Abstract
Chronic rhinosinusitis (CRS) is a complex, heterogenous condition that is likely associated with infectious and inflammatory causative factors. Renewed interest in the role that microbes play in this condition has stemmed from advancements in microbe identification and parallel research implicating the microbiome as having a role in other chronic inflammatory conditions. This clinical commentary provides a review of the current literature relevant to chronic rhinosinusitis. Particular focus is placed on factors specific to investigation of the sinonasal microbiome, evidence for the role of dysbiosis in the disease state, and influences that may affect the microbiome. Possible mechanisms of disease and therapeutic implications through microbial manipulation are also reviewed, as are deficiencies and limitations of the current body of research.
Keywords: Microbiome, chronic rhinosinusitis, probiotic, next-generation sequencing, host-microbial interactions, microbiome manipulation, therapeutic intervention
The etiopathogenesis of chronic rhinosinusitis (CRS) is multifactorial and has aroused renewed interest in the role of microbes and, in particular, bacteria. The diverse bacteria colonizing the sinonasal cavity are essential for physiologic host-microbial interactions that prime the innate immune system in preparation for exposure to exogenous insult. As with many other chronic inflammatory conditions, a consistent finding among patients with CRS is an ecologic breakdown of the local microbiome characterized by an expansion of pathogenic bacteria and a collapse of the commensal population.1,2 As such, changes in the sinonasal microbiome may contribute to the chronic inflammation of the sinonasal mucosa that characterizes this condition.
Although microbiome research in CRS is increasing, the exact role of microbes in the pathogenesis of CRS at the community and individual species levels remains poorly understood. This clinical commentary reviews the research to date in this area and discusses current clinical and research challenges. Specifics of investigation of the sinonasal microbiome, influencing factors, possible mechanisms, and potential therapeutic options are also discussed.
INVESTIGATING THE SINONASAL MICROBIOME: CONSIDERATIONS FOR SAMPLING, SEQUENCING, AND MICROBIOME BIOINFORMATICS
Specimen collection: Specific considerations for collection of sinonasal specimens
Colonizing microbiota can, at least in part, be a function of the local microenvironmental factors such as pH, airflow, humidity, and epithelial barrier function.3 Disease features, such as phenotype, endotype, and severity, may influence composition of the sinonasal microbiome and perhaps even presence of tissue invasion. As a consequence, disease status, site of sampling, and sample type are important variables to be considered in respiratory microbiome analysis.
Sample site is an important confounding factor in studies of CRS and has been given major consideration across studies. Most published studies have relied on samples taken intraoperatively for patient comfort and to facilitate access to the sinus mucosa. This creates a possible selection bias for patients with more severe disease, who require surgery. Studies of multiple sampling sites within a single patient demonstrate variability in composition of the microbiome across sinuses, but in general, interpersonal variability greatly outweighs intrapersonal variability.4–6 A recent study of 225 patients with CRS and 100 controls found that the anterior nares and middle meatus could represent the sinus microbiome,7 confirming the findings of prior smaller studies comparing the middle meatus with the maxillary and ethmoid sinus microbiome signatures.4,6 These findings support sampling at more accessible sinonasal sites during community visits or in the clinic.
Sampling techniques and sample type vary across studies, ranging from guarded swabs and/or brushings, unguarded endoscopically guided swabs, mucosal biopsy, and nasal lavage (Fig 1). Guarded or carefully performed endoscopically guided swabs may reduce the risk of anterior nares contamination, which is particularly important in studies of a specific sinonasal niche, where contamination may influence the interpretation of results. A small study comparing mucosal biopsy samples and mucosal swabs from patients with CRS demonstrated similar bacterial diversity and compositional profiles between the 2 sample types; once again, interpersonal variation was a stronger driver of bacterial composition.8 In light of these potential confounding factors, it is important for the investigator to practice consistency in sample collection methods across his or her study.
FIG 1.
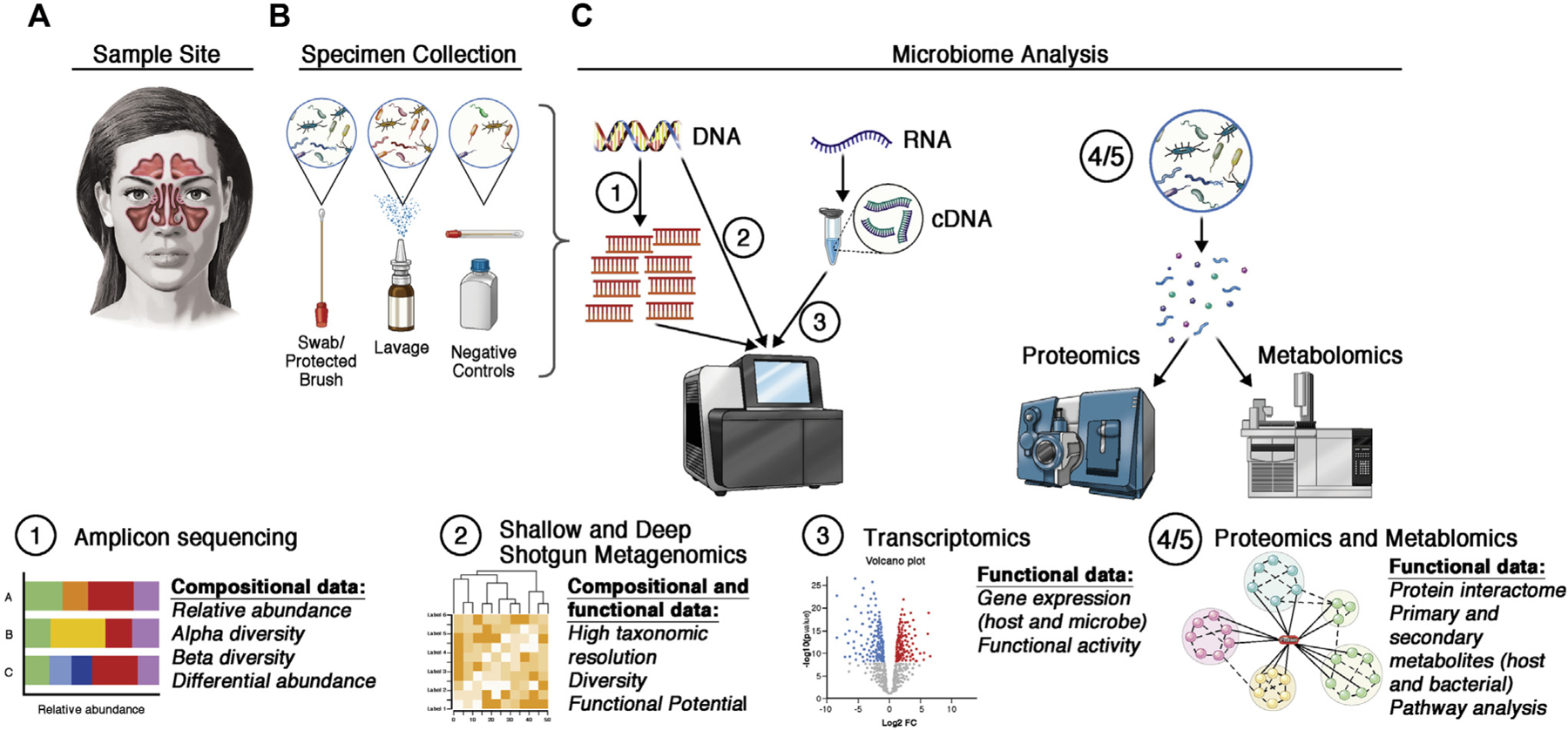
Considerations for analysis of the sinonasal microbiota. A, Investigators must consider the clinical feasibility, benefits, and limitations of the site in the sinonasal cavity. B, Different sampling techniques must also be considered carefully. Negative controls (extraction blanks and blank swabs) can help to identify and reduce contamination. C, Commonly used techniques for microbiome analyses can be used to answer distinct research questions. Amplicon sequencing (eg, 16S rRNA gene sequencing) can yield insights into community composition. Shallow shotgun metagenomics allows for high resolution of the microbial species and strains, whereas deep shotgun metagenomics can yield insights into the putative functions in a community. Gene expression can be analyzed via RNA sequencing (of reverse-transcribed cDNA) to determine which microbial and host genes are expressed in a given condition. Functional analyses can quantify protein production (proteomics) or host- and microbiome-derived metabolites (metabolomics).
