Abstract
Substance use disorders are associated with disruptions to both circadian rhythms and cellular metabolic state. At the molecular level, the circadian molecular clock and cellular metabolic state may be interconnected through interactions with the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase, Sirtuin 1 (SIRT1). In the nucleus accumbens (NAc), a region important for reward, both SIRT1 and the circadian transcription factor neuronal PAS domain protein 2 (NPAS2) are highly enriched, and both are regulated by the metabolic cofactor NAD+. Substances of abuse, like cocaine, greatly disrupt cellular metabolism and promote oxidative stress; however, their effects on NAD+ in the brain remain unclear. Interestingly, cocaine also induces NAc expression of both NPAS2 and SIRT1, and both have independently been shown to regulate cocaine reward in mice. However, whether NPAS2 and SIRT1 interact in the NAc and/or whether together they regulate reward is unknown. Here, we demonstrate diurnal expression of Npas2, Sirt1, and NAD+ in the NAc, which is altered by cocaine-induced upregulation. Additionally, co-immunoprecipitation reveals NPAS2 and SIRT1 interact in the NAc, and cross-analysis of NPAS2 and SIRT1 chromatin immunoprecipitation sequencing reveals several reward-relevant and metabolic-related pathways enriched among shared gene targets. Notably, NAc-specific Npas2 knock-down or a functional Npas2 mutation in mice attenuates SIRT1-mediated increases in cocaine preference. Together, our data reveal an interaction between NPAS2 and SIRT1 in the NAc which may serve to integrate cocaine’s effects on circadian and metabolic factors, leading to regulation of drug reward.
Keywords: Circadian, Reward, NPAS2, SIRT1, Nucleus Accumbens, Cocaine
Graphical Abstract
Both the metabolic cofactor SIRT1 and the circadian transcription factor NPAS2 are highly enriched in the nucleus accumbens (NAc) and regulate reward in mice. Here we demonstrate that NPAS2 and SIRT1 interact in the NAc and share several reward-and metabolic relevant gene targets. Moreover, the increase in preference for cocaine seen with SIRT1 activation is abolished with disruption of NPAS2. These studies shed light on how NPAS2 and SIRT1 in the NAc regulate metabolic state and drug reward.
INTRODUCTION
Substance use disorders, or SUDs, are widely prevalent and debilitating psychiatric disorders with devastating social and economic burdens. While the exact mechanisms contributing to the development of SUDs are still unknown, emerging evidence suggests disruptions in both circadian rhythms and redox metabolism as key contributors (Logan et al., 2014; Uys et al., 2014). For example, drugs of abuse are known to disrupt circadian rhythms and sleep (Hasler et al., 2012), as well as redox homeostasis (Cunha-Oliveira et al., 2013). Conversely, disruption to circadian genes directly alters reward regulation in rodent models (Parekh et al., 2015), and pharmacologically targeting redox regulation has shown promise as a treatment option for SUDs in both clinical and preclinical animal studies (Womersley et al., 2019). Interestingly, few studies have looked at how crosstalk between circadian rhythms and redox homeostasis may contribute to SUD vulnerability, despite the growing understanding of how the circadian and metabolic systems are interconnected at the molecular level (Eckel-Mahan & Sassone-Corsi, 2013).
Nearly all cells in mammalian organisms express a circadian molecular clock, consisting of several transcription and translation feedback loops (Partch et al., 2014). At its core, Circadian Locomotor Output Cycles Kaput (CLOCK) or its homolog, Neuronal PAS Domain 2 (NPAS2), hetero-dimerize with Brain and Muscle ARNT-like 1 (BMAL1) to drive transcription of many output genes, including Period and Cryptochrome genes. Throughout the day, translated Period (PER) and Cryptochrome (CRY) proteins accumulate in the cytoplasm and eventually shuttle back into the nucleus as a heterodimer within a complex to inhibit their own transcription. An additional auxiliary feedback loop involves the transcriptional regulation of Bmal1 and Npas2 by the clock-controlled nuclear receptors RAR Related Orphan Receptor A (RORα) and REV-ERBα (or NR1D1), with RORα promoting their transcription and REV-ERBα repressing (Preitner et al., 2002; Sato et al., 2004; Crumbley et al., 2010). Together, these negative feedback loops cycle roughly every 24 hours and control cellular physiology across the day, with nearly half of all protein-coding genes being regulated by the molecular clock (Partch et al., 2014; Zhang et al., 2014). Notably, NPAS2 is structurally and functionally similar to CLOCK (DeBruyne et al., 2007; Landgraf, Wang, et al., 2016), but the two proteins differ in their expression across the brain; while CLOCK is ubiquitously expressed, NPAS2 is enriched in specific regions, including the forebrain (Reick et al., 2001). Unlike CLOCK, NPAS2 is highly enriched in the striatum and nucleus accumbens (NAc) (Garcia et al., 2000), regions integral to processing reward and motivated behaviors (Volkow & Morales, 2015). Previous work from our lab has demonstrated that mutations in Clock increase cocaine and alcohol intake (McClung et al., 2005; Ozburn et al., 2012, 2013); however, mutations or NAc knock-down of Npas2 reduce cocaine preference (Ozburn et al., 2015; Parekh et al., 2019). This differential regulation of reward may arise through NAc-specific interactions between NPAS2 and cellular metabolic state.
In addition to NPAS2’s direct regulation of key metabolic pathways (O’Neil et al., 2013), NPAS2 can integrate metabolic/redox information itself. Work from the McKnight lab and others have demonstrated NPAS2’s ability to bind the heme cofactor and act as a gas-responsive redox sensor (Dioum et al., 2002; Uchida et al., 2005). Moreover, NPAS2’s DNA binding activity is also influenced by changes in pH and the mitochondrial cofactor nicotinamide adenine dinucleotide (NAD), whereby transcriptional activity is promoted by higher pH and/or NADH levels (Rutter et al., 2001; Yoshii et al., 2015). NAD exists in both an oxidized (NAD+) and reduced form (NADH) that both respond to changes in redox state and help maintain cellular metabolic homeostasis (Ying, 2008; Houtkooper et al., 2010). While levels of NAD have been shown to modulate NPAS2 function under ultra-physiological conditions in vitro (Rutter et al., 2001), a more likely mechanism in vivo may be through interactions with the NAD+-dependent deacetylase, sirtuin 1 (SIRT1).
SIRT1 is a histone and protein deacetylase whose activity is directly coupled to redox balance via NAD+ levels, and it regulates transcriptional control of the cellular metabolic state (Cantó & Auwerx, 2012). SIRT1 has also been shown to play an integral role in regulating NAc function and reward in mice. Work from the Nestler lab and colleagues has found that SIRT1 regulates drug-associated synaptic plasticity, such that overexpression of SIRT1 increases NAc dendritic spine density and plasticity-related gene expression (Ferguson et al., 2013). Furthermore, both SIRT1 overexpression in the NAc and activation via the SIRT1 agonist resveratrol increase cocaine preference in mice, while antagonists decrease preference (Renthal et al., 2009; Ferguson et al., 2013). One mechanism that may underlie these reward-related findings is the known integration of SIRT1 metabolic signaling with the circadian molecular clock. Extensive work from Sassone-Corsi, Schibler, and others illustrates how SIRT1 in the liver and in cell culture models directly regulates the molecular clock through both NAD+ dependent PER2 deacetylation and/or repression of CLOCK/BMAL1 transcription (Asher et al., 2008; Nakahata et al., 2008). This crosstalk is thought to be governed by a transcriptional-enzymatic feedback loop through circadian control of the nicotinamide phosphoribosyltransferase (NAMPT) NAD+ salvage pathway by CLOCK and SIRT1 (Nakahata et al., 2009; Ramsey et al., 2009). While previous studies have characterized rhythmicity of NAD+ in the liver (Ramsey et al., 2009) and rhythmicity of related metabolic factors FAD and NAD(P)H in the hippocampus and suprachiasmatic nucleus (Wang et al., 2012; Naseri Kouzehgarani et al., 2020), characterization of NAD+ in the brain has yet to be done outside of our previous study investigating the ventral tegmental area (VTA) (Logan et al., 2019). Interestingly, in this study, we demonstrated in the VTA that CLOCK and SIRT1 regulate dopaminergic activity and cocaine reward, mediated by NAD+ cellular redox (Logan et al., 2019). This work is further supported by recent findings in the VTA showing NAMPT-mediated NAD biosynthesis directly promotes cocaine reward in a SIRT1 dependent manner (Kong et al., 2018). However, it remains unclear whether similar mechanisms hold true in the NAc and/or with CLOCK’s homolog NPAS2. No studies to date have investigated whether SIRT1 and NPAS2 interact in the NAc or whether together they may be important for regulating reward.
In the present study, we find diurnal variation in expression of Npas2 and Sirt1 in the mouse NAc that is upregulated by cocaine exposure, as well as an altered diurnal variation of Nampt expression and NAD+ levels via upregulation. Most notably, we demonstrate NPAS2 and SIRT1 interact in the NAc and share several reward-relevant and metabolic-related gene targets, and this interaction is complimented by our findings further demonstrating NPAS2 and SIRT1 promote reward behavior through their expression in the NAc.
MATERIALS & METHODS
Animals
Experiments utilized either male C57BL/6J mice (The Jackson Laboratory; Bar Harbor, ME; IMSR Cat# JAX:000664, RRID: IMSR_JAX:000664) or Npas2 mutant and wild-type (WT) littermates (maintained on a C57BL/6J background) obtained from the Weaver lab (Dallmann et al., 2011), ages 8–12 weeks. Npas2 mutant mice carry a mutation resulting in NPAS2 lacking its functional domain, rendering it incapable of binding BMAL1 and carrying out its core circadian functions (Garcia et al., 2000). All mice were maintained on a 12:12 light-dark cycle (lights on at 0700, zeitgeber time (ZT) 0, and lights off at 1900, ZT12) with irradiated food and autoclaved water provided ad libitum (Standard Chow: ProLab IsoPro RMH 3000 5P76 irradiated; LabDiet, St. Louis, MO), housed at 22 ± 1 °C with standard coarse aspen Sani-Chips bedding (P.J. Murphy; Montville, NJ) and Nestlets nesting pad (Ancare; Bellmore, NY). Separate cohorts of mice were used for each of the experiments described below unless otherwise indicated. Animal use was conducted in accordance with the National Institute of Health guidelines, and all procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Drug Administration
Cocaine hydrochloride was provided by the National Institute on Drug Abuse (NIDA) and dissolved with 0.9% saline (Fisher Scientific). Mice were injected intraperitoneally (i.p.) with cocaine (5 mg/kg for CPP or 20 mg/kg for molecular experiments), resveratrol (20 mg/kg; SelleckChem, Houston, TX, USA; dissolved in 5% hydroxypropyl β-cyclodextrin; Sigma-Aldrich, St. Louis, MO), or 0.9% saline – all at 10 ml/kg volume. Systemic administration of resveratrol has been shown to readily cross the blood-brain barrier (Baur & Sinclair, 2006). For molecular experiments, mice were administered saline (10 ml/kg i.p.), chronic cocaine (20 m/kg i.p. for 14 days), or acute cocaine (saline for 13 days and cocaine for on the last day at 20 m/kg i.p.). Lower cocaine dosage was used for CPP to avoid ceiling effects in behavior; 5 mg/kg was previously determined to be optimal by our lab (Ozburn et al., 2015). Higher cocaine dosage (20 mg/kg) for molecular experiments was used, aligning with previous SIRT1 studies (Renthal et al., 2009; Ferguson et al., 2013). Drugs were administered between ZT4–8, as previously published (Falcon et al., 2013).
