Abstract
Applying methods in natural language processing on electronic health records (EHR) data is a growing field. Existing corpus and annotation focus on modeling textual features and relation prediction. However, there is a paucity of annotated corpus built to model clinical diagnostic thinking, a process involving text understanding, domain knowledge abstraction and reasoning. This work introduces a hierarchical annotation schema with three stages to address clinical text understanding, clinical reasoning, and summarization. We created an annotated corpus based on an extensive collection of publicly available daily progress notes, a type of EHR documentation that is collected in time series in a problem-oriented format. The conventional format for a progress note follows a Subjective, Objective, Assessment and Plan heading (SOAP). We also define a new suite of tasks, Progress Note Understanding, with three tasks utilizing the three annotation stages. The novel suite of tasks was designed to train and evaluate future NLP models for clinical text understanding, clinical knowledge representation, inference, and summarization.
Keywords: clinical natural language processing, electronic health record, clinical reasoning, corpus
1. Introduction
Patients in the hospital have a multidisciplinary team of physicians, nurses, and support staff who attend to their care. As part of this care, providers input daily progress notes to update the diagnoses and treatment plan, and to document changes in the patient’s health status. The electronic health record (EHR) contains these daily progress notes, and they are one of the most frequent note types that carry the most relevant and viewed documentation of a patient’s care (Brown et al., 2014). While EHRs are intended to provide efficient care, they are still riddled with problems of note bloat (copying and pasting), information overload (automatically embedded data and administrative documentation), and poorly organized notes that overwhelm physicians and lead to burnout and, ultimately, inefficient care (Shoolin et al., 2013).
Applying methods in natural language processing to the EHR is a growing field with many potential applications in clinical decision support and augmented care. Corpus and annotation on EHR data are created to model semantic features and relation through linguistic cues, including relation extraction (Mowery et al., 2008), named entity recognition (Wang, 2009; Patel et al., 2018; Lybarger et al., 2021), question answering (Pampari et al., 2018; Raghavan et al., 2021), natural language inference (Romanov and Shivade, 2018), etc. However, few corpora have been built to model clinical thinking, especially about clinical diagnostic reasoning, a process involving clinical evidence acquisition, generating hypothesis, integration and abstraction over medical knowledge and synthesizing a conclusion in the form of a diagnosis and treatment plan (Bowen, 2006). In this work, we introduce a hierarchical annotation with three stages addressing clinical text understanding, reasoning and abstraction over evidence, and diagnosis summarization. The annotation guidelines were designed and developed by physicians with clinical informatics expertise in conjunction with computational linguistic experts to model the healthcare provider decision making process. Our annotations were built on top of the Medical Information Mart for Intensive Care-III (MIMIC-III), a publicly available English-language EHR (Johnson et al., 2019).1
Meanwhile, there exists only a handful of language benchmark tasks in medicine such as the National Library of Medicine BLUE Benchmark (Peng et al., 2019), Biomedical Language Understanding and Reasoning Benchmark (Gu et al., 2021), and shared tasks hosted through Harvard’s National NLP Clinical Challenges (N2C2).2. Some of these benchmarks contain non-EHR data (Peng et al., 2019; Gu et al., 2021). Many of the EHR-based benchmarks are time-insensitive, such as discharge summaries (Mullenbach et al., 2021; Uzuner et al., 2008), radiology reports (Abacha et al., 2021; Peng et al., 2018). They also have a strong focus on modeling clinical language instead of potentials for clinical applications with a practitioner-derived focus (Hultman et al., 2019). Recent advances in large scale language modeling enables pre-training on massive corpora and fine-tuning for in-domain tasks, such as transfer learning for BERT (Devlin et al., 2019; He et al., 2020) with ClinicalBERT (Alsentzer et al., 2019; Hao et al., 2020). These pre-trained models are evaluated on existing benchmarks with a great emphasis on domain knowledge representation, but few are tested for clinical reasoning and inference. Better benchmarks for clinical use are needed to evaluate clinical NLP systems that model clinical reasoning and thinking.
The goal of the proposed research is to build a suite of tasks for the following: (1) identify the relevant sections of the daily progress note - Subjective, Objective, Assessment and Plan format (SOAP Note); (2) identify problems/diagnoses from a set of daily progress notes; and (3) accurately discriminate related plans for an assessment in the Assessment and Plan section (A/P of the SOAP note) of daily progress notes. We refer to our suite of tasks as Progress Note Understanding, which covers automatic section segmentation, Assessment and Plan reasoning, and diagnoses summarization. The three tasks are built from different levels of text units and annotation stages.
Our contribution includes: (1) a novel and hierarchical annotation protocol jointly designed by NLP researchers and physicians; (2) annotated data for training and evaluating NLP models on clinical text understanding, reasoning and text generation; (3) a new suite of tasks proposed to develop and evaluate NLP models for clinical applications that help to improve efficiency of bedside care and reduce medical errors.
2. Background
The daily progress note follows a specific format with four major components: Subjective, Objective, Assessment, and Plan (SOAP). SOAP note documentation is engrained in medical school curriculum as well as other training curricula, developed by Larry Weed, MD, known as the father of the problem-oriented medical record and inventor of the ubiquitous SOAP daily progress note (Weed, 1964). It provides an easily recognizable template and pattern for systematic documentation, which lends itself also to reliable annotation. The main purpose of SOAP documentation is to record the patients’ information, including recent events in their care and active problems in a readable and structured way so the patients’ diagnoses are readily identified. This is separate from other note types, such as the admission note (history and physical, also known as H&P) or discharge summary, which are not documented daily. The progress note is a time-series data collection with a reproducible format on all hospitalized patients, and it is the most frequently viewed note by care teams (Hripcsak et al., 2011). As a patient’s condition becomes more severe with an increasing number of interactions with providers, the progress note may also increase in length.
Figure 1 shows an example of a daily progress note with the sections and their corresponding subsections labeled. Every subsection in the progress note belongs to a section of SOAP. Subjective includes sections of free text describing patients’ symptoms, conditions, daily changes in care, etc. Objective contains sections of structured data such as lab results, vital sign, radiology reports. Assessment and Plan sections are considered by providers as the most important components in SOAP note, synthesizing evidences from Subjective and Objective and concluding the diagnoses and treatment plans. Specifically, Assessment is the section describing the patient and establishing the main symptoms or problems for their encounter, and Plan addresses each differential diagnosis/problem with an action plan or treatment plan for the day, so called Plan Subsection. Figure 1 contains two Plan Subsection, marked by purple color. Text “UGIB … daily transfusions” is a different Plan Subsection than “EtOH abuse … to years plus”. Each Plan Subsection starts with a summary of a problem/diagnosis, e.g. UGIB in the second subsection summarizing from admit with upper GI bleed in the Assessment section. The section headings are not always available in the raw progress notes. Depending on the patients’ conditions, some sections are not always required to be present; hence, the number of sections in the progress note may vary. In the end, the SOAP note is reflective of the provider’s effort to collect the most recent and relevant data and synthesize the collected information into a coherent understanding of patient’s condition for decision-making and to ensure coordination of care. This skill in documentation requires clear reasoning to link symptoms, lab and imaging results, and other observations into temporally relevant and problem-specific treatment plans.