Sinonasal microbiome samples can be problematic to process and analyze owing to low microbial biomass and high host DNA contamination. Propidium monoazide can be used to deplete host DNA before extraction by selectively depleting exogenous DNA.9 Clustered regularly interspaced short palindromic repeats (CRISPR)-based approaches can be used to specifically deplete host mitochondrial DNA after DNA extraction before PCR.10 Alternatively, host DNA can be removed bioinformatically by using alignment-based approaches. Low–microbial biomass samples may also be disproportionately affected by exogenous contaminants, including extraction reagents, swabs, or laboratory spaces. Therefore, it is critical that negative controls be included to assess laboratory contamination. In silico approaches, such as use of the R package decontam, are also useful to identify true contaminant sequences in a low–microbial biomass data set.
Recent advances in technology and the bioinformatics pipeline for microbiome research have the potential to alleviate many prior concerns (Fig 1).
In summary, for sampling patients, the authors recommend use of guarded flocked swabs taken from a consistent location representative of the sinus microecology, such as the middle meatus. The site of sampling should be consistent with the hypothesis to be tested. In terms of analysis, the authors currently recommend amplicon sequencing of the hypervariable region of the 16S ribosomal RNA (rRNA) gene for bacteria and the 18S rRNA gene or internal transcribed spacer region for fungi. However, it is likely that as next-generation sequencing technology evolves, these recommendations will change.
SINONASAL MICROBIOME IN HEALTH AND CRS
The core microbiome is defined as organisms or genes that are shared across unrelated individuals.11 Identifying a core sinonasal microbiome offers insight into both the stability of microbial communities and potential drivers of disease. A finding of the International Sinonasal Microbiome Study, the largest sinonasal microbiome study to date, was that Corynebacterium, Staphylococcus, Moraxella, Streptococcus, and Haemophilus were the most abundant genera within the middle meatus of patients with or without CRS.12 Although similar bacterial genera exist between health and disease states, disturbances resulting in changes to the relative and absolute abundances of these taxa may be an etiologic risk factor or potential disease modifier in CRS.
Functional gene predictions from amplicon data using phylogenetic investigation of communities by reconstruction (PIC-RUSt) bioinformatic software and in vitro studies of bacterial isolates from patients with CRS suggest that the CRS microbiome is altered.13 These studies suggest the CRS microbiome is significantly richer in genes involved in antigen processing and presentation, nucleotide binding and oligomerization domain (NOD)-like receptor signaling pathways, mismatch repair, peptidoglycan biosynthesis, peptidases, and DNA replication proteins. Another study found that CRS samples were less functionally diverse than healthy samples and were significantly enriched in bacterial virulence pathways and antimicrobial metabolite production.14 A further study found that CRS samples were significantly enriched in mucin degrading genes15 and that the core taxa driving mucin degradation were Prevotella, Fusobacterium, and Streptococcus. Degradation of mucin by these genera augmented the growth of Staphylococcus aureus. These studies have provided useful insights into the functional capacity of the CRS microbiome, but no consistent core of CRS-associated genes have been described.
EVIDENCE FOR BACTERIAL DYSBIOSIS IN CRS AND FACTORS THAT MAY INFLUENCE THE MICROBIOME
CRS-associated bacterial dysbiosis is typically described relative to a healthy cohort, and it is characterized by a reduction in diversity, an increase in overall bacterial load, loss of keystone species, fragmentation between networks, and an increase in pathobionts. Accepted criteria for dysbiosis have not been defined. Nor is it known whether dysbiosis can be remediated by treatment or whether remediation of dysbiosis translates to resolution of disease. Nonetheless, research has begun by identifying factors that may influence the composition, stability, and resilience of the sinonasal microbiota.
