Abstract
Seven Fusarium species complexes are treated, namely F. aywerte species complex (FASC) (two species), F. buharicum species complex (FBSC) (five species), F. burgessii species complex (FBURSC) (three species), F. camptoceras species complex (FCAMSC) (three species), F. chlamydosporum species complex (FCSC) (eight species), F. citricola species complex (FCCSC) (five species) and the F. concolor species complex (FCOSC) (four species). New species include Fusicolla elongata from soil (Zimbabwe), and Neocosmospora geoasparagicola from soil associated with Asparagus officinalis (Netherlands). New combinations include Neocosmospora akasia, N. awan, N. drepaniformis, N. duplosperma, N. geoasparagicola, N. mekan, N. papillata, N. variasi and N. warna. Newly validated taxa include Longinectria gen. nov., L. lagenoides, L. verticilliforme, Fusicolla gigas and Fusicolla guangxiensis. Furthermore, Fusarium rosicola is reduced to synonymy under N. brevis. Finally, the genome assemblies of Fusarium secorum (CBS 175.32), Microcera coccophila (CBS 310.34), Rectifusarium robinianum (CBS 430.91), Rugonectria rugulosa (CBS 126565), and Thelonectria blattea (CBS 952.68) are also announced here.
Citation: Crous PW, Sandoval-Denis M, Costa MM, Groenewald JZ, van Iperen AL, Starink-Willemse M, Hernández-Restrepo M, Kandemir H, Ulaszewski B, de Boer W, Abdel-Azeem AM, Abdollahzadeh J, Akulov A, Bakhshi M, Bezerra JDP, Bhunjun CS, Câmara MPS, Chaverri P, Vieira WAS, Decock CA, Gaya E, Gené J, Guarro J, Gramaje D, Grube M, Gupta VK, Guarnaccia V, Hill R, Hirooka Y, Hyde KD, Jayawardena RS, Jeewon R, Jurjević Ž, Korsten L, Lamprecht SC, Lombard L, Maharachchikumbura SSN, Polizzi G, Rajeshkumar KC, Salgado-Salazar C, Shang Q-J, Shivas RG, Summerbell RC, Sun GY, Swart WJ, Tan YP, Vizzini A, Xia JW, Zare R, González CD, Iturriaga T, Savary O, Coton M, Coton E, Jany J-L, Liu C, Zeng Z-Q, Zhuang W-Y, Yu Z-H, Thines M (2022). Fusarium and allied fusarioid taxa (FUSA). 1. Fungal Systematics and Evolution 9: 161–200. doi: 10.3114/fuse.2022.09.08
Keywords: Longinectria, multi-gene phylogeny, Nectriaceae, Neocosmospora, new taxa, systematics, typification
INTRODUCTION
Several initiatives in recent years have addressed problems that face contemporary fungal taxonomy. The Fungal Planet series was launched to overcome the reluctance of most mycology journals to publish single new species descriptions (Crous et al. 2011). The Genera of Fungi (GoF) project facilitated the application of fungal generic names through the re-collection of generic types and the designation of epitypes or neotypes (Kirk et al. 2013, Crous et al. 2014). The Fungal Systematics and Evolution (FUSE) series allowed the effective combination of molecular phylogenetic data with phenotypic data to link sexual, asexual and synasexual morphs to known or newly described taxa following the end of the dual nomenclatural system (Crous et al. 2015). Finally, the Genera of Phytopathogenic Fungi (GOPHY) project was introduced to stabilize the taxonomy of fungal phytopathogens at generic and species levels, coupled with biological information about host distribution, pathogenicity, disease symptomatology and DNA barcodes for accepted species (Marin-Felix et al. 2017). The aforementioned publication series inspired other similar initiatives worldwide, such as Fungal Biodiversity Notes (Liu et al. 2015), Fungal Biodiversity Profiles (Adamčík et al. 2015), Mycosphere Notes (Thambugala et al. 2017), and the more recent New and Interesting Fungi (Crous et al. 2018). With an average of 10 to more than 100 new taxa per issue, these publications have become valuable tools for the description of new fungal families, genera and species, as well as for the dissemination of knowledge about the world’s fungal diversity.
In FUSA we introduce a new series of specialised papers focusing on the taxonomy, phylogeny, systematics, ecology and pathogenicity of known and novel Fusarium and allied fusarioid taxa. Fusarium (F.) and related genera are globally distributed fungi, found in diverse substrates, although most commonly in soil, living and dead plant material, air and water (Nelson et al. 1994, Leslie & Summerell 2006, Aoki et al. 2014, Leslie & Summerell 2011). Much of the historical importance of these fungi is based on the economically impactful of plant pathogenic species that infect a wide spectrum of crops inducing cankers, dieback, dry rot of roots and seeds, scab and wilt diseases (Booth 1971, Summerell et al. 2003); as well as numerous mycotoxigenic species endanger animal and human health (Nelson et al. 1994, O’Donnell et al. 2018). Nevertheless, in the last decade several taxa have gained importance as opportunistic human and animal pathogens, particularly members of Neocosmospora (formerly the Fusarium solani species complex), Bisifusarium (formerly the Fusarium dimerum species complex) and members of at least five species complexes of Fusarium sensu stricto (van Diepeningen et al. 2014, Lombard et al. 2015, Sandoval-Denis et al. 2018, 2019, Crous et al. 2021b).
The main goal of FUSA is to publish modern diagnoses of fusarioid taxa, based on multilocus phylogenies, ideally accompanied by genomic data, morphological descriptions, as well as physiological and ecological data. These data will subsequently be placed in an online database, www.fusarium.org, linked to the fusarioid-ID database, which aims to provide a stable, regularly updated, and user-friendly platform for the identification of Fusarium and other fusarioid genera and species through advanced BLASTn queries of well-curated DNA sequences.
Contributors are encouraged to use FUSE as an instrument for typification events to stabilise the application of names by designating accurate lectotypes, epitypes and neotypes; proposing taxonomic novelties such as new combinations and replacement names; and publishing undescribed morphologies for known taxa (asexual/sexual-morph connections). The selection of culture media, culture conditions and the morphological treatment must be based on standardised fusarioid laboratory protocols, as outlined in Crous et al. (2021b); fungal descriptions must be standardised and follow given examples; description of new species should be accompanied by a brief, comprehensive taxonomic discussion; all taxonomic novelties must be registered in MycoBank and ex-type or ex-isotype strains should be deposited in the CBS collection if possible (hosted in the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands). Mycologists and other researchers wishing to contribute to future issues of FUSA are encouraged to contact the Editor-in-Chief (p.crous@wi.knaw.nl).
MATERIALS AND METHODS
Methods, media, protocols and molecular analyses follow guidelines as outlined by Crous et al. (2021b). Sequences derived in this study were deposited in GenBank (Table 1), alignments and phylogenetic trees in Figshare (www.figshare.com; doi identifier 10.6084/m9.figshare.20076044), and taxonomic novelties in MycoBank (www.MycoBank.org; Crous et al. 2004). Alignments composition and evolutionary models are summarized in Table 2.
Table 1.
Collection details and GenBank accession numbers of isolates treated in this study.
| Species | Strain1 | Country and substrate/host |
GenBank accession number2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| acl1 | ITS | LSU | rpb1 | rpb2 | tef1 | tub2 | |||
| Fusarium abutilonis | NRRL 66737T | Canada, Abutilon theophrasti | JAJJWN010000057† | JAJJWN010000064† | JAJJWN010000135† | ||||
| Fusarium aconidiale | CBS 147772T | France, Triticum aestivum | - | - | - | MZ078192 | MZ078218 | MZ078246 | - |
| Fusarium algeriense | CBS 142638T | Algeria, Triticum durum | - | - | - | MF120488 | MF120499 | MF120510 | - |
| Fusarium anguioides | LC7240 | China, bamboo | - | - | - | MW024433 | MW474388 | MW580442 | - |
| NRRL 25385 | China, bamboo | - | - | - | JX171511 | JX171624 | MH742689 | - | |
| Fusarium atrovinosum | CBS 445.67T | Australia, Triticum aestivum | - | - | - | MN120713 | MW928822 | MN120752 | - |
| CBS 130394 | USA, human leg | - | - | - | MN120714 | MN120734 | MN120753 | - | |
| NRRL 13444 | Australia, corn soil | - | - | - | JX171454 | JX171568 | GQ505403 | - | |
| NRRL 34013 | USA, human toe nail | - | - | - | - | GQ505472 | GQ505408 | - | |
| NRRL 34016 | USA, human leg | - | - | - | HM347170 | GQ505475 | GQ505411 | - | |
| Fusarium austroafricanum | NRRL 66741T | South Africa, Pennisetum clandestinum | - | - | - | MH742537 | MH742616 | MH742616 | - |
| NRRL 66742 | South Africa, Pennisetum clandestinum | - | - | - | MH742538 | MH742617 | MH742688 | - | |
| Fusarium aywerte | NRRL 25410T | Australia, soil | - | - | - | JX171513 | JX171626 | JABCQV010000336† | - |
| Fusarium bambusarum | CGMCC 3.20820T | China, bamboo | - | - | - | MW024434 | MW474389 | MW580443 | - |
| LC7187 | China, bamboo | - | - | - | MW024435 | MW474390 | MW580444 | - | |
| Fusarium beomiforme | CBS 100160T | Australia, soil | - | - | - | MF120485 | MF120496 | MF120507 | - |
| Fusarium buharicum | CBS 178.35ET | Uzbekistan, Gossypium herbaceum | - | - | - | KX302920 | KX302928 | KX302912 | - |
| CBS 796.70 | Iran, Hibiscus cannabinus | - | - | - | JX171449 | JX171563 | - | - | |
| Fusarium burgessii | CBS 125537T | Australia, soil | - | - | - | MT409440 | HQ646393 | HQ667148 | - |
| Fusarium camptoceras | CBS 193.65ET | Costa Rica, Theobroma cacao | - | - | - | MW928800 | MN170383 | AB820706 | - |
| Fusarium celtidicola | MFLUCC 16-0526T | Italy, Celtis australis | - | - | - | MH576579 | ON759296 | ON745620 | - |
| Fusarium chlamydosporum | CBS 145.25NT | Honduras, Musa sapientum | - | - | - | MN120715 | MN120735 | MN120754 | - |
| CBS 615.87 | Cuba, Colocasia esculenta | - | - | - | JX171526 | GQ505469 | GQ505405 | - | |
| CBS 677.77 | Solomon Islands, soil | - | - | - | MN120716 | GQ505486 | GQ505422 | - | |
| NRRL 34019 | USA, human eye | - | - | - | - | GQ505478 | GQ505414 | - | |
| NRRL 43633 | USA, human sinus | - | - | - | - | GQ505493 | GQ505429 | - | |
| Fusarium citricola | CBS 142421T | Italy, Citrus reticulata | - | - | - | LT746290 | LT746310 | LT746197 | - |
| CPC 27067 | Italy, Citrus limon | - | - | - | LT746287 | LT746307 | LT746194 | - | |
| Fusarium concolor | CBS 183.34T | Uruguay, Hordeum vulgare | - | - | - | MH742492 | MH742569 | MH742650 | - |
| CBS 677.94 | South Africa, soil | - | - | - | MH742503 | MH742580 | MH742660 | - | |
| Fusarium convolutans | CBS 144207T | South Africa, Kyphocarpa angustifolia rhizosphere | - | - | - | LT996193 | LT996141 | LT996094 | - |
| CBS 144208 | South Africa, Kyphocarpa angustifolia rhizosphere | - | - | - | LT996194 | LT996142 | LT996095 | - | |
| Fusarium guadeloupense | CBS 102302T | Guadeloupe, soil | JAJJWL010000373† | JAJJWL010000322† | JAJJWL010000221† | ||||
| NRRL 66743 | USA, human blood | JAJJWM010000272† | JAJJWM010000096† | JAJJWM010000091† | |||||
| Fusarium humicola | CBS 124.73T | Pakistan, soil | - | - | - | MN120718 | MN120738 | MN120757 | - |
| Fusarium juglandicola | CBS 147773T | France, Juglans regia | - | - | - | MZ078190 | MZ078215 | MZ078243 | - |
| CBS 147775 | France, Juniperus sp. | - | - | - | MZ078191 | MZ078217 | MK034341 | - | |
| Fusarium kotabaruense | InaCC F963T | Indonesia, Musa sp. | - | - | - | LS479875 | LS479859 | LS479445 | - |
| Fusarium lateritium | NRRL 13622 | USA, Ulmus sp. | - | - | - | JX171457 | JX171571 | JAAVTZ000000000† | - |
| Fusarium microconidium | CBS 119843T | Unknown | - | - | - | MN120721 | - | MN120759 | - |
| Fusarium nelsonii | CBS 119876T | South Africa, plant debris | - | - | - | MN120722 | GQ505468 | GQ505404 | - |
| CBS 119877 | Unknown | - | - | - | MN120721 | MN120741 | MN120759 | - | |
| Fusarium neosemitectum | CBS 189.60T | Congo, Musa sapientum | - | - | - | - | MN170422 | MN170489 | - |
| CBS 190.60 | Congo, Musa sapientum | - | - | - | - | MN170423 | MN170490 | - | |
| Fusarium peruvianum | CBS 511.75T | Peru, Gossypium sp. | - | - | - | MN120728 | MN120746 | MN120767 | - |
| Fusarium salinense | CBS 142420T | Italy, Citrus sinensis | - | - | - | LT746286 | LT746306 | LT746193 | - |
| CPC 26403 | Italy, Citrus sinensis | - | - | - | LT746304 | LT746191 | LT746284 | - | |
| Fusarium sp. (FCSC9) | NRRL 13338 | Australia, soil | - | - | - | JX171447 | JX171561 | GQ505402 | - |
| Fusarium sp. 1 | NRRL 66179 | USA, Hibiscus moscheutos | - | - | - | KX302921 | KX302929 | KX302913 | - |
| NRRL 66180 | USA, Hibiscus moscheutos | - | - | - | KX302922 | KX302930 | KX302914 | - | |
| NRRL 66181 | USA, Hibiscus moscheutos | - | - | - | KX302923 | KX302931 | KX302915 | - | |
| NRRL 66182 | USA, Hibiscus moscheutos | - | - | - | KX302924 | KX302932 | KX302916 | - | |
| NRRL 66183 | USA, Hibiscus moscheutos | - | - | - | KX302925 | KX302933 | KX302917 | - | |
| NRRL 66184 | USA, Hibiscus moscheutos | - | - | - | KX302926 | KX302934 | KX302918 | - | |
| Fusarium sp. 2 | NRRL 66739 | China, unknown | JAJJWO010000055† | JAJJWO010000203† | JAJJWO010000256† | ||||
| Fusarium spinosum | CBS 122438T | Brazil, Cucumis melo | - | - | - | MN120729 | MN120747 | MN120768 | - |
| NRRL 43631 | USA, human leg | - | - | - | HM347187 | GQ505491 | GQ505427 | - | |
| Fusarium sporodochiale | CBS 220.61T | South Africa, soil | - | - | - | MN120731 | MN120749 | MN120770 | - |
| Fusarium stilboides | NRRL 20429 | Nyasaland, Coffea sp. | - | - | - | JX171468 | JX171582 | - | - |
| Fusarium sublunatum | CBS 189.34T | Costa Rica, soil | - | - | - | JX171451 | JX171565 | - | - |
| CBS 190.34 | Costa Rica, soil | - | - | - | KX302927 | KX302935 | KX302919 | - | |
| Fusarium tjaynera | NRRL 66246T | Australia, Triodia microstachya | - | - | - | KP083268 | KP083279 | EF107152 | - |
| Fusicolla acetilerea | BBA 63789T | Japan, polluted soil | HQ897839 | HQ897790 | U88108 | - | HQ897701 | - | - |
| Fusicolla aquaeductuum | CBS 268.53 | Netherlands, rubber tubing | - | MH857190 | MH868728 | - | - | - | - |
| CBS 837.85ET | Germany, plug in water tap | - | KM231823 | KM231699 | - | - | - | KM232094 | |
| Fusicolla betae | BBA 64317ET | Germany, Triticum aestivum | HQ897917 | - | - | - | HQ897781 | - | - |
| Fusicolla bharatavarshae | NFCCI 4423T | India, Avicennia marina | - | MK152510 | MK152511 | - | MK157022 | - | MK376462 |
| Fusicolla cassiae-fistulae | MFLUCC 19-0318T | Thailand, Cassia fistula | - | MT215497 | MT215549 | - | - | - | - |
| Fusicolla elongata | CBS 148934T | Zimbabwe, soil | ON759286 | ON763203 | ON763200 | - | ON759297 | - | ON745628 |
| CBS 148935 | Zimbabwe, soil | ON759287 | ON763204 | ON763201 | - | ON759298 | - | ON745629 | |
| Fusicolla epistroma | BBA 62201ET | UK, Diatrypella sp., on Betula sp. | HQ897901 | - | AF228352 | - | HQ897765 | - | - |
