A landscape of UPL3-ubiquitinated proteins by ubiquitome profile indicates UPL3 interacting with UBP12 as a hub of regulator on proteolysis-independent regulation and proteolysis-dependent degradation.
Abstract
The HECT-type UPL3 ligase plays critical roles in plant development and stress protection, but understanding of its regulation remains limited. Here, the multi-omics analyses of ubiquitinated proteins in upl3 mutants were performed. A landscape of UPL3-dependent ubiquitinated proteins is constructed: Preferential ubiquitination of proteins related to carbon fixation represented the largest set of proteins with increased ubiquitination in the upl3 plant, including most of carbohydrate metabolic enzymes, BRM, and variant histone, whereas a small set of proteins with reduced ubiquitination caused by the upl3 mutation were linked to cysteine/methionine synthesis, as well as hexokinase 1 (HXK1) and phosphoenolpyruvate carboxylase 2 (PPC2). Notably, ubiquitin hydrolase 12 (UBP12), BRM, HXK1, and PPC2 were identified as the UPL3-interacting partners in vivo and in vitro. Characterization of brm, upl3, ppc2, gin2, and ubp12 mutant plants and proteomic and transcriptomic analysis suggested that UPL3 fine-tunes carbohydrate metabolism, mediating cellular senescence by interacting with UBP12, BRM, HXK1, and PPC2. Our results highlight a regulatory pattern of UPL3 with UBP12 as a hub of regulator on proteolysis-independent regulation and proteolysis-dependent degradation.
Introduction
Cell senescence, including developmental senescence and stress-induced cell senescence, is triggered by internal and external factors and often involves degradation and remobilization of cellular components. During plant senescence, a visible change of leaf yellowing is an indication of chloroplast damage and chlorophyll degradation. At the molecular level, catabolism of macromolecules is a major event in senescent cells, especially involving proteolysis. It has been documented that during Arabidopsis leaf senescence, alterations in transcriptional regulation, histone-associated epigenetic processes, posttranslational modification (PTM), and macromolecule/organelle degradation are genetically determined and developmentally programmed (Buchanan-Wollaston et al, 2005; Woodson et al, 2015; Yolcu et al, 2017). Pathways for protein degradation include the 26S proteasome, the organelle degradation, autophagy processes, and the monoubiquitination- or short ubiquitin chain–dependent proteinases (Vierstra, 2009; Miller et al, 2010; Berndsen & Wolberger, 2014; Lan & Miao, 2019). The targeting for 26S proteasome degradation is a sequential process that starts with the ubiquitin activation by the E1 (ubiquitin-activating) enzyme in an ATP-dependent manner. The activated ubiquitin is then transferred from the E1 to the E2 (ubiquitin-conjugating) enzyme that acts as an intermediate. Finally, the E3 (ubiquitin ligase) enzyme mediates the deposition of the activated ubiquitin to, normally, a lysine residue of the target protein. Depending on the E3 type, protein ubiquitination may be a direct or indirect process (Berndsen & Wolberger, 2014; Zheng & Shabek, 2017).
The HECT E3 ligase family contains seven members (UPL1-UPL7) in Arabidopsis, which plays important roles in protein fate and protein function during the senescence process (Lan & Miao, 2019). Although animal studies have revealed diverse mechanisms in the functions and regulation of HECT E3s, their plant counterparts are less explored. So far, the Arabidopsis UPL3 and UPL5 are known to be involved in plant development and response to stress (Downes et al, 2003; Miao & Zentgraf, 2010; Patra et al, 2013; Bensussan et al, 2015; Furniss et al, 2018; Miller et al, 2019). Although the upl5 mutant shows a premature aging phenotype, UPL3 functions in trichome, vascular, and seed development, as well as in immune response. The UPL5 protein is able to target the transcription factor WRKY53 for ubiquitination and degradation, playing an antagonist role in leaf senescence (Miao et al, 2004; Miao & Zentgraf, 2010). UPL3 may target GLABROUS 3 (GL3) and ENHANCER OF GL3 (EGL3), two bHLH transcription factors, and positively regulate trichome development and flavonoid biosynthesis in Arabidopsis (Patra et al, 2013). The upl3 mutant shows larger stem diameter than that of the WT plant, indicating a role of UPL3 in regulation of vascular development (Bensussan et al, 2015). UPL3 is required for development of plant immunity (Furniss et al, 2018). Recently, Miller et al (2019) showed that UPL3 controls the protein stability of LEAFY COTYLEDON2 (LEC2), a key transcriptional regulator of seed maturation, and regulates the seed size and crop yields (Miller et al, 2019). Thus, UPL3 emerges as a critical player with pleiotropy traits in Arabidopsis thaliana. However, the underlying mechanisms are still limited understood.
In this study, we conducted globally proteomic and ubiquitomic analyses by using a label-free mass spectrometry-based analysis of protein ubiquitination with the di-Gly-Lys–remnant antibody enrichment approach in the upl3 mutant relative to the wild type to construct a landscape of UPL3-dependent ubiquitylated proteins, and analysis of ubiquitin footprint provides direct evidence of the ubiquitination of proposed target proteins. GFP nanotrap–mass spectrometry and yeast two-hybrid assay further confirmed direct UPL3 targets. Further UPL3-interacting proteins combined with their mutants’ phenotyping and analysis of transcriptome dataset suggested that UPL3 targeted deubiquitination enzymes (DUBs) and acted as regulators impacting on metabolism-related cell senescence.
Results
Mutation of UPL3 affects plant development
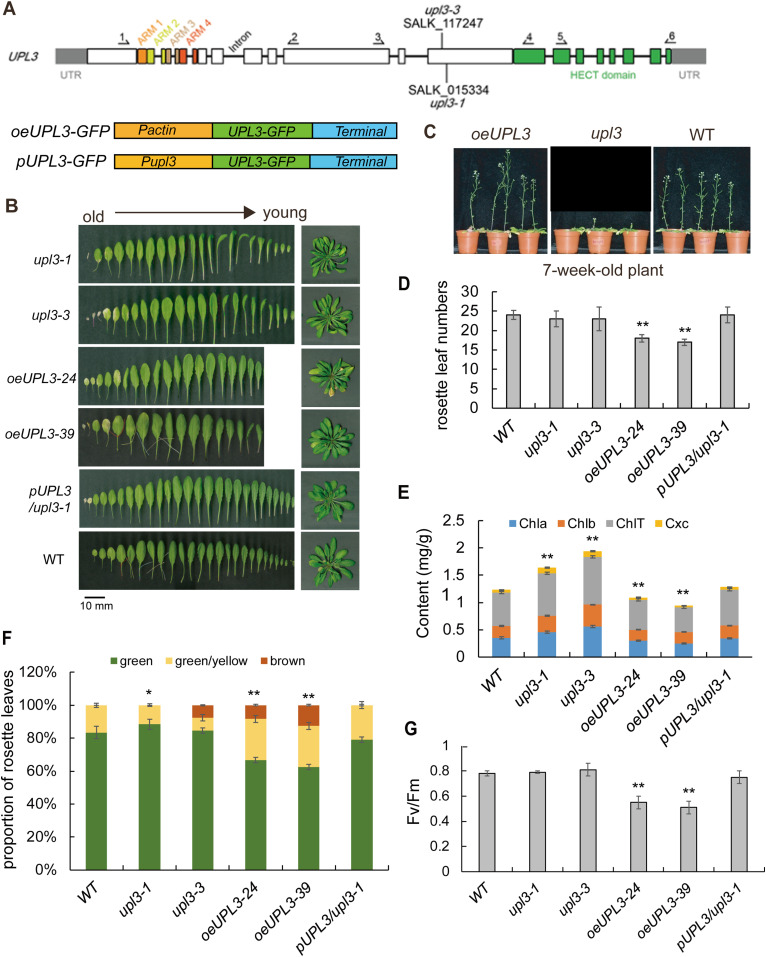
To address more defined roles of UPL3, we systematically analyze the phenotype of upl3 lines; two knockout lines (upl3-1, upl3-3), two overexpressing lines (oeUPL3-24, oeUPL3-39), and two complementation lines (comUPL3-1, comUPL3-2) were produced and confirmed at the transcriptional and protein levels (Figs 1A and S1). Plants of the upl3 lines exhibited apparently downward-curled leaves at the late stage and a 2-wk delay in bolting and leaf senescence, compared with the wild-type (WT) plant (Figs 1B–D and S2C). In contrast, the overexpressing UPL3 lines displayed premature leaf aging and 1-wk earlier bolting (Figs 1B–D and S2C), and lower number of rosette leaves are observed in UPL3 overexpressing plants (Fig 1D). Complementation by the full-length UPL3 rescued the upl3 mutants’ phenotypes (Fig 1B–D). Consistently, chlorophyll content (Fig 1E), photosystem II fluorescent activity (Fv/Fm) (Fig 1G), and green leaves/yellow leaves ratio (Fig 1F) increased significantly in the upl3 lines and decreased in the oeUPL3 line, compared with the WT. And the transcript level of early senescence marker gene (WRKY53) and floral transition marker gene (FT) declined significantly in the upl3 lines and increased significantly in the oeUPL3 line (Fig S2D). Thus, UPL3 functions in organ development, aging, and flowering by accelerating leaf senescence, under natural developmental conditions.
Figure 1. Phenotype of representative 6-wk-old wild-type and upl3 plants.
(A) Organization of the UPL3 gene and plasmid constructs for transgenic plants. (B) Phenotype of representative 6-wk-old wild-type, upl3, and oeUPL3 transgenic plants, as well as complemented line (pUPL3/upl3-1). Leaf senescence and curled leaves are indicated. Bars = 10 mm. (C) Representative images showing the bolting of plants in the seventh week after germination. (D) Rosette leaf numbers of the wild-type, upl3-1, oeUPL3-24, and pUPL3/upl3-1 plants (n > 20), indicating flowering time. (E) Chlorophyll content and carotenoid content measured in the rosettes of 6-wk-old wild-type, upl3-1, and oeUPL3-24 plants. Chla, chlorophyll a; Chlb, chlorophyll b; ChlT, total chlorophyll; Cxc, carotenoid. (F) Proportion of green and yellow leaves of whole rosette of 8-wk-old plants (n = 12). Error bars represent the SD of six biological replicates. Asterisks denote statistically significant differences from the WT, calculated using t test: *P < 0.05; **P < 0.01; and ***P < 0.001. (G) Photosystem II fluorescence activity (Fv/Fm) of the fifth rosette leaf of 6-wk-old plants (n = 9).
Figure S1. Transcript level of UPL3 detected in the upl3 mutants by semi-qRT-PCR.
(A) Schematic of T-DNA insertion site of UPL3 gene in genome. (B) Transcript level of the upl3 mutants detected by semi-qRT-PCR. GAPC2 is used as a control. (A) Primer (P) pairs’ position is indicated in (A). Primer sequences are listed in Table S2. (C) Protein level of the upl3 mutants (upl3-1, upl3-3) and oeUPL3 lines (oe-39, oe-24) and complement line (pUPL3/upl3-1) detected by Western blot using an antibody against GFP. The predicted size of the entire protein is ∼250 kD appeared in oeUPL3-24/-39 lines, along with an additional 100-kD band (possible proteolysis statue). Yellow arrowhead presents UPL3-GFP.
Figure S2. The expression pattern of UPL3 in different tissue during plant development.
(A) Expression level of UPL3 from the Arabidopsis Information Resource (TAIR) eFP Browser dataset (https://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). (B) GUS staining of PUPL3:GUS plants. (a) Three days after germination (dag), (b) 4 dag, (c) 52 dag, (d) and (g) 10 dag, (e) and (h) 21 dag, (f) and (i) 35 dag, (j) 1 dag seedlings. Bar = 5 mm. (C) Flowering rate of the upl3 and the oeUPL3 plants relative to WT. Hundred plants are used to calculate. (D) Transcript level of early senescence marker gene (WRKY53) and floral transition marker gene (FT) in the upl3 and the oeUPL3 plants relative to WT. Asterisk indicates significant differences (*P < 0.05, **P < 0.01) based on the t test compared with WT plants.
