of a double-crossover event. Confirmation of these mutations was obtained by Southern blot analyses.
To generate large amounts of NolA for antibody production, a polyhistidine tag system was used to express NolA as a fusion protein from the T7 promoter of the vector pRSETB (Invitrogen, San Diego, Calif.). The plasmid used for the expression of NolA was constructed as follows. Based on the nolA sequence, oligonucleotide primers containing BglII (5′-GGAGATCTGAACAGAGCTACACCAA-3′) or EcoRI (5′-TAGAATTCGTCAGTAAGGCTGATCC-3′) restriction sites were used to PCR amplify the entire nolA coding region. The amplified fragment was isolated, blunt ended with Klenow DNA polymerase, phosphorylated with T4 polynucleotide kinase, and blunt-end ligated with T4 ligase to form concatemers. Following digestion with BglII and EcoRI, the nolA fragment was cloned into the BamHI-EcoRI site of pRSETB. The resulting plasmid (pBGT7A-2) was transformed into E. coli HMS174(DE3)(pLysS), which harbors a chromosomal T7 RNA polymerase under the control of the lac promoter (49). The in-frame translation fusion in pBGT7A-2 was confirmed by DNA sequence analysis, using the dideoxynucleotide chain termination method of Sanger et al. (42).
β-Galactosidase activity assays.
β-Galactosidase activity in B. japonicum was assayed as described by Yuen and Stacey (56). β-Galactosidase activity was measured 12 h after induction with soybean seed extract (SSE), genistein, or soybean seedling extract (SSG). SSE was prepared as described by Smit et al. (47) and was added at 20 μl/ml. Genistein was added at a final concentration of 2 μM. SSG was prepared from soybean seedlings as follows. Soybean seeds were germinated in the dark for 5 to 7 days at 30°C. The seedlings were then blended in a Waring blender and incubated with 95% ethanol (2 ml of ethanol/g of seedling) for 5 h at 25°C in a rotary shaker, and the mixture was centrifuged at 10,000 × g. The supernatant containing the seedling extract was concentrated by rotary evaporation and mixed with ethyl acetate at a ratio of 1.5 volumes of ethyl acetate per volume of seedling extract. The top layer containing ethyl acetate was concentrated by rotary evaporation and resuspended in methanol. This extract was used in β-galactosidase assays at 2.5 μl/ml.
The β-galactosidase activity of E. coli cells was assayed as follows. Cells were grown overnight in M9 medium (41) supplemented with 0.5% (wt/vol) Casamino Acids (M9-CA medium), 20 μg of tryptophan per ml, and the appropriate antibiotics. Overnight cultures were harvested by centrifugation and diluted 1:20 into fresh M9-CA medium in the absence or presence of tryptophan. β-Galactosidase activity was measured 2 h after subculture.
Primer extension.
The transcriptional start sites of nolA and nodD2 were determined by primer extension as described by Chun and Stacey (9). The following primers were used in the reactions: for nolA, primer 1 (5′-GCGACTTGGACTTCTATGCG-3′), primer 2 (5′-CGAATCTGATGAACCCGTTGCC-3′), primer 3 (5′-GTGTGCTCATAATGGTGCAGCGT-3′), and primer 4 (5′-GTTACTCCGGTCGCCTCTGCAA-3′), which are complementary to bases +275 to +296, +160 to +182, +76 to +96, and +47 to +68, respectively; and for nodD2, 5′-GCTAATTGGTCTTGCCGGTTCCG-3′ and 5′-GCAGATCAGCCCAGTGTTCGTCA-3′), which are complementary to bases −221 to −199 and −280 to −258, respectively. The bases are numbered such that +1 is the first base in the nolA or nodD2 coding region. Size standards were obtained with the same primers in a dideoxy sequencing (42) with plasmids containing the nolA or nodD2 regions as templates.
Protein purification.
E. coli cells harboring pBGT7A-2 were grown to an absorbance at 600 nm (A600) of 0.5 and induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Induced cells were grown for an additional 3 h, harvested by centrifugation at 5,000 × g for 5 min, and lysed by sonication (450 sonifier; Branson, Danbury, Conn.). The cell lysate was centrifuged at 31,000 × g for 20 min, and the proteins in the resultant supernatant (soluble fraction) and pellet (insoluble fraction) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel. Coomassie blue staining of the polypeptides revealed that most of the fusion protein was found in the insoluble fraction. Given this observation, the following steps were used to purify the protein. The insoluble fraction containing protein inclusion bodies was washed once with phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 2 M urea, three times with PBS containing 1% Triton X-100, and once with PBS. These washes removed most of the contaminating proteins (as analyzed by SDS-PAGE) and resulted in a preparation that was predominantly (approximately 90%) polyhistidine-tagged NolA fusion protein. The washed inclusion bodies were solubilized in SDS sample buffer (65 mM Tris-Cl [pH 6.8], 10% glycerol, 1% SDS, 150 mM β-mercaptoethanol, 0.005% bromophenol blue) and separated by SDS-PAGE on a preparative 12% acrylamide gel. This gel was lightly stained with Coomassie blue, and the band corresponding to the NolA fusion protein was excised. The protein was electroeluted from the gel slice by the method described by Harlow and Lane (21) and concentrated by ultrafiltration with a Centricon-10 cartridge (Amicon, Inc., Beverly, Mass.). Protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce Inc., Rockford, Ill.).
Antibody generation.
Female New Zealand White rabbits were immunized by subcutaneous and intramuscular injections of approximately 500 μg of gel-purified protein emulsified in Freund’s complete adjuvant. Booster injections were administered at 5-week intervals with 200 μg of gel-purified NolA emulsified in Freund’s incomplete adjuvant. Blood samples were collected 7 to 10 days after each booster injection, and the serum was processed by standard methods (25). NolA-specific antibodies were then affinity purified by the method described by Gu et al. (20). Briefly, washed inclusion bodies containing His-tagged NolA protein were solubilized in 6 M guanidine-HCl and applied to a Sepharose 6B column (Sigma, St. Louis, Mo.) that had been activated with Ni2+ by the method recommended by Novagen Inc. (Madison, Wis.). The column was washed with 15 volumes of wash buffer A (20 mM imidazole, 500 mM NaCl, 20 mM Tris · Cl [pH 7.9]) followed by 15 volumes of equilibration buffer (150 mM NaCl, 50 mM Tris · Cl [pH 7.4]). Crude antiserum was then applied to the column, and the column was allowed to sit at room temperature for 30 min. The column was washed with 5 column volumes of equilibration buffer and 5 volumes of wash buffer B (2 M NaCl, 50 mM Tris · Cl [pH 7.4]). Anti-NolA antibody was eluted from the column by incubating the column with 1 column volume of 4 M MgCl2 for 15 min and then adding a second column volume, after which the eluate was collected. The eluate, containing the affinity purified antibody, was dialyzed against water for 1 h and then against PBS exhaustively at 4°C. Prior to use, the affinity-purified antibody was absorbed with acetone extracts (21) that were made from extracts of the nolA deletion mutant BjB3.
Western blots.
