Abstract
In this study, we sequenced a bacteria isolate Pandoraea sp. 892iso isolated from a Phytophthora rubi strain which is an important plant pathogenic oomycete, identified through genome and combined the data with existing genomic data from other 28 the genus of Pandoraea species. Next, we conducted a comparative genomic analysis of the genome structure, evolutionary relationships, and pathogenic characteristics of Pandoraea species. Our results identified Pandoraea sp. 892iso as Pandoraea sputorum at both the genome and gene levels. At the genome level, we carried out phylogenetic analysis of single-copy, gene co-linearity, ANI (average nucleotide identity) and AAI (average amino acid identity) indices, rpoB similarity, MLSA phylogenetic analysis, and genome-to-genome distance calculator calculations to identify the relationship between Pandoraea sp. 892iso and P. sputorum. At the gene level, the quorum sensing genes ppnI and ppnR and the OXA-159 gene were assessed. It is speculated that Pandoraea sp. 892iso is the endosymbiont of the Oomycetes strain of Phytophthora rubi.
Introduction
The genus Pandoraea, originating from the term “Pandora’s box”, refers to the source of all evil in Greek mythology and was established by Coenye et al. in 2000 [1]. The species are characterized as nonspore-forming, catalase-positive, aerobic, gram-negative rods with polar flagella. Some species in this genus were once identified closest to Burkholderia cepacia complex (Bcc), Ralstonia pickettii, or Ralstonia paucula based on phenotype. [1–3]. The genus Pandoraea includes 28 named species (Pandoraea anapnoica, P. anhela, P. apista, P. aquatica, P. bronchicola, P. capi, P. captiosa, P. cepalis, P.commovens, P. communis, P. eparura, P. faecigallinarum, P. fibrosis, P. horticolens, P. iniqua, P. morbifera, P. norimbergensis, P. nosoerga, P. oxalativorans, P. pneumonica, P. pnomenusa, P. pulmonicola, P. soli, P. sputorum, P. terrae, P. terrigena, P. thiooxydans and P. vervacti [3–5]. Pandoraea sp. types have been predominantly isolated from patients with septicemia or respiratory tract infections (mostly cystic fibrosis), as well as from food, water, soil, and food [2, 4, 6–9].
Clinical manifestations of this terrorizing pathogen revolve around nosocomial infections and its ability to deteriorate lung function and even cause multiple organ impairment [10–12]. These organisms appear to be potential pathogens for individuals with cystic fibrosis as well for cross-infection [13]. Further, Pandoraea spp. isolated from environmental samples have considerable potential for biotechnological application given various beneficial degradation abilities, such as removing isomers of 1,2,3,4,5,6-hexachlorocyclohexane (HCH) [13], catalyzing the aerobic transformation of biphenyl and various polychlorinated biphenyls (PCBs) [14, 15], catalyzing the decarboxylation of 2,6-dihydroxybenzoate and regioselective carboxylation of 1,3-dihydroxybenzene to 2,6-dihydroxybenzoate, catalyzing the regioselective carboxylation of phenol and 1,2-dihydroxybenzene [16], degrading kraft lignin without any cosubstrate under high alkaline conditions [17], degrading chlorobenzene [18], biodegrading endosulfan classified as an organochlorine pesticide [19], treating malachite green [20], and metabolizing oxalate [21].
Reflecting on previous research, Pandoraea spp. have frequently been misidentified in many clinical laboratories, leading to a lack of clinical documentation on their virulence potential. Therefore, it is important to accurately identify Pandoraea spp.. Earlier classification of prokaryotes was based solely on phenotypic similarities [22], but modern prokaryote characterization has been strongly influenced by advances in genetic methods. One criterion to be considered a species is to be essentially a collection of types that are characterized by at least one diagnostic phenotypic trait and to have purified DNA molecules that show at least 70% cross-hybridization (DNA-DNA hybridization, DDH) [22–25]. This is pragmatic and universally applicable within the bacterial domain, while the lack of this standard has been increasingly found when it comes to reliable diagnosis of infectious disease agents, international regulations for transport, quarantine, and so on [26–28]. Subsequently, this parameter has been applied most frequently in species identification at the whole genome level [29–32]. Genome Blast Distance Phylogeny (GBDP) [33], the core and pangenome [32], and the genomic-distance index based on DNA maximal unique matches (MUM) [34] are used to identify new species. Unfortunately, our understanding of Pandoraea spp. at the genomic level is relatively superficial, whereby the majority of the literature focuses principally on the usage of genotypic data to facilitate accurate genus- and species-level identification and secondarily on biotechnological potential [1, 2, 18, 21].
In the present study, suspected bacteria isolated from an oomycete strain was identified through whole genome sequencing. The taxonomic status of this isolate was verified at the genome and gene levels, and its phylogenetic relationship with similar species was explored using indices, such as ANI/AAI, MLSA (Multi-locus Sequence Analysis) phylogenetic analysis, genome-to-genome distance calculations, quorum sensing, and oxacillinase gene analysis.
Materials and methods
Strains, cultures, and DNA extraction
When we performed morphological observations on the hyphae of a Phytophthora rubi strain (No. 109892) from Westerdijk Fungal Biodiversity Institute, we inadvertently discovered the structure of suspected bacteria present in the mycelia. The structure still existed after the isolation by monofilament isolation and monospore isolation of the fungus. After isolation and culture, we obtained an analytical strain of bacteria, so that part of the name of which is called “892iso isolate”. Separation, purification, and culture were carried out on beef extract peptone medium plates at 30°C for 48 h. A TIANamp Bacteria DNA Kit (Tiangen, China) was used for genomic DNA.
Sequencing, assembly, and annotation
The whole genome was sequenced and assembled by a strategy that combined paired-end and mate-paired libraries. One targeted insert size of 500 bp was constructed using the TruSeq Nano DNA LT Library Prep Kit (Illumina, USA). One mate-paired library (2 kb) was constructed by the Nextera Mate Pair Sample Prep Kit (Illumina, FC-132–1001, USA) on the Illumina HiSeq 2500 platform. SOAPdenovo (v2.04) was used for de novo assembly. The assembled genome was annotated with a web-based tool called RAST (http://rast.nmpdr.org). RAST can identify repeat sequences in the genome, protein-encoding rRNA and tRNA genes, and assign functions to the genes.
Whole genome alignment and some indices calculation
Mauve (version 2.3.1) was used to align genomes for synteny analysis. The calculation of ANI and AAI was based on BLAST alignment results using a Perl script. The genome-to-genome distance calculator calculations were based on a web server (https://ggdc.dsmz.de/) that uses multi-FASTA files as input. The ppnI/ppnR genes of P. pnomenusa were download from NCBI (accession ID KF887500.1 and KF900148.1), then aligned with all Pandoraea gene sets, all matches with the identity greater than 0.3 and score greater than 100 were retained. The ppnI candidates should contain PF00765 domain and ppnR candidates contain PF03472 domain, and the candidate pairs should be adjacent to each other. An intrinsic Carbapenem-Hydrolyzing Oxacillinases gene of Pandoraea sp. HD7676 was download from NCBI (accession ID: KP771987.1). BLAST was employed to identify homolog genes in the 28 Pandoraea species.
Comparative genome analysis
All protein sequences in reference genomes were downloaded and set as the query for all-vs-all BLASTP. OrthoMCL (version 2.0.8) was used to identify single-copy genes with I (inflation) set at 1.5. Next, MUSCLE (version 3.8.425) was used to align the sequences of the associated proteins. PAL2NAL (version 14.0) was used to convert the protein alignment to codon alignment. Gblock (version 0.91b) was used to remove the alignment results that were deemed unreliable. The phylogenetic tree was built by single-copy genes, with Burkholderia cepacia strain LO6 as the outgroup. MCMCTree software in PAML (version 4.7) was used to estimate the divergence time. CAFÉ (version 4) was used to calculate the expansion and contraction of these gene families.