| Fusicolla gigantispora | HKAS 101990 | Thailand, Bruguiera sp. | - | MN047106 | MN017870 | - | - | - | - |
| MFLU 16-1206T | Thailand, Avicennia marina | - | MN047105 | MN017876 | - | - | - | - | |
| Fusicolla gigas | CGMCC 3.20680 | China, soil | - | OK465362 | OK465449 | - | - | - | - |
| Fusicolla guangxiensis | CGMCC 3.20679 | China, rotten twig | - | OK465363 | OK465450 | - | - | - | - |
| Fusicolla matuoi | CBS 581.78 | Japan, Albizzia julibrissin | HQ897858 | KM231822 | KM231698 | - | HQ897720 | - | KM232093 |
| Fusicolla melogrammae | CBS 141092T | UK, Melogramma campylosporum on Carpinus sp. | - | KX897140 | KY092489 | - | HQ897720 | - | MW834305 |
| Fusicolla meniscoidea | CBS 110189T | Australia, soil | MW834043 | MW827613 | MW827654 | - | MW834010 | - | MW834306 |
| CBS 186.34 | Germany, Acer sp. | - | MH855482 | MH866963 | - | - | - | - | |
| Fusicolla ossicola | CBS 140161T | Belgium, bone of wild boar | - | MF628022 | MF628021 | - | MW834011 | - | MW834307 |
| Fusicolla quarantenae | CBS 141541T | Brazil, Melocactus zehntneri | MW834044 | MW553789 | MW553788 | - | MW556626 | - | MW556624 |
| Fusicolla septimanifiniscientiae | CBS 144935T | Netherlands, soil | - | MK069422 | MK069418 | - | - | - | MK069408 |
| Fusicolla siamensis | MFLUCC 17-2577T | Thailand, Cassia fistula | - | MT215498 | MT215550 | - | - | - | - |
| Fusicolla sporellula | CBS 110191T | South Africa, soil | MW834044 | MW827614 | MW827655 | - | MW834012 | - | MW834308 |
| Fusicolla violacea | CBS 634.76T | Iran, Quadraspidiotus perniciosus | - | KM231824 | U88112 | - | HQ897696 | - | KM232095 |
| Geejayessia atrofusca | NRRL 22316 | USA, Staphylea trifolia | - | AF178423 | - | JX171496 | EU329502 | AF178361 | - |
| Geejayessia cicatricum | CBS 125552 | Slovenia, dead twig | - | HQ728145 | - | - | HQ728153 | HM626644 | - |
| Macroconia leptosphaeriae | CBS 100001 | Netherlands, Leptosphaeria sp. | HQ897891 | HQ897810 | HQ897755 | MW834203 | HQ728164 | - | KM232097 |
| Neocosmospora acutispora | CBS 145461T | Guatemala, Coffea arabica | - | LR583700 | - | MW834210 | LR583814 | LR583593 | - |
| Neocosmospora akasia | CBS 146880T | Indonesia, Euwallacea perbrevis | - | MN954357 | - | - | MT009931, MT010011 | MT009971 | - |
| CMW52865 | Indonesia, Acacia crassicarpa | - | MN954330 | - | - | MT009904, MT009984 | MT009943 | - | |
| Neocosmospora ambrosia | CBS 571.94ET | India, Euwallacea fornicatus | - | EU329669 | - | MW834211 | EU329503 | FJ240350 | - |
| NRRL 62942 | Sri Lanka, Camellia sinensis | - | KM406631 | - | KM406638 | KM406638, KM406645 | KM406624 | - | |
| Neocosmospora awan | CBS 146882T | Indonesia, Acacia crassicarpa | - | MN954345 | - | - | MT009919, MT009999 | MT009973 | - |
| CBS 146884 | Indonesia, Acacia crassicarpa | - | JQ038014 | - | - | JQ038028 | JQ038007 | - | |
| Neocosmospora brevis | CBS 144387T | Belgium, soil-water | - | LR583708 | - | MW834214 | LR583822 | LR583601 | - |
| CPC 27191 | Italy, Citrus sinensis | - | LT746248 | - | - | LT746313 | LT746200 | - | |
| YJ1 | China, Rosa chinensis | - | MW724816 | - | - | MW795356 | MW795357 | - | |
| YJ2 | China, Rosa chinensis | - | MW724817 | - | - | MW795358 | MW795359 | - | |
| Neocosmospora cryptoseptata | CBS 145463T | French Guiana, bark | - | AF178414 | - | MW834215 | EU329510 | AF178351 | - |
| Neocosmospora drepaniformis | NRRL 62941T | Singapore, unknown | - | KM406633 | - | JAALXN000000000† | KM406640, KM406647 | KM406626 | - |
| Neocosmospora duplosperma | NRRL 62583T | USA, Euwallacea fornicatus | - | KC691581 | - | KC691611 | KC691642, KC691671 | KC691553 | - |
| NRRL 62585 | USA, Euwallacea fornicatus | - | KC691577 | - | KC691607 | KC691638, KC691667 | KC691549 | - | |
| Neocosmospora euwallaceae | CBS 135854T | Israel, Euwallacea sp. | - | JQ038014 | - | JQ038021 | JQ038028 | JQ038007 | - |
| NRRL 62626 | USA, Euwallacea sp. | - | KC691560 | - | KC691590 | KC691621, KC691650 | KC691532 | - | |
| Neocosmospora floridana | NRRL 62608 | USA, Boxelder tree infested with Euwallacea interjectus | - | KC691562 | - | KC691592 | KC691623, KC691652 | KC691534 | - |
| NRRL 62628T | USA, Euwallacea interjectus | - | KC691563 | - | KC691593 | KC691624, KC691653 | KC691535 | - | |
| Neocosmospora geoasparagicola | CBS 148936 | Netherlands, soil | - | ON763206 | - | ON759289 | ON759300 | ON745621 | - |
| CBS 148937T | Netherlands, soil | - | ON763207 | - | ON759290 | ON759301 | ON745622 | - | |
| CPC 39931 | Netherlands, soil | - | ON763208 | - | ON759291 | ON759302 | ON745623 | - | |
| CPC 39932 | Netherlands, soil | - | ON763209 | - | ON759292 | ON759303 | ON745624 | - | |
| CPC 40571 | Netherlands, soil | - | ON763210 | - | ON759293 | ON759304 | ON745625 | - | |
| CPC 40579 | Netherlands, soil | - | ON763211 | - | ON759294 | ON759305 | ON745626 | - | |
| CPC 40628 | Netherlands, soil | - | ON763212 | - | ON759295 | ON759306 | ON745627 | - | |
| Neocosmospora illudens | CBS 147303 | New Zealand, Beilschmiedia tawa | - | AF178393 | - | JX171488 | JX171601 | AF178326 | - |
| Neocosmospora kuroshio | CBS 142642T | USA, Euwallacea sp. gallery | - | LR583723 | - | KX262236 | KX262256 | KX262216 | - |
| NRRL 62946 | USA, Platanus racemosa | - | KM406637 | - | KM406644 | KM406650 | KM406630 | - | |
| Neocosmospora kurunegalensis | CBS 119599T | Sri Lanka, recently cut tree | - | JF433036 | - | MW834228 | LR583838 | DQ247511 | - |
| Neocosmospora lichenicola | CBS 623.92ET | Germany, human | - | - | - | LR583845 | LR583620 | - | |
| Neocosmospora mahasenii | CBS 119594T | Sri Lanka, unknown tree | - | JF433045 | - | MW834231 | LT960563 | DQ247513 | - |
| Neocosmospora mekan | CBS 146885T | Indonesia, Euwallacea similis | - | MN954342 | - | - | MT009916, MT009996 | MT009956 | - |
| CBS 146886 | Indonesia, Acacia crassicarpa infested with Euwallaceae spp. | - | MN954335 | - | - | MT009909, MT009989 | MT009962 | - | |
| Neocosmospora nirenbergiana | CBS 145469T | French Guiana, Bark | - | AF178403 | - | - | EU329505 | AF178339 | - |
| Neocosmospora obliquiseptata | NRRL 62610 | Australia, Euwallacea sp. gallery | - | KC691575 | - | KC691605 | KC691636, KC691665 | KC691547 | - |
| NRRL 62611T | Australia, Euwallacea sp. gallery | - | KC691576 | - | KC691606 | KC691637, KC691666 | KC691535 | - | |
| Neocosmospora oligoseptata | CBS 143241T | USA, Euwallacea validus | - | KC691566 | - | KC691596 | LR583854 | KC691538 | - |
| NRRL 62582 | USA, Ailanthus sp. | - | KC691569 | - | KC691599 | KC691630, KC691659 | KC691541 | - | |
| Neocosmospora papillata | NRRL 62943T | Sri Lanka, Camellia sinensis | - | KM406635 | - | KM406642 | S24402* | KM406628 | - |
| NRRL 62944 | Sri Lanka, Euwallaceae sp. on Camellia sinensis | - | KM406634 | - | KM406641 | KM406648 | KM406627 | - | |
| Neocosmospora phaseoli | CBS 265.50 | USA, Phaseolus sp. | - | LR583750 | - | - | KJ511278 | FJ919464 | - |
| Neocosmospora plagianthi | NRRL 22632 | New Zealand, Hoheria glabrata | - | AF178417 | - | JX171501 | JX171614 | AF178354 | - |
| Neocosmospora rectiphora | CBS 125726 | Sri Lanka, dead tree | - | JF433043 | - | MW834248 | MW834028 | JF433026 | - |
| CBS 125727T | Sri Lanka, dead tree | - | JF433034 | - | MW834249 | LR583871 | DQ247509 | - | |
| Neocosmospora rekana | CMW53690 | Indonesia, Euwallacea fornicatus | - | MN249098 | - | - | MN249141, MN249112 | MN249155 | - |
| CMW52862T | Indonesia, Euwallacea perbrevis | - | MN249094 | - | - | MN249137, MN249108 | MN249151 | - | |
| Neocosmospora robusta | CBS 145473T | Venezuela, bark | - | AF178405 | - | MW834251 | EU329507 | AF178341 | - |
| Neocosmospora samuelsii | CBS 114067T | Guyana, bark | - | LR583764 | - | MW834252 | LR583874 | LR583644 | - |
| Neocosmospora sp. (AF-6) | NRRL 62590 | USA, Euwallacea fornicatus gallery | - | KC691574 | - | KC691604 | KC691635, KC691664 | KC691546 | - |
| NRRL 62591 | USA, Euwallacea fornicatus gallery | - | KC691573 | - | KC691603 | KC691634, KC691663 | KC691545 | - | |
| Neocosmospora sp. (AF-9) | NRRL 22643 | Costa Rica, Xyleborus ferrugineus | - | KC691583 | - | KC691613 | KC691644, KC691673 | DQ247628 | - |
| NRRL 66088 | USA, Delonix regia | - | KM406632 | - | KM406639 | KM406646 | KM406625 | - | |
| Neocosmospora sp. (AF-13) | UCR4674 | Taiwan, Euwallacea sp. | - | KX262208 | - | KX262248 | KX262268 | KX262228 | - |
| UCR4675 | Taiwan, Euwallacea sp. | - | KX262209 | - | KX262249 | KX262269 | KX262229 | - | |
| Neocosmospora sp. (AF-14) | UCR4672 | Taiwan, Euwallacea sp. | - | KX262206 | - | KX262246 | KX262266 | KX262226 | - |
| UCR4681 | Taiwan, Euwallacea sp. | - | KX262215 | - | KX262255 | KX262275 | KX262235 | - | |
| Neocosmospora sp. (AF-15) | UCR4679 | Taiwan, Euwallacea sp. | - | KX262213 | - | KX262253 | KX262273 | KX262233 | - |
| Neocosmospora sp. (AF-16) | UCR4673 | Taiwan, Euwallacea sp. | - | KX262207 | - | KX262247 | KX262267 | KX262227 | - |
| UCR4678 | Taiwan, Euwallacea sp. | - | KX262212 | - | KX262252 | KX262272 | KX262232 | - | |
| Neocosmospora sp. (AF-17) | UCR4676 | Taiwan, Euwallacea sp. | - | KX262210 | - | KX262250 | KX262270 | KX262230 | - |
| UCR4680 | Taiwan, Euwallacea sp. | - | KX262214 | - | KX262254 | KX262274 | KX262234 | - | |
| Neocosmospora sp. (AF-18) | UCR4677 | Taiwan, Euwallacea sp. | - | KX262211 | - | KX262251 | KX262271 | KX262231 | - |
| Neocosmospora tuaranensis | NRRL 22231T | Malaysia, Hevea brasiliensis | - | KC691570 | - | KC691600 | KC691660, KC691631 | KC691542 | - |
| NRRL 46519 | Malaysia, beetle on Hevea brasiliensis | - | KC691572 | - | KC691602 | KC691633 | KC691544 | - | |
| Neocosmospora variasi | CBS 146888T | Indonesia, Acacia crassicarpa infested with E. perbrevis | - | MN954356 | - | - | MT009913, MT009993 | MT009967 | - |
| CBS 146889 | Indonesia, Acacia crassicarpa infested with E. perbrevis | - | MN954357 | - | - | MT009914, MT009994 | MT009968 | - | |
| Neocosmospora vasinfecta | NRRL 22166ET | USA, Gossypium sp. | - | DQ094319 | - | SSHR01002742† | EU329497 | AF178350 | - |
| NRRL 43467 | USA, human eye | - | EF453092 | - | HM347178 | EF469979 | EF452940 | - | |
| Neocosmospora warna | CBS 146891T | Indonesia, Euwallacea perbrevis | - | MN954346 | - | - | MT009920, MT010000 | MT009955 | - |
| CBS 146893 | Indonesia, Euwallacea perbrevis | - | MN954351 | - | - | MT009925, MT010005 | MT009958 | - | |
| Scolecofusarium ciliatum | CBS 148938 | Ukraine, Peniophora rufomarginata | ON759288 | ON763205 | ON763202 | - | ON759299 | - | ON745630 |
1 CBS: Westerdijk Fungal Biodiverity Institute (WI), Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection Centre, Beijing, China. CMW: Culture collection at the FABI, University of Pretoria, South Africa; CPC: Collection of P.W. Crous, held at WI; HKAS: Herbarium of Cryptogams, Kunming Institute of Botany, Kunming, China; InaCC: Indonesian Culture Collection, Cibinong, Indonesia; LC: Collection of Lei Cai, held at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NFCCI; National Fungal Culture Collection of India, Pune, India; NRRL: Agricultural Research Service Culture Collection, National Center for Agricultural Utilization Research, USDA, Peoria, USA; UCR: collection of the University of California, Riverside, USA; YJ: Pathology Laboratory, Nanjing Forestry University, Nanjing, China. ET: Ex-epitype; NT: Ex-neotype; T: Ex-type.
2 acl1: partial ATP citrate lyase gene; ITS: internal transcribed spacer regions with intervening 5.8S nrRNA gene; LSU: 28S large subunit of the nrDNA; rpb1: partial DNA-directed RNA polymerase II largest subunit gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene, two accession numbers refer to two non-contiguous fragments; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene. †: sequences extracted from full genome sequences; *: sequence available at TreeBASE (study number); sequences generated in this study are shown in bold.
Table 2.
Summary of phylogenetic information for the different analyses in this study.
| Genus | Locus1 |
Number of sites (including gaps)2
|
Model selection3
|
|||||
|---|---|---|---|---|---|---|---|---|
| Total | Conserved | Variable | Informative | BI unique site patterns | IQ-TREE (BIC) | BI (AIC) | ||
| Fusarium | rpb1 | 1 774 | 1 134 | 639 | 568 | 713 | TNe+I+G4 | SYM+I+G |
| rpb2 | 1 657 | 1 085 | 572 | 535 | 592 | TIM2e+I+G4 | SYM+I+G | |
| tef1 | 517 | 217 | 285 | 245 | 348 | TIM2e+G4 | GTR+G | |
| Combined | 3 948 | 2 436 | 1 496 | 1 348 | 1 653 | - | - | |
| Neocosmospora | ITS | 464 | 333 | 128 | 99 | 180 | TNe+R3 | GTR+I+G |
| rpb1 | 1 588 | 1 151 | 437 | 319 | 435 | TIM3e+I+G4 | GTR+I+G | |
| rpb2 | 1 465 | 1 057 | 408 | 336 | 454 | TNe+I+G4 | GTR+I+G | |
| tef1 | 688 | 394 | 283 | 200 | 342 | TIM2+F+G4 | GTR+I+G | |
| Combined | 4 205 | 2 935 | 1 256 | 954 | 1 411 | - | - | |
| Fusicolla | acl1 | 866 | 454 | 382 | 201 | 298 | TNe+G4 | GTR+G |
| ITS | 516 | 391 | 110 | 56 | 123 | TIM2e+G4 | GTR+G | |
| LSU | 474 | 423 | 50 | 28 | 56 | K2P+I | GTR+G+I | |
| rpb2 | 1 702 | 1 220 | 482 | 290 | 415 | TIM2e+G4 | GTR+G+I | |
| tub2 | 482 | 299 | 175 | 109 | 177 | K2P+G4 | HKY+G | |
| Combined | 4 040 | 2 787 | 1 199 | 684 | 1 069 | - | - | |
1 acl1: ATP citrate lyase large subunit; LSU: 28S large subunit of the nrDNA; ITS: Internal transcribed spacer region of the nrDNA; tef1: partial translation elongation factor 1-alpha gene; rpb1: partial DNA-directed RNA polymerase II largest subunit gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene; tub2: partial beta-tubulin gene.
2 BI: Bayesian inference.
3 BIC: Evolutionary model selected by ModelFinder in IQ-TREE; AIC: Evolutionary model selected by MrModeltest under the Akaike Information Criterion
Genome assembly
DNA was extracted from mycelium grown on SAM (Kruse et al. 2017) culture plates as described earlier (Mishra et al. 2018). Library construction and short-read sequencing was done by a commercial sequencing provider (BGI, Hongkong, PRC). Pair-end reads (150 bp, 400 bp insert) were cleaned with Trimmomatic v. 0.39 (Bolger et al. 2014) with the following settings: remove leading and trailing low quality (< 3) or N bases; cutting when the average quality per base dropped below 15 in a 4-base sliding window; Illumina adaptor removal; removing reads shorter than 70 bp. Cleaned reads were used to assemble genomes with velvet v. 1.2.10 (Zerbino & Birney, 2008) using a k-mer value of 93. Assembly statistics were obtained using the stats.sh script of the BBTools package (Bushnell 2021). The assembly quality was evaluated with BUSCO v. 5.2.2 against the fungi_odb10 library (Manni et al. 2021). Genome annotation was done with maker v. 3.01.03 (Cantarel et al. 2008) for gene prediction using the protein sequences of Fusarium oxysporum from the UniProt database as reference. All genomes were submitted to GenBank (see Table 3 for details).
Table 3.