B. japonicum cells were cultured in RDY medium to an A600 of approximately 0.8. The cells were inoculated into HM medium to obtain an A600 of 0.05. The bacterial cells were then grown to an A600 of approximately 0.6 in the presence or absence of SSG. The cells were harvested by centrifugation, washed with PBS buffer, and resuspended in the same buffer. They were lysed by sonication, and the cell lysates were centrifuged at 31,000 × g for 30 min. Proteins contained in the supernatant were separated by SDS-PAGE on a 12% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, Calif.) with a Hoefer Scientific Instruments Inc. (San Francisco, Calif.) electrophoretic transfer apparatus. The filters were blocked for 2 h in TBS (20 mM Tris · Cl [pH 7.5], 0.5 M NaCl) containing 5% bovine serum albumin (BSA) (TBS-BSA). They were then incubated overnight with a 1:500 dilution of anti-NolA, washed three times with TTBS (TBS containing 0.05% Tween 20 [Sigma]), and incubated for 1 h in alkaline phosphatase-conjugated goat anti-rabbit antibodies (Bio-Rad Laboratories) in TBS-BSA. The membrane was washed three times with TTBS, and immunoreactive bands were visualized with nitroblue tetrazolium (Bio-Rad Laboratories) and 5 bromo-4-chloro-3-indolyl phosphate (Bio-Rad Laboratories) as substrates.
Plant nodulation assays.
Glycine max (soybean) cv. Essex and Vigna unguiculata (cowpea) cv. Caloona seeds were surface sterilized as described by Nieuwkoop et al. (35). Following germination, the seedlings were transferred into sterile Leonard jars containing 2 parts vermiculite and 1 part perlite. The plants were grown in a Conviron 4030 plant growth chamber (Conviron, Winnipeg, Canada) at 25°C under 16 h of daylight per 24-h period. For plant tests involving G. max PI 377578 and G. max cv. Kasota, the seeds were prepared as previously reported (40). The plants were incubated with a photoperiod of 18 h per 24-h period and a constant temperature of 20°C. They were watered, alternately, with nitrogen-free nutrient solution and water as needed. Nitrogen fixation activity was detected by acetylene reduction assays (53) with a Shimadzu GC-8A gas chromatograph equipped with a 6-ft Poropak R column. The detector and column were maintained at 100 and 75°C, respectively.
RESULTS
NolA can be translated from three ATGs.
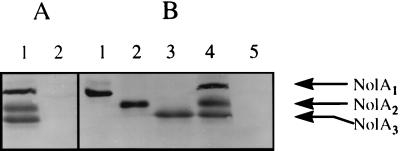
Examination of the nolA sequence (Fig. 1) revealed the presence of three possible ATG start sites from which NolA can be translated. Each of these initiation codons is preceded by a putative ribosome-binding site. Translation of these proteins from the initiation sites ATG1, ATG2, and ATG3 would result in the synthesis of proteins of 25, 22, and 19 kDa, respectively. These proteins have identical C-terminal ends, since they have the same translational reading frame. Consistent with this, immunoblotting of cell extracts of B. japonicum cells induced with SSG with anti-NolA antibody revealed the presence of three cross-reacting bands (Fig. 2A, lane 1). These polypeptides, designated NolA1, NolA2, and NolA3, migrated on SDS-PAGE gels with the molecular masses (i.e., 25, 22, and 19 kDa) expected for polypeptides translated from ATG1, ATG2, and ATG3, respectively. To facilitate further studies on the translational initiation at each of the ATGs, we altered the nolA gene by site-directed mutagenesis of the individual ATG initiation codons. These mutations resulted in the replacement of ATG1, ATG2, and ATG3 with codons encoding valine, alanine, and leucine, respectively. In addition, they created restriction sites within the nolA sequence (i.e., HpaI, NdeI, and XhoI) that allowed the selection of the desired ATG mutation. To analyze the protein products resulting from these mutations, the nolA gene was cloned into plasmid pTE3 harboring a trp promoter and introduced into the chromosomal nolA deletion mutant BjB3. In plasmid pTE3A12, mutations to ATG1 and ATG2 block translation so that only NolA3 can be made. Similarly, plasmids pTE3A23 and pTE3A13 contain mutations that would allow, respectively, only NolA1 or NolA2 to be expressed. Consistent with this, when extracts of BjB3 harboring these plasmids were analyzed, cells carrying pTE3A12 produced only NolA3 whereas BjB3 cells carrying pTE3A13 and pTE3A23, respectively, expressed exclusively NolA2 or NolA1 (Fig. 2B). In contrast, Western blot analysis of BjB3 harboring a plasmid mutated in all three ATGs (i.e., pTE3A123) revealed no labeling of NolA.
FIG. 1.
Promoter and 5′ region of the nolA gene. The two transcriptional start sites, P1 and P2, are shown. The consensus −10 and −35 regions 5′ of P2 are boxed. A region of dyad symmetry between these two boxes is shown by the solid arrows. The translational start sites ATG1, ATG2, and ATG3 are indicated by the shaded boxes. Each is preceded by a putative ribosome-binding site (RBS, underlined). A possible stem-loop region encompassing ATG2 is shown by the reversed arrows. Sequence identity between the nolA and nodD2 promoters is also shown (D2, broken underline).
FIG. 2.
Western blot analysis of cell extracts with a polyclonal antibody against NolA. The three immunoreactive bands are designated NolA1, NolA2, and NolA3. (A) Cell extracts of B. japonicum USDA 110 uninduced or induced with SSG. Lanes: 1, SSG-treated sample; 2, uninduced sample. (B) BjB3 (nolA mutant) complemented with pTE3A23 (lane 1), pTE3A13 (lane 2), pTE3A12 (lane 3), pTE3A (lane 4), and pTE3A123 (lane 5).
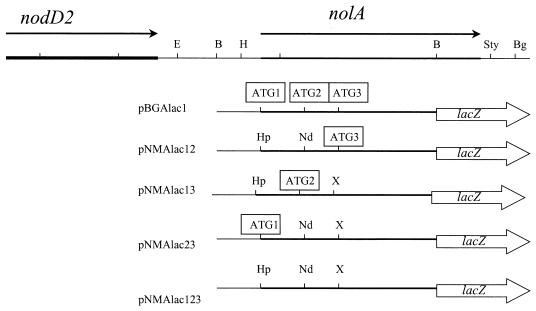
The notion that NolA can be translated from the three putative initiation sites was further tested by fusing the wild-type or mutated nolA genes to a promoterless lacZ gene, generating the translational fusions shown in Fig. 3. Each of these fusions contained mutations in at least two of the ATG start sites in the nolA coding sequence. Therefore, plasmids pNMAlac12, pNMAlac13, and pNMAlac23 would yield fusion proteins NolA1-LacZ, NolA2-LacZ, and NolA3-LacZ, respectively. To assay the translational fusions in B. japonicum, the nolA-lacZ plasmids were conjugated into B. japonicum. Previously, we reported that treatment of B. japonicum cells harboring pBGAlac1 (a wild-type nolA-lacZ fusion) with genistein, a known nod gene inducer of B. japonicum, resulted in little or no induction of nolA-lacZ expression (16). Other isoflavones (e.g., diadzein and biochainin) known to induce nod gene expression, as well as all other flavonoids (e.g., luteolin) tested, were unable to induce nolA expression from pBGAlac1. Treatment with SSE resulted in only a twofold induction. However, nolA expression was greatly induced by ethanol extracts of 5-day-old etiolated soybean seedlings (SSG) (Table 2). Moreover, this induction was observed in B. japonicum USDA 110 harboring each of the plasmids encoding the nolA1-lac, nolA2-lac, or nolA3-lacZ fusion. The greatest levels of induction (approximately 20-fold) were observed with both the NolA1 and NolA3 fusions. NolA2 expression in B. japonicum was consistently lower than NolA1 or NolA3 expression, with only a 10-fold induction observed for SSG treatment. Strains containing pNMAlac123, with all three ATG initiation sites deleted, showed little or no expression when treated with SSG (data not shown).