Loss of UPL3 induces global ubiquitin enrichment in 6-wk-old upl3 plants
UPL3 was highly expressed in the senescent leaf (after 6 wk) (Winter et al, 2007; Fig S2A and B); therefore, we performed a label-free mass spectrometry (MS)–based analysis of protein ubiquitination using the di-Gly-Lys–remnant antibody enrichment approach using 6-wk-old upl3 and WT plants (Figs 2A and S3). Identified proteins based on their tandem mass spectra matching against the UniProt A. thaliana Columbia database (current total of 39,211 reads) using the MaxQuant software are listed in Supplemental Data 1 (418.1KB, xlsx) . Label-free quantification, with a false discovery rate (FDR) adjusted to <0.01 and a minimum score for modified peptides set as >40, resulted in a set of over 1,310 potential ubiquitinated targets. This was further refined to a subset of 1,155 targets by at least two PTMs (Fig 2B and Supplemental Data 2 (62.5KB, xlsx) ).
Figure 2. Identification of ubiquitin conjugates in 6-wk-old upl3 plants compared with the wild type.
(A) A label-free mass spectrometry–based analysis procedure of protein ubiquitination using K-epsilon-GG remnant antibody enrichment approach. (B) Venn diagrams showing the enrichment of differentially ubiquitinated proteins in the upl3 background. (C) Immunodetection of global ubiquitinated proteins in plants of WT and mutants using an antibody against ubiquitin. The upl5 plant is included for a comparison. WB of β-tubulin is used as protein-loading controls. The gray intensity of three replicates was calculated by ImageJ. (D) Distribution of enriched proteins within differential fold-change levels by upl3 relative to WT. (E) Volcano plot of individual ubiquitination site showing their P-value and the log2FC. Dark-gray points are conjugates considered to be “abundant” by their detection in three biological replicates in either background (upl3 and/or the wild type). Proteins with a significant decrease or increase in ubiquitination in the upl3 mutant compared with the wild type (P-value < 0.05) are highlighted in red and blue, respectively. Ubiquitinated targets identified in all wild-type biological replicates and never or only once in the upl3 mutant but that were above the significance threshold of P-value > 0.05 are in yellow. The dashed line represents the theoretical situation, where conjugate abundance in the wild type and upl3 is equal. The horizontal dashed line highlights a P-value = 0.05. The vertical dashed lines highlight a 1.2-fold (log2FC = 0.26) increase or decrease.
Source data are available for this figure.
Figure S3. Quality detection of the tryptic peptides of ubiquitin conjugates in the 6-wk-old upl3 plants and wild-type plants.
(A) Detection of the mass error of all identified peptides (mass error). The quality error takes 0 as the central axis and is concentrated in the range of less than 10 ppm, indicating that the quality error meets the requirements. (B) The length of most peptide segments is distributed between 8– and 20–amino acid residues, which is in line with the law of trypsin-digesting peptide segments, indicating that the sample preparation meets the standard. (C) Detection of statistically significant similarity of two replicates by heatmap of Pearson’s correlation coefficient. rep: replicate, A: WT, B: upl3. (D) Detection of statistically significant similarity of two replicates by scatter diagram. rep: replicate, A: WT, B: upl3-1. This coefficient is a measure of the degree of linear correlation between the two groups of data: when the Pearson coefficient is closer to −1, it is negative correlation; closer to 1, it is positive correlation; and closer to 0, it is irrelevant.
Spectrum identification information for upl3 and WT. LSA-2022-01492_Supplemental_Data_1.xlsx (418.1KB, xlsx)
Differential ubiquitination targets of upl3/WT (FC 1.3 and 1.5). LSA-2022-01492_Supplemental_Data_2.xlsx (62.5KB, xlsx)
To identify the alteration of ubiquitin conjugates associated with the UPL3 mutation, a fold-change greater than 1.2 or less than 1/1.2 and P-value < 0.05 of two replicates were used to filter conjugate targets in the library, whose ubiquitination was up-regulated or down-regulated. All the differentially ubiquitin conjugates (DUCs) data in upl3/WT (|FC| > 1.2, P-value < 0.05) were shown in Fig 2B and Supplemental Data 2 (62.5KB, xlsx) . Among them, ubiquitination of 545 sites (356 proteins) was found to be up-regulated, and ubiquitination of 198 sites (189 proteins) was down-regulated in the upl3, compared with the WT plant (Fig 2B). These unexpected results were verified by global ubiquitination immunodetection using an anti-polyubiquitin antibody, in which deletion of UPL3 led to enhanced signals of global levels of protein ubiquitination, whereas loss of its homolog UPL5 did not, as a control (Fig 2C, Miao & Zentgraf, 2010). Overexpression of UPL3 retained comparable global levels of protein ubiquitination with the WT (Fig 2C). Indeed, more proteins were found in the up-regulated category than in the down-regulated set, in term of modified protein sites (Fig 2D). Specifically, 188 conjugates had enhanced abundance over WT fold-change by greater than 1.5-fold, whereas the levels of only 79 conjugates were reduced fold-change by less than 1/1.5-fold (0.67) (Fig 2D). Fold-change levels relative to the statistic P-value of individual ubiquitinated site were plotted to give more details on the dataset (Fig 2E). Of 267 DECs (|FC| > 1.5) was the fact that ubiquitin-conjugating enzyme 35 (UBC35) and ubiquitin-activating enzyme 1 (UBA1) were among the targets with significantly enhanced ubiquitination levels (Fig 2E), and RPN10, a known target of UPL3 (Furniss et al, 2018), was appeared in dataset of ubiquitome with a down-regulated ubiquitination level, although several conjugates including HXK1, PPC2, VAMP714 etc. were indicated in P-value > 0.05 (yellow dots) of two replicates; they were both significantly down-regulated in two replicates (Supplemental Data 2 (62.5KB, xlsx) ), indicating that the quality of ubiquitomic datasets were properly sound. Together, this surprising rise in protein ubiquitination caused by loss of UPL3 perhaps means that UPL3 has function on deubiquitinating pathway or UPL3 promotes deubiquitinases (DUBs) function.
Differential ubiquitomic enrichment reveals that metabolic enzymes are among the processes significantly affected by UPL3
GO term enrichment showed that the molecular functions of assembled ubiquitin conjugates were related to protein–protein interactions (47%), catalytic activity (34%), transport activity (7%), structural molecule activity (5%), and other (7%). For the DUCs (|FC| > 1.2, P-value < 0.05) of upl3 relative to WT, the protein–protein interaction function enrichment was slightly increased to 49% (Fig 3A), implying a major involvement in molecular interaction for the UPL3-regulated targets. Further assignment to biological processes showed that the DUCs included mainly proteins involved in the response to metal ion stresses and enzymes related to carbon metabolism and nucleotide metabolism (Fig S4). Hence, it is plausible that UPL3 maintains the ubiquitination status of enzymes and regulatory factors to fine-tune cellular metabolism.
Figure 3. GO term and KEGG pathway enrichments for the UPL3-associated ubiquitin conjugates (|FC| > 1.2, P-value < 0.05).
(A) GO term enrichment in categories of molecular functions of UPL3-associated ubiquitin conjugates. (B) Bubble plots showing the KEGG pathway of UPL3-associated ubiquitin conjugate differentially ubiquitin conjugates. (C) The UPL3-regulated ubiquitin conjugates of the upl3/WT mapped in the carbon fixation pathway. (D) The UPL3-ubiquitinated conjugates of the upl3/WT mapped in cysteine and methionine metabolism pathway. (E) Distribution of UPL3-associated differentially ubiquitin conjugates associated with cellular components. (F) Subcellular localization of UPL3 in the nucleus of Arabidopsis leaves transiently expressing a C-terminal GFP fusion. The GFP-alone expressing leaves sample is used as the control. The nucleus is counter-stained with DAPI. Bars = 20 μm.
Figure S4. The UPL3-related ubiquitin conjugates.
(A, B) The list of the UPL3-influenced ubiquitin conjugates (A) and UPL3-dependent ubiquitin conjugates (B) in the upl3/WT. Pink color indicates ubiquitin enriched in the upl3 relative to the wild type, and blue color indicates ubiquitin down-regulated in the upl3 relative to the wild type.
This notion was further validated via KEGG pathway analysis (Figs 3B and S4). A large subset of the DUCs was enzymes related to biosynthesis of secondary metabolites, carbon fixation, carbon metabolism, and amino acid metabolism. The ubiquitinated forms of the Calvin–Benson enzymes, such as ribulose-1,5-P2-carboxylase (RuBisCO), phosphoglycerate kinase (PGK), GAPDH, and the aldoketo reductase family members (ALDOs), as well as the CAM enzymes malate dehydrogenases MDH1 and MAEB homolog, were enriched in the upl3 plants (Fig 3C). On the other hand, a small subset of declined ubiquitin conjugates was cysteine and methionine metabolism–related enzymes, such as 5-methyltetrahydropteroyl-triglutamate, homocysteine methyltransferase 2 (MS2), S-adenosylmethionine synthase 1 (SAM1), cysteine synthase 1 (OASA1), methionine aminotransferase (BCAT4), SNARE-like superfamily protein (YKT61), and vesicle-associated membrane protein 714 (VAMP714) in the upl3 plants (Fig 3D and Supplemental Data 2 (62.5KB, xlsx) ). Interestingly, carbon metabolism was also represented by a reduction in ubiquitin conjugates of phosphoenolpyruvate carboxylase (PPC2) and hexokinase 1 (HXK1) in the upl3 background (Fig 3C and Supplemental Data 2 (62.5KB, xlsx) ).
Our Arabidopsis ubiquitome proteins can be classified as cytoplasmic (36%), nuclear (20%), chloroplast (20%), membrane (14%), and others (10%). Among these, the percent ratio of the UPL3-related DUCs (|FC| > 1.2, P-value < 0.05) was higher in the cytoplasmic and the nuclear sections (Figs 3E and S4). The Cytoscape protein–protein interaction network of the 545 UPL3-related DUCs generated distinctly clustered interaction nodes (Fig S5) (Shannon et al, 2003). The UPL3-regulated conjugates were dispersed throughout the network, suggesting that UPL3 was likely involved in broad control of stress or stimulate response (Fig S5). UPL3 fused GFP transformed transiently in tobacco leaves showed that UPL3 was clearly localized in the nucleus (Fig 3F). Therefore, nuclear proteins such as those regulatory functioning in chromatin remodeling, chromosome regulation, and transcriptional complexes related to stresses are likely primary UPL3-dependent targets.
Figure S5. Gene Ontology (GO) enrichment of genes with differential ubiquitinated conjugates (fold-change > 1.5) of upl3/WT.
(A) Ubiquitin-enriched targets. (B) Ubiquitin–down-regulated targets. Cytoscape analysis exhibited a biological network of differentially ubiquitin conjugates in many clustered interaction network with broadly distinct hubs. Functionally clustered network with terms as nodes linked based on their kappa score level (≥0.3). Edge weight indicates intensity of clustered interaction. The red gradient of nodes represents the term enrichment significance. Cluster without any interaction partners within the network indicate singletons. The size of node indicates the gene proportion of each cluster associated with the term.
