Abstract
Because the rod structure of the flagellar basal body crosses the inner membrane, the periplasmic space, and the outer membrane, its formation must involve hydrolysis of the peptidoglycan layer. So far, more than 10 genes have been shown to be required for rod formation in Salmonella typhimurium. Some of them encode the component proteins of the rod structure, and most of the remaining genes are believed to encode proteins involved in the export process of the component proteins. Although FlgJ has also been known to be involved in rod formation, its exact role has not been understood. Recently, it was suggested that the C-terminal half of the FlgJ protein has homology to the active center of some muramidase enzymes from gram-positive bacteria. In this study, we showed that the purified FlgJ protein from S. typhimurium has a peptidoglycan-hydrolyzing activity and that this activity is localized in its C-terminal half. Through oligonucleotide-directed mutagenesis, we constructed flgJ mutants with amino acid substitutions in the putative active center of the muramidase. The resulting mutants produced FlgJ proteins with reduced enzymatic activity and showed poor motility. These results indicate that the muramidase activity of FlgJ is essential for flagellar formation. Immunoblotting analysis with the fractionated cell extracts revealed that FlgJ is exported to the periplasmic space, where the peptidoglycan layer is localized. On the basis of these results, we conclude that FlgJ is the flagellum-specific muramidase which hydrolyzes the peptidoglycan layer to assemble the rod structure in the periplasmic space.
The bacterial flagellum is a supramolecular structure which originates from the cell envelope and extends into the extracellular space. The individual flagellum consists of at least three substructures: a basal body, a hook, and a filament (1, 5). The basal body is embedded in the cell membrane and consists of a rod with an inner ring (MS ring) and two outer rings (L and P rings). The rod crosses the inner membrane, the periplasmic space, and the outer membrane. The MS ring is associated with the inner membrane, whereas the L ring connects with the outer membrane. The P ring resides in the periplasmic space and is believed to interact with the peptidoglycan layer (6). The MS ring is composed of a single species of protein, FliF, and is believed to be built first during the assembly of the basal body (1, 20, 36). Four proteins, FlgB, FlgC, FlgF, and FlgG, comprise the rod and are postulated to be transported from the cytoplasm into the periplasmic space via a flagellum-specific protein export pathway (12). These proteins assemble onto the MS ring to form the MS ring-rod structure. Subsequently, the FlgI and FlgH proteins, which are transported via the conventional signal sequence-dependent export pathway, assemble around the rod to form the P and L rings, respectively (16). Therefore, the rod is the earliest structure which should penetrate and assemble through the peptidoglycan layer. Because the peptidoglycan is a mechanically rigid structure (11, 31), it has been postulated that breakdown of peptidoglycan should be a prerequisite for rod formation (7, 9). Peptidoglycan hydrolases such as muramidase and amidase (10, 33) are presumed to be responsible for this process.
In addition to the structural genes for the rod subunit proteins, more than 10 genes are known to be required for rod formation (19, 35). Most of them are believed to encode the component proteins of the flagellum-specific export apparatus. They include flhA, fliI, and fliH (37). Mutants defective in any one of these genes produce the MS ring lacking the rod and other flagellar substructures (19). Because flgJ mutants also produce the MS ring lacking the rod (19), FlgJ must be essential for the rod assembly. Through the comparative study of the primary structures of proteins, Joris et al. (17) demonstrated that the C-terminal half of FlgJ has homology to the catalytic domain of two species of muramidase, autolysin from Streptococcus faecalis and muramidase 2 from Enterococcus hirae. Recently, the same region of FlgJ was also shown to be homologous to the Lactococcus lactis AcmA protein, which acts as a muramidase essential for cell separation (4). Especially, two amino acids, aspartic acid and glutamic acid, which are conserved in the active center of the enzymes of the muramidase family, are also present in the corresponding positions of FlgJ (Glu-223 and Asp-248) (Fig. 1A). According to these observations, Dijkstra and Keck (7) proposed a hypothesis that FlgJ may be responsible for the penetration of the peptidoglycan by the rod structure. However, this remained to be proven experimentally. This work was carried out to address this issue.
FIG. 1.
