Abstract
C4 and CAM photosynthesis have repeatedly evolved in plants over the past 30 million years. Because both repurpose the same set of enzymes but differ in their spatial and temporal deployment, they have long been considered as distinct and incompatible adaptations. Portulaca contains multiple C4 species that perform CAM when droughted. Spatially explicit analyses of gene expression reveal that C4 and CAM systems are completely integrated in Portulaca oleracea, with CAM and C4 carbon fixation occurring in the same cells and CAM-generated metabolites likely incorporated directly into the C4 cycle. Flux balance analysis corroborates the gene expression findings and predicts an integrated C4+CAM system under drought. This first spatially explicit description of a C4+CAM photosynthetic metabolism presents a potential new blueprint for crop improvement.
Portulaca oleracea has a rare metabolism derived from the integration of C4 and CAM photosynthesis.
INTRODUCTION
C4 and crassulacean acid metabolism (CAM) photosynthesis are two important adaptations that have evolved multiple times in terrestrial plants (1). Both act as carbon concentration mechanisms (CCMs) that alleviate energy losses caused by photorespiration, which can occur when atmospheric CO2 levels are low, internal leaf temperatures are high, or plant stomatal conductance is reduced, for instance, because of water stress (2). Both CCMs have repurposed a shared set of core metabolic enzymes that are present in all plants, but they differ in how they isolate and create a CO2-enriched environment around ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO), the enzyme that fixes atmospheric CO2 into sugars via the Calvin cycle. In C4 metabolism, phosphoenolpyruvate (PEP) carboxylase (PEPC) first interacts with dissolved CO2 in mesophyll cells to form a temporary four-carbon molecule, typically malate or aspartate. These molecules are then transported to inner bundle sheath cells where they are decarboxylated, elevating CO2 concentrations to levels that saturate RuBisCO. There, CO2 enters the Calvin cycle and is assimilated into carbohydrates (3). Thus, C4 is essentially a temporally synchronous two-cell photosynthetic system, with separate compartments for PEPC and RuBisCO, which results in C4 plants achieving the highest rates of photosynthesis (4). In CAM plants, stomata open and CO2 fixation by PEPC occurs during the night. The four-carbon malate is stored overnight in the form of malic acid in mesophyll cell vacuoles. During the day, stomata close, malate exits the vacuole for decarboxylation, and CO2 is released and assimilated by the Calvin cycle in the same cell (5). Thus, CAM is a temporally asynchronous, single-cell photosynthetic system, with initial carbon capture by PEPC occurring at night but eventual RuBisCO assimilation and carbohydrate production occurring during the day in the same cell. The CAM inverted stomatal behavior provides increased water use efficiency by avoiding water loss through stomata during the hottest daytime hours (6). There is also notable variability in the degree to which CAM is expressed: Many species use CAM as their primary metabolism but possibly more common is a facultative CAM system, where plants operate a C3 metabolism but exhibit facultative CAM as a stress response [“C3+CAM” sensu (1)]. Both CCMs exhibit remarkable evolutionary convergence. C4 has evolved at least 60 times, and C4 species include some of the most productive plants on Earth, comprising upward of 30% of gross terrestrial primary productivity and including several essential crops such as maize and sorghum (7). While the number of CAM origins is less well known, it is speculated to be higher than C4, and some form of CAM metabolism is dominant in a variety of ecosystems (8).
Despite the large number of independent origins of both C4 and CAM, for the most part, plant lineages tend to evolve one CCM or the other. Over 40 years ago, Portulaca oleracea was identified as the first known C4 plant that also operates a facultative CAM cycle (C4+CAM) in response to drought or changes in photoperiod (9). Full integration of C4 and CAM cycles, whereby both operate in the same population of photosynthetic mesophyll cells, seemed implausible on multiple fronts: (i) The shared set of enzymes would need to be expressed at different times of day and in different cells, creating substantial regulatory conflict. (ii) The activity of RuBisCO (for CAM) and PEPC (for C4) in the same mesophyll cells during the day would result in futile competition for CO2 and metabolic cycling. (iii) The anatomical requirements for optimizing each CCM are distinct and potentially antagonistic. (iv) Each CCM might evolve under distinct selection pressures, as the benefits to CAM are primarily to increase water use efficiency, whereas C4 plants take advantage of high light environments and seasonally available water to achieve fast growth and high productivity (10). Only a handful of immunolocalization studies have pursued the spatial resolution of C4+CAM in Portulaca (11, 12), and results were equivocal because of lack of C4-specific versus CAM-specific molecular markers. Regardless, the most accepted hypothesis has been that CAM and C4 cycles operate independently in Portulaca, with C4 in bundle sheath and mesophyll cells and CAM in specialized water storage cells. Only (11) suggested a possible integration: that malate generated from CAM could theoretically be processed in the C4 bundle sheath.
Recent transcriptomic information for Portulaca has provided an opportunity to resolve the spatial configuration of C4 and CAM reactions. Multiple studies revealed that PEPC is encoded by multiple gene copies in Portulaca and its relatives. This redundancy of PEPC-encoding genes allowed one copy to be specifically used for C4 (PPC-1E1a′) and another for CAM (PPC-1E1c). On the basis of gene expression estimates in P. oleracea, PPC-1E1a′ shows a diurnal peak in expression during C4 and C4+CAM states, while the expression of PPC-1E1c peaks during the day-night transition in droughted plants expressing C4+CAM (13–15). C4- and CAM-specific orthologs for these and other CCM-affiliated enzymes provide key markers of the cell populations where CAM and C4 CO2 fixation occur. The lack of significant up-regulation of distinct homologs of CAM decarboxylation genes in multiple Portulaca species is notable (15) and suggests that some elements of C4 and CAM biochemistry may be shared. However, these analyses were performed on whole-leaf transcriptomes and lacked the spatial resolution needed to identify where C4 and CAM processes were occurring. We performed two different spatially explicit gene expression analyses of P. oleracea leaves under well-watered and drought conditions that isolated mesophyll and bundle sheath cell populations. Moreover, we used constraint-based modeling to identify the most efficient biochemical model of C4 and CAM integration across mesophyll and bundle sheath cells under a variety of scenarios. These analyses of P. oleracea reveal a previously unknown type of plant photosynthesis: an integrated C4+CAM metabolism.
RESULTS
Assessment of CAM induction
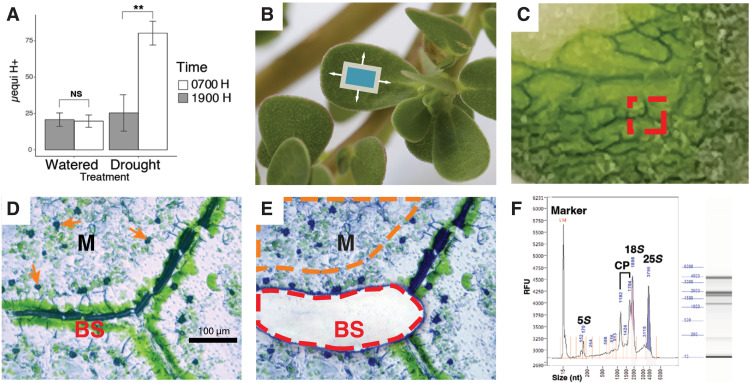
Titratable acidity analysis of well-watered P. oleracea leaves at 7 and 19 hours did not result in significant accumulation of acids overnight (two-sample t test, P = 0.62, t = −0.33, df = 5.95; Fig. 1A and table S1). This confirmed little or no accumulation of malic acid from CAM activity under well-watered conditions. After 7 days of total water withholding, significant accumulation of acids was detected in leaves collected early in the morning compared to those collected at the end of the photoperiod (t test, P = 0.00032, t = 7.34, df = 5.17; Fig. 1A and table S1), confirming CAM induction.
Fig. 1. Drought induction of CAM and laser capture microdissection.
(A) Diel fluctuation of titratable acidity from whole leaves of well-watered plants and after a 7-day drought treatment. NS, not significant. (B) P. oleracea, illustrating the orientation of paradermal sections (blue box). (C) Fresh, flash-frozen paradermal leaf section in cryosection block. The red square indicates an area used for tissue dissection. (D) Microphotograph of a 12-μm-thick leaf paradermal section indicating bundle sheath (BS) and mesophyll (M) tissues before (D) and after (E) BS cell capture. Orange arrows in (D) indicate calcium oxalate crystals in M cells. The red line in (E) indicates a laser-cut area of BS tissue, and the orange line illustrates an area of M tissue for laser capture. (F) RNA profile by electrophoresis for quality control. CP, chloroplast RNAs; RFU, relative fluorescence units.
Laser microdissection–RNA sequencing and read alignments
To obtain cell-specific gene expression, we made paradermal sections from well-watered and droughted P. oleracea leaves (Fig. 1, A and B, and table S1) and captured groups of mesophyll and bundle sheath cells using laser microdissection (LCM) (Fig. 1, C to E). We isolated mRNA from populations of each tissue for short read sequencing. We sequenced 28 libraries that represented 1.05 billion 100–base pair (bp) paired-end reads with a mean of 37.49 million (SD = 3.85 million) reads per library (Fig. 2 and table S2; see Materials and Methods). After quality filtering and trimming, a mean of 68.97% of the reads was kept. We used the P. oleracea de novo assembled transcriptome from (15) as the reference for read mapping and abundance quantification; this transcriptome consisted of 444,307 contigs and 230,895 Trinity “unigenes.” Here, we used the software TransDecoder v5.5.0 (http://transdecoder.github.io) to predict coding sequences in 152,530 of the contigs and CD-HIT v4.8.1 (16) to reduce redundant contigs to a final set of 54,241 contigs of 37,518 unigenes (online table S1). The reduced dataset of mRNA contigs represented 83.20 Mbp with a 2101-bp N50. The mean reads mapped to the reduced set of P. oleracea mRNA contigs by Kallisto v-0.45.0 (17) were 75.30% per library (SD = 3.65%), and a mean of 59.96% (SD = 3.48%) of mapped reads had unique alignments (table S2). Reads mapping to CCM-related genes constituted 10.97% (SD = 2.49%) of daytime mesophyll cell libraries and 12.18% (SD = 2.94%) of the daytime bundle sheath cell libraries. We considered CCM-related genes to include all members of gene families with known roles in C3, C4, and CAM photosynthesis, including those involved with starch metabolism, light sensing and circadian rhythm, and photorespiration. On average, 5.20% (SD = 1.83%) of reads from nocturnal libraries mapped to CCM-related genes, while 11.62% (SD = 2.70%) of reads mapped to CCM-related genes in daytime libraries.