The UPL3-dependent conjugates are enriched in histone H1/H5 and stress-related protein domains with a noncanonical lysine pattern
Scanning protein domains of the DUCs using InterProScan identified several UPL3-ubiquitinated (reduced ubiquitination in the upl3 background) protein domains including the histone H1/H5 domain, jacalin-like lectin domain, GST domain, S15/NS1 RNS binding domain, and heavy-metal–associated domain (Fig 4A blue), whereas the UPL3-regulated ubiquitin conjugates (enhanced ubiquitination in the upl3 background) contained leucine-rich repeat, histone H2A/H2B/H3, ribosomal protein S5, double-stranded RNA-binding domain, and histone fold domains (Fig 4A orange). For protein domains of the individual DUC within each fold-change range (Q1, Q2, Q3, and Q4), the results were shown in Fig S4 and Supplemental Data 2 (62.5KB, xlsx) . Again, it was noted that protein domains associated with RNA binding, protein translation, and amino acid synthesis–related proteins were overrepresented in proteins showing reduced ubiquitination in the upl3 plants, followed by heavy-metal–associated domain and remorin- and jacalin-like lectin domain of abiotic (metal)/biotic stress–responsive proteins. In contrast, protein domains associated with protein binding, transport ATPase, ubiquitin carboxyl-terminal hydrolase, and metabolism enzyme were the most significant domains in proteins showing enhanced ubiquitination in the upl3 plants, followed by domains contained in the ubiquitination/26S proteasome system (UPS) regulatory complex (Fig S6 and Supplemental Data 2 (62.5KB, xlsx) ). It suggested that proteins containing heavy-metal–associated domains, GST domains, jacalin-like lectin domains, and histone H1/H5 domains were likely UPL3-ubiquitinated conjugates. Correspondingly, UPL3-regulated conjugates contain chromatin remodeling ATPase BRM, histone H2A/H2B/H3, histone fold, and SPK1/BTB/POZ–binding domain, most of which have functions related to protein binding activity and gene transcriptional control (Fig S6). In this scenario, UPL3-regulated conjugates in the nucleus are likely mediators of chromatin accessibility and transcriptional processes that control downstream gene expression.
Figure 4. Protein domains and lysine footprints of UPL3-associated differentially ubiquitin conjugates (|FC| > 1.2, P-value < 0.05).
(A) GO terms enrichment analysis of enriched protein domains of differentially ubiquitin conjugates (|FC| > 1.2, P-value < 0.05) via InterProScan. (B) The consensus ubiquitin attachment motif identified by the MEME Suite. (C) List of apparent ubiquitin attachment sites and motif in identified proteins (|FC| > 1.5, P-value < 0.05). R1 and R2 are two replicates. Those up-regulated and down-regulated in upl3 relative to WT were highlighted in red and blue, respectively.
Figure S6. The protein domain analysis of differential ubiquitinated conjugates within different fold-change regions by the upl3 relative to WT by heatmap.
Red color indicates up-regulated, and blue color indicates down-regulated.
Ubiquitin can be attached to substrate proteins as a single moiety or polymeric chains and adopt distinct conformations and lead to different functions in cells (Komander & Rape, 2012). To get insight ubiquitinated lysine site pattern of UPL3, the H89R substitution in the tagged ubiquitinated assay was used here to enable the detection of ubiquitination sites (“footprints”) and identify a consensus ubiquitin attachment sequence (Xu et al, 2010). By scanning all generated datasets, we identified 2,778 ubiquitinated sites in total (Supplemental Data 1 (418.1KB, xlsx) ). Among these, 2,359 sites were quantified, 1,641 of the 2,359 sites were in the upl3 plants, and 414 sites were differentially displayed relative to WT plant with a cutoff log2FC of 1.5.
We identified 110 ubiquitinated modification sites on 77 differentially ubiquitinated proteins in upl3/WT (log2FC > 2, P-value < 0.05) (Supplemental Data 2 (62.5KB, xlsx) ). Motif analysis around the modified lysine using MEME identified a consensus ubiquitin attachment sequence in 44 of the 110 sites (Fig 4B) that strongly matched the c-K-x-E/D/G ubiquitination motif (where c and x represent a hydrophobic and any amino acid, respectively), which was a prevalent motif in yeast and animal ubiquitinated targets (Xu et al, 2010). However, the remaining 66 sites (60%) were unrelated to this motif, indicating that noncanonical sites were also common. In addition, referring to the GPS-SUMO algorithm (Zhao et al, 2014), one or more copies of this consensus sequence were detected in ubiquitinated targets accounting for 74%, 66%, and 59% of the three enriched ubiquitination categories, the UPL3-ubiquitinated, abundant, and total ubiquitinated proteins, respectively. Among the 44 sites with a consensus sequence, of the 110 sites, 14 of the 21 mapped attachment sites on 18 UPL3–up-regulated targets belonged to the canonical c-K-x-E/D/G motif, with the remainder had alternative sequences (Supplemental Data 3 (236.7KB, xlsx) ), including GAPC1, GAPC2, CASA1, NADP-ME2, RuBisCO, AAT1, FBAB, PGK3, MDH1, GLO1, and SHM4 (Fig 4C). Specifically, the ubiquitinated sites identified here for the UPL3-ubiquitinated targets, namely, ALDH, OASA1, HXK1, and PPC2, were within a noncanonical x-A-K-x- motif or x-K-A-x motif (Fig 4C).
Analysis of ubiquitin footprints (K sites) of upl3/WT.LSA-2022-01492_Supplemental_Data_3.xlsx (236.7KB, xlsx)
Identifying the potential ubiquitylated targets in UPL3-bound proteins
To evaluate the direct connection between UPL3 and conjugates, we assessed protein–protein interaction using a GFP nanotrap–assisted pulldown-MS assay (Fig S7), in which the GFP-tagged UPL3 was a bait. To the end, the rosette leaves of 6-wk-old stable transgenic plants expressing either the UPL3-GFP or the control GFP driven by the ACTIN3 promoter were used for total protein isolation, purification, and immunodetection (Fig 5A and B). The trypsin-digested proteins were subjected to mass spectrometry analysis with high-energy collisional dissociation quantum efficiency mass spectrometry (QE-MS). Tandem mass spectra were searched against UniProt A. thaliana Columbia (89247_20181227) database via Mascot2.2 software. A total of 81 putative proteins were identified after subtracting the GFP control resulted from the UPL3-GFP candidate list (Supplemental Data 4 (29.3KB, xlsx) ). With these putative UPL3-interacting patterns, we identified 29 (|FC| > 1.5, P-value < 0.05) or 11 under more stringent conditions (|FC| > 1.5, P-value < 0.01) overlapping proteins in the UPL3-ubiquitome DUCs (Fig 5C).
Figure S7. Preparation and quality detection of the GFP-binding protein (GBP) bead.
GFP protein (∼30 kD) isolated from ACTIN3:GFP plants was immunodetected by an antibody against GFP (left), and the protein was stained by Coomassie (right). M: protein marker, lane1: GFP input, lane2: GFP-IP, lane3: washing solution (supernatant after immunoprecipitation); ∼30-kD signal indicated GFP protein.
Figure 5. UPL3-interacting proteins overlapping with differentially ubiquitin conjugates (DUCs) identify potential UPL3-ubiquitinated substrates ABCG36, MS2, OEP6, PPC2, and LOS1.
(A) Immunodetection of protein samples isolated from GFP- and UPL3-GFP–expressing plants with an antibody against GFP. (B) Nano-trapped proteins for MS analysis. (C) Overlapping proteins between UPL3-interacting candidates and the DUCs. Eighty-one putative interacting proteins were identified as specific to UPL3-GFP, which contained 29 and 11 DUCs with a fold-change |FC| > 1.2 or 1.5, respectively. (D) Enrichment of the KEGG pathway in the overlapping genes highlights an involvement of potential UPL3-ubiquitinated substrates in the IP3K pathway and in ribosomal activity. Red color indicates up-regulated, and blue color indicates down-regulated. (E) List of most notable interacting candidates of UPL3 with their respective fold-change of ubiquitination level in upl3 versus wild-type plants. Orange indicates up, and blue color indicates down. The fold-change is indicated with the data.
Source data are available for this figure.
The dataset for GFP alone and UPL3-GFP nanotrap-pulldown mass spectrum analysis.LSA-2022-01492_Supplemental_Data_4.xlsx (29.3KB, xlsx)
These 29 proteins were clustered in four categories based on the KEGG pathway database. Consistently, proteins involved in the carbon fixation pathway have enhanced ubiquitination in the upl3 mutant, whereas those in the inositol-1,4,5-trisphosphate-3-kinase (IP3K) signaling pathway have reduced ubiquitination in the upl3 plants versus WT (Fig 5D and E), which included six proteins, namely, ABCG36, MS2, PPC2, LOS1, HXK1, and AT3G63160 (Fig 5E, fold-change depicted in blue). 14 other interacting candidates showing upl3 mutation–enhanced ubiquitination in the upl3 background were H2AXb, RPS2B, PHOT1, ERD14, the SWI-SNF chromatin-remodeling ATPase BRAHMA (BRM), and SWIS3C, as well as carbon metabolism–related enzymes (Fig 5E, fold-change depicted in orange). Notably, UBP12, UBP13, and UBP26 are also among the UPL3-interacting candidates (Fig 5E). UBP12 is an ubiquitin hydrolase, with a demonstrated deubiquitination activity in vitro and localization both in the cytoplasm and the nucleus (Derkacheva et al, 2016; Kralemann et al, 2020).
UPL3 interacting with UBP12, HXK1, and PPC2 are formed a complex
To confirm these putative UPL3 interacting partners, we carried out a yeast two-hybrid assay by selecting 16 candidates. The self-interacting N-terminal fragment (470 aa) of UPL3 containing armadillo repeats was shown interacting with most of the selected candidates except for UPL5 and PHOT1; however, the full-length UPL3 only showed a strong interaction with UBP12 and very weak interaction with BRM, HXK1, PPC2, and UBC35 (Figs 6A and B and S8A), but the full-length UPL3 bait was not able to interact with its N-terminus (Fig 6B). Next, the interaction of UPL3 with UBP12 was confirmed using BD-UBP12 as a bait, which in turn showed a weak interaction with PPC2 and BRM, but not HXK1 (Fig 6C).
Figure 6. Confirmation and characterization of UPL3 interaction with its targets.
(A) Domain structure of the UPL3 protein showing the full-length and the N-terminus used in Y2H assay. (B) Confirmation of UPL3 interaction with its targets in a yeast two-hybrid assay. The AD or BD empty vector was used as a negative control, and the interacting pair UPL5-WRKY53 was used as a positive control. SD/-LT: SD media minus Leu and Trp, SD/-ALTH: SD media minus Ade, Leu, Trp, and His. Bars = 10 mm. (C) UBP12 interacts with BRM, PPC2, but not HXK1 in a yeast two-hybrid assay. SD/-LT: SD media minus Leu and Trp, SD/-LTH: SD media minus Leu, Trp, and His, SD/-ALTH: SD media minus Ade, Leu, Trp, and His. Bars = 10 mm. (D, E) The detection of bimolecular fluorescence complementary assay; an empty vector was used as a negative control (Fig S8B). Bars = 20 μm. (F) CoIP detection of ubiquitin conjugates (representation: HXK1 and PPC2) in the overexpression UPL3-GFP and UBP12-GFP plants compared with wild-type plants. Antibodies against HXK1 and PPC2 (Agrisera) and anti-GFP (Rothe) are used.
Source data are available for this figure.
Figure S8. Interaction of UPL3 with its putative interacting partners.
(A) Interaction of UPL3 with its putative interacting partners by yeast two-hybrid. Bars = 10 mm. (B) Interaction of UPL3 or UBP12 with an empty vector of BiFC is used as negative controls. Bars = 20 μm. (C) Detection of antibodies against HXK1, PPC2, and BRM in the gin2, the ppc2, and the brm mutants compared with WT by Western blotting; Ponceau staining is used as loading controls.