Results
Genome assembly, annotation, and validation of protein-coding genes
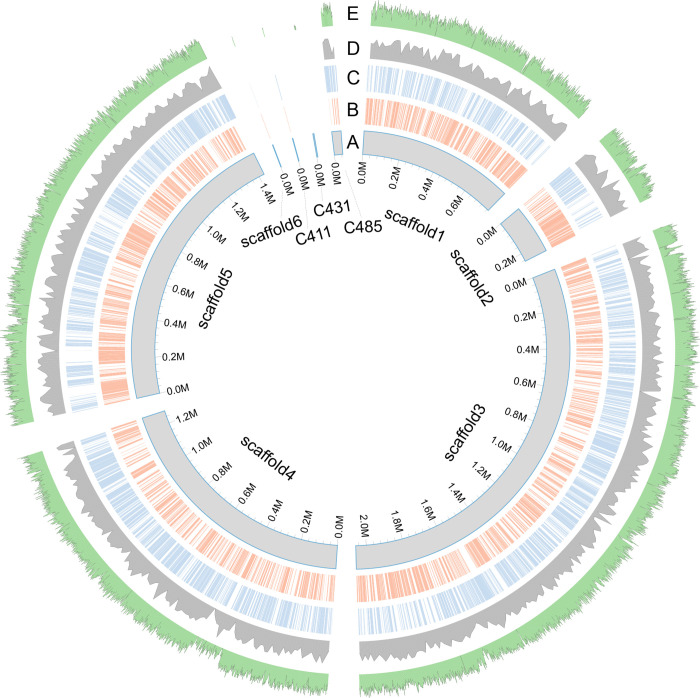
The genome of the Pandoraea sp. 892iso isolate was assembled from sequencing data generated by HiSeq 2000 by SOAPDenovo2 assembler. The total length of the top 48 longest scaffolds was 5.83 Mb, representing approximately 82.6-fold genome sequence coverage. The N50 and maximum lengths of scaffolds was 1.43 kb. Most of the length was concentrated on 12 scaffold sequences over 1,000 bp, of which the longest sequence was 2.06 MB (Fig 1). A total of 5,367 protein-coding genes were predicted from the genome assembly, 5,131 (95.60%) of which were supported by the RNA-seq data (coverage > = 90%). Within these protein-coding genes, 4,274 (79.63%) were assigned a biological function. Among the 1,093 ORFs without known function, 736 showed similarity to other database entries. For Pandoraea sp. 892iso isolate, the coding regions from the predicted genes constituted 88.61% of the genome (total length of all genes divide the genome size) and the average gene density was 919 genes per 1 Mb (total number of all genes divide the genome size, times with 100000bp), which were more or fewer than most of other sequenced Pandoraea species. The GC content of the genome, coding sequences, and repetitive elements were 62.66%, 63.32%, and 57.52%, respectively. A total of 63 tRNA genes were predicted from the assembly. The genome characteristics of Pandoraea sp. 892iso and other Pandoraea species are shown in Table 1.
Fig 1. Structure of the genome assembly.
Table 1. Genome and gene comparison of Pandoraea sp. 892iso and other Pandoraea species.
| Content | Pandoraea sp. 892iso | Pandoraea anapnoica | Pandoraea anhela | Pandoraea apista | Pandoraea aquatica | Pandoraea bronchicola | Pandoraea capi | Pandoraea captiosa | Pandoraea cepalis | Pandoraea commovens | Pandoraea communis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession number | GCF_902459765.1 | GCF_902459655.1 | GCF_001465595.2 | GCF_902459565.1 | GCF_902459805.1 | GCF_902459735.1 | GCF_902459775.1 | GCF_902459625.1 | GCF_902459615.1 | GCF_902459745.1 | |
| Genome | |||||||||||
| Scaffold Number | 48 | 48 | 61 | 2 | 17 | 34 | 31 | 36 | 32 | 26 | 17 |
| Total Length (Mb) | 5.83 | 6.13 | 6.05 | 5.57 | 5.96 | 5.35 | 5.85 | 6.14 | 5.16 | 6.04 | 5.71 |
| GC Content (%) | 62.7 | 62.4 | 63.35 | 62.63 | 62.89 | 62.97 | 63.44 | 63.3 | 63.54 | 62.63 | 62.57 |
| N50 Length | 1,430,084 | 278,466 | 256,277 | - | 442,715 | 323,330 | 401,082 | 280,257 | 286,967 | 434,611 | 434,208 |
| N90 Length | 768,040 | 128,458 | 71,214 | - | 244,118 | 147,078 | 151,211 | 137,159 | 88,384 | 144,451 | 240,155 |
| Longest scaffold | 2,057,907 | 677,688 | 731,537 | - | 1,296,496 | 977,860 | 812,744 | 1,307,560 | 1,062,234 | 1,441,918 | 1,422,275 |
| Gene | |||||||||||
| Gene Number | 5367 | 5,348 | 5,178 | 4,969 | 5,197 | 4,734 | 5,049 | 5,328 | 4,602 | 5,246 | 5,051 |
| Gene Length (bp) | 5,175,057 | 5,334,275 | 5,189,856 | 4,830,549 | 5,211,988 | 4,656,675 | 5,074,754 | 5,326,122 | 4,478,447 | 5,288,517 | 4,979,502 |
| GC Content in Gene Region (%) | 63.32 | 63.11 | 63.92 | 63.21 | 63.6 | 63.52 | 64.13 | 63.89 | 64.03 | 63.33 | 63.13 |
| Gene Length/Genome (%) | 88.61 | 87.07 | 85.84 | 86.7 | 87.48 | 87.02 | 86.72 | 86.75 | 86.8 | 87.6 | 87.23 |
| Gene Average Length (bp) | 964 | 997 | 1,002 | 972 | 1,003 | 984 | 1,005 | 1,000 | 973 | 1,008 | 986 |
| Intergenic Region Length (bp) | 665,083 | 792,413 | 856,156 | 740,711 | 746,139 | 694,448 | 777,390 | 813,460 | 681,119 | 748,432 | 729,101 |
| GC Content in Intergenic Region (%) | 57.52 | 57.63 | 59.87 | 58.9 | 57.95 | 59.27 | 58.97 | 59.45 | 60.3 | 57.73 | 58.74 |
| Intergenic Region Length/Genome (%) | 11.39 | 12.93 | 14.16 | 13.3 | 12.52 | 12.98 | 13.28 | 13.25 | 13.2 | 12.4 | 12.77 |
| Content | Pandoraea eparura | Pandoraea faecigallinarum | Pandoraea fibrosis | Pandoraea horticolens | Pandoraea iniqua | Pandoraea morbifera | Pandoraea norimbergensis | Pandoraea nosoerga | Pandoraea oxalativorans | Pandoraea pneumonica | Pandoraea pnomenusa |
| Accession number | GCF_902459725.1 | GCF_001029105.3 | GCF_000807775.2 | GCF_902459555.1 | GCF_902459685.1 | GCF_902459575.1 | GCF_001465545.3 | GCF_902459585.1 | GCF_000972785.3 | GCF_902459645.1 | GCF_000504585.2 |
| Genome | |||||||||||
| Scaffold Number | 35 | 3 | 1 | 68 | 17 | 47 | 1 | 41 | 5 | 12 | 1 |
| Total Length (Mb) | 5.21 | 5.73 | 5.59 | 6.01 | 6.34 | 5.23 | 6.17 | 4.86 | 6.5 | 5.85 | 5.39 |
| GC Content (%) | 63.68 | 63.45 | 62.82 | 62.31 | 63.06 | 64.65 | 63.06 | 66.13 | 63.08 | 62.45 | 64.89 |
| N50 Length | 259,402 | - | - | 290,798 | 382,973 | 316,192 | - | 229,370 | - | 265,947 | - |
| N90 Length | 102,841 | - | - | 73,897 | 241,289 | 80,719 | - | 91,075 | - | 5,636 | - |
| Longest scaffold | 893,217 | - | - | 787,753 | 1,308,188 | 801,833 | - | 664,052 | - | 2,096,772 | - |
| Gene | |||||||||||
| Gene Number | 4,615 | 5,027 | 4,855 | 5,322 | 5,499 | 4,652 | 5,356 | 4,297 | 5,648 | 5,168 | 4,759 |
| Gene Length (bp) | 4,496,889 | 4,932,939 | 4,868,583 | 5,167,287 | 5,558,312 | 4,536,412 | 5,418,712 | 4,198,421 | 5,522,745 | 5,131,811 | 4,684,824 |
| GC Content in Gene Region (%) | 64.17 | 63.98 | 63.39 | 62.98 | 63.76 | 65.1 | 63.72 | 66.51 | 63.59 | 63.06 | 65.36 |
| Gene Length/Genome (%) | 86.39 | 86.05 | 87.06 | 86 | 87.68 | 86.68 | 87.86 | 86.35 | 84.96 | 87.8 | 86.98 |
