Abstract
Neuroinflammation leads to neuronal stress responses that contribute to neuronal dysfunction and loss. However, treatments that stabilize neurons and prevent their destruction are still lacking. Here, we identify the histone methyltransferase G9a as a druggable epigenetic regulator of neuronal vulnerability to inflammation. In murine experimental autoimmune encephalomyelitis (EAE) and human multiple sclerosis (MS), we found that the G9a-catalyzed repressive epigenetic mark H3K9me2 was robustly induced by neuroinflammation. G9a activity repressed anti-ferroptotic genes, diminished intracellular glutathione levels, and triggered the iron-dependent programmed cell death pathway ferroptosis. Conversely, pharmacological treatment of EAE mice with a G9a inhibitor restored anti-ferroptotic gene expression, reduced inflammation-induced neuronal loss, and improved clinical outcome. Similarly, neuronal anti-ferroptotic gene expression was reduced in MS brain tissue and was boosted by G9a inhibition in human neuronal cultures. This study identifies G9a as a critical transcriptional enhancer of neuronal ferroptosis and potential therapeutic target to counteract inflammation-induced neurodegeneration.
Enhanced G9a activity during neuroinflammation facilitates neuronal loss by the regulation of ferroptosis-related genes.
INTRODUCTION
Neuroinflammation is involved in several neurological and psychiatric diseases, in which activation of brain-resident microglia (1) and infiltration of peripheral immune cells cause neuronal stress and injury (2). As a prototypic neuroinflammatory disease, multiple sclerosis (MS) is characterized by inflammatory lesions that cause damage to the central nervous system (CNS) and lead to neuronal dysfunction and clinical impairment (3). Although inflammatory activity declines over the course of MS, persistent low-grade inflammation and inflammation-independent progression of neurodegeneration lead to continuous worsening of disability (4). In this regard, chronic progressive MS is reminiscent of primary neurodegenerative diseases that are accompanied by low-grade inflammation, such as Parkinson’s or Alzheimer’s disease (5). However, at the stage of progressive neurodegeneration, immunosuppressive treatments that are effective in the early inflammatory phase of MS mostly lose their efficacy. Therefore, to deliver long-term benefits to patients suffering from neuroinflammation, novel neuroprotective therapeutics with direct action on neurons are urgently needed (6).
Neurodegeneration in MS has been attributed to the generation of reactive oxygen and nitrogen species (7), mitochondrial dysfunction with impaired energy production (8), and glutamate excitotoxicity that causes influx of calcium into neurons (9), resulting in ionic dyshomeostasis and cell death (2). Similar pathways have been picked up in recent single-cell transcriptomics analysis of MS postmortem brain samples. Affected neurons showed a strong up-regulation of signatures associated with oxidative stress, mitochondrial dysfunction, and cell death pathways (10). However, the extraction of master regulators of neuronal maladaptation remains challenging (11). As postmortem studies are inherently complicated by the fact that available samples mostly represent late chronic stages of the disease, we recently set out to investigate the neuronal stress response to inflammation under tightly controlled conditions in the MS animal model experimental autoimmune encephalomyelitis (EAE). To generate a comprehensive inventory of neuronal adaptation during inflammatory stress, we exploited translating ribosome affinity purification of spinal cord motor neurons that are directly affected in EAE (12). As part of this study, we found inflammation-induced dysregulation of neuronal proteostasis with toxic aggregation of the presynaptic protein bassoon and we devised a therapeutic approach to clear aggregates by proteasome activation (12). This complements other strategies to ameliorate inflammatory neurodegeneration by modulation of mitochondrial function (13), neuronal ion channels (14–16), or glutamate receptors (17, 18). However, besides the pharmacological appeal of targeting specific molecular pathways, neurodegeneration emerges as a multimodal process with many detrimental cascades running in parallel (19). Therefore, shutting down the underlying maladaptive neuronal stress response as a whole may provide greater and more consistent benefit, as it depends less on specific pathways and their pathological contribution in a given situation.
There is growing recognition of an intrinsic neuronal stress response that orchestrates neuronal adaptation and sets the course for cellular survival or death (20). To reinstall neuronal homeostasis, targeting transcriptional regulators is a highly promising approach because of their broad impact on diverse regulatory cascades with the potential to counteract pathophysiological deregulation in a holistic sense (21). Aberrant gene transcription is a hallmark of neurodegeneration and involves several factors that modulate neuronal gene expression (22–24). These factors target various levels of transcriptional regulation including chemical DNA modification, local recruitment of the transcriptional machinery, and modulation of chromatin accessibility, for example, by histone modifications (25). Of these examples, epigenetic modifiers with enzymatic activity are of particular interest because of their direct accessibility to modulation by small molecules (21). Targeting epigenetic regulators might represent a viable approach to rebalance transcriptional stress responses of neurons and improve clinical outcome.
Another point of convergence in neurodegenerative cascades is the involvement of shared downstream pathways of programmed cell death that provide additional opportunities for intervention. Neurons can undergo diverse forms of cell death depending on the nature and severity of the stressor (19). Under inflammatory conditions, neuronal apoptosis has been described in retinal ganglion cells during EAE (26) and tumor necrosis factor–α (TNF-α)–driven necroptosis was found as an important mechanism of neuronal loss in MS (27, 28). More recently, ferroptosis, an iron-dependent form of regulated cell death (29), has been investigated in cancer development and neurodegenerative disorders (30). Hallmarks of ferroptosis are excessive peroxidation of phospholipid membranes rich in polyunsaturated fatty acids, inactivation of glutathione peroxidase 4 (GPX4), and reduced levels of the intracellular antioxidant glutathione (31). Currently, only circumstantial evidence connects ferroptosis to neuronal injury in neuroinflammation. It has been shown that iron deposits and oxidized phospholipids are increased in MS brains (32, 33), and reduced GPX4 levels have been reported in MS brains and in spinal cord neurons of EAE mice (34). Also, secretion of ferritin heavy chain by oligodendrocytes has been recently identified as an antioxidative defense system to counteract neuronal ferroptosis (35). Thus, it is conceivable that ferroptosis plays an important role in neuronal loss during inflammation of the CNS.
In this study, we identified the inflammation-induced epigenetic regulator euchromatic histone lysine N-methyltransferase 2 (EHMT2; also known as G9a) as a druggable candidate for neuroprotective intervention. We demonstrate that the repressive dimethylation of histone H3 lysine 9 (H3K9me2), which is catalyzed by G9a, is strongly induced in neurons during neuroinflammation in EAE mice and MS patients. Pharmacological inhibition of G9a revealed a robust neuroprotective effect in neuronal cultures and during CNS inflammation in vivo. Mechanistically, G9a dictates neuronal fate by driving ferroptosis via epigenetic repression of anti-ferroptotic genes. Our findings reveal G9a as a stress-induced epigenetic amplifier of neuronal ferroptosis and highlight G9a inhibition as a neuroprotective therapy.
RESULTS
G9a-dependent H3K9me2 is induced by inflammation and excitotoxicity
Because of their ability to regulate broad transcriptional programs, epigenetic modifiers are an emerging class of therapeutic targets with potential value for neuroprotective treatment. To identify epigenetic modifiers implicated in CNS inflammation, we leveraged a neuronal translatome profiling dataset of inflamed spinal cord motor neurons of EAE mice (12) (GSE104899) and ranked deregulated candidates by statistical significance (Fig. 1A). Among the top candidates, we focused on the histone methyltransferase G9a, encoded by the gene Ehmt2 (Fig. 1B), because of its specific induction in inflamed motor neurons (fig. S1A) and the availability of specific inhibitors with in vivo efficacy (36, 37), enabling translational application. We validated the induction of Ehmt2 by quantitative polymerase chain reaction (qPCR) of sorted neuronal nuclei from inflamed spinal cords of EAE mice (fig. S1, B and C). As an epigenetic regulator, a central function of G9a is to catalyze the dimethylation of lysine-9 on core histone 3 (H3K9me2), which results in local transcriptional repression. To assess the catalytic activity of G9a in the context of CNS inflammation, we measured H3K9me2 levels in neuronal nuclei in EAE in comparison to healthy spinal cords by flow cytometry. We found an increase of H3K9me2 levels in neuronal nuclei during acute (day 15) and chronic (day 30) EAE but not before disease onset (day 11) (Fig. 1C). This inflammation-dependent induction of H3K9me2 was accompanied by neuronal loss, as quantified by the percentage of NeuN+ neuronal nuclei in spinal cords of acute and chronic EAE mice (Fig. 1D and fig. S1D). To assess the translatability of our mouse findings to human disease, we investigated the induction of H3K9me2 in MS brain tissue. Of note, we observed a significant increase in H3K9me2 immunofluorescence signal in normal-appearing gray matter (NAGM) as well as in cortical MS lesions compared to control brain tissue (Fig. 1E and table S1), supporting an induction of G9a activity in MS brains.
Fig. 1. G9a-depedent H3K9me2 is induced by inflammation and excitotoxicity.
(A) Gene expression heatmap of up- and down-regulated epigenetic modifiers in inflamed motor neurons during acute EAE on day 15 (n = 5 per group each pooled from three mice). (B) Normalized RNA sequencing expression of Ehmt2 in healthy and inflamed motor neurons (DESeq2 false discovery rate; n = 5 per group). (C) Flow cytometry of H3K9me2 levels in spinal cord neuronal nuclei from healthy and EAE mice at indicated days after immunization [one-way analysis of variance (ANOVA) with Dunnett’s post hoc test; n = 6 per group]. (D) Relative neuronal loss in spinal cords from healthy and EAE mice at indicated days after immunization (one-way ANOVA with Dunnett’s post hoc test; n = 6 per group). (E) H3K9me2 staining of brain sections from MS NAGM, cortical MS lesions and control individuals with quantification in HuC/D cells (one-way ANOVA with Tukey’s post hoc test; controls: n = 10, MS NAGM: n = 4, MS lesion: n = 5). Scale bar, 80 μm. (F) H3K9me2 staining of primary neurons 8 hours after 20 μM glutamate stimulation with quantification (two-tailed Student’s t test; n = 8 control versus n = 7 Glu). Scale bar, 50 μm. (G) Cell viability time course of primary neurons stimulated with 20 μM glutamate with quantification of area under the curve (AUC) 19 hours after glutamate application (left, two-way ANOVA; right, two-tailed Student’s t test; n = 4 independent experiments). Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
To explore whether H3K9me2 induction might be a general feature of stress-induced neurodegeneration, we applied glutamate to primary cortical cultures as a model of excitotoxicity during neuroinflammation (38, 39). Resembling the in vivo situation, glutamate exposure of primary cortical cultures led to an up-regulation of H3K9me2 (Fig. 1F) that was associated with decreased cell viability (Fig. 1G). Notably, stimulation with other inflammatory mediators such as TNF-α or interferon-γ (IFN-γ) did not increase H3K9me2 levels (fig. S1E). Moreover, glutamate stress did not result in significant changes of other repressive (H3K9me1, H3K9me3, and H3K27me3) or permissive (H3K27ac) epigenetic marks, nor did it change the levels of DNA-methyltransferase 3a (DNMT3a). Only H3K9me1, which is also catalyzed by G9a, showed a trend toward being induced by glutamate exposure (fig. S1F).
