ABSTRACT
Non-small cell lung carcinoma (NSCLC) is a type of lung cancer with the highest prevalence and mortality rate worldwide. Many cases of this type of cancer are overexpression on epidermal growth factor receptor (EGFR). The use of currently available EGFR inhibitors as one of the treatment options for NSCLC still shows various shortcomings, especially the high failure rate of therapy due to resistance. It is important to find NSCLC drug candidates with EGFR inhibitory activity. There are various published articles and it is prominent to draw evidence-based scientific conclusions as a basis of decision-making to select potential compounds for further research. Polymer matrix composites and ScienceDirect are used as a database for article screening. Research using molecular docking method targeted to EGFR with parameters of Gibbs energy and amino acid interactions between ligands and drug targets are included in inclusion criteria. Compounds that achieve docking parameters and have comparable activity to NSCLC guideline drugs are conscientiously ranked. There are only 11 compounds that achieved the docking parameters and had comparable EGFR inhibitory potential. Top-rated compounds include 1,3,5-trisubstituted pyrazoline (3c), 1,3,5-trisubstituted pyrazoline (6c), 1,3,5-trisubstituted pyrazoline (8d), N-(3,4-Dimethylphenyl)-2-[(4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydrobenzo[g] quinazolin-2-yl) thio] acetamide. The top-rated compounds can be used and considered for further research processes.
Keywords: Epidermal growth factor receptor, in silico, lung cancer, non-small cell lung cancer
INTRODUCTION
Lung cancer is one of the most frequent cancer worldwide with the mortality cases about 1.8 million deaths.[1] There are two types of lung cancer, namely non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma.[2] NSCLC dominates the incidence of lung cancer for approximately 85% of all lung cancer cases.[3]
Chemotherapy is the most common therapy for patients. One of the mechanisms of action of chemotherapy drugs is to inhibit epidermal growth factor receptor (EGFR). The EGFR, also called HER1 or erbB-1, is a transmembrane glycoprotein receptor tyrosine kinase. EGFR has a role in activating various signal transduction pathways that play a role in the cell cycle. EGFR activation induces dimerization and subsequent phosphorylation of tyrosine kinase, then a cascade of intracellular signaling occurs which helps various important cellular responses such as increased angiogenesis, tumor survival, cell proliferation, invasion and increased metastasis and decreased apoptosis [Figure 1].[4] Many cases of cancers result from EGFR overexpression. There is an overexpression of EGFR (40%–80%) in NSCLC.[5] The first-line anticancer used in NSCLC with EGFR overexpression is erlotinib. The use of erlotinib causes serious side effects such as liver damage and lesions formation in the digestive system.[6,7]
Figure 1.
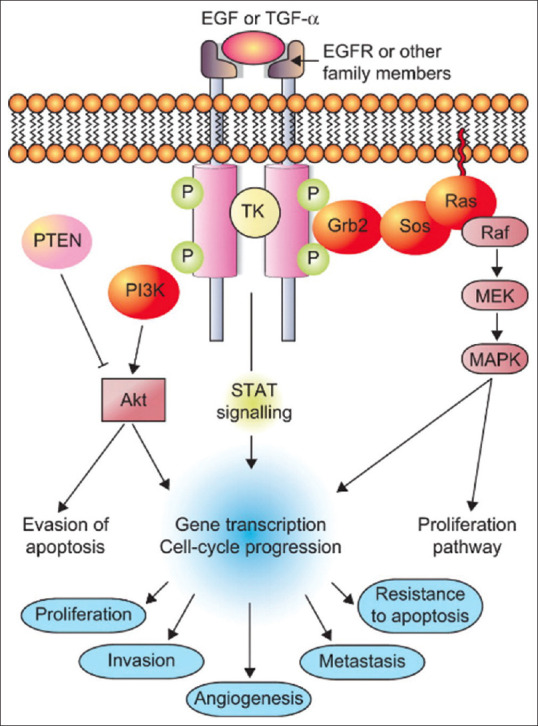
Epidermal growth factor receptor signaling pathway in lung cancer[4]
In the last 10 years, the use of the in silico method as an initial step of anticancer drug discovery and development has increased especially the use of the molecular docking simulation method. The high number of docked molecules through in silico studies requires further research (in vitro, in vivo, and clinical trials) to obtain a new drug that has a better performance than current therapy.
