Abstract
The alk genes are located on the OCT plasmid of Pseudomonas oleovorans and encode an inducible pathway for the utilization of n-alkanes as carbon and energy sources. We have investigated the influence of alternative carbon sources on the induction of this pathway in P. oleovorans and Escherichia coli alk+ recombinants. In doing so, we confirmed earlier reports that induction of alkane hydroxylase activity in pseudomonads is subject to carbon catabolite repression. Specifically, synthesis of the monooxygenase component AlkB is repressed at the transcriptional level. The alk genes have been cloned into plasmid pGEc47, which has a copy number of about 5 to 10 per cell in both E. coli and pseudomonads. Pseudomonas putida GPo12 is a P. oleovorans derivative cured of the OCT plasmid. Upon introduction of pGEc47 in this strain, carbon catabolite repression of alkane hydroxylase activity was reduced significantly. In cultures of recombinant E. coli HB101 and W3110 carrying pGEc47, induction of AlkB and transcription of the alkB gene were no longer subject to carbon catabolite repression. This suggests that carbon catabolite repression of alkane degradation is regulated differently in Pseudomonas and in E. coli strains. These results also indicate that PalkBFGHJKL, the Palk promoter, might be useful in attaining high expression levels of heterologous genes in E. coli grown on inexpensive carbon sources which normally trigger carbon catabolite repression of native expression systems in this host.
When bacteria are confronted with a mixture of carbon sources, many species preferentially utilize one carbon source until depletion and only then begin to utilize the remaining carbon sources. The genes encoding the carbon catabolic enzymes are often regulated in a similar manner; during growth on the preferred carbon source, other carbon catabolic routes are repressed. Carbon catabolite repression is well described for enteric bacteria and some gram-positive bacteria (for a review, see reference 38). In Escherichia coli, the uptake of its preferred carbon source, glucose, takes place via the phosphotransferase system, which indirectly regulates gene expression by regulating the enzyme adenylate cyclase. The product of this enzyme, cyclic AMP (cAMP), together with its cognate pleiotropic transcriptional regulatory protein, the cAMP receptor protein (CRP), modulates the expression of a multitude of catabolic operons (4).
In Pseudomonas species, the preferred carbon source is usually an organic acid such as lactate or a tricarboxylic acid cycle intermediate like citrate or succinate (29). In the presence of these substrates, the expression of catabolic pathways for terpenes such as camphor (24); for aromatic compounds like benzene (31), styrene (37), aniline (25), protocatechuate (47), phenol (34), and toluene (13, 26, 30); for chloroaromatic compounds (32); and for other nonaromatic compounds and carbohydrates (29) is subject to carbon catabolite repression.
The soil bacterium Pseudomonas oleovorans can utilize n-alkanes as sole carbon and energy sources by sequential oxidation of the terminal methyl group of the substrate into n-alkanols, n-alkanals, and n-alkanoic acids. The latter compounds are fed into the β-oxidation pathway for fatty acid degradation. The genes encoding the n-alkane degradation pathway (the alk genes) are clustered in two regions on the catabolic OCT plasmid (6), the alkBFGHJKL operon (16) and the alkST region (14) (see Fig. 1). The genetics and biochemistry of alkane oxidation by P. oleovorans have been reviewed elsewhere (42).
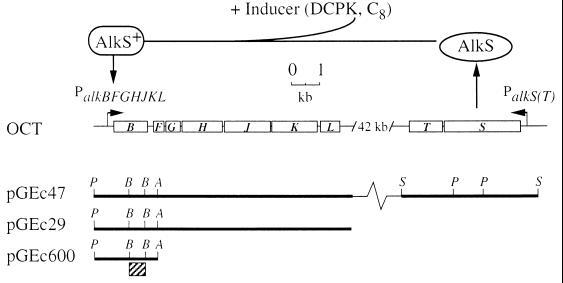
FIG. 1.
Genetic organization and induction of the alk genes on the OCT plasmid. (Top) In response to the presence of an inducer (e.g., DCPK or octane), AlkS initiates transcription of the alkBFGHJKL operon. (Bottom) Plasmids used in this study containing part of or all of the alk genes. The bar below pGEc600 denotes the DNA fragment used as the alkB probe. Restriction enzyme recognition sites: A, Asp718; B, BamHI; P, PstI; S, SalI.
Transcription activation of the alkBFGHJKL operon is mediated by the transcriptional activator AlkS (14) in response to a diverse range of inducers, including substrates like n-alkanes, gratuitous inducers such as dicyclopropylketone (DCPK) (22), and other aliphatic compounds (46) (see Fig. 1).
In the present paper, we show that induction of the PalkBFGHJKL promoter (Palk promoter) is repressed at the transcriptional level in P. oleovorans grown on certain organic acids and glucose. In contrast, when the cloned alk system is expressed in E. coli alk+ recombinants, there is no carbon catabolite repression of the Palk promoter. We present data which suggest that carbon catabolite control of the Palk promoter is a negative control.
MATERIALS AND METHODS
Chemicals.
The digoxigenin (DIG)-DNA-labeling kit, yeast total RNA, RNase-free DNase I, RNase inhibitor, positively charged nylon membrane, RNA molecular weight marker II, anti-DIG-alkaline phosphatase conjugate (Fab fragments), and disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo-[3.3.1.13,7] decan}-4-yl)phenyl phosphate (CSPD) were all from Boehringer Mannheim; X-ray film (Curix) was from Agfa. Amersham supplied 14C-labeled methylated molecular mass markers and l-[35S]methionine. Casamino Acids were from Difco, DCPK was purchased from Sigma, and the RNeasy kit was supplied by Qiagen. All other chemicals were supplied by Fluka.
