Abstract
The loss of detectable hepatitis B surface antigen (HBsAg) is considered a functional cure in chronic hepatitis B. Naturally, HBsAg can be incorporated into the virion envelope or assembled into subviral particles (SVPs) with lipid from host cells. Until now, there has been no detailed structure of HBsAg, and the published SVP structures are controversial. Here, we report the first subnanometer-resolution structures of spherical SVP from hepatitis B virus (HBV) and the related woodchuck hepatitis virus (WHV) determined by cryo–electron microscopy in combination with AlphaFold2 prediction. Both structures showed unique rhombicuboctahedral symmetry with 24 protruding spikes comprising dimer of small HBsAg with four helical domains. The lipid moiety in the SVP is organized in a noncanonical lipid patch instead of a lipid bilayer, which can accommodate the exposed hydrophobic surface and modulate particle stability. Together, these findings advance our knowledge of viral membrane organization and the structures of HBV and WHV spherical SVPs.
High-resolution structures of hepadnavirus spherical subviral particles show unique lipid and protein organization.
INTRODUCTION
Hepatitis B virus (HBV) infection is a substantial global public health concern. Although there is a vaccine to prevent new infection, approximately 240 million people have chronic HBV infections, which results in approximately 887,000 people dying from HBV-related complications, including fibrosis, cirrhosis, and hepatocellular carcinoma (1). Patients may be treated with interferon-α and/or reverse transcriptase (RT) inhibitors. Both treatments have a low probability of clearing the virus. RT inhibitors substantially decrease viral load and reverse liver damage. However, cessation of treatment can result in life-endangering viral flares. Once therapy is initiated, patients can expect lifelong treatment. There is an unmet medical need for new therapeutics that could lead to sustained inhibition of viral replication and the loss of hepatitis B surface antigen (HBsAg) (2), an important serological marker for HBV infection.
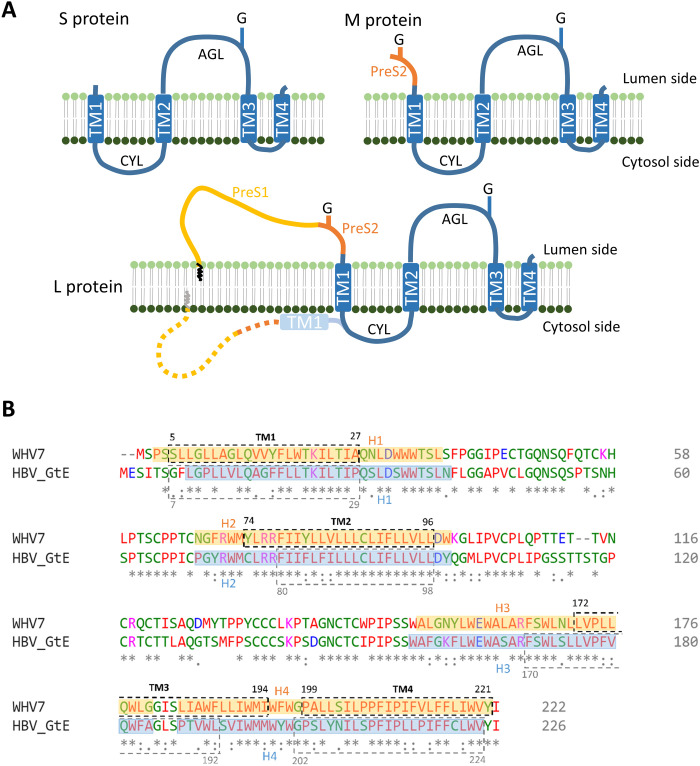
There are four major types of particles found in the sera of chronically HBV-infected patients, including infectious virions, genome-free empty virions, spherical subviral particles (SVPs), and tubular SVPs (3). The highly immunogenic spherical SVPs typically outnumber virions by 103 to 105 fold (4) and are the basis of prophylactic vaccines used against HBV infection to date (5). The HBV virion contains an icosahedral nucleocapsid surrounded by a lipid envelope that is composed of three different surface proteins (HBs) termed L (large), M (medium), and S (small). They are all translated from the same open reading frame but are variable in length at the N-terminal end (4). The S-HBs contains the S domain that is predicted to have four transmembrane helices (TM) (Fig. 1A). M-HBs is extended by an additional PreS2 domain at the N terminus, while L-HBs further adds the PreS1 domain to M-HBs (Fig. 1A). The S domain of all three surface proteins is glycosylated (6, 7). M-HBs also harbors the second N-glycosylation site at its N-terminal region, but it is not used in L-HBs because of cotranslational retention of the PreS (PreS1 + PreS2) domain in the cytosol. The TM1 and TM2 are separated by the cytosolic loop (CYL), which directs the S-HBs for intracellular trafficking (8) and is essential for HBV SVP secretion (9) and virion formation (10). The residues between TM2 and TM3 form the major antigenic loop (AGL) at the surface of the viral particle (11, 12). The ratio of surface proteins L, M, and S distributed on the membrane of virion (3:2:5) is quite different to the ratio of 1:1:4 in tubular SVP (13). Spherical SVP consists almost exclusively of S-HBs, ~10% M-HBs, with only a trace of L-HBs (4).
Fig. 1. Proposed surface protein topology and sequence alignment of WHV and HBV.
(A) Current purposed models for WHV and HBV S, M, and L protein topologies. The blue boxes indicate transmembrane helices (TM). The zigzag line in L protein indicates the N-terminal myristoylation. G, N-glycosylation; CYL, cytosolic loop; AGL, antigenic loop. (B) Sequence alignment of WHV7 and HBV genotype E (GtE) small surface proteins. The TMs were predicted by the TMHMM server, which are indicated by the dashed boxes with their designated position. The helix assignment from this study was presented in transparent colored boxes (orange, WHV; blue, HBV) with their designated position. Identical (*), conserved (:), and semiconserved (.) amino acids are indicated. Blue and purple indicate negatively or positively charged residues, respectively. Red indicates amino acids with hydrophobic side chains.
Now, there are no high-resolution structures for SVPs, although some models at moderate resolution have been obtained by cryo–electron microscopy (cryo-EM) (14–17), atomic force microscopy (AFM) (18), and x-ray scattering data (19). In particular, the particle geometry and membrane organization of spherical SVPs are still under debate. Cyro-EM reconstruction of spherical SVPs produced in transgenic mice showed two predominant sizes with octahedral symmetry and no clear lipid bilayer at 12 Å (14). Cao et al. (17) recently reported that the spherical SVPs obtained from sera of HBV carriers were irregular and contained a lipid bilayer by cryo-EM. Moreover, the yeast-derived HBs virus-like particles (VLP) show a roughly spherical morphology that consists of 96 recombinant S proteins. Although the resolution is limited at 15 Å, the authors proposed that the lipid membrane comprises two discrete regions, an outer uniform lipid monolayer and an inner amorphous layer composed of nonpolar triglycerides (16). In addition, the tubular SVPs purified from human serum also revealed spike-like protrusions across the lipid bilayer packed in a helical lattice. The spikes are modeled with external regions of S domain and PreS regions from L-HBs and M-HBs (15). Last, biochemical analysis of SVP revealed that the lipids in the particles are closely intercalated with the proteins (20–22); however, contradictory results by solid-state nuclear magnetic resonance, light scattering, and cryo-EM suggest that the SVP displays a moderately rigid lipid bilayer (23). It appears that these SVPs are morphologically different depending on their source and sample preparation.