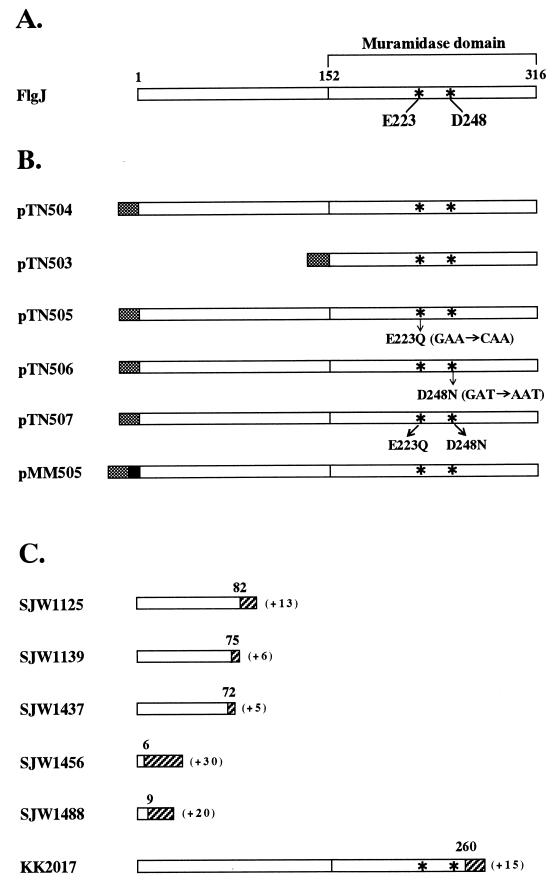
Structures of the wild-type (A), His-tagged (B), and mutant (C) FlgJ proteins. Asterisks indicate the amino acids which are conserved in the putative active center of the enzymes of the muramidase family. Numbers above the bars indicate amino acid residues from the N terminus. Shaded and solid boxes indicate the His10 and FLAG tags, respectively, in the FlgJ proteins encoded by the recombinant plasmids. Hatched areas represent amino acids created by the frameshift mutations, and the numbers of amino acids added are shown in parentheses. Because all the Tn10-induced flgJ mutants (KK strains) carry Tn10 inserted in the identical site, only the structure of the FlgJ protein encoded by KK2017 is shown.
We showed that the purified FlgJ protein has a peptidoglycan-hydrolyzing activity in its C-terminal half. The flgJ mutants with amino acid substitutions in the putative active center of muramidase produced FlgJ proteins with reduced enzymatic activity and showed poor motility. We showed further that FlgJ is exported into the periplasmic space. These results suggest that FlgJ may be the flagellum-specific muramidase which hydrolyzes the peptidoglycan layer to construct the rod structure in the periplasmic space. On the basis of this and other available information, we propose a model of rod assembly in the periplasmic space.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in the present study are listed in Table 1. SJW strains are spontaneously induced flgJ or flhA mutants isolated by S. Yamaguchi, Meiji University, whereas KK strains are Tn10-induced flgJ mutants isolated by one of us (K.K.). Luria broth, Luria agar, motility agar plate, and minimal medium were prepared as described previously (21). Unless otherwise specified, ampicillin was used at 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| HMS174(DE3) | T7 expression host | 34 |
| S. typhimurium | ||
| KK2017 | flgJ::Tn10 | 25 |
| KK2057 | flgJ::Tn10 | 25 |
| KK2063 | flgJ::Tn10 | 25 |
| KK2083 | flgJ::Tn10 | 25 |
| KK2802 | flgJ::Tn10 | 25 |
| KK2813 | flgJ::Tn10 | 25 |
| KK2942 | flgJ::Tn10 | 25 |
| SJW1125 | flgJ1669 | 35 |
| SJW1139 | flgJ1683 | 35 |
| SJW1364 | flhA2014 | 24 |
| SJW1437 | flgJ2076 | 20 |
| SJW1456 | flgJ | 19 |
| SJW1488 | flgJ | 19 |
| Plasmids | ||
| pET19b | T7 expression vector, His tag, Apr | Novagen |
| pET-FLAG-19b | pET19b, His-FLAG tag | 8 |
| pLysS | T7 lysozyme, Cmr | 34 |
| pTrc99A | tac expression vector, Apr | 2 |
| pKK1490 | pBR322 flgIJKL+, Apr | 23 |
| pTN503 | pET19b His-FlgJΔN | This study |
| pTN504 | pET19b His-FlgJ | This study |
| pTN505 | pET19b His-FlgJ(E223Q) | This study |
| pTN506 | pET19b His-FlgJ(D248N) | This study |
| pTN507 | pET19b His-FlgJ(E223Q-D248N) | This study |
| pMM502 | pET-FLAG-19b His-FLAG-FlgJ | This study |
| pMM505 | pTrc99A His-FLAG-FlgJ | This study |
Motility assay.
Motility phenotypes of the cells were detected by observing the formation of spreading colonies (swarms) on motility agar plates (21).
DNA manipulations.