Viral-Placement Surgery and Verification
Npas2 knock-down stereotaxic surgery was performed as previously described (Ozburn et al., 2015). Briefly, male C57BL/6J mice were given bilateral stereotaxic 1 uL injections into the NAc (Bregma: angle 10°, AP +1.5, ML ± 1.5, DV −4.4 mm) of purified high titer adeno-associated virus (AAV2) containing a short hairpin RNA (shRNA) for Npas2 or a scrambled sequence, both expressing a GFP reporter (UNC Vector Core; Chapel Hill, NC, USA; RRID: SCR_002448). The viruses were constructed, packaged, and validated in our previous study (Ozburn et al., 2015). Briefly, the shRNA was constructed against Npas2 with a sequence in the 3’-untranslated region (5′-GAACACTGGATTCTTCCTGTTAAC-3′), and the scramble control consists of a random 24 base sequence (5′-CGGAATTTAGTTACGGGGATCCAC-3′) with no similarities to any known genes or messenger RNA (mRNA). Sirt1 overexpression was performed similarly, though with a purified high titer AAV2-CMV-eGFP-2A-SIRT1 or eGFP control. Sirt1 and control viruses were constructed, packaged, and validated by the manufacturer (Vector Biolabs; Malvern, PA, USA; Cat# AAV-272007). For the injection surgery, so as to minimize pain and discomfort, surgeries were quickly performed under isoflurane anesthesia, and mice were given pain treatment up to 2 days following surgery (Rimadyl, 5mg/kg; Zoetis; Parsippany-Troy Hills, NJ). Mice recovered for at least two weeks to allow for sufficient viral expression before behavioral testing. Following testing, viral placement and spread were verified using immunofluorescence (IF). Mice were perfused with ice-cold phosphate-buffered saline (1x PBS) followed by paraformaldehyde in PBS (4% PFA at pH 7.4). Brains were post-fixed for 24 hours and then transferred to 30% sucrose for another 24 hours. Brains were cryo-sectioned at 40-um sections before processing for IF. Green Fluorescent Protein (GFP) signal was enhanced using a primary anti-GFP antibody (Abcam, Cambridge, UK; Cat# ab13970; RRID: AB_300798) before sections were mounted on slides with VECTASHIELD mounting medium-plus DAPI (Vector Laboratories; Burlingame, CA, USA; Cat# H-1500, RRID: AB_2336788). Sections were imaged at 4x magnification using an Olympus epifluorescence microscope (Olympus Corporation; Shinjuku, Tokyo, Japan). Additionally, virus placement was also verified using a fluorescent protein flashlight (Nightsea; Lexington, MA; Cat# DFP-1) with a fresh tissue section to allow for NAc tissue collection and downstream applications. Mice not expressing virus bilaterally and within the NAc were excluded from analysis.
Behavioral Testing
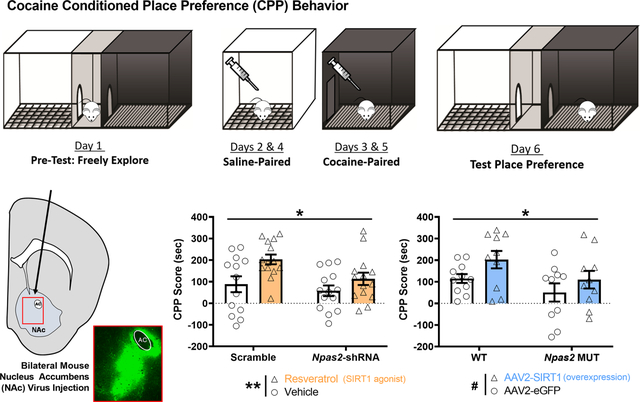
Cocaine conditioned place preference (CPP) was performed as previously published (Parekh et al., 2019). A biased conditioning protocol was utilized whereby cocaine pairing is performed on the mouse’s least preferred side, as determined during the pre-test. Behavioral testing occurred between ZT2–5 with at least 30 minutes of habituation to the room, as previously published (Parekh et al., 2019). Briefly, mice were first allowed to freely explore a 3-chambered apparatus (Med Associates; Fairfax, VT) for 20 minutes to test for inherent bias. Mice exhibiting greater than 10 minutes in any chamber during the pre-test were excluded. On days 2 and 4 of CPP, mice were injected with saline (i.p) and placed in their initially preferred chamber of the apparatus for 20 minutes. On days 3 and 5 of CPP, mice were injected with cocaine (5 mg/kg, i.p.) and placed in their initially non-preferred chamber for 20 minutes. On the final day (Day 6), mice were placed into the 3-chambered apparatus for 20 minutes and allowed to explore freely. Time spent in each chamber was recorded. CPP score was calculated by subtracting the pre-conditioning time spent in the cocaine-paired side from the test-day time on that side (i.e., Test cocaine-paired time minus Pre-Test cocaine-paired time).
Gene Expression: RNA isolation and RT-qPCR
Mice were sacrificed across four times of day (ZT 5, 10, 17, 22) starting 24 hours after the last injection, as previously published (Falcon et al., 2013). Brains were rapidly extracted and the NAc was quickly micro-dissected before being snap-frozen on dry ice. Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen; Hilden, Germany) and converted to complementary DNA (cDNA) using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific; Waltham, MA, USA). Gene expression was measured using quantitative polymerase chain reaction (qPCR) with primers for circadian and metabolic genes of interest, performed in a CFX96 Touch Real-Time PCR detection system (Bio-Rad; Hercules, CA, USA). The following primers were utilized: Mouse 18s forward: 5’-ACCGCAGCTAGGAATAATGGA-3’, Mouse 18s reverse: 5’-GCCTCAGTTCCGAAAACCA-3’; Mouse Npas2 forward: GACACTGGAGTCCAGACGCAA, Mouse Npas2 reverse: AATGTATACAGGGTGCGCCAAA; Mouse Arntl (Bmal1) forward: CTTGCAAGCACCTTCCTTCC, Mouse Arntl (Bmal1) reverse: GGGTCATCTTTGTCTGTGTC; Mouse Nr1d1 (Rev-erbα) forward: GCAAATCAGATCTCAGGTGC, Mouse Nr1d1 (Rev-erbα) reverse: AGGTGATAACACCACCTGTG; Mouse Nr1f1 (Rorα) forward: ACGCCCACCTACAACATCTC, Mouse Nr1f1 (Rorα) reverse: ACATATGGGTTCGGGTTTGA; Mouse Sirt1 forward: TCTGTCTCCTGTGGGATTCC, Mouse Sirt1 reverse: GATGCTGTTGCAAAGGAACC; Mouse Nampt forward: TACAGTGGCCACAAATTCCA, Mouse Nampt reverse: AATGAGCAGATGCCCCTATG. The comparative cycle threshold (ct) method (i.e., 2-ΔΔCT method) was used to calculate relative gene expression (Schmittgen & Livak, 2008), normalized to the 18s reference gene. Relative gene expression values were reported as the mean normalized Ct value ± SEM. Rose plots were manually generated in Adobe Illustrator (Adobe Inc.; San Jose, CA) using observed peaks in expression to weight the rose petals.
Co-Immunoprecipitation (Co-IP) Assay
Mice were sacrificed at two times of day (ZT 5 and 17), brains were rapidly extracted, and the NAc was quickly micro-dissected before being snap-frozen on dry ice. NAc punches were homogenized mechanically and with sonication in pierce immunoprecipitation (IP) lysis buffer (Thermo Fisher Scientific) plus protein phosphatase and protease inhibitor cocktails. A detergent compatible (DC) protein assay (Bio-Rad; Hercules, CA, USA) was performed in an Epoch Microplate Spectrophotometer (BioTek; Winooski, VT, USA) to determine total protein concentrations. Interaction between NPAS2 and SIRT1 was assessed using the Dynabead Co-Immunoprecipitation Kit (Invitrogen; Carlsbad, CA, USA), as per manufacturer instructions. Briefly, NAc samples (500 μg input per sample) were incubated with anti-NPAS2 (Santa Cruz; Dallas, TX, USA; Cat# sc-28708, RRID: AB_2282741) or anti-SIRT1 (Abcam; Cambridge, UK; Cat# ab12193, RRID: AB_298923) coupled M-270 Epoxy Dynabeads, and the protein complex was eluted. Detection of SIRT1 or NPAS2 in the eluted protein complex was assessed using SDS-PAGE followed by immunoblotting for NPAS2 or SIRT1, respectively. Briefly, eluted IP samples were run in a 7.5% mini-gel at 100V (Bio-Rad) and transferred overnight at 4C. Blotting for NPAS2 or SIRT1 was achieved using the LI-COR Odyssey Imaging System (LI-COR Biosciences; Lincoln, NE, USA) with antibodies for SIRT1 (Santa Cruz; Cat# sc-15404, RRID: AB_2188346) or NPAS2 (Invitrogen; Carlsbad, CA, USA; Cat# PA5–95462, RRID: AB_2807265) and GAPDH (Millipore; Burlington, MA; Cat# MAB374, RRID: AB_2107445). Both Goat Anti-Rabbit IgG polyclonal (IRDye 800CW; LI-COR Biosciences, Cat# 926–32211, RRID:AB_621843) and Goat Anti-Mouse IgG polyclonal (IRDye 680RD; LI-COR Biosciences, Cat# 926–68070, RRID:AB_10956588) secondary antibodies were used for detection of NPAS2 or SIRT1 and GAPDH, respectively. Precision Plus Protein Dual Color Standard was used for molecular weight estimation (Bio-Rad; Cat# 1610374). Quantification of blots was performed using Image Studio Lite (LI-COR), normalizing band signal intensity to the ZT5 group’s average band signal intensity. GAPDH was utilized as a negative control and was not detected.