Demographics
The effects of age, sex, and ethnicity on the CRS bacterial community composition and CRS-associated dysbiosis are unclear.3 As is the case in adults, infants and children who are colonized with Dolosigranulum spp and Lactobacillus spp tend to have better overall respiratory health and responses to respiratory syncytial viral illness.16–19 However infant upper respiratory tract (URT) profiles that are dominated by H influenzae or Streptococcus are at risk of developing both acute and chronic URT illnesses.20–22 A recent reanalysis of the Human Microbiome Project data set based on sex found significant differences in composition of the bacterial community of the anterior nares, which the authors hypothesized could influence autoimmune-related diseases.23 Despite these findings, no significant microbiome-demographic associations were observed in analysis of the diverse large cohort of the international sinonasal microbiome study.12
Geographic, environmental, and temporal factors
Data from the collaborative International Microbiome Study of more than 400 patients from 13 centers in 5 different continents indicated that that although the bacterial composition of the core microbiota was preserved across the different sites, significant differences in both alpha and beta diversity occurred according to geography.12 Furthermore, bacterial composition also varied between centers. The most distinct microbial distribution was observed in samples collected from The Netherlands, with depletion of the relative abundances of Corynebacterium and overrepresentation of Staphylococcus. Interestingly, antibiotic use in The Netherlands is among the lowest in the Developed World, with tight regulation of the prescription of antistaphylococcal antibiotics possibly explaining this finding.24
Seasonal changes and temporal effects can affect composition and load of the sinonasal bacterial, fungal, and viral communities,25 although few data exist. There are no published studies describing changes in the viral composition and load of patients with CRS at multiple time points. However, it is well known that exposure to and infection with viruses such as influenza and rhinovirus are strongly influenced by season,26–28 and URT viral infections are known to modulate the bacterial community.29 During the winter and autumn months, significant increases in viral load and detection are noted in both patients with CRS and patients with acute seasonal viral illnesses.30
Most of our knowledge about microbial stability is derived from studies of healthy patients. Longitudinal studies have shown that bacterial composition in healthy participants remains reasonably constant over time, but each subject maintains an identifiable microbial pattern that is very different between subjects.31 The relative abundance and presence of some bacterial taxa such as Acidocella, Hyphomicrobiaceae, Asticcacaulis, Staphylococcus epidermidis, and Cutibacterium acnes can change over time.25,32 These shifts are unpredictable and are likely linked to many factors. Fungal groups are much more dynamic and heavily influenced by changes in seasonal factors such as precipitation, humidity, temperature, and atmospheric pressure.25
Clinical factors and phenotypes
In addition to being a well-known risk factor for the development of CRS, firsthand and secondhand smoking can also affect the sinonasal microbiome. Smoking directly alters the sinonasal epithelium, decreases mucociliary clearance, and increases inflammatory markers.33,34 In 1 study, patients with CRS without nasal polyps (CRSsNP) who were smokers at the time of sampling or who had ever smoked had significantly reduced bacterial richness and complexity.35 Smokers with CRS were colonized by significantly more pathogenic bacteria than were nonsmokers with CRS, and smokers with CRSsNP also had increased relative abundances of potentially pathogenic Bacteroides and Fusobacteria.35 An in vitro study demonstrated that bacterial isolates from smokers with CRS readily produced biofilms in response to exposure to whole tobacco smoke36; however, in another study biofilm formation was not associated with smoking status of patients with CRS.37 Together, these studies suggest that smoking significantly alters CRS disease severity and may promote dysbiosis.
Comorbid conditions such as asthma, atopy, eczema, aspirin-exacerbated respiratory disease, and cystic fibrosis may worsen the bacterial dysbiosis already associated with CRS. A common theme of these studies is that patients with CRS and comorbid respiratory conditions are characterized by an additional increased prevalence and relative abundance of pathogenic microbes, increased bacterial load, decreased bacterial diversity and richness, and decreased abundances of commensals such as Cutibacterium.2,38 For example, when compared with patients with CRS without asthma, patients with CRS and comorbid asthma have been observed to experience decreases in the relative abundances of Corynebacterium,39 Peptoniphilus,2 and Anaerococcus,2 as well as increases in Staphylococcus, Ralstonia, and Acinetobacter.40 Notably, the bacterial communities in patients with CRS with cystic fibrosis tend to be dominated by a single pathogen such as S aureus or Pseudomonas aeruginosa.15,25,39
Therapeutics
The impact of pharmaceutical and surgical interventions on the sinonasal microbial community is unpredictable40 and variable.41 Furthermore, a recently performed randomized controlled trial demonstrated that clinical outcomes following treatments with different medical agents were independent of the microbial outcomes.42 In a recent meta-analysis, short-term and cumulative antibiotic use did not contribute significantly to variation in composition of the sinonasal bacterial community, and few changes were observed versus in patients to whom antibiotics were not prescribed.43 These observations mirror the results from smaller randomized controlled trials showing only modest effects on individual bacterial sequence variants and no significant changes in richness or diversity in patients with CRS who received 7-day courses of doxycycline or roxithromycin versus in controls with CRS.44,45 The effect of topical corticosteroid therapy on the sinonasal microbiome has been less well studied. In their randomized controlled trial, Cherian et al did, however, demonstrate a significant but transient increase in bacterial diversity in patients treated with topical budesonide.42
Composition of the bacterial community may affect surgical outcomes, and surgery may alter composition of the bacterial community. In 1 study, patients with better postoperative outcomes had significantly increased diversity and richness and increased relative abundances of Corynebacterium before surgery.46 The presence of S aureus biofilms and intracellularly localized S aureus are strong predictors of recalcitrant disease and poorer postoperative outcomes.47–49
The effect of endoscopic sinus surgery on bacterial community composition, richness, and diversity is less clear. Some studies have suggested that bacterial diversity and richness increase, whereas others report that endoscopic sinus surgery results in decreased bacterial diversity.50,51 These inconsistencies may be due to interpersonal differences in patients with CRS, postoperative management, and time from surgery to sampling.
POTENTIAL MECHANISMS OF DISEASE
Numerous microbiota-regulated processes exist to maintain the stability of the sinonasal environment (Fig 2). The most intuitive mechanism to consider is pathogen exclusion by commensal populations. Increased microbial diversity is often a hallmark of health, with commensal microbes acting as niche occupiers that symbiotically ingest local nutrients and exclude overpopulation of pathogenic organisms. This theory is supported by the observed low abundance of pathobionts in healthy subjects52 and the reduced diversity with pathogen dominance in patients with CRS.53,54 Further evidence includes the network instability that results from depletion of particular commensal bacteria and observation of direct commensal-pathogen interaction.1 Yan et al55 studied the relationship between S aureus and Corynebacterium in the human nasal cavity and demonstrated that Corynebacteria can be involved in either mutualistic or inhibitory interactions with S aureus. Direct interactions between these 2 microbes have also been demonstrated by other groups researching CRS.56 This supports the findings of Bassiouni et al,57 who observed 3 main sinonasal “microbiotypes” in their analysis of the large International Microbiome Study cohort: the first is Corynebacterium-dominated, the second is Staphylococcus-dominated, and the third is dominated by the other core genera of the sinonasal microbiome (Streptococcus, Haemophilus, Moraxella, and Pseudomonas). Prior work has also demonstrated negative correlation between S aureus and S epidermidis in the nasal cavity,58 and recent work has uncovered direct inhibition by an S epidermidis–derived metabolite.59 Although the aforementioned findings support the dysbiosis hypothesis, causal demonstration in CRS still remains elusive.
FIG 2.