Here, we used cryo-EM to study the spherical SVPs from woodchuck hepatitis virus (WHV), a closely related hepadnavirus to HBV in regard to the nucleic acid sequence, genome organization, virion and capsid morphology, and mechanisms of infection and replication (6, 24). The 6.5-Å three-dimensional (3D) model revealed that the spherical SVP exhibits a novel rhombicuboctahedral symmetry with 24 protruding spikes at the particle surface. Each protrusion is a dimer of S surface proteins (WHs) with four helices. We also found that the spherical SVP contains a noncanonical lipid organization, the density at the outer layer is composed of discrete amorphous lipid, and the density at the inner layer is characterized by protein from the S domain. To see whether the spherical SVPs from HBV also exhibited a similar architecture, we used cryo-EM to solve the structure of spherical SVPs purified from the serum of an HBV carrier to 6.3-Å resolution. In combination with predicted atomic models from AlphaFold version 2 (AlphaFold2), we depicted the first detailed structural models of SVP for both WHV and HBV and provide a mechanistic model for subunit-subunit interactions in the spherical SVP. Together, these results deepen our current understanding of spherical SVP morphogenesis that can improve the efficacy for SVP-based anti-HBV antibodies and enhance the efficiency in using SVP as platforms for delivery of medically relevant antigenic sequences in the future.
RESULTS
WHV SVP purification and reconstruction
The WHV-infected Eastern woodchuck is a critical experimental model for studying immunopathogenesis of HBV infection, liver cancer development, and antiviral therapy against HBV (25). Similar to HBV, WHV also encodes three envelop proteins that contain the common C-terminal S domain (6). The S domain shares 63% amino acid sequence identity to that of HBV and was predicted to have four transmembrane helices, each consisting of an average size of 22 ± 2 amino acid residues (Fig. 1B). The high degree of identity makes the S domain interchangeable between WHV and HBV with minimal effects on the SVP formation (26) and presumably leads to substantial structural-functional conservations.
WHV SVPs were purified from the serum of a woodchuck chronically infected with WHV strain 7 (WHV7) woodchuck by isopycnic CsCl density gradient. Selected fractions were resolved by native agarose gel electrophoresis (NAGE), analyzed by Western blot, and prepared for negative-stain transmission electron microscopy (TEM). The majority of the surface protein located in F16 (fraction 16) to F18 (Fig. 2A, lanes 7 to 9). TEM images showed that F16 had the vast majority of spherical SVPs, whereas F17 contained more tubular SVPs, and virion-like particles started to become detectable in F18 (Fig. 2B). To further investigate the structure of spherical SVPs, F16 was selected, freed from cesium salts, and subjected to cryo-EM single-particle analysis.
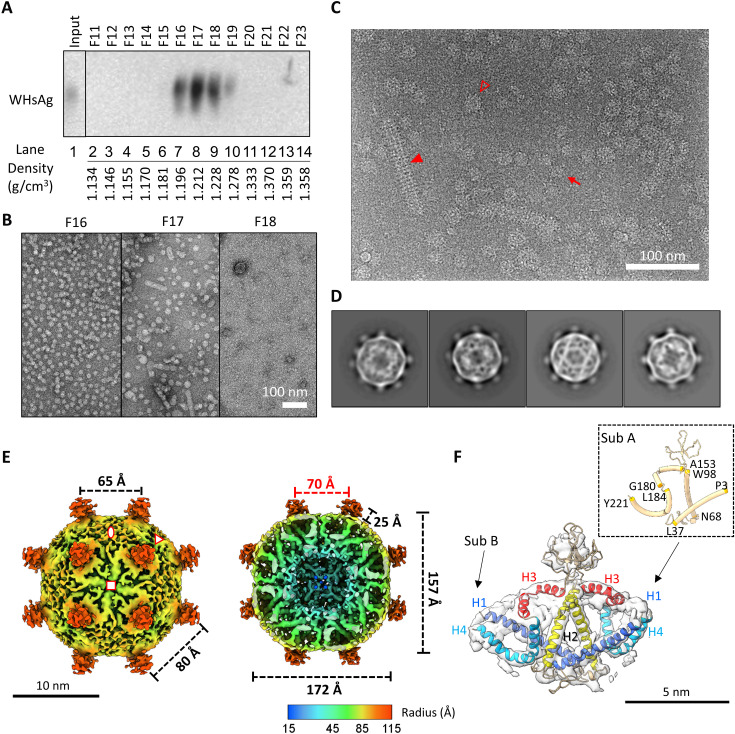
Fig. 2. Structural analysis of WHV SVP.
(A) Western blot of fractions probed for WHV surface proteins. WHV7-infected woodchuck serum was fractionated on CsCl density gradients. Selected fractions (F11 to F23) were resolved by NAGE and analyzed by immunoblotting using a polyclonal antibody (pAb) against HBsAg (ViroStat). (B) Negative-stain images of selected fractions from the CsCl gradient. Scale bar, 100 nm. (C) Cyro-EM micrograph of purified WHV SVP from F16. Small and large spherical particles are indicated by a red arrow or an empty triangle, respectively. Tubular SVP is indicated by red solid triangle. (D) 2D class averages of WHV small spherical SVP. The particle diameter is around 23 nm. (E) Cryo-EM 3D reconstruction of WHV small spherical SVP at 6.5 Å. The map was contoured at ~2.5 σ. Front half (left) and bottom half (right) of radially colored surface representations were viewed at fourfold axis. The asymmetric unit is encompassed by three symbols. Twofold, threefold, and fourfold axes were indicated by oval, triangle, and square, respectively. Red number indicated the particle inner core diameter. (F) Fitting of S-WH dimer. The segmented cryo-EM map is shown as transparent surface. Two subunits and helices 1 to 4 are indicated. The inset shows the residues’ position in the helices in subunit A.
Frozen-hydrated cryo-EM images revealed that WHV SVPs were morphologically heterogeneous (Fig. 2C). Successive runs of reference-free 2D classification were performed to discard classes with blurry and ice-contaminated density. Despite some classes having large diameter with incomplete closed shells, they all had two common features: (i) multiple protruding spikes projected from the particle surface and (ii) two uncommon layers of density where the outer layer seemed to be amorphous and the inner layer contained a knitted mesh-like network (Fig. 2D and fig. S1). We selected the classes with complete closed shell and ordered density for further 3D refinement structural analysis (Fig. 2D). Note that in some of the class images, the particles show clear features of rotational twofold (Fig. 2D, left, 1 to 3) and fourfold (Fig. 2D, right) density. An initial 3D model was reconstructed asymmetrically using Relion software (27). This map exhibited an overall symmetric feature of twofold, threefold, and fourfold spatial arrangement—a characteristic of an octahedral symmetry (fig. S2A). We then applied octahedral symmetry and refined the 3D model to 6.5 Å (Fig. 2E and fig. S2C).
Overall structure of WHV SVP
The 3D reconstruction of WHV spherical SVP resembles a rhombicuboctahedral geometry with a total of 24 spikes protruded from the symmetrical vertices (Fig. 2E and fig. S3A). The height of the spike is approximately 2.5 nm measured from the surface, and the distances between each spike at threefold and fourfold axes are 8 and 6.5 nm, respectively. This results in 8 triangular, 6 squared, and 12 rectangular facets—an elongated derivative of a rhombicuboctahedron. From the outer surface, we observed two different types of density: thick wide density projected sideways from the bottom of the spike and a patch of thin short string-like density filled in the gap at each twofold location. The former density appears as two short tubes arranged in a V shape and laid flat on the particle surface. The short string-like density is comparable to the unspecific density of the lipid observed in the membrane of enveloped viruses (28). From the bottom half view (Fig. 2E, right), multiple folded tubular densities are connected together to form a thickness of 50-Å inner layer. The boundaries of each tube were visible at the high contour level. One curved tubular density comes down from the surface and turns outward to form a U-shape morphology. The other straight tubular density, on the other hand, spans through the inner layer and joins at the ends to form a density layer at the lower radii. Because S protein has been found to have a majority of alpha helices according to the previous prediction (29), we hypothesized that the observed tubular density represents helical regions of the protein.
Together, no conventional lipid bilayer morphology was observed in the WHV spherical SVP. The outer layer contains density of lipid patch mixed with protein density, and the inner layer is formed by complicated but ordered protein structure.