To assess the upstream mechanism of H3K9me2 induction during glutamate toxicity, we applied AP5 or EGTA to block N-methyl-d-aspartate (NMDA) receptors or prevent calcium influx from the extracellular space, respectively. Both substances potently suppressed the induction of H3K9me2 in response to glutamate (fig. S1G), indicating NMDA receptor–mediated calcium influx from the extracellular space as an inducer of increased G9a activity. Besides H3K9me2 levels, NMDA receptor blockage by AP5 also rescued cellular survival (fig. S1H). Together, both CNS inflammation in vivo and glutamate excitotoxicity in neuronal cultures resulted in increased G9a-dependent H3K9me2 levels and neuronal loss.
G9a promotes neuronal damage during glutamate-induced excitotoxicity
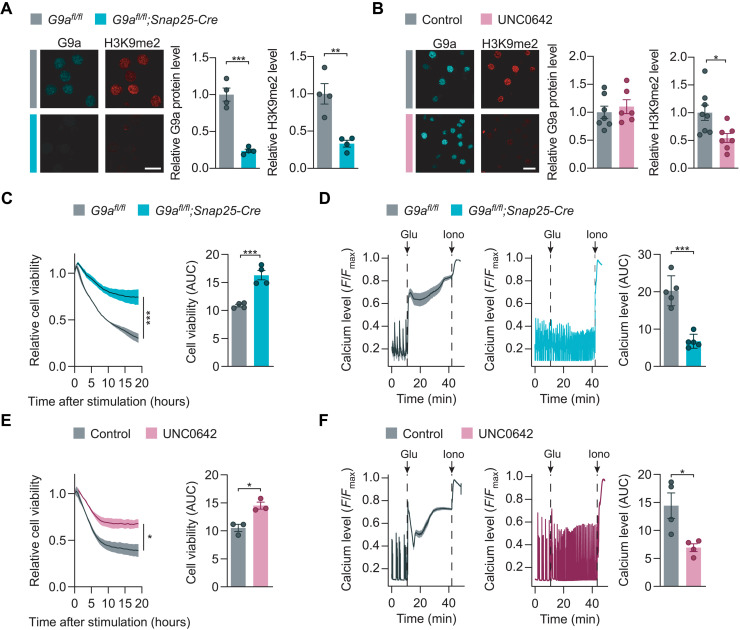
To further explore the impact of G9a and H3K9me2 on neuronal survival during excitotoxicity, we established two strategies for G9a perturbation, namely, genetic disruption in G9afl/fl;Snap25-Cre neurons and pharmacological inhibition by the small-molecule UNC0642 (40). Genetic disruption of G9a in primary cortical neuron cultures resulted in 90% reduction of Ehmt2 mRNA (fig. S2A), 76% reduction of G9a protein, and 67% reduction of H3K9me2 signal (Fig. 2A). Inhibition of the catalytic domain of G9a by UNC0642 did not change the G9a protein level but led to 45% reduction of H3K9me2 signal (Fig. 2B). Next, we investigated the potential of both perturbations to modulate glutamate-mediated neuronal cell death. As experimental readouts, we used real-time measurements of cell viability and toxic calcium overload in glutamate-stimulated primary neuronal cultures. Genetic disruption of G9a in G9afl/fl;Snap25-Cre neurons led to increased neuronal viability (Fig. 2C) and reduced toxic calcium accumulation in comparison to G9afl/fl controls (Fig. 2D) that was comparable to NMDA receptor blockage by AP5 (fig. S2B). To evaluate the potential of pharmacological G9a inhibition, we pretreated neuronal cultures for 24 hours with 1 μM of the G9a-specific small-molecule inhibitor UNC0642 and obtained a similar rescue of cell viability and toxic calcium accumulation as seen for genetic G9a disruption (Fig. 2, E and F, and fig. S2C). Moreover, we treated G9a-deficient neurons from G9afl/fl;Snap25-Cre mice with UNC0642 and observed no additional rescue effect during glutamate stress (fig. S2, D and E), supporting a specific action of UNC0642 on G9a function. Together, G9a activity fostered toxic calcium overload and diminished neuronal survival after glutamate stress.
Fig. 2. G9a promotes neuronal damage during glutamate-induced excitotoxicity.
(A and B) G9a and H3K9me2 stainings of G9afl/fl;Snap25-Cre [(A); two-tailed Student’s t test, n = 4 independent experiments] and UNC0642 pretreated primary neurons compared to respective controls with quantification [(B); two-tailed Student’s t test, G9a: dimethyl sulfoxide (DMSO) n = 7, UNC0642 n = 6, H3K9me2: DMSO n = 8, UNC0642 n = 7]. Scale bars, 20 μm. (C) Cell viability of G9afl/fl versus G9afl/fl;Snap25-Cre primary neurons with time course (left; two-way ANOVA; n = 4 independent experiments) and quantification of AUC (right; two-tailed Student’s t test, n = 4 independent experiments). (D) Cytosolic calcium levels of glutamate-exposed G9afl/fl and G9afl/fl;Snap25-Cre primary neurons with quantification of AUC (two-tailed Student’s t test, n = 5 independent experiments). (E) Cell viability of DMSO- versus UNC0642-treated primary neurons with time course (left; two-way ANOVA; n = 3 independent experiments) and quantification of AUC (right; two-tailed Student’s t test, n = 3 independent experiments). (F) Cytosolic calcium levels of glutamate-exposed primary neurons treated with DMSO or UNC0642 with quantification of AUC (two-tailed Student’s t test, n = 4 independent experiments). Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
G9a potentiates oxidative stress in neurons
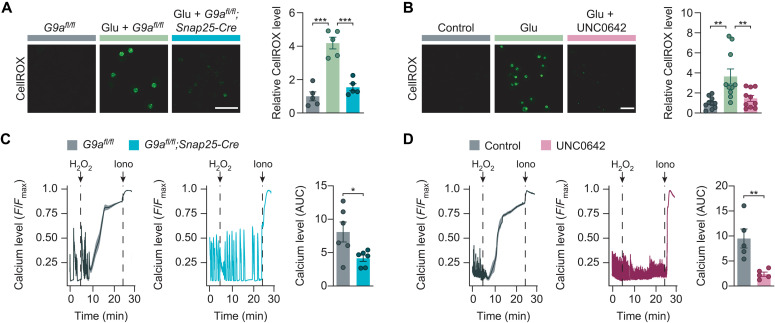
Since neuronal glutamate exposure as well as inflammatory challenges result in an excess of reactive oxygen species (ROS) (7, 41), we next tested whether G9a activity might compromise neuronal resistance to oxidative stress. First, we used CellROX green to measure oxidative stress in primary neuronal cultures after glutamate exposure. While glutamate stimulation of G9afl/fl controls resulted in strong oxidative stress within 1 hour, genetic G9a disruption in G9afl/fl;Snap25-Cre neurons led to a near-complete rescue to the level of cultures without glutamate treatment (Fig. 3A). Similarly, pretreatment with UNC0642 reduced the generation of ROS close to baseline levels as seen without glutamate (Fig. 3B). To test G9a perturbation in a dedicated model with extensive oxidative stress, we performed acute hydrogen peroxide stimulation. We found that both genetic G9a disruption in G9afl/fl;Snap25-Cre neurons and G9a inhibition by UNC0642 rescued hydrogen peroxide–induced calcium overload in comparison to respective controls (Fig. 3, C and D). Notably, hydrogen peroxide treatment itself led to a pronounced up-regulation of H3K9me2, indicating a vicious circle that further drives oxidative stress, while UNC0642 treatment was still able to overcome this effect (fig. S3A). We concluded that G9a exacerbates oxidative stress and consecutive calcium overload.
Fig. 3. G9a potentiates oxidative stress in neurons.
(A) Oxidative stress levels (CellROX green reagent) in G9afl/fl versus G9afl/fl;Snap25-Cre primary neurons with quantification (one-way ANOVA with Tukey’s post hoc test; n = 5 independent experiments). Scale bar, 40 μm. (B) Oxidative stress levels (CellROX green reagent) in DMSO- versus UNC0642-treated primary neurons with quantification (one-way ANOVA with Tukey’s post hoc test; n = 10 independent experiments). Scale bar, 40 μm. (C) Cytosolic calcium levels of H2O2-exposed G9afl/fl and G9afl/fl;Snap25-Cre primary neurons with quantification of AUC (two-tailed Student’s t test, n = 6 independent experiments). (D) Cytosolic calcium levels of H2O2-exposed primary neurons treated with DMSO or UNC0642 with quantification of AUC (two-tailed Student’s t test, n = 5 independent experiments). Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
G9a induces neuronal cell death via ferroptosis
As G9a perturbation supported neuronal viability, we hypothesized a downstream effect of G9a in a central pathway of programmed cell death. Therefore, we used dedicated compounds to induce different forms of programmed cell death that have been implicated in neurodegeneration and oxidative stress and assessed their dependence on G9a function. While neither apoptosis nor necroptosis was affected by G9a perturbation, ferroptosis induced by either erastin or RSL3 was partially rescued in G9a knockout neurons and after G9a inhibition with UNC0642 (Fig. 4, A and B). Moreover, we measured G9a activity by quantification of H3K9me2 immunofluorescence in response to the activation of the different cell death pathways. Again, we did not observe any changes in H3K9me2 signal in apoptosis or necroptosis; however, ferroptosis induction via erastin or RSL3 resulted itself in a significant up-regulation of H3K9me2 (Fig. 4C). Similar to our finding after hydrogen peroxide treatment, this suggests that G9a participates in a self-reinforcing feedback loop, where G9a-dependent H3K9me2 supports ferroptosis and ferroptosis drives H3K9me2. Up-regulation of H3K9me2 in response to ferroptosis induction could be effectively prevented by UNC0642 (fig. S3B).
Fig. 4. G9a induces neuronal cell death via ferroptosis.