Basic statistics of the assembled genomes announced in this publication.
| Species | Strain1 | BioProject ID | Complete BUSCOs [%] | Assembly size [Mbp] | No. of scaffolds | Scaff. N50 [kbp] | Longest scaff. [kbp] | Total no. of CDS |
|---|---|---|---|---|---|---|---|---|
| Fusarium secorum | CBS 175.32 | PRJNA826072 | 99.1 % | 50.5 | 15 085 | 17.3 | 156.3 | 46 001 |
| Microcera coccophila | CBS 310.34 | PRJNA826070 | 98.7 % | 36.7 | 2 725 | 27.3 | 177.9 | 24 411 |
| Rectifusarium robinianum | CBS 430.91T | PRJNA826068 | 98.7 % | 34.7 | 2 358 | 27.4 | 219.8 | 25 210 |
| Rugonectria rugulosa | CBS 126565 | PRJNA826071 | 98.8 % | 46.9 | 2 884 | 56.0 | 353.8 | 30 877 |
| Thelonectria blattea | CBS 952.68T | PRJNA826075 | 98.9 % | 38.9 | 3 001 | 34.8 | 221.9 | 26 348 |
1 T = Ex-type.
RESULTS
Phylogeny
For this study, three multilocus analyses were carried out. The datasets were analysed using IQ-TREE v. 2.1.3 (Nguyen et al. 2015, Minh et al. 2020) and MrBayes v. 3.2.7 (Ronquist & Huelsenbeck 2003) as indicated in Crous et al. (2021b).
An overview of currently accepted taxa in Fusarium species complexes treated in this study is shown in a phylogeny constructed from combined rpb1, rpb2 and tef1 data of 62 strains, encompassing eight species complexes i.e., Fusarium aywerte (FASC), F. buharicum (FBSC), F. burgessii (FBURSC), F. camptoceras (FCAMSC), F. chlamydosporum (FCSC), F. citricola (FCCSC), and F. concolor (FCOSC), including the outgroup taxa (F. lateritium NRRL 13622 and F. stilboides NRRL 20429, both species belonging to the F. lateritium species complex) (Fig. 1). IQ-TREE best tree (log-likelihood -26203.881) was found after 102 iterations. Bayesian analysis lasted for 235 000 generations and recovered 472 trees from which 354 where sampled. The phylogeny resolved all the treated species complexes with high statistical support. Thirty species are recognised (two in FASC, three each in FBURSC and FCAMSC, five each in FCCSC, and FBSC; eight in FCSC, and four in FCOSC). Additionally, three phylogenetic species awaiting formal description were found, of which one resolved in FCSC (Fusarium sp. FCSC 9) and two in the FBSC (clades Fusarium sp. 1, and Fusarium sp. 2)
Fig. 1.
IQ-TREE phylogeny inferred from the combined rpb1, rpb2 and tef1 sequences of currently accepted species belonging to seven species complexes (SC) of Fusarium i.e., F. aywerte (FASC), F. buharicum (FBSC), F. burgessii (FBURSC), F. camptoceras (FCAMSC), F. chlamydosporum (FCSC), F. citricola (FCCSC), and F. concolor (FCOSC). Numbers at the nodes correspond to IQ-TREE bootstrap values ≥ 95 % followed by Bayesian posterior probabilities ≥ 0.95, and IQ-TREE gene concordance factors. The tree is rooted to F. lateritium NRRL 13622 and F. stilboides NRRL 20429 (FLSC). The scale bar indicates the expected number of nucleotide substitutions per site. Species complexes are indicated on the right and highlighted with coloured blocks. Ex-epitype, ex-neotype, and ex-type strains are indicated with ET, NT, and T, respectively.
Fusicolla: A phylogeny was constructed using combined acl1, ITS, LSU, rpb2 and tub2 sequences of 23 strains representing 18 species of Fusicolla (Fu.), plus two outgroup taxa (Macroconia leptosphaeriae CBS 10001 and Scolecofusarium ciliatum CBS 148938) (Fig. 2). IQ-TREE best tree (log-likelihood -15164.779) was found after 117 iterations. Bayesian analysis lasted for 1 535 000 generations and recovered 3 072 trees from which 2 304 where sampled. Two strains obtained from soil in Zimbabwe (MUCL 58143, 58144) are formally described below as the novel species Fusicolla elongata. Sequence data from additional Fusicolla species known from culture (Fu. gigas, Fu. hughesii, Fu. guangxiensis) or sequenced from fungarium specimens (Fu. reyesiana) were initially included in the phylogenies and later removed from the final analyses due to their incomplete datasets (nrDNA or only ITS1 and ITS2 sequences available). Two species recently invalidly published i.e., Fu. gigas and Fu. guangxiensis are re-validated here based on the original protologue (Liu et al. 2022).
Fig. 2.
IQ-TREE phylogeny inferred from the combined acl1, ITS, LSU, rpb2 and tub2 sequences of Fusicolla spp. Numbers at the nodes correspond to IQ-TREE bootstrap values ≥ 95 % followed by Bayesian posterior probabilities ≥ 0.95, and IQ-TREE gene concordance factors. The tree is rooted to Macroconia leptosphaeriae CBS 10001 and Scolecofusarium ciliatum CBS 148938. The scale bar indicates the expected number of nucleotide substitutions per site. Novel taxa are indicated in bold. Ex-epitype and ex-type strains are indicated with ET and T, respectively.
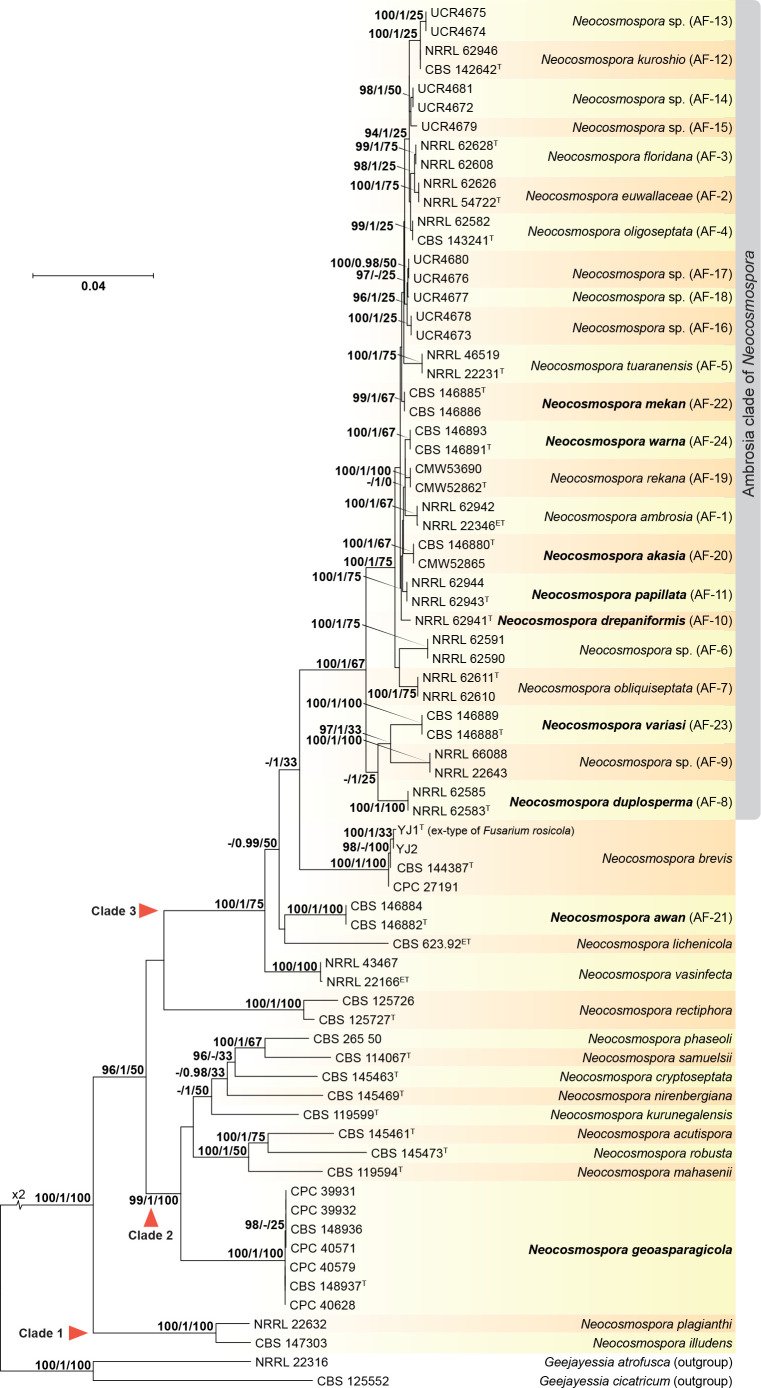
Neocosmospora: A combined alignment was built including ITS, rpb1, rpb2, and tef1 sequences from 73 strains representing the known species diversity of the Ambrosia Clade (Kasson et al. 2013) and close relatives from Clades 1, 2 and 3 of Neocosmospora (O’Donnell 2000) (Fig. 3). IQ-TREE best tree (log-likelihood -20219.033) was found after 103 iterations. Bayesian analysis lasted for 480 000 generations and recovered 962 trees from which 722 where sampled. The Ambrosia Clade was found to encompass 23 phylogenetic species (AF 1-23), 15 of which have been formally described to date. Fusarium species are recombined in Neocosmospora including seven species in the Ambrosia Clade (N. akasia, N. drepaniformis, N. duplosperma, N. mekan, N. papillata, N. variasi, and N. warna) and the distantly related although ecologically similar N. awan. The ex-type of F. rosicola (YJ1) clustered with N. brevis, and the former is synonymised under the latter. A previously undescribed, phylogenetically well-differentiated clade composed of seven soil isolates obtained from different asparagus (Asparagus officinalis) fields, formed a basal lineage in Clade 2. This lineage is formally proposed below as the novel species N. geoasparagicola.
Fig. 3.
IQ-TREE phylogeny inferred from the combined ITS, rpb1, rpb2 and tef1 sequences of representative Neocosmospora spp. Numbers at the nodes correspond to IQ-TREE bootstrap values ≥ 95 % followed by Bayesian posterior probabilities ≥ 0.95, and IQ-TREE gene concordance factors. The tree is rooted to Geejayessia atrofusca NRRL 22316 and G. cicatricum CBS 125552. The scale bar indicates the expected number of nucleotide substitutions per site. New combinations and species are indicated in bold. Numbers between parenthesis indicate former phylogenetic species nomenclature. The ‘Ambrosia clade’ of Neocosmospora is indicated on the right. Ex-epitype and ex-type strains are indicated with ET and T, respectively.
TAXONOMY
Fusarium aywerte species complex (FASC)
Fusarium aywerte (Sangal. & L.W. Burgess) Benyon & L.W. Burgess, Mycol. Res. 104: 1171. 2000. MB 466154. Fig. 4.
Fig. 4.

Fusarium aywerte (CBS 395.96). A. Sporodochium on CLA. B, C. Sporodochia on SNA. D–G, J. Aerial conidiophores with monophialides. H, I. Sporodochial conidiophores. K, L. Macroconidia. Scale bars = 10 μm.
Basionym: Fusarium avenaceum subsp. aywerte Sangal. & L.W. Burgess, Mycol. Res. 99: 287. 1995. MB 363513.
Holotypus: DAR 69501 (dried culture).
Ex-type culture: DAR 69501 = F10108 = NRRL 25410.
Type locality: Australia, Northern Territory, Deep Well.
Type substrate: Soil (from a depth of 5–10 cm) associated with roots of Triodia basedowii.
Descriptions and illustrations: See Sangalang et al. (1995a), Benyon et al. (2000) and Leslie & Summerell (2006).
Reference culture: Australia, Northern Territory, Little Palm Creek, soil under Plectrachne sp. (Poaceae), 1992, D. Backhouse, CBS 395.96 = F 10989.
Diagnostic features: Colonies with greyish rose mycelium and red pigment on PDA, having optimal growth at 25 °C; microconidia not observed; sporodochia with monophialides give rise to long, thin, flexuous, 6–8-septate macroconidia with a long tapering apical cell and a well-developed, elongated foot-shaped basal cell; chlamydospores absent (Sangalang et al. 1995a, Leslie & Summerell 2006).
Notes: Fusarium aywerte was initially described as a subspecies of F. avenaceum (Sangalang et al. 1995b), later to be recognised as a distinct species (Benyon et al. 2000). Besides the molecular differences, there are morphological, physiological and ecological differences between F. aywerte and F. nurragi. Fusarium aywerte has longer macroconidia and a faster growth rate than those of F. nurragi. Further, F. aywerte occurs in the rhizosphere of tussock-forming grasses (Plectrachne, Triodia) in arid tropical regions in northern Australia, while F. nurragi occurs in the rhizosphere of coastal heathland plants (Kunzea ambigua, Banksia serrata, Allocasuarina paradoxa) in temperate regions in southern Australia (Sangalang et al. 1995a, b).
Fusarium tjaynera J.L. Walsh et al., Fungal Diversity 77: 361. 2015. MB 812309. Fig. 5.
Fig. 5.

Fusarium tjaynera (NRRL 66246). A, B. Sporodochia on CLA. C, D. Aerial conidiophores with monophialides giving rise to microconidia. E. Microconidia. F, G. Aerial conidiophores with monophialides giving rise to macroconidia. H. Sporodochial conidiophores. I–K. Macroconidia. Scale bars = 10 μm.
Holotypus: RBG 5367 (metabolically inactive and dried culture).
Ex-type culture: NRRL 66246 = RBG 5367.
Type locality: Australia, Northern Territory, Litchfield National Park.
Type substrate: Triodia microstachya.
Description and illustrations: See Laurence et al. (2016).
Diagnostic features: Colonies with white to greyish rose aerial mycelium and red to burgundy reverse on PDA; mono- to polyphialides give rise to oval, 0–1-septate microconidia in false heads (*1-septate, subcylindrical mesoconidia also present); orange sporodochia give rise to falcate, slender, parallel dorse-ventral sides, (4–)5(–7)-septate macroconidia with a tapering, curved apical cell and well-developed, foot-shaped basal cell; chlamydospores absent (*emended from Laurence et al. 2016).
Notes: Fusarium tjaynera has been isolated from soil as well as from Triodia macrostachya, Sorghum interjectum and S. intrans in northern Australia (Laurence et al. 2016). Fusarium tjaynera is considered endemic to Australia. Fusarium tjaynera resembles F. aywerte, but can be distinguished by the production of microconidia [described as oval, but illustrated as subcylindrical; figs 47, 48 in Laurence et al. (2016)] and red pigmentation on PDA. Compared to F. longipes (distinctly notched basal cell), F. tjaynera has an indistinctly notched basal cell, and a less prominently elongated whip-like apical cell (Burgess et al. 1994, Laurence et al. 2016).
Fusarium buharicum species complex (FBSC)
Fusarium abutilonis Gräfenhan, Nirenberg & Seifert, Mycologia DOI: 10.1080/00275514.2022.2071563 [7]. 2022.
Holotypus: BPI 924391, dried culture of NRRL 66737.
Ex-type culture: NRRL 66737 = DAOMC 213370.
Type locality: Canada, Ontario.
Type substrate: On Abutilon theophrasti.
Descriptions and illustrations: O’Donnell et al. (2022).
Diagnostic features: Colonies reverse orange, sometimes turning greyish brown or greyish blue in the centre; surface smooth or slightly mealy, orange, sometimes turning greyish brown in the centre, aerial mycelium white, sparse to slightly lanose to cottony, margin transparent or white on PDA, having optimal growth at 25 °C; aerial conidia 1–3-septate, sparse to absent, from monophialides; sporodochia pale orange, with monophialides giving rise to almost straight to curved, walls parallel in the centre, (4–)5(–6)-septate macroconidia with a conical and slightly hooked apical cell and well-developed foot-shaped basal cell; chlamydospores sparse, single or in chains of up to six, intercalary or terminal, hyaline, globose (O’Donnell et al. 2022).
Notes: Under some conditions sporodochial conidia of F. abutilonis may appear blue, as reported for F. buharicum (Gerlach & Nirenberg 1982). Fusarium abutilonis is a putative leaf, stem, and root rot pathogen of some Malvaceae and Fabaceae, and has also been isolated from soil (O’Donnell et al. 2022).
Fusarium buharicum Jacz. ex Babajan & Teterevn.-Babajan, Mater. Mikol. Fitopat. Ross.: 216. 1929. MB 314210.
Holotypus: LEP 127667.
Epitypus: Uzbekistan, Tashkent, on Gossypium herbaceum, 1928, A.I. Raillo, CBS 178.35 (preserved as metabolically inactive culture, designated by Crous et al. 2021b).
Ex-epitype culture: CBS 178.35 = DSM 62166 = IMB 11176 = NRRL 25488.
Descriptions and illustrations: See Gerlach & Nirenberg (1982).
Diagnostic features: Colonies pinkish brown, ochraceous to salmon, partly aeruginous, greyish to dark blue or nearly black on PDA, having optimal growth at 25 °C; microconidia not observed; sporodochia with monophialides give rise to straight, subcylindrical, (3–)5(–8)-septate macroconidia with a short, hooked apical cell and well-developed foot-shaped basal cell; chlamydospores in intercalary chains and terminal, in aerial mycelium and especially in conidia (Gerlach & Nirenberg 1982).