Modeling of small WHs
To understand the molecular building block of WHV spherical SVP at the current resolution, we traced the helix density, defined the boundary between closely associated protein density, and segmented one spike from the entire spherical SVP (fig. S3B; see Material and Methods for details) (30). The segmentation can be further separated into two units having virtually identical folds (fig. S3B). Therefore, it is possible to assign the segmented density as two independent monomers. Looking at each monomer, we found two straight helices (named helix 1 and helix 2) interacted at the lower radius of the particle. The helix 2 continuously extends outward to form a mass of density at the site where the spike is located. This density is relatively weak and disappears quickly at high contour level, suggesting the protein conformation in this region is more flexible. Continuing, the density forms a V-shaped helix (helix 3), which lays flat at the particle surface, is not a typical transmembrane helix, and is embedded in the lipid patch as described earlier (fig. S3B, middle). Part of this helix was defined as amphipathic (31, 32). Proceeding inward, the density arranges into the last U-shaped helix (helix 4) that turns in halfway and back up to the surface. This helix overlapped with the hydrophobic C-terminal region identified previously. The resulting model is similar to the predicted topology defined by Suffner et al. (31). Together, our cryo-EM density map suggests that a single spike is made of two S-WHs interacting at the AGL, C terminus of helix 2, and the bottom region of helices 1 and 2.
To better understand the detailed structure, we generated in silico 3D molecular models using AlphaFold2 (33) and subsequently used cryo-EM density map to constrain the predicted protein conformation. The Cα protein backbone was flexibly modeled into the cryo-EM density map using ISOLDE and Coot (34, 35). The resulting structure of S-WHs shows four helical segments and two flexible loop regions (Fig. 2F and fig. S3C). Because of the high confidence in the predicted helical segments, we could unambiguously assign each helix to the corresponding cryo-EM density (Fig. 2F). The two loop regions that connected helices 1/2 and helices 2/3 had the lowest confidence score in the prediction (Fig. 2F). In all five models predicted by AlphaFold2, the loop regions showed substantial differences (fig. S4A). The first loop region connecting helices 1 and 2 is the CYL, which has been shown to be important for maintaining oligomerization of the S domain (31). Despite that there is an extensive amount of cryo-EM density connecting helix 1 and helix 2, the overall volume was insufficient to accommodate the approximate 30 residues of the CYL (Fig. 2). The absence of density is the result of averaging at the flexible region of the protein. The second loop, located between helices 2 and 3, is the AGL, which has been shown to independently mediate S domain oligomerization by mutational analysis (31). The AGL has eight cysteine residues that were found to be cross-linked during the S domain dimerization (12, 36). Although it was predicted to contain two short β strands, the location of the strands in the density was unclear because of the weak cryo-EM density and the low local resolution in this region (Fig. 2F and fig. S2D).
Superimposition of subunits A and B revealed a subtle difference in the tilting angle at both helix 1 and helix 4 (fig. S4, C and D). Thus, our results indicated that the dimer of the asymmetric small surface protein is the building block of the spherical SVP.
Does HBV spherical SVPs have similar architecture?
We were surprised by the overall molecular architecture derived from WHV spherical SVP as described above and were wondering whether the HBV spherical SVP also exhibits similar spatial organization despite that different results were reported earlier (14–17). To determine the structure of HBV spherical SVP, we isolated SVPs from the serum of a patient infected with genotype E (GtE) and performed the same analysis as described earlier for WHV SVP. As expected, the majority of the HBsAg located in F16 to F18 with densities of 1.190 to 1.239 g/cm3 (Fig. 3A, lanes 7 to 9). Images of negatively stained samples indicated F16 has more spherical SVPs compared with other fractions (Fig. 3B) and was used for cryo-EM data collection (Fig. 3C). Like WHV, the HBV SVPs were variable in sizes and shapes (Fig. 3C). The observation is consistent with the previous finding (4, 15).
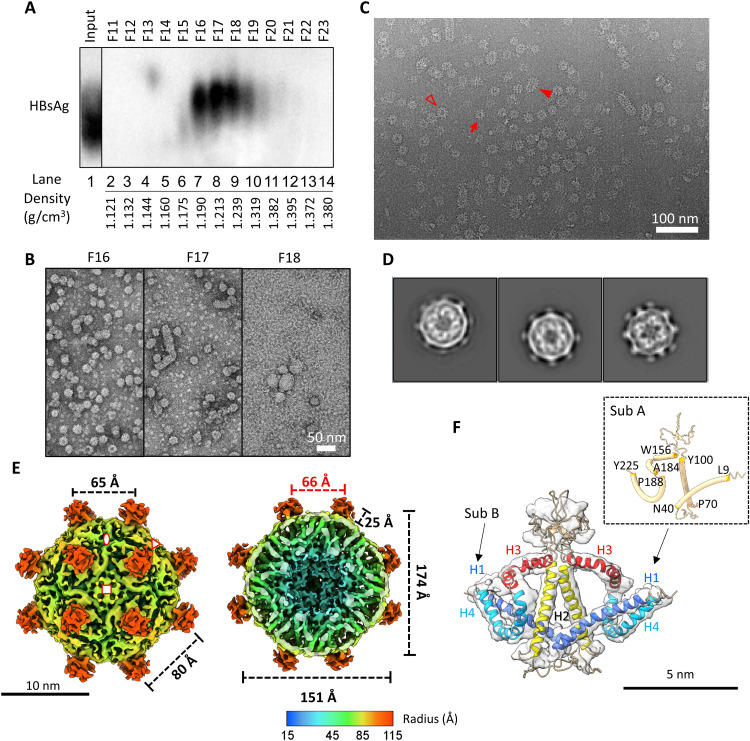
Fig. 3. Isolation of HBV SVP and analysis by Cryo-EM and 3D image processing.
(A) Western blot of fractions probed for HBV surface proteins. HBV-infected human serum was fractionated on isopycnic CsCl density gradients. Selected fractions (F11 to F23) were resolved by NAGE and analyzed by immunoblotting using the pAb against HBsAg (ViroStat). (B) Negative-stain images of selected fractions from CsCl gradient. Scale bar, 50 nm. (C) Cyro-EM micrograph of purified HBV SVP from F16. Small and large spherical particles are indicated by a red arrow or an empty triangle, respectively. Tubular SVP is indicated by red solid triangle. (D) 2D class averages of HBV small spherical SVP. The particle diameter is around 23 nm. (E) Cryo-EM 3D reconstruction of HBV small spherical SVP at 6.3 Å. The map was contoured at ~3.0 σ. Front half (left) and bottom half (right) of radially colored surface representations were viewed at fourfold axis. The asymmetric unit is encompassed by three symbols. Twofold, threefold, and fourfold axes were indicated by oval, triangle, and square, respectively. Red number indicated the particle inner core diameter. (F) Fitting of S-HBs dimer. The segmented cryo-EM map is shown as transparent surface. Two subunits and helices 1 to 4 are indicated. Inset shows the residues position of helices in subunit A.
The 2D class averages exhibited similar features as described above for WHV. All classes showed two layers of densities (Fig. 3D); the density at the outer layer is amorphous, while the density at the inner layer has a complicated protein-protein interaction network. We selected two classes that had complete surface density for further image analysis (Fig. 2D, right two classes). Using asymmetric reconstruction, we found that HBV spherical SVPs also have twofold, threefold, and fourfold characteristics. The four spikes at the twofold facet appeared in a rectangular arrangement instead of a square organization (fig. S2B); therefore, it is also having a rhombicuboctahedron-like symmetry. We further applied octahedral symmetry during model refinement and yielded a final 3D reconstruction at 6.3-Å resolution (fig. S2, C and E).
Like WHV, the HBV spherical SVP also has a diameter of 23 nm and contains 24 protruding spikes (Fig. 3E). Each spike is a dimer of S-HBs encompassing four helical segments. Using AlphaFold2 prediction together with the cryo-EM density map, we were able to generate the first atomic model for S-HBs (Fig. 3F and fig. S4B). The unassigned surface density for the lipid moiety is noticeably lower in HBV than in WHV spherical SVPs (Figs. 2E and 3E and fig. S5).