We further examined the interaction of UPL3 or UBP12 and its interacting partners by bimolecular fluorescence complementation (BiFC) assays and coimmunoprecipitation (CoIP). UPL3 or UBP12 and partners were fused to the YN vector (pCAMBIA1300-N1-YFPN) and YC vector (pCAMBIA1300-N1-YFPC), respectively, and then were cotransformed to Arabidopsis epidermal cells by Agrobacterium-mediated injection. GFP signals in the nucleus were strongly observed in UPL3-GFPn and its interacting partners BRM-GFPc, HXK1-GFPc, PPC2-GFPc, and UBP12-GFPc-co-delivered Arabidopsis epidermal cells (Figs 6D and S8B), as well as in UBP12-GFPn and BRM-GFPc and PPC2-GFPc but not in UBP12-GFPn- and HXK1-GFPc-co-delivered Arabidopsis epidermal cells (Fig 6E); the empty vector was used a negative control (Fig S8B). Next, we detected the interaction of UPL3 with UBP12 or HXK1 or PPC2 in vivo by Co-IP with the oeUPL3-GFP or oeUBP12-GFP transgenic plants. An anti-GFP antibody was used for immunoprecipitation and anti-HXK1, anti-BRM, and anti-PPC2 antibodies were then used for IP Western-blotting detection. UPL3-GFP was coimmunoprecipitated with endogenous HXK1 and PPC2 in 6-wk-old plants, and UBP12-GFP was coimmunoprecipitated with PPC2 but not HXK1 (Figs S8C and 6F), indicating that UPL3 is able to interact with HXK1 and PPC2; UBP12 is able to interact with PPC2, and thus, UPL3, UBP12, BRM, PPC2, and HXK1 may form a complex.
The UPL3 and UBP12 affect HXK1 and PP2C protein stability and carbohydrate contents
To examine whether UPL3 altered the protein level of the bound candidates of UPL3 between upl3 and WT, to the end, we compared the total proteomes of 6-wk-old wild-type and the upl3 plants by tandem MS using the precursor ion intensity of the MS1 scans for quantification. Altogether, 3,557 Arabidopsis proteins could be reproducibly identified and quantified in both samples by our liquid chromatography–mass spectrometry (LC–MS) regime analyzed in triplicate (Fig 7A and Supplemental Data 5 (266KB, xlsx) ). 492 proteins were up-regulated, and 236 proteins were down-regulated in differentially expressed proteins (DEPs, |FC| > 1.5, P-value < 0.05) of proteome dataset from upl3/WT (Fig S9 and Supplemental Data 5 (266KB, xlsx) ). When above DUCs (|FC| > 1.5, P-value < 0.05) was normalized to DEPs (|FC| > 1.5, P-value < 0.05), 105 ubiquitination sites (74 proteins) were enriched and only seven sites (five proteins) were down-regulated (Fig 7B) from upl3/WT. Among them, several UPL3-bound proteins such as UBC35, UBC7, BRM, UBP12, and UPL13, as well as other UBP members (e.g., UBP1C, UBP6, UBP26) were slightly up-regulated 1–1.5 fold in the upl3 mutant relative to WT; the rest UPL3-bound proteins HXK1, PPC2, ABCG36, MS2, LOS1 exhibited a down-regulated 1.5–3-fold in the upl3 mutant relative to WT (Supplemental Data 5 (266KB, xlsx) ). Furthermore, when the ubiquitination enrichment was normalized to the protein level, the level of ubiquitinated UBP12, UBP13, and UBP26 was not significantly changed between the upl3 and WT; however, the ubiquitin level of ubiquitinated UBC7, UBC35, and BRM maintained an up-regulated potential. HXK1, PPC2, ABCG36, MS2, and LOS1 maintained a down-regulated trend (Fig 7C). This suggests that UBP12 and other UBP members are interacting partners but were not altered their ubiquitin level by UPL3.
Figure 7. The protein levels of representation: BRM, HXK1, and PPC2 in the UPL3 or UBP12 mutants.
(A) A total of 3,557 Arabidopsis proteins could be reproducibly identified and quantified in both samples by our liquid chromatography–mass spectrometry (LC–MS) regime analyzed in total triplicate, 3,445 of which were described here as targets of UPL3-associated DEPs (cutoff, P-value < 0.05). (B) Numbers of differentially expression proteins (DEPs) (|FC| > 1.5, P-value < 0.05) of upl3 relative to WT (Left). Numbers of differentially ubiquitinated conjugates (|FC| > 1.2, P-value < 0.05) normalized to DEPs (|FC| > 1.5, P-value < 0.05) (Right). Orange, up-regulated; blue, down-regulated. (C) The fold-change of UBP proteins and BRM, PPC1, PPC2, UBC2, UBC7, HXK1 from proteome dataset of upl3/WT (Supplemental Data 5 (266KB, xlsx) ). (D) Immuno-detection of ubiquitin conjugates (representation: BRM, HXK1, and PPC2) in the upl3 and the ubp12; overexpression UPL3 and UBP12 plants compared with wild-type plants. Antibodies against BRM (provided by Dr. Rongcheng Lin), PPC2, and HXK1 (Agrisera) and polyubiquitin (Cell Signaling Tech) are used, and the tubulin level is used as protein-loading controls. (E) Immuno-detection of ubiquitin conjugates (representation: BRM, HXK1, and PPC2) in the upl3 and the ubp12; overexpression UPL3 and UBP12 plants compared with wild-type plants after treatment of ubiquitination inhibitor MG132. (F, G, H) Starch staining with KI and quantification of starch and sucrose in 18–21-d-old rosettes (n = 5). Error bar represents the SD of triplicates. Asterisks denote the statistically significant level different with the wild type, as verified via t test: *P, 0.05; **P, 0.01, n = 5.
Source data are available for this figure.
Figure S9. Quality detection of the tryptic peptides of proteomes in the 6-wk-old upl3 plants and wild-type plants.
(A) Detection of the mass error of all identified peptides (mass error). The quality error takes 0 as the central axis and is concentrated in the range of less than 10 ppm, indicating that the quality error meets the requirements. (B) The length of most peptide segments is distributed between 8– and 20–amino acid residues, which is in line with the law of trypsin digesting peptide segments, indicating that the sample preparation meets the standard. (C) Detection of statistically significant similarity of two replicates by heatmap of Pearson’s correlation coefficient. rep: replicate, A: WT, B: upl3. (D) Detection of statistically significant similarity of two replicates by scatter diagram. rep: replicate, A: WT, B: upl3. This coefficient is a measure of the degree of linear correlation between the two groups of data: when the Pearson coefficient is closer to −1, it is negative correlation; closer to 1, it is positive correlation; and closer to 0, it is irrelevant.
Differential accumulation protein of upl3/WT (FC 1.2).LSA-2022-01492_Supplemental_Data_5.xlsx (266KB, xlsx)
To examine whether the interaction between UPL3 and UBP12 might contribute to the protein level of ubiquitinated targets in planta, the loss- or gain-of-UPL3 and -UBP12 plants were used. We first detected the protein level of BRM, HXK1, and PPC2 in the ubp12 and the upl3-1 mutant background and overexpressing UBP12 and UPL3-24 transgenic plants relative to WT using antibodies against BRM, HXK1, PPC2, and polyubiquitin. The protein levels of PPC2 and HXK1 were increased by UPL3 mutation and in overexpressing UBP12 plant and decreased by UPL3 overexpression and by UBP12 mutation (Fig 7C). The total ubiquitination level in the upl3 or the ubp12 was comparable or slightly stronger than in the WT, whereas it declined considerably in the UPL3- or UBP12-overexpressing line (Fig 7C), In contrast, the BRM protein level was increased in both upl3 and oeUPL3 but however was down-regulated in the oeUBP12 and did not change in the ubp12. To avoid an influence of UPL3 or UBP12 on BRM, PPC2 and HKX1 gene expression at the transcriptional level, the transcript levels of BRM, PPC2, and HXK1 in loss- or gain-of-UPL3 and -UBP12 mutants were detected by RT-qPCR and showed there were no significantly different of PPC2 and HXK1, but the transcript level of BRM significantly up-regulated in the oeUBP12 and down-regulated in the ubp12 (Fig S10). It suggested that UPL3 affected PPC2 and HXK1 protein levels in the UPS degradation pathway, although UPL3 unexpectedly declined globally polyubiquitination enrichment (Figs 2 and 7C). However, UBP12 and UPL3 displayed the opposite effect on HXK1 and PPC2 protein levels when their genes were mutated or overexpression. Loss of UBP12 or gain of UPL3 reduced but loss of UPL3 or gain of UBP12 increased the protein accumulation relative to WT (Fig 7C). The BRM level was regulated by UBP12 in transcriptional and protein levels. Furthermore, during treatment with inhibitor MG132 of protein ubiquitination in the upl3 and the oeUPL3 plants, the immunodetection of the protein level of PPC2 and HXK1 showed that two protein levels were pronouncedly accumulated in the upl3 and the oeUPL3 after treatment, although PPC2 and HXK1 levels were higher in the upl3 and lower in the oeUPL3 plants relative to WT (Fig 7D). The BRM level was not affected by MG132 treatment in the upl3 and ubp12 background but significantly up-regulated in oeUBP12 and oeUPL3 plants after MG132 treatment. Notably, the polyubiquitin level was apparently increased in the upl3 lines but decreased in the oeUPL3 lines; however, its level was enhanced in the upl3 and declined in the oeUPL3 lines after the treatment of MG132, suggesting that UPL3 has transferring ubiquitin activity, but it did not directly affect the BRM protein level. We concluded that UPL3 has a major effect on PPC2 and HXK1 protein degradation, whereas UBP12 antagonistically affect the HXK1 and PPC2 protein levels and BRM level in the transcriptional level and protein level. These results suggest a complicated molecular interaction network between UPL3 and these proteins, probably in terms of homeostasis in protein ubiquitination.
Figure S10. The transcript level of BRM, PPC2, and HXK1 in the in loss- or gain-of-UPL3 and -UBP12 mutants.
The value represents average value of three replicates.
Because PPC2 is a key enzyme for primary metabolism, the carbohydrate contents of the upl3 and ubp12 mutant plants was monitored to check whether the opposite phenotypes observed in upl3 and ubp12 were related to carbon metabolism in the ppc2 and the gin2, a HXK1 mutant gin2 (glucose-insensitive2) (Cho et al, 2006). The upl3 mutant showed a decreased starch and sucrose accumulation compared with WT, whereas complementation or overexpression of UPL3 restored the starch and sucrose content to the WT level (Fig 7E–H). In contrast, the ubp12 mutant displayed significantly stronger starch and sucrose accumulation than that in WT, whereas effect of UBP12 overexpression was similar to that of the upl3 mutant, with a much lower starch and sucrose content in the plants (Fig 7E–H). The contents of starch and sucrose were significant accumulated in the ppc2 and the gin2 mutant compared with WT. Therefore, starch accumulation is positively correlated with mutations in the two antagonistic ubiquitination/deubiquitination pathway genes.
The UPL3 interacting UBP12 regulates carbohydrate metabolism related leaf aging and flowering
We further sought insights from phenotypic analysis of the upl3-1, brm, ubp12, gin2-1, and ppc2 mutant plants. The results showed that brm, ubp12, gin2, and ppc2 plants shared a common phenotype of lower numbers of rosette leaves and early bolting and flowering, but BRM and PPC2 caused the curled leaves and premature leaf aging phenotype only found in the ppc2 and the brm plants (Fig 8A and B). Summarily, upl3 plants displayed a late-senescence phenotype with more numbers of rosette leaves and delayed flowering time (more than 1 wk); the effect of UPL3 on plant development contrasted those of both PPC2 and HXK1 (Figs 8C and D and S11), thereby a mild curled-leaves phenotype was also observed in aging leaves of the upl3 plants at a later stage of development (Figs 1A and 8A), consistent with previous reports of phenotype of brm, ubp12, and ppc2 mutants (Cui et al, 2013; Xu et al, 2016; Li et al, 2016a, 2016b; Archacki et al, 2017; Park et al, 2019).
Figure 8. The brm, ppc2, ubp12, and oeUPL3 plants show similar early leaf senescence and early-flowering phenotypes.