FIG. 3.
Restriction map of the B. japonicum chromosome showing the location of nolA and nodD2. Only the pertinent restriction sites are shown: H, HindIII; B, BamHI; N, NheI; S, SalI; Sty, StyI; Bg, BglII; E, EagI. Below the restriction map are shown the various nolA derivatives generated by site-directed mutagenesis of the ATG start sites. These mutant derivatives are represented in the figure as nolA-lacZ fusions used in the study. The open arrowhead rectangles represent the pNM480 vector and in-frame C-terminal fusion. The open squares indicate the start sites from which translation of nolA can occur. The restriction sites generated during the mutagenesis are shown: Hp, HpaI; Nd, NdeI; X, XhoI.
TABLE 2.
Expression of plasmid-borne nolA-lacZ fusions in B. japonicum wild-type (USDA 110) and nolA mutant strains
| Strain | Phenotype (chromosomal) | β-Galactosidase activity (U)a
|
|
|---|---|---|---|
| Uninduced | + SSG | ||
| nolA1-lacZ fusion on pNMAlac23 | |||
| USDA 110 | Wild type | 15 ± 3 | 292 ± 15 |
| BjB3 | NolA− | 18 ± 4 | 250 ± 18 |
| BJL823 | NolA1 | 16 ± 7 | 180 ± 15 |
| BJL813 | NolA2 | 17 ± 6 | 238 ± 16 |
| BJL812 | NolA3 | 26 ± 4 | 71 ± 10 |
| BJL83 | NolA1, NolA2 | 19 ± 5 | 255 ± 12 |
| BJL82 | NolA1, NolA3 | 19 ± 4 | 140 ± 12 |
| BJL81 | NolA2, NolA3 | 20 ± 4 | 94 ± 3 |
| BJL123 | Null (Δ123) | 19 ± 10 | 220 ± 20 |
| nolA2-lacZ fusion on pNMAlac13 | |||
| USDA 110 | Wild type | 20 ± 4 | 150 ± 18 |
| BjB3 | NolA− | 25 ± 6 | 52 ± 4 |
| BJL823 | NolA1 | 18 ± 2 | 149 ± 3 |
| BJL813 | NolA2 | 23 ± 6 | 62 ± 6 |
| BJL812 | NolA3 | 17 ± 5 | 45 ± 5 |
| BJL83 | NolA1, NolA2 | 16 ± 5 | 80 ± 8 |
| BJL82 | NolA1, NolA3 | 16 ± 5 | 88 ± 11 |
| BJL81 | NolA2, NolA3 | 18 ± 2 | 56 ± 13 |
| BJL123 | Null (Δ123) | 19 ± 4 | 42 ± 5 |
| nolA3-lacZ fusion on pNMAlac12 | |||
| USDA 110 | Wild type | 25 ± 6 | 250 ± 19 |
| BjB3 | NolA− | 32 ± 7 | 56 ± 8 |
| BJL823 | NolA1 | 16 ± 6 | 228 ± 3 |
| BJL813 | NolA2 | 17 ± 6 | 73 ± 10 |
| BJL812 | NolA3 | 26 ± 4 | 53 ± 8 |
| BJL83 | NolA1, NolA2 | 16 ± 6 | 270 ± 20 |
| BJL82 | NolA1, NolA3 | 19 ± 4 | 87 ± 8 |
| BJL81 | NolA2, NolA3 | 20 ± 5 | 71 ± 5 |
| BJL123 | Null (Δ123) | 11 ± 3 | 57 ± 20 |
Units with CPRG as a substrate. Values are the means of two independent determinations. The standard deviation is indicated.
NolA2 and NolA3 are regulated by NolA1.
Previously, we had shown that the nolA gene was positively autoregulated, requiring the presence of NolA for its expression (16). Our present observation that the nolA gene encodes three proteins raises the question whether all three forms of NolA are autoregulated. To address this, we mobilized the various nolA-lacZ constructs into the B. japonicum nolA mutant BjB3 and compared the translational efficiencies of each fusion. As shown in Table 2, SSG treatment of B. japonicum wild-type or BjB3 cells harboring the nolA1-lacZ plasmid significantly induced β-galactosidase expression. The fact that nolA1-lacZ is induced in BjB3 indicates that NolA1 expression is independent of the presence of NolA. In contrast, little or no β-galactosidase activity was observed in strain BjB3 harboring either a nolA2-lacZ or nolA3-lacZ fusion. These data indicate that only the expression of NolA2 and NolA3 is positively autoregulated. A similar result was also obtained in E. coli cotransformants harboring pTE3A and one of the nolA-lacZ plasmids pNMAlac12, pNMAlac13, pNMAlac23, or pNMAlac123. Compared to NolA1-lacZ (57 ± 4 U), the expression of nolA from the trp promoter of pTE3A resulted in elevated expression of only NolA2-LacZ (342 ± 18 U) and NolA3-LacZ (309 ± 6 U).
A limitation of the above experiments is the fact that the enzymatic activities of the nolA-lacZ fusions were analyzed in the presence of a wild-type nolA gene capable of expressing all three NolA proteins. To further characterize the role of each NolA protein in nolA autoregulation, B. japonicum nolA chromosomal mutants that contained each of the specific mutations to the individual translational initiation sites were generated. These chromosomal mutants possess the exclusive capacity to express NolA1, NolA2, or NolA3 singly or a combination of these proteins. To test the function of NolA proteins, plasmid-borne nolA1-lacZ, nolA2-lacZ, or nolA3-lacZ fusions were transformed into the B. japonicum chromosomal mutants containing the site-directed mutations and the resultant transconjugants were tested for NolA-LacZ induction by SSG. As shown in Table 2, NolA1-LacZ expression was significantly induced in a mutant strain (i.e., BJL123) containing chromosomal mutations to all three ATG start sites. Low levels of NolA2 or NolA3 were observed in BJL123. Therefore, removal of the three translational codons results in a nolA phenotype that is similar or equivalent to the BjB3 nolA deletion. Interestingly, when NolA1-LacZ expression was analyzed in BJL812, BJL82, or BJL81, the enzymatic activity of the fusion was found to be consistently lower than that observed in BJL123 (Table 2). This suggests that NolA3 represses NolA1-LacZ expression. As mentioned above, very little induction of NolA2-LacZ or NolA3-LacZ expression was observed in the null mutant BJL123. Low levels of both fusions were also observed in BJL81 and BJL812, with significant induction of these fusions observed in the presence of NolA1 (e.g., in BJL823). However, the induction of NolA2 and NolA3 was lower in strains expressing, in addition to NolA1, either NolA2 (i.e., BJL83) or NolA3 (i.e., BJL82). These data suggest that all three proteins interact in subtle ways that affect their regulation.