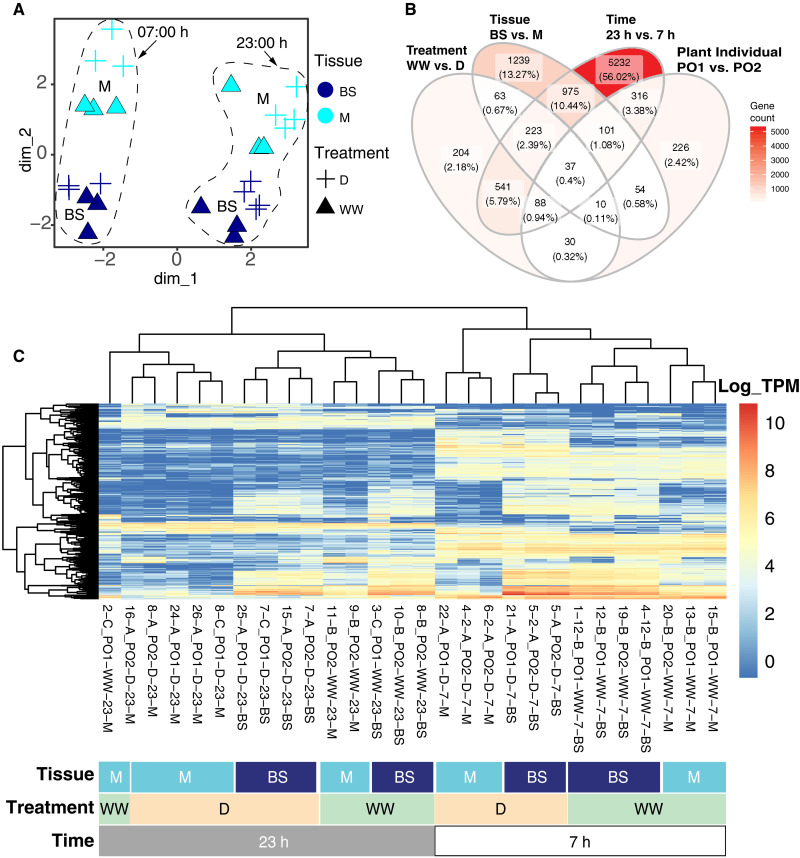
Fig. 2. Transcriptome-wide gene expression patterns across M and BS samples.
(A) All laser microdissection (LMD) samples projected into the first two dimensions of a transcriptome-wide multidimensional scaling analysis of log-transformed expression. Treatments: D, drought; WW, well-watered. (B) Venn diagram with number of DE genes across variables. (C) Heatmap with the log of gene expression of the 500 most variable genes across samples. Dendrograms cluster mRNA libraries (on the top) and genes (on the left) on the basis of gene expression similarities.
Global transcriptional differences across experimental variables
Multidimensional scaling of transcriptome-wide gene expression clustered time points and tissue replicates (n ≥ 3) along dimensions 1 and 2, respectively (Fig. 2A). Multidimensional scaling further separated samples by water status within each time and tissue cluster. Of the 22,509 unigenes that passed the low read mapping filter, 2702 (12.00%) were differentially expressed (DE) between mesophyll and bundle sheath (Padj < 0.05; Fig. 2B and online table S2). Of those, 1030 (38.12%) were more highly expressed in mesophyll and 1672 (61.88%) in bundle sheath. A total of 1196 unigenes (5.31%) were significantly DE across watering treatments; of those, 745 (62.29%) were more abundant in well-watered plants, and 451 (37.71%) were more abundant during drought. We found 7513 DE unigenes (33.37%) across time points, with 3553 (47.29%) and 3960 (52.71%) more abundant during the day and night, respectively.
Using all samples, gene DE analyses revealed that most detected gene expression shifts occurred between day and night, followed by cell type and water status (Fig. 2B). Gene ontology (GO) analyses of DE genes showed mesophyll enrichment of nonphotosynthetic carbon-related GO terms (carbon utilization GO:0015976) while Calvin cycle (GO:0019253) and photorespiratory terms (GO:0019264 and GO:0009853) were enriched in bundle sheath cells (fig. S1A). Genes up-regulated under drought were enriched in stress response GO terms as response to salt and abscisic acid (GO:0009651 and GO:0009737), while genes more highly expressed when well-watered were enriched in photosynthesis and carbon utilization–related terms (GO:0009768 and GO:0015976; fig. S1B).
Transcriptional changes in CCM-related genes across cell types
We annotated 629 trinity unigenes into CCM-related gene families with roles including C3, C4, and CAM photosynthesis; photorespiration; starch metabolism; circadian rhythm; and associated transcription factors (TFs) as identified in other studies of P. oleracea (online tables S3 and S4) (13–15). Most core CCM (C4 and CAM) and starch metabolism unigenes were annotated to known eudicot and Portulaca gene lineages using reference sequences as in (15, 16, 18–20) (see Materials and Methods). When available, gene lineage nomenclature is indicated after gene identifiers followed by a dash. A total of 480 unigenes were kept after filtering lowly abundant unigenes; of those, 170 (35.42%) were DE across cell types, with 83 (48.82%) more abundant in bundle sheath and 87 (51.18%) more abundant in mesophyll. We found that 307 (63.05%) unigenes included in our curated set of gene families were DE over time, with 230 (74.92%) more abundant at 7 hours and 77 (25.08%) more abundant at 23 hours (online table S3). Last, 75 (14.79%) unigenes were DE across water regimen, with 61 (81.33%) more abundant in well-watered plants and 10 (13.33%) more abundant in droughted plants.
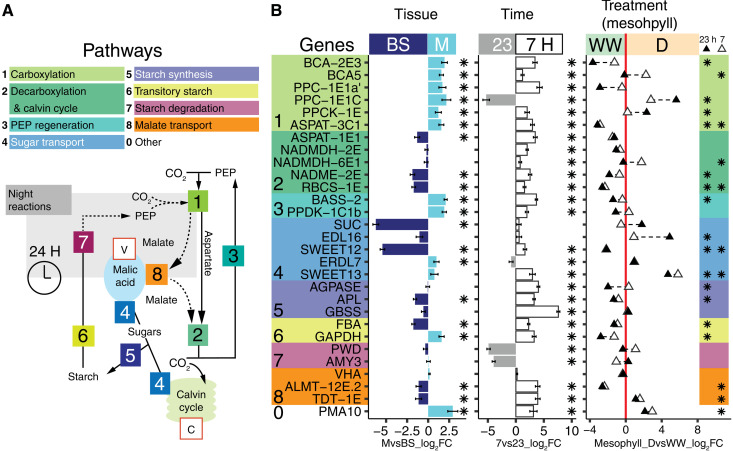
Portulaca C4/CAM-specific orthologs were previously identified by high transcript abundances in comparison to non-CCM homolog genes and by matching to expected transcription patterns across time points and watering treatments using whole leaves (13–15). Here, DE analyses of C4-related genes were consistent with expectations of C4 mesophyll versus bundle sheath expression. Typically, in the C4 carboxylation subpathway, β carbonic anhydrases (BCAs) dissolve CO2 in the form of bicarbonate (HCO3), which is added to PEP by PEPC and affiliated enzymes to form oxalacetate (OAA) in mesophyll cells. OAA is then converted to malate by malate dehydrogenases (MDHs) or to aspartate by aspartate amino transferases (ASPATs). These molecules enter the decarboxylation pathway, where they are decarboxylated by malic enzymes [NAD(P)ME], and the released CO2 is lastly assimilated by the Calvin cycle. Our analyses show that, in well-watered and droughted plants, the initial C4 carbon fixation module that included a carbonic anhydrase (BCA-2E3), PPC-1E1a′, and an aspartate aminotransferase (ASPAT-3C1) occurred in mesophyll cells (mesophyll versus bundle sheath; MvsB log2 fold change (log2FC) > 1.5; Padj < 0.01; module 1; Fig. 3, figs. S2 and S3, and table S3). We identified a second bundle sheath–specific [MvsB log2FC = −1.31; Padj < 0.01] ASPAT homolog (ASPAT-1E1) as a candidate gene representing the entry point for C4 metabolites into the CO2 decarboxylation/assimilation module in the bundle sheath that includes malic enzyme (NADME-2E) and the Calvin cycle (MvsB log2FC > −1.4; Padj < 0.01; module 2; Fig. 3). We uncovered two malic enzymes (MEs) with C4-like expression, e.g., biased toward the morning and restricted to the bundle sheath cells. In addition to the primary P. oleracea ME used for C4-acid decarboxylation, NAD-ME (19), we found a chloroplastic ME (NADPME-1E1b) that may suggest an accessory decarboxylation route (Fig. 4 and fig. S4). However, previous Western blot analyses showed no detectable labeling for NADP-ME when P. oleracea is well-watered (19, 20), suggesting that NADP-ME activity may be posttranscriptionally regulated. In the PEP regeneration pathway, the pyruvate resulting from decarboxylation is used to regenerate PEP via alanine aminotransferase, pyruvate, phosphate dikinase, and affiliated enzymes. In our analyses, the expression of genes involved in the regeneration of PEP was restricted to the mesophyll (MvsB log2FC > 1.4; Padj < 0.01; module 3; Fig. 3 and fig. S2).