Strains and plasmids.
P. putida (oleovorans) TF4-1L (ATCC 29347) is a prototroph strain and carries the OCT plasmid (40). P. putida GPo12 is a variant of P. oleovorans cured of the OCT plasmid (7). E. coli HB101 is Δ(gpt-proA)62 leuB6 thi-1 lacY1 hsdSB20 recA23 rpsL20 ara-14 galK2 xyl-5 mtl-1 supE44 mcrBB (5). E. coli W3110 [F− λ− IN (rrnD-rrnE)1] is a prototrophic K-12 strain (3), and E. coli GEc137 (15) is a fadR derivative of DH1, which is recA1 endA1 supE44 hsdR17 gyrA96 thi-1 relA1 (23). Plasmid pGEc600 carries the alkB gene in pUC19 (36). Plasmid pGEc29 consists of plasmid pLAFR1 and the alkBFGHJKL operon (17), and plasmid pGEc47 consists of the alkBFGHJKL operon and the alkST region in plasmid pLAFR1 (15). The plasmids used in this study are depicted in Fig. 1.
Growth and induction conditions.
Cells were grown on minimal medium E2 agar plates supplemented with MT microelement solution (27). Liquid cultures were grown in M9 medium (33) supplemented with FeSO4 to a final concentration of 10 μM (E. coli) or 30 μM (Pseudomonas) in Erlenmeyer flasks, at 30°C (Pseudomonas) or 37°C (E. coli) in a gyratory shaker set at 200 rpm. Various carbon and energy sources were added to the medium at 0.5% (wt/vol) as indicated in the text. Ampicillin and tetracycline were used at concentrations of 100 and 12.5 μg/ml, respectively. For E. coli GEc137, the growth medium was supplemented with 2 μg of thiamine hydrochloride per ml and 10 g of Casamino Acids per liter. For E. coli HB101(pGEc47), the growth medium was supplemented with 92 μg of l-proline per ml, 80 μg of l-leucine per ml, and 2 μg of thiamine hydrochloride per ml.
Precultures were inoculated from frozen stocks and were used to inoculate assay cultures to a cell density of about 0.05 g (dry weight) of cells per liter, and the alk system was induced (when appropriate) in exponentially growing cultures at a cell density of 0.1 g (dry weight) of cells per liter or higher by addition of DCPK to a concentration of 0.05% (vol/vol). Growth of cultures was monitored by measuring their optical density at 450 nm. Biomass was estimated as described previously (45).
Nucleic acid techniques. (i) DNA manipulations.
Transformation of E. coli, isolation of plasmid and chromosomal DNA, restriction digestions, agarose gel electrophoresis, and Southern transfer of DNA were carried out according to standard protocols (2, 39).
(ii) Conjugative transfer.
Mobilization of pGEc47 to P. putida GPo12 was performed according to the triparental mating procedure of Ditta et al. (12). Exconjugants were selected for simultaneous tetracycline resistance and growth on octane (supplied in vapor phase) on minimal medium agar plates.
(iii) DNA probe.
A 528-bp BamHI fragment from plasmid pGEc600, which represents an internal fragment of alkB, was isolated from an agarose gel by phenol extraction (2). The fragment was labeled nonradioactively with DIG11-dUTP with the DIG-DNA-labeling kit according to the manufacturer’s protocol.
(iv) RNA isolation.
Cells were harvested from exponentially growing cultures and spun down at 4°C (8,000 × g, 5 min). Cell pellets were immediately frozen in liquid nitrogen and kept at −70°C until use. Total RNA was isolated from approximately 0.25 mg (dry weight) of cells with the RNeasy kit according to the manufacturer’s protocol. The RNA samples were treated with RNase-free DNase I in the presence of RNase inhibitor (2). Purity and RNA concentration of the samples were determined by measuring the A260 and A280, with the GeneQuant photospectrometer (Pharmacia). RNA samples were stored at −70°C.
(v) RNA-agarose gel electrophoresis, Northern blotting, and detection.
Total RNA samples, accompanied by RNA molecular weight markers, were separated on a 1% (wt/vol) agarose gel containing formaldehyde as a denaturant (28). Prior to capillary Northern transfer onto a nylon membrane, the gel was subjected to a partial alkaline hydrolysis to improve the transfer of high-molecular-weight RNA fragments (39). After transfer, the RNA was fixed onto the membrane by UV cross-linking (1 min) on a standard UV transilluminator. Prehybridization and hybridization conditions (the alkB probe was used at ±40 ng/ml) for Northern blots were as specified by Boehringer Mannheim, with 50 μg of yeast total RNA per ml added to the (pre)hybridization solutions to reduce background signals. Bound probes were detected with anti-DIG-alkaline phosphatase conjugate (Fab fragments) and CSPD as chemiluminescent substrate according to Boehringer Mannheim’s protocol. Signals were recorded on X-ray film and analyzed in a Molecular Dynamics densitometer.
Pulse-chase labeling, immunoprecipitation, SDS-polyacrylamide gel electrophoresis, fluorography, and quantification of immunoprecipitates.