(A and B) Quantification of dead primary neurons with CellTox green reagent in response to induction of different cell death pathways in G9afl/fl versus G9afl/fl;Snap25-Cre cultures [(A); two-tailed paired Student’s t test, n = 5 independent experiments] and cultures ± UNC0642 [(B); two-tailed paired Student’s t test, n = 6 independent experiments]. (C) H3K9me2 immunostaining of primary neurons after treatment with indicated compounds (repeated-measures one-way ANOVA with Dunnett’s post hoc test; n = 8 independent experiments). (D) COX-2 immunostaining after glutamate stimulation of primary neurons ± UNC0642 with quantification (one-way ANOVA with Tukey’s post hoc test: n = 7 independent experiments). Scale bar, 40 μm. (E) Cytosolic calcium levels after glutamate treatment ± UNC0642 with quantification. Primary neurons were stimulated 10 min before glutamate treatment with ferrostatin-1 (two-way ANOVA with Bonferroni’s post hoc test, n = 6 per group). (F and G) Glutathione (GSH) levels in G9afl/fl versus G9afl/fl;Snap25-Cre [(F); two-tailed Student’s t test, n = 3 independent experiments] or UNC0642-treated primary neurons [(G); two-tailed Student’s t test, n = 4 independent experiments] in response to glutamate. Data are normalized to untreated controls. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001. STS, staurosporine; TSZ, TNF-α + LCL-161 + Z-VAD-FMK.
To further elucidate the involvement of ferroptosis in glutamate-induced cell death, we chose cyclooxygenase 2 (COX-2) as a widely used biomarker of ferroptosis (42). Glutamate excitotoxicity led to a markedly higher percentage of COX-2–positive cells, whereas pharmacological G9a inhibition almost completely abolished its induction (Fig. 4D). As another way to explore the involvement of ferroptosis in glutamate excitotoxicity, we investigated the potential of ferroptosis inhibitors to rescue glutamate-induced calcium overload. Besides the antioxidant ferrostatin-1, the iron chelator α-difluoromethylornithine (DFMO) and the lipophilic radical-trapping antioxidant coenzyme Q10 rescued glutamate-induced calcium overload (Fig. 4E and fig. S3C), substantiating ferroptosis as a contributor to glutamate excitotoxicity along with previously described pathways including apoptosis (43) and necroptosis (44). Moreover, pretreatment with the G9a inhibitor UNC0642 did not further potentiate the rescue effects of ferroptosis inhibition, suggesting a shared pathway of both interventions (Fig. 4E and fig. S3C).
Last, we assessed whether G9a perturbation could increase the levels of glutathione, a key anti-ferroptotic effector molecule with antioxidant function. While glutamate treatment reduced the levels of glutathione in primary neuronal cultures, both genetic G9a disruption in G9afl/fl;Snap25-Cre neurons and pharmacological G9a inhibition by UNC0642 led to a partial rescue of glutathione levels (Fig. 4, F and G). In summary, these findings suggest a pro-ferroptotic function of G9a that can be counteracted by pharmacological inhibition with UNC0642.
G9a amplifies neurodegeneration and represses anti-ferroptotic genes in CNS inflammation
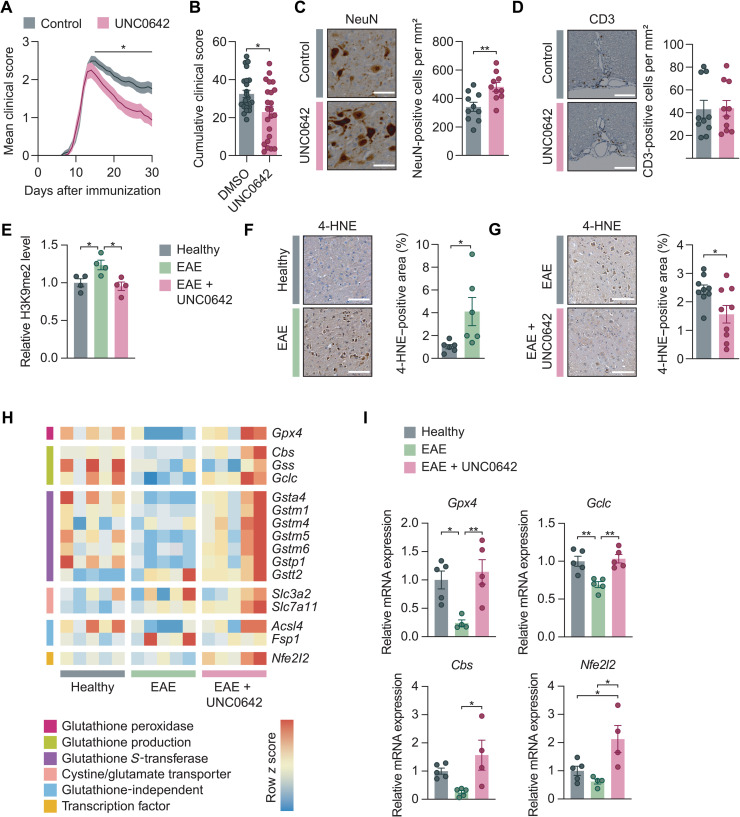
To explore the potential of pharmacological G9a inhibition as a neuroprotective treatment strategy in vivo, we injected EAE animals once daily with UNC0642 (5 mg kg−1 body weight) or vehicle control, starting at symptom onset. This dose was previously reported to effectively reach the CNS without eliciting toxic side effects (40). In accordance with our in vitro findings, treatment with UNC0642 resulted in a significantly ameliorated EAE disease course compared to vehicle-treated mice, which was particularly prominent in the chronic phase of EAE (Fig. 5, A and B). UNC0642 treatment further led to reduced neuronal loss in the spinal cord ventral horn of chronic EAE mice (Fig. 5C), while immune cell infiltration, microglia activation, and body weight changes were not altered (Fig. 5D and fig. S4, A to E).
Fig. 5. G9a amplifies neurodegeneration and represses anti-ferroptotic genes in CNS inflammation.
(A) Disease course of EAE mice treated with DMSO or UNC0642 (5 mg kg−1 body weight). Data are pooled from two independent experiments. DMSO, n = 24; UNC0642, n = 23. (B) Cumulative EAE score from days 15 to 30 (two-tailed Mann-Whitney U test; DMSO, n = 24; UNC0642, n = 23). (C and D) Loss of NeuN+ neurons [(C); two-tailed Mann-Whitney U test; n = 10 per group] and infiltration of CD3+ T cells [(D); two-tailed Mann-Whitney U test; n = 10 per group] in the spinal cord of EAE mice ± UNC0642 at day 30. Scale bars, 100 μm. (E) Flow cytometry of H3K9me2 levels of spinal cord neuronal nuclei from healthy, EAE, and UNC0642-treated EAE mice (one-way ANOVA with Tukey’s post hoc test; n = 4 mice each group). (F and G) 4-HNE immunostaining of cervical spinal cords from healthy versus acute EAE (day 15) [(F); two-tailed Mann-Whitney U test; n = 6 animals per group] and from chronic EAE (day 30) after treatment with vehicle or UNC0642 (5 mg kg−1 body weight) [(G); two-tailed Mann-Whitney U test; n = 12 animals per group]. Scale bars, 100 μm. (H and I) Gene expression data from sorted spinal cord neuronal nuclei of healthy, EAE, and UNC0642-treated EAE mice. n = 5 mice per group (one-way ANOVA with Tukey’s post hoc test; n = 4 to 5 per group). Data are shown as mean values ± SEM. *P < 0.05 and **P < 0.01.
Next, we investigated the effect of UNC0642 in vivo treatment on epigenetic silencing via H3K9me2 and the induction of ferroptosis. The applied UNC0642 treatment regime inhibited H3K9me2 induction and led to a complete normalization of H3K9me2 levels in neuronal nuclei of EAE animals (Fig. 5E). Moreover, we found increased levels of the histopathological ferroptosis marker 4-hydroxy-2-nonenal (4-HNE) in EAE (Fig. 5F), which was significantly reduced by UNC0642 treatment (Fig. 5G). We hypothesized that this effect might be driven by derepression of anti-ferroptotic genes. To test this hypothesis, we sorted NeuN+ neuronal nuclei from the spinal cords of healthy controls, EAE mice, and EAE mice treated with UNC0642 and assessed mRNA expression of ferroptosis-related genes by qPCR. Notably, we observed that many of the genes with anti-ferroptotic function showed a transcriptional suppression during EAE (Fig. 5H and fig. S5A). Among them are Gpx4, encoding the primary cellular defense system against ferroptosis by converting lipid peroxides into nontoxic lipid alcohols (42), and Gclc, encoding the glutamate-cysteine ligase catalytic subunit, the first rate-limiting enzyme of glutathione synthesis (Fig. 5I) (31). Notably, UNC0642 treatment in EAE significantly boosted the expression of Gpx4 and Gclc as well as Cbs, encoding cystathionine β-synthase, the first enzyme of the transsulfuration pathway, an alternative way to fuel glutathione production (45), and Nfe2l2, encoding nuclear factor erythroid 2–related factor 2, a transcription factor that drives several antioxidant genes including Gclc (Fig. 5I) (46). Moreover, multiple glutathione S-transferases followed the same pattern, showing suppression in EAE and derepression upon UNC0642 treatment (fig. S5A). Of note, we also found several necroptosis-related transcripts (Tnf, Ripk1, and Casp8) and proapoptotic Bax significantly induced in neurons during EAE (fig. S5, B and C). Moreover, pro-necroptotic Mlkl and pro-apoptotic Bad were induced in neurons by UNC0642 treatment in EAE (fig. S5, B and C); however, due to their direction, these changes cannot explain the UNC0642-mediated neuroprotection.
Together, these findings exemplify a strong impact of G9a-mediated transcriptional regulation on the glutathione pathway. In contrast, ferroptosis suppressor protein 1 (Fsp1), a glutathione-independent anti-ferroptotic molecule (47), showed no significant regulation (fig. S5A). We concluded that G9a activity led to transcriptional repression of anti-ferroptotic genes, many of which are implicated in the generation of the intracellular antioxidant glutathione. In line with these findings, we found a potent neuroprotective effect of pharmacological G9a inhibition manifesting in increased neuronal survival and improved clinical outcome in CNS inflammation.
Anti-ferroptotic GPX4 is suppressed in MS and induced by G9a inhibition
To translate our findings, we investigated the expression level of the main regulator of ferroptosis, GPX4, in human brain slices of MS patients by RNAscope in situ hybridization. Compared to control brains, we found a reduction of GPX4+ dots per area of SNAP25+ neurons in NAGM and cortical lesions in brain sections of MS patients by comparing single neurons of four subjects per group (Fig. 6A), reflecting the transcriptional repression of Gpx4 in the EAE mouse model (Fig. 5, H and I). By summarizing GPX4 expression levels at a patient level, we observed a trend but no statistical significance due to high interindividual variability and a limited number of subjects (fig. S6A). Furthermore, we detected a prominent increase of H3K9me2 levels in response to erastin-induced ferroptosis in neuronal cultures derived from human induced pluripotent stem cells (hiPSCs), which was rescued by G9a inhibition with UNC0642 (Fig. 6B). This again reflected our observations in murine neuronal cultures (Fig. 4C and fig. S3B) and corroborated induction of G9a activity by ferroptosis and potent suppression by UNC0642 in human cells. Next, we investigated the transcriptional regulation of ferroptosis-related genes by G9a. In line with our findings in UNC0642-treated EAE mice, we found a significant up-regulation of GPX4 and GCLC in response to G9a inhibition via UNC0642 in erastin-treated human neurons (Fig. 6C and fig. S6B). Of note, the expression of SLC7A11, coding for the cystine/glutamate transporter that provides the building blocks for glutathione synthesis, was also highly up-regulated by G9a inhibition (Fig. 6C).