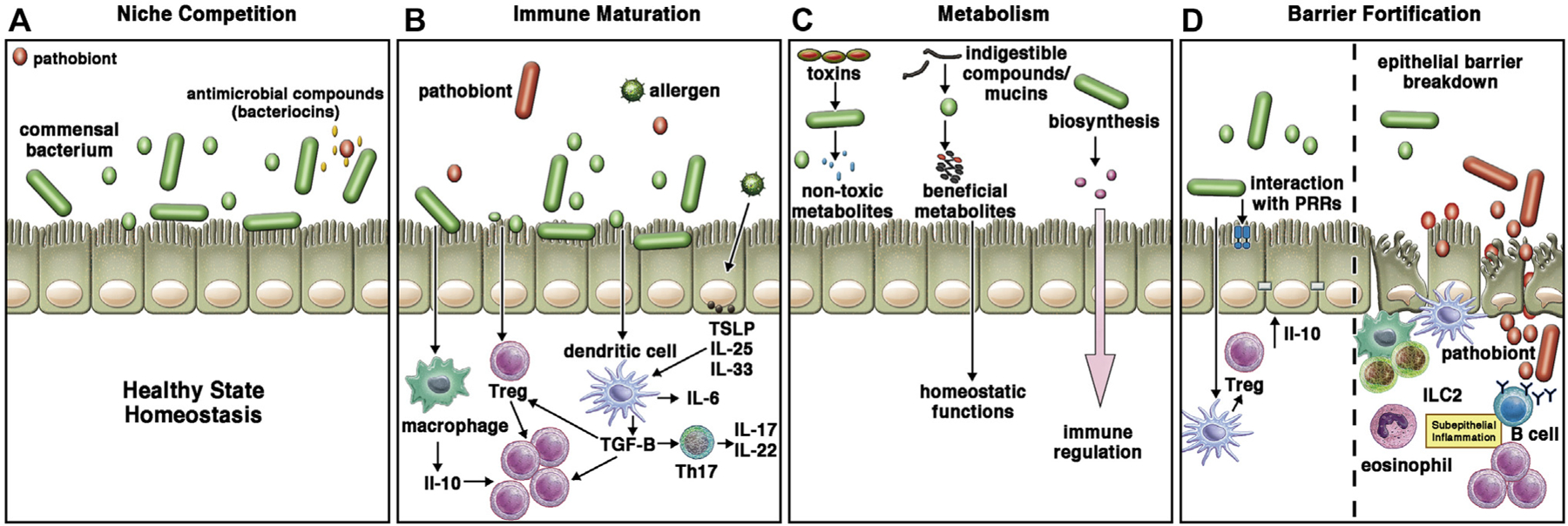
Potential mechanisms for microbiota-regulated processes in CRS. A, Commensal bacteria are niche occupiers precluding colonization or overgrowth of potential pathogens. This can occur directly through the production of bacteriocins, or it may occur indirectly by competition for local resources or induction of various antimicrobial peptides from the apical epithelium. B, Commensal organisms are critical for immune maturation, as has been documented in mouse gut and upper airway studies. Gain of pathogens may direct proinflammatory imbalance over anti-inflammatory homeostatic immune activities. C, Commensal organisms may provide several metabolic actions in the complex apical surface milieu, including biosynthesis of important proteins, degradation of airborne or locally produced toxins to innocuous byproducts, and digestion of mucins into energy sources such as short chain fatty acids. D, Breakdown of the epithelial barrier is a hallmark of CRS, with translocation of microbes into the subepithelial compartment. This can propagate the proinflammatory state. PRR, Pathogen recognition; Treg, regulatory T.
Another mechanism of direct microbiome influence on tissue processes occurs through microbial metabolism. Early attempts applying predictive metagenomics to evaluate the microbiome of samples from patients with CRS and controls identified functionally distinct microbiomes and host immune responses in patients with CRS.14 The various dysfunctional metabolic and pathophysiologic processes identified in these studies included synthesis of ansamycin (an antibiotic metabolite), LPS biosynthesis, tryptophan metabolism, peroxisome proliferator–activated receptor gamma (PPAR-γ) signaling, and invasion of epithelial cell pathways.60
Diverse microbe-derived molecules instruct varied and far-reaching immunologic processes.61,62 Notably, B-cell maturation and production of both IgA and IgG isotypes have recently been reported extranodally within nasal polyps in adults with CRS.63 Consistent with the importance of commensal organisms in gut-associated lymphoid tissue development, histologic observation of the nasal cavity from germ-free mice shows mucosal thinning, loss of goblet cells, increased collagen levels, and loss of nasal-associated lymphoid tissue.64 The influence of pathobionts on immunologic processes is well documented, particularly in the case of toxin release and superantigen production from S aureus.65
Multiple studies have shown correlations between sinonasal microbiota profiles and selected inflammatory markers. Cope et al14 correlated microbiota-based clustering with selected inflammatory markers, associating Corynebacterium-dominant clusters with IL-5 and IFN-γ in patients with CRS with nasal polyps (CRSwNP). Bachert et al66 observed correlations between S aureus presence and IL-5 positivity in patients with CRSwNP and patients with cystic fibrosis with nasal polyps. In another cluster, P aeruginosa colonization was associated with a high TNF-α level in patients with CRSsNP.66 More recently, Vickery et al67 defined disease-specific networks of interacting microbiota and host metabolites through targeted profiling of lipid mediators, suggesting yet another potential mechanism for regulation of local immune and inflammatory processes. These studies remain correlative, and further work is needed to understand the meaning of such associations.
The effect of microbes on epithelial barrier function is well described. Commensal bacteria have been shown to enhance the barrier function in the gut and respiratory tract, whereas numerous pathogenic organisms are known to degrade barrier function.68 A leaky barrier can result in bacterial translocation into the sinus mucosa, with a resulting chronic inflammatory response and tissue damage.69 This effect may be a causative factor in the development of CRS, or it may serve to propagate the disease into its chronic state.70 This phenomenon has been studied the most with S aureus, in which case induction of immunologic and mechanical barrier disruption promotes defective mucociliary clearance leading to bacterial invasion and degradation of intercellular tight junction proteins.71,72 This underlying effect may help explain the repeated observation that commensals inversely associated with Staphylococcus, such as Corynebacterium and Dolosigranulum, are associated with improved healing after sinus surgery in CRS and less disease recalcitrance in CRSwNP.73 Fig 2 summarizes potential mechanisms for microbiota-regulated processes in CRS.
THERAPEUTIC IMPLICATIONS
Microbial manipulation
Probiotics are defined as “live microorganisms that when administered in adequate amounts, confer a health benefit on the host” as proposed by the International Scientific Association for Probiotics and Prebiotics. The consistent finding of a high prevalence of the genus Corynebacterium in the sinuses of healthy patients, a significant reduction in its relative abundance in patients with CRS versus in controls, and a seemingly inverse relationship to the presence of S aureus12,57 supports the probiotic potential for species in this bacterial genus. The presence of 1 species, namely, Corynebacterium accolens, limits the growth of S aureus and other nasal pathobionts. In vitro work by Menberu et al evaluated the effect of CRS-derived isolates of C accolens against methicillin-sensitive S aureus and methicillin-resistant S aureus pathogens that were also isolated from the sinuses of patients with CRS.56 Almost all of the C accolens isolates exhibited antimicrobial activity against methicillin-sensitive S aureus and methicillin-resistant S aureus in both planktonic and biofilm forms.