Pulse-chase-labeling with l-[35S]methionine (specific activity, >1,000 Ci/mmol), immunoprecipitation of AlkB from labeled cell lysates followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, fluorography, and quantification of immunoprecipitates are described in detail elsewhere (41).
Alkane hydroxylase in vivo activity measurement.
DCPK-induced P. oleovorans and E. coli alk+ recombinant cultures were harvested by centrifugation and resuspended to 2 g (dry weight) of cells per liter in assay buffer (0.1 M KPO4 [pH 7.0], 10 mM MgSO4, 0.05% [wt/vol] Triton X-100) containing 1% (wt/vol) of the respective carbon source. Two hundred fifty microliters of the cell suspension was transferred (in duplicate) to glass centrifuge tubes, which could be closed tightly with a screw cap, preventing evaporation of organic solvents. The tubes were placed in a rotary shaker set at 200 rpm at 30°C (P. oleovorans) or 37°C (E. coli alk+ recombinants). After 5 to 10 min of preincubation, 5 μl of n-nonene (99.9% pure) (Wiley Organics, Coshocton, Ohio) was added to the tubes. After incubation for another 15 min, the tubes were cooled down in an ice-water bath, the aqueous phase was saturated with NaCl, and the suspension was extracted by vortexing it for 1 min with 250 μl of n-hexane containing 0.01 or 0.1% (vol/vol) 2-octanol as internal standard. The phases were separated by centrifugation and freezing at −80°C. The hexane phase was analyzed for 1,2-epoxynonane by gas chromatography (Fisons Instruments MFC 800) isothermally at 80°C, by split injection onto a WCOT fused-silica column, CP-sil-5-CB, 25 m by 0.32 mm (Chrompack). Compounds were detected with a flame ionization detector. The specific activity of the alkane hydroxylase system was expressed as international units (IU) per gram (dry weight) of cells (micromoles of 1,2-epoxynonane · minute−1 · gram [dry weight] of cells−1). Control experiments showed that the rate of epoxide formation was linear within the first 20 to 30 min after addition of the substrate. The lower detection limit was about 0.1 U/g (dry weight) of cells. The addition of Triton X-100 to the assay buffer was necessary to obtain maximum epoxidation activities, as already noted by de Smet et al. (11). The assay is not compatible with the presence of n-octane in the growth medium, since n-octane is a competitive substrate for the n-nonene epoxidation reaction.
RESULTS
Influence of the carbon source on alkane hydroxylase activity in P. oleovorans.
Alkane oxidation activity in various P. aeruginosa (10, 43, 44) and P. putida (21, 22) strains has been reported to be subject to carbon catabolite repression by glucose and other carbon sources. We have pursued this line of investigation and have investigated the alkane hydroxylase activity of cultures of P. oleovorans grown on different carbon sources. Exponentially growing cultures were induced with the gratuitous inducer DCPK, and after 2 h, cells were harvested and incubated with n-nonene. The alkane hydroxylase system converts this substrate into 1,2-epoxynonane, which is readily analyzed by gas chromatography.
The results are summarized in Table 1 (left columns) and show that the carbon source on which the cells were grown influenced the epoxidation rates: whereas a high activity was observed for the glycerol-, pyruvate-, and citrate-grown cells, low activity was observed for the succinate- and especially the lactate-grown cells. Thus, expression of the alkane hydroxylase system was subject to carbon catabolite repression in P. oleovorans.
TABLE 1.
Influence of carbon sources on the alkane hydroxylase activities of P. oleovorans and P. putida GPo12(pGEc47) cultures
| Carbon sourceb | Alkane hydroxylase activity (IU · g [dry wt] of cells−1)a
|
|||
|---|---|---|---|---|
|
P. oleovorans
|
P. putida GPo12(pGEc47)
|
|||
| − Ind. | + Ind. | − Ind. | + Ind. | |
| Glycerol | 0.14 ± 0.05 | 9.2 ± 0.4 | 0.17 ± 0.05 | 10.3 ± 0.1 |
| Pyruvate | 0 | 9.6 ± 0.3 | NDc | ND |
| Citrate | 0 | 11.0 ± 0.1 | ND | ND |
| Succinate | 0.15 ± 0.02 | 1.6 ± 0.1 | 0 | 5.1 ± 0.2 |
| Lactate | 0 | 0.1 | 0 | 10.4 ± 0.1 |
Exponentially growing cultures were split into two subcultures; one of these cultures was induced with 0.05% (vol/vol) DCPK (+ Ind.), and the other one was not (− Ind.). The alkane hydroxylase activity of resting cells was measured 2 h after induction of the + Ind. culture and represents the average and the standard deviation thereof of two independent measurements.
All carbon sources were added to the cultures at a concentration of 0.5% (wt/vol).
ND, not determined.
AlkB induction kinetics in P. oleovorans.
To investigate whether the reduced alkane hydroxylase activity reflected reduced expression of the alk genes, we determined the relative synthesis rate of AlkB in P. oleovorans cultures grown on minimal medium with different carbon sources.
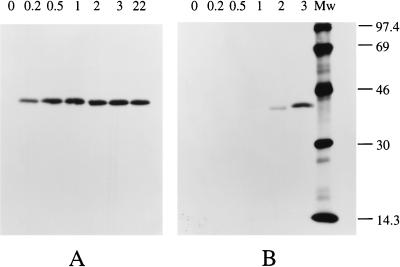
The relative AlkB synthesis rate after induction was determined by pulse-chase-labeling cells with [35S]methionine, followed by immunoprecipitation and quantification of labeled AlkB from cell lysates. Fluorograms of AlkB immunoprecipitated from P. oleovorans grown on glycerol and glucose illustrate the efficacy and specificity of this method (Fig. 2).