A step is required to conclude the results of this computational research to provide references to researchers worldwide regarding candidate compounds that computationally have optimal interaction to EGFR. This systematic review is expected to be an evidence-based scientific conclusion for researchers as a basis for decision-making in selecting compounds to be further investigated in the discovery of anti-NSCLC agents to stimulate the rate of preclinical research anticancer in the drug discovery and development process.
MATERIALS AND METHODS
This systematic review searches followed the preferred reporting items for systematic review and meta-analyses (PRISMA) guideline to increase the credibility of the study and is conducted through polymer matrix composites (PMC) and ScienceDirect databases using three groups of keywords “(((Carcinoma, NonSmall-Cell Lung) AND Molecular Docking Simulation) AND EGFR) NOT Review” in the last 10 years. A total of 355 articles yielded and duplicate articles are eliminated by using the EndNote reference manager yield 350 articles.
Based on the keywords above, the articles collected are selected by several inclusion criteria including:
The research method used must be in silico method, especially molecular docking simulation
The results of the research must contain the Gibbs energy and the interactions between ligands and the amino acid residues from the target protein EGFR
Articles that do not achieve the inclusion criteria were then eliminated while articles that achieve the criteria are then analyzed so that there is a list of compounds capable of interacting with the target protein computationally.
In this stage, 18 articles achieve the inclusion criteria and were suitable to be further analyzed.
In analyzing compound data obtained from inclusion articles, several parameters of compound selection for further analysis have been determined. The determination of the parameters of these compounds is intended to obtain a list of compounds that could be compared to each other for the study process. These parameters include:
The compounds should have a more negative than − 5.50 kcal/mol Gibbs energy value. The determination of the score is based on a theory that indicates that a compound has inhibition activity if it has Gibbs energy value <−5.50 kcal/mol[8]
The compounds should have the ability to form hydrogen bond interactions with the active sites on the target protein
Compounds capable of forming hydrogen bond interactions with catalytic amino acids on the active site of the target protein will be ranked higher.
At this final stage, 9 articles could be further analyzed to find the most potent compound that could be a promising EGFR inhibitor for NSCLC treatment.
RESULT AND DISCUSSION
The scheme of article selection
The article selection scheme based on the PRISMA guideline of systematic review using the keyword of “(((Carcinoma, Non-Small-Cell Lung) AND Molecular Docking Simulation) AND EGFR) NOT Review.” The results showed that 9 articles are reviewed over 355 articles obtained [Figure 2].
Figure 2.