NAD+ Measurement
Mice were sacrificed across four times of day (ZT 5, 10, 17, 22) starting 24 hours after the last injection, as previously published (Falcon et al., 2013). Brains were rapidly extracted and the NAc was quickly micro-dissected before being snap frozen using liquid nitrogen. High-Performance Liquid Chromatography (HPLC) quantification of NAD+ was performed as previously published (Yoshino & Imai, 2013; Stromsdorfer et al., 2016). Briefly, NAc punches were rapidly homogenized in perchloric acid and then neutralized in potassium carbonate. NAD+ concentrations were determined using an HPLC system (Prominence; Shimadzu Scientific Instruments; Columbia, MD, USA) and a SUPELCOSIL LC-18-T column (Sigma Aldrich; St. Louis, MO, USA). NAD+ concentrations were normalized to weights of the frozen tissue samples.
Chromatin Immunoprecipitation sequencing (ChIP-seq) Pathway Analyses
Previously, we performed ChIP-seq for NPAS2 in the NAc to identify novel DNA binding targets (Ozburn et al., 2015). NAc SIRT1 ChIP-seq data from Ferguson et al. (2015) was utilized for comparative analyses investigating shared binding targets between NPAS2 and SIRT1 in the NAc. Here, both Ingenuity Pathway Analysis (IPA) software (QIAGEN; Hilden, Germany; https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis; RRID: SCR_008653) (Krämer et al., 2014) and the online bioinformatics database Metascape (https://metascape.org/; RRID: SCR_016620) (Zhou et al., 2019) were used to identify enriched pathways and processes among the shared binding targets gene list between NPAS2 and SIRT1 at ZT2, the shared time point between the two datasets. For both analyses, shared genes were analyzed as follows: (1) The 3189 genes bound by SIRT1 (e.g., gene body and/or promoter; Saline; ZT 2) and the 2347 genes bound by NPAS2 at ZT2 (e.g., gene body and/or promoter) were first input through the Bioinformatics Evolutionary Genomics Venn Diagram Tool to identify 390 shared gene binding targets (http://bioinformatics.psb.ugent.be/webtools/Venn/); (2) the 390 shared genes bound were then used as an input list for both IPA and Metascape analyses, with 11,005 total identified genes from both datasets used as a reference gene set; and (3) for reporting of significant pathways and processes, a significance threshold of p<0.05 (or a -log10(p-value) of 1.3 in figures, depicted by a red line) was utilized. IPA software was used to identify enriched canonical molecular pathways and top disease and biological functions, while Metascape was used to identify enriched biological processes using only Gene Ontology (GO) Biological Processes as the ontology source. Within Metascape, all statistically enriched terms, accumulative hypergeometric p-values, and enrichment factors were automatically calculated and used for filtering. Remaining significant terms were then hierarchically clustered into a tree based on Kappa-statistical similarities among their gene memberships, with a 0.3 kappa score applied as the threshold to cast the tree into term clusters. Process enrichment was detected using the default settings of a p<0.01 cutoff, minimum overlap of 3, and a minimum enrichment of 1.5. A subset of representative terms from this cluster were then converted into an enrichment network using Cytoscape (v3.1.2; RRID:SCR_003032) (Shannon et al., 2003), with each term represented by a circle node colored by cluster identity and terms with a similarity score > 0.3 linked together (the thickness representing the similarity score). (Zhou et al., 2019)
Statistical Analysis
For all statistical analyses, GraphPad Prism 9 software was used (GraphPad Software; San Diego, CA, USA; RRID: SCR_002798). Statistical analysis of more than two groups was performed using either a one-way or two-way analysis of variance (ANOVA) with significant interactions followed by Bonferroni post-hoc tests corrected for multiple comparisons. Otherwise, statistical analysis of just two groups was performed using a Student’s t-test. Across all data sets, outliers were tested for using the Grubbs’ test (α = 0.05), and no more than one outlier was removed if it exceeded the critical value of Z. Data throughout are expressed as mean ± SEM with α = 0.05 considered statistically significant.
RESULTS
The Molecular Clock and Regulators of Cellular Metabolic State Show Diurnal Variation in the NAc
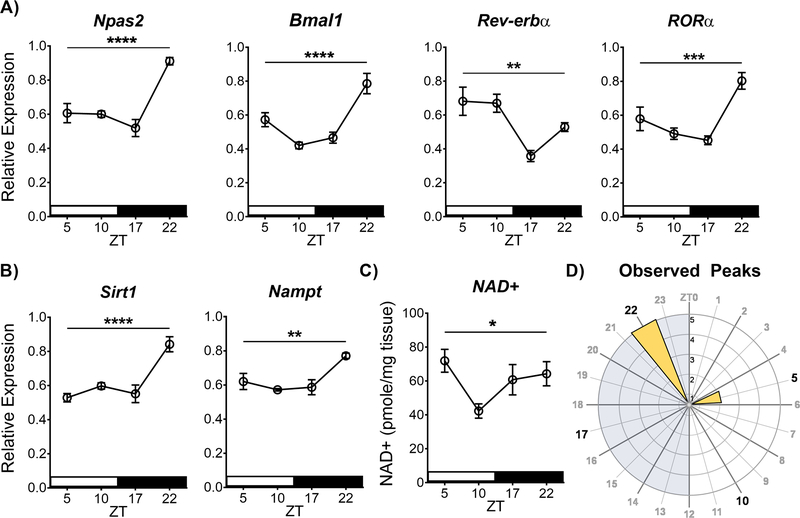
Accumulating evidence suggests circadian molecular clock function may be coupled with the cellular metabolic state through a transcriptional-enzymatic feedback loop via SIRT1 and the NAD+ salvage pathway (Eckel-Mahan & Sassone-Corsi, 2013). However, this phenomenon has largely been characterized in the liver and primarily in the context of SIRT1 and CLOCK/BMAL1. To investigate whether this relationship exists in the NAc and/or with NPAS2/BMAL1, we first sought to confirm and expand our previous findings that core molecular clock genes vary over the 24 hr cycle in the NAc (Falcon et al., 2013). We performed reverse transcriptase quantitative PCR (RT-qPCR) with whole NAc tissue that was collected across four times of day (ZT 5, 10, 17, and 22) from saline-injected C57BL/6J mice (10 ml/kg, i.p., 14 days) starting 24 hours after the last injection, utilizing primers for core circadian genes Npas2, Bmal1, Rev-erbα, and RORα (normalized to 18s) to measure mRNA across time of day. While not all clock genes were assessed, Npas2 and Bmal1 were chosen as central members of the core feedback loop, and Rev-erbα and RORα as members of a key auxiliary feedback loop that regulates both Npas2 and Bmal1 (Preitner et al., 2002; Sato et al., 2004; Crumbley et al., 2010). Much like our previous findings, Npas2 showed diurnal variation in expression (Figure 1A), with a highly significant main effect of time (F(3, 20) = 18.34, p<0.0001, peak: ZT22). Expanding on our previous findings, Bmal1 and the auxiliary feedback loop members Rev-erbα and RORα also showed diurnal variation in the NAc, all with significant main effects of time (F(3, 20) = 15.72, p<0.0001, peak: ZT22; F(3, 20) = 7.94, p=0.001, peak: ZT5; F(3, 20) = 10.94, p=0.0002, peak: ZT22; respectively).
Figure 1. The molecular clock and regulators of cellular metabolic state show diurnal variation in the NAc.
(A) Core molecular clock gene expression of Npas2, Bmal1, Rev-erbα, and RORα show diurnal variation in the NAc, as well as the (B) metabolic genes Sirt1 and Nampt – measured using RT-qPCR. Relative expression normalized to the reference gene 18s. (C) HPLC analysis of NAD+ levels also shows diurnal variation in the NAc. (D) Rose plot of the observed peaks of expression illustrates peaks are primarily at ZT22 during the dark phase. Asterisks and bar indicate a significant main effect of time measured in a one-way ANOVA (* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001). NAc tissue was collected from saline-treated animals (14 days, i.p. at 10 ml/kg, between ZT4–8). Data represented as Mean ± SEM (n=5–6 / ZT). White stripe indicates lights on, black stripe indicates lights off. Grey shading on the rose plot indicates lights off, and the inset axis shows the number of genes. ZT 0 = 7 am.
While SIRT1 is known to be expressed in the NAc (Renthal et al., 2009; Ferguson et al., 2015), no studies to date have investigated its expression across time of day. Here we find Sirt1 expression shows a diurnal variation in the NAc (Figure 1B), with a highly significant main effect of time (F(3, 19) = 14.32; p<0.0001, peak: ZT22). Moreover, we find Nampt, a clock-controlled enzyme in the NAD+ salvage pathway and proposed transcriptional-enzymatic feedback loop (Ramsey et al., 2009), also display diurnal variation in the NAc (Figure 1B), with a significant main effect of time (F(3, 18) = 7.5; p=0.001, peak: ZT22). Most notably, we also investigated the diurnal variation of NAD+ in the NAc. Importantly, using HPLC, we find NAD+ shows a diurnal variation in the NAc (Figure 1C), with a significant main effect of time (F(3, 20) = 3.26, p<0.05, peak: ZT5). Together, Npas2, Sirt1, and the NAD+ salvage pathway are circadian regulated in the NAc, with peaks of expression generally around ZT22 during the dark phase (Figure 1D).