Fig. 6. Anti-ferroptotic GPX4 is suppressed in MS and induced by G9a inhibition.
(A) RNAscope fluorescence in situ hybridization of GPX4 transcripts in brain sections of control individuals and MS NAGM and cortical MS lesions (one-way ANOVA with Dunnett’s post hoc test; control, n = 99,202 neurons; MS NAGM, n = 229,718 neurons; MS lesion, n = 131,294 neurons). Scale bar, 100 μm. (B) H3K9me2 immunostaining in response to ferroptosis induction with erastin in hiPSC-neurons with quantification (repeated-measures one-way ANOVA with Tukey’s post hoc test; n = 3 independent experiments). Scale bar, 40 μm. (C) mRNA expression levels of indicated genes from hiPSC-neurons treated with erastin ± UNC0642 versus control (one-way ANOVA with Tukey’s post hoc test; n = 3 to 4 per group). Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
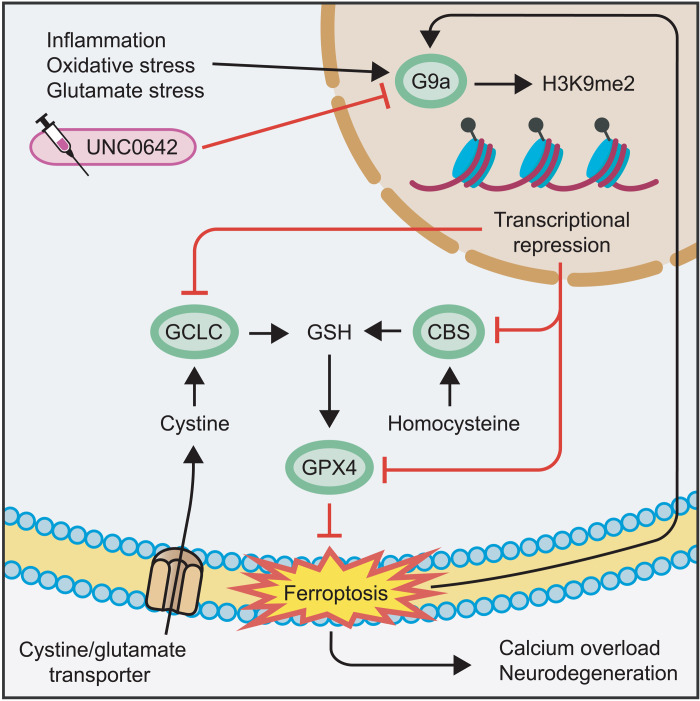
In summary, these results showed an induction of H3K9me2 and repression of GPX4 in neurons of MS patients. Both effects could be reversed by pharmacological G9a inhibition in human neuronal cultures. Together with the evidence from EAE and murine neuronal cell culture, our findings indicate a deleterious induction of neuronal G9a activity during neuroinflammation (Fig. 7). The resulting increase of repressive H3K9me2 leads to epigenetic silencing of key anti-ferroptotic genes including Gpx4, Gclc, and Cbs, causing a depletion of glutathione, accumulation of ROS, and ultimately ferroptotic cell death. Epigenetic reset of neurons by G9a inhibition counteracts this maladaptive cascade and improves neuronal survival and clinical outcome (Fig. 7).
Fig. 7. G9a propagates neurodegeneration by enhancing ferroptosis.
CNS inflammation enhances oxidative stress and increases extracellular glutamate levels, together inducing G9a activity in neurons. G9a catalyzes the repressive mark H3K9me2 that suppresses the expression of Gclc, Cbs, and Gpx4. Thereby, G9a limits the availability of GSH, induces lipid peroxidation, and finally leads to ferroptosis, calcium overload, and neuronal loss. Moreover, ferroptosis itself enhances G9a activity in a self-reinforcing feedback loop. Pharmacological inhibition of G9a by UNC0642 counteracts these effects and results in neuroprotection.
DISCUSSION
Inflammation in the CNS puts neurons at risk for dysfunction and degeneration. Paradoxically, neurons partly support their own demise by mounting maladaptive stress responses that further compromise their function and accelerate their destruction (20). Moreover, stable epigenetic encoding of this maladaptive state might render neurons more prone to subsequent challenges and incapable of autonomous recovery (48). In this study, we specifically searched for epigenetic modifiers that are affected by inflammation and ranked them by statistical significance, listing Ehmt2 second after Kdm1b. We focused on the histone methyltransferase G9a, encoded by Ehmt2, as the most significant candidate with available specific and potent small-molecule inhibitors, allowing to reset the deregulated transcriptional state of inflamed neurons. This complements previous investigations into deregulated neuronal candidates from other biological pathways, which we reported earlier (12, 13). We found G9a to reshape neuronal gene expression in a way that promotes ferroptosis, neuronal loss, and clinical disability. Pharmacological inhibition of G9a rescued these detrimental effects and ultimately resulted in improved clinical outcome. Our findings have profound therapeutic implications for diseases involving inflammation-induced neurodegeneration. This includes diseases with florid or low-grade neuroinflammation like MS (49), infectious encephalitis (50), Alzheimer’s disease (51), Parkinson’s disease (52), and neuropsychiatric diseases like autism (53).
Uncovering the driving forces behind progressive neurodegeneration remains a major scientific challenge. An illustrative example is MS, where the early inflammatory stage of the disease responds well to immunomodulatory treatment, but once patients enter the less inflammatory progressive stage, they elude therapeutic access (11). One reason might be persistent, locally structured inflammation in or near the CNS that is not reached by immunomodulatory drugs (54). However, another possibility is that neuronal stress responses triggered by inflammation do not completely resolve but persist, making these neurons more vulnerable to future damage or destruction (12, 55). In this context, transcriptional reprogramming via stable epigenetic marks like histone modifications (56) or DNA methylation (23) is a plausible underlying molecular mechanism. Epigenetic dysregulation has been implicated in several neurodegenerative diseases including Huntington’s disease (57), Alzheimer’s disease (58), and MS (25). As a result, histone methyltransferases have become promising therapeutic targets in neurodegenerative diseases (48). More broadly, epigenetic mechanisms are suspected to shape acute and long-term gene expression dynamics to allow cellular adaptation to environmental challenges and toxic stimuli (59). It has been proposed that neurons can develop an epigenetic transcriptional memory, whereby a transcriptional response to a stimulus is predefined by a prior exposure (60). In this study, we detected a consistent induction of the G9a-dependent epigenetic mark H3K9me2 in inflamed neurons in EAE mice and MS patients. This H3K9me2 induction appeared to be persistent, as it was maintained even in the chronic stage of EAE, in which inflammation already declined. Moreover, in MS patients, H3K9me2 induction affected neurons across the whole CNS tissue including NAGM, suggesting a generalized shift in the neuronal epigenetic state that is not limited to neurons that are currently directly affected by inflammation. Similarly, broad induction of H3K9me2 was found in the prefrontal cortex of Alzheimer’s disease patients, possibly also driven by inflammation (37). The widespread nature of H3K9me2 induction suggests that large parts of the CNS could benefit from epigenetic normalization by G9a inhibition. However, like our own exploratory analysis of human autopsy material, this study relied on limited cohort sizes requiring confirmation in larger numbers of individuals.
The potential of epigenetic therapy to modulate the transcriptional state of cells is increasingly explored to counteract diverse pathologies (61). The most prominent example is cancer therapy, where various agents with effects on DNA methylation (62), histone methylation (63), and histone acetylation (64) have provided benefit in hematological malignancies and solid tumors in preclinical and clinical trials. But also, in neurological and psychiatric diseases, epigenetic-targeted therapies including G9a inhibition showed promising results. For example, in a mouse model of Alzheimer’s disease, G9a inhibition led to recovery of glutamate receptor expression and excitatory synaptic function and rescued impaired memory performance (37). In Shank3-deficient mice, a model of autism spectrum disorders, G9a inhibition rescued autism-like social deficits and restored NMDA receptor–mediated synaptic function (36). In both studies, the pathology resulted in elevated H3K9me2 levels and disturbed transcriptional signatures, both of which were counteracted by inhibiting G9a. Among several G9a inhibitors that have been used for CNS treatment (36, 37), UNC0642 was shown to have high potency, low cell toxicity, excellent selectivity, and favorable pharmacokinetic properties for in vivo applications (40). However, due to the vibrant drug development activity of epigenetic modifiers for tumor therapy (65), it can be assumed that neurological and psychiatric diseases will profit from repurposing of newly developed agents with further improved pharmacological characteristics (66). This might also include enhanced kinetics to cross the blood-brain barrier, especially since a subgroup of UNC0642-treated mice only showed limited phenotypic benefit in EAE, most probably due to a protracted buildup of effective drug levels in the CNS when starting the treatment at disease onset. However, we chose this setup to focus on benefits in neuronal resilience and minimize the risk of potential immunosuppressive effects during immune cell priming. Of note, numbers and composition of the infiltrating immune cells were not affected by UNC0642 treatment.
In previous reports, beneficial effects of neuronal G9a inhibition were mainly attributed to the restoration of synaptic protein expression (36, 37). Unexpectedly, in this study, we uncovered an additional role for G9a as a potent regulator of the nonapoptotic cell death pathway ferroptosis (29). This notion is supported by improved neuronal survival associated with reduced levels of ferroptotic markers, including 4-HNE (67) and COX-2 (42), upon interference with G9a function. Furthermore, G9a inhibition rendered neurons more resistant against oxidative stress induced by hydrogen peroxide and ferroptosis induced by RSL3 or erastin. Last, G9a inhibition reduced ROS and elevated the level of the intracellular antioxidant glutathione. Thus, we found G9a to be tightly involved in regulating cellular redox homeostasis and ferroptotic cell death. These findings allude to an epigenetic regulatory framework that modulates ferroptosis susceptibility and contributes to ferroptosis-related pathology. A pathological contribution of ferroptosis has been proposed in several neurological diseases including neurodegenerative disorders (30), stroke (68), and traumatic brain injury (69). Moreover, characteristic iron deposits, oxidized phospholipids (32, 33), and reduced GPX4 levels have also been reported in MS and EAE (34). Thus, besides anti-ferroptotic agents with predominantly antioxidant properties that are currently under development (70), targeting the upstream epigenetic regulation of ferroptosis might represent an alternative therapeutic approach to modulate ferroptosis in a wide range of pathologies.