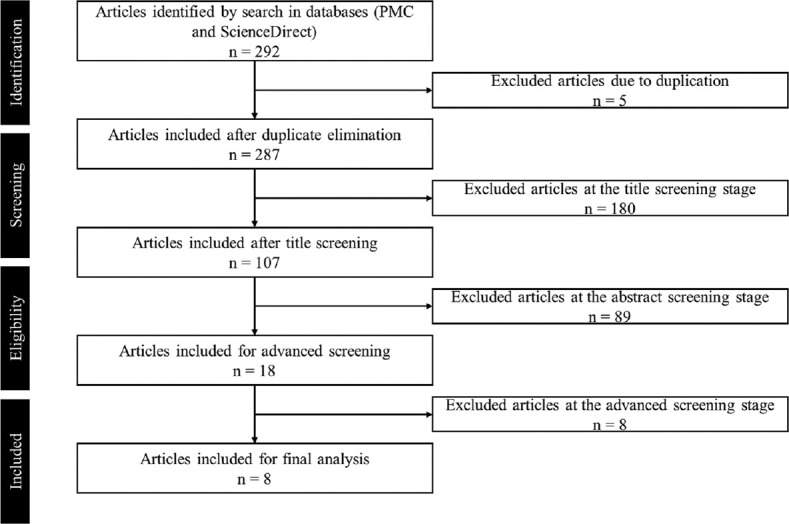
Article selection scheme based on the PRISMA Guideline of Systematic Review using the keyword of “(((Carcinoma, Non-Small-Cell Lung) AND Molecular Docking Simulation) AND Epidermal growth factor receptor) NOT Review.” The results showed that 8 articles are reviewed over 292 articles obtained
Description of the research
Based on the search results on the PMC and ScienceDirect databases using three predetermined keywords, “(((Carcinoma, Non-Small-Cell Lung) AND Molecular Docking Simulation) AND EGFR) NOT Review” with the total was 292 articles [Figure 2]. However, most of the articles did not meet the inclusion criteria that had been determined. There were 180 articles excluded due to their disability to fulfill the inclusion criteria that had been determined. In addition, at the abstract screening stage, 89 articles were excluded due to the unsuitability of the research method. Entering the advance stage, articles that are lack of information regarding the interaction between the test ligands and amino acids interaction in the EGFR protein were excluded. Therefore, 8 articles were included in the inclusion criteria for a more in-depth analysis in this systematic review.
Each test compound discussed in the included article is ranked based on their docking performance by referring to Gibbs free energy data (the more negative the Gibbs free energy, the better) and its ability to interact with key amino acids in EGFR protein through hydrogen bonds. Based on the results, 28 compounds were successfully docked to the EGFR protein consisting of one comparison compound (erlotinib, Food and Drug Administration -approved drug as EGFR inhibitor) and 27 test compounds.
The ideal criteria of molecular docking simulation against epidermal growth factor receptor protein
Ideally, the quality of the molecular docking simulation is determined by three important parameters, i.e., the number of clusters, Gibbs free energy, and the ability of a ligand to interact with its receptor through a stable binding mode. Therefore, to make a ranking list of the docking results, it is necessary to refer to these three data. First, the docking data taken from the molecular docking results must represent at least 75% of the docking population (showed by the number of the cluster) so that the Gibbs energy value and the amino acid interaction mode can be representative of a large number of the experimental population [Table 1].
Table 1.
The ideal parameter of molecular docking simulation against epidermal growth factor receptor protein
| Number of clusters | Gibbs energy (kcal/mol) | Hydrogen bond interaction with amino acid residues | References |
|---|---|---|---|
| 75% from population | −5.5 | Cys773, Met769, Thr766, Lys721, Thr830, Gly772 and Leu694 | [9] |
The second parameter to look at is the Gibbs free energy value. Based on Eleftheriou et al.(2020), a compound can stably interact with its receptors if it has a value of Gibbs free energy lower than 5.5 kcal/mol. Therefore, the compound that will be referred to in the next stage of research must have a Gibbs free energy value lower than 5.5 kcal/mol to avoid problems related to the pharmacodynamic properties of the drug [Table 1].
The third parameter that must be considered in molecular docking simulation is the mode of interaction between ligands and their receptors [Table 1]. EGFR is a targeted receptor in the treatment of lung cancer. Therefore, it is very important to analyze the structure of these proteins atomically to characterize the important properties of these proteins. So that we can find out strategies for inactivating protein function through the inhibitory mechanism that will be carried out by the test ligands. There are five main regions in the EGFR-ATP binding pocket, i.e., a sugar pocket, phosphate-binding pocket, adenine binding pocket, and two hydrophobic regions. The Cys773 residue in the sugar region is unique to EGFR which provides potency and selectivity. The phosphate-binding region subjected to high solvent exposure offers a little opportunity for inhibitor-binding to this region. Electrostatic interaction between the amino group of the adenine ring and the Met769 residue occurs at the adenine binding site. Thr766, Lys721, and Thr830 residues which comprise the back hydrophobic region play a pivotal role in the inhibitor selectivity while the front hydrophobic region is formed by Gly772 and Leu694 residues. Therefore, those amino acids were considered to play the key role in EGFR and need to be inhibited by the ligand through a stable interaction (hydrogen Interaction).