Fig. 3. Differential gene expression of M and BS across experimental conditions.
(A) Schematic of C4, CAM, and accessory biochemical pathways. Solid and dotted lines indicate C4 and CAM routes of carbon concentration, respectively; the gray box contains night reactions, and key substrates are shown (PEP). Intracellular compartments are indicated within red boxes (V, vacuole; C, chloroplast). (B) Differential transcript abundance (measured in log2FC) of selected genes in M relative to BS tissue (left) and at 7 hours relative to 23 hours (middle ) across LMD samples. Gene color backgrounds reflect pathways in (A). In the third panel, white triangles indicate log2FC of 7-hour M samples in D relative to 7-hour M samples WW. Black triangles indicate log2FC of 23-hour M samples in drought compared to 23-hour M samples WW. A triangle in the WW region (negative log2FC, left to the red line) indicates higher expression during WW, while triangles within the D region (right to the red line) indicate higher expression in D. In all panels, asterisks indicate DE significance (Padj < 0.05).
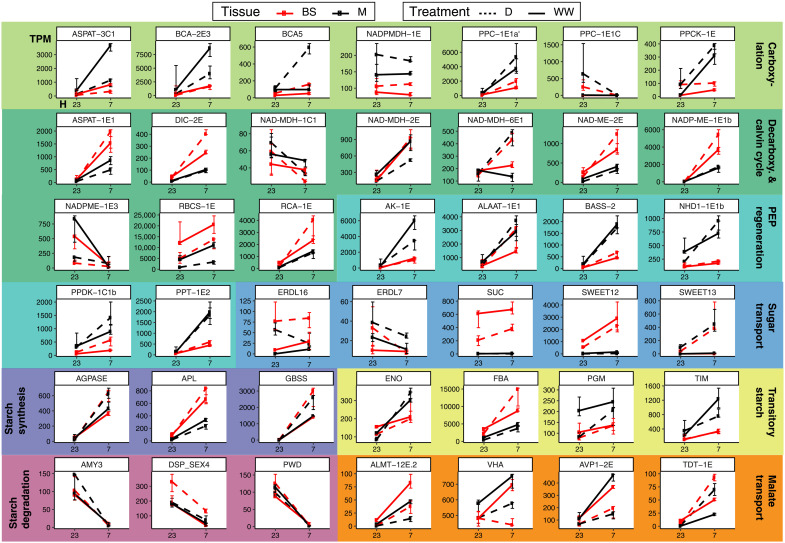
Fig. 4. Transcription abundance of selected CCM-related genes.
Median of transcripts per million (TPM; y axis) across time points (7 and 23 hours, x axis) in LMD mRNA libraries. Black and red lines indicate M or BS samples, respectively. Plain lines indicate watered, and dotted lines indicate drought. Error bars represent the interquartile range of expression.
The ortholog identity and tissue localization of core C4 cycle genes generally remained the same in well-watered and drought-induced C4+CAM P. oleracea. Typically, in CAM, the carboxylation module composed of BCA, PEPC, and MDH operates at the end of the light period in mesophyll cells to store CO2 in the form of malate during the night. In P. oleracea, when CAM was induced, different homologs of carbonic-anhydrase (BCA-2), PEPC (PPC-1E1c), and MDH (NADMDH-6E1) markedly increased their mesophyll abundance (mesophyll, drought versus well-watered; M_DvsWW log2FC > 1.7; Padj < 0.01; Figs. 3 and 4). In the mesophyll, the CAM-specific paralog PPC-1E1c exhibited a 5.6 log2FC at night under drought, compared to daytime samples, which had near-zero abundance (Padj < 0.01; Figs. 3 to 4). In parallel, transcripts of the PEPC-activating protein, PEPC kinase (PPCK-1E), increased in abundance at night in drought samples (M_DvsWW log2FC > 2.3; Padj < 0.01; Figs. 3 and 4). PEPC is inhibited by malate (21); thus, increased PPCK activity might be a requirement to maintain PEPC activity, while malate is highly abundant during the dark period of CAM.
Malate produced at night is purportedly actively pumped into the vacuole by an aluminum-activated malate transporter (ALMT) (22), which is driven by cation currents created by a vacuolar-type proton adenosine triphosphatase (ATPase; VHA) (23), although neither showed genes increasing in expression during drought in any cell type (module 8 and fig. S3; see table S3 and online tables S2 and S3 for GO information). However, we found a sharp up-regulation of the gene encoding the plasma membrane proton pump ATPase 10 (PMA10) in drought samples (M_DvsWW log2FC = 3; Padj < 0.05; module 0; Fig. 3) restricted to the mesophyll (MvsBS log2FC = 2.9; Padj < 0.01; Fig. 3), but a role in CAM is only speculative. The mechanisms responsible for vacuolar malate efflux in CAM remain unknown (24), but the most plausible is the tonoplast dicarboxylate transporter (TDT-1E), whose expression increased during drought, particularly in mesophyll (M_DvsWW log2FC = 1.6; Padj < 0.01; module 8; Fig. 3).
Unlike C4 plants, CAM plants generally produce PEP via degradation of starch and sugars (25); however, our samples only showed significant up-regulation of amyloplastic phosphoglucan phosphatase DSP4 (DSP_SEX4) among starch and sugar degradation genes during CAM induction (modules 6 and 7; Figs. 3 and 4). Temporally, starch degradation genes were more abundant at night (module 7; Fig. 3 and fig. S2). Spatially, most of the transitory starch-related genes (module 6; Fig. 3 and fig. S2) were more abundant in mesophyll during the day, with the exception of chloroplastic fructose-bisphosphate aldolase (FBA gene family, ALFP lineage; orthology in table S3), which was restricted to bundle sheath (MvsBS log2FC = 1.8; Padj < 0.01; module 6; Fig. 3). This aldolase degrades fructose 1,6-bisphosphate into glyceraldehyde 3-phosphate (G3P) before G3P is further degraded to PEP in a series of glycolytic reactions whose genes were mainly expressed in the mesophyll during the day (module 6; Fig. 3 and fig. S2). Tonoplast and chloroplast sugar transporters showed the highest expression specificity to bundle sheath (module 5; Fig. 3 and fig. S2), suggesting a role in the Calvin cycle of C4 and C4+CAM Portulaca. Other sugar transporters, such as ERD6-like (EDL16) and the mesophyll-restricted SWEET13, also increased in abundance with CAM induction (M_DvsWW log2FC > 5,8; Padj < 0.01; Fig. 3 and fig. S3). Our data suggest an increase in sugar transport linked to CAM induction, with G3P originating in the bundle sheath but the final steps of PEP regeneration mainly occurring in mesophyll.
We also found up-regulated genes related to cell wall architecture at night in CAM-induced plants: EXPA2 and SAU32 in the mesophyll and NAP2 in bundle sheath (table S4 and fig. S4). In relation to other transporters, mesophyll-specific up-regulation of the ion transporters probable magnesium transporter (NIPA7) and plasma membrane–type ATPase 10 (PMA10) and bundle sheath–specific mitochondrial uncoupling protein 4 (PUMP4) (table S4, fig. S5, and online table S2). A gene involved in vacuole processes, vacuolar-processing enzyme β-isozyme 1 (VPE1), is also up-regulated at night during CAM induction in bundle sheath. We detected expression shifts in transcription factors (TFs) including increased diurnal expression of the transcription repressor MYB4 in mesophyll and phenylacetaldehyde reductase PAR1 in bundle sheath and decreased diurnal abundance of TF BH062 in both cell types. During the night, we observed up-regulation in both cell types of the TFs scarecrow-like protein 15 (SCL15), protein nuclear fusion defective 4 (NFD4), homeobox-leucine zipper protein ATHB7, probable WRKY TF7 (WRKY7), and protein light-dependent short hypocotyls 3 (LSH3) in the mesophyll specifically (table S4 and fig. S5).
Visium spatial gene expression
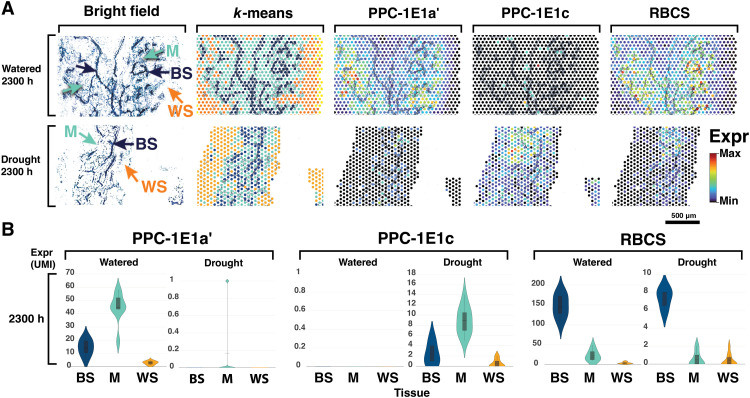
To confirm laser microdissection (LMD) results, we captured and spatially tagged mRNA directly on leaf paradermal sections using the 10x Genomics Visium spatial transcriptomics platform (10x Genomics, Pleasanton, CA) across watering treatments (table S5). K-means clustering of total gene expression grouped areas corresponding to mesophyll, bundle sheath, and water storage tissues (Fig. 5A). Genes with transcription estimates of less than 200 transcripts per million (TPM) in the LMD analysis were poorly represented in the Visium libraries. Transcript abundance mapped onto leaf-section microphotographs revealed that CCM and Calvin cycle genes were lowly abundant or undetected in water storage tissue, including both C4- and CAM-specific PEPC paralogs (Fig. 5 and fig. S6). Our Visium analyses confirmed the near absence of CAM PPC-1E1c expression in well-watered samples and the day-night alternation of abundance of C4 and CAM PEPC paralogs in drought samples. Both C4 and CAM PEPC paralogs were mostly restricted to the mesophyll, with decarboxylation genes and RuBisCO confined to the bundle sheath (Fig. 5 and fig. S6).