Transcriptional start sites of nolA and nodD2.
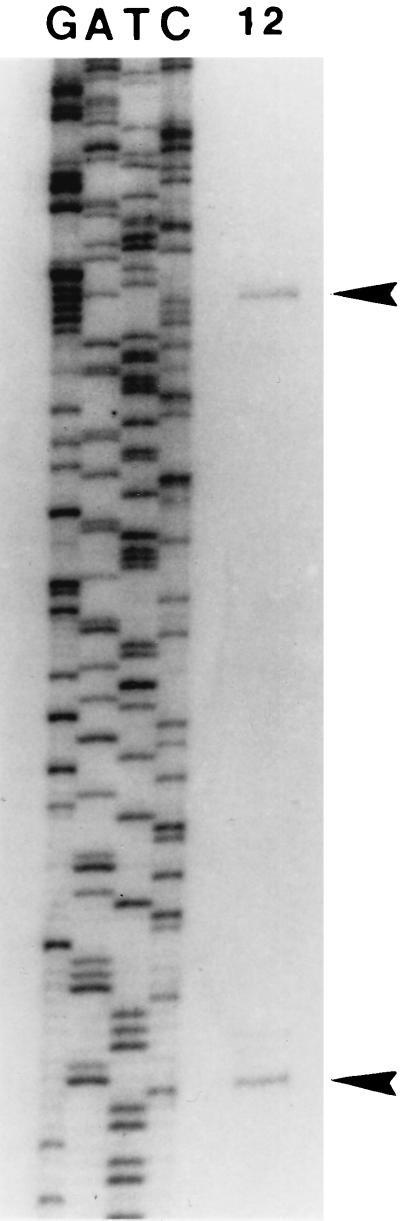
The differential expression of NolA1-LacZ, NolA2-LacZ, and NolA3-LacZ, as well as results of Western blot analyses, clearly indicates that nolA encodes three proteins via translation from three alternative ATG start codons. Therefore, we examined whether the expression of all three proteins could be controlled via transcription from different promoters. Primer extension analysis was performed to identify the transcriptional start site(s) of nolA. Using four independent primers in the extension reactions, we identified two transcriptional start sites (Fig. 4). The first transcriptional start site (derived from promoter P1) is found 82 bases upstream of ATG1 (Fig. 1). The second transcriptional start site (from promoter P2) is found 7 bases upstream of ATG1 and is immediately downstream of a putative NolA-binding site (Fig. 1). This putative NolA-binding site has similar characteristics to the DNA target sites of the MerR-type regulatory proteins (i.e., conserved −10 and −35 hexamers separated by 19 bp, with an inverted repeat contained in the 19-bp intervening sequence).
FIG. 4.
Determination of the transcriptional start sites of nolA. Results of primer extension studies with 50 μg of RNA extracted from uninduced B. japonicum cells (lane 1) and from cells which were induced with soybean seedling extract (lane 2) are shown. A DNA-sequencing ladder is shown for comparison. The two nolA transcripts are indicated (arrowheads). The two nolA transcripts were detected with four different primers (see Materials and Methods). The results shown were obtained with primer 2.
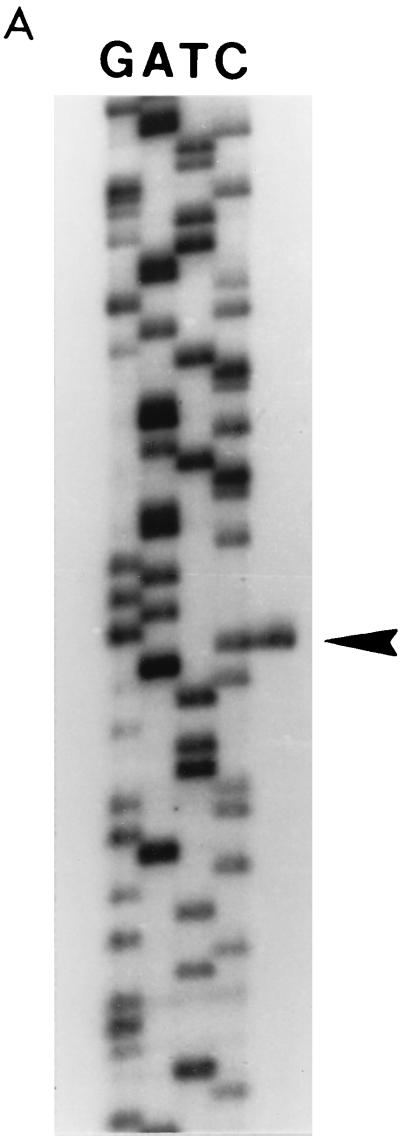
Previously we reported that in addition to the nolA gene, nodD2 expression requires the nolA gene product (16). Examination of the DNA sequence upstream of the predicted nodD2 ATG start codon revealed a putative NolA binding site which has significant homology to the P2 upstream DNA of nolA (i.e., 10 of the 3′ bases within the 19-bp intervening region are identical). Therefore, we determined if nodD2 is transcribed immediately downstream of the putative NolA-binding site (Fig. 5A). The results of the primer extension with two independent primers showed that nodD2 transcription starts 7 bp downstream of the putative NolA-binding site (Fig. 5B). These results further support the idea that NolA1 is the molecular form of NolA that acts as a positive transcriptional regulator of nodD2, as well as that of nolA.
FIG. 5.
Determination of the transcriptional start site of nodD2. (A) nodD2 transcript (arrowhead) as determined by primer extension experiments with 50 μg of RNA extracted from uninduced B. japonicum cells. A DNA-sequencing ladder is shown for comparison. (B) The nodD2 promoter region showing the location of the transcriptional start site and the proposed nodD2 ATG start codon. The putative NolA1-binding site found immediately upstream of the nodD2 transcriptional initiation site is also shown. The putative −10 and −35 hexamers are boxed.
Biological significance of NolA1, NolA2, and NolA3.