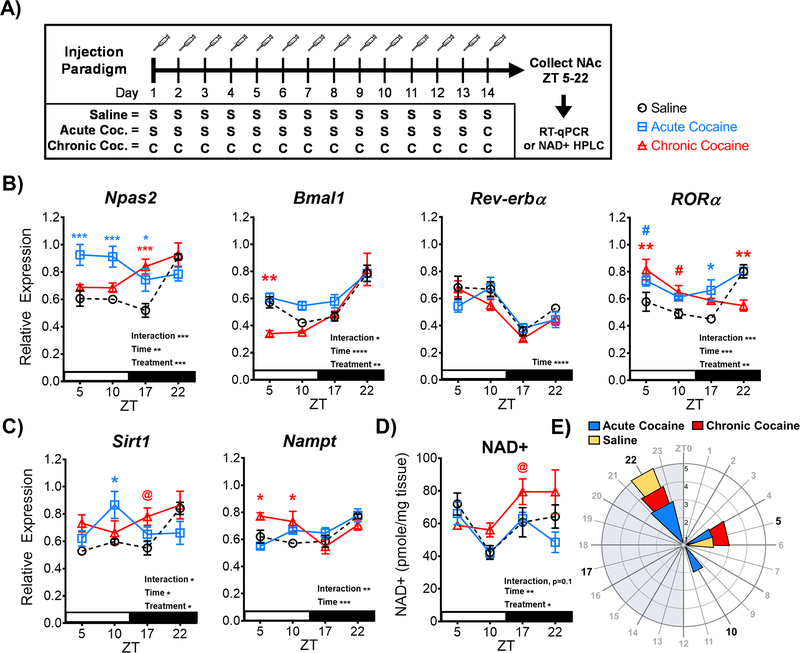
Diurnal Variations of the Molecular Clock, SIRT1, and NAD+ are Affected by Cocaine
Previous studies have found that NPAS2 and SIRT1 expression are induced by cocaine when measured at a single time of day (Renthal et al., 2009; Falcon et al., 2013). We next looked to extend these findings by investigating how the expression is affected by cocaine across time of day in the NAc. Mice were injected with either saline (10 ml/kg, i.p., 14 days; from Figure 1) or cocaine (acute: 20 mg/kg, i.p., 1 day; chronic: 14 days) and sacrificed across four times of day (ZT 5, 10, 17, 22; starting 24hr after the last injection) for NAc tissue to be used in gene expression analyses (Figure 2A). Notably, diurnal expression of Npas2 is upregulated by both acute and chronic cocaine exposure (Figure 2B), with a significant interaction and main effects of time and treatment (Interaction: F(6, 59) = 5.134, p=0.0003; Time: F(3, 59) = 5.331, p=0.002; Treatment: F(2, 59) = 10.47, p=0.0001). Acute cocaine significantly increases Npas2 expression across ZT5, 10, and 17 (p<0.05), leading to a phase-advance in observed peak expression to the light phase (new peak: ZT5). Chronic cocaine significantly increases Npas2 expression only at ZT 17 (p<0.001), relative to saline. Diurnal expression is also altered by cocaine exposure for both Bmal1 (Interaction: F(6, 59) = 2.293, p<0.05; Time: F(3, 59) = 29.46, p<0.0001; Treatment: F(2, 59) = 6.768, p=0.002) and RORα (Interaction: F(6, 60) = 5.297, p=0.0002; Time: F(3, 60) = 7.912, p=0.0002; Treatment: F(2, 60) = 6.148, p=0.003), but not Rev-erbα, which only shows a main effect of time (F(3, 60) = 21.85, p<0.0001). Chronic cocaine significantly reduced expression of Bmal1 at ZT5 (p=0.005), along with a significant upregulation of RORa at ZT5 (p=0.003) and a significant downregulation of RORa at ZT22 (p=0.001) – advancing RORa’s observed peak expression to the light phase (new peak: ZT5) (Figure 2B). While previous studies suggest NAc Sirt1 expression is induced by cocaine exposure (Renthal et al., 2009), no studies have investigated how cocaine affects Sirt1 expression across time of day. Here we find that diurnal variation of Sirt1 expression is altered by both acute and chronic cocaine exposure (Figure 2C), with a significant interaction and main effects of time and treatment (Interaction: F(6, 59) = 2.808, p=0.01; Time: F(3, 59) = 3.179, p<0.05; Treatment: F(2, 59) = 3.628, p<0.05). Much like Npas2, acute cocaine upregulates Sirt1 expression at ZT10 (p<0.05), leading to a phase advance in peak expression to the light phase (new peak: ZT10), while chronic cocaine increases Sirt1 only at ZT 17 (@, p=0.0511). Moreover, the gene encoding the NAD+ salvage pathway enzyme, Nampt, also shows significant upregulation of expression (Figure 2C), with a significant interaction and a main effect of time (Interaction: F(6, 57) = 4.084, p=0.001; Time: F(3, 57) = 7.639, p=0.0002). Chronic cocaine’s upregulation of Nampt expression at ZT5 and ZT10 (p<0.05) also leads to a phase-advancing of observed peak expression to the light phase (new peak: ZT5).
Figure 2. Cocaine exposure alters diurnal variation of the circadian molecular clock and cellular metabolic state in the NAc.
(A) Schematic outlining the 14-day injection paradigm. Mice were given either saline (S; 10 ml/kg, i.p.), chronic cocaine (C; 20 mg/kg, i.p.), or acute cocaine (A; 13 days x saline, 1-day x cocaine). NAc tissue was collected beginning 24 hours after the last injection for molecular assays. (B) Following cocaine exposure, diurnal variation of core molecular clock gene expression of Npas2, Bmal1, and ROR, as well as (C) the metabolic genes Sirt1 and Nampt, were significantly altered in the NAc – as indicated by significant interactions and main effects of time and/or treatment by 2-way ANOVA. (ROR blue #, p=0.0751; ROR red #, p=0.0732; Sirt1 @, p=0.0511). (D) HPLC analysis revealed cocaine exposure also significantly disrupts diurnal variation of NAD+ levels in the NAc, with significant main effects of time and treatment. (NAD+ @, p=0.0572). (E) Rose plot illustrates observed peaks in expression are primarily at ZT22 during the dark phase, but cocaine exposure advances more peaks of expression to the light phase. Saline data from Figure 1 are depicted as dotted lines for comparison purposes. Injections were administered between ZT4–8. White stripe indicates lights on, black stripe indicates lights off. Grey shading on rose plot indicates lights off, and the inset axis shows the number of genes. Asterisks, pounds, and at-signs above the plotted lines indicate a statistical difference relative to saline, revealed in a Bonferroni post-hoc analysis. Color of asterisk, pound, or at-sign corresponds to the treatment group compared. Data represented as Mean ± SEM (n=5–6 / ZT). (* p<0.05, ** p<0.01, *** p<0.001). ZT 0 = 7 am.
Extending our finding that NAD+ shows diurnal variation in the NAc (Figure 1C), we next wanted to investigate how cocaine exposure may affect NAD+ levels across time of day (Figure 2D). Interestingly, diurnal variation of NAD+ levels in the NAc are significantly altered by cocaine exposure, with significant main effects of time and treatment (Time: F(3, 54) = 6.157, p=0.001; Treatment: F(2, 54) = 3.895, p=0.02) and no significant interaction (F(6, 54) = 1.884, p=0.1). Expanding on the simple main effects, relative to saline, chronic cocaine increased NAD+ at ZT 17 (@, p=0.057) and delayed the observed peak expression to the dark phase (new peak: ZT22). Together, cocaine significantly alters diurnal variation of Npas2, Sirt1, and related metabolic regulators in the NAc through upregulation of expression and a phase advancing of observed peak expression, resulting in more peaks in expression during the light phase (Figure 2E).
NPAS2 and SIRT1 are Important for Cocaine Conditioned Place Preference
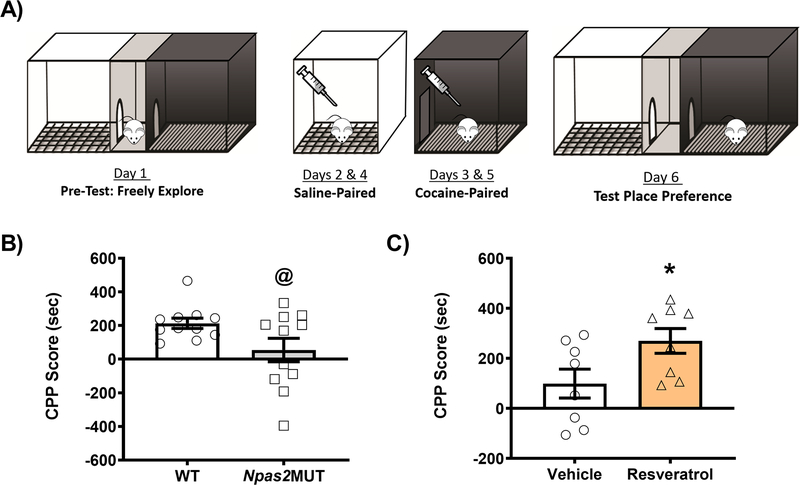
Previous work from our lab has demonstrated NPAS2 is important for cocaine preference, in that mice carrying a functional mutation in Npas2 (Npas2 mutant mice) show decreased cocaine CPP (Ozburn et al., 2015). Additionally, work from our lab and others has demonstrated that SIRT1 is important for cocaine preference, in that activation of SIRT1 via resveratrol increases cocaine CPP (Renthal et al., 2009; Logan et al., 2019). Before determining whether together NPAS2 and SIRT1 are important for cocaine preference through their expression in the NAc, we first wanted to reconfirm the global/systemic findings from previous studies. To accomplish this, Npas2 mutant mice were run through the cocaine CPP paradigm (Figure 3A) and conditioned to both saline and 5 mg/kg cocaine (i.p.) paired chambers. Consistent with our previous findings, Npas2 mutant mice show a decrease in cocaine CPP, as compared to WT littermates (t(20)=2.082, @ p=0.0504; Figure 3B). To test the effects of resveratrol, C57BL/6J mice were run through the cocaine CPP paradigm and conditioned to both saline and 5 mg/kg cocaine paired chambers; however, during conditioning days, mice were injected with either saline or 20 mg/kg resveratrol (i.p) 30 minutes before conditioning. As previously published, mice injected with the SIRT1 agonist resveratrol show a significant increase in cocaine CPP (Figure 3C), relative to saline controls (t(14)=2.243, p<0.05).
Figure 3. NPAS2 and SIRT1 are important for cocaine preference.
(A) Schematic showing the cocaine conditioned place preference (CPP) paradigm. Mice were conditioned to saline (10 ml/kg, i.p.) or cocaine (5 mg/kg, i.p.) on conditioning days. (B) Npas2 mutant mice show decreased cocaine preference relative to wild-type (WT) littermates. (C) C57BL/6J mice were administered vehicle or resveratrol (20 mg/kg; i.p.) 30 minutes before conditioning. Mice administered resveratrol showed a significant increase in cocaine preference relative to vehicle controls. Data represented as Mean ± SEM (n= 8–11). CPP score is calculated by subtracting the initial pre-test time in the cocaine paired chamber from the same chamber’s test-day time (i.e., Test cocaine-paired time minus Pre-Test cocaine-paired time). Asterisk indicates significance relative to controls (* p<0.05). At-sign indicates a trend towards significance (@, p=0.0504).