Because of their pathophysiological relevance, molecular pathways involved in ferroptosis are of high interest for clinical translation. As a new player, FSP1 has been recently found to replenish reduced ubiquinone (Q10), which counteracts ferroptosis by scavenging lipid peroxyl radicals and reducing lipid peroxidation (47). This pathway complements the reduction of lipid peroxides by GPX4, which is considered the main mechanism of cellular defense against ferroptosis (30). The glutathione pool needed for GPX4 function is regenerated from cysteine that has been either imported from the extracellular space via the cystine/glutamate transporter or converted from intracellular homocysteine via the transsulfuration pathway (45). Both the generation of glutathione and the conversion of homocysteine critically depend on the function of the rate-limiting enzymes GCLC and CBS, respectively. In this study, we found a robust transcriptional suppression of all three anti-ferroptotic key enzymes (GPX4, GCLC, and CBS) in nerve cells during neuroinflammation in vivo, leaving neurons more prone to degeneration. Notably, treatment with the G9a inhibitor UNC0642 completely rescued the transcriptional repression of GPX4, GCLC, and CBS, as well as the transcription factor NRF2 that supports the cellular antioxidant response (46). Corroborating this finding for potential translational application, we also found an induction of GPX4 and GCLC upon G9a inhibition in human neuronal culture. Furthermore, G9a inhibition induced the anti-ferroptotic gene SLC7A11 encoding the cystine/glutamate transporter. Reassuringly, G9a can also be extracted as a driver of ROS and lipid peroxidation from a recent genome-wide CRISPR screen in human neurons that aimed to uncover pathways of chronic oxidative stress (71). Together, this evidence supports the potential of G9a inhibition to suppress ferroptosis also in human cells.
When thinking about the clinical application of G9a inhibition, a cautionary note needs to be considered. A big advantage of G9a is that its inhibition boosts the expression of several key anti-ferroptotic genes and potentially other neuroprotective pathways without the need to directly target them. On the other hand, this intended broad effect on the transcriptional landscape can also confer unwanted side effects, especially since pharmacological inhibitors of G9a are not cell type specific and might affect gene expression in various parts of the body. However, in contrast to cell culture experiments, G9a inhibition in vivo never completely suppressed the levels of H3K9me2. In contrast, even at high doses, we did not detect H3K9me2 reduction in spinal cord, brain, or spleen. Only after pathological induction of H3K9me2 in the EAE model, G9a inhibition resulted in a reduction of H3K9me2 to healthy baseline levels. This might indicate that G9a inhibition is sufficient to restore but not unphysiologically dampen H3K9me2 levels, representing a desirable pharmacological feature. One possible explanation might be a higher stability of H3K9me2 marks at “physiological” positions in the genome, which might be favored by the local epigenetic chromatin landscape. Nevertheless, further studies are needed to mechanistically explore this observation and explicitly investigate potential organism-wide side effects, especially since genetic G9a deletion has been shown to impair learning and memory (72), potentially limiting the applicability of this broad epigenetic intervention in clinical care. Moreover, dosing schemes including optimal dosage, time point, and interval of application need to be explored to identify a therapeutic setup with potent neuroprotective effects but limited side effects. In an ideal setting, the inhibitor would be able to restore physiological H3K9me2 levels in neurons after a short-term induction therapy, rendering neurons more resistant to upcoming challenges. Such a therapy might then be repeated at a given time interval.
The urgent clinical demand to counteract neurodegeneration by genuine neuroprotective treatments continues unabated (11). Here, we characterize G9a as a key driver of neuronal loss in inflammatory neurodegeneration. By genetic and pharmacological perturbation of G9a, we found that it destabilizes cellular redox homeostasis and promotes ferroptosis through epigenetic repression of anti-ferroptotic pathways. The G9a inhibitor UNC0642 showed beneficial effects on neuronal survival in glutamate excitotoxicity and oxidative stress and reduced neuronal loss and clinical impairment in the preclinical MS model EAE. In conclusion, our findings highlight the potential value of G9a inhibition to reset the maladaptive state of neurons by erasing their epigenetic memories of inflammation. This approach might prove valuable to deliver neuroprotection in inflammation-induced neurodegeneration and other ferroptosis-dependent pathologies.
MATERIALS AND METHODS
Study design
The current study focuses on the investigation of the epigenetic regulator G9a in inflammation-induced neurodegeneration and its downstream effects on the regulation of ferroptosis in MS and its mouse model EAE. We first investigated the regulation of G9a during neuroinflammation in vivo and in vitro and tested the effect of G9a on neuronal viability in glutamate-stressed primary neuronal cultures. To identify the underlying pathway that regulates neuronal viability, we examined different programmed cell death pathways and identified ferroptosis as regulated by G9a. Next, we probed the therapeutic efficacy of pharmacological G9a inhibition in the EAE model and confirmed the regulation of ferroptosis by neuronal gene expression analysis and immunohistochemistry. Last, we translated our findings into human MS by applying immunohistochemistry and RNAscope in situ hybridization to MS autopsy brain samples and testing the regulation of ferroptosis-related genes by G9a inhibition in hiPSC-derived neurons.
Mice
All mice, C57BL/6J wild-type (The Jackson Laboratory), G9afl/fl (73), and G9afl/fl;Snap25-Cre (Strain #023525 from The Jackson Laboratory) were kept under specific pathogen–free conditions in the central animal facility of the University Medical Center Hamburg-Eppendorf (UKE). We used adult female mice in the age of 6 to 16 weeks. All animal experimental procedures were in accordance with institutional guidelines and conformed to the requirements of the German Animal Welfare Act. Ethical approvals were obtained from the State Authority of Hamburg, Germany (approval no. 122/17).
Experimental autoimmune encephalomyelitis
Mice were immunized subcutaneously as previously described (17) with 200 μg of MOG35–55 peptide (Schafer-N) in complete Freund’s adjuvant (CFA) (Difco, catalog no. DF0639-60-6) containing Mycobacterium tuberculosis (4 mg ml−1) (Difco, catalog no. DF3114-33-8). In addition, 300 ng of pertussis toxin (Calbiochem, catalog no. CAS70323-44-3) was injected intraperitoneally on the day of immunization and 48 hours later. Animals were scored daily for clinical signs by the following system: 0, no clinical deficits; 1, tail weakness; 2, hindlimb paresis; 3, partial hindlimb paralysis; 3.5, full hindlimb paralysis; 4, full hindlimb paralysis and forelimb paresis; 5, premorbid or dead. Animals reaching a clinical score of ≥4 were euthanized according to the regulations of the local Animal Welfare Act (17). Where indicated, animals were injected intraperitoneally with UNC0642 (5 mg kg−1) (Sigma-Aldrich, catalog no. SML 1037) starting from the day of disease onset. We used littermate controls in all EAE experiments. UNC0642 was prediluted in dimethyl sulfoxide (DMSO), and the final injection contained 10% DMSO ± UNC0642 and 90% Dulbecco’s phosphate-buffered saline (DPBS; PAN-Biotech). The investigators were blind to the treatment in the EAE experiments.
Candidate gene identification
We previously generated an RNA sequencing dataset of spinal cord motor neuronal translatomes during CNS inflammation in the EAE model (Gene Expression Omnibus: GSE104899) (12). We reanalyzed this dataset focusing on known epigenetic regulators (dbEM—a database of epigenetic modifiers) that were differentially expressed in inflamed versus healthy motor neurons. Differential expression analysis was performed with DESeq2 (v.1.14.1), calling genes with a minimal twofold change and false discovery rate–adjusted P < 0.05 differentially expressed.
hIPSC-derived neurons
hiPSCs (ZIPi013-B) (74) were maintained under feeder-free conditions on Matrigel (Corning)–coated plates in mTeSR1 medium (STEMCELL Technologies, catalog no. 85850). For neuronal induction, iPSCs were dissociated with Accutase and seeded at a density of 3 × 106 cells per well on AggreWell800 plates [10,000 cells per embryoid body (EB), STEMCELL Technologies] in SMADi Neural Induction Medium (SMADi NIM; STEMCELL Technologies, catalog no. 08582) supplemented with 10 μM Y-27632 (STEMCELL Technologies, catalog no. 72302). On day 6, EBs were harvested and cultivated on Matrigel-coated plates in SMADi NIM for 12 days. Newly formed neural rosettes were manually picked and cultured for another 4 days. To release neural precursor cells (NPCs), neural rosettes were dissociated with Accutase and maintained for several passages at high density in Neural Progenitor Medium (STEMCELL Technologies, catalog no. 05833) on Matrigel-coated plates. hiPSC-derived NPCs were differentiated into neurons as previously described (75, 76) with some modifications. Briefly, NPCs were seeded at a density of 1 × 105 cells/cm2 in Neural Progenitor Medium onto Matrigel-coated plates. After 24 hours, the medium was replaced by neural differentiation medium (day 0 of differentiation) composed of Neurobasal Plus medium (Gibco, catalog no. A3582901) containing 1× B-27 Plus Supplement (Gibco, catalog no. A3582801), 1× N2 Supplement-A (STEMCELL Technologies, catalog no. 07152), 1× MEM nonessential amino acids (Gibco, catalog no. 11140050), laminin (1 μg ml−1) (Sigma-Aldrich, catalog no. L2020), 1 μM dibutyryl-cAMP (adenosine 3′,5′-monophosphate) (STEMCELL Technologies, catalog no. 73882), l-ascorbic acid (10 ng ml−1 ) (STEMCELL Technologies, catalog no. 72132), brain-derived neurotrophic factor (10 ng ml−1) (STEMCELL Technologies, catalog no. 78005), and glial-derived neurotrophic factor (10 ng ml−1) (STEMCELL Technologies, catalog no. 78058). To promote a glutamatergic neuronal cell type, 5 μM cyclopamine (STEMCELL Technologies, catalog no. 72072) was additionally added to the medium during the first week of differentiation. On day 14, the cells were detached using Accutase and reseeded onto 96-well poly-l-ornithine-/laminin–coated plates suited for confocal microscopy. Thereafter, cells were maintained in neural differentiation medium supplemented with CultureOne (Gibco, catalog no. A3320201) for up to 30 weeks to increase maturity.
Primary mouse neuronal cultures
Primary neuronal cultures were prepared as previously described (17). Briefly, pregnant C57BL/6J or G9afl/fl;Snap25-Cre mice were euthanized. To ensure comparability between genotypes, only embryos from heterozygous breedings were used. Brainstem tissue of each embryo for genotyping was reserved, cortices were isolated and dissociated, and cells were plated at a density of 1 × 105 cells per 1 cm2 on poly-d-lysine–coated wells (5 μM; Sigma-Aldrich, catalog no. A-003-M). If not stated otherwise, cells were maintained in Neurobasal Plus medium (supplemented with B-27 Plus, penicillin, streptomycin, and l-glutamine; Gibco, catalog no. A3582901) at 37°C, 5% CO2, and a relative humidity of 98% and treated with 1 μM cytarabine (Sigma-Aldrich, catalog no. BP383) at 1 day in vitro (div) to inhibit glial cell proliferation. If no cytarabine was applied, cells were maintained in Primary Neuron Basal Medium (supplemented with neural cell survival factor, gentamycin, and l-glutamine; Lonza). Throughout, cultures after 14 to 23 div were used for experiments. The G9a inhibitor UNC0642 was used in a concentration of 1 μM, and cultures were pretreated 24 hours before further experimental procedures.