Figure 1:
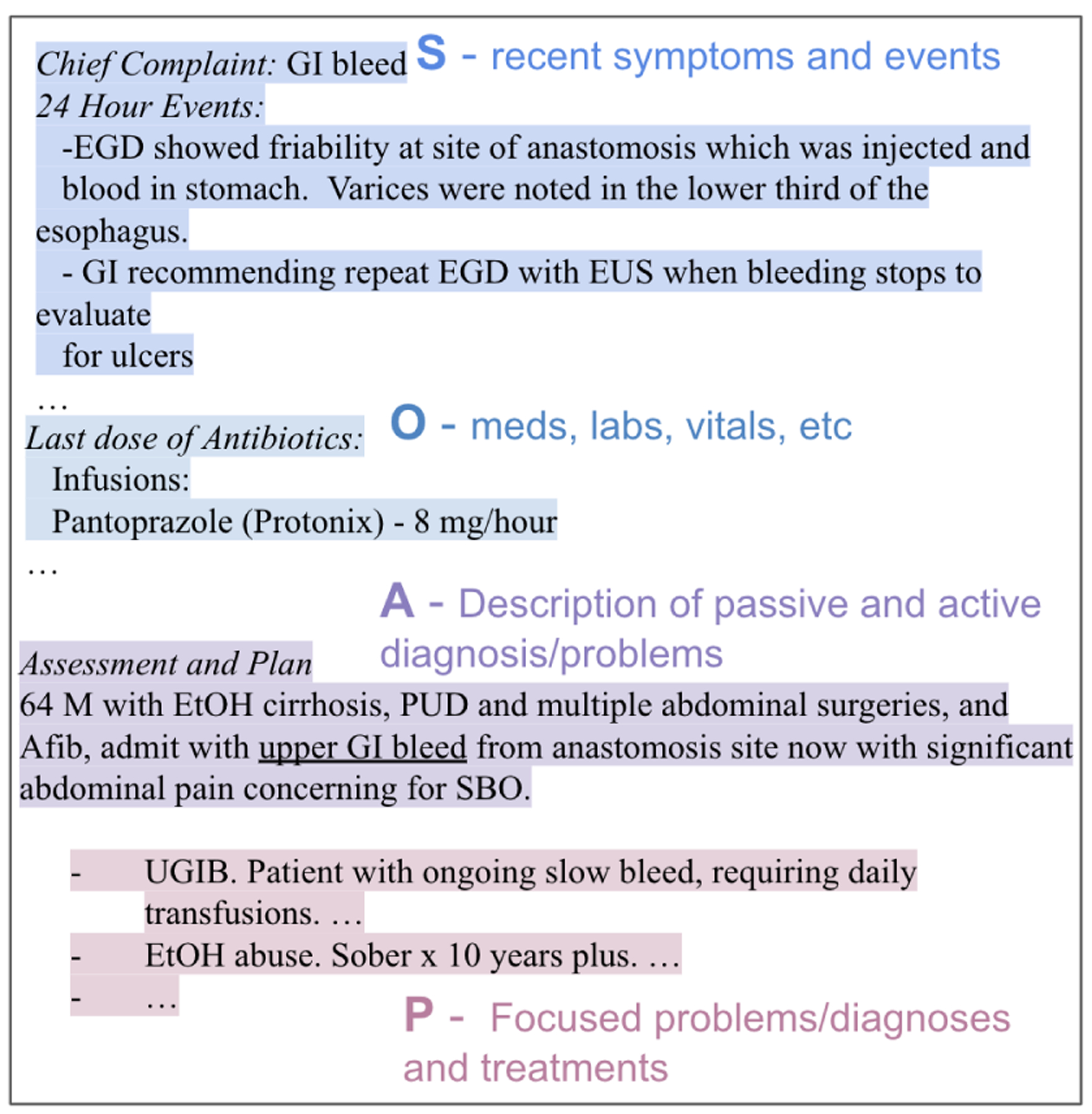
An example progress note with SOAP sections annotated. Line breaks are preserved from the raw note.
3. Data and Preparation
3.1. Annotation Tool and Data Source
All annotation procedures were performed using a dedicated software tool called INCEpTION3 (Klie et al., 2018). INCEpTION is an open-source software tool that serves as a semantic annotation platform and offers intelligent assistance and knowledge management. A sampling of 5,000 unique daily progress notes using the method described below were imported into the INCEpTION data corpus from the MIMIC-III notes file.
The annotations performed using INCEpTION were designed to provide labels and metadata in the daily progress notes extracted from MIMIC-III. MIMIC-III is an openly available dataset developed by the MIT Lab for Computational Physiology, comprising de-identified health data associated with approximately 60,000 intensive care hospitalizations. MIMIC includes demographics, vital signs, laboratory tests, medications, and over ten years of clinical notes collected across all the intensive care units at Beth Israel Hospital in Boston, Massachusetts, USA. To build the corpus of notes, a uniform random sample of progress notes across unique patient encounters were extracted from the NOTEVENTS table of MIMIC-III, which contained surgery, medicine, cardiovascular, neurology, and trauma daily progress notes. The annotations included an oversampling of medical progress notes because that is the largest service unit in the hospital.
3.2. Note Type Selection
The progress note types from MIMIC-III included a total of 84 note types (DESCRIPTION header) including the following: Physician Resident Note, Intensivist Note (SICU, CVICU, TICU), PGY1 Progress Note, PGY1/Attending Daily Progress Note MICU, MICU Resident/Attending Daily Progress Note. Other note types were excluded such as Nursing Progress Note and Social Worker Progress Note because these are not commonly structured in the SOAP format. Further, history and physical notes (admission notes) and discharge summaries were excluded as these are not daily progress notes and preclude analytics at the hospital encounter level in a time-series manner.
3.3. Ineligible Progress Notes
After the sampling of notes was finalized, initial review by annotators was to assess appropriate listing of problems/diagnoses in the Assessment and Plan section. Progress notes with a Plan section that did not contain problems or diagnoses were excluded for annotation. In particular, many progress notes may be written with a systems-oriented Plan section which only lists organ systems with related details but without a diagnosis or problem identified (i.e., general systems-oriented note only include mentions such as “Neurology”, “Cardiovascular”, “Renal”, “Hematology”, etc.). These types of progress notes occurred in 741 (49%) of the reviewed notes from the sampled corpus and were excluded for the following reasons: (1) they do not follow the problem-oriented view that is needed for clinical decision support; and (2) they are not the medical documentation best practices endorsed by medical school curriculum. The corpus of ineligible notes may be used to develop NLP tools in clinical training on proper documentation of health status and the development of diagnoses for medical trainees.
4. Annotation Protocol
We propose a hierarchical annotation schema consisting of three stages: SOAP Section Tagging organizing all sections of the progress note into a SOAP category; Assessment and Plan Relation Labeling specifying causal relations between symptoms and problems mentioned in the Assessment and diagnoses covered in each Plan Subsection; Problem List Identification highlighting the final diagnoses. Every stage of the annotation builds on top of the previous annotation. Figure 3 illustrates the flow of the annotation. The input to the flow is the entire SOAP progress note. The first stage of annotation, SOAP Section Tagging, produces text segments labeled with different sections (example presented in Figure 1). Assessment and Plan sections are segmented at stage 1, and will be used as input to stage 2 Assessment and Plan Relation Labeling. The Plan section containing multiple subsections will be input to stage 3 for Problem List Identification, producing a list of diagnoses/problems.