(A) Phenotypes of 5-wk-old plants. Bars = 5 mm. (B) Bolting and flowering phenotype of the 5-wk-old plants. Bars = 40 mm. (C) Rosette leaf numbers of different genotypes at indicated time (n > 20). (D) Chlorophyll content in rosette leaves of 6-wk-old plants (n = 3). (E) Photosystem II fluorescence activity (Fv/Fm) of rosette leaves of 6-wk-old plants (n = 5). (F) Proportion of green and yellow leaves of the whole rosette of 8-wk-old plants (n = 12). (G) Quantitative real-time PCR of gene expression from 6-wk-old upl3 and wild-type plants for 13 selected plant aging and senescence-related transcription factors. Error bars represent the SD of three biological replicates. (H) GO terms enrichment analysis of 1,467 differential display genes (DEGs, |FC| > 2, P-value < 0.05) of upl3/WT.
Figure S11. Flowering rates of upl3, brm, gin2, ppc2, and ubp12 lines compared with wild type.
A total of 120 plants were used to calculate.
Based on a delaying senescence phenotype of upl3 mutant and premature senescence and early bolting phenotype of ubp12 mutant, we further selected 13 genes coding for transcription factors such as WRKYs and NACs related to leaf aging (e.g., WRKY53, WRKY75, and ORE1) and salicylic acid responsive senescence (e.g., WRKY38, WRKY63, and WRKY51 etc.), as well as floral transition genes (e.g., FT and FLC) for RT-qPCR analysis (Fig 8G and H). The results showed that the expression level of WRKY53, WRKY75, WRKY38, WRKY63, and WRKY51, as well as NACs was significantly decreased either in the upl3 (Fig 8G and H) or in the oeUBP12, in contrast significantly increased either in the oeUPL3 or in the ubp12. These data confirmed the notion that UPL3 had a profound functional involvement in stress-responsive cell senescence and developmental cell senescence (aging) via ubiquitination of their regulators either directly or indirectly.
Because UPL3 interacts with its partners in the nucleus, we compared transcriptome data of the upl3 and WT plants, which released from Furniss et al (2018) (Supplemental Data 6 (267.2KB, xlsx) ). A total of 1,467 differentially expressed genes between the upl3 and WT seedlings were identified (Figs S12A and Supplemental Data 6 (267.2KB, xlsx) ). Of these, genes related to stress responses (drug, hypoxia, oxidative, and UV) were most significantly up-regulated, followed by genes associated with cellular glucan metabolic processes, anthocyanin-containing compound biosynthesis, and cellular polysaccharide catabolic processes. A third set of up-regulated genes was involved in protein transport and leaf senescence. In contrast, genes for response to (a)biotic stress (metal ion salt stress and fungus) and plant aging were most significantly down-regulated, followed by genes related to secondary metabolic processes, MAPK signaling pathway, leaf cell death, and phenylpropanoid biosynthesis (Figs 8H and S12B).
Figure S12. Co-analysis of UPL3-dependent transcriptome highlights stress-related genes.
(A) Venn diagrams showing overlaps of differentially ubiquitin conjugates (fold-change > 1.5) and transcriptome DEGs (log2FC > 1.3) of upl3 relative to WT, RNA-seq data released from the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-7374 (Furniss et al, 2018). (A, B) List of the 26 overlapping genes of differentially ubiquitin conjugates and DEGs, as in (A). Text in blue and orange denotes a down- or up-regulation in the upl3 plants, respectively.
Differential display expression targets of upl3/WT transcriptome dataset.LSA-2022-01492_Supplemental_Data_6.xlsx (267.2KB, xlsx)
We analyzed DUCs and DEGs datasets of upl3/WT and showed that 26 proteins were identified as the overlapping proteins of DEGs and DUCs, with an altered ubiquitinated protein level (|FC| > 1.5, P-value < 0.05) and gene expression level (|FC| > 1.3, P-value < 0.05) by the upl3 mutation (Fig S12A). Among these proteins, four proteins (Fig S12B, fold-change depicted with two orange values) with increased ubiquitin conjugates were up-regulated in gene expression in the upl3 background, including glucomannan 4-β-mannosyltransferase 9 (CSLA9), inositol-3-phosphate synthase isozyme 1 (MIPS1), calcium-binding protein 16 (CML16), and the Patellin-2 (PATL2) (Fig S12B, fold-change depicted with two orange values). Two other proteins (Fig S12B, fold-change depicted with a blue and an orange value) with reduced ubiquitin conjugates but with an increased transcript level in the upl3 mutant were cysteine lyase (CORI3) and cinnamyl alcohol dehydrogenase 7 (CAD7), both of which had reported functions in the amino acid metabolic process (Tsuwamoto & Harada, 2011; Tanaka et al, 2018). On the other hand, 9 proteins (Fig S12B, fold-change depicted with an orange and a blue value) showed an enhanced ubiquitin conjugates level but a down-regulated expression level in the upl3 mutant plants. These included calmodulin-like protein10 (CML10), aquaporin (PIP1-5), triacylglycerol lipase-like 1 (TLL1), leucine-rich repeat ser/thr protein kinase (LRR-RLK), the jacalin-related lectin 23 (JAL23), ABCB transporter member 19 (ABCB19), methionine aminotransferase (BCAT4), and the ankyrin repeat-containing protein (BDA1). The remaining 11 proteins (Fig S12B, fold-change depicted in blue) were those with both a reduced ubiquitin conjugates level and down-regulated gene expression in the upl3 mutant. Their functions seemed to be related to glycoside metabolism and transport pathways. From these data, it is obvious that the UPL3-centered molecular network involves both feed-forward and feed-back regulatory pathways and mostly impacts on cellular metabolism, stress-responsive cell death, and aging.
Discussion
During cell senescence, the predominant regulation is protein degradation, which is critical not only in signal transduction but also for the execution of the senescence syndrome (Buchanan-Wollaston et al, 2005; Woodson et al, 2015; Yolcu et al, 2017; Guo et al, 2021). Our multi-omics analysis provides an overview of UPL3 downstream targets that were either directly or indirectly affected at the protein level and the transcript level. An unexpected finding is that loss of UPL3 results in a globally enhanced ubiquitination of metabolism-related proteins. The unveiling of physical interactions with the ubiquitin-specific proteases UBP12, BRM, HXK1, and PPC2 is of particular interest, suggesting a potential action model of UPL3 interacting with UBP12 in BRM, HXK1, and PPC2, mediating carbohydrate metabolic–related protein turnover during plant development (Fig 9).
Figure 9. A noncanonical working model of UPL3 in regulating cell senescence of Arabidopsis by multi-omics and genetics analysis.
Based on integrative datasets of ubiquitome, proteome, and transcriptome. Preferential ubiquitination of proteins related to carbon fixation represented the largest set of proteins with increased ubiquitination in the upl3 plant, whereas a small set of proteins with reduced ubiquitination caused by the upl3 mutation were linked to cysteine/methionine synthesis processes. Notably, ubiquitin hydrolase 12 (UBP12), BRM, HXK1, and PPC2 were among the UPL3-interacting partners identified as the UPL3-interacting partners by both GFP nanotrap–mass spectrometry analyses and yeast two-hybrid assay; characterization of upl3-, brm-, ppc2-, gin2-, and ubp12-mutant plants and transcriptome analysis suggested that UPL3 fine-tunes carbon metabolism, mediating cellular senescence via proteolysis-independent regulation and proteolysis-dependent degradation on metabolism-mediated cell senescence. Red color represents up-regulated, blue color represents down-regulated, thick frame represents more numbers of ubiquitinated proteins.
Globally, the loss of the HECT-type E3 ubiquitin ligase UPL3 leads to the depletion of ubiquitin conjugates of specific proteins, accounting for ∼1/3 of the total proteins whose ubiquitination status is significantly altered, with a ratio |FC| > 1.2 (Fig 2). And the proteome of the upl3, accounting for 1,737 proteins, is significantly altered with a fold-change |FC| > 1.2 (Supplemental Data 5 (266KB, xlsx) ). Although two combined proteomic and ubiquitomic datasets of three replicates are used to analyze and the variant P-value is not so low about 0.6, we harvested more 500 DUCs (|FC| > 1.2, P-value < 0.05). After normalized to proteome dataset, these proteins are considered as putative candidates regulated by the UPL3 enzyme, although not all proven experimentally yet. And some of these are demonstrated UPL3 targets, for example, GLS3 (Saracco et al, 2009; Patra et al, 2013; Kim et al, 2015; Gao et al, 2017). It did not appear in our dataset; however, other common known ubiquitinated targets included in the list are AHA1, CDC48A, ERD4, LEC2, and RPN10 proteins (Fig S4, and Ref. in Downes et al [2003], Park et al [2008], Finley [2009], Rai et al [2012], Besche et al [2014], Yamauchi et al [2016], Furniss et al [2018], and Kumari et al [2019]), playing roles in cell division, cell senescence, and stress-induced cell senescence.
UPL3 alters the levels of ubiquitin conjugates involved in metabolism and heavy-metal stress-induced cell senescence
UPL3 has several new putative targets, such as MS2, OASA1, ALDHA3, HXK1, and PPC2. The protein level of these five putative target proteins were accumulated by proteome analysis (Fig 7C and Supplemental Data 5 (266KB, xlsx) ), and the ubiquitin level of them were down-regulated in the upl3 mutant relative to WT by ubiquitome analysis (Figs 2 and 3 and Supplemental Data 2 (62.5KB, xlsx) ); the ubiquitinated sites were identified as a noncanonical pattern (Fig 4); two of them: HXK1 and PPC2 were detected to be direct interacting partners of UPL3 (Fig 6). This evidence supposed that UPL3 recruited them and affects their ubiquitin level and protein level in the ubiquitination/26S proteasome system (UPS) degradation pathway. It has been reported that these five putative proteins play important roles in promoting cell proliferation and expansion during early leaf development (HXK1, Van Dingenen et al, 2019), delaying flowering and leaf senescence (ALDHA3, Stiti et al, 2011), reducing starch accumulation (PPC2, Shi et al, 2015; You et al, 2020), and lowering cadmium tolerance, defense response against abiotic stresses such as salicylic acid, salinity, heavy metal, and leaf cell death (OASA1, Dominguez-Solís et al, 2001; Shirzadian-Khorramabad et al, 2010; Birke et al, 2013), which is consistent with the upl3 phenotype (Figs 1 and 8; Furniss et al, 2018). Therefore, HXK1 and PPC2 might be direct targets of UPL3 to affect metabolite flux in carbon fixation in C3 plant such as Arabidopsis. In most nonphotosynthetic tissues and the photosynthetic tissues of C3 plants, the fundamental function of PPC2 is to anaplerotically replenish tricarboxylic acid cycle intermediates, which can be activated by its positive effector, Glc-6-P, and inhibited by its negative effectors, such as malate, Asp, and Glu, as well as the levels of glycine and serine (O’Leary et al, 2011; You et al, 2020). Our result showed that UPL3 also affected the ubiquitin level of MS2 and OASA1 in methionine and cysteine biosynthesis pathways (Fig 3), which feedbacks perhaps negatively regulates the PPC2 protein level through carbohydrate and amino acid biosynthetic process.