Procedures for manipulation of DNA in vitro and transformation were performed as described previously (26). Restriction enzymes and T4 DNA ligase were purchased from Takara, Toyobo, Nippon Gene, or New England Biolabs. PCR amplification was carried out with DNA thermal cycler model 480 (Perkin-Elmer) as specified by the manufacturer. Taq DNA polymerase was purchased from Promega or Boeringer Mannheim. The DNA sequence was determined by the dideoxy chain termination method with the ABI 373S DNA sequencing system (Perkin-Elmer). Oligonucleotide primers used in the present study were purchased from Amersham-Pharmacia or synthesized with a model 393 DNA/RNA synthesizer (Perkin-Elmer). Their sequence designs are summarized in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea |
|---|---|
| PGJ1 | GGCATATGATCGGAGACGGTAAATTGC |
| PGJ2 | GGGGATCCCGACTACGTGGACTTGAGCA |
| PGJ3 | GCTCTCCCATATGAGTAAAGACTTTCTGGC |
| PGJ4 | GCCATTTTCGTATTGAGTGGTGGTG |
| PGJ5 | CACCACCACTCAATACGAAAATGGC |
| PGJ6 | CAGCGCGACATAATTCGATAATGCC |
| PGJ7 | GGCATTATCGAATTATGTCGCGCTG |
| PGJ8 | CTACAGAGCGTGCGTTCCAG |
| PGJ9 | GGAAATCATCATATGATCGGAGAC |
| PGJ10 | AGCGACTACGGATCCTTGAGCAAT |
The sequence is written in the 5′-to-3′ direction. The underlined nucleotides are the substitution sites used in the oligonucleotide-directed mutagenesis.
Plasmid construction and oligonucleotide-directed mutagenesis.
The T7 expression vector pET19b was used for overproduction of the wild-type and mutant FlgJ proteins fused to a His10 tag at their N terminus. Plasmid pKK1490 contains the flgIJKL genes of Salmonella typhimurium (23). The flgJ gene was amplified by PCR with pKK1490 as a template and two oligonucleotides, PGJ1 and PGJ2, as primers. The amplified product was digested with NdeI and BamHI and inserted into the corresponding site of pET19b to obtain pTN504. This plasmid specifies the entire FlgJ protein fused to the His tag at its N terminus (Fig. 1B). The 3′ half of the coding region of the flgJ gene was also amplified with the PGJ3 and PGJ2 primers and inserted in the same way as above into pET19b to obtain pTN503. This plasmid specifies the C-terminal half of the FlgJ protein fused to the His tag at its N terminus (Fig. 1B).
To obtain mutant plasmids which specify FlgJ proteins with amino acid substitutions in the putative active center of muramidase (Fig. 1B), oligonucleotide-directed mutagenesis was performed by multiple rounds of PCR. Plasmid pTN505 specifying the E223Q mutant FlgJ protein in which Glu-223 was replaced with Gln was constructed as follows. Plasmid pKK1490 was used as a template for the first round of PCR. By using two sets of primers, PGJ1 plus PGJ4 and PGJ5 plus PGJ2, 0.69- and 0.33-kb DNA fragments, which correspond to N-terminal and C-terminal regions of FlgJ, respectively, were amplified. These two fragments were mixed and used as templates for the second round of PCR with the PGJ1 and PGJ2 primers. The amplified 0.99-kb fragment was digested with NdeI and BamHI and inserted into the corresponding site of pET19b to yield pTN505. Plasmid pTN506 specifying the D248N mutant FlgJ protein in which Asp-248 was replaced with Asn was constructed in the same way as above, except that primers PGJ4 and PGJ5, used for the construction of pTN505 in the first round of PCR, were replaced with primers PGJ6 and PGJ7. Plasmid pTN507, which specifies the E223Q-D248N mutant FlgJ protein, was constructed the same way as pTN506, except that pTN505 was used as a template for the first round of PCR.
To obtain a hybrid gene specifying the FLAG-tagged FlgJ protein, the wild-type flgJ gene was amplified by PCR with primers PGJ9 and PGJ10. The amplified product was digested with NdeI and BamHI and inserted into the corresponding site of pET-FLAG-19b to obtain pMM502. A 1-kb NcoI-BamHI fragment was excised from pMM502 and inserted into the corresponding site of pTrc99A to yield pMM505 (Fig. 1B). This plasmid carries an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible gene encoding the entire FlgJ protein fused to the His-FLAG tag at its N terminus.
Overexpression and purification of the His-tagged FlgJ protein.