NolA was first identified as a genotype-specific nodulation gene that could extend the host range of B. japonicum USDA 123 strains to include certain soybean genotypes that restrict nodulation by these strains. For example, USDA 438, a B. japonicum serogroup 123 strain, can nodulate G. max cv. Williams but not G. max PI 377578 (40). Complementation of USDA 438 with the nolA gene conferred upon transconjugants the ability to nodulate the restricted PI 377578 genotype. To test the importance of each NolA protein in this nodulation process, two separate sets of USDA 438 transconjugants were examined for their capacity to nodulate the restricting PI 377578 genotype and the nonrestrictive cultivar Kasota. The first set of transformants were generated by cloning the nolA gene into the broad-host-range vector pRK290 and mobilizing the resultant plasmids into USDA 438. In these cases, each NolA protein was expressed from a wild-type promoter. As shown in Table 3, all the USDA 438 transconjugants nodulated the nonrestrictive soybean genotype Kasota. When the same strains were applied to the roots of PI 377578, only strains expressing NolA1 (strain designations are given in Table 3) were able to nodulate this plant. Strains harboring the vector control (pRK290) or plasmids whose nolA gene allowed the expression of NolA from only ATG2 or ATG3 were unable to nodulate cultivar PI 377578. The inability of the last two mutants to nodulate PI 377578 may be explained by the fact that NolA2 and NolA3 require NolA1 for its expression. Given this possibility, USDA 438 cells were transformed with a second set of plasmids expressing singly NolA1 (i.e., pTE3A23), NolA2 (i.e., pTE3A13), or NolA3 (i.e., pTE3A12) or no NolA protein (i.e., pTE3A123) from the constitutive trp promoter of pTE3. The control comprised a vector-only sample. As shown in Table 3, soybean genotype Kasota was nodulated normally by these strains. In contrast, only transconjugants expressing NolA3 were able to nodulate PI 377578. The nodules formed on these plants were capable of reducing acetylene (Fix+, data not shown). The other transconjugants (containing pTE3A123, pTE3A13, or the vector pTE3) failed to nodulate the restricting PI 377578 genotype. Interestingly, the constitutive expression of NolA1 from the trp promoter in USDA 438(pTE3A23) resulted in no nodule formation on soybean PI 377578. This result is in contrast to that obtained previously with USDA438 complemented with pJLDA23, where NolA1 is expressed from its own promoter.
TABLE 3.
Nodulation phenotype of B. japonicum USDA 438 and its transconjugants on G. max PI 377578 and G. max cv. Kasota
| Strain | NolA expressed | No. of nodules/planta
|
|
|---|---|---|---|
| PI 377578 | cv. Kasota | ||
| nolA from wild-type promoter | |||
| USDA 438 | |||
| + pRK290 | None | 2 ± 1 | 50 ± 11 |
| + pJLDA | NolA1, NolA2, NolA3 | 46 ± 6 | 53 ± 17 |
| + pJLDA23 | NolA1 | 27 ± 7 | 48 ± 9 |
| + pJLDA13 | NolA2 | 1 ± 1 | 52 ± 5 |
| + pJLDA12 | NolA3 | 2 ± 1 | 51 ± 5 |
| + pJLDA123 | Null (Δ123) | 2 ± 0 | 39 ± 6 |
| nolA from trp promoter | |||
| USDA 438 | |||
| + pTE3 | None | 1 ± 1 | 54 ± 7 |
| + pTE3A | NolA1, NolA2, NolA3 | 23 ± 5 | 37 ± 11 |
| + pTE3A23 | NolA1 | 0 | 32 ± 4 |
| + pTE3A13 | NolA2 | 0 | 45 ± 4 |
| + pTE3A12 | NolA3 | 27 ± 1 | 52 ± 13 |
| + pTE3A123 | Null (Δ123) | 0 | 42 ± 1 |
Nodule numbers per plant were compared 30 days after inoculation. Values are means for three plants ± standard error of means.
In addition to being necessary for the nodulation of certain soybean genotypes, nolA is essential for the nodulation of cowpea plants (16). For example, the nolA mutant strain BjB3 exhibited significantly lower nodulation and nitrogen fixation on cowpea but was not affected in these traits when inoculated on soybean. Given this observation, we tested the ability of NolA1, NolA2, or NolA3 to complement the nodulation-deficient phenotype observed with this bacterial strain. Similar to results observed in the nodulation of soybean cultivar PI 377578 by USDA 438 transconjugants, only B. japonicum USDA 110 strains expressing NolA3 from pTE3A12 were able to enhance the nodulation and nitrogen fixation of cowpea plants (data not shown). In contrast, expression of NolA1 from pTE3A23 or NolA2 from pTE3A13 resulted in a nodulation phenotype similar to that observed with the BjB3 vector control (data not shown). The role of NolA in the nodulation process was also tested by using chromosomal nolA mutants that contained mutations to the individual ATG initiation sites. When inoculated onto cowpea plants, only strain BJL823 (expressing NolA1) and BJL82 (expressing NolA1 and NolA3) were capable of effective nodulation of cowpea plants (Table 4). Expression of NolA2 (e.g., BJL83) appeared to counteract the effects of NolA1, resulting in decreased nodulation efficiency of cowpea plants. The soybean control (i.e., G. max cv. Essex) revealed little or no difference in nodulation when inoculated with these chromosomal mutants.
TABLE 4.
Nodulation and nitrogen fixation phenotypes of B. japonicum nolA mutants harboring mutations to the translation initiation sites of nolA
| Strain | Phenotype | No. of nodules/planta
|
Nitrogen fixation (nmol/plant/h)b
|
||
|---|---|---|---|---|---|
| Cowpea | G. max cv. Essex | Cowpea | G. max cv. Essex | ||
| BJL823 | NolA1 | 17 ± 4 | 18 ± 4 | 3,000 ± 500 | 2,350 ± 114 |
| BJL813 | NolA2 | 8 ± 2 | 14 ± 2 | 200 ± 50 | 2,000 ± 110 |
| BJL812 | NolA3 | 8 ± 1 | 16 ± 5 | 766 ± 25 | 2,485 ± 250 |
| BJL83 | NolA1, NolA2 | 10 ± 3 | 15 ± 1 | 399 ± 30 | 2,000 ± 500 |
| BJL82 | NolA1, NolA3 | 20 ± 3 | 20 ± 4 | 3,333 ± 20 | 3,050 ± 300 |
| BJL123 | Null (Δ123) | 9 ± 3 | 18 ± 3 | 1,009 ± 273 | 3,259 ± 359 |
| USDA 110 | Wild type | 23 ± 2 | 23 ± 7 | 3,200 ± 172 | 3,500 ± 450 |
Nodule number determined 28 days after inoculation. Results are the means for 18 plants assayed over two trials (standard errors are shown).
Acetylene reduction assay performed 28 days after inoculation.
DISCUSSION
Our understanding of gene expression was initially governed by the one-gene–one-enzyme concept (24). Since then, many exceptions to this rule have been found, predominantly among eukaryotic and viral genes. The regulation of these multiple gene products can be controlled at the transcriptional (e.g., the Dfer gene of Drosophila melanogaster [38]), posttranscriptional (e.g., RNA processing of bacteriophage T4 terminase genes [14]), and translational (e.g., the Sp3 retinoblastoma gene [15]) levels. In contrast, examples of multiple proteins derived from one prokaryotic gene have rarely been reported. There are a few examples (e.g., tipA, infB, clpA, clpB, and fbcH) where two prokaryotic polypeptides are derived from one gene (23, 33, 37, 44, 52). These products can be regulated translationally (e.g., clpA and clpB) via the use of alternative translational start sites on the same mRNA or posttranslationally via proteolytic processing (e.g., fbcH). To our knowledge, only two examples detailing the expression of three proteins derived from one gene have been reported for prokaryotes. These are the celA gene of Ruminococcus albus (3) and PPL3316 of Peptostreptococcus albus 3316 (34). For celA, transcriptional control has been proposed to account for the expression of three polypeptides. In contrast, the presence of three translational start sites is thought to account for the expression of the PPL proteins.