UBP12 is an UPL3-interacting protein, which in-turn might act as an important downstream player in protein ubiquitination
As described in previous review, the HECT E3 N-terminus not only simply serves as an adapter for direct binding but it also provides various ways to regulate HECT E3s’ substrate recruitment and catalytic activity (Lan & Miao, 2019; Wang & Spoel, 2022). In this study, we showed that UPL3 seemed to interact with histone variants, histone modifiers (BRM), PPC2, HXK1, several U-box proteins (UBC35), and DUBs (UBP12) in the nucleus (Figs 6 and S8). The 2/3 ubiquitin conjugates (DUCs) including BRM, histone variants, and UBC35 were all enhanced in the upl3 plants (Fig 2). If we only look at canonical function of UPL3 E3 ligase in the UPS pathway, it seems to be hard to explain the UPL3 affecting the polyubiquitination pattern. Furniss et al (2018) suggested that UPL3 might have E4 function, recruiting polyubiquitin to targets (Furniss et al, 2018); however, it could not explain our issue yet. This study showed that UPL3 could interact with UBP12, UBP13, and UBP26 (Figs 5 and 6); however, the ubiquitin level of UBP12, UBP13, UBP26, and other members were not significantly altered in the upl3 (Fig 7). Thus, we supposed that UBP12, UBP13, and UBP26 might be recruited by UPL3 for nuclear access or playing their roles together. In fact, it has reported that UBP12 has function in the protein deubiquitination and is able to interact in vivo with a Polycomb G protein (LHP1) and EMBRYONIC FLOWER1 complex (EMF1c) to form a complex and make removal of H2A ubiquitin, being necessary for transcriptional regulation such as BRM, several WRKYs, and NACs transcription factors in plant senescence and flowering and repress genes involved in stimulus response (Figs 8 and S10) (Li et al, 2016a; Miao & Zentgraf, 2007; Guo et al, 2017; Zhang et al, 2017; Kralemann et al, 2020). In this study, similar phenotypes among oeUPL3, brm, ubp12, ppc2, and gin2 mutants or between upl3 and oeUBP12 are helpful to explain UPL3 and UBP12 playing their roles together to determine ubiquitination status of histone variants or candidate targets for correct biological function in plant senescence and flowering and organ development. oeUPL3 had more similar phenotype with the ubp12 ubp13 double-mutant and brm mutant: early flowering (Cui et al, 2013; An et al, 2018), premature leaf aging phenotype (Park et al, 2019; Vanhaeren et al, 2020), and response to SA (Furniss et al, 2018) and JA (Ewan et al, 2011; Jeong et al, 2017), as well as plant defense response (Ewan et al, 2011), starch and sucrose accumulation (Figs 7 and 8), exhibiting a delayed senescence, and flowering phenotype (Cao et al, 2008; Kotliński et al, 2017; Rutowicz et al, 2019; Kralemann et al, 2020), but the transcript level of BRM, HXK1, and PPC2 was not regulated by UPL3, although a series of transcription factors, such as WRKYs, ORE1, and NACs, as well as CML10, PIP1-5, TLL1, LRR-RLK, JAL23, ABCB19, BCAT4, and BDA1, whose functions were involved in response to (a)biotic stress and in plant development and plant aging in response to light, auxin, JA, or calcium (Wu et al, 2010; Yang et al, 2012, 2013; Debernardi et al, 2014), are parts of common downstream genes of UPL3 and UBP12/13 (Figs 8 and S12; Lee et al, 2019). The functions of UPL3 interacting with UBP12 might have two possibilities: (1) recruiting UBP12 in enhancing the deubiquitination of a large set of proteins, such as carbon metabolism–related enzymes; (2) recruiting UBP12 in chromatin accessing and activating downstream gene expression. The detailed insight into the regulatory mechanism of ubiquitination dynamic between UPL3 and UBP12 with their substrate complex would be the next challenge in the field.
The UPL3 active site is dependent on the noncanonical ubiquitination motif
Based on the analysis of the Kub site information in this study, the canonical c-K-x-E/G motif sites are highly enriched in the ubiquitin conjugates in the upl3, whereas the noncanonical motif –x-A-K-x- sites show low enrichment for HXK1 and PPC2 (Fig 4), because HXK1 at K77 was previously reported to be ubiquitinated at a site of noncanonical linkages in animal cell (Huang et al, 2018). Thus, our omics results confirm the assembly of polyubiquitin chains in plants. By scanning our ubiquitome datasets for ubiquitination sites using footprints containing ubiquitin remnants after trypsin cleavage, modifications by SUMO1 at K23 and K42 were detected in addition to polyubiquitin linked via K48 linkages. Furthermore, according to acetylation of lysine (Kac) sites of GAPDH protein both in human and in rice under stress condition is required for protein translocation into the nucleus (Boukouris et al, 2016; Li et al, 2016a, 2016b; Huang et al, 2018). Our data presented here indicate that one lysine (K76 in AtGAPC) was ubiquitinated in the upl3 plants. Perhaps, it is competed this lysine site for acetylation or phosphorylation (Swaney et al, 2013; Elia et al, 2015; Ohtake et al, 2015). However, different extracellular stimuli and their intracellular signaling messengers (e.g., heat, salt, osmotic stress, heavy metal, reactive oxygen species, and lipid mediators) might differentially and specifically induce ubiquitination of GAPC for nuclear access via the Kub site pattern (Li et al, 2016a, 2016b; Kim et al, 2020). From this study, several proteins such as BRM, BCAT4, and SAM1 and different lysine sites showed various signs of up- or down-regulated ubiquitination (Fig 2E). Therefore, we speculate that the Kub site preference pattern of UPL3 mediates their function and regulation. It can be explained that 80% ubiquitinated proteins are enriched outside of nucleus.
Materials and Methods
Materials
The seeds of Arabidopsis (A. thaliana) Col-0 and transgenic plants were germinated, and seedlings were grown on half-strength Murashige–Skoog (MS) medium supplemented with 0.7% (wt/vol) agar. Ten-d-old seedlings were transplanted to the vermiculite, watering with half-strength MS medium in climate rooms under controlled conditions (22.5°C, 13/11 h of light/dark photoperiod with a light intensity of 80 μmol photons m−2 s−1, 60% relative humidity). The mutant seeds of upl3-1 (SALK_015334) and upl3-3 (SALK_117247) were obtained from the Nottingham Arabidopsis Stock Centre (NASC).
UPL3 or UBP12 overexpression (oeUPL3) and complementation plants (comUPL3, pUPL3/upl3) were constructed by cloning the UPL3 or UBP12 coding sequence into pCAMBIA3301 vector using SpeI and Smal under ACTIN3 promoter and its own promoter PUPL3, which contains 2 kb upstream of the ATG start codon) with the primers described in Table S1. Arabidopsis transformation was performed by the floral-dip method (Clough & Bent, 1998). The homozygous upl3 T-DNA insertion knockout mutants were obtained by genome insertion screening and RNA-level screening (Fig S1) with the primers described in Table S2. The brm, gin2, ppc2, ubp12, ubp12 ubp13, and oeUBP12 seeds were kindly provided by other scientists.
Table S1 Primers used in the construction of binary vectors. (19.4KB, docx)
Table S2 Primers used in mutant plants identification. (18.3KB, docx)
Starch and glucose measurement and staining
Whole rosettes of 18–21-d ubp12 and upl3 transgenic plants were either harvested or covered with black trays at 10 am after 2 h. At 10 am of the next day, rosettes of covered plants were harvested. Rosettes were cleared in 80% (vol/vol) ethanol plus 5% (vol/vol) formic acid at room temperature, stained in I2-KI solution (5 g I2 and 10 g KI per 100 ml sterile water), and washed three times in water (Huang, et al, 2020). The contents of starch and sucrose were measured according to the method described by Huang et al (2020)
Protein extraction, trypsin digestion, HPLC fractionation, affinity enrichment, and LC–MS/MS analysis
The rosette leaves of 10 6-wk-old upl3 and wild-type plants were pooled and ground in liquid nitrogen into cell powder and then transferred to a 5-ml centrifuge tube, collected in three tubes for one biological replicates; a total of 30 plants were collected for three biological replicates. After that, the method described according to the experimental manual of proteomic/ubiquitinomics determination in the “Jingjie Bio” company was followed.
Four volumes of lysis buffer (8 M urea, 1% Triton-100, 10 mM dithiothreitol, and 1% Protease Inhibitor Cocktail [Roche]) was added to the cell powder, followed by sonication three times on ice using a high-intensity ultrasonic processor (Scientz). For PTM experiments, inhibitors were also added to the lysis buffer, for example, 3 μM TSA and 50 mM NAM for acetylation. The remaining debris was removed by centrifugation at 20,000g at 4°C for 10 min. Finally, the protein was precipitated with cold 20% TCA for 2 h at −20°C. After centrifugation at 12,000g 4°C for 10 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone for three times. The protein was redissolved in 8 M urea, and the protein concentration was determined with a BCA kit according to the manufacturer’s instructions.
Trypsin digestion
A total of 5 mM dithiothreitol was added to the protein solution for reduction for 30 min at 56°C, and the solution was alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM NH4HCO3 to urea concentration less than 2 M. Finally, trypsin was added at 1:50 trypsin-to-protein-mass ratio for the first digestion overnight and 1:100 trypsin-to-protein-mass ratio for a second 4-h digestion. Approximately 100 mg of protein for each sample was digested with trypsin for the following experiments.
HPLC fractionation
The tryptic peptides were fractionated by high-pH reverse-phase HPLC using the Thermo BetaSil C18 column (5-μm particles, 10 mm ID, 250 mm length). Briefly, peptides were first separated with a gradient of 8–32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into 18 fractions and dried by vacuum centrifugation.
Affinity enrichment
To enrich Kub peptides, tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, and 0.5% Nonidet P-40, pH 8) were incubated with prewashed antibody beads at 4°C overnight with gentle shaking; the di-Gly-Lys specific antibody was used (PTM Biolabs). Then the beads were washed four times with NETN buffer and twice with distilled, deionized water. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. The eluted fractions were combined and vacuum-dried. The resulting peptides were cleaned with C18 ZipTips (Millipore) according to the manufacturer’s instructions, followed by LC–MS/MS analysis.
LC–MS/MS analysis
Three parallel analyses for each fraction were performed. LC–MS/MS analysis was performed according to previously described protocols (Xie et al, 2015). The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a home-made reverse-phase analytical column (15-cm length, 75 μm i.d.). The gradient was from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23–35% in 8 min, and climbing to 80% in 3 min and then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl/min on an EASY-nLC 1000 UPLC system.
The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q Exactive TM Plus (Thermo Fisher Scientific) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350–1,800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using NCE setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500.
A data-dependent procedure that alternated between one mass spectrometry scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 1.5 × 104 in the mass spectrometry survey scan with 30-s dynamic exclusion. The electrospray voltage applied was 2 kV. Automatic gain control was used to prevent overfilling of the ion trap; 5 × 104 ions were accumulated for the generation of MS/MS spectra. For mass spectrometry scans, the mass-to-charge ratio scan range was 350 to 1,800. The fixed first mass was set as 100 mass-to-charge ratios.
Database search
The resulting MS/MS data were processed using the Maxquant search engine (v.1.5.2.8) (https://www.maxquant.org). Tandem mass spectra were searched against the Uniprot database concatenated with a reverse-decoy database. Trypsin/P was specified as cleavage enzyme allowing up to four missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in First search and 5 ppm in Main search, and the mass tolerance for fragment ions was set as 0.02 D. Carbamidomethyl on Cys was specified as fixed modification, and GlyGly on Lys and oxidation on Met were specified as variable modifications. The label-free quantification method (Xu et al, 2010) was employed to calculate the relative abundance of the modified peptides. FDR was adjusted to < 1%, and the minimum score for modified peptides was set to >40.
Ubiquitin footprints
Ubiquitin footprints were identified through Proteome Discoverer (version 2.0.0.802; Thermo Fisher Scientific) by searching the TAIR10 protein database using the variable modification of lysine residues by ubiquitin (Gly–Gly, +114.043 m/z). Peptides were assigned using SEQUEST HT (Thermo Fisher Scientific), with search parameters set to assume trypsin digestion with a maximum of two missed cleavages, a minimum peptide length of 6, precursor mass tolerances of 10 ppm, and fragment mass tolerances of 0.02 D. Carbamidomethylation of cysteine was specified as a static modification, whereas oxidation of methionine and N-terminal acetylation were specified as dynamic modifications. The target FDR of #1% (strict) was used as validation for PSMs and peptides. Proteins that contained similar peptides and which could not be differentiated based on the MS/MS analysis alone were grouped to satisfy the principles of parsimony. MEME Suite 4.11.4 was used to identify the ubiquitin-binding cKxE/D/G motif, whereas the prevalence of these sites was predicted by referring to GPS-ubiquitin (Zhao et al, 2014).