FIG. 2.
Synthesis of AlkB by P. oleovorans during growth on glycerol or glucose. The P. oleovorans cultures, growing exponentially on glycerol (A) or glucose (B), were induced with DCPK at t = 0. At the indicated time points (hours), cell samples were pulse-labeled with [35S]methionine, and AlkB was immunoprecipitated from cell lysates containing 100,000 trichloroacetic acid-precipitable counts. After SDS-polyacrylamide gel electrophoresis analysis and treatment with 1 M sodium salicylate as fluorophore, X-ray films were exposed to the gels at −80°C for 142 h. Molecular mass markers (lane Mw) are indicated in kilodaltons.
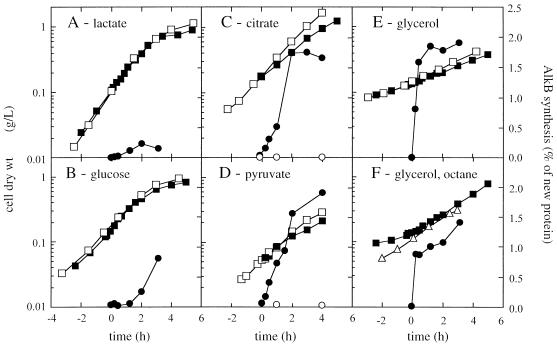
The induction kinetics of AlkB were dependent on the carbon source on which the cells were grown (Fig. 3). After induction, the P. oleovorans culture growing on glycerol immediately started synthesizing AlkB. After about 30 min, a steady-state AlkB level of 1.5 to 1.75% (wt/wt) relative to newly synthesized protein was attained. Citrate and pyruvate showed a slight repressive effect during the first 2 h, whereas glucose and especially lactate significantly repressed the induction of AlkB synthesis.
FIG. 3.
Growth and relative AlkB synthesis rates in P. oleovorans cultures growing on different carbon sources. The cultures were induced at t = 0, after which cell samples were pulse-chase-labeled with [35S]methionine at the indicated time points. Growth of noninduced (□) and induced (■) cultures is expressed as dry weight of cells. AlkB was immunoprecipitated from cell lysates, and its synthesis rate was quantified relative to total protein synthesis as described in Materials and Methods. AlkB synthesis in noninduced (○) and induced (●) cultures was expressed as weight percentage of newly synthesized protein. For induction, 0.05% (vol/vol) DCPK was used for the cultures grown on lactate (A), glucose (B), citrate (C), pyruvate (D), and glycerol (E). The culture in panel F was grown on glycerol and induced with 5% (vol/vol) n-octane. In addition, growth of a P. oleovorans culture with 5% (vol/vol) n-octane as sole carbon source is indicated (▵) in panel F.
P. oleovorans grew on the various C sources with a distinct growth rate, which decreased somewhat after addition of DCPK for the cultures growing on glycerol, citrate, and pyruvate but not for the lactate- or glucose-grown culture. A similar decrease in growth rate after induction of the alk genes in P. oleovorans has been observed by Chen et al. (8).
We also induced the alk system of a P. oleovorans culture growing exponentially on glycerol with 5% (vol/vol) octane instead of DCPK. In this case, however, the growth rate of the culture increased from 0.25 to 0.35 h−1, which corresponds to the growth rate of a P. oleovorans shaking-flask culture growing on octane (Fig. 3F). This change in growth rate suggests that P. oleovorans preferentially used n-octane over glycerol as the main carbon source. The relative AlkB synthesis level of the octane-induced culture was somewhat lower than that of the DCPK-induced culture, reflecting the potency of DCPK as an inducer of the alk system (22). To allow for numerical comparison, the results of the AlkB synthesis induction measurements have been summarized in Table 2.
TABLE 2.
Relative AlkB synthesis rates and specific growth rates in P. oleovorans grown on various carbon sourcesa
| Carbon sourceb | μ (h−1)
|
AlkB synthesisc (% of new protein)
|
||
|---|---|---|---|---|
| − Ind.d | + Ind. | − Ind. | + Ind. | |
| Octane | NAe | 0.35 | NA | 1.1 |
| Lactate | 0.55 | 0.48 | <0.01 | 0.24 |
| Glucose | 0.55 | 0.54 | 0.03 | 0.25 |
| Succinate | 0.60 | 0.54 | NDf | ND |
| Citrate | 0.55 | 0.42 | 0.06 | 1.75 |
| Pyruvate | 0.42 | 0.32 | 0.06 | 1.6 |
| Glycerol | 0.23 | 0.18 | <0.01 | 1.8 |
All cultures except the octane-grown culture were induced by addition of 0.05% (vol/vol) DCPK.
All carbon sources were supplied at a concentration of 0.5% (wt/vol) except octane, which was added to 5% (vol/vol).
The relative AlkB synthesis rate (as weight percentage of newly synthesized protein) is shown.
Ind., inducer.
NA, not applicable.
ND, not determined.
AlkB induction kinetics in E. coli alk+ recombinants.