Fig. 5. Visium spatial gene expression.
(A) Microphotograph of a leaf paradermal section; K-means clustering of total gene expression; and abundance of the main C4, CAM, and Calvin cycle carboxylases using the 10x Genomics Visium platform. K-means clustering of sampling spots corresponds to BS (dark blue), M (light blue), and water storage (WS; orange) tissues; abundances of PPC-1E1a′, PPC-1E1c, and RBCS are shown relative to their observed unique molecule index (UMI) ranges. (B) Violin plots of transcript abundance in UMI across sample spots classified by tissue type in 2300-hour samples.
Flux balance model
To further explore how C4 and CAM might be integrated, we built a two-cell two-phase model adapted from a highly curated plant core metabolic model (26). The metabolic model includes all major metabolic enzymes and reactions found to be highly conserved across a wide array of plant genomes and represents a “core” stoichiometric model that captures the central carbon metabolism in leaves. Our new model represents how C4 and CAM could be integrated in a parsimonious manner from first principles of general plant metabolism. We used parsimonious flux balance analysis (FBA) to model the efficiency of possible C4 and CAM configurations. In our models, metabolic fluxes were constrained by the rates at which metabolites were consumed or produced by each reaction. All reactions were permitted to occur in both cells and at any time, and we considered diel fluxes within mesophyll and bundle sheath cells as distinct compartments. The predicted fluxes for major reactions in the metabolic models were largely consistent with the differential gene expression results (Fig. 6 and fig. S7), and most inconsistencies fell within the predicted variance of flux variability analysis (FVA; fig. S8).
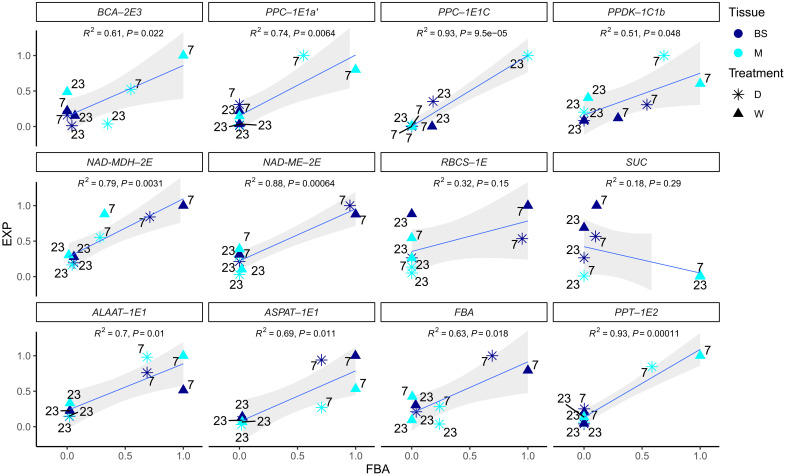
Fig. 6. Correlations between predicted enzymatic fluxes and estimated gene expression in M and BS samples.
Pearson correlation for individual enzymes between predicted reaction fluxes in the most efficient C4+CAM flux balance model (FBA) and the expression (EXP) of their encoding genes as the mean of transcript abundance in the LMD samples (TPM). Each data point corresponds to an experimental group of samples composed of D or wet conditions (W), M or BS tissue, and daytime (7 h) or nighttime (23 h). Time labels are shown next to each point. Expression and flux balance results are normalized by maximal values for each enzyme/gene. Regression lines are shown in blue, and SEs are shown in gray.
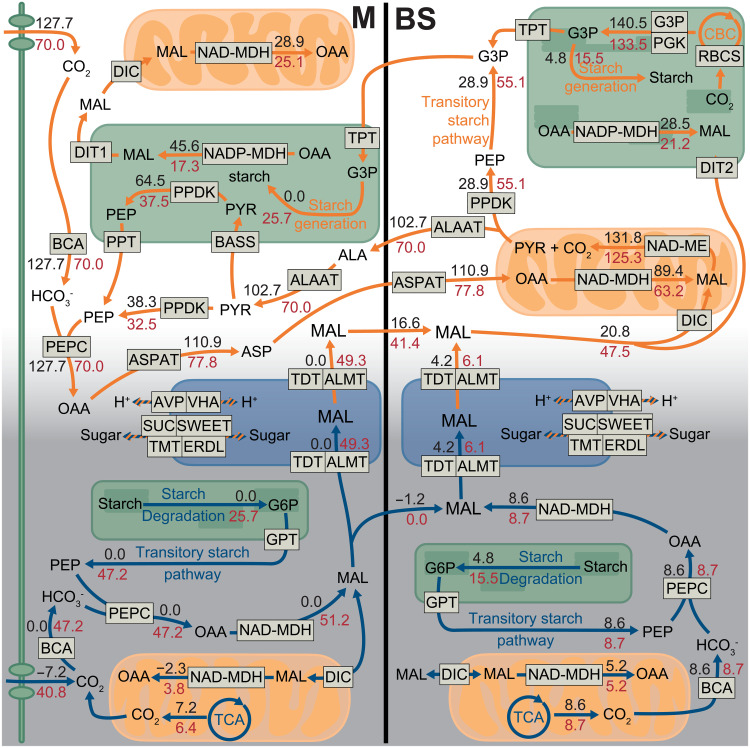
Under well-watered conditions, the flux balance model predicted a C4 photosynthesis pathway with a small amount of CAM cycling in the bundle sheath; under drought conditions, the model predicted the emergence of a C4+CAM pathway (Fig. 7 and table S6). While the model predicted a canonical C4 cycle, the predicted CAM cycle was a two-cell cycle spanning mesophyll and bundle sheath using both atmospheric and respired CO2 and a regular one-cell cycle in bundle sheath using respired CO2. For the two-cell CAM cycle, CO2 was assimilated by PEPC, converted to malate, and stored in the mesophyll vacuole. During the day, malate was transferred to the bundle sheath and decarboxylated to provide CO2 to the Calvin cycle. All the essential C4- and CAM-related reactions (PEP carboxylation, carbonic anhydrase activity, RuBisCO carboxylation, CO2 uptake, NAD-ME activity, and PEP dikinase regulation) had little variation in the FVA, indicating that they were robust processes (fig. S8). Most of the processes related to metabolite turnover (i.e., malate, oxaloacetate, alanine, aspartate, and pyruvate) were quite flexible and exhibited significant variation in FVA without affecting the efficiency of the system. (fig. S7). The predicted leaf area–based photosynthetic rates under well-watered conditions can be converted to 23.2 μmol m−2 s−1 (see Materials and Methods), which is comparable with the gas exchange measurements for P. oleracea (19).
Fig. 7. Biochemical flux results in the FBA model of an integrated C4+CAM photosynthesis.
Schematic of major metabolic fluxes related to C4 and CAM in the M and BS; orange and blue arrows indicate daytime and nighttime reactions, respectively. Black numbers indicate fluxes under WW conditions (no specific CO2 uptake constraint; stomata open responding to WW condition); red numbers indicate fluxes under drought conditions (daytime CO2 uptake constraint as 45% stomata closure compared to WW condition). Minor fluxes are not shown for simplicity, and therefore, not all shown input and output fluxes are equal. The unit for the flux values is mmol gDW−1 day−1.
We also investigated whether blocking intercellular malate transfer and malate storage would affect the efficiency of the C4+CAM system. Blocking malate transfer did not affect the efficiency of the system under either well-watered or drought conditions (table S7) because malate, aspartate, and alanine transfer between mesophyll and bundle sheath were interchangeable; therefore, blocking one metabolite induced conversion to an alternative metabolite for intercellular movement. Furthermore, modeling results indicated that malate transport between mesophyll and bundle sheath might have the smallest enzymatic and metabolic cost and was more efficient than other C4 acids in the C4+CAM system. Blocking malate storage in mesophyll or bundle sheath alone did not affect the efficiency of the system; however, blocking malate storage in both mesophyll and bundle sheath greatly reduced efficiency. Malate storage capability is thus important for C4+CAM metabolism, but it is not necessarily important whether the malate is stored in mesophyll, bundle sheath, or both.
In summary, under well-watered conditions, FBA predicted a C4 pathway with only a very small amount of CAM in the bundle sheath, but under drought, FBA predicted a C4+CAM system, consistent with our gene expression results (Fig. 7 and figs. S7 and S8). Under drought, integrating malate from CAM into the C4 cycle maximized phloem photosynthate output with minimal enzymatic investment compared to models where C4 and CAM cycles ran independently of one another. In the integrated model, most nocturnal CO2 fixation occurred in the mesophyll, and malate was stored in vacuoles of both cells. During the day, fluxes of malate from mesophyll vacuoles moved to the bundle sheath to enter the C4 decarboxylation module. Under drought (45% stomatal closure compared with well-watered conditions), phloem outputs of C4+CAM plants were only reduced by 8% (table S6). However, without CAM, phloem output was reduced by 52% (table S7).