Molecular docking of various small molecules against epidermal growth factor receptor protein
EGFR is a protein that is very important in cancer signaling pathways, especially lung cancer. In transducing the signal, EGFR requires one ATP molecule to bind to the binding site on the protein so that electronic signaling can be continued. The ATP binding pocket contained in the EGFR (as mentioned in part III) can be competitively inhibited by a ligand. This strategy is used by researchers to find candidates for new drug compounds for cancer therapy, one of which is lung cancer.[4]
1,3,5-trisubstituted pyrazoline (3c) is one of the compounds that was docked with the best Gibbs energy value over the last 10 years (ΔG = −11.16) by forming hydrogen bonds in the heterocyclic amine and nitrogen groups to the amino acid residues Gln767 and Met769 [Figure 3 and Table 2].[9] The docking parameter was considered to be much better than the reference value (−5.5 kcal/mol) and the comparison compound that has been approved by FDA as EGFR inhibitor (erlotinib, ΔG = −7.23 kcal/mol). The Gibbs energy comparison and the interaction to the key amino acid residues between the most potential compounds are shown in Figures 4 and 5. Although this compound has the greatest Gibbs energy, its mode of interaction with the EGFR amino acid residue is not yet considered satisfactory. 1,3,5-trisubstituted pyrazoline (3c) is only able to interact with one key amino acid, Met769, which acts as an electrostatic bond between the amino group of the adenine ring of the ATP molecule in the adenine pocket. Meanwhile, to increase the selectivity and potency of the drug, the test ligand must be able to interact with Cys773 which acts as a ribose sugar binder.
Figure 3.
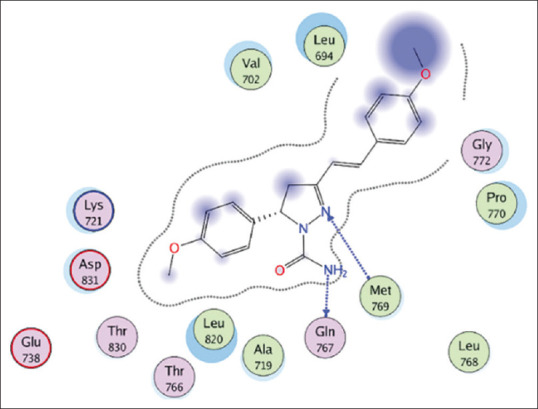
The interaction between 1,3,5-trisubstituted pyrazoline (3c) as the best-docked molecule against epidermal growth factor receptor. The amine and nitrogen at the heterocyclic ring of 1,3,5-trisubstituted pyrazoline (3c) interact with Gln767 and Met 769, respectively through a hydrogen bond[9]
Table 2.