| Gene Average Length (bp) | 974 | 981 | 1,003 | 971 | 1,011 | 975 | 1,012 | 977 | 978 | 993 | 984 |
| Intergenic Region Length (bp) | 708,688 | 799,725 | 723,482 | 841,203 | 780,817 | 696,886 | 748,658 | 663,693 | 977,986 | 713,267 | 701,122 |
| GC Content in Intergenic Region (%) | 60.55 | 60.15 | 58.94 | 58.19 | 58.13 | 61.77 | 58.29 | 63.71 | 60.23 | 58.09 | 61.71 |
| Intergenic Region Length/Genome (%) | 13.61 | 13.95 | 12.94 | 14 | 12.32 | 13.32 | 12.14 | 13.65 | 15.04 | 12.2 | 13.02 |
| Content | Pandoraea pulmonicola | Pandoraea soli | Pandoraea sp. XY-2 | Pandoraea sputorum | Pandoraea terrae | Pandoraea thiooxydans | Pandoraea vervacti | ||||
| Accession number | GCF_000815105.2 | GCF_902459595.1 | GCF_004193915.1 | GCF_900187205.1 | GCF_902459695.1 | GCF_001017775.3 | GCF_000934605.2 | ||||
| Genome | |||||||||||
| Scaffold Number | 1 | 51 | 1 | 1 | 81 | 1 | 2 | ||||
| Total Length (Mb) | 5.87 | 4.96 | 5.06 | 5.74 | 6.18 | 4.46 | 5.74 | ||||
| GC Content (%) | 64.3 | 63.62 | 63.76 | 62.78 | 62.79 | 63.19 | 63.52 | ||||
| N50 Length | - | 370,563 | - | - | 194,136 | - | - | ||||
| N90 Length | - | 61,129 | - | - | 60,237 | - | - | ||||
| Longest scaffold | - | 921,398 | - | - | 456,896 | - | - | ||||
| Gene | |||||||||||
| Gene Number | 4,996 | 4,393 | 4,512 | 4,994 | 5,590 | 4,091 | 4,889 | ||||
| Gene Length (bp) | 5,040,965 | 4,324,589 | 4,386,412 | 5,002,422 | 5,421,742 | 3,998,582 | 4,955,787 | ||||
| GC Content in Gene Region (%) | 65 | 64.13 | 64.26 | 63.5 | 63.31 | 63.76 | 64.11 | ||||
| Gene Length/Genome (%) | 85.91 | 87.15 | 86.75 | 87.1 | 87.78 | 89.57 | 86.39 | ||||
| Gene Average Length (bp) | 1,009 | 984 | 972 | 1,002 | 970 | 977 | 1,014 | ||||
| Intergenic Region Length (bp) | 826,656 | 637,393 | 669,794 | 740,701 | 755,081 | 465,604 | 780,495 | ||||
| GC Content in Intergenic Region (%) | 59.97 | 60.12 | 60.47 | 57.89 | 58.98 | 58.3 | 59.82 | ||||
| Intergenic Region Length/Genome (%) | 14.09 | 12.85 | 13.25 | 12.9 | 12.22 | 10.43 | 13.61 |
Comparative genomics and identification at the genome level
Comparative genomic analysis
A total of genes in Pandoraea sp. 892iso were classified through cluster analysis. The distribution of best hits within the genus Pandoraea is shown in Fig 2. In total, 3,849 orthologous genes were shared in common between Pandoraea sp. 892iso and the other four Pandoraea species. The cluster analysis of Pandoraea sp. 892iso, 28 Pandoraea species, and Burkholderia cepacia as an outgroup was carried out by orthoMCL to obtain the result of a common single-copy gene family. The phylogenetic relationship in view of these single-copy genes is shown in Fig 3 and S1 Table, which shows the closest phylogenetic relationship to be between Pandoraea sp. 892iso and the P. sputorum strain DSM21091. Meanwhile, eight specific gene families, including 21 genes, were clustered, 17 genes were hypothetical proteins, and the other four are shown in Table 2.
Fig 2. Venn diagram of genes common to Pandoraea sp. 892iso and the four other Pandoraea types.
Fig 3. Phylogenetic analyses of the evolutionary relationships between Pandoraea sp. 892iso and Pandoraea types.
A neighbor-joining phylogenetic tree constructed based on single-copy genes common to these nine bacterial genomes. The neighbor-joining method was used in MEGA6, where a bootstrap test (1,000 replicates) is shown next to the branches.
Table 2. Details of the four respective genes of Pandoraea sp. 892iso.
| Gene | Position | direction | Detail |
|---|---|---|---|
| fig|93222.8.peg.1 | C163_3_104 | - | DNA-cytosine methyltransferase |
| fig|93222.8.peg.10 | C237_1_126 | - | DNA-cytosine methyltransferase |
| fig|93222.8.peg.14 | C273_3_104 | - | DNA-cytosine methyltransferase |
| fig|93222.8.peg.2519 | scaffold3_1641978_1640653 | - | DNA-cytosine methyltransferase |
| fig|93222.8.peg.4329 | scaffold5_320715_322067 | + | DNA-cytosine methyltransferase |
| fig|93222.8.peg.4363 | scaffold5_361619_362311 | + | Transcriptional regulator, GntR family |
| fig|93222.8.peg.4364 | scaffold5_363002_362316 | - | Transcriptional regulator, GntR family |
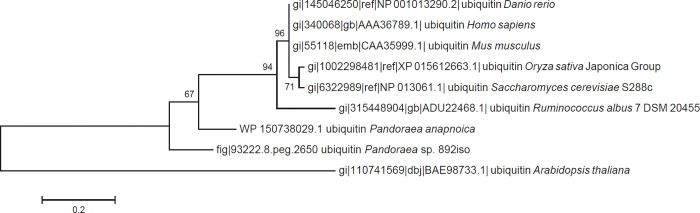
The global genome clustering and alignment of Pandoraea types were complicated by Mummer. The results showed the best gene co-linearity among these Pandoraea types and that rearrangement was almost absent, except for P. thiooxydans and P. sputorum, which were phylogenetically closest to Pandoraea sp. 892iso (S1 Fig). It was speculated that the small external selection pressure of the Pandoraea group and the genome evolution occurred in a similar way. More attention should be given to Pandoraea sp. 892iso and its proximal P. sputorum, both of which rearranged compared to other Pandoraea types. Rearrangements existed in the five largest scaffold alignments, especially in scaffolds 3, 4, and 5, as shown in S2 Fig. A special unique insertion sequence in scaffold3_1802763_1803544 of Pandoraea sp. 892iso contains the gene fig|93222.8.peg.2650 with the function of ubiquitin in the NR database, which may be related to the function of covalent attachment to other cellular proteins associated with stability changing, localization, and activity of the target protein [35]. The ubiquitin gene in Pandoraea sp. 892iso was found to be different from that in human, mouse, zebrafish, rice, Arabidopsis, yeast, or other model organisms by phylogenetic analysis (Fig 4).