Chemicals
Chemicals used in vitro are listed in table S2 including function, supplier, catalog number, and concentration.
Human immunohistochemistry
Human CNS tissue was fixed with 4% paraformaldehyde (PFA) and embedded in paraffin as described previously (77), and 2-μm-thin sections were cut. Tissue sections were deparaffinized, and antigen retrieval [Marmite Pascal tris-EDTA (pH 9.0), 125°C, 30 s] was performed. To prevent unspecific binding of primary antibody, tissues were incubated with normal goat serum (10% in PBS) before overnight incubation with a mouse immunoglobulin G2a (IgG2a) anti-H3K9me2 (1:200; Abcam, ab1220) and a mouse IgG2b anti-HuC/D (1:100; Invitrogen, A-21271) antibody. Autofluorescence was removed (Merck, catalog no. 2160), and primary antibodies were visualized using goat anti-mouse IgG2a Alexa Fluor 488 (Invitrogen, A21131) and goat anti-mouse IgG2b Alexa Fluor 647 (Life, A21242) (see table S3 for antibodies). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, D3571). All steps were performed at room temperature. Stained sections were scanned using the Pannoramic 250 FLASH II (3DHISTECH) Whole Slide Scanner at a resolution of 0.221 μm per pixel. Their use for scientific purposes was in accordance with institutional ethical guidelines and approved by the ethics committee of the University of Geneva (Switzerland). Informed consent was obtained from subjects, in accordance with approval of the local ethical committee.
RNAscope in situ hybridization
We performed RNAscope Fluorescent Multiplex V2 [Advanced Cell Diagnostic (ACD), catalog no. 323100] according to the manufacturer’s standard protocol for PFA-fixed tissue. The human probes Hs-GPX4 (ACD, catalog no. 456851) and Hs-SNAP25 (ACD, 518851) were commercially available from ACD. RNAscope human samples were scanned using the Pannoramic 250 FLASH II (3DHISTECH) Digital Slide Scanner at ×20 magnification. GPX4+ SNAP25+ neurons were quantified by a blinded experimenter using a custom-made script, which was based on Cognition Network Language (Definiens Cognition Network Technology; Definiens Developer XD software).
Mouse immunohistochemistry
For histopathology of EAE mice, spinal cord tissue was fixed, decalcified, dehydrated, cast in paraffin, and stained according to the standard procedures of the UKE Mouse Pathology Facility as previously described (17). The tissue was stained with hematoxylin (blue color) for orientation, followed by immunolabeling, that was visualized using the avidin-biotin complex technique with 3,3′-diaminobenzidine (brown stain). Slides were analyzed with a NanoZoomer 2.0-RS digital slide scanner and NDP.view2 software (Hamamatsu). CD3-positive cells were quantified in the white matter tract of the spinal cord using a customized counting mask with Fiji (ImageJ). NeuN+ cells (neurons) were manually counted in the ventral horn outflow tract of the spinal cord. Area of 4-HNE and Iba1 stainings were analyzed with a customized mask using Fiji (ImageJ). See table S3 for antibody specifications. Analysis was standardized across all conditions. At least three images were analyzed per animal, and the mean per animal was used for subsequent statistical comparisons.
Immunocytochemistry
Immunocytochemistry was performed as described previously (14). hiPSC neurons and mouse neurons were cultivated on 12-mm-diameter coverslips or in 96-well plates suited for confocal microscopy, fixed with 4% PFA, and incubated in 10% normal donkey serum (NDS) containing 0.25% Triton X-100, and subsequently, immunolabeling was performed. Immunostainings were imaged with a Zeiss LSM 700 confocal microscope.
RealTime-Glo cell viability assay
RealTime-Glo (Promega, catalog no. G9711) MT cell viability assay was performed as previously described (17). Briefly, the substrate and NanoLuc Enzyme were mixed together, added to neuronal cultures, and then incubated for 5 hours for equilibration of luminescence signal before respective treatments were applied. Luminescence was acquired with a Spark 10M multimode microplate reader (Tecan) at 37°C and 5% CO2 every 30 min over a total time period of 24 hours. At least five technical replicates per condition were used. For analysis, every well’s data point was normalized to its last value before the stressor was added and then normalized to the mean of the control wells for every time point using a customized MATLAB script. Thereby, normalization accounted for well-to-well seeding variability.
Cell death induction
Primary neurons were subjected to dedicated compounds initiating either apoptosis (staurosporine, STS), necroptosis (TNF-α + LCL-161 + Z-VAD-FMK, or shikonin), or ferroptosis (RSL3, or erastin) for 48 hours in G9afl/fl;Snap25-Cre cultures, or cultures were pretreated for 24 hours with 1 μM UNC0642. Together with the cell death–inducing compounds, CellTox Green Cytotoxicity Assay (Promega) was added to primary neurons 1:2000. Before fixation with 4% PFA, cells were counterstained with Hoechst 3342. Neurons were imaged on a Zeiss LSM 700 confocal microscope using ×20 magnification. Dyeing cells were identified with Fiji (ImageJ) by their bright green nuclear fluorescence and quantified as percentage of total cells, measured by the number of Hoechst-positive nuclei.
Real-time PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen) and subsequently reverse-transcribed to complementary DNA with the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Gene expression was analyzed by real-time PCR performed in the ABI Prism 7900 HT Fast Real-Time PCR System (Applied Biosystems) using TaqMan Gene Expression Assays (Thermo Fisher Scientific; listed in table S4). Gene expression was calculated as 2−ΔCt relative to Tbp as the endogenous control.
Calcium imaging
Calcium recordings were performed as previously described (17). Primary neuronal cultures were seeded on Ibidi 60 μ-Dish Quad (catalog no. 80411) plates. Cultures were infected with an adeno-associated virus 7 (AAV7) containing pAAV-Syn-GCamp6f-WPRE-SV (Addgene 100837) at 7 to 10 div with a 20,000-fold multiplicity of infection. AAV particles were produced according to the standard procedures of the UKE vector facility. Images were acquired with a confocal LSM 700 laser scanning confocal microscope (Zeiss) every 0.48 s with a ×20 magnification in an imaging chamber maintaining 37°C and 5% CO2. Infected cultures were imaged in the respective culture medium. Before applying the indicated chemicals, 5 to 10 min of baseline activity was recorded. At the end of the recording, 8 μM ionomycin was applied to induce the maximum cellular calcium response that was used for normalization. Specific assay details and concentrations can be found in the respective figure legends or table S2. For data analysis, mean fluorescence values of every cell were measured using Fiji software [National Institutes of Health (NIH)] and normalized to the maximal calcium response after ionomycin challenge. For each cell, mean area under the curve (AUC) of the calcium response was calculated using a custom R script.
Oxidative stress detection
For the detection of oxidative stress in response to glutamate treatment, CellROX green reagent (Thermo Fisher Scientific) was used. Neuronal cultures were stimulated with 20 μM glutamate for a total incubation time of 1 hour. CellROX reagent was added to the wells in a 5 μM final concentration after 30 min of glutamate stress. Hoechst 33342, a cell-permeant nuclear counterstain, was added for 10 min. Cells were washed two times with PBS before image acquisition. The cells were subsequently imaged on a Zeiss LSM 700 confocal microscope using ×20 magnification. Immunofluorescence of nuclear CellROX dye was quantified using Fiji (ImageJ).
Glutathione measurement
GSH-Glo Glutathione assay (Promega) was used as described by the manufacturer. Primary cortical neurons were cultured in a 96-well plate and stimulated with glutamate for 2 hours. Cells were washed with PBS and GSH-Glo reagent was added to the wells for 30 min at room temperature, followed by the addition of luciferin detection reagent. After 15 min, luminescence was detected using a Spark 10M multimode microplate reader (Tecan).
Neuronal nuclei isolation and cell sorting
Nuclei of mouse spinal cords were isolated with the Nuclei Isolation Kit (Sigma-Aldrich, catalog no. NUC101) according to the manufacturer’s protocol with minor modifications. Briefly, mice were sacrificed with CO2 and perfused with cold PBS. Whole spinal cords were removed and mechanically dissociated with a scalpel on a petri dish placed on a cooled metal block. Tissue was added to 2 ml of EZ buffer (Sigma-Aldrich, catalog no. NUC101) and further dissociated using a glass douncer (Sigma-Aldrich, D9063). After 5-min incubation on ice, the homogenate was centrifuged (500g, 5 min, 4°C) and the pellet was washed in 2 ml of EZ buffer, followed by two washing steps in nuclei incubation buffer (340 mM sucrose, 2 mM MgCl2, 25 mM KCl, 65 mM glycerophosphate, 5% glycerol, 1 mM EDTA, 1% bovine serum albumin). Nuclei were filtered using a 30-μm filter and directly stained with primary labeled NeuN antibody (ab190565, Abcam) and propidium iodide. NeuN+ nuclei were sorted using a BD FACSAria III cell sorter (BD Biosciences). RNA for real-time PCR was processed as described above.
Neuronal nuclei isolation and flow cytometry
Nuclei of mouse spinal cords were isolated as described above. After filtering of the homogenate, nuclei were fixed with methanol for 10 min on ice, followed by two washing steps with nuclei incubation buffer. For intranuclear staining, nuclei were permeabilized with nuclei incubation buffer supplemented with 0.5% Triton X-100 for 15 min on ice. After blocking with nucleus incubation buffer and 10% NDS, primary antibody was added for 1 hour at room temperature, followed by a washing step and subsequent incubation in secondary antibody for 30 min at room temperature. Immunofluorescence was acquired on an LSR II FACS analyzer (BD Biosciences). Data analysis was performed with the FlowJo v.10 analysis software (FlowJo LLC).
Isolation of CNS-infiltrating immune cells and flow cytometry
Following procedures as previously described (12), mice were perfused intracardially with ice-cold PBS immediately after sacrifice to remove blood from the intracranial vessels. The brain and spinal cord were prepared with sterile instruments, minced with a scalpel, and incubated with agitation in RPMI 1640 (PAA) containing collagenase A (1 mg ml−1) (Roche) and recombinant ribonuclease-free deoxyribonuclease I (0.1 mg ml−1) (Roche) for 45 min at 37°C. Tissue was triturated through a 100-μm cell strainer and washed with PBS (pellet was centrifuged at 300g for 10 min at 4°C). The homogenized tissue was resuspended in 30% isotonic Percoll (GE Healthcare) and carefully underlaid with 78% isotonic Percoll. After gradient centrifugation (1500g for 30 min at 4°C), CNS-infiltrating immune cells were recovered from the gradient interphase and washed in ice-cold PBS. Single-cell suspensions were stained in the presence of 123count eBeads (Thermo Fisher Scientific). The antibodies and the respective antigen, host species, supplier, catalog number, clone, and dilution are listed in table S3. Data were acquired on an LSR II FACS analyzer (BD Biosciences). Data analysis was performed with the FlowJo v.10 analysis software (FlowJo LLC).