The docked small molecules against epidermal growth factor receptor (PDB ID: 1M17)
| Rank | Molecules | Gibbs energy (kcal/mol) | Hydrogen bond interaction with amino acid residues | References |
|---|---|---|---|---|
| Reference value | -5.5 | Cys773, Met769, Thr766, Lys721, Thr830, Gly772 and Leu694 | [9] | |
| 1 | Erlotinib | −7.23 | Met769, Cys773 | [10] |
| 2 | 1,3,5-trisubstituted pyrazoline (3c) | −11.16 | Gln767, Met769 | [9] |
| 3 | 1,3,5-trisubstituted pyrazoline (6c) | −10.86 | Lys721, Met769 | [9] |
| 4 | 1,3,5-trisubstituted pyrazoline (8d) | −10.52 | Met769, Cys773 | [9] |
| 5 | N-(3,4-Dimethylphenyl)-2-[(4-oxo-3- (4-sulfamoylphenyl)-3,4- dihydrobenzo[g] quinazolin-2-yl) thio] acetamide | −9.88 | Met769, Cys773, Phe699 | [11] |
| 6 | 7-Cyano-N-phenyl-6-(3,4,5-trimethoxybenzamid0) -2,3-dihydro-1H-pyrrolizine-5-carboxamide | −9.07 | Thr830, Asp831 | [12] |
| 7 | Doxorubicin | −8.94 | Lys721, Thr766, Met769, Thr830, Asp831 | [10] |
| 8 | Steroidal D-homo lactones16) | −8.7 | Lys721, Glu738, Thr766, Met769, Thr830 | [13] |
| 9 | Steroidal D-homo lactones9) | −8.3 | Lys721, Glu738, Cys733, Thr830 | [13] |
| 10 | Steroidal D-homo lactones5) | −7.8 | Lys731, Glu738, Cys773, Thr830 | [13] |
| 11 | Sarcophine | −7.17 | Lys721 | [10] |
| 12 | Sinulolide B | −5.82 | Thr766, Thr830 | [10] |
| 13 | [Zn (ANA) 2Cl2] | −5.79 | Lys721, Arg817, Asn818, Asp831 | [14] |
| 14 | 1,3,5-trisubstituted pyrazoline (7d) | −9.4 | Thr766, Gln767 | [9] |
| 15 | 1,3,5-trisubstituted pyrazoline (8b) | −9.55 | - | [9] |
| 16 | Sardisterol | −8.13 | Thr766, Asp776 | [10] |
| 17 | 1,3,4-Thiadiazoline−coumarin (7-OC2H5) | −6.69 | Lys745 | [15] |
| 18 | Matrine | −6.19 | Arg836 | [16] |
| 19 | Sinulolide A | II−6.14 | Met769, Asp831 | [10] |
| 20 | (E)-2-(1-(3-aminophenyl) ethylidene) hydrazinecarboxamide | −5.8 | Arg836, Tyr869 | [17] |
| 21 | (E)-2-(2-aminobenzylidene) hydrazinecarboxamide | −5.7 | Asp916 | [17] |
| 22 | 1,3,4-Thiadiazoline−coumarin (6-OnC5H11) | −5.7 | Ala722 | [15] |
| 23 | Triptolide | −5.69 | Lys757, Asp761 | [16] |
| 24 | 1,3,4-Thiadiazoline−coumarin (6-OnC4H9) | −5.57 | Lys745, Ser720 | [15] |
| 25 | [Hg (ANA) 2Cl2] | −5.43 | Lys721, Asp831 | [14] |
| 26 | [Cd (ANA) 2Cl2] | −5.13 | Asp813, Arg817 | [14] |
| 27 | 1,3,4-Thiadiazoline−coumarin (7-OiC5H11) | −5.11 | Lys745, Lys875 | [15] |
| 28 | 1,3,4-Thiadiazoline−coumarin (7-OiC4H9) | −3.978 | Ser720, Arg803 | [15] |
| 29 | Hydroxyjolkinolide B | −3.19 | Phe856, Ala859 | [16] |
All the testing ligands are compared to erlotinib (FDA-approved drug for EGFR inhibitor). Eleven compounds have a better docking parameter (green) compared to erlotinib and the reference value (blue). Besides that, sixteen compounds are excluded as a drug candidate due to their worse docking parameter (red). EGFR: Epidermal growth factor receptor, FDA: Food and Drug Administration
Figure 4.