Fig 4. Phylogenetic analyses of evolutionary relationships of ubiquitin genes among Pandoraea sp. 892iso and Pandoraea types.
A neighbor-joining phylogenetic tree constructed based on single-copy genes common to these nine bacterial genomes. The neighbor-joining method was used in MEGA6, where a bootstrap test (1,000 replicates) is shown next to the branches.
ANI and AAI
ANI (average nucleotide identity), as the new method for bacterial species definition, provides several benefits, avoids misplacement based on phenotypic similarities or chemical characteristics, provides a scalable and uniform approach that works for both culturable and nonculturable species, is faster and cheaper than traditional taxonomic methods, and, most importantly, falls in line with Darwin’s vision of classification [30]. AAI (average amino acid identity), a method that compares all conserved protein-coding genes present in a given set of genomes, clusters types into groups that share more than 95% AAI [36]. ANI and AAI characteristics have been used to evaluate the accuracy of these genotypic methods in the identification of Pandoraea species. Given the availability of whole genome sequence data and Pandoraea sp. 892iso nucleotide and amino acid data as query, Blastn by CDS sequence coverage was ≥ 50% and tblastn by protein coverage was ≥ 70%. We performed sequence-based genotypic microbial identification analysis using the RefSeq database by genome comparison between Pandoraea sp. 892iso and Pandoraea sputorum and generated an ANI value of 98.81% and an AAI value of 91.18%; genome comparison with other in-house sequenced Pandoraea species provided an ANI value of less than 93.34% and an AAI value of 84.90% (Table 3). Based on previous results using the ANI value for species definition, ANI and AAI values of ≥ 95% corresponded to the traditional 70% DNA-DNA. Using the ANI and AAI values of Pandoraea sp. 892iso, it can be unequivocally stated that Pandoraea sp. 892iso is phylogenetically close to P. sputorum.
Table 3. Average nucleotide identity (ANI) and average amino acid identity (AAI) analyses.
Genome comparisons of Pandoraea sp. 892iso and other Pandoraea-type species.
| Species | ID | ANI | AAI | ||
|---|---|---|---|---|---|
| value | percent | value | percent | ||
| Pandoraea anapnoica | GCF_902459765.1 | 94.10 | 83.55 | 93.57 | 89.88 |
| Pandoraea anhela | GCF_902459655.1 | 87.56 | 61.88 | 85.97 | 84.01 |
| Pandoraea apista | GCF_001465595.2 | 86.43 | 54.74 | 84.98 | 83.38 |
| Pandoraea aquatica | GCF_902459565.1 | 92.99 | 83.01 | 93.06 | 89.25 |
| Pandoraea bronchicola | GCF_902459805.1 | 86.63 | 55.10 | 84.51 | 81.14 |
| Pandoraea capi | GCF_902459735.1 | 87.77 | 67.77 | 88.02 | 87.59 |
| Pandoraea captiosa | GCF_902459775.1 | 87.24 | 61.15 | 86.49 | 85.41 |
| Pandoraea cepalis | GCF_902459625.1 | 86.48 | 48.39 | 82.57 | 77.98 |
| Pandoraea commovens | GCF_902459615.1 | 94.51 | 85.62 | 94.43 | 90.55 |
| Pandoraea communis | GCF_902459745.1 | 86.60 | 53.85 | 83.93 | 82.50 |
| Pandoraea eparura | GCF_902459725.1 | 86.58 | 48.85 | 81.94 | 77.51 |
| Pandoraea faecigallinarum | GCF_001029105.3 | 87.43 | 60.07 | 85.94 | 83.10 |
| Pandoraea fibrosis | GCF_000807775.2 | 86.50 | 57.54 | 85.87 | 83.29 |
| Pandoraea horticolens | GCF_902459555.1 | 86.53 | 53.29 | 83.58 | 82.45 |
| Pandoraea iniqua | GCF_902459685.1 | 85.58 | 54.26 | 83.99 | 86.77 |
| Pandoraea morbifera | GCF_902459575.1 | 86.21 | 51.70 | 83.57 | 82.34 |
| Pandoraea norimbergensis | GCF_001465545.3 | 85.45 | 53.68 | 83.84 | 86.68 |
| Pandoraea nosoerga | GCF_902459585.1 | 86.24 | 50.42 | 82.72 | 78.67 |
| Pandoraea oxalativorans | GCF_000972785.3 | 93.57 | 77.73 | 90.99 | 86.06 |
| Pandoraea pneumonica | GCF_902459645.1 | 85.60 | 52.62 | 83.58 | 85.52 |
| Pandoraea pnomenusa | GCF_000504585.2 | 86.27 | 53.21 | 83.90 | 82.50 |
| Pandoraea pulmonicola | GCF_000815105.2 | 86.28 | 53.72 | 83.78 | 82.39 |
| Pandoraea soli | GCF_902459595.1 | 86.52 | 48.07 | 82.37 | 77.75 |
| Pandoraea sp. XY-2 | GCF_004193915.1 | 86.49 | 48.39 | 80.31 | 74.75 |
| Pandoraea sputorum | GCF_900187205.1 | 99.29 | 88.49 | 97.03 | 90.91 |
| Pandoraea terrae | GCF_902459695.1 | 82.67 | 25.17 | 72.62 | 73.04 |
| Pandoraea terrigena | GCF_902459705.1 | 86.36 | 48.80 | 82.03 | 79.21 |
| Pandoraea thiooxydans | GCF_001017775.3 | 80.09 | 10.79 | 67.18 | 62.49 |
| Pandoraea vervacti | GCF_000934605.2 | 87.33 | 60.28 | 86.32 | 83.73 |
rpoB similarity and MLSA phylogenetic analysis
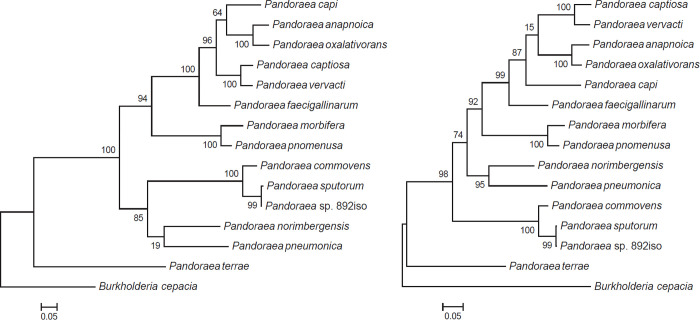
The rpoB gene, encoding the β-subunit of RNA polymerase, has emerged as a core gene candidate for phylogenetic analyses and identification of bacteria; it is a single-copy gene, belongs to the common set of genes, and is long enough to contain phylogenetically useful information for some bacterial declination [37–40]. Multilocus sequence analysis (MLSA) is a currently widely used method for prokaryotic taxonomy, which utilizes internal fragments of several protein-coding genes. It was introduced by Gevers et al. and is increasingly being applied to obtain higher resolution power among species within a genus [39, 41]. As a typing technique for type characterization that shows variation in multiple housekeeping genes, a concatenation of five housekeeping genes, shikimate dehydrogenase (aroE), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL), was recommended for our bacterial delineation, as well as for clarifying the taxonomic situation within the Pandoraea family [39, 41]. The phylogenetic tree topologies of Pandoraea sp. 892iso and other Pandoraea spp. by rpoB similarity (Fig 5A) and MLSA analysis (Fig 5B) revealed Pandoraea sp. 892iso to have the closest phylogenetic relationship with Pandoraea sputorum strain DSM21091.