Examination of the nolA coding region has revealed that in addition to the ATG translational start codon (ATG1) proposed by Sadowsky et al. (40), two start codons (ATG2 and ATG3) preceded by putative ribosome-binding sites are present within the coding region. Translation from ATG1 would give a full-length protein (NolA1) which contains the N-terminal helix-turn-helix DNA-binding motif that has sequence similarity to the DNA-binding domains of the MerR family of regulatory proteins (40). Translation from ATG2 and ATG3 would result in the N-terminally truncated proteins NolA2 and NolA3, which do not possess the DNA-binding domain. However, since all three ATGs have the same open reading frame, NolA1, NolA2, and NolA3 have identical carboxyl termini. To investigate the possibility that three molecular forms of NolA could be derived from one gene, Western blot analysis was performed on cell extracts of B. japonicum by using a NolA-specific antibody. Three polypeptides whose molecular weights matched those predicted from translation of the nolA sequence at the different ATG start sites were identified. Mutations to the individual translational start sites resulted in a concomitant removal of the polypeptide whose ATG site had been removed, supporting the notion that nolA can be translated from three separate ATG start sites. Consistent with this, when the nolA gene and its mutant derivatives were fused with a β-galactosidase gene to generate nolA-lacZ translational fusions, functional fusion proteins could be generated from the mutated nolA constructs as long as at least one of the three translational sites was retained within the nolA coding region.
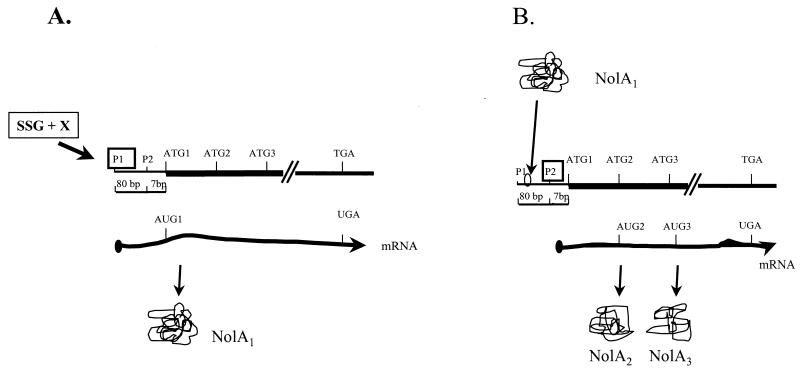
The expression of NolA1, NolA2, and NolA3 is differentially regulated, as evidenced by the data using the various nolA-lacZ fusions. For example, the expression of NolA2 and NolA3 requires NolA1, while the expression of NolA1 is activated by SSG. Several possibilities exist to account for the regulation of these proteins. For instance, the expression of NolA could be regulated either transcriptionally (e.g., by the translation of three mRNAs) or posttranscriptionally (e.g., by the translation of a single mRNA transcript via alternate ATG start codons). In this regard, results of primer extension studies showed that nolA is transcribed from two promoters, designated P1 and P2. Transcription from P1 starts 82 bases upstream of ATG1, whereas transcription from P2 starts 7 bases from the ATG1 codon. Importantly, P2 is located immediately downstream of a putative NolA-binding site characteristic of the MerR-type promoters. A possible model describing the expression of NolA1, NolA2, and NolA3 is shown in Fig. 6. This model proposes that initiation of transcription from P1 results in the production of NolA1 by translation from ATG1. Transcription from P1 is not regulated by NolA1, as indicated by the results of the nolA1-lacZ studies, as well as by the absence of a putative NolA-binding site upstream of P1. Therefore, transcription from P1 may be regulated by some unknown cellular factor(s) (X) in B. japonicum, which requires a compound in SSG. Once NolA1 is produced, this protein can bind to the P2 promoter and initiate transcription from P2. The mRNA made from P2 would have only 7 bases before ATG1, probably resulting in inefficient translation from this start site but favoring translation from ATG2 or ATG3. This model predicts that NolA2 and NolA3 production would require NolA1. Indeed, the results of the nolA-lacZ studies indicate that NolA2 and NolA3 expression is significantly reduced in the absence of NolA1. Although somewhat speculative, the model in Fig. 6 will be helpful in designing future experiments.
FIG. 6.
Proposed model for the transcriptional control of NolA1, NolA2, and NolA3 expression. (A) Transcription from P1 is regulated by an unknown factor, X, in B. japonicum, which is activated by SSG. (B) NolA1 produced from this transcript then binds to the P2 promoter to activate transcription from P2. NolA2 and NolA3 are translated from AUG2 and AUG3 on this transcript.
In addition to the regulation of NolA2 and NolA3 by NolA1, additional fine-tuning of NolA expression is apparent. For example, a region of secondary structure (ΔG = −10.4 kcal/mol) surrounding ATG2 may explain why the NolA2-LacZ expression is consistently lower than the NolA3-LacZ activity. The formation of a stem-loop structure in the mRNA surrounding this region could sequester the ATG2 initiation codon and impair ribosome binding to the initiation region. Examples of such regulation include mcrA, infC, trmD, and arsA of E. coli (27, 45, 51, 55). Additional fine-tuning of NolA expression is also observed in the ability of NolA3 to affect the expression of NolA1, as well as its own expression. Analysis of NolA1-LacZ expression, for instance, showed the levels of NolA1 expression to be higher when the fusion was expressed in strains lacking the capacity to express NolA3 (e.g., compare BJL82 and BJL823). One possible explanation for this observation is that NolA3 could interact with the inducer compound, reducing the levels of active inducer available for NolA1 activation. By modulating the levels of NolA1, the NolA3 protein could prevent the uncontrolled amplification of nolA transcription and NolA production.
Functionally, NolA1, NolA2, and NolA3 also appear to play different roles in the nodulation process. This observation was made by testing the ability of the NolA proteins to allow either B. japonicum USDA 110 or USDA 438 to nodulate cowpea or soybean. The proteins were expressed from either the wild-type promoter (e.g., in the chromosomal nolA mutants or on multicopy plasmids harboring nolA) or the constitutive trp promoter of the vector pTE3. The latter constructs are important since they allowed the study of the function of NolA2 and NolA3 independently of the need for NolA1. Of the various combinations tested, complementation was noted only when NolA1 was expressed from its own promoter and when NolA3 was expressed either from the trp promoter of pTE3 or from its own promoter in the presence of NolA1. Expression of NolA2, on the other hand, either from the wild-type promoter or from pTE3 did not promote nodulation efficiency but, rather, appeared to decrease the ability of B. japonicum to nodulate cowpea. The ability of both NolA1 or NolA3 to complement nodulation in these strains is puzzling since these two proteins appear to be redundant in function. As mentioned above, NolA1 is required for the expression of NolA3, which we have demonstrated to be sufficient to cause effective nodulation. The observation that NolA1 alone is sufficient to allow nodulation to occur may suggest that the role of NolA1 extends beyond the regulation of NolA3. For example, NolA1 could be regulating genes, in addition to nolA, that when expressed are sufficient to allow nodulation to occur. A possible target gene is nodD2, which requires nolA for its expression (16). Examination of the B. japonicum nodD2 promoter revealed a putative NolA1 DNA-binding site which was similar to the promoters of genes controlled by MerR-type regulators (1, 22, 23, 36). Using primer extension experiments, we showed that the transcriptional start site of nodD2 lay immediately downstream of this putative promoter, further supporting a role of NolA1 in nodD2 expression. This transcriptional start site is unusual in that it is 328 bp upstream of the predicted ATG start codon for NodD2. Such a situation has been found in Rhizobium meliloti, where the syrM gene is transcribed from a start site 499 bp 5′ of the ATG start codon and nodD3 is transcribed from a site 659 bp upstream of the translational start codon (6). However, the involvement of nodD2 or as yet unidentified genes is speculative and will have to be supported experimentally.