The results of the study by Menberu et al56 were in agreement with those of a previously published clinical trial showing a 71% eradication of S aureus from the nasal cavity of human carriers after topical implantation of a strain of Corynebacterium spp.74 The exact mechanism of antibiotic action remains unknown but is thought to be due to the release of a yet to be characterized protein. C accolens has similar activity against Streptococcus pneumonia, another common nasal pathobiont.75,76 The mechanism of action against S pneumonia is mediated through degradation of triacylglycerol by C accolens to produce free fatty acids, in particular oleic acid, which interferes with the growth of S pneumoniae in the nasal cavity. In vivo studies evaluating the utility of these probiotic candidates in patients with established CRS has yet to be confirmed.
The only published human studies evaluating probiotic rinses for the treatment of established CRS have been performed by using Lactococcus lactis W136. This bacterium, although not highly prevalent in healthy sinuses, has an established safety profile for oral consumption in humans. In a small, nonblinded, and noncontrolled study, Endam et al77 reported the safety and feasibility of topical application of L lactis W136 in patients with CRS. Although they reported a clinical benefit in symptom reduction in these patients, few significant changes to bacterial profiles were observed following treatment. Lambert et al78 also evaluated a probiotic solution containing L lactis W136. In their nonblinded, controlled, crossover study comparing saline, xylitol, and L lactis W136 solutions, no significant difference in symptom scores were observed between the groups and no meaningful difference in the sinus microbiome was seen. Although their study was not limited to this particular strain of Lactococcus, Martensson et al79 similarly reported no significant changes to clinical outcomes, bacterial community composition, or inflammatory profiles following irrigation fluid containing lactic acid–producing bacteria in patients with CRS.79 Interestingly, despite these findings, commercial preparations of L lactis are now available for CRS “treatment.” The release of such products into the commercial market may be premature, not only given their lack of objective benefit but also given the possibility of their promoting growth of pathogenic strains of bacteria, such as Paeruginosa, as was observed in a coculture study by Cho et al.80
Animal models of sinusitis have also been used to explore the role of probiotic candidates, although similar human trials have not yet been performed. Cleland et al50 investigated the development of sinusitis in mice that had been inoculated with S aureus alone or in the presence of the known commensal S epidermidis. In mice coinoculated with the commensal and pathogenic bacteria, there were significantly fewer markers of inflammation typically associated with CRS than in those receiving S aureus alone, supporting the probiotic effect of S epidermidis. In another murine model for sinusitis, Abreu et al81 utilized Lactobacillus sakei, a commensal identified as being depleted in human patients with CRS, to defend against Corynebacterium tuberculostearicum sinus infection in the context of a depleted sinus bacterial community.
The aforementioned studies highlight the difficulties in identifying promising probiotic microbes and the need for large, well-controlled clinical trials that demonstrate significant improvements in clinical, microbiologic, and inflammatory outcomes in CRS. As evidenced from previous sections of this review, high-fidelity resolution of the microbiome at the species and strain levels is still lacking. Poor taxonomic resolution combined with such high interpersonal variation and the numerous endogenous and exogenous influences exerted on the sinonasal microbiome further complicate progress in this area.
CURRENT LIMITATIONS OF RESEARCH
The main challenge facing the field of CRS microbiome–focused research, and other human microbiome–related research in general, is moving beyond taxonomic associations and toward the establishment of causality.82 Awareness of causal effects, even if only in a few specific scenarios83 such as a particular microbe, mechanism, and disease phenotype or endotype, will be requisite before attempting to intervene with novel therapeutic approaches (Table I).
Table I.
Key questions, unresolved issues, and next steps
|
As a heterogeneous human disease with an unclear etiopatho-genesis, variable and fluctuating natural history, soft boundaries for phenotyping and novel endotyping, susceptibility to infection and local immune dysfunction, and numerous trials of varied therapies before diagnosis, sorting out a possible microbiome role in CRS is a large task.
To date, most work related to the CRS microbiome has utilized the case-control study design, which cannot be used to determine whether the microbiome drives the disease process, whether the disease is altering the microbiome, or whether both are influenced by other confounders. Additionally, the microbiome may not be causative of the disease; rather, it could serve as disease modifier in some critical aspect. Although the varied temporal aspect of disease development and response to therapies has limited the opportunity for longitudinal study of pathogenesis before disease onset, it has afforded the potential to study the microbiome’s role in disease evolution and response to interventions.
Cross-sectional sinonasal microbiome studies are also limited by large intersubject variation and the numerous confounding variables that may exist in human subjects with CRS or atopic disease. Before study enrollment, patients recruited for these studies have been subjected to repeated disease interventions that are known to alter the microbiome (eg, antibiotic or steroid therapies), and they exhibit deficiencies in local epithelial barrier and immune function.69,84 Additionally, the high degree of interpersonal variability is a challenge to overcome, given the relatively small cohorts included in studies. CRS is a broad disease that is clinically defined by a constellation of particular symptoms with an objective finding of inflammation, and it has recently been further characterized by disease phenotype and molecular approaches to endotyping, much like what has been done for asthma. It is apparent that different immunologic process may predominate in certain patients at certain times (ie, type 1, 2, or 3 inflammation or diminished regulatory T–cell function). Direct and indirect host-microbial interactions are present in each of these CRS subcategories, and although little is known at this point, it is certainly possible that the importance of the microbiome and mechanistic insights will be different in at least 1 group. Studies characterizing microbiota relevance by disease endotype are under way, and the introduction of targeted biologics to type 2–dominant CRS offers exciting an opportunity for an intervention study. As we continue to define CRS subtypes beyond the simple phenotype differences, larger sample sizes and narrowly defined disease categories will be required for studies so as to limit the potential for type II error, as in recent trials delineating the role for surgery and biologics in CRS.84 Teasing apart the intricacies within the complex set of features known to exert influences on the microbiome is key to understanding whether, and to what degree, observed changes represent bystander effects.
These limitations associated with in situ studies in patients are further compounded by deficiencies of the small animal models created for study of CRS.85 Compelling microbiome research has utilized animal experiments that include antibiotic depletion, gnotobiotic mice, and microbial transfer approaches. Although acute airway responses can be ascertained in animal models, establishing chronic upper airway inflammation representative of CRS remains elusive. As described in the European Position Paper on Rhinosinusitis and Nasal Polyps,86 “Currently available animal models are either allergic models or genetically manipulated animals that artificially generate an inflammatory response and again, do not answer the cause of the disease.” In rodents, the surface area of the nasal cavity is small and there are no true sinuses. Additionally, nearly half of the nasal epithelium is olfactory epithelium, and rodents express different antimicrobial peptides and mucins than humans do.87 Unsurprisingly, the baseline colonizing microbes are distinct from those of the human upper airway. Furthermore, immune responses in mice are notably different from those in humans.88 Alternatively, the rabbit maxillary sinus model was developed for study of biofilm formation by use of obstruction of sinus outflow by a foreign body,89 and it was more recently utilized for microbiome study.80 Other accepted in vitro approaches such as cell culture with microbial exposure carry limitations, including absence of multicellular interaction (ie, epithelial-immune interaction), absent tissue architecture, inability to maintain cells and many microbes under the same culture conditions, and limited duration of live microbe and cellular coexistence.