DISCUSSION
P. oleracea is a rare C4 species capable of expressing CAM photosynthesis as a stress response to water deficit. This metabolic flexibility allows P. oleracea not only to achieve high rates of photosynthesis when resources are plenty but also to tolerate extreme drought for a relatively long period, breaking a fundamental productivity-stress tolerance trade-off in plant physiology (27). Previous studies identified candidate enzymes linked to C4 and CAM, but how the two pathways were spatially organized within Portulaca leaves remained unknown. We compared well-watered and drought-induced P. oleracea plants using recent advances in spatial gene expression analysis and metabolic modeling to describe a previously unknown biochemical pathway that integrates C4 and CAM cycles into a single photosynthetic metabolism.
Spatial gene expression analyses confirmed CAM and C4 operating in the same cells
The spatial description of gene expression across different tissues via in situ hybridization is necessarily limited to a handful of genes. Here, we estimated the expression of tens of thousands of genes in mesophyll and bundle sheath cells using laser capture microdissection. Furthermore, using the Visium (10x Genomics) spatial gene expression workflow, we mapped and visualized the expression of C4- and CAM-related genes over paradermal anatomical sections of leaves. Spatially explicit gene expression analysis at the cellular scale is still quite rare in plants (28) and holds promise for addressing a diverse set of problems in plant molecular biology and function. On the basis of K-means clustering of total gene expression, our Visium spatial gene expression analysis predicted mesophyll, bundle sheath, and water storage cells across samples. Gene expression maps overlapping microphotographs of leaf sections were congruent with findings in the laser captured cell samples: Genes related to C4 and CAM PEPC activity were more abundant in mesophyll, while decarboxylation and Calvin cycle genes were largely restricted to bundle sheath. Metabolic activity in water storage cells appeared very low, and almost no transcripts were captured for any CCM-related genes. Future work using spatial proteomics and metabolomic profiling would confirm that protein location matches transcription location.
A new photosynthetic pathway: CAM metabolic fluxes are connected to the C4 pathway for decarboxylation
We built a two-cell two-phase diel plant metabolic model that integrates C4 and CAM on the basis of first principles of general plant metabolism. We adapted the highly curated model of (26) and used flux balance analysis (FBA) to test for the most efficient biochemical pathways in which C4 and CAM could operate and correlated the results with observed transcript abundance. Congruent with empirical observations of Portulaca, our model predicted that a purely C4 system was more efficient when well-watered but that C4+CAM was more efficient under drought. Our FBA models further predicted cellular compartmentalization of the major C4 and Calvin Cycle reactions in agreement with our expression data, although FBA slightly underpredicted the transcript abundances of RuBisCO, NAD-MDH, and NAD-ME in mesophyll. These inconsistencies could be explained by an incomplete segregation of enzymes; the imperfect nature of laser microdissection for mesophyll and bundle sheath isolation; or more complex relationships among mRNA, active enzymes, and metabolites. Under drought conditions, the integration and processing of malate from CAM into the C4 cycle maximizes phloem output of photosynthates with the smallest enzyme cost compared to models without integration of cycles. This model predicts that most nocturnal CO2 fixation occurs in mesophyll, with a small fraction of respired CO2 fixed in the bundle sheath, and that malate is stored in vacuoles of both cells. During the day, fluxes of malate from mesophyll reach bundle sheath to enter the C4 decarboxylation module. Together, we present a two-cell CAM system operating in P. oleracea, where CO2 is fixed by both C4 and CAM machinery in the same mesophyll cells but at distinct times, with CAM fluxes of malate entering the C4 cycle during the day in bundle sheath cells to be decarboxylated and assimilated into sugars by RuBisCO and the Calvin cycle.
The evolutionary assembly of a C4+CAM metabolism in Portulaca
Our analyses confirm that P. oleracea has an integrated C4+CAM photosynthesis, where initial C4 and CAM carbon fixation occurs in the same mesophyll cells over a 24-hour period, with decarboxylation and final CO2 assimilation restricted to the bundle sheath. Only a subset of core enzymes recruited distinct homologs for C4 versus CAM expression. Thus, the regulatory constraints that were proposed to prevent this coordination appear easily surmountable; not all genes require a temporal or spatial shift in gene expression, and those that do simply use alternative homologs. So, why is the co-occurrence of C4 and CAM apparently rare, having only been identified in Portulaca, and quite recently, Trianthema (29)? It is possible that many C4+CAM species exist and have not been identified, as most known C4 species have not been investigated for CAM activity. At the same time, it is helpful to look at what Portulaca and Trianthema have in common. Both are mildly succulent plants that are unusual C4 members of clades with predominantly C3+CAM or CAM metabolism. Portulaca was most likely an ancestrally C3+CAM species that evolved C4 in parallel several times (13, 15), and we predict that Trianthema will be shown to have followed a similar evolutionary trajectory. Portulaca achieved the high bundle sheath:mesophyll ratio needed for efficient C4 photosynthesis by evolving a three-dimensional leaf venation system while still maintaining some degree of tissue succulence and leaf thickness (30), thus traversing a potential antagonism of C4 and CAM co-occurrence. We predict that we are more likely to find additional C4+CAM species in mildly succulent C4 clades [e.g., (31)] or C4 clades closely related to CAM-evolving lineages [e.g., (32)].
C4+CAM metabolism and global food security
Facultative CAM cycles in C3 plants help maintain a positive carbon balance during stress (typically drought) (33). Our FBA modeling results were consistent with this expectation, demonstrating how CAM maintained productivity in a C4 plant under drought conditions. P. oleracea is a fast-growing annual weed that achieves high rates of carbon assimilation similar to other highly productive C4 plants (14, 19). In combination with C4, a facultative CAM cycle provides elevated drought tolerance in a plant that can also achieve exceptionally high rates of photosynthesis, essentially circumventing the fundamental productivity-tolerance trade-off that constrains plant function (27). Future work should generate more robust quantifications of the levels of contribution to net carbon gain and survival of facultative CAM in Portulaca experimentally via gene editing. Portulaca is highly amenable to development as a model system for photosynthesis research (34): It has a short life cycle and is self-compatible, and a high-quality genome of Portulaca amilis is already available (15).
An integrated C4+CAM photosynthesis inspires future avenues for crop improvement and food security, as there has been an ongoing global initiative to engineer C4 or CAM pathways into C3 crops (35, 36). Predictions of increasing evapotranspiration with warming climate point to the urgency of developing drought tolerant crops (37), but in maize, a C4 plant, decades of selection for more robust growth and higher yields resulted in significantly higher drought sensitivity (38). Because C4 crops such as maize already have the ability to process C4-originated malate for sugar production during the day, engineering a facultative CAM biochemistry where CAM-produced malate enters the C4 cycle may require fewer changes than a full C3-to-CAM or C3-to-C4 transition would require. One caveat is our uncertainty with regard to the anatomical requirements of a C4+CAM system; we note that the CAM cycle was not detectable in the large water storage cells of Portulaca but in, rather, the considerably smaller mesophyll cells. This suggests that this level of facultative CAM may not require extensive anatomical specialization, as has been hypothesized in recent models of CAM evolution (1). We speculate that engineering a C4+CAM metabolism in a C4 crop is feasible and could confer substantial drought tolerance without sacrificing productivity and yield.
MATERIALS AND METHODS
Methods I: Spatial gene expression
Plant material, drought experiment, and sampling
Two P. oleracea plants were grown at the greenhouse facilities at the Marsh Botanical Garden, Yale University, New Haven, CT, USA. Plants were grown from (voucher accession PO-russ2018) in a greenhouse in soil mix FAFARD GROWING MIX #2 3.8CF, composed of 70% peat moss, 20% perlite, 10% vermiculite, starter nutrients, limestone, and a wetting agent (Sun Gro Horticulture, MA, USA). During the experiment, temperature was maintained at an average of 27° and 22°C during the 15 hours of light and 8 hours of dark a day, respectively. Natural light intensity at plant level peaked around 1600 mol m−2 s−1 (photosynthetically active radiation) at the center of the light period. The light period began at ~5 hours (ending ~20 hours), and leaf samples were taken at 7, 19, and 23 hours under well-watered conditions and again after 7 days of complete water withholding (drought). Leaves used for titratable acidity assays were placed in 1-ml Eppendorf tubes and flash-frozen in liquid nitrogen; for cryosectioning, another three to four whole leaves were immersed in optimal cutting temperature (OCT) compound in plastic histology molds and rapidly placed on an aluminum block partially submerged in liquid nitrogen to flash freeze. All samples were stored at −80°C.
To assess nocturnal accumulation of malate, leaves from 7- and 19-hour samples from both well-watered and drought plants were boiled in 60 ml of 20% ethanol. After half of the volume evaporated, distilled water was added to return the initial 60 ml; this process was repeated twice. The final 30-ml solutions were cooled to room temperature and titrated to a pH of 7.0 using 0.002 M NaOH. Acidity, measured as μEq H+ per gram of fresh mass, was calculated as volume titrant (in μl) × titrant molarity (in M)/fresh tissue mass (in g).
Cell-specific laser microdissection, RNA isolation, and sequencing
LMD relies on cell identification for cell-specific capture. Here, we targeted mesophyll cells and bundle sheath cells for RNA sequencing. We chose to use fresh-frozen tissue, as opposed to fixed tissue and/or embedded in paraffin, to avoid potential RNA degradation. Although freezing can distort the leaf anatomy, bundle sheath cells are easily identified by their dense chloroplasts and proximity to veins, and mesophyll cells are easily identified by sparse chloroplasts and calcium oxalate crystals (Fig. 1D).