Notes: Fusarium buharicum was initially described as a pathogen of cotton (Gossypium) from the cotton plantations near Bukhara city in Uzbekistan (at that time – the Uzbek Soviet Socialist Republic) on which it induced collar rot symptoms, leading to plant death. With the introduction of resistant and more high yielding varieties of cotton, however, the disease lost its economic significance (Booth 1971). Fusarium buharicum was also found to be an important pathogen of kenaf (Hibiscus cannabinus) in Iran (CBS 796.70), on which it caused root, crown and stem rot (Gerlach & Sharif 1970). Sandoval-Denis et al. (2018b) described F. convolutans as a new soil-borne species occurring in South Africa, which is closely related to F. buharicum but distinct in that it has by its shorter, less septate and less curved macroconidia, and forms sterile hyphal coils in culture. Booth (1971) mentioned that older cultures of F. buharicum form intercalary globose chlamydospores in hyphae or in macroconidial cells, being pale brown, smooth-walled 10–14 μm diam at maturity. Gerlach & Nirenberg (1982) designated CBS 178.35 as neotype of F. buharicum as they were unable to locate the type specimen. However, A. Jaczweski did deposit a specimen in LEP, and therefore, CBS 178.35 was retained as epitype for the species (Crous et al. 2021b).
Fusarium convolutans Sand.-Den. et al., MycoKeys 34: 77. 2018. MB 825102.
Holotypus: CBS H-23495 (dried OA culture).
Ex-type culture: CBS 144207 = CPC 33733.
Type locality: South Africa, Kruger National Park, Skukuza, Granite Supersite.
Type substrate: Rhizosphere soil under Kyphocarpa angustifolia.
Description and illustrations: See Sandoval-Denis et al. (2018b).
Diagnostic features: Colonies white to cream coloured on surface, reverse white, with straw to yellow diffusible pigment on PDA, having optimal growth at 30 °C; aerial monophialides giving rise macroconidia in false heads, lunate to falcate, curved to somewhat straight, (1–)3-septate, with a blunt to conical apical cell and papillate to distinct foot-shaped basal cell; sporodochia absent; chlamydospores abundant, in hyphae or conidia, intercalary or terminal, single or in clumps; sterile, coiled, sometimes branched hyphal projections abundantly formed laterally from the substrate and aerial mycelium (Sandoval-Denis et al. 2018b).
Notes: Fusarium convolutans is characterised by forming sterile, coiled hyphal projections, similar to structures observed in F. circinatum, F. pseudocircinatum and F. sterilihyphosum. The three latter species, however, are genetically unrelated to F. convolutans, being members of the FFSC. Furthermore, they are distinct in that they have microconidia, and lack chlamydospores (Leslie & Summerell 2006).
Fusarium guadeloupense Gräfenhan, Nirenberg & Seifert, Mycologia DOI: 10.1080/00275514.2022.2071563 [9]. 2022.
Holotypus: BPI 924391, dried culture of NRRL 36125.
Ex-type culture: NRRL 36125 = CBS 102302 = BBA 70872.
Type locality: Guadeloupe.
Type substrate: From soil.
Descriptions and illustrations: O’Donnell et al. (2022).
Diagnostic features: Colonies reverse orange with greyish brown; surface white to reddish grey, aerial mycelium white to reddish grey, dense, cottony on PDA, fast growing, having optimal growth at 25 °C; microconidia absent; sporodochia pale to greyish orange, with monophialides giving rise to almost straight to slightly curved, dorsal surface more curved than ventral surface, broadest at or slightly above the centre, 5(–6)-septate macroconidia with a conical and slightly bent apical cell and poorly developed foot-shaped basal cell; chlamydospores single or in chains, intercalary or terminal, hyaline, mostly globose (O’Donnell et al. 2022).
Notes: Fusarium guadeloupense is presently known from two strains, one collected from soil in Guadeloupe, and the other from human blood in Texas, USA. The latter isolate was also able to grow at 37 °C, suggesting that it might be able to infect humans and animals, although this remains to be proven (O’Donnell et al. 2022).
Fusarium sublunatum Reinking, Zentralbl. Bakteriol., Abt. 2, 89: 510. 1934. MB 279278. Fig. 6.
Fig. 6.

Fusarium sublunatum (CBS 189.34). A, B. Sporodochia on CLA. C–F. Sporodochial conidiophores. G. Chlamydospores. H. Macroconidia. Scale bars = 10 μm.
Synonyms: Fusarium sambucinum var. sublunatum (Reinking) Bilaĭ, Mikrobiol. Zhurn. (Kiev) 49: 6. 1987. MB 346814.
Fusarium elongatum Reinking, Zentralbl. Bakteriol. Parasitenk., Abt. 2, 89: 511. 1934. MB 263929.
Fusarium sublunatum var. elongatum Reinking, Die Fusarien, ihre Beschreibung, Schadwirkung und Bekämpfung: 82. 1935. MB 434115.
Authentic material: B 70 0100189.
Lectotypus: Costa Rica, Limón, soil from Musa sapientum plantation, 1933, O.A. Reinking, CBS 189.34 (preserved as metabolically inactive culture, designated by Crous et al. 2021b).
Ex-type culture: BBA 62431 = CBS 189.34 = DSM 62431 = IMB 5238 = NRRL 13384 = NRRL 20840.
Descriptions and illustrations: See Reinking (1934), Gerlach & Nirenberg (1982).
Diagnostic features: Colonies pale beige, rose to cinnamon on PDA, having optimal growth at 25 °C; microconidia not observed; sporodochia with monophialides give rise to falcate, inequilaterally curved, (3–)5(–8)-septate macroconidia with a hooked apical cell and well-developed foot-shaped basal cell; chlamydospores abundant in aerial hyphae and conidia, in pairs, chains or clusters (Gerlach & Nirenberg 1982).
Notes: Fusarium sublunatum was described from soil samples collected in a Musa plantation in Costa Rica. No holotype specimen could be located for F. sublunatum and therefore the metabolically inactive culture CBS 189.34 (= IMB 5238), which represents the ex-type culture (Gerlach & Nirenberg 1982), was designated as lectotype (Crous et al. 2021b). Fusarium sublunatum var. elongatum (original culture CBS 190.34 = NRRL 20897), also described from soil collected in a banana plantation in Costa Rica, proved to be a synonym of F. sublunatum (Raillo 1950, Gerlach & Nirenberg 1982).
Fusarium burgessii species complex (FBURSC)
Fusarium algeriense Laraba & O’Donnell, Mycologia 109: 944. 2017 (2018). MB 820565. Fig. 7.
Fig. 7.
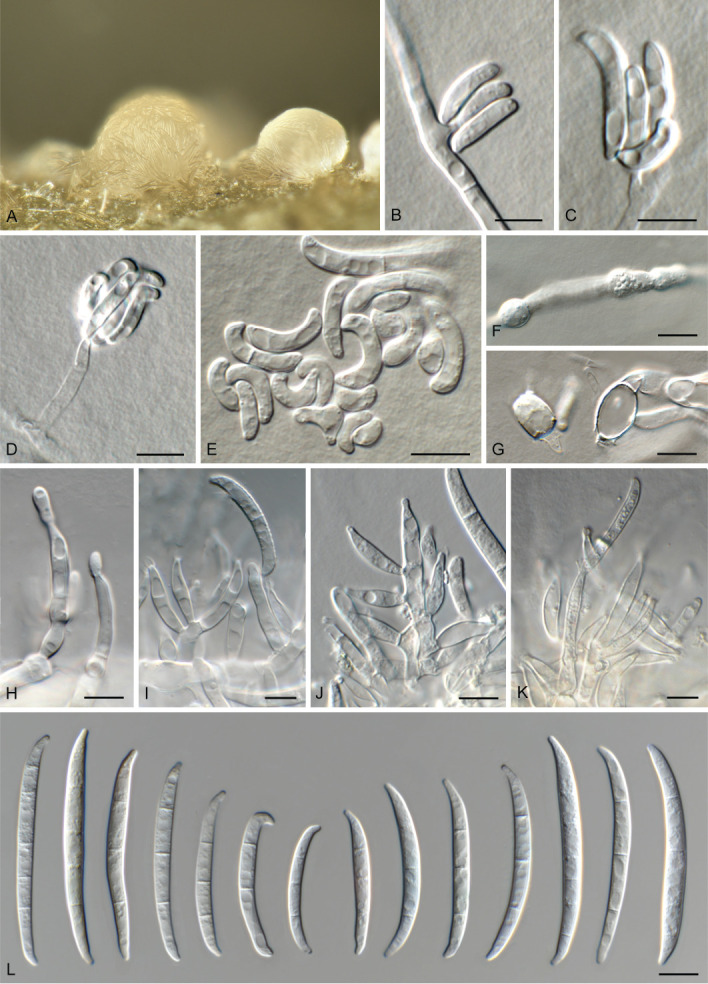
Fusarium algeriense (CBS 142638). A. Sporodochium on CLA. B–D. Aerial conidiophores with monophialides. E. Microconidia. F, G. Chlamydospores. H–K. Sporodochial conidiophores. L. Macroconidia. Scale bars = 10 μm.
Holotypus: BPI 910347 (dried culture).
Ex-type culture: CBS 142638 = IL-79 = KOD 1247 = NRRL 66647.
Type locality: Algeria, Guelma Province, Djeballah Khemissi.
Type substrate: Triticum durum.
Description and illustrations: See Laraba et al. (2017).
Diagnostic features: Colonies reddish orange, brownish grey, yellowish white to purplish grey on PDA, having optimal growth at 25 °C; *microconidia developing in false heads, on superficial and immersed mycelium, subcylindrical, straight to curved, 0–1-septate; sporodochia with monophialides give rise to straight to falcate, slender, 1–3(–4)-septate macroconidia with a hooked apical cell and well-developed foot-shaped basal cell; *chlamydospores intercalary, globose to subglobose, in chains, sparse, hyaline (*emended from Laraba et al. 2017).
Notes: Fusarium algeriense represents a species within the F. burgessii species complex causing crown rot of durum wheat in Algeria (Laraba et al. 2017). Following its description, crown rot symptoms of bread wheat in two provinces of Azerbaijan were also attributed to F. algeriense (Özer et al. 2020).
Morphologically, F. algeriense needs to be compared to F. burgessii and F. beomiforme, which have an optimal growth at 30 °C, and produce abundant chlamydospores. Isolates of F. algeriense had an optimal growth at 25 °C, lacked chlamydospore production in culture, and produced monophialides, with reniform or ellipsoidal, mostly aseptate microconidia. In contrast, F. burgessii has polyphialides, and F. beomiforme has monophialides, but with globose-to-napiform, 0–1-septate microconidia (Laraba et al. 2017).
Fusarium beomiforme P.E. Nelson et al., Mycologia 79: 886. 1987. MB 122057. Fig. 8.
Fig. 8.

Fusarium beomiforme (CBS 100160). A, B. Sporodochia on SNA. C–E. Microconidia. F, G. Chlamydospores developing in macroconidia. H–L. Sporodochial conidiophores. M. Macroconidia. Scale bars = 10 μm.
Holotypus: DAOM 196987 (dried culture).
Ex-type culture: ATCC 64067 = CBS 100160 = DAOM 196987 = DAR 58880 = F 5759 = FRC M-1425 = IMI 316127 = MRC 4593 = NRRL 13606.
Type locality: Australia, Queensland, Rockhampton.
Type substrate: Plant debris in soil.
Descriptions and illustrations: See Nelson et al. (1987) and Leslie & Summerell (2006).
Diagnostic features: Colonies pale orange to white, with orange red to red-brown pigmentation on PDA; optimal growth at 30 °C; monophialides produce false heads with 0–1-septate napiform to globose microconidia in aerial mycelium; sporodochia with monophialides giving rise to long falcate, 3–4(–5)-septate macroconidia with a slightly curved apical cell and notched basal cell, and slow to form, abundant, intercalary, single to chains of chlamydospores in aerial and submerged hyphae (Nelson et al. 1987).
Notes: Fusarium beomiforme was described from soil and plant debris collected in the Markham Valley of Papua New Guinea (where sorghum had been cultivated), from grassland areas in the vicinity of Rockhampton, Emerald, Longreach, and Boulia along the Tropic of Capricorn in Queensland, Australia, and from Hluhluwe, KwaZulu-Natal, South Africa (Nelson et al. 1987). Since then, F. beomiforme has also been recovered from Thailand (from soil where previously sorghum had been cultivated; Mohamed Nor et al. 2019), though to date, F. beomiforme has not been reported to be pathogenic, and is probably a saprobe.
Fusarium burgessii M.H. Laurence et al., Fungal Diversity 49: 109. 2011. MB 519216. Fig. 9.
Fig. 9.

Fusarium burgessii (CBS 125537). A. Sporodochium on CLA. B. Aerial conidiophores with monophialides giving rise to micro- and macroconidia. C–F. Microconidia. G–J. Sporodochial conidiophores. K. Macroconidia. Scale bars = 10 μm.
Holotypus: CBS 125537 (preserved as metabolically inactive culture).
Ex-type culture: CBS 125537 = NRRL 66654 = RBG 5315.
Type locality: Australia, Queensland, Idalia National Park.
Type substrate: Soil.
Description and illustrations: See Laurence et al. (2011).
Diagnostic features: Colonies white to yellow with yellow pigmentation on PDA, having optimal growth at 30 °C; mono- to polyphialides produce false heads with 0–1-septate oval, elliptical or reinform microconidia in aerial mycelium; sporodochia with mono- to polyphialides produce short to medium length, falcate, 3-septate macroconidia with a slightly curved to hooked apical cell and notched to well-developed, foot-shaped basal cell; chlamydospores in both aerial and submerged hyphae, terminal and intercalary, solitary or in chains (Laurence et al. 2011).
Notes: Fusarium burgessii was described from Australia, and is known to occur in soils from Longreach, Queensland, to Finke Gorge National Park, Northern Territory (Laurence et al. 2011). Morphologically, it is allied to F. algeriense and F. beomiforme (see discussion under F. algeriense), and morphotype B (isolated from the rhizosphere of indigenous Gossypium spp.), which presently still represents an undescribed species (Laurence et al. 2011).
Fusarium camptoceras species complex (FCAMSC)
Fusarium camptoceras Wollenw. & Reinking, Phytopathology 15: 158. 1925. MB 259537. Fig. 10.
Fig. 10.

Fusarium camptoceras (CBS 193.65). A. Aerial conidiophores with monophialides. B, C. Chlamydospores. D, E. Macroconidia. Scale bars = 10 μm.
Neotypus: CBS H-24077, designated in Xia et al. (2019).
Ex-neotype culture: ATCC 16065 = ATCC 24364 = BBA 9810 = CBS 193.65 = DSM 62167 = IMB 9810 = IMI 112500 = NRRL 20716 = NRRL 36344.
Neotype locality: Costa Rica.
Neotype substrate: Cushion gall of Theobroma cacao.
Descriptions and illustrations: See Wollenweber & Reinking (1935), Booth (1971), Gerlach & Nirenberg (1982), Marasas et al. (1998) and Leslie & Summerell (2006).
Diagnostic features: Colonies brown on PDA, having optimal growth at 25 °C; microconidia not observed; aerial polyphialides formed on loosely branched conidiophores giving rise to av. 3–4-septate mesoconidia, and macroconidia; sporodochia with monophialides give rise to falcate, 3–5(–7)-septate macroconidia with a pointed apical cell and obtuse to well-developed, foot-shaped basal cell; intercalary chains, pairs or clusters of chlamydospores in aerial and submerged hyphae, never in terminal pairs (Marasas et al. 1998, Leslie & Summerell (2006).
Notes: Fusarium camptoceras was described from subtropical and tropical regions (Costa Rica, Ecuador, Honduras, Angola), recovered from decaying Coffea, Musa and Theobroma spp. (Marasas et al. 1998). Reports prior to 1998 could represent two species separated from F. camptoceras, namely F. musarum and F. nelsonii, which differ regarding their red pigmentation on PDA, size and septation of their mesoconidia (F. musarum av. 5–6-septate; F. nelsonii av. 3-septate), sporodochia (absent in F. musarum; present in F. nelsonii), and the pattern in which chlamydospores are formed (in terminal pairs in F. nelsonii, solitary or chains in F. camptoceras and F. musarum) (Marasas et al. 1998). Further studies are needed to confirm the role of F. camptoceras as plant pathogen.
Fusarium kotabaruense Maryani et al., Persoonia 43: 65. 2019. MB 828964.
Holotypus: InaCC F963 (preserved as metabolically inactive culture).
Ex-type culture: InaCC F963 = Indo172.
Type locality: Indonesia, South Kalimantan, Kota Baru, Kecamatan Pamukan Barat, Desa Sungai Birah.
Type substrate: Infected pseudostem of Musa var. Pisang Hawa (ABB).
Description and illustrations: See Maryani et al. (2019).
Diagnostic features: Colonies rosy buff on PDA, having optimal growth at 25 °C; aerial hyphae and orange sporodochia with mono- and polyphialides give rise to macroconidia, falcate, (2–)3–5(–7)-septate, with blunt apical cell and poorly-developed, foot-shaped basal cell; chlamydospores not observed (Maryani et al. 2019).
Notes: Fusarium kotabaruense represents a fast-growing species which clustered basal to the FIESC, and was shown to be better accommodated in the Fusarium camptoceras species complex (Xia et al. 2019, Crous et al. 2021b). Although assumed to lack sporodochia, isolates on CLA incubated under nuv-light did produce orange sporodochia. This species is characterised by its mono- to polyphialides, fast-growing cultures and multiseptate conidia (Maryani et al. 2019).
Fusarium neosemitectum L. Lombard et al., Persoonia 43: 214. 2019. MB 831845.
Holotypus: CBS H-24067.
Ex-type culture: CBS 189.60.
Type locality: Democratic Republic of the Congo.
Type substrate: Musa sapientum.
Description and illustrations: See Xia et al. (2019).
Diagnostic features: Colonies white, felty to velvety on PDA, with abundant aerial mycelium; aerial mono- to polyphialides giving rise to macroconidia, ellipsoid to falcate, curved dorsiventrally, (1–)2–4(–5)-septate; blunt, conical to slightly papillate apical cell and blunt to poorly-developed, foot-shaped basal cell; sporodochia and chlamydospores not observed (Xia et al. 2019).
Notes: Fusarium neosemitectum can be distinguished from closely related species, such as F. kotabaruense and F. camptoceras, by the presence of short phialidic pegs on the aerial mycelium, not observed for the latter two species. All three species in FCAMSC appear to be tropical species due to their origins and they also share a mutual host genus, Musa (Marasas et al. 1998, Maryani et al. 2019).