NPAS2 and SIRT1 Interact in the NAc and Share Reward-Relevant and Metabolic-Related Gene Targets
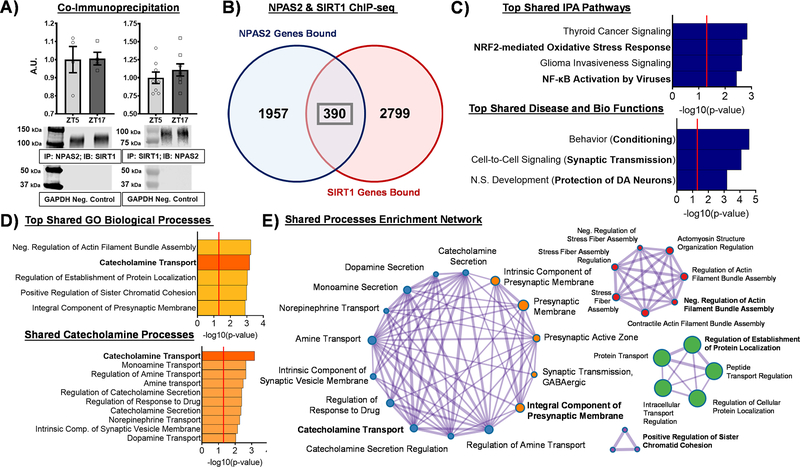
While studies performed in the liver suggest SIRT1 interacts with CLOCK/BMAL1 to integrate metabolic information and regulate molecular clock function (Nakahata et al., 2008, 2009), no studies have investigated whether this interaction holds true with CLOCK’s homolog NPAS2 and/or within the NAc. Given the previous findings indicating both NPAS2 and SIRT1 are induced in the NAc by cocaine exposure (Renthal et al., 2009; Falcon et al., 2013) and they each play a role in regulating cocaine reward (Renthal et al., 2009; Ferguson et al., 2013; Ozburn et al., 2015), we wanted to investigate whether NPAS2 and SIRT1 interact in the NAc. First, utilizing BioGrid (Stark et al., 2006; Hunter et al., 2009) and InterPro (Hunter et al., 2009), we assessed the domains and families common between NPAS2 and SIRT1 interactors (Supp. Figure 1). Interestingly, NPAS2 is structurally similar to its paralog, CLOCK, and it shares BHLH and PAS domains with multiple known SIRT1 interactors. Additionally, known SIRT1 interactors CLOCK, ARNTL (BMAL1), and EPAS1, along with NPAS2, make up 4 of 7 proteins belonging to the Nuclear Translocator Family, which could imply a similar binding pattern with SIRT1 for the group (Supp. Figure 1). Taken together, these common domains and families suggest structurally an interaction between NPAS2 and SIRT1 is plausible.
Next, to assess a potential interaction experimentally, both co-immunoprecipitation (Co-IP) of NPAS2 followed by western blot analysis for SIRT1 and Co-IP of SIRT1 followed by western blot analysis for NPAS2 were performed in NAc tissue from C57BL/6J mice. Across two times of day, SIRT1 was detected in NPAS2 pulldown (Figure 4A, Left) and NPAS2 was detected in SIRT1 pulldown (Figure 4A, Right), together indicating that NPAS2 and SIRT1 interact in the same protein complex in the NAc. Notably, GAPDH was not detected in either Co-IP (Figure 4A, Bottom), illustrating the specificity of the assay. Previous work from our lab and Ferguson et al. utilizing chromatin immunoprecipitation sequencing (ChIP-seq) in NAc tissue demonstrated NPAS2 and SIRT1 regulate transcription of reward-relevant genes (Ferguson et al., 2015; Ozburn et al., 2015); however, an investigation into potential shared gene targets has yet to be studied. Cross analysis of the NPAS2 and SIRT1 ChIP-seq datasets at ZT2, the time shared between the two datasets, revealed 2,347 and 3,189 genes bound (e.g., promoter or gene body) by NPAS2 and SIRT1, respectively, with 390 bound genes shared between the two (Figure 4B). To investigate the functional importance of these 390 shared gene targets, we performed Ingenuity Pathway Analysis (IPA) and Metascape analysis (Zhou et al., 2019) to uncover molecular pathways and biological processes enriched among the shared geneset. Interestingly, IPA revealed NRF2-mediated oxidative stress response and NF-kB activation to be among the top shared canonical pathways (Figure 4C), both pathways known to mitigate oxidative stress and promote metabolic homeostasis (Mauro et al., 2011; Ma, 2013; Vomund et al., 2017; Lingappan, 2018). Moreover, top biological functions enriched among the shared genes include behavioral conditioning, synaptic transmission, and protection of dopamine neurons (Figure 4C) – all relevant to the regulation of reward and reward-related behavior. These results are further supported by Metascape (GO) Biological Processes analysis revealing enriched catecholamine and dopamine-related processes, as well as regulation of synaptic transmission (Figure 4D); notably, when plotted as an enrichment network visualization, these synaptic transmission, catecholamine, and dopamine-related processes are highly interconnected (Figure 4E). These findings not only expand upon previous data implicating NPAS2 and SIRT1 in the regulation of reward, but also suggest an interaction in the NAc that may facilitate co-regulation of reward-relevant genes/processes.
Figure 4. NPAS2 and SIRT1 interact in the NAc and share reward-relevant gene targets.
(A) Co-immunoprecipitation (Co-IP) of NPAS2 from whole NAc tissue and immunoblotting for SIRT1 revealed a band for SIRT1 at ~120kDa across time of day (left), while Co-IP of SIRT1 from whole NAc tissue and immunoblotting for NPAS2 revealed a band for NPAS2 at ~ 92kDa across time of day (right). Data represented as Mean ± SEM (n= 4–8). GAPDH was used as a negative control and was not detected (~37 kDa). (B) Cross analysis of chromatin immunoprecipitation sequencing (ChIP-seq) for NPAS2 and SIRT1 at ZT2, the time shared between the two datasets, revealed 2,347 and 3,189 genes bound (e.g., promoter or gene body) by NPAS2 and SIRT1, respectively, with 390 genes bound in common. (C) The top pathways enriched among these 390 shared genes include metabolic and reward-relevant biological functions. (D) Synaptic transmission, catecholamine transport, and other dopamine-relevant mechanisms are among the top biological processes enriched among NPAS2 and SIRT1’s shared gene targets. (E) Among the enriched shared biological processes, the catecholamine nodes and synaptic transmission nodes show a high degree of interconnectivity. Red line in bar graphs indicates significance threshold of p<0.05 or -log10(p-value) > 1.3.
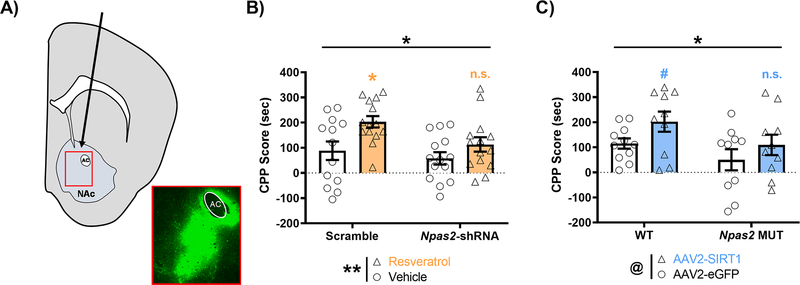
Both NPAS2 and SIRT1 Regulate Cocaine CPP through Expression in the NAc
Given that expression of Npas2 and Sirt1 are both induced in the NAc following cocaine exposure (Figure 2B, C) and NPAS2 and SIRT1 interact in the NAc to regulate reward-relevant genes (Figure 4), we next wanted to determine if NPAS2 in the NAc is necessary for the increase in CPP seen with the SIRT1 agonist resveratrol (Figure 3C). C57BL/6J mice were first injected bilaterally into the NAc with an AAV2-Npas2-shRNA or scramble control (Figure 5A). Mice given the shRNA have a NAc specific knockdown of NPAS2, previously shown in our lab to result in a 4-fold decrease in expression of Npas2 (Ozburn et al., 2015). Mice were then run through the cocaine CPP paradigm and conditioned to both saline and 5 mg/kg cocaine paired chambers, with an administration of either saline or 20 mg/kg resveratrol 30 minutes before conditioning. Notably, both a significant main effect of Npas2-shRNA virus decreasing CPP and a significant main effect of resveratrol treatment increasing CPP were seen (Figure 5B), while no significant interaction was detected (Virus: F(1, 51) = 4.513, p=0.038; Treatment: F(1, 51) = 9.003, p=0.004; Interaction: F(1, 51) = 1.100, p=0.299). Expanding on the simple main effects, resveratrol significantly increased cocaine CPP in the scramble group (*, p<0.05), but not in the Npas2-shRNA group (n.s., p=0.5094) (Figure 5B). Next, we alternatively wanted to determine whether NPAS2 is necessary for the increase in CPP seen with Sirt1 overexpression in the NAc (Ferguson et al., 2013). To investigate this, Npas2 mutant mice or WT controls were injected bilaterally into the NAc with an AAV expressing Sirt1 or eGFP under the CMV promoter, then similarly run through the cocaine CPP paradigm (5 mg/kg). Further confirming our findings, we find a significant main effect of Npas2 genotype decreasing CPP and a trending main effect of Sirt1 virus increasing CPP (Figure 5C), but no significant interaction was detected (Genotype: F(1, 37) = 4.675, p=0.037; Virus: F(1, 37) = 4.061, p=0.051; Interaction: F(1, 37) = 0.148, p=0.702). Expanding on the simple main effects, Sirt1 overexpression increased cocaine CPP in the WT group at trend level significance (#, p=0.0943), but failed to increase CPP in the Npas2 MUT group (n.s., p=0.2691) (Figure 5C).
Figure 5. Both NPAS2 and SIRT1 regulate cocaine preference through their expression in the NAc.
(A) C57BL/6J mice were injected bilaterally into the NAc with either AAV2-Npas2-shRNA-GFP or scramble control, while Npas2 mutant or wild-type mice were injected with either AAV2-CMV-eGFP-2A-Sirt1 or eGFP control. The inset box depicts a representative image of virus placement and spread, as measured by GFP immunofluorescence. AC indicates anterior commissure, while arrow indicates virus injection path. (B) Following two weeks of recovery, C57BL/6J mice injected with either Npas2-shRNA or scramble were conditioned to saline or cocaine (5 mg/kg) and injected with vehicle or resveratrol (20 mg/kg, i.p.) 30 minutes before conditioning. Viral-mediated knock-down of Npas2 in the NAc attenuates resveratrol mediated increases in cocaine CPP – with both main effects of virus and treatment measured by a 2-way ANOVA. Expanding on the simple main effects, resveratrol significantly increased cocaine CPP in the scramble group (*, p<0.05), but not in the Npas2-shRNA group (n.s., p=0.5094). (C) Npas2 mutant mice overexpressing Sirt1 in the NAc conditioned to saline and cocaine (5 mg/kg) show an attenuated preference for cocaine in the CPP task relative to controls – with both a main effect of genotype and a trending effect of virus measured by a 2-way ANOVA (@, p=0.0512). Expanding on the simple main effects, Sirt1 overexpression increased cocaine CPP in the WT group (#, p=0.0943), but not in the Npas2 MUT group (n.s., p=0.2691). Horizontal or vertical bars indicate main effect comparisons. Symbols above plotted bars indicate within group statistical comparisons. Throughout, asterisks indicate significance (* p<0.05, ** p<0.01). Data represented as Mean ± SEM (n= 10–15).