Frozen OCT molds containing fresh leaves were moved from −80° to −20°C inside a cryomicrotome (CM 3050S; Leica Biosystems, MA, US) to acclimate for 30 min. We obtained paradermal sections by cutting leaves parallel to the longitudinal axis with the cryomicrotome. Paradermal sectioning results in larger areas of bundle sheath and mesophyll tissues compared to cross-sectioning. Sections 12 μm across were laid on PEN membrane glass slides for ultraviolet laser microdissection (Thermo Fisher Scientific, MA, USA) and stored at −80°C. At the time of laser microdissection, slides were brought to room temperature inside a desiccant box containing silica gel for ~10 min. Room temperature dry slides were laser cut using an LMD scope (LMD7000; Leica, Leica Biosystems, MA, USA) with the following laser parameters: power, 50; aperture, 9; speed, 4; and ×20 microscope magnification. This relatively wide cutting laser ensured clear separation of bundle sheath and mesophyll tissues. More than 10 areas containing tens of cells each were captured and accumulated in each sample of each tissue type. Mesophyll or bundle sheath samples were captured into separate caps of 0.5-ml microtubes filled with 20 μl of extraction buffer (Arcturus PicoPure RNA Isolation Kit; Thermo Fisher Scientific; MA, USA).
RNA was isolated using PicoPure kits (Arcturus PicoPure RNA Isolation Kit, Thermo Fisher Scientific, MA, USA), with additional on-column deoxyribonuclease (DNase) digestion (RNase-free DNase, QIAGEN, Germany). Extraction quality was evaluated using a TapeStation DNF-472T33 (Agilent Technologies, CA, USA) with the high sensibility assay HS Total RNA, analysis mode: Plant RNA. Only extractions with RNA profiles clearly exhibiting the two plant cell ribosomal RNA peaks and the two chloroplasts ribosomal RNA peaks were sequenced (fig. S1A). We used SMART-Seq v4 Ultra Low Input RNA kits for sequencing (Takara Bio Group, Japan), which first generated high-quality cDNA from ultralow amounts of total RNA and then high-quality Illumina sequencing-ready libraries. The SMARTer anchor sequence and polyadenylate (poly-A) sequence served as universal priming sites for end-to-end cDNA amplification of mRNA. We used 250 pg of total RNA input with 14 polymerase chain reaction (PCR) cycles for cDNA synthesis and 10 PCR cycles for library amplification, following the manufacturer’s guidance. mRNA libraries were pooled and sequenced on a NovaSeq 6000 system (Illumina, CA, USA) to generate ~25 million 100-bp paired-end reads per library. Library preparation and sequencing were done at the Yale Center for Genome Analysis, New Haven, CT, USA.
Spatial whole-transcriptome sequencing
To obtain near-cellular resolution of gene expression across entire leaf paradermal sections, we used the Visium Spatial Gene Expression platform (10x Genomics, CA, USA) following the manufacturer’s directions. The Visium platform captures mRNA released from a tissue section fixed to a slide, spatially tagging each mRNA molecule before sequencing to later map their position and abundance over a corresponding microphotograph of the tissue section. We followed the Tissue Optimization workflow to confirm the compatibility of our leaf tissues with the solution and to optimize the permeabilization conditions to release the maximum mRNA in the shortest time. First, using a cryomicrotome, 12-μm paradermal sections were obtained from flash-frozen leaves and each placed within the frames of one of the eight, 6.5 mm–by–6.5 mm capture areas on an optimization slide. Sections were fixed with methanol and stained with hematoxylin and eosin. The slide was scanned under a bright field using a microscope (Axio Imager.M1; Zeiss, Germany) with a magnification of ×10 per tile for comparison with final fluorescent images. We used permeabilization test times of 3, 6, 12, 18, 24, and 30 min in each capture area, including negative (tissue section not exposed to permeabilization reagents) and positive controls (stock isolated plant mRNA). cDNA was generated from the mRNA that bound to probes on the slide during permeabilization using fluorescently labeled nucleotides. Tissue was then enzymatically removed from the slide, and the remaining fluorescently labeled cDNA was analyzed to select the optimal permeabilization time based on the fluorescence of each test area. Using the fluorescence capacity of the microscope (TRITC filter cube; filter Rhod; 800-ms exposure time), we observed maximum fluorescence in the corresponding sample area after 12 min of permeabilization. The software ZEN 2.6 (Zeiss, Germany) was used to control the microscope and stitch the microphotograph tiles.
Once the tissue permeabilization conditions were optimized, a total of eight sections, comprising two technical replicates at 7 and 23 hours under well-watered and drought conditions, were used for spatial mRNA sequencing. Each replicate was placed on one of two slides containing four capture areas. Each area contained 5000 barcoded capture spots that were 55 μm in diameter to 100 μm center to center between spots. Spots were populated with primers that included Illumina sequencing primers, unique spatial barcodes per spot, unique molecular identifiers (UMIs), and 30-nt of repeating deoxythymidines poly(dT) sequences to capture poly-A mRNA. Tissue sections were processed as in the optimization steps: cryosectioning, followed by fixation, staining, slide imaging, and permeabilization. The mRNA released from overlying cells was captured by the primers on the spots and incubated with a mix containing reverse transcription reagents, producing spatially barcoded cDNA. After the second strand was synthesized, a denaturation step released it from the slide, and cDNA was transferred from each capture area to a corresponding tube for amplification and library construction following the manufacturer’s specifications. Amplification, library preparation, and sequencing were done at the Yale Center for Genome Analysis, following the manufacturer’s directions, with a sequencing depth of at least 50,000 read pairs per spot in the capture area.
P. oleracea lacks genomic resources and only transcriptome sequencing data have been produced to date. The closest relative reference genome available is that of P. amilis. However, on average, only the 32% of our LMD-RNA reads align to the P. amilis genome, preventing further guide for transcriptome assembly or gene expression analyses. Our laboratory previously de novo assembled a P. oleracea transcriptome using Trinity (39) and using more than 1 billion 100-bp pair-end reads from whole leaves (15), and here, we used that assembly to map reads from both LCM and Visium experiments. Before that, we reduced the dataset to contigs containing coding sequences. For that, we first used the software TransDecoder v5.5.0 (code was obtained from http://transdecoder.github.io and accessed in May 2020) to predict coding sequences within the transcripts, providing the software with precomputed BLASTX (40) alignments to the UniProt protein sequences database (41) to improve prediction. Next, we used CD-HIT v4.8.1 (16) to cluster coding sequences sharing 95% identity, retaining one representative. The indexes of the remaining sequences were used to subset the initial transcriptome assembly, yielding a reduced transcriptome for read mapping. Transcripts were functionally annotated using the software Trinotate v3.2.1 (code was obtained from http://trinotate.github.io and accessed in May 2020), selecting the best Blast hit (lowest e-value score) of all the possible alignments. Annotation of transcripts into gene lineages for gene families with a previously known role in any plant lineage in CCMs and starch metabolism was performed as with gene lineage notations as in (13–15, 18, 42). For that, we used the original set of sequences per gene family annotated in those studies to retrieve homolog coding sequences from our dataset using Blast (e-value, 0.01). For each gene family, the retrieved sequences were aligned on the basis of codon positions to the reference sequences using MAFFT alignment software (43) and manual curation. Maximum likelihood gene family trees were inferred from the alignments using IQ-TREE (44) with automatic model selection and 100 bootstraps. Query sequences were assigned to gene lineages on the basis of the annotation of its sister reference sequence on the tree. Our gene lineage annotations matched those in (15).
mRNA reads from mesophyll and bundle sheath tissues captured using laser microdissection were filtered and trimmed using Trimmomatic-0.39 (45). With this tool, ten base pairs were removed from the beginning of the read; Illumina adapters, poly-A tails, and SMART-Seq primers were removed from the reads; a 5-bp sliding window trimming approach was used to clip the read at the 5′ end where the average quality score within the window fell below 20; and single, low-quality bases from the beginnings and ends were also clipped. Only reads longer than 18 bp were kept after filtering. To quantify abundance, reads were mapped to the transcriptome assembly using the pseudo-aligner Kallisto v-0.45.0 (17) with 100 bootstrap replicates.
We used DEseq2 (46) to test for gene DE across experimental variables. To generate Trinity unigene-level counts (as opposed to transcript-level counts), we used Sleuth (47) in “gene_mode” to compile the Kallisto transcript counts into unigene counts. The resulting matrix of estimated counts was used to measure DE with DEseq2 in R (48). To estimate the magnitude and direction of the change in expression (measured as log2FC) and to statistically test for DE unigenes across cell types, time points, and water status, we included those variables and controlled for sequencing batch and plant individuals using all samples for DEseq2 normalization. In addition, to specifically measure the effect of drought on each cell type at each time point, we separated our counts matrix by 7 and 23 hours and ran the DEseq2 function for each partition, including, in these cases, a cell type and water status interaction term for normalization. The P values obtained from each test were adjusted (P-adjusted) for multiple testing by means of false discovery rate using the Benjamini and Hochberg method (49). We then tested for enrichment of GO terms in the lists of significant DE unigenes from each test (Padj < 0.05). First, we extracted the corresponding GO terms from the Trinotate analysis and then used Fisher’s exact test, as implemented in TopGO (50), using the GO terms from all unigenes as the background “gene universe.”