The alkane hydroxylase system was introduced into E. coli on plasmid pGEc47, allowing controlled and functional expression of the alk genes in E. coli (15). The induction kinetics of AlkB in two E. coli alk+ recombinants, HB101(pGEc47) and W3110(pGEc47), were investigated as described above for P. oleovorans. As carbon sources for the E. coli alk+ recombinants, glycerol (a prototype nonrepressive carbon source) and glucose (a prototype repressive carbon source) were chosen.
As in P. oleovorans, induction of the alk genes reduced the growth rate of the E. coli alk+ recombinants, confirming earlier observations (18, 36).
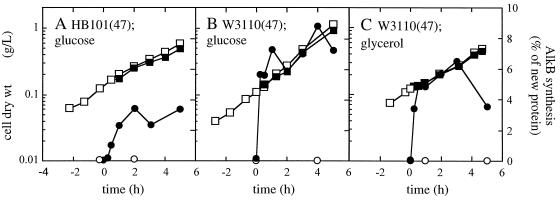
Glucose-grown E. coli HB101(pGEc47) and W3110(pGEc47) cultures showed a rapid increase in their relative AlkB synthesis rates, to approximately 3.5 and 7 to 8% of newly synthesized protein, respectively (Fig. 4). In the glycerol-grown E. coli W3110(pGEc47) cultures, AlkB accounted for approximately 6% of newly synthesized protein under steady-state conditions.
FIG. 4.
Growth and relative AlkB synthesis levels in E. coli alk+ recombinant cultures growing on different carbon sources. The cultures were induced with 0.05% (vol/vol) DCPK at t = 0, after which cell samples were pulse-chase-labeled with [35S]methionine at the indicated time points and analyzed for newly synthesized AlkB. Growth of noninduced (□) and induced (■) cultures is expressed as dry weight of cells, and AlkB synthesis in noninduced (○) and induced (●) cultures was expressed as weight percentage of newly synthesized protein. (A) E. coli HB101(pGEc47) grown on glucose; (B) E. coli W3110(pGEc47) grown on glucose; (C) E. coli W3110(pGEc47) grown on glycerol.
Thus, in contrast to P. oleovorans, no carbon catabolite repression of alkB expression was observed in induced E. coli alk+ recombinants. In fact, glucose-grown E. coli W3110(pGEc47) showed a slightly higher relative AlkB synthesis rate than did glycerol-grown cells.
Expression from the Palk promoter.
The specificity of the DIG-labeled alkB probe (consisting of an internal alkB sequence [Fig. 1]) was verified on Southern blots of chromosomal and plasmid DNA from various strains (data not shown). No alkB DNA sequences were found in chromosomal DNA samples of P. putida GPo12, thereby confirming the absence of the alk genes from this strain.
We investigated the influence of different carbon sources on the induction of the Palk promoter in more detail. To this end, cultures of P. oleovorans and E. coli alk+ recombinants were grown on different carbon sources and the alk system was induced with DCPK. After 2 h, cells were harvested from induced and noninduced control cultures. Total RNA was isolated from the cells, and equal amounts of RNA per sample were analyzed by agarose gel electrophoresis and Northern blotting. The results of the Northern blotting experiments are shown in Fig. 5.
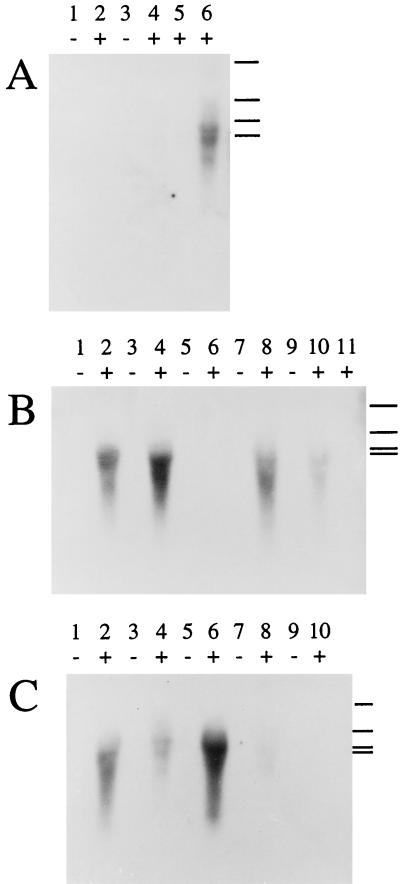
FIG. 5.
Northern blot analysis of total RNA isolated from Pseudomonas and E. coli recombinant cultures with or without 2-h induction of the alk genes with DCPK. Bound DIG-labeled alkB DNA probes were detected with a chemiluminescent substrate. The signal of the blots in panels A, B, and C was recorded on X-ray film by exposure for 5, 2, and 20 min, respectively. The plus and minus signs above the lanes indicate samples taken from induced and uninduced cultures, respectively. RNA molecular size markers of 5.3, 2.8, 1.9, and 1.6 kb (top to bottom, respectively) are indicated in the right margin of each panel. The alkB signal was detected at about 1.5 kb. (A) Lanes 1 and 2, P. putida GPo12 grown on glycerol; lanes 3 and 4, P. oleovorans grown on succinate; lane 5, P. oleovorans grown on glucose; lane 6, P. oleovorans grown on glycerol. (B) Lanes 1 and 2, E. coli W3110(pGEc47) grown on glucose; lanes 3 and 4, E. coli HB101(pGEc47) grown on glucose; lanes 5 and 6, P. oleovorans grown on glucose; lanes 7 and 8, E. coli W3110(pGEc47) grown on pyruvate; lanes 9 and 10, P. oleovorans grown on pyruvate; lane 11, P. oleovorans grown on succinate. (C) Lanes 1 and 2, E. coli W3110(pGEc47) grown on glycerol; lanes 3 and 4, P. oleovorans grown on glycerol; lanes 5 and 6, E. coli HB101(pGEc47) grown on lactate; lanes 7 and 8, P. oleovorans grown on lactate; lanes 9 and 10, E. coli GEc137(pGEc29) grown on glucose.