DISCUSSION
Emerging evidence suggests disruptions in both circadian rhythms and redox metabolism as key contributors underlying SUDs (Logan et al., 2014; Uys et al., 2014). Interestingly, circadian molecular clock function is integrated with the cellular metabolic state through a transcriptional-enzymatic feedback loop involving SIRT1 and the clock-controlled NAD+ salvage pathway (Nakahata et al., 2009; Ramsey et al., 2009). However, this interaction has yet to be explored in the NAc and/or in the context of the circadian transcription factor NPAS2. Results from this study illustrate how cocaine exposure affects the circadian expression of core circadian and metabolic genes, namely Npas2 and Sirt1, and how together these two may interact in the NAc to regulate NAc function. Core circadian genes Npas2, Bmal1, and their regulators Rev-erbα and RORα all show diurnal variation in the NAc. Expression of metabolic regulators Sirt1, Nampt, and levels of the mitochondrial coenzyme NAD+ also showed diurnal variation in the NAc. Interestingly, the diurnal variation in expression of the molecular clock and regulators of metabolic state were all shown to be altered in the NAc following cocaine exposure. Specifically, Npas2, its transcriptional activator RORα, and the metabolic regulators Sirt1, and Nampt were all phase-advanced in response to cocaine exposure, primarily driven by upregulation and shifting of observed peak expression to the light phase. NAD+ was also upregulated by cocaine exposure, but showed a delaying of observed peak expression to the dark phase. Most notably, in addition to reconfirming past findings that global/systemic modulation of NPAS2 and SIRT1 regulate cocaine CPP, we found NPAS2 and SIRT1 interact in the mouse NAc and their NAc expression is important in the regulation of cocaine reward. Through co-immunoprecipitation in the NAc followed by western blot analysis, we found that NPAS2 and SIRT1 interact in a shared protein complex across time of day. Furthermore, cross-analysis of NPAS2 and SIRT1 ChIP-seq datasets revealed several metabolic and reward-relevant pathways to be enriched among their shared gene targets; these pathways and processes included NRF2-mediated oxidative stress response and NF-kB activation, as well as many processes important for transport of dopamine, synaptic transmission, regulation of response to drug, and behavioral conditioning.
Finally, in line with previous studies, we find that systemic treatment with the SIRT1 activating drug, resveratrol significantly increases cocaine CPP and we produce a similar increase in cocaine CPP with Sirt1 knock-down in the NAc. Interestingly, when we knock-down Npas2 in the NAc and then treat mice with resveratrol, or knock-down Sirt1 in the NAc of Npas2 mutant mice, either way we fail to produce the SIRT1 mediated increase in CPP. These results highlight the important role of these proteins in the NAc in the modulation of drug reward, though future, more specifically targeted studies are necessary to determine if it is indeed the NPAS2/SIRT1 protein complex in the NAc that modulates reward or if they regulate reward via parallel pathways.
Our gene expression findings presented here are consistent with the previous literature from our lab and others illustrating that the NAc is under circadian control. Previous studies from our lab have found in mice that core clock genes Npas2, Clock, and Period all show diurnal variation in the NAc (Falcon et al., 2013), as well as rhythmicity measured by Per2:luciferase luminescence assays (Logan et al., 2015). This is further supported by studies from the Welsh group demonstrating NAc Per2:luciferase rhythms (Landgraf, Long, et al., 2016; Porcu et al., 2020), as well as diurnal variation in CRYPTOCHROME protein levels in the NAc (Porcu et al., 2020). Interestingly, this is also seen in time of death analyses of gene expression in human post-mortem tissue, where the canonical clock genes (e.g., Bmal1, Npas2, Period, Cryptochrome, Rev-erbα) are all highly rhythmic in the NAc of healthy donors (Li et al., 2013; Ketchesin et al., 2021). Here we further support these findings and expand upon them by showing Npas2, Bmal1, and their transcriptional regulators Rev-erbα and RORα all have significant diurnal variation in the NAc. This circadian regulation also extends to the regulation of Sirt1, Nampt, and levels of the mitochondrial coenzyme NAD+, which all display diurnal variation in the NAc. While these metabolic genes and cofactors have previously been shown to be diurnally regulated in the liver (Nakahata et al., 2008, 2009; Ramsey et al., 2009), our findings are the first to suggest this relationship also exists in the NAc. Future investigation with more time points may be necessary to fully capture the rhythmicity across time of day, especially considering the potential for bimodal circadian oscillations as seen with NAD+ in the liver (Ramsey et al., 2009). Furthermore, future investigation may also explore these changes in a cell-type-specific manner to fully appreciate the regulation of circadian rhythms in the NAc.
Through decades of research, aberrant functioning of the brain’s reward circuitry, including dopamine signaling in the NAc, has been linked to the manifestation of SUDs (Koob & Le Moal, 2001; Koob & Volkow, 2010; Pierce et al., 2020). The dopamine hypothesis posits that substances of abuse act directly on the brain’s reward system to increase mesolimbic DA release and promote both reward and subsequent reward-seeking behavior (Koob, 1992; Willuhn et al., 2010; Volkow & Morales, 2015). In fact, psychostimulants like cocaine or amphetamine act directly at NAc synapses to increase the concentration of extracellular DA available either by blocking dopamine transporter (DAT) mediated DA reuptake or promoting greater DA release, respectively (Kalivas, 2007). Interestingly, through our NPAS2 and SIRT1 ChIP-seq cross-analysis, we found catecholamine/dopamine transport, catecholamine secretion, regulation of response to drugs, and regulation of presynaptic membrane synaptic transmission to all be significantly enriched biological processes among shared gene targets between NPAS2 and SIRT1 in the NAc. This is further evidenced by IPA pathway analyses revealing behavioral conditioning, synaptic transmission, and protection of dopamine neurons to all be significantly enriched biological functions among shared gene targets. One gene target in particular, glial cell-derived neurotrophic factor (GDNF), is shared among all the enriched processes and pathways and has been extensively shown to be important for dopamine neuron survival and function (Airaksinen & Saarma, 2002). GDNF and its signaling pathways have also been associated with reward physiology relevant for the development of SUDs - specifically shown to regulate dopamine neuronal excitability, synaptic transmission, synaptic plasticity, and sensitivity to psychostimulants (Ron & Janak, 2005; Carnicella & Ron, 2009; Ghitza et al., 2010). Though originally characterized as a neuronal pas domain protein (Hogenesch et al., 1997; Zhou et al., 1997), notably, NPAS2 has since been shown to be among the top expressed genes in glial cell development and to play a role in regulating astrocyte-mediated glutamate uptake (Cahoy et al., 2008; Beaulé et al., 2009); thus, regulation of GDNF is not all too surprising. However, while particularly interesting in the context of our current findings, the relationship between NPAS2, SIRT1, and GDNF and how this shared gene target may contribute to reward regulation remains to be studied. Taken together, the many dopamine-related processes enriched among shared gene targets between NPAS2 and SIRT1 in the NAc further highlights their functional relevance for reward processing and warrants future mechanistic investigation. Moreover, while not performed in this study, future follow-up studies will employ gene expression analyses to validate and investigate specific gene targets of interest.
An abundance of evidence suggests substances of abuse disrupt both circadian rhythms and metabolic redox state of the cell (Hasler et al., 2012; Cunha-Oliveira et al., 2013; Uys et al., 2014). Here, our findings further support these observations showing the diurnal variation of both core clock genes (e.g., Npas2, Bmal1, and RORα) and regulators of metabolic state (e.g., Sirt1, Nampt, and NAD+ levels) are significantly altered by exposure to cocaine. The disruption in diurnal variation in expression of Npas2, Sirt1, and the NAD+ salvage pathway may point toward a mechanism in which cocaine alters both circadian and metabolic regulation of the NAc. In both human and animal studies, cocaine has been shown to produce increased oxidative stress/damage and subsequent compensatory upregulation of glutathione antioxidant signaling (Cunha-Oliveira et al., 2013; Uys et al., 2014). This is further supported by the human and rodent literature suggesting therapeutic targeting of redox signaling through antioxidants (e.g., n-acetylcysteine) may prove beneficial in treating SUDs (Womersley et al., 2019). Interestingly, NRF2-mediated oxidative stress response and NF-kB activation are highly enriched pathways among NPAS2 and SIRT1’s gene targets. Both pathways are known to mitigate oxidative stress and promote redox homeostasis through antioxidant expression (Mauro et al., 2011; Ma, 2013; Vomund et al., 2017; Lingappan, 2018). Levels of NAD+ are directly correlated with the redox state of the cell (Ying, 2008) and can directly impact circadian molecular clock function, either through direct actions on CLOCK or NPAS2/BMAL1 (Rutter et al., 2001) or through SIRT1 (Asher et al., 2008; Nakahata et al., 2008). Our findings that cocaine exposure significantly upregulates and disrupts diurnal variation of NAD+ levels and members of the SIRT1/NAMPT transcriptional-enzymatic feedback loop might suggest a mechanism by which cocaine directly modulates molecular clock function through NPAS2. However, while our findings suggest NPAS2 and SIRT1 do interact in the same protein complex in the NAc, future investigation would be needed to determine whether this upregulation of NAc NAD+ levels leads to altered molecular clock function through direct impacts on the clock, through SIRT1, or both. Nevertheless, our findings underscore the complexity of the bidirectional relationship between cellular metabolic state and the circadian molecular clock, while also revealing this interaction may be important in the NAc.
In addition to the growing body of evidence suggesting a bidirectional link between disruptions in circadian rhythms / metabolic state and the development or perpetuation of SUDs (Logan et al., 2014; Uys et al., 2014), interestingly, pharmacologically targeting redox regulation has shown promise as a treatment option for SUDs in both clinical and preclinical animal studies (Womersley et al., 2019). Of the most promising clinical studies, treatment with N-acetylcysteine, a precursor to the primary endogenous antioxidant glutathione, has shown potential across several large scale clinical trials; e.g., SUDS patients given oral doses of N-acetylcysteine show reductions in self-reported cravings, drug motivation, and total consumption (reviewed in: (Womersley & Uys, 2016; Womersley et al., 2019). While the exact therapeutic mechanism has not been determined, interestingly, N-acetylcysteine can directly attenuate SIRT1 repression, increase Sirt1 mRNA, and decrease SIRT1’s protein degradation (Zhao et al., 2018). Along with the clinical and preclinical studies implicating SIRT1 in SUDs and other comorbid psychiatric disorders (Abe et al., 2011; Ferguson et al., 2013, 2015; Liu et al., 2019; Kapoor et al., 2020), our findings presented here suggest targeting of SIRT1 may have therapeutic implications in regulating reward through its role in integrating circadian and metabolic processes. However, future clinical investigation into the therapeutic potential of SIRT1 modulation for the development of novel treatments for SUDs is still necessary.