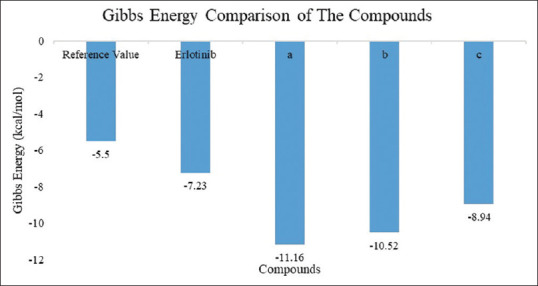
Gibbs Energy Comparison of The Compounds (a) 1,3,5-trisubstituted pyrazoline (3c) (b) 1,3,5-trisubstituted pyrazoline (8d) (c) Doxorubicin
Figure 5.
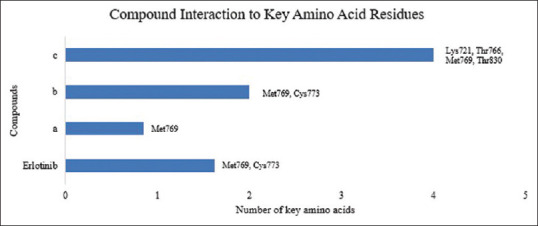
Compound Interaction to Key Amino Acid Residues (a) 1,3,5-trisubstituted pyrazoline (3c) (b) 1,3,5-trisubstituted pyrazoline (8d) (c) Doxorubicin
Therefore, 1,3,5-trisubstituted pyrazoline (3c) is considered not selective but still able to inhibit EGFR. On the other hand, 1,3,5-trisubstituted pyrazoline (8d) has a similar Gibbs energy value (ΔG = −10.52) but has a better interaction mode with EGFR amino acid residues and is considered very similar to its native ligand (erlontinib) through inhibition of Met769 and Cys 773 by forming hydrogen bonds [Figure 6 and Table 2]. The binding of the test ligand with Cys773 was successfully formed on this ligand so that it is predicted that the interaction mode between 1,3,5-trisubstituted pyrazoline (8d) and ATP binding pocket on the EGFR structure will have high specificity and potential.
Figure 6.
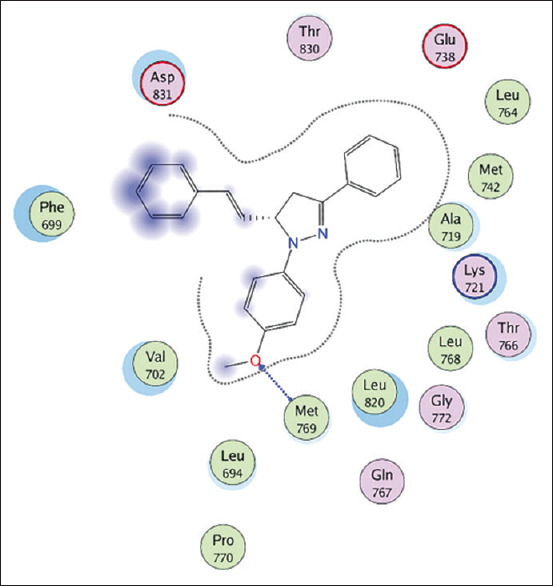
The interaction between 1,3,5-trisubstituted pyrazoline (8d) as the third best-docked molecule against epidermal growth factor receptor. The Oxygen of the hydroxyl group of 1,3,5-trisubstituted pyrazoline (8d) interacts with Gln767 and Met 769, respectively through a hydrogen bond[9]
However, in our findings, there is a very interesting point where doxorubicin, antibiotic that inhibit topoisomerase-II is able to interact well with ATP-binding pocket in EGFR (ΔG = −8.94 kcal/mol). Doxorubicin can interact through hydrogen bonding on four key amino acid residues simultaneously, i.e., Lys721, Thr766, Met769, Thr830 [Figure 7 and Table 2]. Blockade in the adenine region (Met769) as well as Thr766, Lys721, and Thr830 residues which comprise the back hydrophobic region play a pivotal role in the inhibitor selectivity against EGFR.