Structural comparison between WHV and HBV spherical SVPs
The structure of S-HBs is virtually identical to S-WHs at the monomer and dimer levels except that the angles that govern curvatures of the helices are slightly different (fig. S4C). The building block consists of a dimer of two subunits; each subunit contains an AGL (WHV: Lys99-Trp152; HBV: Gln101-Ser155), a CYL (WHV: Ser38-Cys67; HBV: Phe41-Cys69), and four helical domains (WHV: Pro3-Ser38, Asn68-Lys99, Ala153-Gly181, and Leu184-Tyr221; HBV: Leu9-Asn40, Pro70-Tyr100, Trp156-Ala184, and Pro188-Tyr225) (WHV monomers A and B have one residue difference; Fig. 1B). Both AGL and CYL are highly hydrophilic (figs. S6 and S8).
In the monomer, the interactions between H1 and H2 occur inside the particle at the lower radius via salt bridges, hydrogen bonds, hydrophobic interactions, and van der Waals forces bonding (table S1 and fig. S7). The H3, which lies flat at the particle surface, has the hydrophilic residues facing outside of the particle (fig. S6). The interactions found between H2 and H3 could help to maintain the location of H3 on the surface (fig. S7), and the exposed hydrophobic residues at the side are stabilized by the isolated lipid moiety. A recent report suggests that part of the H3 (Trp156 to Arg169) plays a critical role in the morphogenesis of HBV SVPs (32). Last, the H4 forms a hydrophobic core at the center of the U shape and turns the CTD to the particle surface (figs. S6 and S7). Overall, we found that S-WHs have more protein-protein interactions at the monomer level as compared to HBV, and these interactions are asymmetrically within the two subunits (table S1).
Dimeric hydrophobic interactions were found at the H1/H4 and H2/H2 helices. According to the literature, the disulfide bonds formed at AGL and CYL also contribute to the intermolecular homo-oligomerization (9, 10, 31, 37). However, because of the low local resolution and the high flexibility at these two loop regions, we were unable to model the interactions confidently. The orientation of the dimer in WHV and HBV spherical SVPs was different. In WHV, the tip of each H3 (Gly180–181) faces to threefold and fourfold centers, while in HBV, it faces to the twofold center (fig. S5). There is an approximately 70° rotation between WHV and HBV. Consequently, the resulting interactions between dimers at different interfaces were quite distinct between WHV and HBV spherical SVPs.
In WHV, surface exposed hydrophobic residues located at the twofold, threefold, and fourfold axes where the lipid density was observed in the cryo-EM density map (Figs. 2E and 4A and fig. S6A). Likewise, the hydrophobic residues were mostly exposed at the twofold and threefold axes in HBV, which was coincidental with the observed lipid moiety (Figs. 3E and 4B and fig. S8). The extra density around threefold and fourfold centers was the result of averaging N-terminal residues of S-HBs (Figs. 1B and 3E). Overall, the hydrophobic interactions at different dimer-dimer interfaces were the main force for holding the particle together (fig. S8, right most). The hydrophobic buried surface area without considering loop regions was most dominant at the twofold area (fig. S8 and table S2). Together, WHV spherical SVPs have more discrete lipid density and pose a different quaternary structural arrangement than HBV spherical SVPs.
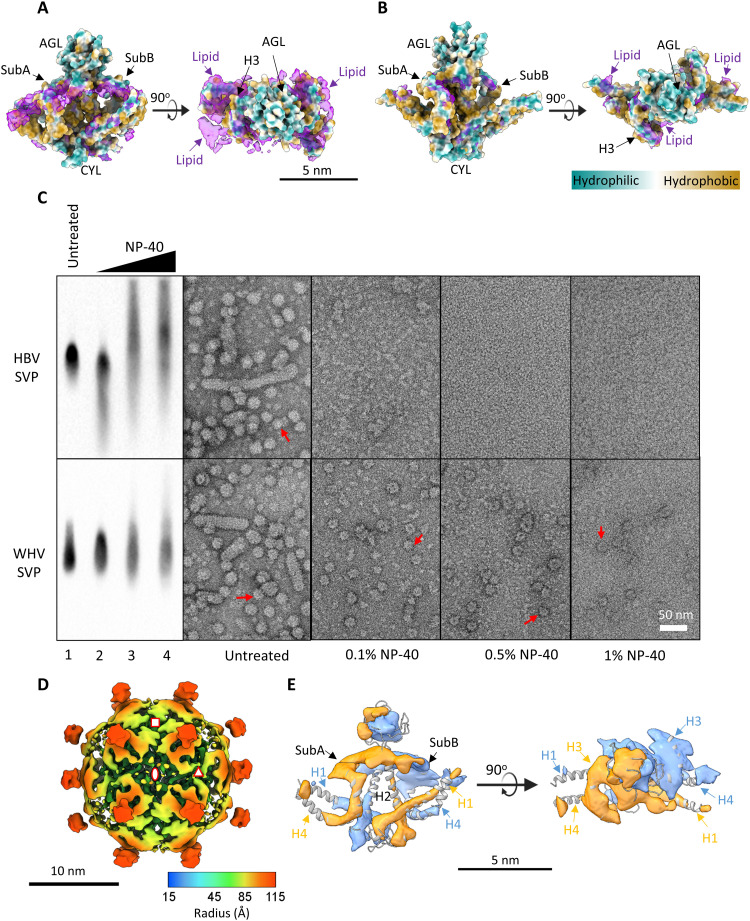
Fig. 4. Effects of lipids on the maintenance of the WHV and HBV SVP integrity.
Location of lipid density in the WHV (A) and HBV (B) SVPs. Superimposing side (left) and top (right) views of S protein dimer (colored by local hydrophobicity) with segmented cryo-EM density map of dimers (purple, excluding AGL region) showed that the WHV SVP contains more lipid density. (C) Effects of lipid removal on WHV and HBV SVP stability. The dialyzed SVP-containing CsCl F16 from WHV and HBV, without any further treatment (untreated) or treated with increasing concentrations of NP-40 (0.1, 0.5, and 1%) for 30 min at room temperature, before further analysis. The samples were resolved by 1% NAGE and immunoblotting by the pAb against HBsAg (ViroStat). The samples were visualized by negative-stain EM. Scale bar, 50 nm. Intact particles are indicated by red arrows. (D) Cryo-EM 3D reconstruction of the WHV small spherical SVP under 0.1% NP-40 treatment. The 9.7-Å cryo-EM density map was radially colored and viewed at twofold axis. The asymmetric unit is encompassed by three symbols. Octahedral symmetry axes are indicated with red symbols: twofold (oval), threefold (triangle), and fourfold (square). The 3D reconstruction was contoured at the level where the electron density covers the atomic structure. (E) Segmented cryo-EM density map of S-WH dimer was flexibly fitted with atomic structure.
Lipid in the SVP of both WHV and HBV
Structurally, we observed a different amount of lipid distribution in WHV and HBV spherical SVPs through cryo-EM reconstructions. Our data indicated that the particles were held by hydrophobic interactions (fig. S8). Each dimer was arranged to have the hydrophilic residues exposed at the outmost and inmost surfaces of the particles (fig. S8). We wondered what would happen to the particle if the lipids were removed.