Figure 3:
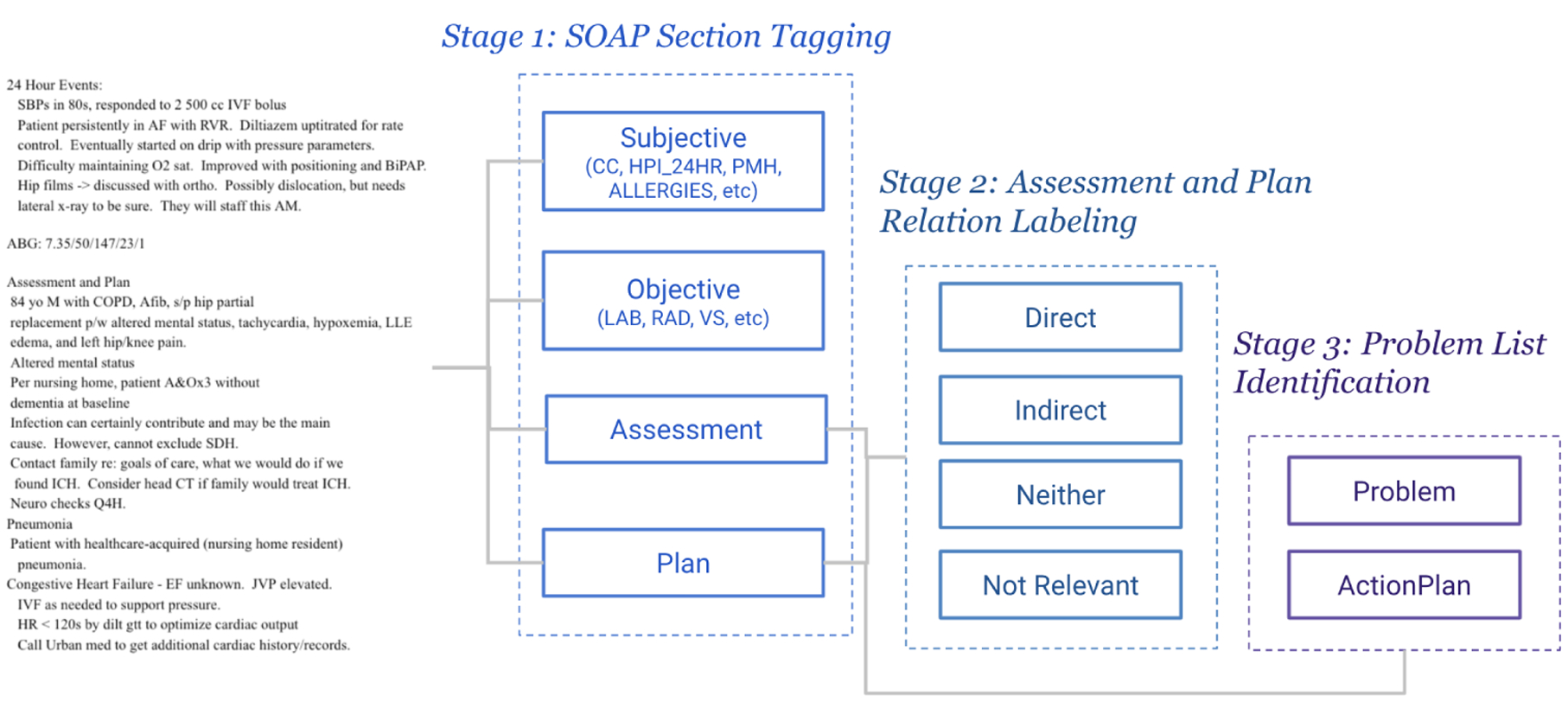
Annotation flow diagram for Progress Note Understanding. Dotted line boxes indicate different stages. Square text boxes are the labels produced at each stage.
4.1. Progress Note Section Tagging
The first stage of annotation is to segment the progress notes into sections, where each section belongs to a type of SOAP. Table 1 presents a list of sections and headers that may appear in the progress notes and their corresponding soap types, following the definitions in (Weed, 1964). Given a progress note, the annotator will mark each line of text using one of the attributes, as shown in Figure 2. Most of the sections start with a section header, indicating the lines below it falling into the same category of information until next section. When there is no section header, the annotator should mark the attributes by the content expressed in the lines (e.g. the line 14 Last dose of Antibiotics belonging to Medications). We post-process the attribute labels and further categorize them as one of the SOAP type. A full description of the functionality of each section in SOAP notes could be found in (Podder et al., 2020).
Table 1:
Section headings in the progress notes, annotated tag attributes and corresponding SOAP types.
| Sections | Attributes | SOAP Type |
|---|---|---|
| Chief Complaint | CC | S |
| History of Present Illness | HPL_24HR | S |
| Past Medical History | PMH | S |
| Allergies | ALLERGIES | S |
| Patient Surgical History | PSH | S |
| Social History | SH | S |
| Family History | FH | S |
| Review of System | ROS | S |
| Physical Exam | PE | O |
| Medications | MED | O |
| Laboratory | LAB | O |
| Radiology | RAD | O |
| Vital Sign | VS | O |
| Assessment | PROBLEMLIST | A |
| Plan | PLAN | P |
| Addendum | ADDENDUM | OTHERS |
Figure 2:

A screenshot of INCEpTION interface. The line numbers are indicated at left hand side. This piece of text are labeled with CC, HPI_24HR, ALLERGIES, MED, indicating four sections.
4.2. Assessment and Plan Relation Labeling
The Assessment and Plan (A&P) sections are highlighted through the first stage of annotation. Recall that the Plan section contains multiple Plan Subsection, each of which lists a detailed plan for one specific diagnosis/problem. The second stage of annotation is to label Plan Subsection for its relation to the assessment: (1) directly related; (2) indirectly related; (3) not related; and (4) not relevant. These relations indicate whether a Plan Subsection addresses the primary diagnoses or problems related to the primary diagnosis (direct), an adverse event or consequence from the primary diagnosis or comorbidity mentioned in the Assessment (indirect), a problem or diagnosis not mentioned in the progress note (neither), or a Plan subsection without a problem or diagnosis listed (not relevant). Figure 4 shows the guidelines for marking the relations between the Assessment and each subsection of the Plan. Figure 5 presents an example of linking Plan Subsection with the Assessment using INCEpTION interface.