Fusarium chlamydosporum species complex (FCSC)
Fusarium atrovinosum L. Lombard & Crous, Fungal Syst. Evol. 4: 190. 2019. MB 831559.
Holotypus: CBS H-24015.
Ex-type culture: BBA 10357 = CBS 445.67 = DSM 62169 = IMB 10357= IMI 096270 = NRRL 26852 = NRRL 26913.
Type locality: Australia.
Type substrate: Triticum aestivum.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies on the surface greyish rose to vinaceous to buff in the centre, with abundant aerial mycelium, and livid red to dark vinaceous in reverse on PDA; aerial polyphialides giving rise to false heads with fusiform to ellipsoidal to obovoid, 0–1(–2)-septate microconidia; chlamydospores abundant, globose to subglobose, thick-walled, smooth to slightly verrucose, formed terminally or intercalarily in chains of three or more (Lombard et al. 2019).
Notes: Fusarium atrovinosum is closely related to F. chlamydosporum, F. spinosum and F. sporodochiale and can be distinguished from these three species by the lack of monophialides on aerial mycelium, the lack of sporodochia, and abundant chlamydospores.
Fusarium chlamydosporum Wollenw. & Reinking, Phytopathology 15: 156. 1925. MB 260522.
Synonyms: Fusarium chlamydosporum var. chlamydosporum, Phytopathology 15: 156. 1925. MB 429587.
Fusarium sporotrichioides var. chlamydosporum (Wollenw. & Reinking) Joffe, Mycopathol. Mycol. Appl. 53: 211. 1974. MB 348165.
Dactylium fusarioides Gonz. Frag. & Cif., Bol. Real Soc. Esp. Hist. Nat. 27: 280. 1927. MB 265606.
Fusarium fusarioides (Gonz. Frag. & Cif.) C. Booth, The genus Fusarium: 88. 1971. MB 314214.
Pseudofusarium purpureum Matsush., Microfungi of the Solomon Islands and Papua-New Guinea: 47. 1971. MB 321785.
Neotypus: CBS 145.25 (preserved as metabolically inactive culture), designated in Lombard et al. (2019).
Ex-neotype culture: CBS 145.25 = NRRL 26851 = NRRL 26912.
Neotype locality: Honduras, Tela.
Neotype substrate: Pseudostem of Musa sapientum.
Descriptions and illustrations: See Booth (1971), Gerlach & Nirenberg (1982) and Leslie & Summerell (2006).
Diagnostic features: Colonies with white mycelium and greyish rose to burgundy pigment on PDA; microconidia abundant, straight to reniform, 0(–2)-septate, arising from aerial mono- and polyphialides; sporodochia rare, with monophialides give rise to thick-walled, unequal dorsiventrally curved, 3–5-septate macroconidia with a short, curved, pointed apical cell and poorly to well-developed, foot-shaped basal cell; chlamydospores abundant, formed rapidly in aerial mycelium, submerged hyphae and on agar surface, verruculose and pale brown, in chains or clusters (Marasas et al. 1998, Leslie & Summerell 2006).
Notes: Fusarium chlamydosporum (FCSC) is common in soils and grains from arid and semi-arid regions (Burgess & Summerell 1992, Kanaan & Bahkali 1993, Sangalang et al. 1995a), and from plant material displaying disease symptoms that include crown rot (Du et al. 2017), blight (Satou et al. 2001), damping-off (Engelbrecht et al. 1983, Lazreg et al. 2013) and stem canker (Fugro 1999). It has also been implicated in human and animal fusarioses (O’Donnell et al. 2009). Records prior to Lombard et al. (2019) need to be interpreted with care, as this was shown to be a species complex O’Donnell et al. (2009, 2018). Subsequent to these studies, five of these taxa were named, with several additional species in the FCSC still awaiting formal description. Furthermore, F. chlamydosporum var. fuscum was raised to species level, as F. coffeatum, in the F. incarnatum-equiseti species complex (FIESC) (Lombard et al. 2019).
Fusarium humicola L. Lombard & Crous, Fungal Syst. Evol. 4: 191. 2019. MB 831561.
Holotypus: CBS H-24016.
Ex-type culture: ATCC 24372 = CBS 124.73 = IMI 128101 = NRRL 25535.
Type locality: Pakistan.
Type substrate: Soil.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies fulvous to ochreous in the centre becoming vinaceous to livid red towards the margin, reverse dark vinaceous to vinaceous on PDA; aerial mono- to polyphialides giving rise to microconidia in false heads, ellipsoidal to obovoid, 0–3-septate; sporodochia pale luteous to pale salmon, with monophialides give rise to falcate, mostly straight with dorsiventrally curved apical and basal cells 3–5-septate macroconidia with a curved, blunt to papillate apical cell and well-developed, foot-shaped basal cell; chlamydospores not observed (Lombard et al. 2019).
Note: Fusarium humicola is closely related to F. nelsonii, which has smaller, more strongly curved sporodochial conidia, and abundant chlamydospores.
Fusarium microconidium L. Lombard & Crous, Fungal Syst. Evol. 4: 192. 2019. MB 831562.
Holotypus: CBS H-24017.
Ex-type culture: CBS 119843 = KSU 11396 = MRC 8391.
Type locality: Unknown.
Type substrate: Unknown.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies rose to rosy vinaceous to pale luteous on surface, with abundant aerial mycelium, and livid red to dark vinaceous in reverse on PDA; aerial mono- or polyphialides giving rise to microconidia, fusoid to ellipsoidal to obovoid, 0–1-septate; sporodochia and chlamydospores not observed (Lombard et al. 2019).
Notes: Fusarium microconidium is distinguished from other species in the FCSC based on the production of predominantly aseptate microconidia and lack of sporodochia and chlamydospores.
Fusarium nelsonii Marasas & Logrieco, Mycologia 90: 508. 1998. MB 443596.
Holotypus: BPI 802927; isotypi DAOM 225260 and PREM 55396.
Ex-type culture: ATCC 201410 = CBS 119876 = FRC R-8670 = ITEM 1229 = MRC 4570 = NRRL 28505 = NRRL 53945.
Type locality: South Africa, Western Cape Province, Malmesbury.
Type substrate: Plant debris in wheat field soil.
Descriptions and illustrations: See Marasas et al. (1998) and Leslie & Summerell (2006).
Diagnostic features: Colonies with white floccose mycelium and red pigmentation on PDA, having optimal growth at 30 °C; aerial polyphialides giving rise to mesoconidia, fusoid to lanceolate, straight to curved, (0–)3-septate; sporodochia cream coloured, with monophialides giving rise to straight or falcate, 3(–5)-septate macroconidia with a curved, blunt apical cell (beak-like) and poorly-developed, foot-shaped basal cell; chlamydospores abundant and rapidly formed in aerial and submerged hyphae, intercalary or terminal, single, in pairs, chains or clumps (Marasas et al. 1998, Leslie & Summerell 2006).
Notes: Fusarium nelsonii was described from South Africa, where it was isolated from Triticum soil, plant debris, Medicago roots, Sorghum malt and Zea mays kernels (Marasas et al. 1998). It has been reported from Triticum in Iran (Chehri et al. 2010), Sorghum in India (Lincy et al. 2011), fruit blight of Cucumis sativus var. sativus and stalk rot of Zea mays in China (Ahmad et al. 2020, Zhang et al. 2021).
Fusarium nelsonii produces macro- and mesoconidia (aerial mycelium), which distinguishes it from F. musarum (macroconidia absent), and has shorter meso- and macroconidia than F. camptoceras.
Fusarium peruvianum L. Lombard & Crous, Fungal Syst. Evol. 4: 194. 2019. MB 831564.
Holotypus: CBS H-24019.
Ex-type culture: CBS 511.75.
Type locality: Peru.
Type substrate: Seedlings of Gossypium sp.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies fulvous to ochreous in the centre becoming coral to vinaceous towards the margin, with abundant aerial mycelium, and livid red to dark vinaceous in reverse on PDA; aerial phialides mostly polyphialidic, giving rise to micro- and macroconidia; microconidia ellipsoid to obovoid, 0–3(–4)-septate, macroconidia fusoid to falcate, straight or gently dorsiventrally curved, with a blunt apical cell and indistinct papillate to poorly-developed, foot-shaped basal cell; chlamydospores abundant, intercalary or terminal, single or in pairs; sporodochia not observed (Lombard et al. 2019).
Note: Fusarium peruvianum can be distinguished from other species in the FCSC by having falcate aerial macroconidia and 4-septate obovoid microconidia.
Fusarium spinosum L. Lombard et al., Fungal Syst. Evol. 4: 195. 2019. MB 831565.
Holotypus: CBS H-24020.
Ex-type culture: CBS 122438.
Type locality: Brazil.
Type substrate: Galia melon imported into the Netherlands.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies rose to rosy vinaceous to pale luteous in the centre, with abundant aerial mycelium, reverse fulvous to ochreous with rosy vinaceous flames on PDA; aerial mono- to polyphialides giving rise to micro- and macroconidia in false heads; microconidia fusoid to ellipsoidal to obovoid, straight to curved, 0–3-septate; macroconidia falcate, slightly dorsiventrally curved, 3-septate, apex blunt, with an indistinct papillate to poorly-developed foot-shaped basal cell; chlamydospores abundant, intercalary or terminal, single or in chains; sporodochia not observed (Marasas et al. 1998, Leslie & Summerell 2006).
Note: Fusarium spinosum is distinguished from other species in the FCSC by only forming 3-septate, falcate macroconidia.
Fusarium sporodochiale L. Lombard & Crous, Fungal Syst. Evol. 4: 196. 2019. MB 831566.
Holotypus: CBS H-12681.
Ex-type culture: ATCC 14167 = CBS 220.61 = MUCL 8047 = NRRL 20842.
Type locality: South Africa, Gauteng Province, Johannesburg.
Type substrate: Soil.
Description and illustrations: See Lombard et al. (2019).
Diagnostic features: Colonies rose to rosy vinaceous to sulphur yellow, with abundant aerial mycelium, reverse livid red to dark vinaceous on PDA; aerial phialides mostly polyphialidic, giving rise to microconidia in false heads, fusoid to ellipsoidal to obovoid, (0–)1-septate; sporodochia pale luteous to pale orange, with monophialides giving rise to falcate, slightly to strongly dorsiventrally curved macroconidia, tapering towards both ends, with an elongated, strongly curved apical cell and a blunt and distinct foot-shaped basal cell, (1–)5–6(–10)-septate; chlamydospores not observed (Lombard et al. 2019).
Notes: Fusarium sporodochiale is unique within the FCSC, producing up to 10-septate sporodochial macroconidia. Additionally, the apical cell of macroconidia is more elongated and hooked than those of other species in this complex.
Fusarium citricola species complex (FCCSC)
Fusarium aconidiale L. Lombard & Crous, Persoonia 46: 523. 2021. MB 839622.
Holotypus: CBS H-24769.
Ex-type culture: CBS 147772 = CPC 37959 = UBOCC-A-109005.
Type locality: France.
Type substrate: Triticum aestivum.
Description and illustrations: See Crous et al. (2021a).
Diagnostic features: Colonies white to rosy buff, flat, woolly to cottony with radial patches of white aerial mycelium, reverse white to pale rosy buff on PDA; aerial phialides monophialidic, but microconidia not observed; sporodochia crystalline to pale cream, with monophialides giving rise to falcate, straight to moderately curved macroconidia, tapering towards the basal part, apical cell more or less equally sized than the adjacent cell, curved to hooked; basal cell well-developed, foot-shaped, rarely papillate, 3(–5)-septate; chlamydospores not observed (Crous et al. 2021a).
Notes: Fusarium aconidiale is similar to F. juglandicola but does not produce red pigment under continuous white light nor any chlamydospores or aerial microconidia, distinguishing it from other members of the FCCSC. Furthermore, F. aconidiale produces predominantly 3-septate sporodochial conidia and much less frequently 4- and 5-septate sporodochial conidia compared to F. juglandicola. (Crous et al. 2021a).
Fusarium celtidicola Q.J. Shang et al., Phytotaxa 361: 255. 2018. MB 553845. Figs 11, 12.
Fig. 11.

Fusarium celtidicola (MFLUCC 16-0526). A. Sporodochium on CLA. B. Aerial conidiophore. C–I. Sporodochial conidiophores with monophialides. J. Chlamydospore. K. Macroconidia. Scale bars = 10 μm.
Fig. 12.

Fusarium celtidicola (MFLUCC 16-0526). A. Perithecial ascomata on host surface. B, C. Vertical section through perithecia. D–G. Hamathecial catenophyses, and asci. H. Germinating ascospore. (F, G in Melzer’s reagent). Scale bars: A = 100 μm, B, C = 30 μm, D–H = 10 μm (Photos from Shang et al. 2018).
Holotypus: MFLU 15-3646; isotypus HKAS 95020.
Ex-type culture: KUMCC 16-0019 = MFLUCC 16-0526; ex-isotype culture KUMCC 16-0019 = MFLUCC 16-0526.
Type locality: Italy, Forlì-Cesena Province, Forlì, Viale dell’Appennino.
Type substrate: Dead branch of Celtis australis.
Description and illustrations: See Shang et al. (2018).
Diagnostic features: Colonies on the surface white, reddish at the centre, and reddish white in reverse on PDA; aerial monophialides giving rise to micro- and macroconidia; microconidia oblong to naviculate, straight or curved, 1–3-septate; macroconidia naviculate to falcate, 3–5-septate with a curved, blunt apical cell and poorly-developed, foot-shaped basal cell; chlamydospores intercalary in aerial hyphae, in pairs or chains (Shang et al. 2018).
Notes: Fusarium celtidicola is distinct from other members of the FCCSC in that it produces chlamydospores, and has a sexual morph with blue-black to dark purple perithecia, and ellipsoid to obovoid to fusoid, 0–3-septate, smooth-walled ascospores (Shang et al. 2018).
Fusarium citricola Guarnaccia et al., Persoonia 40: 12. 2017 (2018). MB 820246.
Holotypus: CBS H-23020 (dried SNA/CL culture).
Ex-type culture: CBS 142421 = CPC 27805.
Type locality: Italy, Cosenza, Rocca Imperiale.
Type substrate: Crown of Citrus reticulata ‘Caffin’.
Description and illustrations: See Sandoval-Denis et al. (2018a).
Diagnostic features: Colonies pale luteous to pale yellow on surface (orange to red when incubated in light), reverse pale luteous to straw (diffusible pigment absent in the dark, an orange to red pigment sometimes present when incubated in the light) on PDA; aerial monophialides giving rise to microconidia, ellipsoidal to falcate, 0–3-septate; sporodochia bright orange, with monophialides giving rise to falcate, dorsiventrally curved macroconidia with almost parallel sides, tapering slightly towards both ends, with a blunt to papillate, curved apical cell and poorly to well-developed, foot-shaped basal cell, (1–)2–4(–6)-septate; chlamydospores absent (Sandoval-Denis et al. 2018a).
Notes: Fusarium citricola was shown to be the cause of cankers on diverse Citrus spp. in Apulia and Calabria in southern Italy. Fusarium citricola resembles F. salinense, but can be distinguished in having slightly smaller sporodochial conidia, often with a gentle and symmetrical dorsiventral curvature, and 0–3-septate microconidia (vs the often asymmetrically curved macroconidia and 0–1(–2)-septate microconidia in F. salinense) (Sandoval-Denis et al. 2018a).
Fusarium juglandicola L. Lombard & Crous, Persoonia 46: 521. 2021. MB 839621.
Holotypus: CBS H-24770.
Ex-type culture: CBS 147773 = CPC 37962 = UBOCC-A-119001.
Type locality: France, Rhone-Alps region.
Type substrate: Bud of Juglans regia.
Description and illustrations: See Crous et al. (2021a).
Diagnostic features: Colonies white to pale luteous on surface and reverse on PDA; aerial monophialides giving rise to macroconidia; microconidia absent; sporodochia with monophialides giving rise to falcate, moderately dorsiventrally curved macroconidia with almost parallel sides, tapering towards both ends, with a blunt to slightly hooked, somewhat curved apical cell and papillate to well-developed, foot-shaped basal cell, (1–)3–4(–5)-septate; chlamydospores absent (Crous et al. 2021a).
Notes: Fusarium juglandicola was isolated from walnut, Juniperus sp., and eggs from an unknown species in southeast France. Fusarium juglandicola is unique within the FCCSC by lacking microconidia and red pigments, even when incubated under continuous white light (Crous et al. 2021a).
Fusarium salinense Sand.-Den. et al., Persoonia 40: 15. 2017 (2018). MB 820245.
Holotypus: CBS H-23019 (dried SNA/CL culture).
Ex-type culture: CBS 142420 = CPC 26973.
Type locality: Italy, Sicily, Messina, Leni.
Type substrate: Twigs of Citrus sinensis.
Description and illustrations: See Sandoval-Denis et al. (2018a).
Diagnostic features: Colony surface pale luteous to sulphur yellow with white to pale luteous margins, reverse pale luteous to orange toward the centre of the colony. Yellow diffusible pigment sometimes present, while red colonies and diffusible pigments occur when incubated in light on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia, ovoid, ellipsoid to falcate, 0–1(–2)-septate; sporodochia flesh, salmon to orange coloured, with monophialides give rise to falcate, (2–)3–4(–5)-septate, slender macroconidia, with a gentle curvature and nearly parallel dorsiventral lines or an unequal curvature, slightly more pronounced in the upper part of the spore, tapering slightly towards the basal end, with a papillate and curved apical cell and a poorly-developed, foot-shaped basal cell; chlamydospores absent, but rounded, thin-walled hyphal swellings sometimes present in old cultures. (Sandoval-Denis et al. 2018a).