To test the role of lipid in particle stability, we used nonionic detergent Nonidet P-40 (NP-40) at different concentrations to remove the lipid from SVPs without denaturing the proteins (Fig. 4C). Remarkably, NP-40 decreased the SVP integrity of both WHV and HBV as judged by NAGE Western blot analysis and negative-stain TEM (Fig. 4C). The HBV SVPs were completely collapsed into smaller density under only 0.1% NP-40 treatment (Fig. 4C, top). The protein band migrated slightly faster compared to the untreated sample (Fig. 4C, top, lanes 1 and 2). Increasing NP-40 concentrations further broke these structures into smaller globular density with different sizes, resulting in slower and broader migration on NAGE (Fig. 4C, top, lanes 3 and 4). A possible explanation is that the small surface antigen contains overall positive net charges, which could slow down the protein migration if totally exposed in the disassembled intermediates. Nevertheless, the surface antigen signals were noticeably reduced with the amount of added NP-40 (Fig. 4C), which is consistent with an earlier finding (38).
In contrast to HBV, some WHV SVPs maintained the particle integrity even under 1% NP-40 treatment (Fig. 4C, bottom), suggesting qualitatively that the amount of lipid content was higher in the WHV SVPs or that the lipid was more difficult to be extracted. We attempted to reconstruct WHV SVP treated with 0.1% NP-40 using cryo-EM; however, the resolution was limited to 9.7 Å. The low particle number and the possible partially defected surface organization could account for the reduction of the final resolution (Fig. 4D). Discernable differences on the helical region were observed. First, weaker densities appeared at H1, H2, and H4 helices (Fig. 4E). The weaker densities suggest that parts of the helical domain at these regions (H1, amino acids 3 to 21; H2, amino acids 80 to 93; and H4, amino acids 209 to 222) correspond to a transiently occupied conformation. Second, the helices at the threefold axis (A subunit: H3 and H4) have less lipid content compared to the equivalent helices at the fourfold axis (B subunit: H3 and H4). Once these lipids were removed, the helical domains became more flexible (39). Together, our results indicated that lipids play an important role in stabilizing the structure of SVPs.
DISCUSSION
For many years, the reported structures of HBV spherical SVP were substantially conflicting (14–17). None of the 3D reconstructions have shown clear density of the helices, although it has been long proposed on the basis of sequence, mutational, and biophysical analysis (Fig. 1). The lack of high-resolution structure has hampered our understanding of the spatial organization of the surface antigen and the SVPs. In this study, we used cryo-EM single-particle analysis to examine WHV and HBV spherical SVPs isolated from the serum samples from infected woodchucks and patients, respectively. To our knowledge, these are the first 3D models of spherical SVPs resolved to subnanometer resolution (Figs. 2E and 3E). Our cryo-EM structures showed that both SVPs were organized into rhombicuboctahedral-like particles with approximately 23-nm diameter. Each particle contains 24 asymmetric S surface protein dimers assembled with discrete lipid patches, which is different from the canonical lipid bilayer organization. To understand the mechanism of particle formation, we further used cryo-EM density map to model the predicted structures from AlphaFold2 and obtained the first reliable atomic structures for both WHV and HBV S proteins (Figs. 2F and 3F).
S dimerization is the building block of small spherical SVP for both WHV and HBV
Previous studies postulated the presence of S protein dimerization through molecular modeling (14) and mutagenesis (9, 31, 32, 40, 41), yet controversial results were also reported on the basis of fluorescence correlation spectroscopy, AFM, and cryo-EM (16, 18, 42). Our cryo-EM 3D reconstruction and segmentation provided direct visualizing evidence for identifying the oligomerization state of S protein in the SVPs (Figs. 2F and 3F and fig. S3B). Using asymmetric reconstruction, we have revealed that the SVPs arranged into a rhombicuboctahedron-like surface lattice with 24 protruding spikes (fig. S2). Similar symmetrical architecture exists in the COPII (coat protein complex II)–coated vesicular transportation machinery at the endoplasmic reticulum (ER) (43). Whether the SVPs utilize COPII vesicles for trafficking to the ER-Golgi intermediate compartment needs further investigation (44). By imposing octahedral symmetry, we were able to increase the final resolution and identified that each spike is composed of an asymmetric S protein dimer. Therefore, a total of 48 S proteins were found in the spherical SVP. Our result is markedly different to the number of 70 to 100 copies of S protein per particle reported earlier (14, 16, 18, 41, 42, 45). This discrepancy can be explained by different level of heterogeneity in the SVP samples. We have observed a wide range of particle sizes (Figs. 2D and 3D and fig. S1). The particles with larger diameter have noticeable defects at the surface (fig. S1) and were excluded from the image analysis. Likewise, the tubular SVPs, which have more surface proteins, were also excluded. These particles could contribute to the calculations of the S protein number in these earlier studies.
Combining cryo-EM density map and predicted AlphaFold2 structure, we identified that H1/H4, H2/H2, AGL, and CYL were involved in the S protein dimerization contact. Mutagenesis analysis on S-HBs showed the replacement of residues Phe158, Trp165, and other residues involved in the hydrophobic surface of the amphipathic helix (amino acids 156 to 169 in H3), as well as Arg79 and part of the H2 (amino acids 80 to 98) significantly reduced SVP secretion (10, 31, 32), which were also found to be involved in multiple protein interactions at intrasubunit (monomer) or intersubunit (dimer) (fig. S7). Moreover, our structure confirmed the unique epitope sequence (amino acids 178 to 186) recognized by anti-HBsAg monoclonal antibody (Mab) 4-7B is surface exposed at H3 (46). Consistent with our 3D model, mutational analysis at amino acids 8 to 22 (H1), deletion of amino acids 24 to 28 (H1), and deletion of amino acids 178 to 226 (H3 and H4) still allowed SVP formations, suggesting that these regions are not essential for S protein oligomerization (31). Despite that AlphaFold2 predicted models with high precision for the four helical domains, including a horizontal H3 helix laid parallelly on the particle surface, the results for nonsecondary structural regions need to be viewed with great caution (figs. S3, S4, and S7 and table S1). Among all predicted models, the spatial organization for the loop regions was highly divergent and the angles for the preceding and following helices were different. In WHV and HBV, the orientation of the S protein dimer residing within the particles was different (figs. S4 and S5). We speculate that this difference originates from the distinct protein folding in the AGL region. Although the protein sequence at AGL is variable (Fig. 1B), the eight cysteine residues in this region are highly conserved and directly involved in disulfide bridge formation and/or particle secretion (9, 47). It is possible that the disulfide isomerase at the ER catalyzes a different level of intra- and intermolecular disulfide bond formation between human and woodchuck surface proteins, which allows proteins to fold differently and supports the rotation of the S dimer observed here (Figs. 2E and 3E and fig. S5). Our cryo-EM density maps do not have a resolution high enough to allow directly modeling the loop region, which needs to be clarified in further research.
Noncanonical membrane organization of WHV and HBV spherical SVPs
We did not observe a typical lipid bilayer structure in both WHV and HBV spherical SVPs (Figs. 2 and 3 and fig. S6). Although 2D class averages and 3D reconstruction vaguely showed two layers of density, our cryo-EM analysis revealed that the outer layer density was composed of H3 laid parallelly to the plane of the particle surface together with discrete lipid patches that were located between different helices. The inner-layer density, which originally faced the cytosolic side when inserted at the ER membrane, was formed by the C-terminal region of H1, CYL, and the N-terminal region of H2 to give rise to a highly curved hydrophilic surface (Figs. 2 and 3and fig. S8). The tubular-shaped helical domain has much higher density than the mass of lipid moiety, which makes it more apparent when contoured at the higher density threshold (fig. S3B).
The orientation of the surface antigens in the spherical SVP allowed the innermost and outermost surface to be hydrophilic and enclosed the hydrophobic helices in the middle (fig. S8). This spatial arrangement and the protein-protein interaction make SVPs quite different from low-density lipoproteins (LDLs). Unlike LDLs, spherical SVPs have a higher protein-to-lipid ratio (table S3) and an ordered inner core (fig. S2) (23, 42). The exposed hydrophobic areas were covered by the lipid patches (Fig. 4, A and B, and fig. S6), presumably to stabilize the particles. We have found that removing the lipid content by NP-40 destabilized the particles (Fig. 4C) and unwound part of the helix domains (Fig. 4, D and E). Furthermore, electron spin resonance, AFM, and time-resolved fluorescence spectroscopy experiments suggest that the lipid molecules are in tight association with the S protein, resulting in an immobilized state (14, 22, 41, 42), different from the fluid mosaic lipid molecules in the traditional bilayer conformation. On the basis of the protein arrangements and the noncanonical lipid patches revealed in both WHV and HBV SVPs, our results indicate that spherical SVPs are lipid-stabilized “protein-like” particles (Figs. 2E and 3E).