Figure 4:

Guidelines for annotating the four relations (direct, indirect, neither, not relevant) between Assessment and each subsection of Plan
Figure 5:
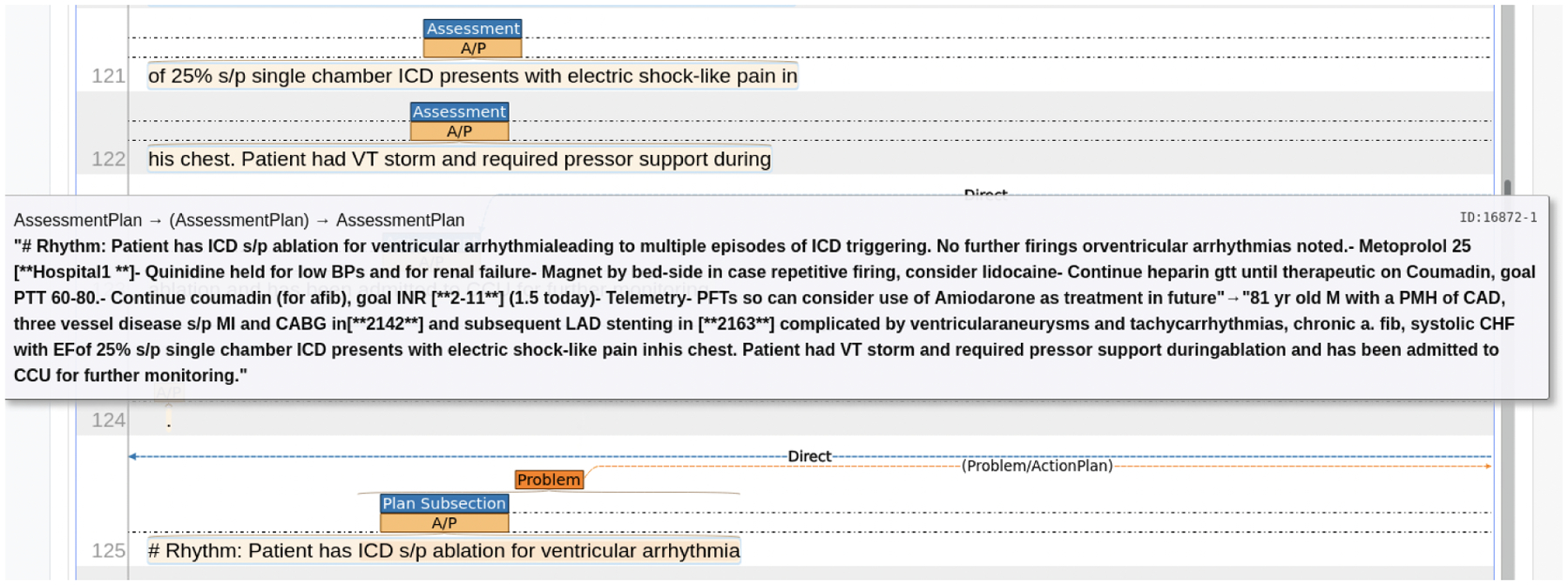
A screenshot of INCEpTION interface on A&P Labeling. The Plan Subsection (line 125) contains a Problem ICD sp ablation for ventricular arrythmia. A direct link connects the Plan Subsection and the Assessment, indicating that the problem is a major diagnosis. Once the link establishes, a text window pops up showing the text in the Plan Subsection (“Rhythm: Patient has ICD … treatment in future”, and text in Assessment (“81 yr old M with a PMH … further monitoring.”), separated by a dash.
direct, indirect, and neither relations all indicate that a diagnosis/problem is found in the progress note. When the label is not relevant, the plan subsection has no mention of a problem. Instead, it might describe quality improvement/administrative details such as physical therapy, occupational therapy, nutrition, prophylaxis (stress ulcer and gastric ulcer), or disposition.
4.3. Problem List Identification
The relevant plan subsections include a problem/diagnosis with an associated treatment or action plan, stating how the provider will address the problem. At the third stage of the annotation, the goal is to highlight the problems/diagnoses mentioned in the Plan subsections separately from the treatment or action plans for the day. In identifying problems/diagnoses, the annotators only labeled the text spans covering the problem in each Plan subsection, using the label problem. Once the problem/diagnosis was labeled then annotators labeled the accompanying actionplan for that problem and link the two attributes indicated by problemapproach. Figure 6 shows an example plan subsection where the problem is “Ventricular Arrhythmia”, and the actionplan indicating the medical treatment for the ventricular arrhythmia.
Figure 6:
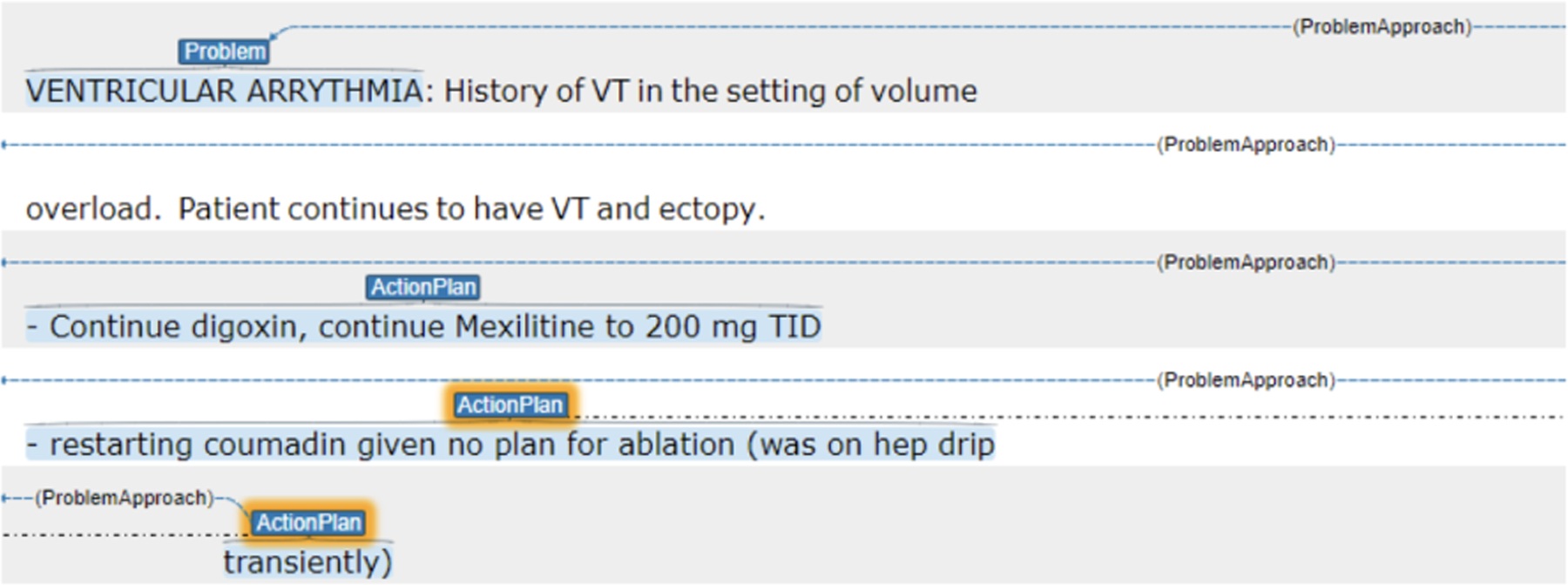
A Plan Subsection annotated with the problem (“Ventricular Arrythmia”) and ActionPlan.
Problems sometimes include the syndrome (i.e., sepsis syndrome, acute respiratory distress syndrome) along with the underlying diagnosis (i.e, urinary tract infection, COVID-19 pneumonia). We ask the annotators to mark the entire span of text where the text contained both syndrome and/or diagnosis (e.g. “sepsis likely due to urinary tract infection”, where sepsis is the syndrome, and urinary tract infection is the diagnosis). We include both syndrome and diagnosis because the same diagnosis may lead to different medical treatment pathways depending on the syndrome. For instance, antibiotics are needed for the urinary tract infection and then there are resuscitation efforts and additional monitoring needed for the sepsis syndrome. Phrases connecting between syndrome and diagnosis were included as part of the problem label, such as “caused by” or “likely due to” were included prior and after for diagnosis. These are strong linguistic cues showing the causal relations between syndrome and diagnosis, and reflecting a physicians’ reasoning process. However, additional details about the diagnoses were not needed. For example, “Myocardial infarction involving the LAD s/p PCI” should be labeled only for the diagnosis/problem of “myocardial infarction”. Another example, “SVT with hypotension: This occurred in setting of having RA/RV manipulated with wire” should be labeled as just “SVT with hypotension”. These were descriptions of characteristics of the diagnosis.