Fig 5. Phylogenetic tree highlighting the position of Pandoraea sp. 892iso relative to the other Pandoraea species.
The tree was aligned with the characteristics of the rpoB gene (a) and MLSA (b) under the maximum likelihood (ML) criterion.
Genome-to-genome distance calculator
In silico genome-to-genome comparison to obtain an estimate of the overall similarity between the genomes of two types has enabled the taxonomist to perform genome-based species delineation and genome-based subspecies delineation. These distance functions can also cope with heavily reduced genomes and repetitive sequence regions. The Genome-to-Genome Distance Calculator (GGDC) calculates the distances by comparing genomes to obtain HSPs (high-scoring segment pairs) and interfering distances from a set of formulas: 1) HSP length/total length; 2) identities/HSP length; and 3) identities/total length [42]. An estimated GGDC of the overall similarity between Pandoraea sp. 892iso and other Pandoraea species is shown in Table 4. In probability DDG ≥70% index analysis, the pairwise comparison of the genome with P. sputorum was found to be 98.49%, 96.97%, and 99.88% for the HSP length/total length, identities/HSP length, and identities/total length ratios, respectively. Thus, the close relationship of Pandoraea sp. 892iso and P. sputorum was verified.
Table 4. Pairwise comparison of Pandorarae sp. 892iso and Pandoraea species using the GGDC.
| Query | Reference | ID | HSP length/total length | identities/HSP length | identities/total length | G+C difference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Distance | DDH estimate (GLM-based) | Prob. DDH>70% | Prob. DDH>79% | Distance | DDH estimate (GLM-based) | Prob. DDH>70% | Prob. DDH>79% | Distance | DDH estimate (GLM-based) | Prob. DDH>70% | Prob. DDH>79% | ||||
| Pandoraea sp. 892iso | Pandoraea anapnoica | GCF_902459765.1 | 78.3 | [74.3–81.8%] | 0.1411 | 89.13 | 52.1 | [49.5–54.8%] | 0.0669 | 25.38 | 74.9 | [71.4–78.1%] | 0.1985 | 88.31 | 0.26 |
| Pandoraea sp. 892iso | Pandoraea anhela | GCF_902459655.1 | 48.1 | [44.7–51.5%] | 0.3215 | 8.92 | 29 | [26.6–31.5%] | 0.1476 | 0.07 | 42.4 | [39.4–45.4%] | 0.4217 | 0.31 | 0.69 |
| Pandoraea sp. 892iso | Pandoraea apista | GCF_001465595.2 | 48 | [44.6–51.4%] | 0.322 | 8.82 | 26.6 | [24.3–29.1%] | 0.1626 | 0.02 | 41.3 | [38.3–44.3%] | 0.4323 | 0.21 | 0.03 |
| Pandoraea sp. 892iso | Pandoraea aquatica | GCF_902459565.1 | 80.7 | [76.8–84.1%] | 0.1286 | 91.75 | 46.5 | [43.9–49.1%] | 0.0803 | 10.85 | 74.9 | [71.4–78.1%] | 0.1985 | 88.3 | 0.23 |
| Pandoraea sp. 892iso | Pandoraea bronchicola | GCF_902459805.1 | 48.6 | [45.2–52%] | 0.3173 | 9.79 | 27 | [24.6–29.5%] | 0.1603 | 0.03 | 41.8 | [38.9–44.9%] | 0.4268 | 0.26 | 0.31 |
| Pandoraea sp. 892iso | Pandoraea capi | GCF_902459735.1 | 61 | [57.3–64.6%] | 0.2337 | 45.76 | 29.3 | [27–31.8%] | 0.1458 | 0.08 | 51.6 | [48.5–54.7%] | 0.3455 | 4.22 | 0.78 |
| Pandoraea sp. 892iso | Pandoraea captiosa | GCF_902459775.1 | 48.7 | [45.3–52.2%] | 0.3164 | 9.99 | 28.4 | [26–30.9%] | 0.1515 | 0.05 | 42.6 | [39.6–45.6%] | 0.42 | 0.32 | 0.64 |
| Pandoraea sp. 892iso | Pandoraea cepalis | GCF_902459625.1 | 38.3 | [34.9–41.8%] | 0.4098 | 1.11 | 26.9 | [24.6–29.4%] | 0.1605 | 0.03 | 34.5 | [31.6–37.6%] | 0.5045 | 0.02 | 0.88 |
| Pandoraea sp. 892iso | Pandoraea commovens | GCF_902459615.1 | 85.3 | [81.6–88.4%] | 0.1056 | 95.14 | 54.2 | [51.5–56.9%] | 0.0625 | 32.2 | 81.4 | [78–84.3%] | 0.1615 | 96.5 | 0.03 |
| Pandoraea sp. 892iso | Pandoraea communis | GCF_902459745.1 | 43.9 | [40.5–47.4%] | 0.3559 | 4.04 | 26.8 | [24.4–29.3%] | 0.1614 | 0.02 | 38.5 | [35.6–41.6%] | 0.4599 | 0.08 | 0.09 |
| Pandoraea sp. 892iso | Pandoraea eparura | GCF_902459725.1 | 36.8 | [33.4–40.3%] | 0.426 | 0.75 | 27.5 | [25.1–30%] | 0.1571 | 0.03 | 33.6 | [30.6–36.7%] | 0.5161 | 0.01 | 1.02 |
| Pandoraea sp. 892iso | Pandoraea faecigallinarum | GCF_001029105.3 | 49.8 | [46.4–53.2%] | 0.3082 | 11.94 | 28.8 | [26.4–31.3%] | 0.1492 | 0.06 | 43.5 | [40.5–46.5%] | 0.4115 | 0.44 | 0.79 |
| Pandoraea sp. 892iso | Pandoraea fibrosis | GCF_000807775.2 | 52.1 | [48.6–55.5%] | 0.2918 | 16.86 | 26.7 | [24.4–29.2%] | 0.1618 | 0.02 | 44.1 | [41.1–47.1%] | 0.4064 | 0.52 | 0.16 |
| Pandoraea sp. 892iso | Pandoraea horticolens | GCF_902459555.1 | 41.6 | [38.3–45.1%] | 0.3767 | 2.47 | 26.8 | [24.5–29.3%] | 0.1612 | 0.02 | 36.9 | [33.9–40%] | 0.4771 | 0.04 | 0.35 |
| Pandoraea sp. 892iso | Pandoraea iniqua | GCF_902459685.1 | 39.5 | [36.2–43%] | 0.397 | 1.51 | 25.6 | [23.3–28.1%] | 0.1697 | 0.01 | 35 | [32–38%] | 0.4994 | 0.02 | 0.41 |
| Pandoraea sp. 892iso | Pandoraea morbifera | GCF_902459575.1 | 43.2 | [39.9–46.7%] | 0.3621 | 3.49 | 26.5 | [24.1–29%] | 0.1636 | 0.02 | 37.9 | [34.9–40.9%] | 0.4665 | 0.06 | 2 |
| Pandoraea sp. 892iso | Pandoraea norimbergensis | GCF_001465545.3 | 39.3 | [36–42.8%] | 0.3992 | 1.44 | 25.5 | [23.2–28%] | 0.1702 | 0.01 | 34.8 | [31.8–37.9%] | 0.5015 | 0.02 | 0.4 |
| Pandoraea sp. 892iso | Pandoraea nosoerga | GCF_902459585.1 | 40.8 | [37.4–44.2%] | 0.3849 | 2.03 | 26.8 | [24.4–29.3%] | 0.1615 | 0.02 | 36.3 | [33.3–39.3%] | 0.4842 | 0.03 | 3.47 |
| Pandoraea sp. 892iso | Pandoraea oxalativorans | GCF_000972785.3 | 62.6 | [58.9–66.2%] | 0.2242 | 51.57 | 49.4 | [46.8–52%] | 0.073 | 17.48 | 61 | [57.7–64.2%] | 0.2809 | 29.7 | 0.43 |