FUTURE DIRECTIONS
Sinonasal microbiome research is still largely in a descriptive phase. It is likely that the microbial profiles that have been described to date are the culmination of the complex interplay between the environment, microbiota, and the host immune system. In vitro and in vivo studies examining microbial interaction of known pathobionts and commensals in patients with CRS, as well the interaction between microbes and the host’s local immune cells, are clearly needed. Furthermore, without researching and understanding the early and intermediary stages of the disease process, resolving whether the microbiome changes observed are a cause, a disease modifier, or a result of disease is impossible. Ideally, generational, longitudinal cohort studies documenting the microbiome from health to disease could be performed to help resolve this question.
Short sequences have offered limited insights into function, and they limit important taxonomic assignments to species or strain. Long-read sequencing may be adapted to confidently distinguish the species- and strain-level differences between the microbiota of healthy subjects and patients with CRS. Future studies should also focus on distinguishing changes between healthy individuals and patients with CRS in the core genetic components transcriptome and proteome. Although some small, cross-sectional studies in each of these areas have been performed, larger and more definitive studies will be required.
Disclosure of potential conflict of interest:
A. J. Psaltis reports receiving fees for consulting from ENT technologies, Tissium, Fusetec, and Medtronic; receiving honoraria from Sequiris for speakers bureau participation; and being a shareholder of Chitogel. V. R. Ramakrishnan reports fees for consulting from Medtronic, Inc, and Optinose, US, and honoraria from Genentech, AstaZeneca, Regeneron, GlaxoSmithK-line, and MicroGen DX for serving on scientific advisory boards. The rest of the authors declare that they have no relevant conflicts of interest.
Supported in part by the Auckland Medical Research Foundation (grant 1120008 [to B.W.M.]); the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award R15AI147148 [to E.K.C.]); the Ludeman Family Center for Women’s Health Research at the University of Colorado Anschutz Medical Campus (to V.R.R.); and the National Institutes of Health/National Institute on Deafness and Other Communication Disorders (grant K23DC014747 [to V.R.R.]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding organizations.
Abbreviations used
- CRS
Chronic rhinosinusitis
- CRSsNP
Chronic rhinosinusitis without nasal polyps
- CRSwNP
Chronic rhinosinusitis with nasal polyps
- rRNA
Ribosomal RNA
- URT
Upper respiratory tract
REFERENCES
- 1.Wagner Mackenzie B, Waite DW, Hoggard M, Douglas RG, Taylor MW, Biswas K. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol 2017;19:381–92. [DOI] [PubMed] [Google Scholar]
- 2.Hoggard M, Biswas K, Zoing M, Wagner Mackenzie B, Taylor MW, Douglas RG. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int Forum Allergy Rhinol 2017;7:230–9. [DOI] [PubMed] [Google Scholar]
- 3.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017;15:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joss TV, Burke CM, Hudson BJ, Darling AE, Forer M, Alber DG, et al. Bacterial communities vary between sinuses in chronic rhinosinusitis patients. Front Microbiol 2015;6:1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lal D, Keim P, Delisle J, Barker B, Rank MA, Chia N, et al. Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int Forum Allergy Rhinol 2017;7:561–9. [DOI] [PubMed] [Google Scholar]
- 6.Ramakrishnan VR, Gitomer S, Kofonow JM, Robertson CE, Frank DN. Investigation of sinonasal microbiome spatial organization in chronic rhinosinusitis. Int Forum Allergy Rhinol 2017;7:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Boeck I, Wittouck S, Martens K, Claes J, Jorissen M, Steelant B, et al. Anterior nares diversity and pathobionts represent sinus microbiome in chronic rhinosinusitis. mSphere 2019;4:e00532–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassiouni A, Cleland EJ, Psaltis AJ, Vreugde S, Wormald PJ. Sinonasal microbiome sampling: a comparison of techniques. PLoS One 2015;10:e0123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marotz CA, Sanders JG, Zuniga C, Zaramela LS, Knight R, Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 2018; 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu W, Crawford ED, O’Donovan BD, Wilson MR, Chow ED, Retallack H, et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol 2016;17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 2009;19:1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paramasivan S, Bassiouni A, Shiffer A, Dillon MR, Cope EK, Cooksley C, et al. The international sinonasal microbiome study: a multicentre, multinational characterization of sinonasal bacterial ecology. Allergy 2020;75:2037–49. [DOI] [PubMed] [Google Scholar]
- 13.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cope EK, Goldberg AN, Pletcher SD, Lynch SV. Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 2017;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas SK, Feddema E, Boyer HC, Hunter RC. Diversity of cystic fibrosis chronic rhinosinusitis microbiota correlates with different pathogen dominance. J Cyst Fibros 2021;20:678–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015;17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 2012;78: 6262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014;190: 1283–92. [DOI] [PubMed] [Google Scholar]
- 19.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J Allergy Clin Immunol 2018;142:1447–56.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016;48:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulraiz F, Bellinghausen C, Bruggeman CA, Stassen FR. Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB J 2015;29:849–58. [DOI] [PubMed] [Google Scholar]
- 23.Ma ZS, Li W. How and why men and women differ in their microbiomes: medical ecology and network analyses of the microgenderome. Adv Sci (Weinh) 2019;6: 1902054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goossens H, Ferech M, Coenen S, Stephens P, European Surveillance of Antimicrobial Consumption Project Group. Comparison of outpatient systemic antibacterial use in 2004 in the United States and 27 European countries. Clin Infect Dis 2007;44:1091–5. [DOI] [PubMed] [Google Scholar]