Statistical analysis
The statistical analyses applied during the bioinformatics analysis are detailed in the respective sections of the article. Flow cytometric data were analyzed by using FlowJo (LLC). Images were analyzed using Fiji software (NIH). Experimental data obtained from RealTime-Glo cell viability assay were analyzed with a customized MATLAB script, and calcium imaging data were processed using a customized R script. Further experimental data were analyzed with the Prism 9 software (GraphPad Software) for Mac and are presented as means ± SEM. Unless otherwise stated, differences between two experimental groups were determined using an unpaired, two-tailed Student’s t test; differences between three or more experimental groups were determined using one-way analysis of variance (ANOVA) with Tukey’s post hoc test; differences between two or more experimental groups under multiple conditions or over time were analyzed using two-way ANOVA with Tukey’s post hoc test. Statistical analysis of the clinical scores in the EAE experiments was performed by applying a Mann-Whitney U test to the cumulative score of each individual animal. Significant results are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We thank the UKE vector facility for supplying viral vectors and for technical advice and the UKE Mouse Pathology Facility for histopathology of EAE mice. We thank A. Tarakhovsky, Rockefeller University, New York, for providing the G9afl/fl mice. We thank M. Kreutzfeldt for helping with RNAscope analysis.
Funding: This work was supported by the Deutsche Forschungsgemeinschaft (grant no. FR1720/11-2 to M.A.F.), the Deutsche Multiple Sklerose Gesellschaft (to M.A.F.), the Gemeinnützige Hertie-Stiftung (Hertie Network of Excellence in Clinical Neuroscience Fellowship to J.B.E.), the Forschungsförderungsfonds of the Universitätsklinikum Hamburg-Eppendorf (postdoc fellowship to N.R. and startup funds to J.B.E.), the Swiss National Science Foundation (grant nos. 310030B_201271 and 310030_185321 to D.M.), and the ERC (grant no. 865026 to D.M.).
Author contributions: N.R. conducted most of the experiments and analyzed the data. N.R. and J.B.E. performed EAE experiments with UNC0642 treatment. M.S.W. helped with immunocytochemistry and calcium imaging. L.B.-L. helped with neuronal cell culture and viability assays. S.B. helped with mouse preparation and qPCR. G.D.L., K.E., I.W., and D.M. conducted human histopathology and RNAscope experiments. U.H. and O.P. established and characterized hiPSC-neurons. N.R., J.B.E., and M.A.F. designed the experiments and wrote the initial version of the manuscript. M.A.F. and J.B.E. conceived, supervised, and funded the study.
Competing interests: N.R., J.B.E., and M.A.F. are inventors on a patent application related to this work filed by Universitätsklinikum Hamburg-Eppendorf (no. EP22169820.2, filed on 25 April 2022). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Publicly available datasets used in this study are available from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104899).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S4
REFERENCES AND NOTES
- 1.Prinz M., Masuda T., Wheeler M. A., Quintana F. J., Microglia and central nervous system-associated macrophages—From origin to disease modulation. Annu. Rev. Immunol. 39, 251–277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friese M. A., Schattling B., Fugger L., Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 10, 225–238 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Dendrou C. A., Fugger L., Friese M. A., Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15, 545–558 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Luchetti S., Fransen N. L., van Eden C. G., Ramaglia V., Mason M., Huitinga I., Progressive multiple sclerosis patients show substantial lesion activity that correlates with clinical disease severity and sex: A retrospective autopsy cohort analysis. Acta Neuropathol. 135, 511–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ransohoff R. M., How neuroinflammation contributes to neurodegeneration. Science 353, 777–783 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Dangond F., Donnelly A., Hohlfeld R., Lubetzki C., Kohlhaas S., Leocani L., Ciccarelli O., Stankoff B., Sormani M. P., Chataway J., Bozzoli F., Cucca F., Melton L., Coetzee T., Salvetti M., Facing the urgency of therapies for progressive MS—A progressive MS alliance proposal. Nat. Rev. Neurol. 17, 185–192 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Haider L., Fischer M. T., Frischer J. M., Bauer J., Hoftberger R., Botond G., Esterbauer H., Binder C. J., Witztum J. L., Lassmann H., Oxidative damage in multiple sclerosis lesions. Brain 134, 1914–1924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell G. R., Ziabreva I., Reeve A. K., Krishnan K. J., Reynolds R., Howell O., Lassmann H., Turnbull D. M., Mahad D. J., Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann. Neurol. 69, 481–492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macrez R., Stys P. K., Vivien D., Lipton S. A., Docagne F., Mechanisms of glutamate toxicity in multiple sclerosis: Biomarker and therapeutic opportunities. Lancet Neurol. 15, 1089–1102 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Schirmer L., Velmeshev D., Holmqvist S., Kaufmann M., Werneburg S., Jung D., Vistnes S., Stockley J. H., Young A., Steindel M., Tung B., Goyal N., Bhaduri A., Mayer S., Engler J. B., Bayraktar O. A., Franklin R. J. M., Haeussler M., Reynolds R., Schafer D. P., Friese M. A., Shiow L. R., Kriegstein A. R., Rowitch D. H., Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faissner S., Plemel J. R., Gold R., Yong V. W., Progressive multiple sclerosis: From pathophysiology to therapeutic strategies. Nat. Rev. Drug Discov. 18, 905–922 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Schattling B., Engler J. B., Volkmann C., Rothammer N., Woo M. S., Petersen M., Winkler I., Kaufmann M., Rosenkranz S. C., Fejtova A., Thomas U., Bose A., Bauer S., Träger S., Miller K. K., Brück W., Duncan K. E., Salinas G., Soba P., Gundelfinger E. D., Merkler D., Friese M. A., Bassoon proteinopathy drives neurodegeneration in multiple sclerosis. Nat. Neurosci. 22, 887–896 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Rosenkranz S. C., Shaposhnykov A. A., Träger S., Engler J. B., Witte M. E., Roth V., Vieira V., Paauw N., Bauer S., Schwencke-Westphal C., Schubert C., Bal L. C., Schattling B., Pless O., van Horssen J., Freichel M., Friese M. A., Enhancing mitochondrial activity in neurons protects against neurodegeneration in a mouse model of multiple sclerosis. eLife 10, e61798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schattling B., Steinbach K., Thies E., Kruse M., Menigoz A., Ufer F., Flockerzi V., Bruck W., Pongs O., Vennekens R., Kneussel M., Freichel M., Merkler D., Friese M. A., TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 18, 1805–1811 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Friese M. A., Craner M. J., Etzensperger R., Vergo S., Wemmie J. A., Welsh M. J., Vincent A., Fugger L., Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat. Med. 13, 1483–1489 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Yan J., Bengtson C. P., Buchthal B., Hagenston A. M., Bading H., Coupling of NMDA receptors and TRPM4 guides discovery of unconventional neuroprotectants. Science 370, eaay3302 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Woo M. S., Ufer F., Rothammer N., Di Liberto G., Binkle L., Haferkamp U., Sonner J. K., Engler J. B., Hornig S., Bauer S., Wagner I., Egervari K., Raber J., Duvoisin R. M., Pless O., Merkler D., Friese M. A., Neuronal metabotropic glutamate receptor 8 protects against neurodegeneration in CNS inflammation. J. Exp. Med. 218, e20201290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith T., Groom A., Zhu B., Turski L., Autoimmune encephalomyelitis ameliorated by AMPA antagonists. Nat. Med. 6, 62–66 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Andreone B. J., Larhammar M., Lewcock J. W., Cell death and neurodegeneration. Cold Spring Harb. Perspect. Biol. 12, a036434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farley M. M., Watkins T. A., Intrinsic neuronal stress response pathways in injury and disease. Annu. Rev. Pathol. 13, 93–116 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Bhat K. P., Umit Kaniskan H., Jin J., Gozani O., Epigenetics and beyond: Targeting writers of protein lysine methylation to treat disease. Nat. Rev. Drug Discov. 20, 265–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein H. U., McCabe C., Gjoneska E., Sullivan S. E., Kaskow B. J., Tang A., Smith R. V., Xu J., Pfenning A. R., Bernstein B. E., Meissner A., Schneider J. A., Mostafavi S., Tsai L. H., Young-Pearse T. L., Bennett D. A., De Jager P. L., Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 22, 37–46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kular L., Needhamsen M., Adzemovic M. Z., Kramarova T., Gomez-Cabrero D., Ewing E., Piket E., Tegner J., Beck S., Piehl F., Brundin L., Jagodic M., Neuronal methylome reveals CREB-associated neuro-axonal impairment in multiple sclerosis. Clin. Epigenetics 11, 86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh J. L., Garg P., Thin T. H., Yoo S., Dutta R., Trapp B. D., Haroutunian V., Zhu J., Donovan M. J., Sharp A. J., Casaccia P., Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 17, 121–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh J. L., Casaccia P., Epigenetic mechanisms in multiple sclerosis: Implications for pathogenesis and treatment. Lancet Neurol. 12, 195–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer R., Weissert R., Diem R., Storch M. K., de Graaf K. L., Kramer B., Bähr M., Acute neuronal apoptosis in a rat model of multiple sclerosis. J. Neurosci. 21, 6214–6220 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofengeim D., Ito Y., Najafov A., Zhang Y., Shan B., DeWitt J. P., Ye J., Zhang X., Chang A., Vakifahmetoglu-Norberg H., Geng J., Py B., Zhou W., Amin P., Berlink Lima J., Qi C., Yu Q., Trapp B., Yuan J., Activation of necroptosis in multiple sclerosis. Cell Rep. 10, 1836–1849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picon C., Jayaraman A., James R., Beck C., Gallego P., Witte M. E., van Horssen J., Mazarakis N. D., Reynolds R., Neuron-specific activation of necroptosis signaling in multiple sclerosis cortical grey matter. Acta Neuropathol. 141, 585–604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X., Stockwell B. R., Conrad M., Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon S. J., Stockwell B. R., The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 3, 35–54 (2019). [Google Scholar]