Notes: Fusarium salinense is known from Sicily (Italy), and Salina (Aeolian Island), and is associated with canker symptoms on three different Citrus species. It produces sparingly branched conidiophores in the aerial mycelium, especially in young cultures, but its growth soon becomes pionnotal. Fusarium salinense can be distinguished from F. citricola by producing shorter sporodochial phialides and slightly longer and robust macroconidia, often with an unequal dorsiventral curvature (Sandoval-Denis et al. 2018a).
Fusarium concolor species complex (FCOSC)
Fusarium bambusarum M.M. Wang & L. Cai, Persoonia 48: 25. 2022. MB 346784.
Typus: HMAS 351575 (dried SNA/CL culture).
Type locality: China, Jiangxi Province.
Type substrate: From bamboo.
Descriptions and illustrations: See Wang et al. (2022).
Diagnostic features: Colonies white on PDA, with dense aerial mycelium; aerial monophialides giving rise to microconidia in false heads, ovoid to fusoid-ellipsoid, aseptate; sporodochia orange grey on carnation leaf agar, with monophialides give rise to falcate macroconidia, slightly bent with parallel sides, with a papillate to hooked, curved apical cell, and well-developed, foot-shaped basal cell, 3–6-septate; chlamydospores terminal, globose, becoming rough and thick-walled (Wang et al. 2022).
Notes: Fusarium bambusarum is distinguished from other taxa in the FCOSC based on its 3–6-septate macroconidia, and having monophalidic aerial phialides (Wang et al. 2022). Presently this taxon is only known from bamboo collected in Jiangxi Province, China.
Fusarium anguioides Sherb., Mem. Cornell Univ. Agric. Exp. Sta. 6: 169. 1915. MB 159197.
Synonym: Fusarium avenaceum var. anguioides (Sherb.) Bilaĭ, Mikrobiologicheskij Zhurnal (Kiev) 49: 6. 1987. MB 346784.
Typus: ?CUP-007479, BPI 72044 neotype (not Code compliant).
Type locality: USA, New York, Castile.
Type substrate: Solanum tuberosum.
Descriptions and illustrations: See Sherbakoff (1915), Gerlach & Nirenberg (1982) and Nelson et al. (1995).
Diagnostic features: Colonies cream, pink, rose to carmine or yellowish to ochre, becoming yellowish brown or red-brown to brown with age on PDA, having optimal growth at 25 °C; aerial mono- to polyphialides giving rise to microconidia, ovoid to fusoid, 0–3-septate; sporodochia orange to cinnamon or brick coloured, with monophialides give rise to falcate, macroconidia, slightly bent to anguiform, slender, tapering toward both ends, with an elongated, elegantly curved apical cell and well-developed, foot-shaped basal cell, (3–)5–7-septate; chlamydospores absent, but hyphal swellings do occur (Gerlach & Nirenberg 1982).
Notes: Sherbakoff (1915) provided an illustration with the original protologue of F. anguioides and placed material in CUP, as CUP-007479. The neotype (BPI 72044) designated by Nelson et al. (1995) originated from China and was isolated from soil in a bamboo grove, and is thus unsuitable. An isolate from the original locality (USA) and host (Solanum tuberosum) needs to be selected.
Fusarium austroafricanum A. Jacobs et al., Mycologia 110: 1197. 2018. MB 823959. Fig. 13.
Fig. 13.

Fusarium austroafricanum (CBS 120990). A, B. Sporodochia on CLA. C–F. Aerial conidiophores with polyphialides giving rise to microconidia. G. Microconidia. H. Chlamydospore. I–L. Sporodochial conidiophores giving rise to macroconidia. M. Macroconidia. Scale bars = 10 μm.
Holotypus: PREM 62137 (dried culture); paratypi PREM 62138 and PREM 62139 (dried cultures).
Ex-holotype culture: NRRL 66741 = PPRI 10408 = PPRI 23548; ex-paratype cultures: CBS 120990 = DAOM 192987 = FRC M-2406 = NRRL 53441 = PPRI 23546 and NRRL 66742 = PPRI 10412.
Type locality: South Africa, Eastern Cape Province, Humansdorp.
Type substrate: Endophyte of Pennisetum clandestinum.
Description and illustrations: See Jacobs-Venter et al. (2018).
Diagnostic features: Colony surface white to reddish white, reverse pale orange on PDA, having optimal growth at 30 °C; aerial mono- to polyphialides giving rise to microconidia, oval to obovoid, aseptate; sporodochia with monophialides give rise to falcate, (3–)5(–8)-septate macroconidia with a blunt apical cell and poorly-developed, foot-shaped basal cell; chlamydospores singly or in intercalary or terminal clusters (Jacobs-Venter et al. 2018).
Notes: Fusarium austroafricanum is similar morphologically to F. concolor and F. babinda, but forms white to reddish white colonies on PDA, whereas those of F. concolor are white to pale orange, and those of F. babinda are pale orange to violet. Morphologically, F. austroafricanum differs from F. concolor and F. babinda in the shape of the apical cell on the macroconidia, i.e. blunt (F. austroafricanum), papillate (F. concolor) or slightly curved to hooked (F. babinda) (Reinking 1934, Marasas et al. 1986, Jacobs-Venter et al. 2018).
Fusarium concolor Reinking, Zentralbl. Bakteriol., Abt. 2, 89: 512. 1934. MB 261626.
Synonym: Fusarium polyphialidicum Marasas et al., Mycologia 78: 678. 1986. MB 102972.
Holotypus: IMI 112502.
Ex-type culture: BBA 2607 = BBA 63601 = CBS 183.34 = DAOM 225131 = DSM 62179 = IMB 10330 = IMI 112502 = NRRL 13994.
Type locality: Uruguay, Montevideo.
Type substrate: Hordeum vulgare.
Descriptions and illustrations: See Gerlach & Nirenberg (1982) and Marasas et al. (1986).
Diagnostic features: Colonies whitish, reverse white to yellow on PDA, having optimal growth at 25 °C; aerial mono- to polyphialides giving rise to microconidia in false heads, obovoid, fusoid to subclavate, (0–)1(–2-)septate; sporodochia white to pale orange, with mono- to polyphialides give rise to straight or falcate, 3–5(–9)-septate macroconidia with a long and tapered to curved apical cell and well-developed, foot-shaped basal cell; chlamydospores abundant, intercalary and terminal in hyphae and conidia, single, in pairs, chains or clusters (Gerlach & Nirenberg 1982, Marasas et al. 1986).
Notes: Balmas et al. (2010) and Jacobs-Venter et al. (2018) considered that F. polyphialidicum was a synonym of F. concolor, which was originally described based on a single isolate from diseased barley in Uruguay (Reinking 1934). Fusarium concolor has a wide distribution and host range, occurring in Africa (South Africa, Zimbabwe), Australasia (Australia), Europe (Italy, Spain), South America (Uruguay), and North America (USA, Hawaii), and has also been associated with human infections (Jacobs-Venter et al. 2018).
Taxonomic novelties
Novel species are described in Fusicolla and Neocosmospora. Additionally, arguments for recognising distinct genera in the terminal fusarioid clade of the Nectriaceae were presented by Crous et al. (2021b) and Hill et al. (2022). In this regard, several species recently assigned to Fusarium s. str., are herewith allocated to Neocosmospora.
Fusicolla elongata Decock, Crous & Sand.-Den., sp. nov. MycoBank MB 843499. Fig. 14.
Fig. 14.
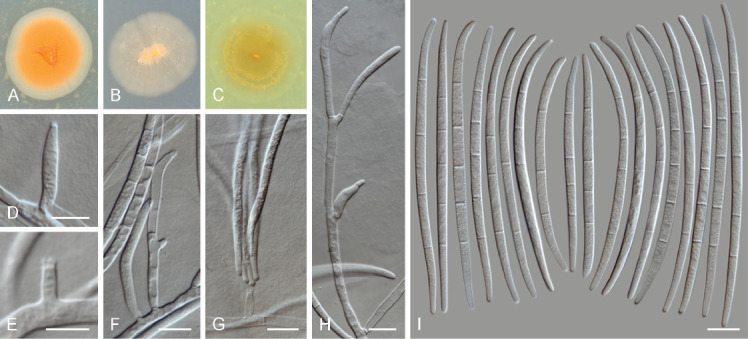
Fusicolla elongata (MUCL 58143 ex-type). A–C. Colony surface on PDA, SNA and OA, respectively. D–H. Conidiophores and conidiogenous cells. I. Conidia. Scale bars: E = 5 μm; all others = 10 μm.
Etymology: From Latin elongare, meaning elongated, in reference to its long conidia.
Description: Conidiophores prostrate, emerging from vegetative hyphae, intermixed and confluent, commonly as single phialides borne laterally on hyphae or reduced to phialidic pegs; rarely and mostly on the colony periphery, conidiophores erect, simple or branched once or twice laterally and irregularly, terminating in a single conidiogenous cell. Conidiogenous cells monophialidic, subcylindrical, (3–)8–31.5(–40) × (1.5–)2–4(–4.5) μm, smooth- and thin-walled, with or without noticeable periclinal thickening, and a minute, non-flared apical collarette. Macroconidia slender to somewhat elongate, almost straight to gently curved, apical cell barely curved with a rounded apex, basal cell obtuse, non-foot-shaped, (3–)4–5-septate, predominantly 5-septate, hyaline, smooth- and thin-walled; 3-septate conidia: 66.5–82 × 2.5–3 μm (av. 73.6 × 2.8 μm); 4-septate, (64–)74.5–92.5(–97) × (2.5–)3–4 μm (av. 83.5 × 3.2 μm); 5-septate, (81.5–)85–96(–100.5) × 2.5–4 μm (av. 90.5 × 3.3 μm). Microconidia, chlamydospores and sexual morph not observed.
Culture characteristics: Colonies at 25 °C after 7 d: On PDA reaching 17–22 mm diam, orange to apricot at centre, white to pale salmon at periphery, flat, slightly folded to cerebriform at centre, membranous to slimy, lacking aerial mycelium, margin entire; reverse white to pale salmon, without diffusible pigments. On SNA reaching 15–22 mm diam, buff to pale salmon, flat, membranous to slimy at centre, aerial mycelium lacking or scattered in irregular, short patches; reverse white to pale saffron without diffusible pigments. On OA reaching 22–28 mm diam, pale luteous, pale amber to ochraceous, flat, membranous, with abundant and confluent sporulation forming slimy masses and concentric rings, lacking aerial mycelium, margin entire to filamentous; reverse pale luteous without diffusible pigments.
Typus: Zimbabwe, Matabeleland North, Victoria Falls area, from soil, Apr. 1996, C. Decock, isol. number 51V (holotype CBS H-24945, culture ex-type MUCL 58143 = CBS 148934).
Additional material examined: Zimbabwe, Matabeleland North, Victoria Falls area, from soil, Apr. 1996, C. Decock, isol. number 52V, culture MUCL 58144 = CBS 148935.
Notes: Fusicolla elongata produces characteristic long 3–5-septate conidia. Other Fusicolla species producing conidia with similar septation include Fu. acetilerea, Fu. violacea. However, Fu. elongata forms exceptionally long conidia which distinguishes this species from every other known species in the genus. Fusicolla elongata can be further distinguished from Fu. acetilerea by the lack of chlamydospores in the former species. Additionally, while both Fu. acetilerea and Fu. violacea have brownish to dark red-brown colony pigmentation, colonies of Fu. elongata are consistently orange to salmon coloured (Gerlach & Nirenberg 1982).
Fusicolla gigas Chang Liu, Z.Q. Zeng & W.Y. Zhuang, sp. nov. MycoBank MB 844496.
Etymology: Name refers to the large-sized macroconidia produced by this species.
Holotypus: CGMCC 3.20680 (permanently preserved in a metabolically inactive state).
Ex-type culture: CGMCC 3.20680.
Type locality: China, Chongqing City, Wushan County, Hongchiba National Forest Park.
Type substrate: Isolated from soil.
Description and illustration: Liu et al. (Phytotaxa 536: 167. 2022).
Diagnostic features: Colonies orange to pale yellow with orange margin and slimy appearance on PDA; aerial monophialides giving rise to micro- and macroconidia; microconidia aseptate, slightly to markedly curved; macroconidia falcate to long-fusiform, (1–)3(–4)-septate, with a hooked apical cell and foot-shaped basal cell; chlamydospores and sexual morph not observed (Liu et al. 2022).
Fusicolla guangxiensis Z.Q. Zeng, C. Liu & W.Y. Zhuang, sp. nov. MycoBank MB 844497.
Etymology: Name refers to the type locality of the type specimen.
Holotypus: CGMCC 3.20679 (permanently preserved in a metabolically inactive state).
Ex-type culture: CGMCC 3.20679.
Type locality: China, Guangxi autonomous region, Fangchenggang City, Shangsi County, Shiwandashan National Forest Park.
Type substrate: Isolated from an unidentified rotten twig.
Description and illustration: Liu et al. (Phytotaxa 536: 169. 2022).
Diagnostic features: Colonies orange with pale luteous margin and slimy appearance on PDA; aerial monophialides giving rise to macroconidia; macroconidia falcate to long-fusiform, (0–)1(–3)-septate, with an acute to hooked apical cell and an acute, non-pedicellate basal cell; microconidia, chlamydospores and sexual morph not observed (Liu et al. 2022).
Notes: Fusicolla gigas and Fu. guangxiensis were invalidly published because the protologue did not explicitly mention the holotypes were preserved in a metabolically inactive state [Art. 40.8 (Shenzhen)]. Both species are validated here.
Longinectria O. Savary, M. Coton, E. Coton & J-L. Jany, gen. nov. MycoBank MB 844395.
Etymology: From the Latin longus = long, “Longi-” refers to the phialides length observed for the Longinectria species and “-nectria” refers to the Nectriaceae family.
Ascomatal morph unknown. Conidiophores with variable-length phialides, sometimes extremely long (e.g. 153–237 μm), lateral, sometimes verticillate, hyaline. Macroconidia straight to slightly curved, apical cell morphology blunt to papillate and a basal cell often notched, 0–3-septate, hyaline. Microconidia ovoid, ellipsoid to allantoid, 0–1 septate, hyaline. Chlamydospores absent to abundant, globose, single, in pairs or chains, intercalary or terminal (from Savary et al. 2021).
Type species: Longinectria lagenoides O. Savary, M. Coton, E. Coton & J-L. Jany
Notes: The genus Longinectria, together with its two known species, L. lagenoides and L. verticilliformis, were invalidly published as two numbers were cited as holotypes for each species [Art. 40.7, 40.8 (Shenzhen)] (Savary et al. 2021). The names were subsequently published in Index Fungorum, but as the type species of the genus was not indicated, the genus was still not validly published [Art. 40.1 (Shenzhen)], and the species also rendered invalid [Art. 35.1 (Shenzhen)]. The genus and species are thus validated here.
Longinectria lagenoides O. Savary, M. Coton, E. Coton & J-L. Jany, sp. nov. MycoBank MB 844396.
Holotypus: UBOCC-A-120039 (permanently preserved in a metabolically inactive state).
Ex-type culture: UBOCC-A-120039 = CBS 147588.
Type locality: France.
Type substrate: Isolated from Swiss cheese.
Description and illustration: Savary et al. (Mycosphere 12: 1089. 2021) .
Etymology: From Latin lagoena = bottle, refers to the observed phialide shape.
Diagnostic features: Colonies brown with folded surface and brown pigmentation and powdery aerial mycelium (sporulation) on PDA, growing between 5 and 25 °C, having optimal growth at 20 °C; aerial monophialides giving rise to micro- and macroconidia; monophialides extremely long or reduced to conidiogenous pegs on hyphae; microconidia 0–1-septate, ovoid to allantoid; macroconidia straight, 0–3-septate, apical cell blunt to papillate, and poorly-developed, foot-shaped basal cell; chlamydospores globose, typically intercalary, or terminal, two or more. No known mycotoxins already described to be produced by Fusarium, Penicillium, Aspergillus or Alternaria spp. were detected (Savary et al. 2021).
Longinectria verticilliformis O. Savary, M. Coton, E. Coton & J-L. Jany, sp. nov. MycoBank MB 844397.
Etymology: Name refers to the subverticillate arrangement of phialides.
Holotypus: UBOCC-A-120043 (permanently preserved in a metabolically inactive state).
Ex-type culture: UBOCC-A-120043 = CBS 147589.
Type locality: France.
Type substrate: Isolated from an Italian cheese (Alpeggio).
Description and illustration: Savary et al. (Mycosphere 12: 1091. 2021).
Diagnostic features: Colonies white to white grey with powdery to cottony aerial mycelium on PDA, growing between 5 and 25 °C, with optimal growth at 20 °C; aerial monophialides giving rise to micro- and macroconidia; microconidia 0–1-septate, straight or curved, reniform; macroconidia straight, ellipsoidal, 1–3-septate, with a blunt to papillate apical cell and foot-shaped basal cell; chlamydospores not observed. No known mycotoxins already described to be produced by Fusarium, Penicillium, Aspergillus or Alternaria spp. were detected (Savary et al. 2021).
Neocosmospora akasia (Lynn & I. Barnes) Crous & Sand.-Den., comb. nov. MycoBank MB 843501.
Basionym: Fusarium akasia Lynn & I. Barnes, Mycologia 113: 544. 2021. MB 834436.
Holotypus: PREM 62607; paratypi PREM 62608 and PREM 62609.
Ex-type culture: CBS 146880 = CMW 54735 = PPRI 27978; ex-paratype cultures CBS 146881 = CMW 54741 = PPRI 27979 and CBS 147161 = CMW 54752 = PPRI 27980.
Type locality: Indonesia, Riau, Pelalawan.
Type substrate: From head (including mycangium) of Euwallacea perbrevis (TSHBa) in stems of Acacia crassicarpa.
Description and illustrations: Lynn et al. (2021).