- 25.Wagner Mackenzie B, Dassi C, Vivekanandan A, Zoing M, Douglas RG, Biswas K. Longitudinal analysis of sinus microbiota post endoscopic surgery in patients with cystic fibrosis and chronic rhinosinusitis: a pilot study. Respir Res 2021;22: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prussin AJ 2nd, Torres PJ, Shimashita J, Head SR, Bibby KJ, Kelley ST, et al. Seasonal dynamics of DNA and RNA viral bioaerosol communities in a daycare center. Microbiome 2019;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipsitch M, Viboud C. Influenza seasonality: lifting the fog. Proc Natl Acad Sci U S A 2009;106:3645–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee WM, Lemanske RF Jr, Evans MD, Vang F, Pappas T, Gangnon R, et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 2012;186:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choe YJ, Smit MA, Mermel LA. Seasonality of respiratory viruses and bacterial pathogens. Antimicrob Resist Infect Control 2019;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lima JT, Paula FE, Proenca-Modena JL, Demarco RC, Buzatto GP, Saturno TH, et al. The seasonality of respiratory viruses in patients with chronic rhinosinusitis. Am J Rhinol Allergy 2015;29:19–22. [DOI] [PubMed] [Google Scholar]
- 31.Koutsourelakis I, Halderman A, Khalil S, Hittle LE, Mongodin EF, Lane AP. Temporal instability of the post-surgical maxillary sinus microbiota. BMC Infect Dis 2018;18:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camarinha-Silva A, Jauregui R, Pieper DH, Wos-Oxley ML. The temporal dynamics of bacterial communities across human anterior nares. Environ Microbiol Rep 2012;4:126–32. [DOI] [PubMed] [Google Scholar]
- 33.Shi JB, Fu QL, Zhang H, Cheng L, Wang YJ, Zhu DD, et al. Epidemiology of chronic rhinosinusitis: results from a cross-sectional survey in seven Chinese cities. Allergy 2015;70:533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamashiro E, Xiong G, Anselmo-Lima WT, Kreindler JL, Palmer JN, Cohen NA. Cigarette smoke exposure impairs respiratory epithelial ciliogenesis. Am J Rhinol Allergy 2009;23:117–22. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishnan VR, Frank DN. Impact of cigarette smoking on the middle meatus microbiome in health and chronic rhinosinusitis. Int Forum Allergy Rhinol 2015;5: 981–9. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein-Daruech N, Cope EK, Zhao KQ, Vukovic K, Kofonow JM, Doghramji L, et al. Tobacco smoke mediated induction of sinonasal microbial biofilms. PLoS One 2011;6:e15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Kofonow JM, Finkelman BS, Doghramji L, Chiu AG, Kennedy DW, et al. Clinical factors associated with bacterial biofilm formation in chronic rhinosinusitis. Otolaryngol Head Neck Surg 2011;144:457–62. [DOI] [PubMed] [Google Scholar]
- 38.Cook KA, Domissy A, Simon RA, White AA, Modena BD. Dysbiosis in aspirin-exacerbated respiratory disease. Int Forum Allergy Rhinol 2021;11:1116–20. [DOI] [PubMed] [Google Scholar]
- 39.Pletcher SD, Goldberg AN, Cope EK. Loss of microbial niche specificity between the upper and lower airways in patients with cystic fibrosis. Laryngoscope 2019; 129:544–50. [DOI] [PubMed] [Google Scholar]
- 40.Hauser LJ, Ir D, Kingdom TT, Robertson CE, Frank DN, Ramakrishnan VR. Investigation of bacterial repopulation after sinus surgery and perioperative antibiotics. Int Forum Allergy Rhinol 2016;6:34–40. [DOI] [PubMed] [Google Scholar]
- 41.Hoggard M, Jacob B, Wheeler D, Zoing M, Chang K, Biswas K, et al. Multiomic analysis identifies natural intrapatient temporal variability and changes in response to systemic corticosteroid therapy in chronic rhinosinusitis. Immun Inflamm Dis 2021;9:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherian LM, Bassiouni A, Cooksley CM, Vreugde S, Wormald PJ, Psaltis AJ. The clinical outcomes of medical therapies in chronic rhinosinusitis are independent of microbiomic outcomes: a double-blinded, randomised placebo-controlled trial. Rhinology 2020;58:559–67. [DOI] [PubMed] [Google Scholar]
- 43.Lux CA, Wagner Mackenzie B, Johnston J, Zoing M, Biswas K, Taylor MW, et al. Antibiotic treatment for chronic rhinosinusitis: prescription patterns and associations with patient outcome and the sinus microbiota. Front Microbiol 2020;11: 595555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain R, Hoggard M, Zoing M, Jiang Y, Biswas K, Taylor MW, et al. The effect of medical treatments on the bacterial microbiome in patients with chronic rhinosinusitis: a pilot study. Int Forum Allergy Rhinol 2018;8:890–9. [DOI] [PubMed] [Google Scholar]
- 45.Siu J, Mackenzie BW, Klingler L, Biswas K, Wang Y, Hung CT, et al. Sinonasal and gastrointestinal bacterial composition and abundance are stable after 1 week of once-daily oral antibiotic treatment for chronic rhinosinusitis. Int Forum Allergy Rhinol 2021;11:1355–66. [DOI] [PubMed] [Google Scholar]
- 46.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol 2015;136:334–42.e1. [DOI] [PubMed] [Google Scholar]
- 47.Psaltis AJ, Weitzel EK, Ha KR, Wormald PJ. The effect of bacterial biofilms on post-sinus surgical outcomes. Am J Rhinol 2008;22:1–6. [DOI] [PubMed] [Google Scholar]
- 48.Singhal D, Psaltis AJ, Foreman A, Wormald PJ. The impact of biofilms on outcomes after endoscopic sinus surgery. Am J Rhinol Allergy 2010;24:169–74. [DOI] [PubMed] [Google Scholar]
- 49.Ou J, Drilling A, Singhal D, Tan NC, Wallis-Hill D, Vreugde S, et al. Association of intracellular Staphylococcus aureus with prognosis in chronic rhinosinusitis. Int Forum Allergy Rhinol 2016;6:792–9. [DOI] [PubMed] [Google Scholar]
- 50.Cleland EJ, Drilling A, Bassiouni A, James C, Vreugde S, Wormald PJ. Probiotic manipulation of the chronic rhinosinusitis microbiome. Int Forum Allergy Rhinol 2014;4:309–14. [DOI] [PubMed] [Google Scholar]
- 51.Jain R, Hoggard M, Biswas K, Zoing M, Jiang Y, Douglas R. Changes in the bacterial microbiome of patients with chronic rhinosinusitis after endoscopic sinus surgery. Int Forum Allergy Rhinol 2017;7:7–15. [DOI] [PubMed] [Google Scholar]
- 52.Ramakrishnan VR, Feazel LM, Gitomer SA, Ir D, Robertson CE, Frank DN. The microbiome of the middle meatus in healthy adults. PLoS One 2013;8:e85507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi EB, Hong SW, Kim DK, Jeon SG, Kim KR, Cho SH, et al. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy 2014;69:517–26. [DOI] [PubMed] [Google Scholar]
- 54.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope 2012;122: 467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal micro-environments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 2013;14:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menberu MA, Liu S, Cooksley C, Hayes AJ, Psaltis AJ, Wormald PJ, et al. Corynebacterium accolens has antimicrobial activity against Staphylococcus aureus and methicillin-resistant S. aureus pathogens isolated from the sinonasal niche of chronic rhinosinusitis patients. Pathogens 2021;10:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassiouni A, Paramasivan S, Shiffer A, Dillon MR, Cope EK, Cooksley C, et al. Microbiotyping the sinonasal microbiome. Front Cell Infect Microbiol 2020;10: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 2010;5: e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010;465:346–9. [DOI] [PubMed] [Google Scholar]