| Pandoraea sp. 892iso | Pandoraea pneumonica | GCF_902459645.1 | 39.9 | [36.6–43.4%] | 0.393 | 1.67 | 25.3 | [23–27.8%] | 0.1718 | 0.01 | 35.1 | [32.2–38.2%] | 0.4973 | 0.02 | 0.21 |
| Pandoraea sp. 892iso | Pandoraea pnomenusa | GCF_000504585.2 | 43.6 | [40.2–47%] | 0.3588 | 3.78 | 26.6 | [24.3–29.1%] | 0.1626 | 0.02 | 38.2 | [35.3–41.3%] | 0.463 | 0.07 | 2.23 |
| Pandoraea sp. 892iso | Pandoraea pulmonicola | GCF_000815105.2 | 41.3 | [37.9–44.8%] | 0.3797 | 2.29 | 26.6 | [24.2–29.1%] | 0.163 | 0.02 | 36.6 | [33.6–39.6%] | 0.4809 | 0.04 | 1.64 |
| Pandoraea sp. 892iso | Pandoraea soli | GCF_902459595.1 | 39.4 | [36–42.9%] | 0.3985 | 1.46 | 27 | [24.7–29.5%] | 0.16 | 0.03 | 35.4 | [32.4–38.4%] | 0.4948 | 0.02 | 0.96 |
| Pandoraea sp. 892iso | Pandoraea sp. XY-2 | GCF_004193915.1 | 39.7 | [36.3–43.1%] | 0.3957 | 1.56 | 27 | [24.6–29.5%] | 0.1603 | 0.03 | 35.5 | [32.6–38.6%] | 0.4926 | 0.03 | 1.1 |
| Pandoraea sp. 892iso | Pandoraea sputorum | GCF_900187205.1 | 94.2 | [91.7–96%] | 0.0565 | 98.49 | 94 | [92.2–95.4%] | 0.0077 | 96.97 | 96.1 | [94.4–97.3%] | 0.0638 | 99.88 | 0.12 |
| Pandoraea sp. 892iso | Pandoraea terrae | GCF_902459695.1 | 18.2 | [15.1–21.7%] | 0.7652 | 0 | 22.6 | [20.3–25%] | 0.194 | 0 | 17.9 | [15.3–20.9%] | 0.8108 | 0 | 0.13 |
| Pandoraea sp. 892iso | Pandoraea terrigena | GCF_902459705.1 | 39 | [35.6–42.4%] | 0.403 | 1.31 | 26.7 | [24.4–29.2%] | 0.1621 | 0.02 | 34.9 | [32–38%] | 0.4998 | 0.02 | 0.82 |
| Pandoraea sp. 892iso | Pandoraea thiooxydans | GCF_001017775.3 | 14.2 | [11.4–17.6%] | 0.9145 | 0 | 20.2 | [18–22.6%] | 0.2177 | 0 | 14.4 | [12–17.2%] | 0.9331 | 0 | 0.54 |
| Pandoraea sp. 892iso | Pandoraea vervacti | GCF_000934605.2 | 50.9 | [47.5–54.4%] | 0.2999 | 14.26 | 28.4 | [26–30.9%] | 0.1512 | 0.05 | 44.1 | [41.1–47.2%] | 0.4058 | 0.53 | 0.87 |
Some special genes among Pandoraea sp
Quorum sensing (QS)
The most studied QS molecule is N-acyl homoserine lactone (AHL), which is secreted by gram-negative proteobacteria. AHLs are secreted by LuxI homologs until a threshold concentration of AHL is attained before they bind to LuxR homologs and subsequently activate a cascade of QS-regulated gene expression [43]. The predicted putative AHL synthase (ppnI) and AHL receptor protein (ppnR) in Pandoraea sp. 892iso and the nine Pandoraea species are shown in Table 5. The phylogenetic trees of putative AHL synthase (ppnI) and AHL receptor protein (ppnR) are shown in Fig 6.
Table 5. The identified ppnI and ppnR genes in Pandoraea sp. 892iso and nine Pandoraea species.
| Species | Accession number | scaffold | gene | start | end | strand |
|---|---|---|---|---|---|---|
| Pandoraea sp. 892iso | fig|93222.8.peg.1246 | scaffold3 | ppnI | 215501 | 216286 | + |
| Pandoraea sp. 892iso | fig|93222.8.peg.1247 | scaffold3 | ppnR | 216253 | 216966 | - |
| Pandoraea oxalativorans | WP_046292715.1 | NZ_CP011253.3 | ppnI | 4024825 | 4025493 | - |
| Pandoraea oxalativorans | WP_046293945.1 | NZ_CP011253.3 | ppnR | 4024031 | 4024732 | + |
| Pandoraea anapnoica | WP_150739377.1 | NZ_CABPSP010000011.1 | ppnI | 57228 | 57914 | - |
| Pandoraea anapnoica | WP_150739515.1 | NZ_CABPSP010000011.1 | ppnR | 56433 | 57134 | + |
| Pandoraea pneumonica | WP_150681584.1 | NZ_CABPSK010000004.1 | ppnI | 583193 | 583867 | - |
| Pandoraea pneumonica | WP_174988328.1 | NZ_CABPSK010000004.1 | ppnR | 582433 | 583146 | + |
| Pandoraea morbifera | WP_150566717.1 | NZ_CABPSD010000005.1 | ppnI | 208906 | 209694 | - |
| Pandoraea morbifera | WP_150566716.1 | NZ_CABPSD010000005.1 | ppnR | 208206 | 208919 | + |
| Pandoraea sputorum | WP_174555901.1 | NZ_LT906435.1 | ppnI | 1348270 | 1349055 | + |
| Pandoraea sputorum | WP_039402529.1 | NZ_LT906435.1 | ppnR | 1349022 | 1349723 | - |
| Pandoraea terrae | WP_150700195.1 | NZ_CABPRZ010000043.1 | ppnI | 19106 | 19732 | - |
| Pandoraea terrae | WP_150700194.1 | NZ_CABPRZ010000043.1 | ppnR | 18360 | 19076 | + |
| Pandoraea vervacti | WP_044456583.1 | NZ_CP010897.2 | ppnI | 4037152 | 4037835 | - |
| Pandoraea vervacti | WP_044458339.1 | NZ_CP010897.2 | ppnR | 4036372 | 4037073 | + |
| Pandoraea captiosa | WP_150627103.1 | NZ_CABPSQ010000011.1 | ppnI | 88267 | 88950 | - |
| Pandoraea captiosa | WP_150627162.1 | NZ_CABPSQ010000011.1 | ppnR | 87492 | 88193 | + |
| Pandoraea pnomenusa | WP_023871914.1 | NC_023018.2 | ppnI | 3778787 | 3779572 | - |
| Pandoraea pnomenusa | WP_080685145.1 | NC_023018.2 | ppnR | 3778087 | 3778800 | + |
| Pandoraea commovens | WP_174985011.1 | NZ_CABPSA010000008.1 | ppnI | 204518 | 205333 | + |
| Pandoraea commovens | WP_150666021.1 | NZ_CABPSA010000008.1 | ppnR | 205300 | 206013 | - |
| Burkholderia cepacia | WP_042976961.1 | NZ_CP045236.1 | ppnI | 471746 | 472354 | - |
| Burkholderia cepacia | WP_021162347.1 | NZ_CP045236.1 | ppnR | 473082 | 473801 | + |
| Pandoraea faecigallinarum | WP_167362711.1 | NZ_CP011807.3 | ppnI | 3690884 | 3691549 | - |
| Pandoraea faecigallinarum | WP_053059408.1 | NZ_CP011807.3 | ppnR | 3690044 | 3690820 | + |
| Pandoraea capi | WP_150721274.1 | NZ_CABPRV010000004.1 | ppnI | 224554 | 225237 | - |
| Pandoraea capi | WP_150721396.1 | NZ_CABPRV010000004.1 | ppnR | 223772 | 224473 | + |
| Pandoraea norimbergensis | WP_157125706.1 | NZ_CP013480.3 | ppnI | 1418662 | 1419441 | + |
| Pandoraea norimbergensis | WP_064675185.1 | NZ_CP013480.3 | ppnR | 1419408 | 1420109 | - |
Fig 6. Phylogenetic tree of ppnI and ppnR.