4.4. Annotator Training
The annotation guidelines and rules were initially developed and trialed by two physicians with board certifications in critical care medicine, pulmonary medicine, and clinical informatics. The physicians practice in the same field as the authors of the source notes. They are also experts in clinical research informatics with an extensive research track in machine learning and natural language processing, one of them serves as the mentor and adjudicator for the trained annotators. Two medical students were recruited and had received training in their medical school curriculum in medical history taking and documentation (including SOAP format), anatomy, pathophysiology, and pharmacology. An additional three week period with orientation and training was provided by one of the critical care physicians to the annotators. Each annotator met an inter-rater reliability with a kappa score of > 0.80 with the adjudicator prior to independent review. The annotators augmented their medical knowledge with a subscription and access to a large medical reference library, UpToDate®4. The adjudicator performed audits of the charts after approximately 200 were completed and if the inter-rater reliability fell below the 0.80 threshold then cases with disagreements were reviewed to consensus and the annotator was re-trained again until threshold kappa agreement was met.
5. Annotation Results
In total, two annotators labeled 768 progress notes with 28,945 labels from all annotation stages. We further split the corpus into train/dev/test, resulting in 608, 76 and 87 notes, respectively. All annotations were stored as XML files. We reported statistics of the labels for each stage.
Statistics for Section Tagging
We collected 3,790, 6,090, 787 and 2,742 labels for SOAP and Others, respectively. Table 2 presents the statistics of labels broken down by each section attribute, showing the total and average number of section tagged and average length of sections by tokens. Laboratory (LAB) was the most frequent section type across our sampled progress notes, with 4 sections per note on average. Often, more than one lab test was required to provide evidences for certain diseases, and every lab tested was included in the progress note as an individual section. Assessment and Plan sections were the longest with 577.29 tokens per note, much more than any other sections. Recall that Assessment and Plan viewed as the most important piece in SOAP note, summarizing the evidence from other sections and listed diagnoses with treatment plans. Physicians tend to spend most of their time reading the Assessment and Plan sections and less than 10 percent of the content in physicians’ verbal handoffs were found outside the A/P section (Brown et al., 2014).
Table 2:
Total and average number of section tags, and average length for each section counting by tokens.
| Attributes | #Labels | Avg #Labels Per Note | Avg Length |
|---|---|---|---|
| CC | 750 | 0.98 | 12.70 |
| HPL_24HR | 807 | 1.51 | 94.37 |
| PMH | 731 | 0.95 | 7.42 |
| ALLERGIES | 764 | 0.99 | 17.24 |
| PSH | 12 | 0.02 | 33.91 |
| SH | 15 | 0.02 | 15.87 |
| FH | 711 | 0.93 | 5.05 |
| ROS | 740 | 0.96 | 21.61 |
| PE | 789 | 1.03 | 131.64 |
| MED | 823 | 1.07 | 76.31 |
| LAB | 3083 | 4.01 | 55.59 |
| RAD | 1395 | 1.82 | 38.75 |
| VS | 974 | 1.27 | 159.92 |
| A&P | 787 | 1.02 | 577.29 |
| Addendum | 727 | 0.95 | 47.62 |
Statistics of A&P Relation Labeling
Recall that A&P relations were labeled between every pair of Plan Subsection and the Assessment. Across the sample set of progress notes, we had 7.73 plan subsections per note on average. Table 3 summarizes the count of labels on all annotated notes and the average per note. The distribution across four relations were relatively balanced, with neither being the most frequently labeled and not relevant being the least frequently labeled.
Table 3:
Total and average number of relation labels.
| Count | direct | indirect | neither | not rel |
|---|---|---|---|---|
| Total | 1404 | 1599 | 1913 | 1018 |
| Per Note | 1.83 | 2.09 | 2.49 | 1.32 |
Statistics of Problem Lists
We collected 4,843 and 4,759 labels for problem and actionplan, respectively. For every note, the average numbers of problem and actionplan were both approximately 6. Recall that the problem was highlighted after the annotation for ap Relation Labeling, we were able to count the problems that were labeled as direct and indirect, which was 2,866 in total. We post-processed the annotation such that for every assessment, there was a list of direct problems and a list of indirect problems. Every list could be regarded as a short summary, with distinct problems delimited by semi-colons. In total we collect 743 and 619 summaries for direct problems and indirect problems, respectively.
Inter-Annotator Agreement
We measured Cohen’s Kappa on the AP relation labeling task, as it was deemed the most difficult by the annotators because it was the only task that required clinical reasoning and medical knowledge. The two annotators achieved a Cohen’s Kappa of 0.74 on 10 randomly sampled notes, which represented good quality given the complexity of the task. Figure 7 presents the agreement and disagreement between the two annotators on four relations. Most of the disagreement occurred between indirect and neither, taking 9.83% of all labels, exposing the difficulties in deciding whether a problem is due to a subsequent event of the main diagnosis (indirect) or a separate disease altogether (neither). Nonetheless, the percentage of labels agreed by both annotators was 81.9%.
Figure 7:
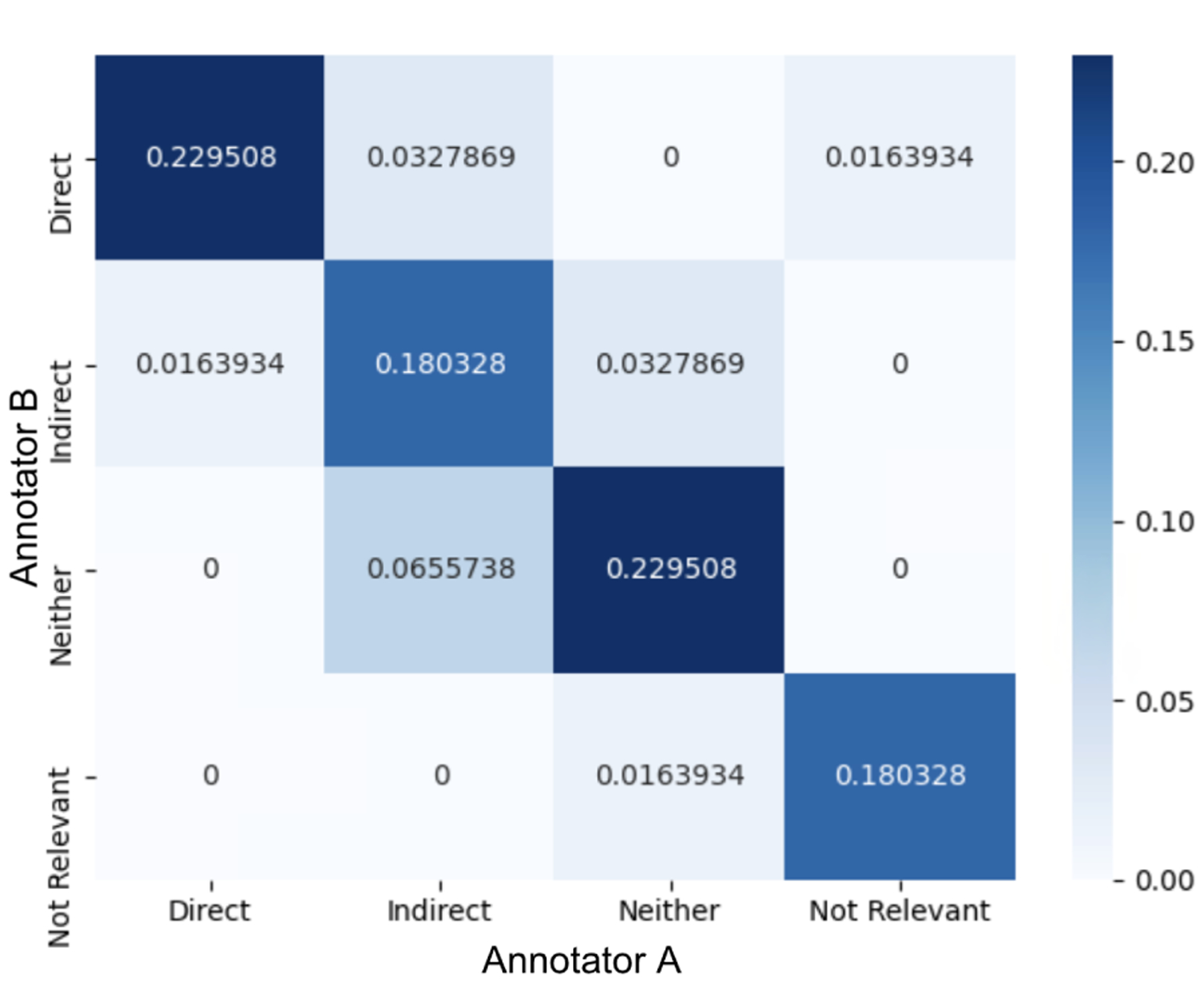
Confusion matrix on the AP relation labels between two annotators.