While work from our lab and others in the VTA suggest SIRT1 interacts with CLOCK to regulate dopaminergic activity via NAMPT driven NAD+ biosynthesis (Kong et al., 2018; Logan et al., 2019), it was unclear if this would hold true with CLOCK’s functional homolog NPAS2 and/or in the NAc, a region downstream of the VTA. The findings presented in this study are the first to show that NPAS2 and SIRT1 interact in the same protein complex and together they may be important for regulating NAc function. However, further mechanistic investigation into how this interaction is modulating reward is still necessary. Interestingly, work from Nestler and colleagues, in addition to a recent study published from our lab, might suggest that regulation of synaptic plasticity is at the root of this interaction. We recently demonstrated the importance of NPAS2 for regulating drug-related synaptic plasticity in the NAc through cell-type-specific modulation of Drd1-medium spiny neurons at the glutamatergic inputs onto the NAc (Parekh et al., 2019). This is particularly relevant, considering SIRT1 is considered a key mediator of drug-related molecular and cellular plasticity through the regulation of numerous synaptic proteins and dendritic spines in the NAc (Ferguson et al., 2013). Supporting this idea, in addition to regulation of dopamine transport and synaptic transmission, regulation of actin filament bundle assembly was the top enriched biological process among the shared gene targets between NPAS2 and SIRT1 revealed by ChIP-seq analysis. Essential to synaptic plasticity, the post-synaptic elements of excitatory synapses consist of actin-rich protrusions from dendrites called dendritic spines that adapt in shape and size in response to synaptic activity; the dynamics of dendritic spine formation and modulation are driven by the regulation of actin filament bundles and actin-binding proteins (Lin & Webb, 2009; Gordon-Weeks & Fournier, 2014; Konietzny et al., 2017; Borovac et al., 2018). It has long been appreciated that drug-induced long-term changes in synaptic plasticity and spine density/morphology in the NAc are associated with addiction pathophysiology and behavior (Kauer & Malenka, 2007; Russo et al., 2010). Studies have shown actin and its cycling between filamentous and depolarized forms is immediately relevant for psychostimulant reward-related behavior (Toda et al., 2006; Shibasaki et al., 2011; Young et al., 2015; Areal et al., 2019). Perhaps together the interaction between NPAS2 and SIRT1 shown in this study may be important for regulating drug-related synaptic transmission and plasticity in the NAc; however, future investigation into this idea is still needed.
While this study focused on NPAS2 and SIRT1 in the NAc, a role for CLOCK in the NAc is still possible. Though NPAS2 is highly enriched in the mouse forebrain and specifically within the striatum and NAc (Garcia et al., 2000), CLOCK is still expressed in these regions (Falcon et al., 2013; Ozburn et al., 2015). Outside of the NAc, complete loss of CLOCK can be compensated by NPAS2 and vice versa due to independent interactions with BMAL1 to regulate the molecular clock (DeBruyne et al., 2007; Bertolucci et al., 2008; Landgraf, Wang, et al., 2016). While CLOCK and NPAS2 may be regulating molecular clock function similarly, their differential contribution to cocaine reward highlights their divergence in functions outside of the transcription-translation feedback loop. For example, Clock mutant mice show increased cocaine reward (McClung et al., 2005; Ozburn et al., 2012), Npas2 mutant mice show decreased cocaine reward that is recapitulated through knock-down of Npas2 in the NAc (Ozburn et al., 2015); knock-down of Clock in the NAc fails to produce an effect on cocaine reward (Ozburn et al., 2015) – further supporting the notion that NPAS2 and CLOCK serve different roles in regulating reward, especially within the NAc. However, despite these differences in cocaine preference, a recent study from our lab found similarities in operant cocaine self-administration. Much like how Clock mutant mice have increased cocaine self-administration and motivation (Ozburn et al., 2012), Npas2 mutant mice also show increased intravenous cocaine self-administration, but in both sex- and time-specific manners (DePoy et al., 2020). Considering these new findings and the limitations associated with our current study utilizing only male mice, future studies will benefit from including both sexes in the continued investigation of NPAS2 in the NAc. Finally, while an interaction between CLOCK and SIRT1 in the VTA is important for regulating dopaminergic activity and reward (Logan et al., 2019), whether CLOCK and SIRT1 interact in the NAc and whether this interaction may be important remains to be investigated. Future studies may continue to tease apart the role of NPAS2 versus CLOCK in the NAc, and how their interaction with SIRT1 may differ.
Ultimately, the results of this study demonstrate that NPAS2 and SIRT1 interact in the NAc and introduce a potential mechanism by which cocaine’s effects on the cellular metabolic state may affect circadian regulation of the NAc and downstream reward function. These proteins together may have important implications for the development of substance use disorders and future investigation into the therapeutic potential of targeting NPAS2 and/or SIRT1 for the development of novel treatments is necessary.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. David Weaver for providing the Npas2 mutant mice. We thank Dr. Jude Samulsky and the University of North Carolina Gene Therapy Vector Core for their work in the preparation of viral constructs. We would also like to thank Mark Brown, Ioannis H. Migias, and Allison Cerwensky for their role in animal care and genotyping. Data analysis was performed using Ingenuity Pathway Analysis software licensed through the Molecular Biology Information Service of the Health Sciences Library System at the University of Pittsburgh. Cocaine HCl was provided by NIDA via the NIH Drug Distribution Center. This work was funded and supported by the National Institutes of Health (NIH): R01DA039865 (PI: McClung CA), K02DA042886 (PI: McClung CA), P50DA039841 (PI: McClung CA), R33DA041872 (PIs McClung CA and Logan RW), R21DA037636 (PI: McClung CA), & T32 NS007433-18 (PI: Sved AF).
ABBREVIATIONS
- AAV
Adeno-Associated Virus
- BMAL1
Brain and Muscle ARNT-like protein 1
- ChIP-Seq
Chromatin Immunoprecipitation Sequencing
- CLOCK
Circadian Locomotor Output Cycles Kaput
- Co-IP
Co-Immunoprecipitation
- CPP
Conditioned Place Preference
- eGFP
enhanced Green Fluorescent Protein
- HPLC
High-Performance Liquid Chromatography
- IPA
Ingenuity Pathway Analysis
- NAc
Nucleus Accumbens
- NAD
Nicotinamide Adenine Dinucleotide
- NAMPT
Nicotinamide Phosphoribosyltransferase
- NPAS2
Neuronal PAS Domain Protein 2
- REV-ERBα
Reverse Strand of ERBα protein
- RORα
RAR Related Orphan Receptor A
- RT-qPCR
Reverse Transcriptase Quantitative Polymerase Chain Reaction
- SIRT1
Sirtuin-1
- SUD
Substance Use Disorder
- VTA
Ventral Tegmental Area
- ZT
Zeitgeber Time
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to declare.
DATA ACCESSIBILITY STATEMENT
Data are available from the corresponding author upon request.
REFERENCES
- Abe N, Uchida S, Otsuki K, Hobara T, Yamagata H, Higuchi F, Shibata T, & Watanabe Y (2011) Altered sirtuin deacetylase gene expression in patients with a mood disorder. J. Psychiatr. Res, 45, 1106–1112. [DOI] [PubMed] [Google Scholar]
- Airaksinen MS & Saarma M (2002) The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci, 3, 383–394. [DOI] [PubMed] [Google Scholar]
- Areal LB, Hamilton A, Martins-Silva C, Pires RGW, & Ferguson SSG (2019) Neuronal scaffolding protein spinophilin is integral for cocaine-induced behavioral sensitization and ERK1/2 activation. Mol. Brain, 12, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, & Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell, 134, 317–328. [DOI] [PubMed] [Google Scholar]
- Baur JA & Sinclair DA (2006) Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug Discov, 5, 493–506. [DOI] [PubMed] [Google Scholar]
- Beaulé C, Swanstrom A, Leone MJ, & Herzog ED (2009) Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS One, 4, e7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolucci C, Cavallari N, Colognesi I, Aguzzi J, Chen Z, Caruso P, Foá A, Tosini G, Bernardi F, & Pinotti M (2008) Evidence for an overlapping role of CLOCK and NPAS2 transcription factors in liver circadian oscillators. Mol. Cell. Biol, 28, 3070–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovac J, Bosch M, & Okamoto K (2018) Regulation of actin dynamics during structural plasticity of dendritic spines: Signaling messengers and actin-binding proteins. Mol. Cell. Neurosci, 91, 122–130. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, & Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci, 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C & Auwerx J (2012) Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol. Rev, 64, 166–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicella S & Ron D (2009) GDNF--a potential target to treat addiction. Pharmacol. Ther, 122, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumbley C, Wang Y, Kojetin DJ, & Burris TP (2010) Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J. Biol. Chem, 285, 35386–35392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Oliveira T, Rego A, & Oliveira C (2013) Oxidative stress and drugs of abuse: an update. Mini Rev. Org. Chem, 10, 321–334. [Google Scholar]
- Dallmann R, DeBruyne JP, & Weaver DR (2011) Photic resetting and entrainment in CLOCK-deficient mice. J. Biol. Rhythms, 26, 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, & Reppert SM (2007) CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci, 10, 543–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePoy LM, Becker-Krail DD, Zong W, Petersen K, Shah NM, Brandon JH, Miguelino AM, Tseng GC, Logan RW, & McClung CA (2020) Circadian- and sex-dependent increases in intravenous cocaine self-administration in Npas2 mutant mice. J. Neurosci,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A, & McKnight SL (2002) NPAS2: a gas-responsive transcription factor. Science, 298, 2385–2387. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K & Sassone-Corsi P (2013) Metabolism and the circadian clock converge. Physiol. Rev, 93, 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon E, Ozburn A, Mukherjee S, Roybal K, & McClung CA (2013) Differential regulation of the period genes in striatal regions following cocaine exposure. PLoS One, 8, e66438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, Renthal W, Neve R, Liu X, Shao N, Sartorelli V, Shen L, & Nestler EJ (2013) Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J. Neurosci, 33, 16088–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Shao N, Heller E, Feng J, Neve R, Kim H-D, Call T, Magazu S, Shen L, & Nestler EJ (2015) SIRT1-FOXO3a regulate cocaine actions in the nucleus accumbens. J. Neurosci, 35, 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, & McKnight SL (2000) Impaired cued and contextual memory in NPAS2-deficient mice. Science, 288, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Ghitza UE, Zhai H, Wu P, Airavaara M, Shaham Y, & Lu L (2010) Role of BDNF and GDNF in drug reward and relapse: a review. Neurosci. Biobehav. Rev, 35, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks PR & Fournier AE (2014) Neuronal cytoskeleton in synaptic plasticity and regeneration. J. Neurochem, 129, 206–212. [DOI] [PubMed] [Google Scholar]