The morphogenesis of spherical SVPs starts with S protein translocation at the ER membrane, dimer formation, transport to the ER-Golgi intermediate compartment, and oligomerization into particles (4, 32, 48). A perplexing and unanswered question is how the particle formation sheds the inner lipid layer. Although the detailed mechanism remains to be further elucidated, we can offer some possible explanations. Because of the lateral interaction between individual S protein dimers, the membrane curvature brings the CYL regions closely together and displaces the cytosolic lipids (21, 22). Our cryo-EM results also support this hypothesis. The spherical SVPs have an inner core of ~7.0 nm in diameter (181 nm3 in volume), which can roughly accommodate all CYL regions from 48 subunits (180 nm3 in volume), leaving almost no space left for the lipid membrane.
Docking the atomic models into cryo-EM density maps, we found that WHV spherical SVPs contain more lipid (~25% v/v) than HBV spherical SVPs (~20% v/v; table S3). Detergent-based lipid removal also showed that WHV spherical SVPs have a higher resistance to NP-40 treatment (fig. S4). Because lipid and protein interactions play an important role in functional and structural integrity of the S protein (41, 49), removal of lipids breaks the lipid-protein interface, decreases the helical content, reduces the antigenic activity of the particles, and affects the assembly of SVPs (13, 38). In WHV, the intrasubunit and intersubunit interactions of the S protein are stronger than in HBV (fig. S7 and table S1). The total hydrophobic interaction between S protein dimers at different symmetrical axes is also higher in WHV than in HBV spherical SVPs (table S2). Together, our results revealed that the WHV spherical SVP is more stable than its HBV counterpart.
Conformational flexibility of spherical SVP
The resolution of our cyro-EM 3D reconstruction was limited to 6.5 and 6.3 Å for WHV and HBV, respectively. There are several potential reasons that hampered the overall resolution achieved here, and it is likely that more than one is a contributing factor. First, the two flexible loop regions CYL and AGL could affect the estimated resolution. The cryo-EM structure at the AGL region had the weakest density (fig. S3B) and had the lowest local resolution (fig. S2, D and E). Likewise, AlphaFold2 also predicted different structures with various secondary structural elements for the AGL and CYL regions at very low confidence score. Because of the poorly resolved density, we could not confidently model structures for these two loop regions into the cryo-EM density (fig. S3C). In addition, it is possible the asymmetry of each spike correlated with only half of the AGLs in the SVP was found to be N-glycosylated; yet the distribution and composition of the N-glycans on the particle remain to be determined (6, 7).
Second, the M protein also participates in the assembly of spherical SVP, albeit at a lesser amount than S protein (50, 51). The additional residues of the PreS2 domain at its N terminus were found to be exposed at the particle surface (52) and may sterically hinder the neighboring subunits. If there were only a few M proteins randomly incorporated in the particle, then their densities would be averaged out by the applied octahedral symmetry during our data processing. It is intriguing to think that the particles with smear surface, as seen in fig. S1, were the result of several M proteins accumulated locally that interrupted the particle lattice. Last, the nonuniform lipid density on the SVP also affected the resolution estimation (fig. S2D). Lipid, whose density is less well defined compared to the helices, is important to stabilize the SVP structures. Although lipid molecules were tightly associated with the S protein (21, 22), they also have a low local resolution (fig. S2), probably because of their variation in fatty acid compositions and orientation related to the hydrophobic surface. Together, the conformational flexibility and compositional heterogeneity of the surface proteins and lipids could prevent the determination of high-resolution structures.
In summary, we have solved the highest resolution of spherical SVP for both WHV and HBV to subnanometer resolution using cryo-EM to date. In combination with predictions from AlphaFold2, we have presented the detailed atomic structure of small surface antigens for WHV and HBV. We have described the structure and assembly mechanisms of the spherical SVP, which are composed of 24 dimers of small surface antigens. As the nanoparticle is a potentially effective platform for modern vaccine design (53), improvements in our understanding of these mechanisms might facilitate the design of new antiviral strategies in the future.
MATERIALS AND METHODS
Purification and identification of SVPs
Infection of woodchucks with the WHV7 has been described, and serum samples from a woodchuck with chronic WHV infection were used (54). Serum of HBsAg-positive patients infected with HBV GtE was obtained from BioCollections Worldwide Inc. (Miami, FL).
Serum sample containing SVPs was fractionated by CsCl gradient ultracentrifugation as described previously (55). Briefly, 1 ml of serum was subjected to CsCl [0.25 g/ml in 10 mM tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), and 100 mM NaCl] at 50,000g for 96 hours at 4°C. The purified fractions or serum samples were resolved on a 1% NAGE. After transferring the viral particles to a nitrocellulose membrane, SVPs were detected by a polyclonal rabbit anti-HB antibody (ViroStat), which cross-react with WHs (56). The refractive index of fractions was measured with a spectronic refractometer (Thermo Fisher Scientific, 334610).
All SVP fractions were negatively stained with 2% uranyl acetate on continuous carbon grids and examined by a JEOL 2100 (Lab6) transmission microscope operating at 200 kV. Selected samples were dialyzed against TNE buffer [10 mM tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA (pH 8.0)] for future use.
Sequence alignment and helix prediction
Protein sequence alignments between WHV7 and HBV GtE were performed using Clustal Omega. The NCBI database accession numbers for WHV7 and HBV GtE surface protein are M18752.1 and KU702884.1, respectively. The transmembrane helix was identified using the TMHMM server (57).
Negative-stain TEM
Samples were applied on a glow-discharged grid coated with continuous carbon film and stained with 2% (w/v) uranyl acetate. Images were acquired using JEM-2100 equipped with Gatan UltraScan 4000. The particle number varied in different regions on the grid.
Cryo–electron microscopy
Grid preparation and data collection followed the standard protocol. Briefly, selected SVP fraction (F16) was dialyzed against TNE buffer [10 mM tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA (pH 8.0)] twice at 4°C to remove cesium salts using Slide-A-Lyzer mini dialysis devices [20 kDa, molecular weight cutoff (MWCO); Thermo Fisher Scientific]. Approximately 4 μl of dialyzed SVP fraction was applied to either ultrathin continuous or Quantifoil R 2/2 holey carbon film coating grids that were glow discharged before use to improve surface hydrophilicity. The grids were then vitrified using Thermo Fisher Scientific Vitrobot (Mark IV; blotting force, 0; blotting time, 4 s; wait time, 15 s) at 18°C under 100% humidity. The cryo-EM micrographs were taken on a Thermo Fisher Scientific Titan Krios operating at 300 kV by EPU software under superresolution and counting modes with nominal defocus range from −1.0 to −3.0 μm (table S4). Low-dose (~30 e−/Å2) images at a magnification of ×64,000 (0.678 Å per pixel) for HBV SVPs and of ×105,000 (0.84 Å per pixel) for WHV SVPs were collected on Gatan BioContinuum using an energy slit of 30 eV (table S4).
Image processing and 3D reconstruction
Dose-weighted motion correction was done with MotionCor2, and contrast transfer function parameters were estimated using ctffind4 in RELION (v3.1) software (27).