Escherichia coli HMS174(DE3) cells harboring pLysS and one of the His-tagged fusion plasmids were grown overnight at 37°C in 1 ml of Luria broth containing 200 μg of ampicillin per ml and 25 μg of chloramphenicol per ml. The whole culture was inoculated into 500 ml of the same medium and incubated at 37°C with gentle shaking. When the cell growth reached mid-log phase, IPTG was added at a final concentration of 1 mM and incubation was continued for a further 3 h. The cells were harvested by centrifugation and stored overnight at −80°C. The cell pellet was thawed, suspended in 40 ml of binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and disrupted by sonication. The inclusion bodies containing the fused proteins were collected by low-speed centrifugation and resuspended in 40 ml of binding buffer containing 6 M urea. After insoluble materials were removed by centrifugation, the supernatant was subjected to affinity chromatography by loading on the His · Bind resin column (Novagen) as specified by the manufacturer. The column was washed with binding buffer containing 6 M urea and then with washing buffer (60 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]) containing 6 M urea. His-tagged proteins were eluted with elution buffer (0.5 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]) containing 6 M urea. Each fraction was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (29). Fractions containing His-tagged proteins were pooled and dialyzed three times against 500 ml of phosphate-buffered saline containing 6 M urea. Protein concentrations were determined with a protein assay kit (Bio-Rad).
Zymogram analysis.
To detect the peptidoglycan-hydrolyzing activities of the purified proteins, a zymogram analysis was performed essentially by the methods described by Potvin et al. (32) and Buist et al. (4). Purified proteins were electrophoretically separated in an SDS-polyacrylamide gel containing 0.2% (wt/vol) autoclaved, lyophilized Micrococcus lysodeikticus ATCC 4698 cells (Sigma). After electrophoresis, the gel was soaked for 48 h with gentle shaking in renaturation buffer (25 mM Tris-HCl [pH 7.5], 1% [vol/vol] Triton X-100) at 37°C with six to eight changes of the buffer to remove SDS and allow the protein to be renatured. To obtain better contrast, the gel was stained with 1% (wt/vol) methylene blue (Nacalai tesque) in 0.01% (wt/vol) KOH. After being destained with deionized water, the gel was photographed. If a protein had peptidoglycan-hydrolyzing activity, an unstained clear band appeared in the blue background at the position corresponding to the protein band.
Immunoblotting analysis of the proteins from the periplasmic contents and the culture supernatants.
The flgJ or flhA mutant cells harboring pMM505 were grown at 37°C in minimal medium containing 100 μg of ampicillin per ml. When the cell density reached an optical density at 600 nm of 1.0 to 1.2, 1.5 ml of the culture was centrifuged and the cells and culture supernatants were collected separately. The cells were washed twice with TN buffer (10 mM Tris-HCl [pH 8.0], 0.3 M NaCl), suspended in 400 μl of sucrose buffer (20% sucrose, 100 mM Tris-HCl [pH 8.0], 0.5 mM EDTA), and incubated at room temperature for 20 min. After centrifugation, the cells were resuspended in 750 μl of 0.5 mM MgCl2 and placed on ice for 10 min to release the periplasmic fraction (30). The proteins in the supernatant and periplasmic fractions were concentrated by precipitation with 10% trichloroacetic acid and separated by SDS-PAGE as described previously (15). After electrophoresis, the proteins in the gel were transferred to a nitrocellulose membrane. For detection of the FLAG-tagged protein, the membrane was probed with anti-FLAG M2 monoclonal antibody (Eastman Kodak) with the ECL immunoblotting detection kit (Amersham-Pharmacia) as specified by the manufacturer. To confirm that the periplasmic contents were successfully obtained, immunoblotting was performed with antibody against β-lactamase (5Prime→3Prime Inc.) (data not shown).
RESULTS
Sequence analysis of the flgJ mutants.
The flgJ gene belongs to the hook-basal body operon (the flgB operon) and is transcribed last in that operon (25). The wild-type flgJ gene encodes a protein of 316 amino acids (16). In this study, we used five spontaneously induced and seven Tn10-induced flgJ mutants. All the spontaneously induced mutants formed compact colonies on motility agar plates (data not shown), indicating that they totally lack the ability to produce flagella. On the other hand, all the Tn10-induced mutants formed bushy colonies on motility agar plates (data not shown), indicating that they retain a residual activity to produce flagella. To determine the sequence changes which result in these mutant phenotypes, we amplified the flgJ genes from the chromosomal DNAs of these mutants by PCR with primers PGJ8 and PGJ2 and determined the DNA sequences of the amplified products. All the spontaneously induced mutants were found to carry 1- to 347-bp deletions near the 5′ end of the coding region, which result in frameshift mutations and create new termination codons just downstream of the mutation sites. These changes may cause premature termination of the FlgJ protein, and thus these mutants may encode only N-terminal small portions of the FlgJ protein which lack the region homologous to muramidase (Fig. 1C). All the Tn10-induced mutants carry Tn10 inserted at an identical site near the 3′ end of the coding region. This insertion disrupts the FlgJ protein near the C terminus, and the resulting truncated FlgJ protein retains the active center of the putative muramidase domain (Fig. 1C). This may account for the leaky phenotype of the Tn10-induced flgJ mutants used here.
Peptidoglycan-hydrolyzing activity of the FlgJ protein.