Visium Spatial Gene Expression analysis
Reads obtained from tissue sections using the Visium Spatial Gene Expression platform were processed using the analysis pipelines implemented in Space Ranger v1.1.0 (10x Genomics; https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/what-is-space-ranger). We used the “spaceranger mkref” to construct a reference transcriptome, parsing the exon unigene information generated by TransDecoder and the sequence of the longest isoform by unigene. We mapped reads to the most highly expressed unigene per Portulaca CCM-related gene lineage when possible or per gene family from the above LCM analysis, with a total of 140 sequences. We manually aligned the bright-field microphotograph based on fiducial markers and selected the areas where capture spots covered tissue. We then ran “spaceranger count” to filter-trim reads and align them to the reference transcriptome, with default parameters. UMIs were later used to correct and estimate counts of aligned reads per unigene per spatial capture spot. Principal components analysis and K-means clustering of spots by expression similarity were performed automatically in Space Ranger and visualized in the Loupe Browser 5.0 (10x Genomics).
Methods II: Flux balance model
To further explore how C4 and CAM might be integrated, we built a two-cell two-phase model adapted from a highly curated plant core metabolic model (26). The core metabolic model includes all major metabolic enzymes and reactions found to be highly conserved across a wide array of plant genomes and represents a “core” stoichiometric model, which captures the central carbon metabolism in leaves. Our new model represents how C4 and CAM could be integrated in a parsimonious manner from first principles of general plant metabolism.
Model construction
Previous models only considered C3 or CAM metabolism in a single-cell model or C4 metabolism in a two-cell model but without diel variation (26, 51, 52). To consider the C4 and CAM cycles together, we built a new metabolic model including both mesophyll and bundle sheath cells and diel-flux changes. Our model is an extension of the model in (26), which considered diel-flux and charge balance in each cellular compartment. The advantage of including charge balance is that it considers the effects of organellar pH on the metabolites’ charge states, which is important for the CAM acidification process. Starch was allowed to accumulate in the plastid, and sugars (glucose, sucrose, and fructose), carboxylic acids (malate, citrate, and isocitrate), proteinogenic amino acids, and nitrate were allowed to accumulate in the vacuole for storage and were used during both day and night. We duplicated all the reactions to represent bundle sheath and mesophyll cell types individually. We then added transport reactions between bundle sheath and mesophyll for daytime and nighttime conditions. We allowed particular metabolites to be transferred between bundle sheath and mesophyll on the basis of previous studies (53–56): malate, aspartate, alanine, pyruvate, G3P, dihydroxyacetone phosphate, PEP, 2-phosphoglyceric acid, sucrose, sulfate, ammonium, nitrate, and CO2. We also added charge and proton balances for these reactions following the method in (26). Our model had the following four first-order compartments: daytime mesophyll, nighttime mesophyll, daytime bundle sheath, and nighttime bundle sheath. Under each of these first-order compartments, there were the following second-order compartments where corresponding biochemical reactions happen: plastid, peroxisome, vacuole, thylakoid, mitochondrion, extracellular, cytoplasm, mitochondrial intermembrane space, and inner membrane spaces. We also made the following revisions to the model: allowed proton and oxygen transfer between day and night, fixed mannan biosynthesis reactions, and allowed protons to be transferred reversibly between cytoplasm and mitochondria, vacuole, and extracellular space. Thus, our model contains all the major biochemical reactions related to C3, C4, and CAM photosynthesis. Our model is available on Dryad (https://datadryad.org; DOI: 10.5061/dryad.931zcrjm6).
Modeling constraints
We used the COBRApy package (57) to perform the flux balance analysis. The non–growth-associated maintenance costs were controlled by constraining an ATPase and three reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase pseudo-reactions (cytoplasmic, mitochondrial, and plastidic) to a 3:1 (adenosine triphosphate hydrolysis:NADPH oxidase) ratio, based on (58) and used by previous studies (26). The maintenance costs, which were represented by ATPase fluxes, had a linear relationship with photon uptake fluxes (26). We used a photon uptake flux similar to light levels in our greenhouse experiments described above and a corresponding non–growth-associated maintenance ATPase flux of 33.66 μmol m−2 s−1. We also set an anatomical constraint that bundle sheath cells occupied 30% of the leaf area and mesophyll cells occupied 70%. All the fluxes in mesophyll and bundle sheath were adjusted according to the above ratios. RuBisCO carboxylation to oxygenation ratio was set to 3:1 in mesophyll but to 10:1 in bundle sheath because of lower diffusion of O2 and the CCM (56). We used a relatively lower RuBisCO carboxylation to oxygenation ratio compared to other studies of C4 models, but sensitivity analysis indicated that our results were robust to varied and higher ratios (table S8). Last, as P. oleracea primarily uses NAD-type C4 (20, 59), we constrained our model to use NAD-ME as the decarboxylation enzyme and accordingly blocked the transfer of pyruvate, PEP, 2PG, and G3P between mesophyll and bundle sheath (60). We did not constrain any other major biochemical reactions related to C3, C4, or CAM.
Modeling scenarios
We parameterized the models with the growth environmental conditions in the greenhouse experiment and performed the FBA. The primary objective function maximized phloem output, which meant letting the model dictate which pathways were most efficient (i.e., generate the highest phloem output) under given environmental conditions. The secondary objective function minimized the absolute sum of fluxes, which was a proxy for reducing enzymatic costs while fulfilling the primary objective function (61). We then performed FVA to examine the potential flux space without affecting the primary objective in FBA analysis. We first let the model freely determine the potential pathways among C3, C4, and CAM with two objectives, without adding additional constraints, under two environmental conditions: well-watered, under which we did not specifically constrain the CO2 uptake to mimic stomata open to well-watered condition, and drought conditions, under which we constrained CO2 uptake to be 55% of well-watered condition to represent a drought condition, stomata closure of 45%. We also modeled another two drought conditions, which represent 25 and 75% stomatal closure, to support the conclusions and results are robust.
We then tested several scenarios related to malate transfer and storage: (i) blocking malate transfer between bundle sheath and mesophyll, (ii) blocking malate storage in mesophyll, (iii) blocking malate storage in bundle sheath, and (iv) blocking malate storage in both mesophyll and bundle sheath.
Last, we performed alternative modeling scenarios related to C4 anatomy. We revised the constraint of CO2-proof bundle sheath cells (typical C4 anatomy) by allowing CO2 to diffuse into bundle sheath but maintained the same atmospheric CO2 as in modeling scenarios with CO2-impermeable bundle sheath. To set a CAM scenario, we set daytime CO2 at 0 and the RuBisCO carboxylation–to–oxygenation ratio to 5.15, following (27). All the other settings and constraints were the same for C3, C4, and CAM scenarios. The unit for the fluxes is “mmol gDW−1 day−1.” “DW” represents “dry weight.” The flux unit can be converted to net CO2 uptake on a leaf area basis as follows
Thus, the net CO2 uptake flux is 127.7 − 7.2 = 120.5 mmol gDW−1 day−1 at well-watered condition, and specific leaf area (SLA) for P. oleracea is 600 cm2 g−1on average (62). s represents 3600 seconds per hour and h represents 24 hours per day. Then, the CO2 uptake predicted by the model is converted to be 23.2 μmol m−2 s−1 on a leaf area basis.
Acknowledgments
We are grateful to E. Guillon at Yale University for sharing with us her expertise with the microscopes. We also thank the Horsley and Wolenski laboratories at the Department of Molecular, Cellular, and Developmental Biology, Yale University, for sharing with us laboratory instruments and to C. Bolick at Yale’s Marsh Botanic Gardens for the support with the experiment. We thank R. Shawn Abrahams, A. Chomentowska, K. Dion, N. Heaphy, J. Randall, and O. Oso for comments on earlier drafts that have improved this manuscript. Last, we thank Ian C. Berriman for the help with the design of the figures.
Funding: This work was funded by the NSF (IOS-1754662 award to E.J.E). This work is supported by the NOAA Climate and Global Change Postdoctoral Fellowship Program (award no. NA18NWS4620043B to H.Z).
Author contributions: Conceptualization: J.J.M.-V., H.Z., and E.J.E. Methodology: J.J.M.-V., H.Z., I.S.G., S.L.T., C.Y.M.C., and E.J.E. Investigation: J.J.M.-V., H.Z., S.L.T., and C.Y.M.C. Visualization: J.J.M.-V., H.Z., I.S.G., and E.J.E. Funding acquisition: E.J.E. and H.Z. Writing (original draft): J.J.M.-V., H.Z., and E.J.E. Writing (review and editing): J.J.M.-V., H.Z., I.S.G., S.L.T., C.Y.M.C., and E.J.E.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Online table S1 to S5 (.xlsx) and Visium loupe files data files S1 to S6 (.loupe) are available on Dryad (https://datadryad.org; DOI: 10.5061/dryad.931zcrjm6). All short-sequencing reads are available in the GeneBank Sequence Read Archive (https://ncbi.nlm.nih.gov/sra) BioProject: PRJNA774250. The scripts used for the data analyses are available on Dryad (https://datadryad.org; DOI: 10.5061/dryad.931zcrjm6).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S8
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
Data files S1 to S6
REFERENCES AND NOTES
- 1.Edwards E. J., Evolutionary trajectories, accessibility and other metaphors: The case of C4 and CAM photosynthesis. New Phytol. 223, 1742–1755 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Sage R. F., Sage T. L., Kocacinar F., Photorespiration and the evolution of C4 photosynthesis. Rev. Plant Biol. 63, 19–47 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Hatch M. D., Slack C. R., Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101, 103–111 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black C. C., Chen T. M., Brown R. H., Biochemical basis for plant competition. Weed Sci. 17, 338–344 (1969). [Google Scholar]
- 5.Osmond C. B., Crassulacean acid metabolism: A curiosity in context. Annu. Rev. Plant Physiol. 29, 379–414 (1978). [Google Scholar]
- 6.S. R. Szarek, I. P. Ting, Photosynthetic efficiency of CAM plants in relation to C3 and C4 plants, in Environmental and Biological Control of Photosynthesis, R. Marcelle, Ed. (Springer, 1975), pp. 289–297. [Google Scholar]
- 7.Sage R. F., Christin P.-A., Edwards E. J., The C4 plant lineages of planet earth. J. Exp. Bot. 62, 3155–3169 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Silvera K., Santiago L. S., Cushman J. C., Winter K., Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the orchidaceae. Plant Physiol. 149, 1838–1847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch K., Kennedy R. A., Characteristics of crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L. Plant Physiol. 65, 193–197 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. F. Sage, in Functional Plant Biology (Funct Plant Biol, 2002), vol. 29, pp. 775–785; https://pubmed.ncbi.nlm.nih.gov/32689525/.