The same data process scheme was applied to the datasets of both WHV and HBV SVPs. In general, particles were semimanually picked and subjected to reference-free 2D classification using RELION (27). After building an initial model for each dataset using C1 symmetry, low-resolution 3D models were obtained and further used as reference templates for automated particle picking from all micrographs in RELION. Multiple times of reference-free 2D classification were performed to discard classes with blurred and ice-contaminated density. The model applied for particle picking was also used for initial 3D classification and 3D refinement using C1 symmetry. Data processing started with downscaled pixel size at bin = 8 and gradually moved up to bin = 2. After defining the symmetry, cryo-EM density maps were subsequently refined using their C1 models aligned to octahedral symmetry from the 3D classification results. The refinement process was continued until no further improvement occurred, which was followed by a postprocessing step using B-factor sharpening. A total number of 34,520 particles were used in the final 3D reconstruction at a resolution of 6.5 Å for WHV SVP, and 36,166 particles were used to build the final 3D reconstructions at a resolution of 6.3 Å for HBV SVP. The resolution of the map is determined by Fourier shell correlation criteria at 0.143 cutoff (fig. S2C).
Visualization is done in UCSF ChimeraX (v1.3) with “hide dust” function (4.3 for WHV and 6.8 for HBV) to remove some noise when composing figures (58). All protein subunit densities were segmented from DeepEMhancer (59) postprocessing maps using ChimeraX. Local resolution was estimated within Relion. We used the average protein density of 1.3 g/cm3 for volume calculation.
Protein structure predictions and atomic model buildup
At the current resolution, the information is insufficient to build a reliable atomic model directly from the density; however, the helices can already be identified (fig. S3B). AlphaFold2 was used to predict small surface protein structures using Google Colab notebook AlphaFold2_advanced.ipynb (33). No template information was used for structure modeling. Fast method from ColabFold was applied for multiple sequence alignment. For all targets, five models were generated. All of them had high confidence scores for helical regions but relative low confidence scores for loops. Despite the length and conformation of the helices from different models were virtually identical, the loops that connected between helices were orientated differently (fig. S4, A and B).
The predicted atomic models of the S surface protein were docked into the segmented cryo-EM density map as one rigid body using ChimeraX to find the best fitting model. Flexible model fitting was performed using ISOLDE (34), accessed through ChimeraX (58). Additional real-space refinement was performed using Coot (35). The side-chain orientation movement and the atom locations were refined locally according to the published data on the protein-protein interactions and used the weighted sum of the electron-density values over the atomic centers. Ramachandran plot, bond geometry, rotamers, and clashes were also optimized to obtain the final 3D models.
The optimized model was used to construct a dimer model according to the segmented dimer density using flexible fitting steps as described above. Side-chain clashes and Ramachandran were detected using MOLPROBITY (60). The dimer model was then rigid body fitted into the complete DeepEMhancer postprocessing map using the “Fit in Map” function in ChimeraX. Full particle model was then generated using octahedral symmetry operation.
Contact and interaction analysis between subunits was performed in ChimeraX. Lipid density calculation was performed by subtracting low-pass–filtered WHV and HBV SVP atomic structures (using molmap to generate maps at 6.5 and 6.3 Å, respectively) from the cryo-EM maps. CYL and AGL loop regions were excluded in the calculation. Buried surface area of hydrophobic residues (Ala, Val, Ile, Leu, Met, Phe, Tyr, and Trp) and surface hydrophobicity rendering were done in ChimeraX.
Removal of lipid from the SVP surface
The dialyzed CsCl fractions rich in SVPs from both HBV and WHV were incubated with NP-40 at room temperature for 30 min. The NP-40 was added before the start of incubation at a final concentration of 0, 0.1, 0.5, and 1%. The mixture was checked by NAGE and negative stain as previously described above. Cryo-EM analysis of 0.1% NP-40–treated WHV spherical SVPs was performed following the same procedure described above (table S4).
Acknowledgments
We thank A. Zlotnick for insightful discussion. We thank The Pennsylvania State University College of Medicine for access to Cryo-EM (RRID: SCR_021178) and the HPC core facilities.
Funding: The work was supported by the startup fund from The Pennsylvania State University College of Medicine and NIH grant R21-AI1641191 to J.C.-Y.W. and R37AI043453 to J.H.
Author contributions: Conceptualization: H.L. and J.C.-Y.W. Methodology: H.L. and J.C.-Y.W. Investigation: H.L., X.H., J.X., and J.C.-Y.W. Visualization: H.L. and J.C.-Y.W. Supervision: S.M., J.H., and J.C.-Y.W. Writing—original draft: H.L. and J.C.-Y.W. Writing—review and editing: H.L., X.H., J.X., S.M., J.H., and J.C.-Y.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Electron Microscopy Data Bank (EMDB) accession code for WHV SVP is EMD-26118, for HBV SVP is EMD-26117, and for WHV SVP with 0.1% NP-40 treatment is EMD-26119. Details describing these reconstructions are in table S4.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S4
REFERENCES AND NOTES
- 1.Revill P. A., Chisari F. V., Block J. M., Dandri M., Gehring A. J., Guo H., Hu J., Kramvis A., Lampertico P., Janssen H. L. A., Levrero M., Li W., Liang T. J., Lim S. G., Lu F., Penicaud M. C., Tavis J. E., Thimme R., Zoulim F., Arbuthnot P., Boonstra A., Chang K. M., Chen P. J., Glebe D., Guidotti L. G., Fellay J., Ferrari C., Jansen L., Lau D. T. Y., Lok A. S., Maini M. K., Mason W., Matthews G., Paraskevis D., Petersen J., Rehermann B., Shin E. C., Thompson A., van Bömmel F., Wang F. S., Watashi K., Yang H. C., Yuan Z., Yuen M. F., Block T., Miller V., Protzer U., Bréchot C., Locarnini S., Peters M. G., Schinazi R. F., A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 4, 545–558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block T. M., Chang K.-M., Guo J.-T., Prospects for the global elimination of hepatitis B. Annu. Rev. Virol. 8, 437–458 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Hu J., Liu K., Complete and incomplete hepatitis B virus particles: Formation, function, and application. Viruses 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seitz S., Habjanič J., Schütz A. K., Bartenschlager R., The hepatitis B virus envelope proteins: Molecular gymnastics throughout the viral life cycle. Annu. Rev. Virol. 7, 263–288 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Eddleston A., Hepatitis. Lancet 335, 1142–1145 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Tolle T. K., Glebe D., Linder M., Linder D., Schmitt S., Geyer R., Gerlich W. H., Structure and glycosylation patterns of surface proteins from woodchuck hepatitis virus. J. Virol. 72, 9978–9985 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson D. L., Nath N., Gavilanes F., Structure of hepatitis B surface antigen. Correlation of subtype with amino acid sequence and location of the carbohydrate moiety. J. Biol. Chem. 257, 10414–10420 (1982). [PubMed] [Google Scholar]
- 8.Zeyen L., Döring T., Stieler J. T., Prange R., Hepatitis B subviral envelope particles use the COPII machinery for intracellular transport via selective exploitation of Sec24A and Sec23B. Cell. Microbiol. 22, e13181 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Mangold C., Streeck R. E., Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J. Virol. 67, 4588–4597 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Löffler-Mary H., Dumortier J., Klentsch-Zimmer C., Prange R., Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270, 358–367 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Carman W. F., Zanetti A. R., Karayiannis P., Waters J., Manzillo G., Tanzi E., Zuckerman A. J., Thomas H. C., Vaccine-induced escape mutant of hepatitis B virus. Lancet 336, 325–329 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Mangold C. M., Unckell F., Werr M., Streeck R. E., Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology 211, 535–543 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Ho J. K.-T., Jeevan-Raj B., Netter H.