6. Suite of Tasks for Progress Note Understanding
In this section, we further propose the suite of clinical NLP tasks: Progress Note Understanding. The suite of tasks was developed from the annotation and designed to train and evaluate models for clinical text understanding in future work. The suite of tasks was set up in the same stream as the annotation, with each task corresponding to an annotation stage and targeted at a different NLP problem. Figure 8 presents a diagram of the suite set up. The level of NLP difficulty in moving from Task 1 to Task 3 were intended to increase from classification to classification with medical knowledge (NLI) to text generation. We consider the SOAP Note Section Segmentation as the easiest task that only requires text understanding, and the Diagnoses Summarization to be the hardest as it investigates clinical text understanding, clinical reasoning and text generation.
Figure 8:
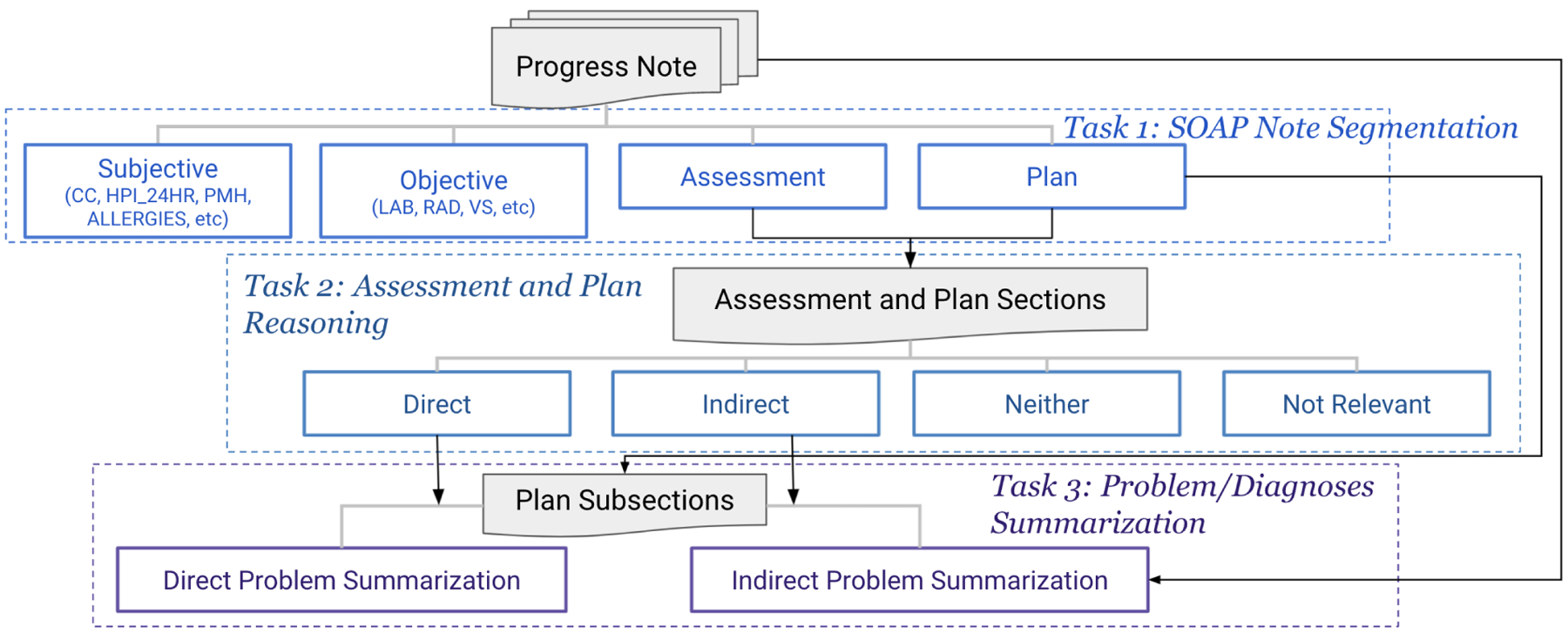
The Progress Note Understanding suite is consisted of three tasks, each of which corresponds to one annotation stage. Task 3, Problem/Diagnoses Summarization, has two subtasks, Direct Problem Summarization and Indirect Problem Summarization, using the problems identified in annotation stage 2 and 3.
6.1. Task 1: SOAP Note Segmentation
The suite of tasks started with a clinical text understanding task where the goal was to segment the entire SOAP note into topic-relevant sections. We formulated this task as labeling each line of the daily progress note by its soap type based on the information contained in the line. This task helped to train and evaluate models that automatically understand the SOAP note structure and topic-level contents, and highlights the problem-oriented sections. We considered this task as a fundamental and essential step for NLP system pipelines for clinical note processing, as previously shown in (Krishna et al., 2021; Cillessen and de Vries Robbé, 2012; Mowery et al., 2012). Previous work also focused on segmenting SOAP notes (Mowery et al., 2012; Ganesan and Subotin, 2014). Mowery et al. (2012) formulated the task as sentence labeling using 50 emergency department reports. Our proposed task is similar to Ganesan and Subotin (2014) where the task is on line-basis instead of sentence, but our work is a larger corpus and is focused on daily progress notes. Lastly, we plan to make our corpus available unlike prior work. Evaluation will be measured using the F1 score and Accuracy as in previous work (Mowery et al., 2012).