- 32.Lassmann H., van Horssen J., Oxidative stress and its impact on neurons and glia in multiple sclerosis lesions. Biochim. Biophys. Acta 1862, 506–510 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Hametner S., Wimmer I., Haider L., Pfeifenbring S., Bruck W., Lassmann H., Iron and neurodegeneration in the multiple sclerosis brain. Ann. Neurol. 74, 848–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu C. L., Nydes M., Shanley K. L., Morales Pantoja I. E., Howard T. A., Bizzozero O. A., Reduced expression of the ferroptosis inhibitor glutathione peroxidase-4 in multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neurochem. 148, 426–439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee C., Kling T., Russo B., Miebach K., Kess E., Schifferer M., Pedro L. D., Weikert U., Fard M. K., Kannaiyan N., Rossner M., Aicher M. L., Goebbels S., Nave K.-A., Krämer-Albers E.-M., Schneider A., Simons M., Oligodendrocytes provide antioxidant defense function for neurons by secreting ferritin heavy chain. Cell Metab. 32, 259–272.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z. J., Zhong P., Ma K., Seo J. S., Yang F., Hu Z., Zhang F., Lin L., Wang J., Liu T., Matas E., Greengard P., Yan Z., Amelioration of autism-like social deficits by targeting histone methyltransferases EHMT1/2 in Shank3-deficient mice. Mol. Psychiatry 25, 2517–2533 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y., Liu A., Wang Z. J., Cao Q., Wang W., Lin L., Ma K., Zhang F., Wei J., Matas E., Cheng J., Chen G. J., Wang X., Yan Z., Inhibition of EHMT1/2 rescues synaptic and cognitive functions for Alzheimer’s disease. Brain 142, 787–807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitt D., Werner P., Raine C., Glutamate excitotoxicity in a model of multiple sclerosis. Nat. Med. 6, 67–70 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Birkner K., Wasser B., Ruck T., Thalman C., Luchtman D., Pape K., Schmaul S., Bitar L., Krämer-Albers E.-M., Stroh A., Meuth S. G., Zipp F., Bittner S., β1-Integrin- and KV1.3 channel-dependent signaling stimulates glutamate release from Th17 cells. J. Clin. Invest. 130, 715–732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F., Barsyte-Lovejoy D., Li F., Xiong Y., Korboukh V., Huang X. P., Allali-Hassani A., Janzen W. P., Roth B. L., Frye S. V., Arrowsmith C. H., Brown P. J., Vedadi M., Jin J., Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J. Med. Chem. 56, 8931–8942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergun O., Sobolevsky A. I., Yelshansky M. V., Keelan J., Khodorov B. I., Duchen M. R., Exploration of the role of reactive oxygen species in glutamate neurotoxicity in rat hippocampal neurones in culture. J. Physiol. 531, 147–163 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., Stockwell B. R., Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Bhavnani B. R., Glutamate-induced apoptosis in neuronal cells is mediated via caspase-dependent and independent mechanisms involving calpain and caspase-3 proteases as well as apoptosis inducing factor (AIF) and this process is inhibited by equine estrogens. BMC Neurosci. 7, 49 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X., Chua C. C., Kong J., Kostrzewa R. M., Kumaraguru U., Hamdy R. C., Chua B. H., Necrostatin-1 protects against glutamate-induced glutathione depletion and caspase-independent cell death in HT-22 cells. J. Neurochem. 103, 2004–2014 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Mosharov E., Cranford M. R., Banerjee R., The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry 39, 13005–13011 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Harvey C. J., Thimmulappa R. K., Singh A., Blake D. J., Ling G., Wakabayashi N., Fujii J., Myers A., Biswal S., Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 46, 443–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doll S., Freitas F. P., Shah R., Aldrovandi M., da Silva M. C., Ingold I., Goya Grocin A., Xavier da Silva T. N., Panzilius E., Scheel C. H., Mourao A., Buday K., Sato M., Wanninger J., Vignane T., Mohana V., Rehberg M., Flatley A., Schepers A., Kurz A., White D., Sauer M., Sattler M., Tate E. W., Schmitz W., Schulze A., O’Donnell V., Proneth B., Popowicz G. M., Pratt D. A., Angeli J. P. F., Conrad M., FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Hwang J. Y., Aromolaran K. A., Zukin R. S., The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthews P. M., Chronic inflammation in multiple sclerosis—Seeing what was always there. Nat. Rev. Neurol. 15, 582–593 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Klein R. S., Garber C., Funk K. E., Salimi H., Soung A. K., Kanmogne M., Manivasagam S., Agner S., Cain M., Neuroinflammation during RNA viral infections. Annu. Rev. Immunol. 37, 73–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nichols M. R., St-Pierre M. K., Wendeln A. C., Makoni N. J., Gouwens L. K., Garrad E. C., Sohrabi M., Neher J. J., Tremblay M. E., Combs C. K., Inflammatory mechanisms in neurodegeneration. J. Neurochem. 149, 562–581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sulzer D., Alcalay R. N., Garretti F., Cote L., Kanter E., Agin-Liebes J., Liong C., McMurtrey C., Hildebrand W. H., Mao X., Dawson V. L., Dawson T. M., Oseroff C., Pham J., Sidney J., Dillon M. B., Carpenter C., Weiskopf D., Phillips E., Mallal S., Peters B., Frazier A., Lindestam Arlehamn C. S., Sette A., T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 546, 656–661 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matta S. M., Hill-Yardin E. L., Crack P. J., The influence of neuroinflammation in autism spectrum disorder. Brain Behav. Immun. 79, 75–90 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Kaufmann M., Evans H., Schaupp A. L., Engler J. B., Kaur G., Willing A., Kursawe N., Schubert C., Attfield K. E., Fugger L., Friese M. A., Identifying CNS-colonizing T cells as potential therapeutic targets to prevent progression of multiple sclerosis. Med 2, 296–312.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheeler M. A., Clark I. C., Tjon E. C., Li Z., Zandee S. E. J., Couturier C. P., Watson B. R., Scalisi G., Alkwai S., Rothhammer V., Rotem A., Heyman J. A., Thaploo S., Sanmarco L. M., Ragoussis J., Weitz D. A., Petrecca K., Moffitt J. R., Becher B., Antel J. P., Prat A., Quintana F. J., MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He H., Hu Z., Xiao H., Zhou F., Yang B., The tale of histone modifications and its role in multiple sclerosis. Hum. Genomics 12, 31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alcala-Vida R., Seguin J., Lotz C., Molitor A. M., Irastorza-Azcarate I., Awada A., Karasu N., Bombardier A., Cosquer B., Skarmeta J. L. G., Cassel J. C., Boutillier A. L., Sexton T., Merienne K., Age-related and disease locus-specific mechanisms contribute to early remodelling of chromatin structure in Huntington’s disease mice. Nat. Commun. 12, 364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nativio R., Donahue G., Berson A., Lan Y., Amlie-Wolf A., Tuzer F., Toledo J. B., Gosai S. J., Gregory B. D., Torres C., Trojanowski J. Q., Wang L. S., Johnson F. B., Bonini N. M., Berger S. L., Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 21, 497–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsankova N., Renthal W., Kumar A., Nestler E. J., Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 8, 355–367 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.-C., Widom J., Brickner J. H., H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLOS Biol. 5, e81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z., Zhang R., Epigenetics in autoimmune diseases: Pathogenesis and prospects for therapy. Autoimmun. Rev. 14, 854–863 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Plummer R., Vidal L., Griffin M., Lesley M., de Bono J., Coulthard S., Sludden J., Siu L. L., Chen E. X., Oza A. M., Reid G. K., McLeod A. R., Besterman J. M., Lee C., Judson I., Calvert H., Boddy A. V., Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin. Cancer Res. 15, 3177–3183 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Segovia C., Jose-Eneriz E. S., Munera-Maravilla E., Martinez-Fernandez M., Garate L., Miranda E., Vilas-Zornoza A., Lodewijk I., Rubio C., Segrelles C., Valcarcel L. V., Rabal O., Casares N., Bernardini A., Suarez-Cabrera C., Lopez-Calderon F. F., Fortes P., Casado J. A., Duenas M., Villacampa F., Lasarte J. J., Guerrero-Ramos F., de Velasco G., Oyarzabal J., Castellano D., Agirre X., Prosper F., Paramio J. M., Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat. Med. 25, 1073–1081 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Insinga A., Monestiroli S., Ronzoni S., Gelmetti V., Marchesi F., Viale A., Altucci L., Nervi C., Minucci S., Pelicci P. G., Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat. Med. 11, 71–76 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Mohammad H. P., Barbash O., Creasy C. L., Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 25, 403–418 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Jan S., Dar M. I., Wani R., Sandey J., Mushtaq I., Lateef S., Syed S. H., Targeting EHMT2/ G9a for cancer therapy: Progress and perspective. Eur. J. Pharmacol. 893, 173827 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Conrad M., Kagan V. E., Bayir H., Pagnussat G. C., Head B., Traber M. G., Stockwell B. R., Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 32, 602–619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alim I., Caulfield J. T., Chen Y., Swarup V., Geschwind D. H., Ivanova E., Seravalli J., Ai Y., Sansing L. H., Ste Marie E. J., Hondal R. J., Mukherjee S., Cave J. W., Sagdullaev B. T., Karuppagounder S. S., Ratan R. R., Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell 177, 1262–1279.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Xie B. S., Wang Y. Q., Lin Y., Mao Q., Feng J. F., Gao G. Y., Jiang J. Y., Inhibition of ferroptosis attenuates tissue damage and improves long-term outcomes after traumatic brain injury in mice. CNS Neurosci. Ther. 25, 465–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang D., Chen X., Kang R., Kroemer G., Ferroptosis: Molecular mechanisms and health implications. Cell Res. 31, 107–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian R., Abarientos A., Hong J., Hashemi S. H., Yan R., Drager N., Leng K., Nalls M. A., Singleton A. B., Xu K., Faghri F., Kampmann M., Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci. 24, 1020–1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaefer A., Sampath S. C., Intrator A., Min A., Gertler T. S., Surmeier D. J., Tarakhovsky A., Greengard P., Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sampath S. C., Marazzi I., Yap K. L., Sampath S. C., Krutchinsky A. N., Mecklenbrauker I., Viale A., Rudensky E., Zhou M. M., Chait B. T., Tarakhovsky A., Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell 27, 596–608 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Tandon R., Brandl B., Baryshnikova N., Landshammer A., Steenpass L., Keminer O., Pless O., Muller F. J., Generation of two human isogenic iPSC lines from fetal dermal fibroblasts. Stem Cell Res. 33, 120–124 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Brennand K. J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D., McCarthy S., Sebat J., Gage F. H., Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Djuric U., Cheung A. Y. L., Zhang W., Mok R. S., Lai W., Piekna A., Hendry J. A., Ross P. J., Pasceri P., Kim D. S., Salter M. W., Ellis J., MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells. Neurobiol. Dis. 76, 37–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kreutzfeldt M., Bergthaler A., Fernandez M., Bruck W., Steinbach K., Vorm M., Coras R., Blumcke I., Bonilla W. V., Fleige A., Forman R., Muller W., Becher B., Misgeld T., Kerschensteiner M., Pinschewer D. D., Merkler D., Neuroprotective intervention by interferon-γ blockade prevents CD8+ T cell-mediated dendrite and synapse loss. J. Exp. Med. 210, 2087–2103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S4