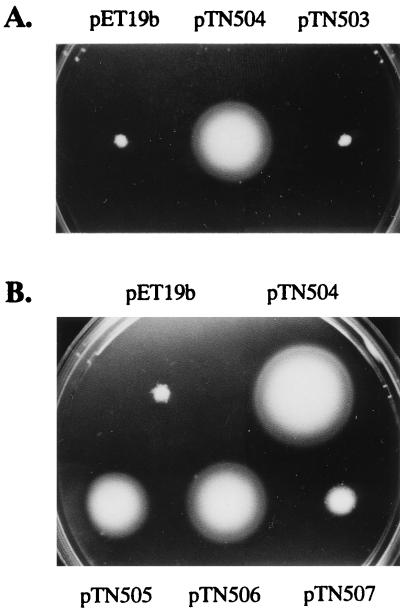
Plasmid pTN504 encodes an entire FlgJ protein fused to the His tag at the N terminus (His-FlgJ). When introduced into the flgJ mutants SJW1488 and KK2017, this plasmid was able to restore motility to the cells (Fig. 2A). This indicates that the fused gene is expressed even in the cells lacking the gene for T7 RNA polymerase and that the fused protein is functional in vivo.
FIG. 2.
Motility recovery of the flgJ mutants by the His-tagged FlgJ-encoding plasmids. (A) Plasmids were introduced into SJW1488 and KK2017 by transformation. Single colonies of the resulting transformants were stabbed onto motility agar plates and incubated for 4 h at 37°C. In this figure, only the result with SJW1488 is shown. The same result was obtained with KK2017. (B) Experiments were carried out as for panel A, but only SJW1488 was used.
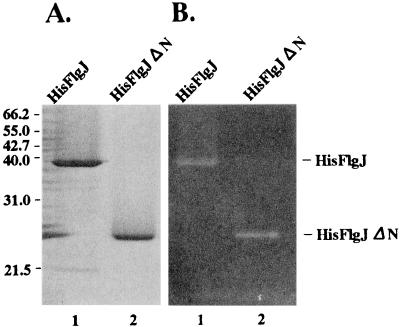
The His-FlgJ fusion protein was overproduced and recovered from inclusion bodies as described in Materials and Methods. After being solubilized with 6 M urea, the protein was purified by affinity chromatography with His · Bind resin. The purified protein gave a single band in SDS-PAGE (Fig. 3A). The estimated molecular mass of the protein in the gel was 37 kDa, which is in good agreement with its calculated molecular mass (37,152 Da). To detect the muramidase activity with the purified protein, we tried to solubilize the protein in the absence of urea. However, we did not obtain enough protein in a soluble form under any conditions we used. Therefore, we decided to apply the zymogram analysis technique to the protein separated in the SDS-polyacrylamide gel containing M. lysodeikticus cells (see Materials and Methods). We found a clear band in the position corresponding to the purified protein on the gel (Fig. 3B). This indicates that the His-FlgJ fusion protein has the activity to hydrolyze peptidoglycan. Because FlgJ has homology to known muramidase enzymes, we believe that the peptidoglycan-hydrolyzing activity of FlgJ is due to its muramidase activity. This is discussed below.
FIG. 3.
SDS-PAGE and zymogram analyses of the His-tagged FlgJ proteins. (A) The purified proteins were separated in an SDS–12% polyacrylamide gel. After electrophoresis, the gel was stained with 0.25% Coomassie brilliant blue. The molecular masses of the marker proteins are indicated in kilodaltons on the left. (B) The purified proteins were separated in an SDS–12% polyacrylamide gel containing M. lysodeikticus cells. After electrophoresis, the gel was treated as described in Materials and Methods for the zymogram analysis. Equal molar amounts of protein (54 pmol) were applied to each lane. The proteins used were His-FlgJ (lane 1) and His-FlgJΔN (lane 2).
As described above, the region homologous to muramidase is localized in the C-terminal half of FlgJ (Fig. 1A). To examine whether the C-terminal half of FlgJ suffices for the peptidoglycan-hydrolyzing activity, we constructed a hybrid plasmid, pTN503, which specifies the C-terminal half of the FlgJ protein fused to the His tag at its N terminus (His-FlgJΔN) (Fig. 1B), and purified the fusion protein by procedures similar to those applied to His-FlgJ. When the purified protein was examined for peptidoglycan-hydrolyzing activity by the zymogram analysis, a clear band was observed at the position corresponding to the purified protein on the gel (Fig. 3B). Therefore, we conclude that the N-terminal half of FlgJ is dispensable for the muramidase activity. When the same molar amounts of the purified proteins were subjected to zymogram analysis, the clear area seemed to be slightly larger for His-FlgJΔN than for His-FlgJ (Fig. 3B).