The alkB probe hybridized with RNA isolated from induced P. oleovorans and E. coli alk+ cells and gave a signal at approximately 1.5 kb, which represents the alkB transcript (35). Noninduced cultures showed no detectable alkB mRNA synthesis, and neither did induced and noninduced cultures of P. putida GPo12 (Fig. 5A).
For P. oleovorans, the intensity of the alkB hybridization signal was generally weak. The low intensity of the alkB mRNA signal can be attributed to the relatively low expression of AlkB in the wild-type host, where this protein never exceeds 1.8% of the total cell protein for any of the carbon sources tested (Fig. 3 and Table 2). The use of glycerol (Fig. 5A, lane 6, and 5C, lane 4) and pyruvate (Fig. 5B, lane 10) as carbon sources allowed detection of some mRNA, while all other carbon sources used (lactate, glucose, and succinate) failed to show an mRNA signal.
When induced, E. coli HB101(pGEc47) and W3110(pGEc47) showed higher alkB mRNA levels than did P. oleovorans on all carbon sources tested, and no repression of alkB transcription was observed on any of the carbon sources tested (Fig. 5B and C), in accord with the higher levels of AlkB synthesis in E. coli alk+ recombinants, as shown in Fig. 4. Rather surprisingly, E. coli HB101(pGEc47) showed somewhat higher alkB mRNA levels than did E. coli W3110(pGEc47), despite the fact that the latter produces two- to threefold more AlkB than does the former recombinant (Fig. 4), indicating that these experiments provide only a rough indication of the presence or absence of alkB mRNA.
Plasmid pGEc29 carries the alkBFGHJKL operon, which cannot be expressed due to the absence of its positive regulator AlkS (Fig. 1). As expected, total RNA from an induced culture of E. coli GEc137(pGEc29) grown on glucose did not show an alkB mRNA signal (Fig. 5C, lane 10).
These results indicate that the regulation of the Palk promoter with respect to carbon catabolite repression in the E. coli alk+ recombinants is fundamentally different from that in P. oleovorans.
Carbon catabolite repression of the Palk promoter in P. putida GPo12(pGEc47) is reduced.
Reintroduction of the alk genes on plasmid pGEc47 restores the ability of P. putida GPo12 to use octane as a carbon source (15). We investigated whether the carbon catabolite repression of alkane hydroxylase activity was controlled identically in P. oleovorans and in P. putida GPo12(pGEc47). Analogously to the experiments performed with P. oleovorans, the alkane hydroxylase activities of induced and noninduced cultures grown on different carbon sources were measured (Table 1, right columns).
The alkane hydroxylase activities of P. putida GPo12(pGEc47) cultures grown on glycerol and on lactate were similar to each other and to that found for P. oleovorans grown on glycerol. The alkane hydroxylase activity of P. putida GPo12(pGEc47) grown on succinate was about 50% lower. Thus, carbon catabolite repression is strongly reduced or abolished in this recombinant strain.
DISCUSSION
Carbon catabolite repression in pseudomonads appears to be regulated differently from that in Bacillus subtilis or E. coli (9). Whereas in the latter microorganism, the cAMP-CRP complex plays an important role in carbon catabolite repression, no such role for cAMP was found in pseudomonads (29). The only known Pseudomonas CRP homolog, Vfr, was found to be involved primarily in quorum sensing, not in carbon catabolite repression control (1). Thus, the molecular mechanisms of carbon catabolite repression in pseudomonads remain elusive to date.
We have studied the carbon catabolite repression of alkane degradation in P. oleovorans, a P. putida strain carrying the OCT plasmid, in some detail. The induction of the alkane hydroxylase activity was eliminated or reduced significantly during growth on succinate and lactate. Analysis of AlkB synthesis induction indicated that during growth on succinate, lactate, or glucose the expression of the alkBFGHJKL operon was repressed. These results were confirmed by alkB mRNA analysis, showing that carbon catabolite repression of the Palk promoter in P. oleovorans takes place at the transcriptional level.
Induction with DCPK during growth of P. oleovorans on glycerol, citrate, and pyruvate, but not on lactate or glucose, led to significant expression of AlkB (>0.5% of new protein within 2 h after induction) and to a decrease in growth rate. As we have found for other alk+ recombinants (8, 18, 36), the higher the expression of the alkane hydroxylase system and of the alkB gene in particular, the greater the difference in specific growth rates between the noninduced and the induced cultures. As a result, repressive carbon sources are good growth substrates for P. oleovorans even in the presence of alk system inducers, as is indicated by their respective growth rates (μ) in Table 2. The contrary statement is not true: citrate is an excellent growth substrate for P. oleovorans, yet it allows full induction of the Palk promoter at the expense of a distinct reduction in growth rate.