Figure 7.
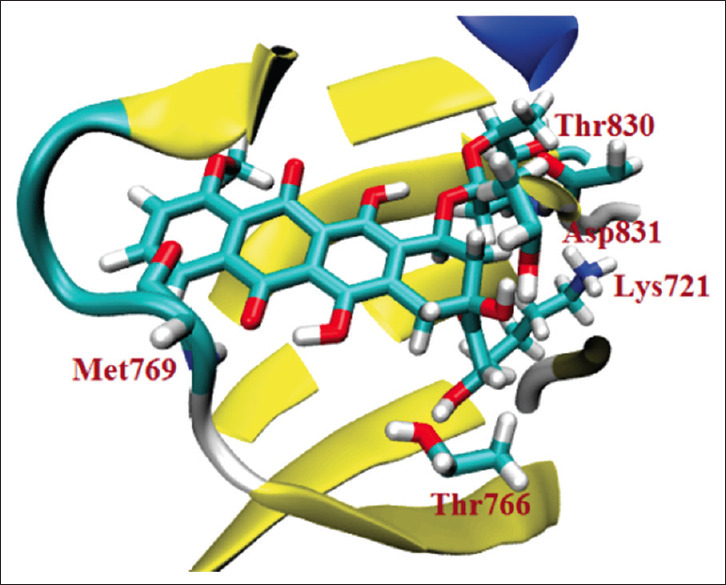
The interaction between doxorubicin against epidermal growth factor receptor. The ligand can interact with the key amino acid of epidermal growth factor receptor (Lys721, Thr766, Met769, Thr830) through a hydrogen bond[10]
CONCLUSION
Based on systematic review, eleven compounds achieve the molecular docking parameters, thus that they were considered worthy to enter the further research stage (in vitro and in vivo). The eleven compounds include 1,3,5-trisubstituted pyrazoline (3c), 1,3,5-trisubstituted pyrazoline (6c), 1,3,5- trisubstituted pyrazoline (8d), N-(3,4-Dimethylphenyl)-2- [(4-oxo-3-(4-sulfamoylphenyl)-3,4 -dihydrobenzo [g] quinazolin-2-yl) thio] acetamide, Doxorubicin, Steroidal D-homo lactones (16), Steroidal D-homo lactones (9), Steroidal D-homo lactones (5), Sarcophine, Sinulolide B, [Zn (ANA) 2Cl2]. This review can be used as a reference for researchers in deciding on the selection of compounds that are suitable for further investigation to find anti-NSCLC drugs so that they can stimulate the rate of preclinical research in the process of finding and developing anti-NSCLC drugs.17
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This research was supported by the Department of Pharmaceutical Analysis and Medicinal Chemistry, Faculty of Pharmacy, Universitas Padjadjaran. The authors were very grateful to Prof. apt. Muchtaridi, M. Si., Ph. D. for his aid in reviewing this article.
REFERENCES
- 1.World Health Organization. Cancer. 2021. [Last accessed on 2021 Apr 28]. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer .
- 2.Kementerian Kesehatan. Jakarta: The National Committee of Cancer Countermeasures; 2017. The Ministry of Health of The Republic of Indonesia. The Guideline of Medical care: Lung Cancer. [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–94. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brambilla E, Gazdar A. Pathogenesis of lung cancer signalling pathways: Roadmap for therapies. Eur Respir J. 2009;33:1485–97. doi: 10.1183/09031936.00014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santosa W. Epidermal Growth Factor Receptor (EGFR) sebagai Target Baru dalam Terapi Kanker. J Indon Med Assoc. 2012;62:125–6. [Google Scholar]
- 6.Gridelli C, De Marinis F, Di Maio M, Cortinovis D, Cappuzzo F, Mok T. Gefitinib as first-line treatment for patients with advanced non-small-cell lung cancer with activating epidermal growth factor receptor mutation: Review of the evidence. Lung Cancer. 2011;71:249–57. doi: 10.1016/j.lungcan.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Research UK. Side Effects of Erlotinib (Tarceva) 2018. [Last accessed on 2021 Apr 28]. Available from: https://www.cancerresearchuk.org/about-cancer/ofgeneral/treatment/cancer-drugs/drugs/erlotinib/side-effects .