When introduced into the flgJ mutants SJW1488 and KK2017, pTN503 was unable to restore motility to the mutants (Fig. 2A). This suggests that the muramidase domain alone does not suffice for FlgJ to support flagellation of the cells.
Mutants with amino acid substitutions at the active center of muramidase.
Next, we performed the following experiments to show that the muramidase activity of FlgJ is actually involved in the flagellar assembly process of the cell. As mentioned above, the active center of the muramidase activity is predicted to be located around Glu-223 and Asp-248 of FlgJ. By using oligonucleotide-directed mutagenesis, these residues were replaced with Gln and Asn, respectively. We constructed three mutant plasmids, pTN505, pTN506, and pTN507, which specify altered FlgJ proteins with substitutions Glu to Gln, Asp to Asn, and both of them, respectively (Fig. 1B). In this study, these mutant proteins were called E223Q, D248N, and E223Q-D248N, respectively.
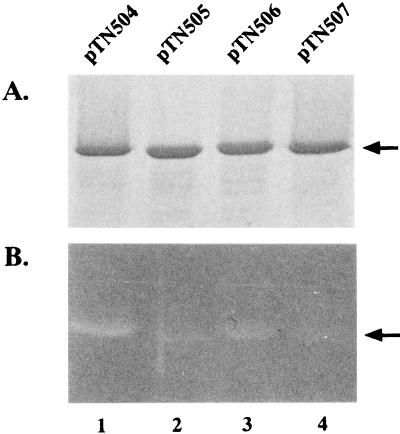
These mutant proteins were all purified as the His-tagged fusion proteins and examined for their peptidoglycan-hydrolyzing activity by zymogram analysis (Fig. 4). They all showed greatly reduced activity of the enzyme, indicating that these amino acids are important for FlgJ to show muramidase activity. However, residual activities of the enzyme were detectable in all the mutant proteins. The observed activity was highest in the D248N protein and lowest in the E223Q-D248N protein.
FIG. 4.
SDS-PAGE (A) and zymogram (B) analyses of the FlgJ proteins with amino acid substitutions in the putative active center of muramidase. The procedures used were the same as those described in the legend to Fig. 3. The arrows indicate the positions of the His-tagged FlgJ proteins. The proteins used were His-FlgJ (lane 1), His-FlgJ(E223Q) (lane 2), His-FlgJ(D248N) (lane 3), and His-FlgJ(E223Q-D248N) (lane 4).
These mutant plasmids were introduced into the flgJ mutant SJW1488 and examined for their ability to restore motility to the cells (Fig. 2B). All the mutant plasmids showed reduced abilities compared with pTN504, which specifies the wild-type FlgJ protein. Especially, pTN507 showed severely attenuated ability to restore motility. The sizes of the swarms formed by the flgJ mutant cells harboring these plasmids were in good agreement with the magnitude of the enzymatic activity remaining in the FlgJ mutant proteins encoded by the plasmids. This strongly suggests that the muramidase activity of FlgJ is responsible for flagellar morphogenesis of S. typhimurium.
Export of FlgJ into the periplasmic space.
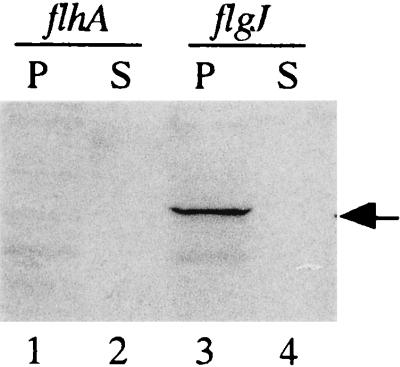
The peptidoglycan layer resides in the periplasmic space. Therefore, if FlgJ actually acts as a muramidase, it must be exported into the periplasmic space. To test this possibility, we examined FlgJ export from IPTG-treated cells of the flgJ mutant, SJW1437, harboring pMM505. Because this plasmid encodes the FLAG-tagged FlgJ protein, this fusion protein can be detected immunologically with the anti-FLAG antibody. Proteins in the periplasm and the culture supernatant were thus analyzed by Western blotting with this antibody as described in Materials and Methods. The FLAG-tagged protein was detected in the periplasmic contents but not in the culture supernatant (Fig. 5). This result indicates that FlgJ is exported into the periplasmic space but not across the outer membrane.
FIG. 5.
Export assay of FlgJ. Proteins from the periplasmic content (P) and culture supernatant (S) were prepared from the 1 mM IPTG-induced culture of the cells harboring pMM505. After separation by SDS-PAGE, the proteins were transferred to a nitrocellulose membrane and visualized immunologically with anti-FLAG antibody. The arrow indicates the position of the His-FLAG-tagged FlgJ protein. The strains used were SJW1364 (flhA) (lanes 1 and 2) and SJW1437 (flgJ) (lanes 3 and 4).