In order to also study the carbon catabolite repression of the Palk promoter-AlkS system in enteric bacteria, we transformed two E. coli strains with broad-host-range plasmid pGEc47 containing the alk genes. The induction of alkB mRNA and that of AlkB synthesis were not repressed in the two E. coli alk+ recombinants growing on glycerol, lactate, or glucose. Thus, carbon catabolite repression of the Palk promoter-AlkS system is completely absent in E. coli strains.
To study the carbon catabolite repression behavior of the cloned alk genes in their original genetic background, plasmid pGEc47 was introduced into P. putida GPo12. The resulting alk+ recombinant displayed high alkane hydroxylase activities not only when grown on glycerol but also when grown on lactate or succinate (Table 1). Thus, carbon catabolite repression of alkane hydroxylase activity was almost completely abolished in this recombinant strain.
There are several possible explanations for these findings. First, a putative negative control factor might reduce alkS expression in P. oleovorans under carbon catabolite repression conditions, thereby reducing transcription of the alkBFGHJKL operon. Second, a putative negative control factor might bind to AlkS and/or to Palk, thus compromising the ability of AlkS to initiate transcription from the Palk promoter.
The OCT plasmid of P. oleovorans has a copy number of 1 to 2 (19). Plasmid pGEc47, an RK2 derivative, has a copy number between 5 and 10 in E. coli and pseudomonads (20). The putative factor exerting negative control over Palk induction appears to be specific for pseudomonads, since it is completely absent in the E. coli alk+ recombinants. The fact that carbon catabolite repression of the Palk promoter is almost completely abolished when the alk genes are reintroduced into P. putida GPo12 suggests that the control factor might be encoded on the OCT plasmid (outside of the two EcoRI fragments that contain the alkST region and the alkBFGHJKL operon [Fig. 1]). Alternatively, the putative control factor (conferring carbon catabolite repression on the Palk promoter in P. oleovorans) could be titrated by the extra copies of the Palk promoter genes present in the P. putida GPo12(pGEc47) recombinants. Additional experimental work will be necessary to clarify these issues.
The finding that the Palk promoter-AlkS system is not subject to carbon catabolite repression in E. coli allows the use of this system for expression of target genes in recombinant E. coli strains. This strategy can combine several virtues: low-cost inducers (e.g., octane and DCPK), the tightly controlled and efficient gene expression of the Palk promoter-AlkS system, and the high cell and product yields and low costs associated with the use of glucose as a carbon source.
ACKNOWLEDGMENTS
We thank Maarten Nieboer for the gift of plasmid pGEc600 and Marcel Wubbolts for useful suggestions.
This research was supported by the Swiss National Science Foundation through the Swiss Priority Program in Biotechnology, grant no. 5002-037023.
REFERENCES
- 1.Albus A M, Pesci E C, Runyen-Janecky L J, West S E H, Iglewski B H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 3.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 1st ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1191–1219. [Google Scholar]
- 4.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarty A M, Chou G, Gunsalus I C. Genetic regulation of octane dissimulation plasmids in Pseudomonas. Proc Natl Acad Sci USA. 1973;70:1137–1140. doi: 10.1073/pnas.70.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Janssen D B, Witholt B. Growth on octane alters the membrane lipid fatty acids of Pseudomonas oleovorans due to the induction of alkB and synthesis of octanol. J Bacteriol. 1995;197:6894–6901. doi: 10.1128/jb.177.23.6894-6901.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Janssen D B, Witholt B. Physiological changes and alk gene instability in Pseudomonas oleovorans during induction and expression of alk genes. J Bacteriol. 1996;178:5508–5512. doi: 10.1128/jb.178.18.5508-5512.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the Pseudomonas. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 10.Dalhoff A, Rehm H J. Studies on regulation of tetradecane oxidation in Pseudomonas aeruginosa. II. The effect of glucose on tetradecane oxidation. Eur J Appl Microbiol. 1976;3:203–211. [Google Scholar]
- 11.de Smet M J, Wijnberg H, Witholt B. Synthesis of 1,2-epoxyoctane by Pseudomonas oleovorans during growth in a two-phase system containing high concentrations of 1-octene. Appl Environ Microbiol. 1981;42:811–816. doi: 10.1128/aem.42.5.811-816.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duetz W A, Marqués S, De Jong C, Ramos J L, Van Andel J G. Inducibility of the TOL catabolic pathway in Pseudomonas putida (pWW0) growing on succinate in continuous culture—evidence of carbon catabolite repression control. J Bacteriol. 1994;176:2354–2361. doi: 10.1128/jb.176.8.2354-2361.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggink G, Engel H, Meijer W G, Otten J, Kingma J, Witholt B. Alkane utilization in Pseudomonas oleovorans: structure and function of the regulatory locus alkR. J Biol Chem. 1988;263:13400–13405. [PubMed] [Google Scholar]
- 15.Eggink G, Lageveen R G, Altenburg B, Witholt B. Controlled and functional expression of the Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J Biol Chem. 1987;262:17712–17718. [PubMed] [Google Scholar]
- 16.Eggink G, van Lelyveld P H, Arnberg A, Arfman N, Witteveen C, Witholt B. Structure of the Pseudomonas putida alkBAC operon. Identification of transcription and translation products. J Biol Chem. 1987;262:6400–6406. [PubMed] [Google Scholar]