GFP nanotrap–MS analysis
The rosette leaves of 10 6-wk-old ACTIN3:UPL3-GFP overexpression plants and ACTIN3:GFP plants were harvested and ground in liquid nitrogen and homogenized in immunoprecipitation (IP) buffer (50 mM Tris–HCl, pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 mM PMSF, 10% glycerol, and 1× protease inhibitor cocktail [Roche]). After centrifugation at 16,000g for 10 min at 4°C, the supernatant of each sample was mixed with 30 μl GBP beads (Fig S6) (Rothbauer et al, 2008) and rotated at 4°C for 2 h. GBP beads were pelleted and washed three times with IP buffer. The immunoprecipitated proteins were eluted from the beads with 2× SDS–PAGE sample buffer by heating at 95°C for 10 min. Triplicate protein samples were separated by 15% SDS–PAGE and then extracted for mass spectrometry analysis according to Deng et al (2016). The antibody against GFP was purchased from Roche company.
Semi-RT-PCR and RT-qPCR
Semi-RT-PCR and RT-qPCR analyses employed the oligonucleotide primers described in Tables S2 and S3. RNA was extracted from rosette leaves of 6-wk-old upl3 and wild-type plants, and the following procedure was performed according to the description (Huang et al, 2020). Semi-qRT-PCR was performed on an ARKTIK thermal cycler (Thermo Fisher Scientific). The GAPC2 (AT1G13440) was chosen as an internal control (25 cycles). PCR products were run on a 1.0% TAE agarose gel. The transcript abundance of RT-qPCR was normalized to that generated with GAPC2 based on the comparative threshold method (Pfaffl, 2001). Three independent biological replicates with three technical repeats were performed.
Table S3 Primers used in RT–qPCR. (21.2KB, docx)
Yeast two-hybrid assay
Yeast two-hybrid assays were performed as described in the manual for the GAL4-based two-hybrid system 3 protocol (Clontech). Full-length or different regions of candidates were cloned into pGADT7-AD vectors to construct prey constructs, and the Bait vector pGBKT7-BD expressed the wild-type UPL3 or UPL3 variants fused to the GAL4 DNA-binding domain (BD). The procedure was according to that described by Huang et al (2020). The primers were listed in Table S4.
Table S4 Primers used in yeast two-hybrid assays. (20.8KB, docx)
Coimmunoprecipitation and Western blot analyses
The oeUPL3-GFP, oeUBP12-GFP, and oeGFP plants from the fifth to eighth rosette leaves of 6-wk-old plants were harvested and ground in liquid nitrogen. Total proteins were extracted from 1 g of materials and performed CoIP according to the description by Huang et al (2022).
To extract soluble proteins from plant tissue of oeUPL3, upl3, ubp12, and oeUBP12 and WT plants, 200 mg of leaf material were batch-frozen in liquid nitrogen, ground into powder, resuspended in 100 μl of extraction buffer (100 mM Tris, pH 7.2, 10% sucrose, 5 mM MgCl2, 5 mM EGTA, protease inhibitor), and centrifuged at 15,000g for 10 min. The supernatant was used for immunoblotting analysis. Antibodies (Agrisera) against polyubiquitin, HXK1, PPC2, and β-tubulin (CW0098, KWBIO) were used. Anti-BRM was kindly provided by Dr Rongcheng Lin (Institute of Botany, Chinese Academy of Sciences). The procedure was performed according to Miao and Zentgraf (2010). Proteins were separated on 6% acrylamide gels and transferred to nitrocellulose membranes using standard protocols.
Bioinformatic analysis
Bioinformatic analysis was performed according to previously described protocols (Xie et al, 2015). The detail procedure described as following:
Enrichment of Gene Ontology analysis: Proteins were classified by GO annotation into three categories: biological process, cellular compartment, and molecular function. For each category, a two-tailed Fisher’s exact test was employed to test the enrichment of the differentially expressed protein against all identified proteins. The GO with a corrected P-value < 0.05 is considered significant.
Enrichment of pathway analysis: Encyclopedia of Genes and Genomes (KEGG) database was used to identify enriched pathways by a two-tailed Fisher’s exact test to test the enrichment of the differentially expressed protein against all identified proteins. The pathway with a corrected P-value < 0.05 was considered significant. These pathways were classified into hierarchical categories according to the KEGG website.
Enrichment of protein domain analysis: For each category proteins, InterPro (a resource that provides functional analysis of protein sequences by classifying them into families and predicting the presence of domains and important sites) database was researched, and a two-tailed Fisher’s exact test was employed to test the enrichment of the differentially expressed protein against all identified proteins. Protein domains with a corrected P-value < 0.05 were considered significant.
Enrichment-based clustering: For further hierarchical clustering based on differentially expressed protein functional classification (such as: GO, Domain, Pathway, Complex), we first collated all the categories obtained after enrichment along with their P-values and then filtered for those categories which were at least enriched in one of the clusters with P-value < 0.05. This filtered P-value matrix was transformed by the function x = −log10 (P-value). Finally, these x values were z-transformed for each functional category. These z scores were then clustered by one-way hierarchical clustering (Euclidean distance, average linkage clustering) in Genesis. Cluster membership was visualized by a heatmap using the “heatmap.2” function from the “gplots” R-package.
Summary
Bioinformatic analysis was performed according to previously described protocols (Xie et al, 2015). GO term association and enrichment analysis were performed using the Database for Annotation, Visualization, and Integrated Discovery. The KEGG database (2019-11-19) was used to annotate protein pathways (Kanehisa & Goto, 2000). The KEGG online service tool KAAS was used to annotate the proteins’ KEGG database descriptions. The annotation results were mapped on the KEGG pathway database using the KEGG online service tool KEGG Mapper. The domain annotation was performed with InterProScan (version_7.0) on the InterPro domain database via Web-based interfaces and services. WoLF PSORT was used to predict subcellular localization (Horton et al, 2007). The CORUM 3.0 database was used to annotate protein complexes. Motif-X software was used to analyze the models of the sequences with amino acids in specific positions of ubiquityl-21-mers (10 amino acids upstream and downstream of the Kub site) in all of the protein sequences. In addition, the IPI Arabidopsis (A. thaliana) proteome was used as the background database, and the other parameters were set to default values. The parameter settings for searching motifs using Motif-X software were occurrences 20 and the Bonferroni-corrected P = 0.005. Protein–protein interaction networks were analyzed with the IntAct database (http://www.ebi.ac.uk/intact/). The protein–protein interaction network map was generated with the Cytoscape software (Shannon et al, 2003).
Data Availability
All datasets have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027037. The description of dataset was shown in Tables S5, S6, and S7.
Table S5 LSA-2022-01492_TableS5.docx (17.2KB, docx) The description of dataset in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027037.
Table S6. List of apparent ubiquitin attachment sites and motif in identified proteins (|FC| > 1.5, P-value < 0.05). R1 and R2 are two replicates. It is also indicated in Fig 4C. LSA-2022-01492_TableS6.xlsx (11.2KB, xlsx)
Table S7. List of most notable interacting candidates of UPL3 with their respective fold-change of ubiquitination level in upl3 versus wild-type plants. It is also indicated in Fig 5E. LSA-2022-01492_TableS7.xlsx (11.2KB, xlsx)
Supplementary Material
Acknowledgements
We thank the Nottingham Arabidopsis Stock Centre (NASC) for T-DNA insertion lines. Dr. Shi Xiao (SUN YATSEN University) provided the gin2 and ppc2 seeds. Dr. Rongcheng Lin (Institute of Botany, Chinese Academy of Sciences) kindly provided the antibody against BRM, and Dr. Keqiang Wu (Taiwan University) provided the brm seeds. Dr Wenqiang Tang (Hebei Normal University) provided the ubp12, ubp12ubp13, and oeUBP12 seeds. We thank Jingjie PTM Biolabs for providing the methods for partial data analysis. This work was supported by a grant from the National Natural Science Foundation of China (grant number 31770318) and grant of excellent PhD candidate program of Fujian Agriculture and Forestry University (No. 324-1122yb049). We thank Dr. Binghua Wu (Fujian Agriculture and Forestry University) for critically reading the manuscript.
Author Contributions
W Lan: resources, data curation, software, formal analysis, investigation, and methodology.
W Ma: data curation, formal analysis, investigation, and methodology.
S Zheng: resources, investigation, and methodology.
Y Qiu: resources, investigation, and methodology.
H Zhang: resources, investigation, and methodology.
H Lu: investigation.
Y Zhang: software, formal analysis, validation, and visualization.
Y Miao: conceptualization, data curation, supervision, validation, visualization, project administration, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- An Z, Liu Y, Ou Y, Li J, Zhang B, Sun D, Sun Y, Tang W (2018) Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci U S A 115: 1123–1128. 10.1073/pnas.1714177115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Yatusevich R, Buszewicz D, Krzyczmonik K, Patryn J, Iwanicka-Nowicka R, Biecek P, Wilczynski B, Koblowska M, Jerzmanowski A, et al. (2017) Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res 45: 3116–3129. 10.1093/nar/gkw1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensussan M, Lefebvre V, Ducamp A, Trouverie J, Gineau E, Fortabat MN, Guillebaux A, Baldy A, Naquin D, Herbette S, et al. (2015) Suppression of dwarf and irregular xylem phenotypes generates low-acetylated biomass lines in Arabidopsis. Plant Physiol 168: 452–463. 10.1104/pp.15.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C (2014) New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Boil 21: 301–307. 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L, et al. (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J 33: 1159–1176. 10.1002/embj.201386906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke H, Heeg C, Wirtz M, Hell R (2013) Successful fertilization requires the presence of at least one major O-acetylserine(thiol)lyase for cysteine synthesis in pollen of Arabidopsis. Plant Physiol 163: 959–972. 10.1104/pp.113.221200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouris AE, Zervopoulos SD, Michelakis ED (2016) Metabolic enzymes moonlighting in the nucleus: Metabolic regulation of gene transcription. Trends Biochem Sci 41: 712–730. 10.1016/j.tibs.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585. 10.1111/j.1365-313x.2005.02399.x [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602. 10.1105/tpc.108.062760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear Hexokinase1 complex in glucose signaling. Cell 127: 579–589. 10.1016/j.cell.2006.09.028 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu Q, Cui X, Zheng S, Liu B, et al. (2013) Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol 162: 897–906. 10.1104/pp.112.213009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF (2014) Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79: 413–426. 10.1111/tpj.12567 [DOI] [PubMed] [Google Scholar]
- Deng X, Lu T, Wang L, Gu L, Sun J, Kong X, Liu C, Cao X (2016) Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc Natl Acad Sci U S A 113: 5447–5452. 10.1073/pnas.1522458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M, Liu S, Figueiredo DD, Gentry M, Mozgova I, Nanni P, Tang M, Mannervik M, Kohler C, Hennig L (2016) H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat Plants 2: 16126. 10.1038/nplants.2016.126 [DOI] [PubMed] [Google Scholar]
- Dominguez-Solís JR, Gutierrez-Alcalá G, Vega JM, Romero LC, Gotor C (2001) The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J Biol Chem 276: 9297–9302. 10.1074/jbc.M009574200 [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742. 10.1046/j.1365-313x.2003.01844.x [DOI] [PubMed] [Google Scholar]
- Elia A, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ (2015) Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell 59: 867–881. 10.1016/j.molcel.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewan R, Pangestuti R, Thornber S, Craig A, Carr C, O’Donnell L, Zhang C, Sadanandom A (2011) Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol 191: 92–106. 10.1111/j.1469-8137.2011.03672.x [DOI] [PubMed] [Google Scholar]
- Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss JJ, Grey H, Wang Z, Nomoto M, Jackson L, Tada Y, Spoel SH (2018) Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog 14: e1007447. 10.1371/journal.ppat.1007447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Li D, Jin C, Duan S, Qi S, Liu K, Wang H, Ma H, Hai J, Chen M (2017) Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem Biophys Res Commun 485: 360–365. 10.1016/j.bbrc.2017.02.074 [DOI] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870. 10.1105/tpc.17.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ren G, Zhang K, Li Z, Miao Y, Guo H (2021) Leaf senescence: Progression, regulation, and application. Mol Hortic 1: 5. 10.1186/s43897-021-00006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: Protein localization predictor. Nucleic Acids Res 35: W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Yu J, Cai Q, Chen Y, Li Y, Ren Y, Miao Y (2020) Triple-localized WHIRLY2 influences leaf senescence and silique development via carbon allocation. Plant Physiol 184: 1348–1362. 10.1104/pp.20.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Lan W, Ma W, Huang R, Lin W, Li M, Chen CY, Wu K, Miao Y (2022) WHIRLY1 recruits the histone deacetylase HDA15 repressing leaf senescence and flowering in Arabidopsis. J Integr Plant Biol 64: 1411–1429. 10.1111/jipb.13272 [DOI] [PubMed] [Google Scholar]
- Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, Qi S, Locasale JW, Roeder RG, Zhao Y, et al. (2018) p300-Mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol Cell 70: 984. 10.1016/j.molcel.2018.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Jung C, Seo JS, Kim J-K, Chua N-H (2017) The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29: 1406–1424. 10.1105/tpc.17.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Oh JE, Noh YS, Noh B (2015) Epigenetic control of juvenileto- adult phase transition by the Arabidopsis SAGA-like complex. Plant J 83: 537–545. 10.1111/tpj.12908 [DOI] [PubMed] [Google Scholar]
- Kim SC, Guo L, Wang X (2020) Nuclear moonlighting of cytosolic glyceraldehyde-3-phosphate dehydrogenase regulates Arabidopsis response to heat stress. Nat Commun 11: 3439. 10.1038/s41467-020-17311-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kotliński M, Knizewski L, Muszewska A, Rutowicz K, Lirski M, Schmidt A, Baroux C, Ginalski K, Jerzmanowski A (2017) Phylogeny-based systematization of Arabidopsis proteins with histone H1 globular domain. Plant Physiol 174: 27–34. 10.1104/pp.16.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralemann LEM, Liu S, Trejo-Arellano MS, Munoz-Viana R, Köhler C, Hennig L (2020) Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable polycomb-mediated gene repression in Arabidopsis. Genome Biol 21: 144. 10.1186/s13059-020-02062-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Chetelat A, Nguyen CT, Farmer EE (2019) Arabidopsis H+-ATPase AHA1 controls slow wave potential duration and wound-response Jasmonate pathway activation. Proc Natl Acad Sci U S A 116: 20226–20231. 10.1073/pnas.1907379116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Miao Y (2019) New aspects of HECT-E3 ligases in cell senescence and cell death of plants. Plants 8: 483. 10.3390/plants8110483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Li MW, Feke A, Liu W, Saffer AM, Gendron JM (2019) GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat Commun 10: 3750. 10.1038/s41467-019-11769-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, et al. (2016. a) SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 7: 12235. 10.1038/ncomms12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, et al. (2016. b) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48: 687–693. 10.1038/ng.3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmerman P, Zentgraf U (2004) Targets of AtWRKY 53 transcription factor and its role during the senescence of Arabidopsis leaf. Plant Mol Biol 55: 853–867. [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63: 179–188. 10.1111/j.1365-313x.2010.04233.x [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830. 10.1105/tpc.106.042705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Wells R, McKenzie N, Trick M, Ball J, Fatihi A, Dubreucq B, Chardot T, Lepiniec L, Bevan MW (2019) Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 levels, seed size and crop yield in Brassica napus. Plant Cell 31: 2370–2385. 10.1105/tpc.18.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A 107: 16512–16517. 10.1073/pnas.1004181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F, Saeki Y, Sakamoto K, Ohtake K, Nishikawa H, Tsuchiya H, Ohta T, Tanaka K, Kanno J (2015) Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep 16: 192–201. 10.15252/embr.201439152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC (2011) The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J 436: 15–34. 10.1042/bj20110078 [DOI] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY (2008) In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol 148: 246–258. 10.1104/pp.108.121897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Jeong JS, Seo JS, Park BS, Chua NH (2019) Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol 223: 1447–1460. 10.1111/nph.15879 [DOI] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L (2013) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74: 435–447. 10.1111/tpj.12132 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Suprasanna P, D’Souza SF, Kumar V (2012) Membrane topology and predicted RNA-binding function of the ‘early responsive to dehydration (ERD4)’ plant protein. PLoS One 7: e32658. 10.1371/journal.pone.0032658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7: 282–289. 10.1074/mcp.m700342-mcp200 [DOI] [PubMed] [Google Scholar]
- Rutowicz K, Lirski M, Mermaz B, Teano G, Schubert J, Mestiri I, Kroteń MA, Fabrice TN, Fritz S, Grob S, et al. (2019) Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biol 20: 157. 10.1186/s13059-019-1767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358. 10.1111/j.1365-313x.2009.03862.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JH, Yi K, Liu Y, Xie L, Zhou Z, Chen Y, Hu Z, Zheng T, Liu R, Chen Y, et al. (2015) Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol 167: 671–681. 10.1104/pp.114.254474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadian-Khorramabad R, Jing HC, Everts GE, Schippers JHM, Hille J, Dijkwel PP (2010) A mutation in the cytosolic O-acetylserine (thiol) lyase induces a genome-dependent early leaf death phenotype in Arabidopsis. BMC Plant Biol 10: 80. 10.1186/1471-2229-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiti N, Missihoun TD, Kotchoni SO, Kirch HH, Bartels D (2011) Aldehydede hydrogenases in Arabidopsis thaliana: Biochemical requirements, metabolic pathways, andf unctional analysis. Front Plant Sci 2: 65. 10.3389/fpls.2011.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10: 676–682. 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ikeda A, Shiojiri K, Ozawa R, Shiki K, Nagai-Kunihiro N, Fujita K, Sugimoto K, Yamato KT, Dohra H, et al. (2018) Identification of a hexenal reductase that modulates the composition of green leaf volatiles. Plant Physiol 178: 552–564. 10.1104/pp.18.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Harada T (2011) The Arabidopsis CORI3 promoter contains two cis-acting regulatory regions required for transcriptional activity in companion cells. Plant Cell Rep 30: 1723–1733. 10.1007/s00299-011-1080-4 [DOI] [PubMed] [Google Scholar]
- Van Dingenen J, Vermeersch M, ·De Milde L, Hulsmans S, ·De Winne N, Van Leene J, Gonzalez N, Dhondt S, De Jaeger G, Rolland F, et al. (2019) The role of HEXOKINASE1 in Arabidopsis leaf growth. Plant Mol Biol 99: 79–93. 10.1007/s11103-018-0803-0 [DOI] [PubMed] [Google Scholar]
- Vanhaeren H, Chen Y, Vermeersch M, De Milde L, De Vleeschhauwer V, Natran A, Persiau G, Eeckhout D, De Jaeger G, Gevaert K, et al. (2020) UBP12 and UBP13 negatively regulate the activity of the ubiquitin-dependent peptidases DA1, DAR1 and DAR2. Elife 9: e52276. 10.7554/elife.52276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397. 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- Wang Z, Spoel SH (2022) HECT ubiquitin ligases as accessory proteins of the plant proteasome. Essays Biochem 66: EBC20210064. 10.1042/EBC20210064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454. 10.1126/science.aac7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Cameron JN, Ljung K, Spalding EP (2010) A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J 62: 179–191. 10.1111/j.1365-313x.2010.04137.x [DOI] [PubMed] [Google Scholar]
- Xie X, Kang H, Liu W, Wang GL (2015) Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J Proteome Res 14: 2017–2025. 10.1021/pr5009724 [DOI] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 28: 868–873. 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Guo C, Zhou B, Li C, Wang H, Zheng B, Ding H, Zhu Z, Peragine A, Cui Y, et al. (2016) Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol 172: 2416–2428. 10.1104/pp.16.01588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S, Takemiya A, Sakamoto T, Kurata T, Tsutsumi T, Kinoshita T, Shimazaki K (2016) The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol 171: 2731–2743. 10.1104/pp.16.01581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Richter GL, Wang X, Młodzińska E, Carraro N, Ma G, Jenness M, Chao Dy, Peer WA, Murphy AS (2013) Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74: 37–47. 10.1111/tpj.12103 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang Y, Ding P, Johnson K, Li X, Zhang Y (2012) The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol 159: 1857–1865. 10.1104/pp.112.197152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolcu S, Li X, Li S, Kim YJ (2017) Beyond the genetic code in leaf senescence. J Exp Bot 69: 801–810. 10.1093/jxb/erx401 [DOI] [PubMed] [Google Scholar]
- You L, Zhang J, Li L, Xiao C, Feng X, Chen S, Guo L, Hu H (2020) Involvement of abscisic acid, ABI5, and PPC2 in plant acclimation to low CO2. J Exp Bot 71: 4093–4108. 10.1093/jxb/eraa148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307. 10.1104/pp.16.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J (2014) GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42: W325–W330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Shabek N (2017) Ubiquitin ligases: Structure, function, and regulation. Annu Rev Biochem 86: 129–157. 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data for Figure 2LSA-2022-01492_SdataF2_F5_F6_F7.pdf (2.5MB, pdf)
Spectrum identification information for upl3 and WT. LSA-2022-01492_Supplemental_Data_1.xlsx (418.1KB, xlsx)
Differential ubiquitination targets of upl3/WT (FC 1.3 and 1.5). LSA-2022-01492_Supplemental_Data_2.xlsx (62.5KB, xlsx)
Analysis of ubiquitin footprints (K sites) of upl3/WT.LSA-2022-01492_Supplemental_Data_3.xlsx (236.7KB, xlsx)
Source Data for Figure 5LSA-2022-01492_SdataF2_F5_F6_F7.pdf (2.5MB, pdf)
The dataset for GFP alone and UPL3-GFP nanotrap-pulldown mass spectrum analysis.LSA-2022-01492_Supplemental_Data_4.xlsx (29.3KB, xlsx)
Source Data for Figure 6LSA-2022-01492_SdataF2_F5_F6_F7.pdf (2.5MB, pdf)
Source Data for Figure 7LSA-2022-01492_SdataF2_F5_F6_F7.pdf (2.5MB, pdf)
Differential accumulation protein of upl3/WT (FC 1.2).LSA-2022-01492_Supplemental_Data_5.xlsx (266KB, xlsx)
Differential display expression targets of upl3/WT transcriptome dataset.LSA-2022-01492_Supplemental_Data_6.xlsx (267.2KB, xlsx)
Table S1 Primers used in the construction of binary vectors. (19.4KB, docx)
Table S2 Primers used in mutant plants identification. (18.3KB, docx)
Table S3 Primers used in RT–qPCR. (21.2KB, docx)
Table S4 Primers used in yeast two-hybrid assays. (20.8KB, docx)
Table S5 LSA-2022-01492_TableS5.docx (17.2KB, docx) The description of dataset in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027037.
Table S6. List of apparent ubiquitin attachment sites and motif in identified proteins (|FC| > 1.5, P-value < 0.05). R1 and R2 are two replicates. It is also indicated in Fig 4C. LSA-2022-01492_TableS6.xlsx (11.2KB, xlsx)
Table S7. List of most notable interacting candidates of UPL3 with their respective fold-change of ubiquitination level in upl3 versus wild-type plants. It is also indicated in Fig 5E. LSA-2022-01492_TableS7.xlsx (11.2KB, xlsx)
Data Availability Statement
All datasets have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027037. The description of dataset was shown in Tables S5, S6, and S7.
Table S5 LSA-2022-01492_TableS5.docx (17.2KB, docx) The description of dataset in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027037.
Table S6. List of apparent ubiquitin attachment sites and motif in identified proteins (|FC| > 1.5, P-value < 0.05). R1 and R2 are two replicates. It is also indicated in Fig 4C. LSA-2022-01492_TableS6.xlsx (11.2KB, xlsx)
Table S7. List of most notable interacting candidates of UPL3 with their respective fold-change of ubiquitination level in upl3 versus wild-type plants. It is also indicated in Fig 5E. LSA-2022-01492_TableS7.xlsx (11.2KB, xlsx)