Because FlgJ lacks a cleavable signal peptide at its N terminus (16), its export is expected to be via the flagellum-specific export pathway. To test this possibility, we examined the export of the FLAG-tagged FlgJ protein in the flhA mutant SJW1364, because FlhA is believed to be one of the components of the flagellum-specific export apparatus (37). As expected, the FLAG-tagged protein was not detected either in the periplasmic contents or in the culture supernatant (Fig. 5).
DISCUSSION
In this study, we demonstrated the peptidoglycan-hydrolyzing activity of the flgJ gene product. Mutations which affected this enzymatic activity also affected the motility of the cell, indicating that this enzymatic activity is involved in the process of flagellar formation. Because FlgJ is essential for the formation of the rod and because the rod must penetrate and assemble through the peptidoglycan layer, our results strongly suggest that the peptidoglycan-hydrolyzing activity of FlgJ is responsible for the penetration of the peptidoglycan layer by the rod.
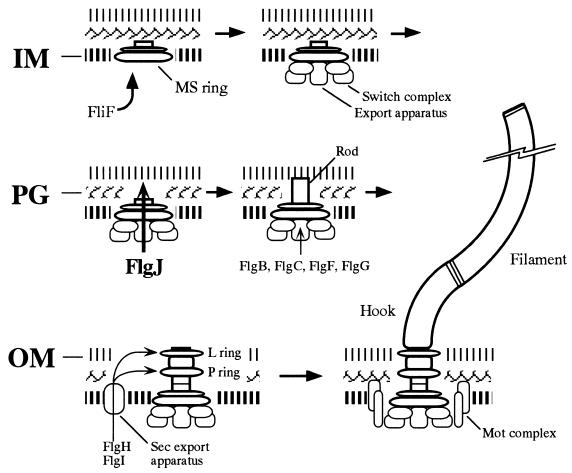
On the basis of the above results and other available information, we propose a model for the flagellar morphogenetic pathway with special emphasis on the processes of crossing three envelope barriers by the rod (Fig. 6). First, the FliF protein integrates into the inner membrane and self-assembles to form the MS ring (20, 36), which together with the flagellum-specific export apparatus makes a selective pore through the inner membrane. Second, the muramidase activity of FlgJ hydrolyzes the peptidoglycan layer to make a hole for rod elongation, which allows the rod to penetrate the peptidoglycan. This process may also be involved in P-ring formation, which is postulated to occur in the peptidoglycan layer (6). Third, formation of the L ring in the outer membrane around the tip of the rod makes a hole through the outer membrane (16). Through these processes, the rod can cross all the envelope barriers. Finally, the hook and the filament can be formed in the extracellular space to make a mature flagellum.
FIG. 6.
Model of the flagellar assembly pathway with special emphasis on the processes involved in the crossing of three envelope barriers by the rod structure. The rod is composed of at least four proteins, FlgB, FlgC, FlgF, and FlgG, which are exported into the periplasmic space via the flagellum-specific export pathway. Rod assembly proceeds by the formation of holes in the inner membrane (IM) by FliF (36), in the peptidoglycan (PG) by FlgJ (this study), and in the outer membrane (OM) by FlgH (16). Details are described in the text. Other assembly processes are drawn on the basis of the model described previously (1, 19).
We showed that the domain for the peptidoglycan-hydrolyzing activity is confined to the C-terminal half, which has homology to known muramidase enzymes. Mutations which replaced amino acids at the putative active center of the muramidase reduced the peptidoglycan-hydrolyzing activity, suggesting that the peptidoglycan-hydrolyzing activity of FlgJ is due to its muramidase activity.
Although the N-terminal half of FlgJ is dispensable for the peptidoglycan-hydrolyzing activity, the truncated FlgJ protein lacking the N-terminal half could not support flagellation of the cells. This suggests that the N-terminal region is also required for FlgJ to function in vivo.
Because the peptidoglycan layer resides in the periplasmic space, FlgJ must be transported across the inner membrane before or at the time of the rod formation. However, unlike other known muramidases (4, 17), FlgJ has no signal sequence at its N terminus (16). Most of the flagellar proteins lack signal sequence and are believed to be exported via the flagellum-specific export pathway which resides within the flagellar structure (12, 13, 14, 18, 22, 27). Their export signals are postulated to exist in their N-terminal regions (15, 28). In this study, we showed evidence suggesting that FlgJ may be exported into the periplasmic space via the flagellum-specific export pathway. Therefore, its N-terminal region is likely to contain information essential for its own export. This may be one of the reasons why the N-terminal half is indispensable. However, we cannot exclude the possibility that the N-terminal half plays an additional essential role in the flagellar assembly process.