- 8.Eleftheriou P, Amanatidou D, Petrou A, Geronikaki A. In silico Evaluation of the effectivity of approved protease inhibitors against the main protease of the novel SARS-CoV-2 virus. Molecules. 2020;25:2529. doi: 10.3390/molecules25112529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George RF, Kandeel M, El-Ansary DY, El Kerdawy AM. Some 1,3,5-trisubstituted pyrazoline derivatives targeting breast cancer: Design, synthesis, cytotoxic activity, EGFR inhibition and molecular docking. Bioorg Chem. 2020;99:103780. doi: 10.1016/j.bioorg.2020.103780. [DOI] [PubMed] [Google Scholar]
- 10.Hegazy MF, Elshamy AI, Mohamed TA, Hamed AR, Ibrahim MA, Ohta S, et al. Cembrene diterpenoids with ether linkages from sarcophyton ehrenbergi: An anti-proliferation and molecular-docking assessment. Mar Drugs. 2017;15:192. doi: 10.3390/md15060192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soliman AM, Alqahtani AS, Ghorab M. Novel sulphonamide benzoquinazolinones as dual EGFR/HER2 inhibitors, apoptosis inducers and radiosensitizers. J Enzyme Inhib Med Chem. 2019;34:1030–40. doi: 10.1080/14756366.2019.1609469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shawky AM, Ibrahim NA, Abdalla AN, Abourehab MAS, Gouda AM. Novel pyrrolizines bearing 3,4,5-trimethoxyphenyl moiety: Design, synthesis, molecular docking, and biological evaluation as potential multi-target cytotoxic agents. J Enzyme Inhib Med Chem. 2021;36:1313–33. doi: 10.1080/14756366.2021.1937618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuzminac IZ, Jakimov DS, Bekić SS, Ćelić AS, Marinović MA, Savić MP, et al. Synthesis and anticancer potential of novel 5,6-oxygenated and/or halogenated steroidal d-homo lactones. Bioorg Med Chem. 2021;30:115935. doi: 10.1016/j.bmc.2020.115935. [DOI] [PubMed] [Google Scholar]
- 14.Mallela R, Konakanchi R, Guda R, Munirathinam N, Gandamalla D, Yellu NR, et al. Zn(II), Cd(II) and Hg(II) metal complexes of 2-aminonicotinaldehyde: Synthesis, crystal structure, biological evaluation and molecular docking study. Inorganica Chim Acta. 2018;469:66–75. [Google Scholar]
- 15.Ngoc Toan V, Dinh Thanh N, Minh Tri N. 1,3,4-Thiadiazoline-coumarin hybrid compounds containing D-glucose/D-galactose moieties: Synthesis and evaluation of their antiproliferative activity. Arab J Chem. 2021;14:103053. [Google Scholar]
- 16.Zhao GF, Huang ZA, Du XK, Yang ML, Huang DD, Zhang S. Molecular docking studies of Traditional Chinese Medicinal compounds against known protein targets to treat non-small cell lung carcinomas. Mol Med Rep. 2016;14:1132–8. doi: 10.3892/mmr.2016.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sennappan M, Skariyachan S, Managutti PB, Gunaga SS. C-demethylation and 1, 2-amino shift in (E)-2-(1-(3-aminophenyl) ethylidene) hydrazinecarboxamide to (E)-2-(2-aminobenzylidene) hydrazinecarboxamide and their applications. Sci Rep. 2020;10:21913. doi: 10.1038/s41598-020-79027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