Interestingly, the levels of NolA1 expression may be critical for efficient nodulation to occur. A high level of constitutive expression of NolA1 from pTE3A23, for instance, leads to essentially no complementation of nodulation efficacy. In contrast, significant nodulation of either the soybean cultivar PI 377578 or cowpea plants is observed only when these plants are inoculated with bacterial strains that allow the expression of nolA from its own promoter. This is reflected in results obtained with both strain USDA 438(pJLDA23), which harbors nolA on a multicopy plasmid, and the chromosomal nolA mutant BJL823. Therefore, effective nodulation appears to require regulated levels of NolA1, which are probably determined by the level of specific plant signals. Such signals are found in 5-day-old etiolated seedlings and do not appear to involve flavonoids such as genistein, a known nod gene inducer in B. japonicum. At present, the nature of the plant signal is unknown. We are working to elucidate the structure of the nolA inducer in order to better understand its physiological relevance to the nodulation process. However, given that NolA belongs to the family of known MerR-type regulatory proteins, it is possible that the inducing compound is similar in nature to those associated with the MerR family (1, 22, 23, 36). For instance, it is known that the MerR regulatory proteins function in the presence of inducer molecules that are toxic. For example, MerR binds mercury, TipA binds the antibiotic thiostrepton, and SoxR responds to superoxide (22, 23, 36). By analogy, one can conjecture that the inducer compound produced by the plant is likely toxic to B. japonicum. The complex regulation of NolA allows the cell to monitor the level of this compound and, in doing so, regulate genes that allow it to withstand or counter the effects of the compound. Indeed, one possible explanation for NolA-dependent genotype-specific nodulation is that the restrictive genotypes of soybean produce this toxin in abundance and therefore only strains possessing the NolA response system can nodulate these genotypes. To date, studies have shown that nod factors, as well as the host-specific bacterial nodulation genes involved in their synthesis (e.g., nodZ in B. japonicum, nodL in Rhizobium leguminosarum, and nodH and nodL in R. meliloti), play a key role in determining host specificity (2, 5, 8, 30). Additional host determinants have also been identified; these include noeD, a negatively acting genotype-specific gene in B. japonicum USDA 110 that controls the level of acetylation of nod factors (29), and the products of the nolBTUVWX genes of Rhizobium fredii USDA 257, which restrict the ability of this strain to nodulate certain soybean cultivars. The biochemical function of the products of these latter genes is unknown (31). In the present case, the possibility that a symbiont is able to detoxify plant secreted toxins would provide an additional mechanism in determining host specific nodulation.
In conclusion, here we report that the nolA gene possesses the rare property of encoding three functionally distinct proteins. Previously, we had shown that nod gene regulation in B. japonicum is regulated by a LysR regulatory protein, NodD1, and a two-component system, NodVW. In light of our present results, it is very clear that nod gene regulation in B. japonicum is surprisingly complex. At present, the biological need for such complexity is unclear but may reflect the need of this bacterial species for developmental and ecological versatility in its interaction with its plant hosts.
ACKNOWLEDGMENTS
This work was supported by grant 96-35305-3627 from the U.S. Department of Agriculture, National Research Initiative, and grant IBN-9728281 from the National Science Foundation.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vasquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Ardourel M, Lortet G, Maillet F, Roche P, Truchet G, Prome J-C, Rosenberg C. In Rhizobium meliloti, the operon associated with nod box n5 comprises nodL, noeA and noeB, three host-range genes specifically required for the nodulation of particular Medicago species. Mol Microbiol. 1995;17:687–699. doi: 10.1111/j.1365-2958.1995.mmi_17040687.x. [DOI] [PubMed] [Google Scholar]
- 3.Attwood G T, Herrera F, Weissenstein L A, White B A. An endo-β-1,4-glucanase gene (celA) from the rumen anaerobe Ruminococcus albus 8: cloning, sequencing, and transcriptional analysis. Can J Microbiol. 1996;42:267–278. doi: 10.1139/m96-039. [DOI] [PubMed] [Google Scholar]
- 4.Banfalvi Z, Kondorosi A. Production of root hair deformation factors by Rhizobium meliloti nodulation genes in Escherichia coli: hsnD (nodH) is involved in the plant host specific modification of the NodABC factor. Plant Mol Biol. 1989;13:1–12. doi: 10.1007/BF00027330. [DOI] [PubMed] [Google Scholar]
- 5.Banfalvi Z, Niewkoop A, Schell M G, Besl L, Stacey G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol Gen Genet. 1988;214:420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- 6.Barnett M J, Rushing B G, Fisher R F, Long S R. Transcription start sites for syrM and nodD3 flank an insertion sequence relic in Rhizobium meliloti. J Bacteriol. 1996;178:1782–1787. doi: 10.1128/jb.178.7.1782-1787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergersen F J. The growth of Rhizobium in synthetic media. Aust Biol Sci. 1961;14:349–360. [Google Scholar]
- 8.Bloemberg G V, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. Nodulation protein NodL of Rhizobium leguminosarum O-acetylates lipo-oligosaccharides, chitin fragments and N-acetylglucosamine in vitro. Mol Microbiol. 1994;11:793–804. doi: 10.1111/j.1365-2958.1994.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 9.Chun J-Y, Stacey G. A Bradyrhizobium gene essential for nodulation competitiveness is differentially regulated from two promoters. Mol Plant-Microbe Interact. 1994;7:248–255. doi: 10.1094/mpmi-7-0248. [DOI] [PubMed] [Google Scholar]
- 10.Cole M A, Elkan G H. Transmissible resistance to penicillin G, neomycin and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother. 1973;4:248–253. doi: 10.1128/aac.4.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dénarié J F, Debellé F, Promé J C. Rhizobium lipo-oligosaccharide nodulation factors. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski D R. A broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egelhoff T, Long S. Rhizobium meliloti nodulation genes: identification of nodABC gene products, purification of NodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol. 1985;164:591–599. doi: 10.1128/jb.164.2.591-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin J L, Mosig G. Expression of the bacteriophage T4 DNA terminase genes 16 and 17 yields multiple proteins. Gene. 1996;177:179–189. doi: 10.1016/0378-1119(96)00299-5. [DOI] [PubMed] [Google Scholar]
- 15.Gallego-M E, Sirand-Pugnet P, Durosay P, Clouet-d’Orval B, d’Aubenton-Carafa Y, Brody E, Expert-Bezancon A, Marie J. Tissue-specific splicing of two mutually exclusive exons of the chicken beta-tropomyosin pre-mRNA: positive and negative regulations. Biochimie. 1996;78:457–465. doi: 10.1016/0300-9084(96)84752-3. [DOI] [PubMed] [Google Scholar]