We would like to consider regulatory roles of the N-terminal half of FlgJ on the muramidase activity. To avoid undesirable cell lysis caused by random hydrolysis of peptidoglycan, muramidases such as autolysin are known to contain information directing themselves to the sites where they should act (3, 17). By analogy to this, the action of FlgJ should be restricted to the area where the flagellum will be formed. In the known muramidases, amino acid sequences conveying this information are located in their C-terminal regions (3, 17). However, FlgJ lacks regions homologous to these amino acid sequences and instead contains a large N-terminal region (17). We suppose that the N-terminal region of FlgJ may contain information for this localized hydrolysis. Specific binding of the N-terminal region of FlgJ to the tip of the MS ring or the growing rod may ensure the localization of the FlgJ protein. Interestingly, the truncated FlgJ protein lacking its N-terminal half showed slightly greater peptidoglycan-hydrolyzing activity than did the intact FlgJ protein in the zymogram analysis (Fig. 3B). This suggests that the N-terminal portion may play some inhibitory role on the muramidase activity. Binding of the N-terminal region of FlgJ to the tip of the MS ring or the growing rod may relieve this inhibition to promote the muramidase activity.
ACKNOWLEDGMENTS
We thank Shigeru Yamaguchi for providing bacterial strains.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, and Science of Japan (to K.K.) and by U.S. Public Health Service grant AI12202 (to R.M.M.).
REFERENCES
- 1.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 2.Amann E, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–314. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17:4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePamphilis M L, Adler J. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971;105:384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePamphilis M L, Adler J. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cytoplasmic membrane. J Bacteriol. 1971;105:396–407. doi: 10.1128/jb.105.1.396-407.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan F, Macnab R M. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 9.Fein J E. Possible involvement of bacterial autolytic enzymes in flagellar morphogenesis. J Bacteriol. 1979;137:933–946. doi: 10.1128/jb.137.2.933-946.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 11.Holtje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab R M. FlgB, FlgC, FlgF, and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 13.Homma M, DeRosier D J, Macnab R M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 14.Iino T. Assembly of Salmonella flagellin in vitro and in vivo. J Supramol Struct. 1974;2:372–384. doi: 10.1002/jss.400020226. [DOI] [PubMed] [Google Scholar]
- 15.Iyoda S, Kutsukake K. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol Gen Genet. 1995;249:417–424. doi: 10.1007/BF00287103. [DOI] [PubMed] [Google Scholar]
- 16.Jones C J, Homma M, Macnab R M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joris B, Englebert S, Chu C-P, Kariyama R, Daneo-Moore L, Shockman G D, Ghuysen J-M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett. 1992;91:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- 18.Joys T M. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J Biol Chem. 1985;260:15758–15761. [PubMed] [Google Scholar]
- 19.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 20.Kubori T, Yamaguchi S, Aizawa S-I. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J Bacteriol. 1997;179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutsukake K, Doi H. Nucleotide sequence of the flgD gene of Salmonella typhimurium which is essential for flagellar hook formation. Biochim Biophys Acta. 1994;1218:443–446. doi: 10.1016/0167-4781(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 23.Kutsukake K, Ide N. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol Gen Genet. 1995;247:275–281. doi: 10.1007/BF00293195. [DOI] [PubMed] [Google Scholar]
- 24.Kutsukake K, Iino T, Komeda Y, Yamaguchi S. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1980;178:59–67. doi: 10.1007/BF00267213. [DOI] [PubMed] [Google Scholar]
- 25.Kutsukake K, Ohya Y, Yamaguchi S, Iino T. Operon structure of flagellar genes in Salmonella typhimurium. Mol Gen Genet. 1988;214:11–15. doi: 10.1007/BF00340172. [DOI] [PubMed] [Google Scholar]
- 26.Kutsukake K, Iyoda S, Ohnishi K, Iino T. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 1994;13:4568–4576. doi: 10.1002/j.1460-2075.1994.tb06778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwajima G, Asaka J-I, Fujiwara T, Fujiwara T, Node K, Kondo E. Nucleotide sequence of the hag gene encoding flagellin of Escherichia coli. J Bacteriol. 1986;168:1479–1483. doi: 10.1128/jb.168.3.1479-1483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwajima G, Kawagishi I, Homma M, Asaka J-I, Kondo E, Macnab R M. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci USA. 1989;86:4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minamino T, Iino T, Kutsukake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nossal N G, Heppel L A. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 31.Park J T. The murein sacculus. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 32.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214:241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 33.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishing Co.; 1994. pp. 131–166. [Google Scholar]
- 34.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki T, Iino T, Horiguchi T, Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978;133:904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno T, Oosawa K, Aizawa S-I. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 37.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]