- 11.Lara M. V., Drincovich M. F., Andreo C. S., Induction of a crassulacean acid-like metabolism in the C4 succulent plant, Portulaca oleracea L.: Study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell Physiol. 45, 618–626 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Guralnick L. J., Edwards G., Ku M. S. B., Hockema B., Franceschi V., Photosynthetic and anatomical characteristics in the C4–Crassulacean acid metabolism-cycling plant Portulaca grandiflora. Funct. Plant Biol. 29, 763–773 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Christin P.-A., Arakaki M., Osborne C. P., Bräutigam A., Sage R. F., Hibberd J. M., Kelly S., Covshoff S., Wong G. K.-S., Hancock L., Edwards E. J., Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. J. Exp. Bot. 65, 3609–3621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari R. C., Bittencourt P. P., Rodrigues M. A., Moreno-Villena J. J., Alves F. R. R., Gastaldi V. D., Boxall S. F., Dever L. V., Demarco D., Andrade S. C. S., Edwards E. J., Hartwell J., Freschi L., C4 and crassulacean acid metabolism within a single leaf: Deciphering key components behind a rare photosynthetic adaptation. New Phytol. 225, 1699–1714 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Gilman I. S., Moreno-Villena J. J., Lewis Z. R., Goolsby E. W., Edwards E. J., Gene co-expression reveals the modularity and integration of C4 and CAM in Portulaca. Plant Physiol. 189, 735–753 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Moreno-Villena J. J., Dunning L. T., Osborne C. P., Christin P.-A., Highly expressed genes are preferentially co-opted for C4 photosynthesis. Mol. Biol. Evol. 35, 94–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voznesenskaya E. V., Koteyeva N. K., Edwards G. E., Ocampo G., Unique photosynthetic phenotypes in Portulaca (Portulacaceae): C3-C4 intermediates and NAD-ME C4 species with pilosoid-type Kranz anatomy. J. Exp. Bot. 68, 225–239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voznesenskaya E. V., Koteyeva N. K., Edwards G. E., Ocampo G., Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3-C4 intermediate in genus Portulaca L. (Portulacaceae). J. Exp. Bot. 61, 3647–3662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimmo H. G., The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 5, 75–80 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., Martinoia E., The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52, 1169–1180 (2007). [DOI] [PubMed] [Google Scholar]
- 23.J. A. C. Smith, J. Ingram, M. S. Tsiantis, B. J. Barkla, D. M. Bartholomew, M. Bettey, O. Pantoja, A. J. Pennington, (1996) Transport across the vacuolar membrane in CAM plants, in Crassulacean Acid Metabolism. Ecological Studies (Analysis and Synthesis), K. Winter, J. A. C. Smith, Eds. (Springer, 1996), vol 114, pp. 53–71. [Google Scholar]
- 24.Borland A. M., Griffiths H., Hartwell J., Smith J. A. C., Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 60, 2879–2896 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Weise S. E., van Wijk K. J., Sharkey T. D., The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J. Exp. Bot. 62, 3109–3118 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Shameer S., Baghalian K., Cheung C. Y. M., Ratcliffe R. G., Sweetlove L. J., Computational analysis of the productivity potential of CAM. Nat. Plants 4, 165–171 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Grime J. P., Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194 (1977). [Google Scholar]
- 28.Giacomello S., Salmén F., Terebieniec B. K., Vickovic S., Navarro J. F., Alexeyenko A., Reimegård J., McKee L. S., Mannapperuma C., Bulone V., Ståhl P. L., Sundström J. F., Street N. R., Lundeberg J., Spatially resolved transcriptome profiling in model plant species. Nat. Plants. 3, 17061 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Winter K., Garcia M., Virgo A., Ceballos J., Holtum J. A. M., Does the C4 plant Trianthema portulacastrum (Aizoaceae) exhibit weakly expressed crassulacean acid metabolism (CAM)? Funct. Plant Biol. 48, 655–665 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Ogburn R. M., Edwards E. J., Repeated origin of three-dimensional leaf venation releases constraints on the evolution of succulence in plants. Curr. Biol. 23, 722–726 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kadereit G., Ackerly D., Pirie M. D., A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc. R. Soc. B Biol. Sci. 279, 3304–3311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Riina R., Morawetz J. J., Haevermans T., Aubriot X., Berry P. E., Molecular phylogenetics and classification of Euphorbia subgenus chamaesyce (Euphorbiaceae). Taxon 61, 764–789 (2012). [Google Scholar]
- 33.Winter K., Ecophysiology of constitutive and facultative CAM photosynthesis. J. Exp. Bot. 70, 6495–6508 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Ferrari R. C., Bittencourt P. P., Nagumo P. Y., Oliveira W. S., Aurineide Rodrigues M., Hartwell J., Freschi L., Developing Portulaca oleracea as a model system for functional genomics analysis of C4/CAM photosynthesis. Funct. Plant Biol. 48, 666–682 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Ermakova M., Danila F. R., Furbank R. T., von Caemmerer S., On the road to C4 rice: Advances and perspectives. Plant J. 101, 940–950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X., Cushman J. C., Borland A. M., Edwards E. J., Wullschleger S. D., Tuskan G. A., Owen N. A., Griffiths H., Smith J. A. C., De Paoli H. C., Weston D. J., Cottingham R., Hartwell J., Davis S. C., Silvera K., Ming R., Schlauch K., Abraham P., Stewart J. R., Guo H.-B., Albion R., Ha J., Lim S. D., Wone B. W. M., Yim W. C., Garcia T., Mayer J. A., Petereit J., Nair S. S., Casey E., Hettich R. L., Ceusters J., Ranjan P., Palla K. J., Yin H., Reyes-García C., Andrade J. L., Freschi L., Beltrán J. D., Dever L. V., Boxall S. F., Waller J., Davies J., Bupphada P., Kadu N., Winter K., Sage R. F., Aguilar C. N., Schmutz J., Jenkins J., Holtum J. A. M., A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 207, 491–504 (2015). [DOI] [PubMed] [Google Scholar]
- 37.DeLucia E. H., Chen S., Guan K., Peng B., Li Y., Gomez-Casanovas N., Kantola I. B., Bernacchi C. J., Huang Y., Long S. P., Ort D. R., Are we approaching a water ceiling to maize yields in the United States? Ecosphere 10, e02773 (2019). [Google Scholar]
- 38.Lobell D. B., Roberts M. J., Schlenker W., Braun N., Little B. B., Rejesus R. M., Hammer G. L., Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science 344, 516–519 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L., BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium T. U., UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christin P.-A. A., Boxall S. F., Gregory R., Edwards E. J., Hartwell J., Osborne C. P., Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5, 2174–2187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K., Standley D. M., A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32, 1933–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L., Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 48.R. Core Team, R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2018).
- 49.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- 50.Alexa A., Rahnenfuhrer J., Lengauer T., Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Blätke M.-A., Bräutigam A., Evolution of C4 photosynthesis predicted by constraint-based modelling. eLife 8, e49305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shameer S., Vallarino J. G., Fernie A. R., Ratcliffe R. G., Sweetlove L. J., Flux balance analysis of metabolism during growth by osmotic cell expansion and its application to tomato fruits. Plant J. 103, 68–82 (2020). [DOI] [PubMed] [Google Scholar]
- 53.de Oliveira Dal’Molin C. G., Quek L.-E., Palfreyman R. W., Brumbley S. M., Nielsen L. K., C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 154, 1871–1885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallmann J., Heckmann D., Bräutigam A., Lercher M. J., Weber A. P. M., Westhoff P., Gowik U., The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3, e02478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arrivault S., Obata T., Szecówka M., Mengin V., Guenther M., Hoehne M., Fernie A. R., Stitt M., Metabolite pools and carbon flow during C4 photosynthesis in maize: 13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 68, 283–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw R., Cheung C. Y. M., A mass and charge balanced metabolic model of Setaria viridis revealed mechanisms of proton balancing in C4 plants. BMC Bioinformatics 20, 357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebrahim A., Lerman J. A., Palsson B. O., Hyduke D. R., COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst. Biol. 7, 1–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung C. Y. M., Williams T. C. R., Poolman M. G., Fell D. A., Ratcliffe R. G., Sweetlove L. J., A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J. 75, 1050–1061 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Gutierrez M., Gracen V. E., Edwards G. E., Biochemical and cytological relationships in C4 plants. Planta 119, 279–300 (1974). [DOI] [PubMed] [Google Scholar]
- 60.Furbank R. T., Evolution of the C4 photosynthetic mechanism: Are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Töpfer N., Braam T., Shameer S., Ratcliffe R. G., Sweetlove L. J., Alternative crassulacean acid metabolism modes provide environment-specific water-saving benefits in a leaf metabolic model. Plant Cell 32, 3689–3705 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Navas M. L., Friess N., Maillet J., Influence of cucumber mosaic virus infection on the growth response of Portulaca oleracea (purslane) and Stellaria media (chickweed) to nitrogen availability. New Phytol. 139, 301–309 (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S8
Tables S1 to S5
Data files S1 to S6