Intrinsic carbapenem-hydrolyzing oxacillinases
Oxacillinases are serine β-lactamases of molecular class D. Many bacterial species could produce OXA-type enzymes, some of them with carbapenem-hydrolyzing activity. The nine Pandoraea-derived oxacillinase genes, named OXA-159, encode 292 amino acids and were found to be new oxacillinase variants [44]. The predicted genes with the function of OXA-159 in Pandoraea sp. 892iso and the nine Pandoraea species are shown in Table 6. The phylogenetic trees of genes with the putative function of OXA-159 are shown in Fig 7.
Table 6. The identified genes with the function of OXA-159 in Pandoraea sp. 892iso and nine Pandoraea species.
| Species | Accession number |
|---|---|
| Pandoraea sp. 892iso | fig|93222.8.peg.176 |
| Pandoraea oxalativorans | WP_052653498.1 |
| Pandoraea nosoerga | WP_150556387.1 |
| Pandoraea morbifera | WP_150567617.1 |
| Pandoraea sputorum | WP_063861062.1 |
| Pandoraea communis | WP_150690981.1 |
| Pandoraea fibrosis | WP_052240481.1 |
| Pandoraea pnomenusa | WP_023872076.1 |
| Burkholderia cepacia | WP_153490194.1 |
| Pandoraea faecigallinarum | WP_053059421.1 |
| Pandoraea capi | WP_150719552.1 |
| Pandoraea norimbergensis | WP_058375744.1 |
| Pandoraea anapnoica | WP_150740206.1 |
| Pandoraea bronchicola | WP_150559740.1 |
| Pandoraea iniqua | WP_150791439.1 |
| Pandoraea apista | WP_048627819.1 |
| Pandoraea pneumonica | WP_150680540.1 |
| Pandoraea cepalis | WP_150607462.1 |
| Pandoraea sp. XY-2 | WP_130026801.1 |
| Pandoraea pulmonicola | WP_052266736.1 |
| Pandoraea soli | WP_150552526.1 |
| Pandoraea vervacti | WP_063389849.1 |
| Pandoraea aquatica | WP_150576315.1 |
| Pandoraea captiosa | WP_150626879.1 |
| Pandoraea commovens | WP_150664304.1 |
| Pandoraea horticolens | WP_150619975.1 |
| Pandoraea anhela | WP_150669648.1 |
Fig 7. Phylogenetic tree of OXA-159 genes.
The neighbor-joining method was used in MEGA7, where a bootstrap test (1,000 replicates) is shown next to the branch.
Conclusions
We sequenced Pandoraea sp. 892iso from the genome of a Phytophthora rubi strain (numbered 109892) and combined the data with existing genomic data for other Pandoraea species. Next, we conducted a comparative genomic analysis of the genome structure, evolutionary relationships, and pathogenic characteristics of Pandoraea species. Our results identified Pandoraea sp. 892iso as Pandoraea sputorum at both the genome and gene levels. At the genome level, we carried out phylogenetic analysis of single-copy, gene co-linearity, ANI and AAI indices, rpoB similarity, MLSA phylogenetic analysis, and genome-to-genome distance calculator calculations to identify the relationship between Pandoraea sp. 892iso and P. sputorum. At the gene level, the quorum sensing genes ppnI and ppnR and the OXA-159 gene were analyzed. It is speculated that Pandoraea sp. 892iso is the endosymbiont of the Phytophthora rubi strain.
Supporting information
(DOC)
Scaffold1, scaffold2, scaffold3, scaffold4, and scaffold5 were the five largest sequences.
(DOC)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Grant funds of National Key R&D Programme of China (No. 2016YFF0203204) and National Key Technology Research and Development Programme of China (No. 2012BAK11B06) for Gui-Ming Zhang are used in study design, data collection and analysis. The funds of Scientific Research Project of General Administration of Customs. P. R. China (No. 2021HK171) for Ying Wang is used for the decision to publish, and preparation of the manuscript.