- 17.Eggink G, van Lelyveld P H, Witholt B. The construction of a gene bank from Pseudomonas oleovorans. Molecular cloning of the alk sequences of the OCT plasmid coding for the alkane oxidizing enzymes. Prog Ind Microbiol. 1984;20:373–380. [Google Scholar]
- 18.Favre-Bulle O, Witholt B. Biooxidation of n-octane by a recombinant Escherichia coli in a two-liquid-phase system: effect of medium components on cell growth and alkane oxidation activity. Enzyme Microb Technol. 1992;14:931–937. [Google Scholar]
- 19.Fennewald M, Prevatt W, Meyer R, Shapiro J. Isolation of Inc P-2 plasmid DNA from Pseudomonas aeruginosa. Plasmid. 1978;1:164–173. doi: 10.1016/0147-619x(78)90036-7. [DOI] [PubMed] [Google Scholar]
- 20.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fish N M, Allenby D J, Lilly M D. Oxidation of n-alkanes: growth of Pseudomonas putida. Eur J Appl Microbiol Biotechnol. 1982;14:259–262. [Google Scholar]
- 22.Grund A, Shapiro J, Fennewald M, Bacha P, Leahy J, Markbreiter K, Nieder M, Toepfer M. Regulation of alkane oxidation in Pseudomonas putida. J Bacteriol. 1975;123:546–556. doi: 10.1128/jb.123.2.546-556.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Hartline R A, Gunsalus I C. Induction specificity and catabolite repression of the early enzymes in camphor degradation by Pseudomonas putida. J Bacteriol. 1971;106:468–478. doi: 10.1128/jb.106.2.468-478.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helm V, Reber H. Investigation on the regulation of aniline utilization in Pseudomonas multivorans strain An1. Eur J Appl Microbiol Biotechnol. 1979;7:191–199. [Google Scholar]
- 26.Holtel A, Marqués S, Möhler I, Jakubzik U, Timmis K N. Carbon sources-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J Bacteriol. 1994;176:1773–1776. doi: 10.1128/jb.176.6.1773-1776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lageveen R G, Huisman G W, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on the formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehrach H, Diamond D, Wozney J M, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions; a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 29.MacGregor C H, Wolff J A, Arora S K, Hylemon P B, Phibbs P V. Catabolite repression control in Pseudomonas aeruginosa. In: Galli E, Silver S, Witholt B, editors. Pseudomonas molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 198–206. [Google Scholar]
- 30.Marqués S, Holtel A, Timmis K N, Ramos J L. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J Bacteriol. 1994;176:2517–2524. doi: 10.1128/jb.176.9.2517-2524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason J R. The induction and repression of benzene and catechol-oxidizing capacity of Pseudomonas putida ML2 studied in perturbed chemostat culture. Arch Microbiol. 1994;162:57–62. doi: 10.1007/BF00264373. [DOI] [PubMed] [Google Scholar]
- 32.McFall S M, Abraham B, Narsolis C G, Chakrabarty A M. A tricarboxylic acid cycle intermediate regulating transcription of a chloroaromatic biodegradative pathway: fumarate-mediated repression of the clcABD operon. J Bacteriol. 1997;179:6729–6735. doi: 10.1128/jb.179.21.6729-6735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 34.Müller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieboer M, Gunnewijk M, van Beilen J B, Witholt B. Determinants for overproduction of the Pseudomonas oleovorans cytoplasmic membrane protein alkane hydroxylase in alk+Escherichia coli W3110. J Bacteriol. 1997;179:762–768. doi: 10.1128/jb.179.3.762-768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieboer M, Kingma J, Witholt B. The alkane oxidation system of Pseudomonas oleovorans: induction of the alk-genes in Escherichia coli W3110 affects membrane biogenesis and results in overexpression of alkane hydroxylase in a distinct cytoplasmic membrane subfraction. Mol Microbiol. 1993;8:1039–1051. doi: 10.1111/j.1365-2958.1993.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor K, Buckley C M, Hartmans S, Dobson A D W. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol. 1995;61:544–548. doi: 10.1128/aem.61.2.544-548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier M H., Jr Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schwartz R D, McCoy C J. Pseudomonas oleovorans hydroxylation-epoxidation system: additional strain improvements. Appl Microbiol. 1973;26:217–218. doi: 10.1128/am.26.2.217-218.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staijen I E, Hatzimanikatis V, Witholt B. The AlkB monooxygenase of Pseudomonas oleovorans: synthesis, stability and level in recombinant Escherichia coli and the native host. Eur J Biochem. 1997;244:462–470. doi: 10.1111/j.1432-1033.1997.00462.x. [DOI] [PubMed] [Google Scholar]
- 42.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 43.van der Linden A C. Epoxidation of α-olefins by heptane-grown Pseudomonas cells. Biochim Biophys Acta. 1963;77:157–159. doi: 10.1016/0006-3002(63)90484-0. [DOI] [PubMed] [Google Scholar]
- 44.van Eyk J, Bartels T J. Paraffin oxidation in Pseudomonas aeruginosa. I. Induction of paraffin oxidation. J Bacteriol. 1968;96:706–712. doi: 10.1128/jb.96.3.706-712.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witholt B. Method for isolating mutants overproducing NAD and its precursors. J Bacteriol. 1972;109:350–364. doi: 10.1128/jb.109.1.350-364.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wubbolts M G. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1994. [Google Scholar]
- 47.Zylstra G J, Olsen R, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase. J Bacteriol. 1989;171:5907–5917. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]