6.2. Task 2: Assessment and Plan Reasoning
Recall that the Assessment and Plan sections contain the most useful information for providers needing to know what is the condition of a patient. Providers spend the most time viewing these sections, where clinical reasoning occurs. Providers conclude the diagnoses/problems from the evidences presented in S and O sections and the symptoms described in A section, then infer the treatment plan for each diagnosis for that day. It is a process of associating the patients’ current condition with clinical problems, and assessing the solutions with medical knowledge. Modeling such understanding and reasoning process greatly benefits the downstream applications that aim at improving efficiency of bedside care and reducing medical errors. We propose task 2, Assessment and Plan Reasoning, a relation classification task that builds on top of the stage 2 annotation. Models trained and evaluated on this task should predict one of four relations. We plan to use F1 score as the evaluation metric for this task.5
6.3. Task 3: Problem/Diagnoses Summarization
Automatically generating a set of diagnoses/problems in a progress note could help providers quickly and efficiently understand and document a patient’s condition, and ultimately reduce effort in document review and augment care during time-sensitive hospital events. Devarakonda et al. (2017) demonstrated that a system that automatically predicts diagnoses captures more problems than human clinicians through a pilot study. They built a two-level classifier that first predicted major category of conditions and then a subsequent prediction on the fine-grained level of problems. Liang et al. (2019) formulated the task as extractive summarization on a disease-specific progress notes. To enrich the research on diagnosis summarization, we propose Problem/Diagnoses Summarization with labels created from annotation stage 2 and 3. Specifically, the task consisted of two subtasks: Direct Problem Summarization and Indirect Problem Summarization. The goal of Direct Problem Summarization is to take the assessment section as input, and predict a list of primary diagnoses. For Indirect Problem Summarization, the goal is to predict a list of adverse events and subsequent diagnoses given the entire progress note. Recall that in Figure 5, indirect problems included diagnoses/problems that were not part of the Assessment or primary diagnoses, which required input from other sections in the progress notes (e.g. LAB, VS) and would make the task harder than Direct Problem Summarization. Different from (Liang et al., 2019), we defined both subtasks as abstractive summarization, since our data covered a larger breadth of diseases/problems and they were not always explicitly mentioned in the progress note. In some instances, the signs or symptoms of a disease were listed, relevant objective data that pointed to a diagnosis, or synonyms and acronyms of the disease or problem were listed in other parts of the note. Recall that the text spans labeled by Problem were concatenated through semi-colons. Figure 9 includes an example Direct Problem Summary with an assessment it derived from.
Figure 9:
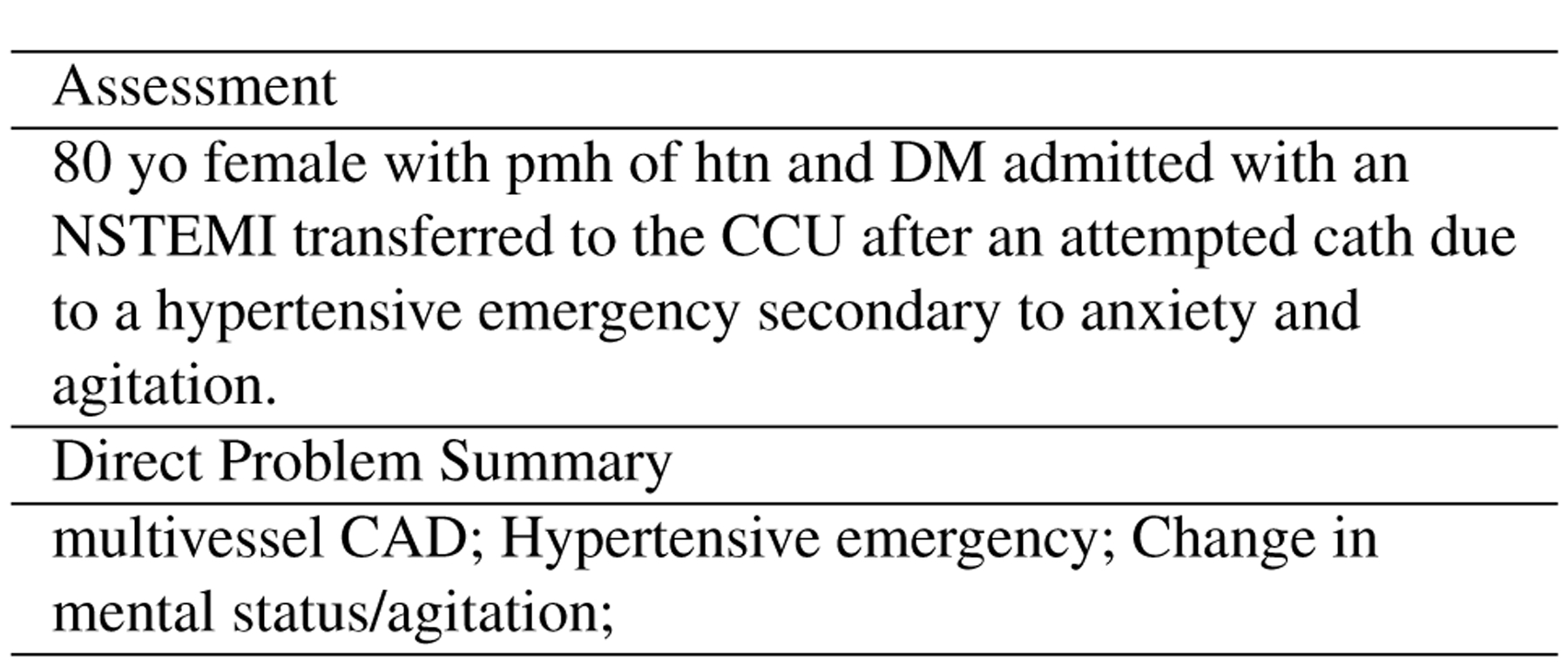
An example Direct Problem Summary given an Assessment section. The diagnosis “Multivessel CAD” is inferred from “DM admitted with an NSTEMI transfered to CCU”; “hypertensive emergency” is mentioned directly in the assessment, and “Change in mental status/agitation” refers to “anxiety and agitation”.
For both summarization subtasks, we plan to follow previous work on EHR summarization (MacAvaney et al., 2019; Adams et al., 2021) and use the standard summarization metrics like ROUGE (Lin, 2004). A diagnosis could be expressed with different lexicons, (e.g. “CAD” and “Coronaries”), hence we also consider post-processing the summaries with the National Library of Medicine’s Unified Medical Language System (UMLS) (Bodenreider, 2004) by mapping the clinical terms to the UMLS metathesaurus of concept unique identifiers (CUIs), and measuring the F1 score for clinical accuracy. Liang et al. (2019) also used this method as their main evaluation.
7. Conclusions
We introduce a novel and hierarchical annotation on a large collection of publicly available EHR data and aim to develop and evaluate models for automated section segmentation, assessment and plan reasoning, and diagnoses summarization. A suite of tasks, Progress Note Understanding, is proposed to utilize the annotation and contains three tasks, each of which corresponds to an annotation stage. Future work will focus on hosting shared tasks for the clinical NLP community and using the tasks to build systems for applications in clinical decision support.
Acknowledgement
We thank Professor Ozlem Uzuner (George Mason University) and Harvard Department of Biomedical Informatics (DBMI) for the feedback and guidance during N2C2 task setup.