Diagnostic features: Colony surface white, buff to saffron or fulvous in dark, buff to honey darkening to red, blood red in ambient daylight, reverse yellowish white to buff ,darkening to isabelline or cinnamon in the dark, saffron to orange, darkening to rust and blood red after 1 mo in ambient daylight on PDA, having optimal growth at 30 °C; aerial monophialides giving rise to microconidia in false heads, ovoid to obovoid, slightly curved, 0–1(–2)-septate; sporodochia buff to pale orange, with monophialides give rise to slightly curved, clavate, with ridged appearance, (0–)1–4(–5)-septate macroconidia with a blunt apical cell and obtuse to poorly-developed, foot-shaped basal cell; chlamydospores sparse, in hyphae and conidia, single or in pairs (Lynn et al. 2021).
Notes: Neocosmospora akasia is associated with the ambrosia beetles, Euwallacea perbrevis and E. similis in plantations of Acacia crassicarpa in Indonesia. It is characterized by clavate conidia which are slightly constricted at the septa, giving it a ridged appearance, and having arched, thick aerial conidiophores that taper slightly at the base (Lynn et al. 2021).
Neocosmospora awan (Lynn & I. Barnes) Crous & Sand.-Den., comb. nov. MycoBank MB 843502.
Basionym: Fusarium awan Lynn & I. Barnes, Mycologia 113: 544. 2021. MB 834437.
Holotypus: PREM 62602; paratypi PREM 62594 and PREM 62604.
Ex-type culture: CBS 146882 = CMW 54719 = PPRI 27973; ex-paratype cultures CBS 146883 = CMW 53705 = PPRI 27957 and CBS 146884 = CMW 54722 = PPRI 27975.
Type locality: Indonesia, Riau, Pelalawan.
Type substrate: From head (including mycangium) of Euwallacea similis in stems of Acacia crassicarpa.
Description and illustrations: Lynn et al. (2021).
Diagnostic features: Colony surface colour white in the dark, white darkening to honey after 1 mo in ambient daylight, in reverse yellowish white to buff in the dark, buff darkening to ochreous after 1 mo in ambient daylight on PDA, having optimal growth at 30 °C; aerial monophialides giving rise to microconidia in false heads, ovoid, 0–1(–2)-septate, and flute-shaped, 1–3-septate macroconidia; sporodochia luteous to ochreous, with monophialides giving rise to curved, cylindrical or slightly clavate or flute-shaped, (0–)2–3(–4)-septate macroconidia with a narrowly papillate to blunt apical cell and obtuse to poorly-developed, foot-shaped basal cell; chlamydospores abundant, intercalary and terminal in hyphae and conidia, single, in pairs or chains (Lynn et al. 2021).
Notes: Neocosmospora awan is associated with ambrosia beetles, Euwallacea perbrevis and E. similis, in plantations of Acacia crassicarpa in Indonesia. It is characterised by having abundant chlamydospores that form in hyphae and mature conidia, having multiseptate aerial macroconidia that are elongated-ovoid in shape, and very narrow sporodochial macroconidia. Furthermore, phylogenetically it groups separate from the Ambrosia Clade within Neocosmospora.
Neocosmospora brevis Sand.-Den. & Crous, Persoonia 43: 119. 2019. MB 831176.
Synonym: Fusarium breve (Sand.-Den. & Crous) O’Donnell et al., Index Fungorum 440: 1. 2020. MB 557673.
New synonym: Fusarium rosicola Lin Huang et al., Pl. Pathol. 70: 2065. 2021. MB 839201.
Holotypus: CBS H-23975.
Ex-type culture: CBS 144387 = MUCL 16108.
Type locality: Belgium, Heverlee.
Type substrate: Soil-water polluted with diethylene glycerol and ethylene glycerol.
Description and illustrations: Sandoval-Denis et al. (2019), He et al. (2021).
Diagnostic features: Colony surface orange to saffron or pale yellow, reverse orange, luteous to amber to pale yellow on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia in false heads, oval, ellipsoidal to subclavate, straight or slightly curved, 0–1(–2)-septate; sporodochia with monophialides give rise to falcate, slightly dorsiventrally curved, 3–5-septate macroconidia, apical cell blunt and rounded, basal cell without a well-developed foot-shaped basal cell; chlamydospores abundant, globose to subglobose, terminal or intercalary on hyphae or conidia, solitary or in chains (Sandoval-Denis et al. 2019).
Notes: Fusarium rosicola was described as a pathogen of Chinese rose (Rosa chinensis) (He et al. 2021). Apparent morphological and physiological differences with its closest relative, N. brevis, in their phylogenetic analysis were not supported in our analysis (Fig. 3). We attribute these differences to intraspecific variability in N. brevis.
Neocosmospora drepaniformis (T. Aoki et al.) Crous & Sand.-Den., comb. nov. MycoBank MB 843503.
Basionym: Fusarium drepaniforme T. Aoki et al., Mycologia 113: 1098. 2021. MB 558018.
Holotypus: BPI 923530 (dried culture), isotypus IMI 351954.
Ex-type culture: NRRL 62941 (= KOD 147) = MAFF 247230.
Type locality: Singapore.
Type substrate: Unknown woody host.
Description and illustrations: Aoki et al. (2021).
Diagnostic features: Colony surface white, yellowish white to pale yellow, becoming pale orange, light orange to greyish orange with age, reverse yellowish white or pale yellow to greyish yellow on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia in false heads, ellipsoidal, oblong-ellipsoidal, fusoid-ellipsoidal to clavate, straight or sometimes curved and reniform or crescent-shaped, some obovate to comma-shaped, 0–1(–3)-septate; sporodochia sparse, with monophialides give rise to clavate and straight (in the dark), to falcate (under nuv-light), (0–)3–7-septate macroconidia, with a papillate apical cell and poorly to well-developed, foot-shaped basal cell; chlamydospores intercalary and terminal in hyphae and conidia, single, in chains or small clusters (Aoki et al. 2021).
Notes: Neocosmospora drepaniformis was originally deposited as “F. bugnicourtii” (on Camellia sinensis: West Bengal) based on IMI 351954. It is characterised by forming multiseptate curved conidia, especially under nuv-light. Some conidia become swollen in the apical part, appearing wedge-shaped (Aoki et al. 2021).
Neocosmospora duplosperma (T. Aoki et al.) Crous & Sand.-Den., comb. nov. MycoBank MB 843504.
Basionym: Fusarium duplospermum T. Aoki et al., Mycologia 113: 1091. 2021. MB 558017.
Holotypus: BPI 923529 (dried culture).
Ex-type culture: NRRL 62583 = MAFF 247220.
Type locality: USA, Florida, Miami-Dade County, Homestead.
Type substrate: From the oral mycangium of Euwallacea perbrevis trapped in a Persea americana grove.
Description and illustrations: Aoki et al. (2021).
Diagnostic features: Colony surface white, yellowish white, pale yellow, light yellow to greyish yellow, becoming pale orange to greyish orange, or reddish white to pale red, reddish grey to greyish red with age in the dark, reverse pigment absent or yellowish white, pale yellow to light yellow, some greyish orange, brownish orange to yellowish brown or brown, sometimes with yellowish pigments in the agar on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia in false heads, ellipsoid, oblong-ellipsoid, fusoid-ellipsoid to short-clavate, straight or sometimes curved, reniform or crescent-shaped, some obovate to comma-shaped, 0–1-septate; sporodochia with monophialides give rise to two distinct conidial types, i) short-clavate to obovate or naviculate, straight or curved, with obtuse apex and truncate base, 0–1(–2)-septate, and ii) straight or curved, wedge-shaped, (1–)3–5(–7)-septate, swollen in the apical region, with a tapering apical cell, base with a poorly to well-developed, foot-shaped basal cell; chlamydospores delayed, intercalary and terminal in hyphae and conidia, single or in chains (Aoki et al. 2021).
Notes: Neocosmospora duplosperma can be distinguished by forming two morphologically distinct types of multiseptate conidia, namely (i) long, slender, and falcate, or (ii) relatively short, apically swollen, curved and wedge-shaped (“dolphin-like”). Furthermore, N. duplosperma is characterised by forming brownish orange colonies on PDA, which differs other species in the Neocosmospora Ambrosia Clade, which typically produce whitish, yellowish, or greyish coloured colonies on PDA (Aoki et al. 2021).
Neocosmospora geoasparagicola Sand.-Den., Crous, de Boer, Katschnig & W. Jonkers, sp. nov. MycoBank MB 843505. Fig. 15.
Fig. 15.

Neocosmospora geoasparagicola (CBS 148937 ex-type). A–D. Sporodochia formed on the surface of carnation leaves. E–H. Aerial conidiophores and conidiogenous cells. I–K. Sporodochial conidiophores and conidiogenous cells. L. Conidia. Scale bars: B–D = 20 μm; J = 5 μm; all others = 10 μm.
Etymology: Named after the substrate from which all the original specimens were collected: soil from Asparagus officinalis fields.
Conidiophores erect or prostrate, borne on the agar substrate and aerial mycelium, 45–190 μm tall, simple or branched laterally and sympodially, bearing terminal single phialides; aerial conidiogenous cells monophialidic, subulate to subcylindrical, smooth- and thin-walled, 21–61 × 2.5–5 μm, with short and flared apical collarettes, periclinal thickening inconspicuous or absent, rarely proliferating laterally and apically. Aerial conidia falcate, smooth- and thick-walled, gently dorsiventrally curved, robust, with a blunt, slightly curved apical cell, basal cell obtuse to poorly-developed, foot-shaped, undistinguishable in shape from sporodochial conidia, 3–4(–5)-septate, predominantly 3-septate, 3-septate conidia: (37–)39–50(–56.5) × (4–)5–6.5 μm (av. 44 × 5.3 μm); 4-septate conidia: (49–)51–63(–67.5) × 5.5–7 μm (av. 56.5 × 6 μm); 5-septate conidia: 54.5 × 5.5 μm (only one element observed); overall: (37–)39–54(–67.5) × 4.5–6.5 μm (av. 46.6 × 5.4 μm), borne at the tip of monophialides and accumulating forming elongated false-heads. Sporodochia pale luteous to pale orange, formed on aerial and substrate mycelium, and on the surface of carnation leaves. Sporodochial conidiophores simple or laterally and irregularly branched bearing terminal monophialides or groups of 2–4 monophialides; sporodochial conidiogenous cells monophialidic, doliiform, subulate to subcylindrical, (13–)14.5–22(–31) × 3.5–6 μm, smooth and thin-walled, with a vasiform apical collarette and inconspicuous to absent periclinal thickening. Sporodochial conidia falcate, gently dorsiventrally curved, robust, with a blunt, slightly curved apical cell, basal cell obtuse to poorly-developed, foot-shaped, 3–5-septate, predominantly 4-septate, hyaline, smooth- and thick-walled; 3-septate conidia: (43.5–)47–55(–60) × 5–7 μm (av. 51 × 6 μm); 4-septate conidia: (46–)52–60(–63) × 5–7 μm (av. 56.1 × 6 μm); 5-septate conidia: (52.5–)55–64(–68) × 5–7 μm (av. 59.2 × 6.1 μm); overall: (43.5–)52–61(–68) × 5–7 μm (av. 56.6 × 6 μm). Chlamydospores and sexual form not observed.
Culture characteristics: Colonies at 25 °C after 7 d: On PDA reaching 38–43 mm diam, white to pale buff, pale vinaceous buff at periphery, flat, dusty to felty with or without cottony patches or concentric rings of short aerial mycelium, membranous at periphery, margin entire to slightly filamentous; reverse white to pale buff, ochreous to umber at centre, without diffusible pigments. On SNA reaching 36–42 mm diam, white to pale buff, flat, membranous to dusty at centre, aerial mycelium scarce; reverse white, without diffusible pigments. On OA reaching 40–48 mm diam, white to pale buff, flat, felty, with concentric rings of short, white aerial mycelium, margin entire to slightly lobate; reverse pale buff without diffusible pigments.
Typus: Netherlands, Limburg, Kessel, from field soil cultured with Asparagus officinalis ‘Guelph Millennium’ field, 19 Nov. 2020, M. Sandoval-Denis & L. Lombard (holotype CBS H-24947, culture ex-type CBS 148937 = CPC 40592).
Additional material examined: Netherlands, Limburg, Kessel, from field soil cultured with Asparagus officinalis field, 2019, W. de Boer (cultures CBS 148936 = CPC 39928, 39931, 39932); from field soil cultured with Asparagus officinalis ‘Cygnus’ field, 13 Nov. 2020, M. Sandoval-Denis & L. Lombard (culture CPC 40579); from field soil cultured with A. officinalis ‘Grolim’ field. 13 Nov. 2020, M. Sandoval-Denis & L. Lombard (culture CPC 40571); from field soil cultured with A. officinalis ‘Schneekopf’ field, 13 Nov. 2020, M. Sandoval-Denis & L. Lombard (culture CPC 40628).
Notes: Neocosmospora geoasparagicola was isolated from soil from several Asparagus officinalis experimental fields (Bejo Zaden, Kessel, Limburg, Netherlands) where diverse Asparagus varieties have been cultivated. Neocosmospora geoasparagicola nested within Clade 2 of Neocosmospora, which contains mostly species from Asia and the Americas, including N. phaseoli, an important root pathogen of Fabaceae (O’Donnell 2000, Nalim et al. 2011, Sandoval-Denis et al. 2019). Subsequent pathogenicity testing, however, showed that N. geoasparagicola is not a pathogen of A. officinalis (data not shown).
Species in Neocosmospora Clade 2 are characterised by forming often large multiseptate macroconidia from aerial and sporodochial phialides, while generally lacking microconidia. While consistent with general morphological features of taxa in Clade 2, N. geoasparagicola clustered basally, and clearly separated phylogenetically and biogeographically from the remaining species in this group. Morphologically, N. geoasparagicola is most similar to N. cryptoseptata and N. nirenbergiana. Neocosmospora geoasparagicola can be differentiated from N. cryptoseptata by it slightly longer conidia and sporodochial phialides. There is considerable morphological overlap between N. geoasparagicola and N. nirenbergiana. However, sporodochial conidia of N. geoasparagicola, which are indistinguishable from aerial macroconidia, are shorter and tend to present longer apical cells than those of N. nirenbergiana. By contrast, aerial conidia of N. nirenbergiana are considerably different from its sporodochial counterparts, being shorter and somewhat pointy. Additionally, N. geoasparagicola lacks reddish pigments, a feature commonly observed in N. nirenbergiana.
Neocosmospora mekan (Lynn & I. Barnes) Crous & Sand.-Den., comb. nov. MycoBank MB 843506.
Basionym: Fusarium mekan Lynn & I. Barnes, Mycologia 113: 547. 2021. MB 834438.
Holotypus: PREM 62600; isotypi PREM 62601 and PREM 62602.
Ex-type culture: CBS 146885 = CMW 54714 = PPRI 27971; ex-paratype cultures CBS 146886 = CMW 53696 = PPRI 27956 and CBS 146887 = CMW 54717 = PPRI 27972.
Type locality: Indonesia, Riau, Pelalawan.
Type substrate: From head (including mycangium) of Euwallacea similis in stems of Acacia crassicarpa.
Description and illustrations: Lynn et al. (2021).
Diagnostic features: Colony surface white, greyish flax blue to greyish violet in the dark, white to pale mouse grey darkening to purple slate and rust after 1 mo in ambient daylight, reverse yellowish white to fawn in the dark, bay darkening to chestnut and blood red after 1 mo in ambient daylight on PDA, having optimal growth at 30 °C; aerial monophialides giving rise to microconidia in false heads, ovoid to obovoid, rarely pyriform, 0–1(–2)-septate, aerial macroconidia long ovoid, apex blunt, basal cell obtuse, 0–3(–4)-septate; sporodochia luteous to ochreous, with monophialides give rise to straight or slightly curved, sub-fusoid, widest in apical third, wedge-shaped, 0–5(–6)-septate macroconidia with a blunt apical cell and obtuse to poorly-developed, foot-shaped basal cell; chlamydospores abundant, intercalary and terminal in hyphae and conidia, single, in pairs or chains, rarely in clusters (Lynn et al. 2021).
Notes: Neocosmospora mekan is associated with Euwallacea perbrevis and E. similis beetles in plantations of Acacia crassicarpa in Indonesia. It is distinguished by its multiseptate (evenly spaced), slightly curved, elongate, subfusoid to wedge-shaped macroconidia, and chlamydospores that tend to form at both the apex and base of mature macroconidia (Lynn et al. 2021).
Neocosmospora papillata (T. Aoki et al.) Crous & Sand.-Den., comb. nov. MycoBank MB 843507.
Basionym: Fusarium papillatum T. Aoki et al., Mycologia 113: 1097. 2021. MB 558019.
Holotypus: BPI 923531 (dried culture).
Ex-type culture: NRRL 62943 (= KOD 796) = MAFF 247228.
Type locality: Sri Lanka, Central Province, Kandy.
Type substrate: From the mycangium of a living female Euwallacea perbrevis beetle from a gallery in a branch of infested Camellia sinensis bush.
Description and illustrations: Aoki et al. (2021).
Diagnostic features: Colony surface white, yellow white to pale yellow, orange white initially, becoming partly pale orange to greyish orange in the dark, reverse pale yellow to light yellow, or greyish yellow on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia in false heads, oblong-ellipsoid, fusoid-ellipsoid to clavate, straight or crescent- or comma-shaped, also sometimes forming swollen clavate to falcate, straight or curved conidia, 0–1(–3)-septate; sporodochia with monophialides give rise clavate to falcate, often gently curved, sometimes crescent-shaped (0–)3–7(–8)-septate macroconidia, often swollen in their upper parts with a papillate apical cell (protrude ventrally), with poorly to well-developed, foot-shaped basal cell; chlamydospores intercalary and terminal in hyphae and conidia, single or in chains (Aoki et al. 2021).