- 60.Mahdavinia M, Engen PA, LoSavio PS, Naqib A, Khan RJ, Tobin MC, et al. The nasal microbiome in patients with chronic rhinosinusitis: analyzing the effects of atopy and bacterial functional pathways in 111 patients. J Allergy Clin Immunol 2018;142:287–90.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ethridge AD, Bazzi MH, Lukacs NW, Huffnagle GB. Interkingdom communication and regulation of mucosal immunity by the microbiome. J Infect Dis 2021; 223(12 suppl 2):S236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim CH. Immune regulation by microbiome metabolites. Immunology 2018;154: 220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song J, Wang H, Zhang YN, Cao PP, Liao B, Wang ZZ, et al. Ectopic lymphoid tissues support local immunoglobulin production in patients with chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 2018;141:927–37. [DOI] [PubMed] [Google Scholar]
- 64.Jain R, Waldvogel-Thurlow S, Darveau R, Douglas R. Differences in the paranasal sinuses between germ-free and pathogen-free mice. Int Forum Allergy Rhinol 2016;6:631–7. [DOI] [PubMed] [Google Scholar]
- 65.Vickery TW, Ramakrishnan VR, Suh JD. The Role of Staphylococcus aureus in patients with chronic sinusitis and nasal polyposis. Curr Allergy Asthma Rep 2019;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chalermwatanachai T, Zhang N, Holtappels G, Bachert C. Association of mucosal organisms with patterns of inflammation in chronic rhinosinusitis. PLoS One 2015; 10:e0136068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vickery TW, Armstrong M, Kofonow JM, Robertson CE, Kroehl ME, Reisdorph NA, et al. Altered tissue specialized pro-resolving mediators in chronic rhinosinusitis. Prostaglandins Leukot Essent Fatty Acids 2021;164:102218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panchatcharam BS, Cooksley CM, Ramezanpour M, Vediappan RS, Bassiouni A, Wormald PJ, et al. Staphylococcus aureus biofilm exoproteins are cytotoxic to human nasal epithelial barrier in chronic rhinosinusitis. Int Forum Allergy Rhinol 2020;10:871–83. [DOI] [PubMed] [Google Scholar]
- 69.Akdis CA. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol 2021;21: 739–51. [DOI] [PubMed] [Google Scholar]
- 70.Kern RC, Conley DB, Walsh W, Chandra R, Kato A, Tripathi-Peters A, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol 2008;22:549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murphy J, Ramezanpour M, Stach N, Dubin G, Psaltis AJ, Wormald PJ, et al. Staphylococcus aureus V8 protease disrupts the integrity of the airway epithelial barrier and impairs IL-6 production in vitro. Laryngoscope 2018;128:E8–15. [DOI] [PubMed] [Google Scholar]
- 72.Altunbulakli C, Costa R, Lan F, Zhang N, Akdis M, Bachert C, et al. Staphylococcus aureus enhances the tight junction barrier integrity in healthy nasal tissue, but not in nasal polyps. J Allergy Clin Immunol 2018;142: 665–8.e8. [DOI] [PubMed] [Google Scholar]
- 73.Gan W, Zhang H, Yang F, Liu S, Liu F, Meng J. The influence of nasal microbiome diversity and inflammatory patterns on the prognosis of nasal polyps. Sci Rep 2021;11:6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect 2000;44:127–33. [DOI] [PubMed] [Google Scholar]
- 75.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 2016;7, e01725–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Speert DP, Wannamaker LW, Gray ED, Clawson CC. Bactericidal effect of oleic acid on group A streptococci: mechanism of action. Infect Immun 1979;26: 1202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Endam LM, Alromaih S, Gonzalez E, Madrenas J, Cousineau B, Renteria AE, et al. Intranasal application of lactococcus lactis W136 is safe in chronic rhinosinusitis patients with previous sinus surgery. Front Cell Infect Microbiol 2020;10:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lambert PA, Gill AL, Gill SR, Allen PD, Man LX. Microbiomics of irrigation with xylitol or Lactococcus lactis in chronic rhinosinusitis. Laryngoscope Investig Otolaryngol 2021;6:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martensson A, Abolhalaj M, Lindstedt M, Martensson A, Olofsson TC, Vasquez A, et al. Clinical efficacy of a topical lactic acid bacterial microbiome in chronic rhinosinusitis: a randomized controlled trial. Laryngoscope Investig Otolaryngol 2017;2:410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho DY, Skinner D, Lim DJ, McLemore JG, Koch CG, Zhang S, et al. The impact of Lactococcus lactis (probiotic nasal rinse) co-culture on growth of patient-derived strains of Pseudomonas aeruginosa. Int Forum Allergy Rhinol 2020;10:444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 2012;4:151ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramakrishnan VR, Frank DN. Microbiome in patients with upper airway disease: moving from taxonomic findings to mechanisms and causality. J Allergy Clin Immunol 2018;142:73–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanage WP. Microbiology: microbiome science needs a healthy dose of scepticism. Nature 2014;512:247–8. [DOI] [PubMed] [Google Scholar]
- 84.Bachert C, Marple B, Hosemann W, Cavaliere C, Wen W, Zhang N. Endotypes of chronic rhinosinusitis with nasal polyps: pathology and possible therapeutic implications. J Allergy Clin Immunol Pract 2020;8:1514–9. [DOI] [PubMed] [Google Scholar]
- 85.Al-Sayed AA, Agu RU, Massoud E. Models for the study of nasal and sinus physiology in health and disease: a review of the literature. Laryngoscope Investig Otolaryngol 2017;2:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, Reitsma S, et al. Executive summary of EPOS 2020 including integrated care pathways. Rhinology 2020; 58:82–111. [DOI] [PubMed] [Google Scholar]
- 87.Casadei E, Tacchi L, Lickwar CR, Espenschied ST, Davison JM, Munoz P, et al. Commensal bacteria regulate gene expression and differentiation in vertebrate olfactory systems through transcription factor REST. Chem Senses 2019;44:615–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004;172:2731–8. [DOI] [PubMed] [Google Scholar]
- 89.Liang KL, Jiang RS, Wang J, Shiao JY, Su MC, Hsin CH, et al. Developing a rabbit model of rhinogenic chronic rhinosinusitis. Laryngoscope 2008;118:1076–81. [DOI] [PubMed] [Google Scholar]