Footnotes
MIMIC is available at https://physionet.org/content/mimiciii/1.4/. Data usage agreement is required. Our annotation is available at https://git.doit.wisc.edu/smph/dom/UW-ICU-Data-Science-Lab/progress-note-understanding-lrec2022.
v0.19.2 (Released on 2021-04-07), available at: https://inception-project.github.io/
The task is part of 2022 National NLP Clinical Challenge (N2C2): https://n2c2.dbmi.hms.harvard.edu/2022-track-3.
Reference
- Abacha AB, M’rabet Y, Zhang Y, Shivade C, Langlotz C, and Demner-Fushman D (2021). Overview of the mediqa 2021 shared task on summarization in the medical domain. In Proceedings of the 20th Workshop on Biomedical Language Processing, pages 74–85. [Google Scholar]
- Adams G, Alsentzer E, Ketenci M, Zucker J, and Elhadad N (2021). What’s in a summary? laying the groundwork for advances in hospital-course summarization. In Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, pages 4794–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsentzer E, Murphy J, Boag W, Weng W-H, Jindi D, Naumann T, and McDermott M (2019). Publicly available clinical bert embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, pages 72–78. [Google Scholar]
- Bodenreider O (2004). The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research, 32(suppl 1):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen JL (2006). Educational strategies to promote clinical diagnostic reasoning. New England Journal of Medicine, 355(21):2217–2225. [DOI] [PubMed] [Google Scholar]
- Brown P, Marquard J, Amster B, Romoser M, Friderici J, Goff S, and Fisher D (2014). What do physicians read (and ignore) in electronic progress notes? Applied clinical informatics, 5(02):430–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillessen F and de Vries Robbé P (2012). Modeling problem-oriented clinical notes. Methods of information in medicine, 51(06):507–515. [DOI] [PubMed] [Google Scholar]
- Devarakonda MV, Mehta N, Tsou C-H, Liang JJ, Nowacki AS, and Jelovsek JE (2017). Automated problem list generation and physicians perspective from a pilot study. International journal of medical informatics, 105:121–129. [DOI] [PubMed] [Google Scholar]
- Devlin J, Chang M-W, Lee K, and Toutanova K (2019). Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 4171–4186. [Google Scholar]
- Ganesan K and Subotin M (2014). A general supervised approach to segmentation of clinical texts. In 2014 IEEE International Conference on Big Data (Big Data), pages 33–40. IEEE. [Google Scholar]
- Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, Naumann T, Gao J, and Poon H (2021). Domain-specific language model pretraining for biomedical natural language processing. ACM Transactions on Computing for Healthcare (HEALTH), 3(1):1–23. [Google Scholar]
- Hao B, Zhu H, and Paschalidis I (2020). Enhancing clinical bert embedding using a biomedical knowledge base. In Proceedings of the 28th international conference on computational linguistics, pages 657–661. [Google Scholar]
- He Y, Zhu Z, Zhang Y, Chen Q, and Caverlee J (2020). Infusing disease knowledge into bert for health question answering, medical inference and disease name recognition. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 4604–4614. [Google Scholar]
- Hripcsak G, Vawdrey DK, Fred MR, and Bostwick SB (2011). Use of electronic clinical documentation: time spent and team interactions. Journal of the American Medical Informatics Association, 18(2):112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman GM, Marquard JL, Lindemann E, Arsoniadis E, Pakhomov S, and Melton GB (2019). Challenges and opportunities to improve the clinician experience reviewing electronic progress notes. Applied clinical informatics, 10(03):446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Pollard T, and Mark III R (2019). Mimic-iii clinical database demo (version 1.4). PhysioNet, 10:C2HM2Q. [Google Scholar]
- Klie J-C, Bugert M, Boullosa B, de Castilho RE, and Gurevych I (2018). The inception platform: Machine-assisted and knowledge-oriented interactive annotation. In Proceedings of the 27th International Conference on Computational Linguistics: System Demonstrations, pages 5–9. [Google Scholar]
- Krishna K, Khosla S, Bigham JP, and Lipton ZC (2021). Generating soap notes from doctorpatient conversations using modular summarization techniques. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 4958–4972. [Google Scholar]
- Liang J, Tsou C-H, and Poddar A (2019). A novel system for extractive clinical note summarization using ehr data. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, pages 46–54. [Google Scholar]
- Lin C-Y (2004). Rouge: A package for automatic evaluation of summaries. In Text summarization branches out, pages 74–81. [Google Scholar]
- Lybarger K, Ostendorf M, and Yetisgen M (2021). Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. Journal of Biomedical Informatics, 113:103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAvaney S, Sotudeh S, Cohan A, Goharian N, Talati I, and Filice RW (2019). Ontology-aware clinical abstractive summarization. In Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval, pages 1013–1016. [Google Scholar]
- Mowery DL, Harkema H, and Chapman W (2008). Temporal annotation of clinical text. In Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing, pages 106–107. [Google Scholar]
- Mowery D, Wiebe J, Visweswaran S, Harkema H, and Chapman WW (2012). Building an automated soap classifier for emergency department reports. Journal of biomedical informatics, 45(1):71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenbach J, Pruksachatkun Y, Adler S, Seale J, Swartz J, McKelvey G, Dai H, Yang Y, and Sontag D (2021). CLIP: A dataset for extracting action items for physicians from hospital discharge notes. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 1365–1378, Online, August. Association for Computational Linguistics. [Google Scholar]
- Pampari A, Raghavan P, Liang J, and Peng J (2018). emrqa: A large corpus for question answering on electronic medical records. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 2357–2368. [Google Scholar]
- Patel P, Davey D, Panchal V, and Pathak P (2018). Annotation of a large clinical entity corpus. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 2033–2042. [Google Scholar]
- Peng Y, Wang X, Lu L, Bagheri M, Summers R, and Lu Z (2018). Negbio: a high-performance tool for negation and uncertainty detection in radiology reports. AMIA Jt Summits Transl Sci Proc, 2017:188–196. [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yan S, and Lu Z (2019). Transfer learning in biomedical natural language processing: An evaluation of bert and elmo on ten benchmarking datasets. In Proceedings of the 2019 Workshop on Biomedical Natural Language Processing (BioNLP 2019). [Google Scholar]
- Podder V, Lew V, and Ghassemzadeh S (2020). Soap notes. StatPearls [Internet]. [PubMed] [Google Scholar]
- Raghavan P, Liang JJ, Mahajan D, Chandra R, and Szolovits P (2021). emrkbqa: A clinical knowledge-base question answering dataset. In Proceedings of the 20th Workshop on Biomedical Language Processing, pages 64–73. [Google Scholar]
- Romanov A and Shivade C (2018). Lessons from natural language inference in the clinical domain. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 1586–1596. [Google Scholar]
- Shoolin J, Ozeran L, Hamann C, and Bria Ii W (2013). Association of medical directors of information systems consensus on inpatient electronic health record documentation. Applied clinical informatics, 4(02):293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner O, Goldstein I, Luo Y, and Kohane I (2008). Identifying patient smoking status from medical discharge records. JOURNAL OF THE AMERICAN MEDICAL INFORMATICS ASSOCIATION, 15(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y (2009). Annotating and recognising named entities in clinical notes. In proceedings of the ACLIJCNLP 2009 Student Research Workshop, pages 18–26. [Google Scholar]
- Weed LL (1964). Medical records, patient care, and medical education. Irish Journal of Medical Science (1926–1967), 39(6):271–282. [DOI] [PubMed] [Google Scholar]