Notes: Neocosmospora papillata frequently forms multiseptate clavate conidia with papillate apical cells that protrude ventrally, especially under nuv-light, which distinguishes it from other species in the Neocosmospora Ambrosia Clade. Morphologically it resembles N. drepaniformis, but is distinct in that macroconidia often possess a papillum protruding ventrally from the apical cells, and their ultimate and penultimate apical cells are often swollen so that they are widest in the terminal half. Macroconidia of N. drepaniformis, however, are often widest at the second to fourth cells from the apex (Aoki et al. 2021).
Neocosmospora variasi (Lynn & I. Barnes) Crous & Sand.-Den., comb. nov. MycoBank MB 843508.
Basionym: Fusarium variasi Lynn & I. Barnes, Mycologia 113: 549. 2021. MB 834439.
Holotypus: PREM 62595; paratypi PREM 62596 and PREM 62597.
Ex-type culture: CBS 146888 = CMW 53734 = PPRI 27958; ex-paratype cultures CBS 146889 = CMW 53735 = PPRI 27959 and CBS 146890 = CMW 54696 = PPRI 27968.
Type locality: Indonesia, Riau, Pelalawan.
Type substrate: From Euwallacea perbrevis in stems of Acacia crassicarpa.
Description and illustrations: Lynn et al. (2021).
Diagnostic features: Colony surface white or livid purple to fawn in the dark, with white to livid purple to bay segments, darkening to dark brick or violate slate or black after 1 mo in ambient daylight, reverse yellowish white to fawn in the dark, with white with rust to umber segments, occasionally entirely darkening to umber or black after 1 mo in ambient daylight on PDA, having optimal growth at 30 °C; aerial monophialides giving rise to microconidia in false heads, ovoid to obovoid, or short-clavate, curved, 0–1(–2)-septate; sporodochia luteous to ochreous or dull green to dark violet, with monophialides that give rise to falcate to clavate, 3–6(–7)-septate macroconidia with a papillate apical cell and poorly to well-developed, foot-shaped basal cell; chlamydospores abundant, intercalary and terminal in hyphae and conidia, single, in pairs, chains or often in clusters (Lynn et al. 2021).
Notes: Neocosmospora variasi is associated with the ambrosia beetle, Euwallacea perbrevis, in plantations of Acacia crassicarpa in Indonesia. It is characterised by having aerial micro- and macroconidia, which vary in size and shape. Furthermore, it produces abundant chlamydospores in clusters, which is unusual for species in the Ambrosia Clade of Neocosmospora. Lynn et al. (2021) were also of the opinion that as presently defined, N. variasi might represent two cryptic taxa.
Neocosmospora warna (Lynn & I. Barnes) Crous & Sand.-Den., comb. nov. MycoBank MB 843509.
Basionym: Fusarium warna Lynn & I. Barnes, Mycologia 113: 551. 2021. MB 834440.
Holotypus: PREM 62603; paratypi PREM 62605 and PREM 62606.
Ex-type culture: CBS 146891 = CMW 54720 = PPRI 27974; ex-paratype cultures CBS 146892 = CMW 54724 = PPRI 27976 and CBS 146893 = CMW 54726 = PPRI 27977.
Type locality: Indonesia, Riau, Pelalawan.
Type substrate: From head (including mycangium) of Euwallacea perbrevis in stems of Acacia crassicarpa.
Description and illustrations: Lynn et al. (2021).
Diagnostic features: Colony surface white to livid purple to vinaceous purple, with white segments, to fawn at margins in the dark, lavender to violet or livid violet with white segments, darkening to livid vinaceous or dark vinaceous to dark purple with sepia margins after 1 mo in ambient daylight, reverse yellowish white to fawn in the dark, pale vinaceous grey white with rust to umber, darkening to dark brick after 1 mo in ambient daylight on PDA, having optimal growth at 25 °C; aerial monophialides giving rise to microconidia in false heads, obovoid to ovoid to short-clavate, rarely curved, 0–3(–4-)septate; sporodochia luteous to ochreous, or dull green to sepia, with monophialides giving rise to short-clavate, wedge-shaped (widest at apical septum), 1–4(–6)-septate macroconidia with a papillate apical cell and obtuse basal cell; chlamydospores sparse, intercalary and terminal in hyphae and conidia, single, in pairs, often clusters (Lynn et al. 2021).
Notes: Neocosmospora warna is associated with Euwallacea perbrevis beetles in plantations of Acacia crassicarpa in Indonesia. It is characterised by multi-septate, thick, short-clavate, wedge-shaped (widest at apical septum), papillate sporodochial conidia that taper toward the obtuse basal cell, and small chlamydospores (Lynn et al. 2021).
Genome announcements
Other than providing illustrations, diagnoses and multilocus phylogenies of fusarioid taxa, a further aim of the FUSA series is to also provide access to genome data of newly sequenced species, the first of which are published here.
The assemblies of Fusarium secorum (CBS 175.32), Microcera coccophila (CBS 310.34), Rectifusarium robinianum (CBS 430.91), Rugonectria rugulosa (CBS 126565), and Thelonectria blattea (CBS 952.68) are announced here. They were obtained from high coverage Illumina data (168–283×). Quality assessment done with BUSCO against 758 genes from the library for Fungi showed a high completeness (> 98 %) and a low duplication level (< 1 %) for the analysed genomes. The genome sizes varied from 34.7 Mbp to 50.5 Mbp. Assemblies of R. rugulosa, M. coccophila, R. robinianum, and T. blattea showed similar number of scaffolds while in the F. secorum genome their amount was significantly increased due to a high number (> 10 k) of scaffolds with sizes smaller than < 1 kbp. The total number of annotated gene models varied from 24 411 in M. coccophila to 46 001 in F. secorum. All assemblies were deposited in GenBank, detailed statistics and BioProject numbers are shown in Table 3.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
REFERENCES
- Adamčík S, Cai L, Chakraborty D, et al. (2015). Fungal Biodiversity Profiles 1–10. Cryptogamie, Mycologie 36: 121–166. [Google Scholar]
- Ahmad A, Akram W, Shahzadi I, et al. (2020). First report of Fusarium nelsonii causing early-stage fruit blight of cucumber in Guangzhou, China. Plant Disease 104: 1542. [Google Scholar]
- Aoki T, Liyanage PNH, Konkol JL, et al. (2021). Three novel Ambrosia Fusarium Clade species producing multiseptate “dolphin-shaped” conidia, and an augmented description of Fusarium kuroshium. Mycologia 113: 1089–1109. [DOI] [PubMed] [Google Scholar]
- Aoki T, O’Donnell K, Geiser D. (2014). Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201. [Google Scholar]
- Balmas V, Migheli Q, Scherm B, et al. (2010). Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 102: 803–812. [DOI] [PubMed] [Google Scholar]
- Benyon FHL, Burgess LW, Sharp PJ. (2000). Molecular genetic investigations and reclassification of Fusarium species in sections Fusarium and Roseum. Mycological Research 104: 1164–1174. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C. (1971). The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, England. [Google Scholar]
- Bushnell B. (2021). BBMap short read aligner. http://sourceforge.net/projects/bbmap/ [Google Scholar]
- Burgess LW, Liddell CM, Summerell BA. (1994). Laboratory manual for Fusarium research, 3rd edn. University of Sydney, Sydney. [Google Scholar]
- Burgess LW, Summerell BA. (1992). Mycogeography of Fusarium: survey of Fusarium species in subtropical and semi-arid grassland soils from Queensland, Australia. Mycological Research 96: 780–784. [Google Scholar]
- Cantarel BL, Korf I, Robb SM, et al. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Research 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehri K, Salleh B, Yli-Mattila T, et al. (2010). Occurrence, pathogenicity and distribution of Fusarium spp. in stored wheat seeds Kermanshah Province, Iran. Pakistan Journal of Biological Sciences 13: 1178–1186. [DOI] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. (2021a). Fungal Planet description sheets: 1182–1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, et al. (2011). Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. (2021b). Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Giraldo A, Hawksworth D, et al. (2014). The Genera of Fungi: fixing the application of type species of generic names. IMA Fungus 5: 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. (2015). Fungal Systematics and Evolution: FUSE 1. Sydowia 67: 81–118. [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. (2018). New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YX, Chen FR, Shi NN, et al. (2017). First report of Fusarium chlamydosporum causing banana crown rot in Fujian Province, China. Plant Disease 101: 1048. [Google Scholar]
- Engelbrecht MC, Smit WA, Knox-Davies PS. (1983). Damping-off of rooibos tea, Aspalathus linearis. Phytophylactica 15: 121–124. [Google Scholar]
- Fugro PA. (1999). A new disease of okra (Abelmoschus esculentus L.) in India. Journal of Mycology and Plant Pathology 29: 264. [Google Scholar]
- Gerlach W, Nirenberg H. (1982). The genus Fusarium – a pictorial atlas. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406. [Google Scholar]
- Gerlach W, Scharif G. (1970). Erreger einer Fusskrankheit an Hibiscus cannabinus in Iran – Fusarium bucharicum Jaczewski. Phytopathologische Zeitschrift 68: 323–333. [Google Scholar]
- He J, Li DW, Zhang Y, et al. (2021). Fusarium rosicola sp. nov. causing vascular wilt on Rosa chinensis. Plant Pathology 70: 2062–2073. [Google Scholar]
- Hill R, Buggs RJA, Vu DT, et al. (2022). Lifestyle transitions in fusarioid fungi are frequent and lack clear genomic signatures. Molecular Biology and Evolution 39: msac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Venter A, Laraba I, Geiser DM, et al. (2018). Molecular systematics of two sister clades, the Fusarium concolor and F. babinda species complexes, and the discovery of a novel microcycle macroconidium–producing species from South Africa. Mycologia 110: 1189–1204. [DOI] [PubMed] [Google Scholar]
- Kanaan YM, Bahkali AH. (1993). Frequency and cellulolytic activity of seed-borne Fusarium species isolated from Sausi Arabian cereal cultivars. Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz 100: 291–298. [Google Scholar]
- Kasson MT, O’Donnell K, Rooney AP, et al. (2013). An inordinate fondness for Fusarium: Phylogenetic diversity of fusaria cultivated by ambrosia beetles in the genus Euwallacea on avocado and other plant hosts. Fungal Genetics and Biology 56: 147–157. [DOI] [PubMed] [Google Scholar]
- Kirk PM, Stalpers JA, Braun U, et al. (2013). A without-prejudice list of generic names of fungi for protection under the International Code of Nomenclature for algae, fungi and plants. IMA Fungus 4: 381–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J, Doehlemann G, Kemen E, et al. (2017). Asexual and sexual morphs of Moesziomyces revisited. IMA Fungus 8: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laraba I, Keddad A, Boureghda , et al. (2017). Fusarium algeriense sp. nov., a novel toxigenic crown rot pathogen of durum wheat from Algeria is nested in the Fusarium burgessii species complex. Mycologia 109: 935–950. [DOI] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, et al. (2011). Fusarium burgessii sp. nov. representing a novel lineage in the genus Fusarium. Fungal Diversity 49: 101–112. [Google Scholar]
- Laurence MH, Walsh JL, Shuttleworth LA, et al. (2016). Six novel species of Fusarium from natural ecosystems in Australia. Fungal Diversity 77: 349–366. [Google Scholar]
- Lazreg F, Belabid L, Sanchez J, et al. (2013). First report of Fusarium chlamydosporum causing damping-off disease on Aleppo pine in Algeria. Plant Disease 97: 1506. [DOI] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. (2006). The Fusarium laboratory manual. Blackwell Publishing Professional, USA. [Google Scholar]
- Leslie JF, Summerell BA. (2011). In search of new Fusarium species. Plant Breeding and Seed Science 63: 94–101. [Google Scholar]
- Lincy SV, Chandrashekar A, Narayan MS, et al. (2011). Natural occurrence of trichothecene-producing Fusaria isolated from India with particular reference to sorghum. World Journal of Microbiology and Biotechnology 27: 981–989. [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. [Google Scholar]
- Liu C, Zhuang WY, Yu ZH, et al. (2022). Two new species of Fusicolla (Hypocreales) from China. Phytotaxa 536: 165–174. [Google Scholar]
- Lombard L, van der Merwe NA, Groenewald JZ, et al. (2015). Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Doorn R, Crous PW. (2019). Neotypification of Fusarium chlamydosporum - a reappraisal of a clinically important species complex. Fungal Systematics and Evolution 4: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn KMT, Wingfield MJ, Durán A, et al. (2021). Novel Fusarium mutualists of two Euwallacea species infesting Acacia crassicarpa in Indonesia. Mycologia 113: 536–558. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. (2021). BUSCO Update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular Biology and Evolution 38: 4647–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA, et al. (1986). Fusarium polyphialidicum, a new species from South Africa. Mycologia 78: 678–682. [Google Scholar]
- Marasas WFO, Rheeder JP, Logrieco A, et al. (1998). Fusarium nelsonii and F. musarum: two new species in section Arthrosporiella related to F. camptoceras. Mycologia 90: 505–513. [Google Scholar]
- Marin-Felix Y, Groenewald JZ, Cai L, et al. (2017). Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology 86: 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. (2019). New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh Q, Schmidt HA, Chernomor O, et al. (2020). IQ-TREE2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Gupta DK, Pfenninger M, et al. (2018). A reference genome of the European beech (Fagus sylvatica L.). GigaScience 7: giy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Nor NMI, Salleh B, Leslie JF. (2019). Fusarium species from Sorghum in Thailand. The Plant Pathology Journal 35: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalim FA, Samuels GJ, Wijesundera RL, et al. (2011). New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia 103: 1302–1330. [DOI] [PubMed] [Google Scholar]
- Nelson PE, Dignani MC, Anaissie EJ. (1994). Taxonomy, biology, and clinical aspects of Fusarium species. Clinical Microbiology Reviews 7: 479–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PE, Toussoun TA, Burgess LW. (1987). Characterization of Fusarium beomiforme sp. nov. Mycologia 79: 884–889. [Google Scholar]
- Nelson PE, Toussoun TA, Marasas WFO. (1995). Neotypification and emended description of Fusarium anguioides. Mycologia 87: 543–546. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, et al. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K. (2000). Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92: 919–938. [Google Scholar]
- O’Donnell K, Gräfenhan T, Laraba I, et al. (2022). Fusarium abutilonis and F. guadeloupense, two novel species in the Fusarium buharicum clade supported by multilocus molecular phylogenetic analyses. Mycologia DOI: 10.1080/00275514.2022.2071563. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Gueidan C, Sink S, et al. (2009). A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genetics and Biology 46: 936–948. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, McCormick SP, Busman M, et al. (2018). Marasas et al. 1984 “Toxigenic Fusarium species: Identity and mycotoxicology” revisited. Mycologia 110: 1058–1080. [DOI] [PubMed] [Google Scholar]
- Özer G, Imren M, Paulitz TC, et al. (2020). First report of crown rot caused by Fusarium algeriense on wheat in Azerbaijan. Plant Disease 104: 582–582. [DOI] [PubMed] [Google Scholar]
- Raillo AI. (1950). Griby roda Fusarium. State publishing house of agricultural literature, Moscow. [Google Scholar]
- Reinking OA. (1934). Interesting new Fusaria. Zentralblatt für Bakteriologie und Parasitenkunde, Abteilung 2. 89: 509–514. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. (2018a). Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Lombard L, Crous PW. (2019). Back to the roots: a reappraisal of Neocosmospora. Persoonia 43: 90–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Swart WJ, Crous PW. (2018b). New Fusarium species from the Kruger National Park, South Africa. MycoKeys 34: 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangalang AE, Burgess LW, Backhouse D, et al. (1995a). Mycogeography of Fusarium species in soils from tropical, arid and mediterranean regions of Australia. Mycological Research 99: 523–528. [Google Scholar]
- Sangalang AE, Summerell BA, Burgess LW, et al. (1995b). Taxonomy of Fusarium: characterization of Fusarium avenaceum subsp. aywerte and Fusarium avenaceum subsp. nurragi. Mycological Research 99: 287–290. [Google Scholar]
- Satou M, Ichinoe M, Fukumoto F, et al. (2001). Fusarium blight of kangaroo paw (Anigozanthos spp.) caused by Fusarium chlamydosporum and Fusarium semitectum. Journal of Phytopathology 149: 203–206. [Google Scholar]
- Savary O, Coton M, Frisvad JC, et al. (2021). Unexpected Nectriaceae species diversity in cheese, description of Bisifusarium allantoides sp. nov., Bisifusarium penicilloides sp. nov., Longinectria gen. nov. lagenoides sp. nov. and Longinectria verticilliforme sp. nov. Mycosphere 12: 1077–1100. [Google Scholar]
- Shang QJ, Phookamsak R, Camporesi E, et al. (2018). The holomorph of Fusarium celtidicola sp. nov. from Celtis australis. Phytotaxa 361: 251–265. [Google Scholar]
- Sherbakoff CD. (1915). Fusaria of potatoes. Memoirs of the Cornell University Agricultural Experimental Station 6: 87–270. [Google Scholar]
- Summerell BA, Salleh B, Leslie JF. (2003). A utilitarian approach to Fusarium identification. Plant Disease 87: 117–128. [DOI] [PubMed] [Google Scholar]
- Thambugala KM, Wanasinghe DN, Phillips AJL, et al. (2017). Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8: 697–796. [Google Scholar]
- van Diepeningen A, Al-Hatmi A, Brankovics B, et al. (2014). Taxonomy and clinical spectra of Fusarium species: Where do we stand in 2014? Current Clinical Microbiology Reports 1: 10–18. [Google Scholar]
- Wang MM, Crous PW, Sandoval-Denis M, et al. (2022). Fusarium and allied genera from China: species diversity and distribution. Persoonia 48: 1– 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber HW, Reinking OA. (1935). Die Fusarien. Verlagsbuchandlung Paul Parey, Berlin, Germany. [Google Scholar]
- Xia JW, Sandoval_Denis M, Crous PW, et al. (2019). Numbers to names – restyling the Fusarium incarnatum-equiseti species complex. Persoonia 43: 186–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo C, Wang C, et al. (2021). First report of maize stalk rot caused by Fusarium nelsonii in China. Plant Disease 105: 4168. [Google Scholar]