- Hasler BP, Smith LJ, Cousins JC, & Bootzin RR (2012) Circadian rhythms, sleep, and substance abuse. Sleep Med. Rev, 16, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, Perdew GH, & Bradfield CA (1997) Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem, 272, 8581–8593. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, & Auwerx J (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev, 31, 194–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJA, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, & Yeats C (2009) InterPro: the integrative protein signature database. Nucleic Acids Res, 37, D211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW (2007) Cocaine and amphetamine-like psychostimulants: neurocircuitry and glutamate neuroplasticity. Dialogues Clin Neurosci, 9, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Chao M, Johnson EC, Novikova G, Lai D, Meyers J, Schulman J, Nurnberger JI, Porjesz B, Liu Y, COGA collaborators, Tatiana Foroud, Edenberg HJ, Marcora E, Agrawal A, & Goate A (2020) Multi-omics integration analysis identifies novel genes for alcoholism with potential link to neurodegenerative diseases. BioRxiv,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer JA & Malenka RC (2007) Synaptic plasticity and addiction. Nat. Rev. Neurosci, 8, 844–858. [DOI] [PubMed] [Google Scholar]
- Ketchesin KD, Zong W, Hildebrand MA, Seney ML, Cahill KM, Scott MR, Shankar VG, Glausier JR, Lewis DA, Tseng GC, & McClung CA (2021) Diurnal rhythms across the human dorsal and ventral striatum. Proc. Natl. Acad. Sci. USA, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Du C, Jiang L, Jiang W, Deng P, Shao X, Zhang B, Li Y, Zhu R, Zhao Q, Fu D, Gu H, Luo L, Long H, Zhao Y, & Cen X (2018) Nicotinamide phosphoribosyltransferase regulates cocaine reward through Sirtuin 1. Exp. Neurol, 307, 52–61. [DOI] [PubMed] [Google Scholar]
- Konietzny A, Bär J, & Mikhaylova M (2017) Dendritic actin cytoskeleton: structure, functions, and regulations. Front. Cell Neurosci, 11, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF (1992) Drugs of abuse: anatomy, pharmacology and function of reward pathways. Trends Pharmacol. Sci, 13, 177–184. [DOI] [PubMed] [Google Scholar]
- Koob GF & Le Moal M (2001) Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology, 24, 97–129. [DOI] [PubMed] [Google Scholar]
- Koob GF & Volkow ND (2010) Neurocircuitry of addiction. Neuropsychopharmacology, 35, 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, & Tugendreich S (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Long JE, & Welsh DK (2016) Depression-like behaviour in mice is associated with disrupted circadian rhythms in nucleus accumbens and periaqueductal grey. Eur. J. Neurosci, 43, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Landgraf D, Wang LL, Diemer T, & Welsh DK (2016) NPAS2 compensates for loss of CLOCK in peripheral circadian oscillators. PLoS Genet, 12, e1005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ, Akil H, & Bunney WE (2013) Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. USA, 110, 9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-H & Webb DJ (2009) Actin and Actin-Binding Proteins: Masters of Dendritic Spine Formation, Morphology, and Function. Open Neurosci. J, 3, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappan K (2018) NF-κB in Oxidative Stress. Curr. Opin. Toxicol, 7, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yan H, Zhou D, Cai X, Zhang Y, Li S, Li H, Li S, Zhou D-S, Li X, Zhang C, Sun Y, Dai J-P, Zhong J, Yao Y-G, Luo X-J, Fang Y, Zhang D, Ma Y, Yue W, Li M, & Xiao X (2019) The depression GWAS risk allele predicts smaller cerebellar gray matter volume and reduced SIRT1 mRNA expression in Chinese population. Transl. Psychiatry, 9, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Edgar N, Gillman AG, Hoffman D, Zhu X, & McClung CA (2015) Chronic Stress Induces Brain Region-Specific Alterations of Molecular Rhythms that Correlate with Depression-like Behavior in Mice. Biol. Psychiatry, 78, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Parekh PK, Kaplan GN, Becker-Krail DD, Williams WP, Yamaguchi S, Yoshino J, Shelton MA, Zhu X, Zhang H, Waplinger S, Fitzgerald E, Oliver-Smith J, Sundarvelu P, Enwright JF, Huang YH, & McClung CA (2019) NAD+ cellular redox and SIRT1 regulate the diurnal rhythms of tyrosine hydroxylase and conditioned cocaine reward. Mol. Psychiatry, 24, 1668–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Williams WP, & McClung CA (2014) Circadian rhythms and addiction: mechanistic insights and future directions. Behav. Neurosci, 128, 387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol, 53, 401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, Moretti M, De Smaele E, Beg AA, Tergaonkar V, Chandel NS, & Franzoso G (2011) NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol, 13, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, & Nestler EJ (2005) Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc. Natl. Acad. Sci. USA, 102, 9377–9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, & Sassone-Corsi P (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell, 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, & Sassone-Corsi P (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science, 324, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseri Kouzehgarani G, Bothwell MY, & Gillette MU (2020) Circadian rhythm of redox state regulates membrane excitability in hippocampal CA1 neurons. Eur. J. Neurosci, 51, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil D, Mendez-Figueroa H, Mistretta T-A, Su C, Lane RH, & Aagaard KM (2013) Dysregulation of Npas2 leads to altered metabolic pathways in a murine knockout model. Mol. Genet. Metab, 110, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozburn AR, Falcon E, Mukherjee S, Gillman A, Arey R, Spencer S, & McClung CA (2013) The role of clock in ethanol-related behaviors. Neuropsychopharmacology, 38, 2393–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozburn AR, Falcon E, Twaddle A, Nugent AL, Gillman AG, Spencer SM, Arey RN, Mukherjee S, Lyons-Weiler J, Self DW, & McClung CA (2015) Direct regulation of diurnal Drd3 expression and cocaine reward by NPAS2. Biol. Psychiatry, 77, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozburn AR, Larson EB, Self DW, & McClung CA (2012) Cocaine self-administration behaviors in ClockΔ19 mice. Psychopharmacology, 223, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh PK, Logan RW, Ketchesin KD, Becker-Krail D, Shelton MA, Hildebrand MA, Barko K, Huang YH, & McClung CA (2019) Cell-Type-Specific Regulation of Nucleus Accumbens Synaptic Plasticity and Cocaine Reward Sensitivity by the Circadian Protein, NPAS2. J. Neurosci, 39, 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh PK, Ozburn AR, & McClung CA (2015) Circadian clock genes: effects on dopamine, reward and addiction. Alcohol, 49, 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, & Takahashi JS (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol, 24, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce RC, Unterwald EM, & Kenny PJ (2020) Addiction, Second. edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Porcu A, Vaughan M, Nilsson A, Arimoto N, Lamia K, & Welsh DK (2020) Vulnerability to helpless behavior is regulated by the circadian clock component CRYPTOCHROME in the mouse nucleus accumbens. Proc. Natl. Acad. Sci. USA, 117, 13771–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, & Schibler U (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell, 110, 251–260. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong H-K, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S-I, & Bass J (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science, 324, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, & McKnight SL (2001) NPAS2: an analog of clock operative in the mammalian forebrain. Science, 293, 506–509. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, & Nestler EJ (2009) Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron, 62, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D & Janak PH (2005) GDNF and addiction. Rev Neurosci, 16, 277–285. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, & Nestler EJ (2010) The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci, 33, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, & McKnight SL (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science, 293, 510–514. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, & Hogenesch JB (2004) A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron, 43, 527–537. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD & Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, & Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki M, Mizuno K, Kurokawa K, Suzuki T, & Ohkuma S (2011) Role of actin depolymerizing factor in the development of methamphetamine-induced place preference in mice. Eur. J. Pharmacol, 671, 70–78. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, & Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res, 34, D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromsdorfer KL, Yamaguchi S, Yoon MJ, Moseley AC, Franczyk MP, Kelly SC, Qi N, Imai S-I, & Yoshino J (2016) NAMPT-Mediated NAD(+) Biosynthesis in Adipocytes Regulates Adipose Tissue Function and Multi-organ Insulin Sensitivity in Mice. Cell Rep, 16, 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S, Shen H-W, Peters J, Cagle S, & Kalivas PW (2006) Cocaine increases actin cycling: effects in the reinstatement model of drug seeking. J. Neurosci, 26, 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Sato E, Sato A, Sagami I, Shimizu T, & Kitagawa T (2005) CO-dependent activity-controlling mechanism of heme-containing CO-sensor protein, neuronal PAS domain protein 2. J. Biol. Chem, 280, 21358–21368. [DOI] [PubMed] [Google Scholar]
- Uys JD, Mulholland PJ, & Townsend DM (2014) Glutathione and redox signaling in substance abuse. Biomed. Pharmacother, 68, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND & Morales M (2015) The brain on drugs: from reward to addiction. Cell, 162, 712–725. [DOI] [PubMed] [Google Scholar]
- Vomund S, Schäfer A, Parnham MJ, Brüne B, & von Knethen A (2017) Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, & Gillette MU (2012) Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science, 337, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willuhn I, Wanat MJ, Clark JJ, & Phillips PEM (2010) Dopamine signaling in the nucleus accumbens of animals self-administering drugs of abuse. Current topics in behavioral neurosciences, 3, 29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womersley JS, Townsend DM, Kalivas PW, & Uys JD (2019) Targeting redox regulation to treat substance use disorder using N-acetylcysteine. Eur. J. Neurosci, 50, 2538–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womersley JS & Uys JD (2016) S-Glutathionylation and Redox Protein Signaling in Drug Addiction. Prog Mol Biol Transl Sci, 137, 87–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W (2008) NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal, 10, 179–206. [DOI] [PubMed] [Google Scholar]
- Yoshii K, Tajima F, Ishijima S, & Sagami I (2015) Changes in pH and NADPH regulate the DNA binding activity of neuronal PAS domain protein 2, a mammalian circadian transcription factor. Biochemistry, 54, 250–259. [DOI] [PubMed] [Google Scholar]
- Yoshino J & Imai S-I (2013) Accurate measurement of nicotinamide adenine dinucleotide (NAD+) with high-performance liquid chromatography. Methods Mol. Biol, 1077, 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EJ, Briggs SB, & Miller CA (2015) The actin cytoskeleton as a therapeutic target for the prevention of relapse to methamphetamine use. CNS Neurol Disord Drug Targets, 14, 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, & Hogenesch JB (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA, 111, 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jin Y, Yang L, Hou Z, Liu Y, Sun T, Pei J, Li J, Yao C, Wang X, & Chen G (2018) Promotion of SIRT1 protein degradation and lower SIRT1 gene expression via reactive oxygen species is involved in Sb-induced apoptosis in BEAS-2b cells. Toxicol. Lett, 296, 73–81. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, & Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun, 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, & McKnight SL (1997) Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA, 94, 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.