- 16.Garcia M L, Dunlap J, Loh J, Stacey G. Phenotypic characterization and regulation of the nolA gene of Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1996;9:625–635. doi: 10.1094/mpmi-9-0625. [DOI] [PubMed] [Google Scholar]
- 17.Gillette W K, Elkan G H. Bradyrhizobium (Arachis) sp. strain NC92 contains two nodD genes involved in the repression of nodA and a nolA gene required for the efficient nodulation of host plants. J Bacteriol. 1996;178:2757–2766. doi: 10.1128/jb.178.10.2757-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göttfert M, Grob P, Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1990;87:2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Göttfert M, Holtzhauser D, Bani D, Hennecke H. Structural and functional analysis of the two different nodD genes in Bradyrhizobium japonicum USDA110. Mol Plant-Microbe Interact. 1992;5:257–265. doi: 10.1094/mpmi-5-257. [DOI] [PubMed] [Google Scholar]
- 20.Gu J, Stephenson C G, Iadarola M J. Affinity purification of antibodies using a 6× His-tagged antigen immobilized ion Ni-NTA. BioTechniques. 1994;17:257–262. [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes D J, Caso J L, Thompson C J. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 1993;12:3183–3191. doi: 10.1002/j.1460-2075.1993.tb05987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horowitz N H. The one gene-one enzyme hypothesis. Genetics. 1948;33:612–613. [PubMed] [Google Scholar]
- 25.Hurn B A L, Chantler S M. Production of reagent antibodies. Methods Enzymol. 1980;70:104–142. doi: 10.1016/s0076-6879(80)70044-7. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lesage P, Chiaruttini C, Graffe M, Dondon V, Milet M, Springer M. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins L35 and L20. J Mol Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 28.Loh J, Garcia M, Stacey G. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohrke S M, Day B, Kumar Kolli V S, Hancock R, Yuen J P-Y, de Souza M L, Stacey G, Carlson R, Tong Z, Hur H-G, Orf J H, Sadowsky M J. The Bradyrhizobium japonicum noeD gene: a negatively activating genotype-specific nodulation gene for soybean. Mol Plant-Microbe Interact. 1998;11:476–488. doi: 10.1094/MPMI.1998.11.6.476. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Lara I M, Blok-Tip L B, Quinto C, Garcia M L, Stacey G, Bloemberg G V, Lamers G E M, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. NodZ of Bradyrhizobium extends the nodulation host range of Rhizobium by adding a fucosyl residue to nodulation signals. Mol Microbiol. 1996;21:397–408. doi: 10.1046/j.1365-2958.1996.00644.x. [DOI] [PubMed] [Google Scholar]
- 31.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 32.Minton N P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 33.Morel-Deville F, Vachon G, Sacerdot C, Cozzone A J, Grunberg-Manago M, Cenatiempo Y. Characterization of the translational start site of IF2β, a short form of Escherichia coli initiation factor IF2. Eur J Biochem. 1990;188:605–614. doi: 10.1111/j.1432-1033.1990.tb15441.x. [DOI] [PubMed] [Google Scholar]
- 34.Murphy J P, Duggleby C J, Atkinson M A, Trowern A R, Atkinson T, Goward C R. The functional units of a peptostreptococcal protein L. Mol Microbiol. 1994;12:911–920. doi: 10.1111/j.1365-2958.1994.tb01079.x. [DOI] [PubMed] [Google Scholar]
- 35.Nieuwkoop A J, Banfavi Z, Deshmane N, Gerhold D, Schell M G, Sirotkin K M, Stacey G. A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J Bacteriol. 1987;169:2631–2638. doi: 10.1128/jb.169.6.2631-2638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Halloran T V, Frantz B, Shin M K, Ralston D M, Wright J G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- 37.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. Site directed mutagenesis of the dual translational initiation sites of clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 38.Paulson R, Jackson J, Immergluck K, Bishop J M. The Dfer gene of Drosophila melanogaster encodes two membrane-associated proteins that can both transform vertebrate cells. Oncogene. 1997;14:641–652. doi: 10.1038/sj.onc.1200875. [DOI] [PubMed] [Google Scholar]
- 39.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 40.Sadowsky M J, Cregan P B, Göttfert M, Sharma A, Gerhold D, Rodriguez-Quinones F, Keyser H H, Hennecke H, Stacey G. The Bradyrhizobium japonicum nolA gene and its involvement in the genotype-specific nodulation of soybeans. Proc Natl Acad Sci USA. 1991;88:637–641. doi: 10.1073/pnas.88.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanjuan J, Grob P, Göttfert M, Hennecke H, Stacey G. NodW is essential for the full expression of the common nodulation genes in Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1994;7:364–369. [Google Scholar]
- 44.Seol J H, Yoo S J, Kim K I, Kang M-S, Ha D B, Chung C H. The 65-kDa protein from the internal translational initiation site of the clpA gene inhibits the ATP-dependent protease Ti in Escherichia coli. J Biol Chem. 1994;269:29468–29473. [PubMed] [Google Scholar]
- 45.Shivapriya R, Prasad R, Naryanan I L, Krishnaswamy S, Dharmalingam K. Expression of the mcrA gene of Escherichia coli is regulated postranscriptionally, possibly by sequestration of the Shine Dalgarno region. Gene. 1995;157:201–207. doi: 10.1016/0378-1119(94)00746-f. [DOI] [PubMed] [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in-vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Smit G, Puvanesarajah V, Carlson R W, Barbour W M, Stacey G. Bradyrhizobium japonicum nodD1 can be specifically induced by soybean flavonoids that do not induce the nodYABSUIJ operon. J Biol Chem. 1992;267:310–318. [PubMed] [Google Scholar]
- 48.So J-S, Hodgson A L M, Haugland R, Leavitt M, Banfalvi Z, Niewkoop A J, Stacey G. Transposon-induced symbiotic mutants of Bradyrhizobium japonicum: isolation of two gene regions essential for nodulation. Mol Gen Genet. 1987;207:15–23. doi: 10.1007/BF00331485. [DOI] [PubMed] [Google Scholar]
- 49.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:61–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 50.Summers A O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki K, Wakao N, Kimura T, Sakka K, Ohmiga K. Expression and regulation of the arsenic resistance operon of Acidiphilim multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl Environ Microbiol. 1998;64:411–418. doi: 10.1128/aem.64.2.411-418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thöny-Meyer L, James P, Hennecke H. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1991;88:5001–5005. doi: 10.1073/pnas.88.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wacek T J, Brill W J. Simple, rapid assay for screening nitrogen-fixing ability in soybean. Crop Sci. 1976;15:519–523. [Google Scholar]
- 54.Wang S-P, Stacey G. Studies of the Bradyrhizobium japonicum nodD1 promoter: a repeated structure for the nod box. J Bacteriol. 1991;173:3356–3365. doi: 10.1128/jb.173.11.3356-3365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wikstrom P M, Lind L K, Berg D, Bjork G R. Importance of mRNA folding and start codon accessibility in the expression of genes in a ribosomal protein operon of Escherichia coli. J Mol Biol. 1992;224:949–966. doi: 10.1016/0022-2836(92)90462-s. [DOI] [PubMed] [Google Scholar]
- 56.Yuen J P-Y, Stacey G. Inhibition of nod gene expression in Bradyrhizobium japonicum by organic acids. Mol Plant-Microbe Interact. 1996;9:424–428. [Google Scholar]