References
- 1.Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan JR, et al. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Micr. 2000; 50(2):887–99. doi: 10.1099/00207713-50-2-887 [DOI] [PubMed] [Google Scholar]
- 2.Schneider I, Queenan AM, Bauernfeind A. Novel carbapenem-hydrolyzing oxacillinase OXA-62 from Pandoraea pnomenusa. Antimicrob Agents Ch. 2006; 50(4):1330–5. doi: 10.1128/AAC.50.4.1330-1335.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stryjewski ME, LiPuma JJ, Messier RH Jr, Reller LB, Alexander BD. Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J Clin Microbiol. 2003; 41(5):2255–7. doi: 10.1128/JCM.41.5.2255-2257.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daneshvar MI, Hollis DG, Steigerwalt AG, Whitney AM, Spangler L, Douglas MP, et al. Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genome species. J Clin Microbiol. 2001; 39(5):1819–26. doi: 10.1128/JCM.39.5.1819-1826.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anandham R, Indiragandhi P, Kwon SW, Sa TM, Jeon CO, Kim YK, et al. Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int J Syst Evol Micr. 2010; 60(1):21–6. doi: 10.1099/ijs.0.012823-0 [DOI] [PubMed] [Google Scholar]
- 6.Peeters C, De Canck E, Cnockaert M, Brandt ED, Snauwaert C, Verheyde B, et al. Comparative genomics of Pandoraea, a genus enriched in xenobiotic biodegradation and metabolism. Front Microbiol, 2019; 10: 2556. doi: 10.3389/fmicb.2019.02556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin N, Tani A, Kotan R, Sedláček I, Kimbara K, Tamer AU, et al. Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int J Syst Evol Micr, 2011; 61(9): 2247–2253. doi: 10.1099/ijs.0.026138-0 [DOI] [PubMed] [Google Scholar]
- 8.Jeong SE, Lee HJ, Jia B, Jeon CO. Pandoraea terrae sp. nov., isolated from forest soil, and emended description of the genus Pandoraea Coenye et al. 2000. Int J Syst Evol Micr, 2016; 66(9): 3524–3530. doi: 10.1099/ijsem.0.001229 [DOI] [PubMed] [Google Scholar]
- 9.Johnson LN, Han J, Moskowitz SM, Burns JL, Qin X, Englund JA. Pandoraea bacteremia in a cystic fibrosis patient with associated systemic illness. The Pediatric infectious disease journal 2004; 23(9):881–882. doi: 10.1097/01.inf.0000136857.74561.3c [DOI] [PubMed] [Google Scholar]
- 10.Caraher E, Collins J, Herbert G, Murphy PG, Gallagher CG, Crowe MJ, et al. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J Med Microbiol. 2008; 57(1):15–20. doi: 10.1099/jmm.0.47544-0 [DOI] [PubMed] [Google Scholar]
- 11.Costello A, Herbert G, Fabunmi L, Schaffer K, Kavanagh KA, Caraher EM, et al. Virulence of an emerging respiratory pathogen, genus Pandoraea, in vivo and its interactions with lung epithelial cells. J Med Microbiol. 2011; 60(3):289–99. doi: 10.1099/jmm.0.022657-0 [DOI] [PubMed] [Google Scholar]
- 12.Jones AM, Webb AK. Recent advances in cross-infection in cystic fibrosis: Burkholderia cepacia complex, Pseudomonas aeruginosa, MRSA and Pandoraea spp. J Roy Soc Med. 2003; 96(Suppl 43):66. doi: 10.1258/jrsm.96.1.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okeke BC, Siddique T, Arbestain MC, Frankenberger WT. Biodegradation of γ-hexachlorocyclohexane (lindane) and α-hexachlorocyclohexane in water and a soil slurry by a Pandoraea species. J Agr Food Chem. 2002; 50(9):2548–55. doi: 10.1021/jf011422a [DOI] [PubMed] [Google Scholar]
- 14.Gómez-Gil L, Kumar P, Barriault D, Bolin JT, Sylvestre M, Eltis LD. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J Bacteriol. 2007; 189(15):5705–15. doi: 10.1128/JB.01476-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liz JAZ, Jan-Roblero J, de la Serna JZ, de León AV, Hernández-Rodríguez C. Degradation of polychlorinated biphenyl (PCB) by a consortium obtained from a contaminated soil composed of Brevibacterium, Pandoraea and Ochrobactrum. World Journal of Microbiology and Biotechnology 2009; 25(1):165–170. doi: 10.1007/s11274-008-9875-3 [DOI] [Google Scholar]
- 16.Matsui T, Yoshida T, Yoshimura T, Nagasawa T. Regioselective carboxylation of 1, 3-dihydroxybenzene by 2, 6-dihydroxybenzoate decarboxylase of Pandoraea sp. 12B-2. Appl Microbiol Biot. 2006; 73(1):95–102. doi: 10.1007/s00253-006-0437-z [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Chai L, Tang C, Yang Z, Zheng Y, Chen Y, et al. Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioproc Biosyst Eng. 2013; 36(12):1957–65. doi: 10.1007/s00449-013-0972-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Liu H, Xu Y, Wang S, Leak DJ, Zhou N. Genetic and biochemical analyses of chlorobenzene degradation gene clusters in Pandoraea sp. strain MCB032. Arch Microbiol. 2009; 191(6):485–92. doi: 10.1007/s00203-009-0476-9 [DOI] [PubMed] [Google Scholar]
- 19.Siddique T, Okeke BC, Arshad M, Frankenberger WT. Biodegradation kinetics of endosulfan by Fusarium ventricosum and a Pandoraea species. J Agr Food Chem. 2003; 51(27):8015–9. doi: 10.1021/jf030503z [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Kuo J, Cheng C, Huang Y, Ho I, Chung Y. Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system. J Hazard Mater. 2009; 172(2):1439–45. doi: 10.1016/j.jhazmat.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 21.Ee R, Yong D, Lim YL, Yin W, Chan K. Complete genome sequence of oxalate-degrading bacterium Pandoraea vervacti DSM 23571 T. J Biotechnol. 2015; 204:5–6. doi: 10.1016/j.jbiotec.2015.03.020 [DOI] [PubMed] [Google Scholar]
- 22.Tindall BJ, Rosselló-Móra R, Busse H, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Micr. 2010; 60(1):249–66. doi: 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- 23.McCarthy BJ, Bolton ET. An approach to the measurement of genetic relatedness among organisms. P Natl Acad Sci Usa. 1963; 50(1):156. doi: 10.2307/71689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schildkraut CL, Marmur J, Doty P. The formation of hybrid DNA molecules and their use in studies of DNA homologies. J Mol Biol. 1961; 3(5):516–95. doi: 10.1016/s0022-2836(61)80024-7 [DOI] [PubMed] [Google Scholar]
- 25.Wayne LG, Brenner DJ, Colwell RR, Grimont P, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Micr 1987; 37(4):463–464. doi: 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- 26.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Micr. 1994; 44(4):846–9. doi: 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- 27.Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000; 38(7):2465–7. doi: 10.1128/JCM.38.7.2465-2467.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosselló-Mora R, Amann R. The species concept for prokaryotes. Fems Microbiol Rev. 2001; 25(1):39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x [DOI] [PubMed] [Google Scholar]
- 29.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philosophical Transactions of the Royal Society B: Biological Sciences 2006; 361(1475):1929–1940. doi: 10.1098/rstb.2006.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan JZ, Halachev MR, Loman NJ, Constantinidou C, Pallen MJ. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. Bmc Microbiol 2012; 12(1):1–11. doi: 10.1186/1471-2180-12-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Micr. 2007; 57(1):81–91. doi: 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- 32.Grim CJ, Kotewicz ML, Power KA, Gopinath G, Franco AA, Jarvis KG, et al. Pan-genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaptation. Bmc Genomics. 2013; 14(1):366. doi: 10.1186/1471-2164-14-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005; 21(10):2329–35. doi: 10.1093/bioinformatics/bth324 [DOI] [PubMed] [Google Scholar]
- 34.Deloger M, El Karoui M, Petit M. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009; 191(1):91–9. doi: 10.1128/JB.01202-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 2004; 1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 36.Thompson CC, Chimetto L, Edwards RA, Swings J, Thompson FL. Microbial genomic taxonomy. Bmc Genomics. 2013; 14(1):1–8. doi: 10.1186/1471-2164-14-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adékambi T, Drancourt M, Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol 2009; 17(1):37–45. doi: 10.1016/j.tim.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 38.Adékambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA–DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Micr 2008; 58(8):1807–1814. doi: 10.1099/ijs.0.65440-0 [DOI] [PubMed] [Google Scholar]
- 39.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol 2005; 3(9):733–739. doi: 10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- 40.Estrada-De Los Santos P, Vinuesa P, Martínez-Aguilar L, Hirsch AM, Caballero-Mellado J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol 2013; 67(1):51–60. doi: 10.1007/s00284-013-0330-9 [DOI] [PubMed] [Google Scholar]
- 41.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 2007; 10(5):504–509. doi: 10.1016/j.mib.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 42.Auch AF, Klenk H, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2010; 2(1):142–148. doi: 10.4056/sigs.541628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001; 35(1):439–68. doi: 10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- 44.Schneider I, Bauernfeind A. Intrinsic carbapenem-hydrolyzing oxacillinases from members of the genus Pandoraea. Antimicrob Agents Ch. 2015; 59(11):7136–41. doi: 10.1128/AAC.01112-15 [DOI] [PMC free article] [PubMed] [Google Scholar]