-J., Hepatitis b virus (HBV) subviral particles as protective vaccines and vaccine platforms. Viruses 12, 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert R. J., Beales L., Blond D., Simon M. N., Lin B. Y., Chisari F. V., Stuart D. I., Rowlands D. J., Hepatitis B small surface antigen particles are octahedral. Proc. Natl. Acad. Sci. U.S.A. 102, 14783–14788 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Short J. M., Chen S., Roseman A. M., Butler P. J. G., Crowther R. A., Structure of hepatitis B surface antigen from subviral tubes determined by electron cryomicroscopy. J. Mol. Biol. 390, 135–141 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Mulder A. M., Carragher B., Towne V., Meng Y., Wang Y., Dieter L., Potter C. S., Washabaugh M. W., Sitrin R. D., Zhao Q., Toolbox for non-intrusive structural and functional analysis of recombinant VLP based vaccines: A case study with hepatitis B vaccine. PLOS ONE 7, e33235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J., Zhang J., Lu Y., Luo S., Zhang J., Zhu P., Cryo-EM structure of native spherical subviral particles isolated from HBV carriers. Virus Res. 259, 90–96 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Milhiet P.-E., Dosset P., Godefroy C., le Grimellec C., Guigner J. M., Larquet E., Ronzon F., Manin C., Nanoscale topography of hepatitis B antigen particles by atomic force microscopy. Biochimie 93, 254–259 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Aggerbeck L. P., Peterson D. L., Electron microscopic and solution X-ray scattering observations on the structure of hepatitis B surface antigen. Virology 141, 155–161 (1985). [DOI] [PubMed] [Google Scholar]
- 20.Gavilanes F., Gonzalez-Ros J. M., Peterson D. L., Structure of hepatitis B surface antigen. Characterization of the lipid components and their association with the viral proteins. J. Biol. Chem. 257, 7770–7777 (1982). [PubMed] [Google Scholar]
- 21.Satoh O., Umeda M., Imai H., Tunoo H., Inoue K., Lipid composition of hepatitis B virus surface antigen particles and the particle-producing human hepatoma cell lines. J. Lipid Res. 31, 1293–1300 (1990). [PubMed] [Google Scholar]
- 22.Satoh O., Imai H., Yoneyama T., Miyamura T., Utsumi H., Inoue K., Umeda M., Membrane structure of the hepatitis B virus surface antigen particle. J. Biochem. 127, 543–550 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Grélard A., Guichard P., Bonnafous P., Marco S., Lambert O., Manin C., Ronzon F., Dufourc E. J., Hepatitis B subvirus particles display both a fluid bilayer membrane and a strong resistance to freeze drying: A study by solid-state NMR, light scattering, and cryo-electron microscopy/tomography. FASEB J. 27, 4316–4326 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Galibert F., Chen T. N., Mandart E., Nucleotide sequence of a cloned woodchuck hepatitis virus genome: Comparison with the hepatitis B virus sequence. J. Virol. 41, 51–65 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suresh M., Menne S., Application of the woodchuck animal model for the treatment of hepatitis B virus-induced liver cancer. World J. Gastrointest. Oncol. 13, 509–535 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerhardt E., Bruss V., Phenotypic mixing of rodent but not avian hepadnavirus surface proteins into human hepatitis B virus particles. J. Virol. 69, 1201–1208 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheres S. H., Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Hardy J. M., Newton N. D., Modhiran N., Scott C. A. P., Venugopal H., Vet L. J., Young P. R., Hall R. A., Hobson-Peters J., Coulibaly F., Watterson D., A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses. Nat. Commun. 12, 3266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonveaux N., Conrath K., Capiau C., Brasseur R., Goormaghtigh E., Ruysschaert J. M., The topology of the S protein in the yeast-derived hepatitis B surface antigen particles. J. Biol. Chem. 269, 25637–25645 (1994). [PubMed] [Google Scholar]
- 30.Pintilie G. D., Zhang J., Goddard T. D., Chiu W., Gossard D. C., Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suffner S., Gerstenberg N., Patra M., Ruibal P., Orabi A., Schindler M., Bruss V., Domains of the hepatitis B virus small surface protein S mediating oligomerization. J. Virol. 92, e02232–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S., Shen Z., Kang Y., Sun L., Viswanathan U., Guo H., Zhou T., Dai X., Chang J., Zhang J., Guo J. T., A putative amphipathic alpha helix in hepatitis B virus small envelope protein plays a critical role in the morphogenesis of subviral particles. J. Virol. 95, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casañal A., Lohkamp B., Emsley P., Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1055–1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangold C., Unckell F., Werr M., Streeck R., Analysis of intermolecular disulfide bonds and free sulfhydryl groups in hepatitis B surface antigen particles. Arch. Virol. 142, 2257–2267 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Prange R., Nagel R., Streeck R. E., Deletions in the hepatitis B virus small envelope protein: Effect on assembly and secretion of surface antigen particles. J. Virol. 66, 5832–5841 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavilanes F., Gomez-Gutierrez J., Aracil M., Gonzalez-Ros J. M., Ferragut J. A., Guerrero E., Peterson D. L., Hepatitis B surface antigen. Role of lipids in maintaining the structural and antigenic properties of protein components. Biochem. J. 265, 857–864 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh J. A., Buried and accessible surface area control intrinsic protein flexibility. J. Mol. Biol. 425, 3250–3263 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Wunderlich G., Bruss V., Characterization of early hepatitis B virus surface protein oligomers. Arch. Virol. 141, 1191–1205 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Zhao Q., Wanga Y., Freedb D., Fub T. M., Gimeneza J. A., Sitrina R. D., Washabaugh M. W., Maturation of recombinant hepatitis B virus surface antigen particles. Hum. Vaccin. 2, 174–180 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Greiner V. J., Egelé C., Oncul S., Ronzon F., Manin C., Klymchenko A., Mély Y., Characterization of the lipid and protein organization in HBsAg viral particles by steady-state and time-resolved fluorescence spectroscopy. Biochimie 92, 994–1002 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Hughson F. M., Both Layers of the COPII Coat Come into View. Cell 134, 384–385 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Zeyen L., Döring T., Prange R., Hepatitis B virus exploits ERGIC-53 in conjunction with COPII to exit cells. Cells 9, 1889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegler V. D., Bruss V., Role of transmembrane domains of hepatitis B virus small surface proteins in subviral-particle biogenesis. J. Virol. 87, 1491–1496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulij W., de Wit P. L. M., Sünnen C. M. G., van Roosmalen M. H., Petersen-van Ettekoven A., Cooreman M. P., Heijtink R. A., Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. J. Gen. Virol. 80, 2121–2126 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Cheong W.-S., Hyakumura M., Yuen L., Warner N., Locarnini S., Netter H. J., Modulation of the immunogenicity of virus-like particles composed of mutant hepatitis B virus envelope subunits. Antiviral Res. 93, 209–218 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Huovila A. P., Eder A. M., Fuller S. D., Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118, 1305–1320 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta K., Donlan J. A. C., Hopper J. T. S., Uzdavinys P., Landreh M., Struwe W. B., Drew D., Baldwin A. J., Stansfeld P. J., Robinson C. V., The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheu S. Y., Lo S. J., Biogenesis of the hepatitis B viral middle (M) surface protein in a human hepatoma cell line: Demonstration of an alternative secretion pathway. J. Gen. Virol. 75 ( Pt. 11), 3031–3039 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Gerlich W. H., Medical virology of hepatitis B: How it began and where we are now. Virol. J. 10, 239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng K. C., Moss B., Selective synthesis and secretion of particles composed of the hepatitis B virus middle surface protein directed by a recombinant vaccinia virus: Induction of antibodies to pre-S and S epitopes. J. Virol. 61, 1286–1290 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen B., Tolia N. H., Protein-based antigen presentation platforms for nanoparticle vaccines. npj Vaccines 6, 70 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suresh M., Czerwinski S., Murreddu M. G., Kallakury B. V., Ramesh A., Gudima S. O., Menne S., Innate and adaptive immunity associated with resolution of acute woodchuck hepatitis virus infection in adult woodchucks. PLOS Pathog. 15, e1008248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi J., Liu H., Hu J., Regulation of hepatitis B virus virion release and envelopment timing by nucleocapsid and envelope interactions. J. Virol. 96, e0130521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong X., Luckenbaugh L., Perlman D., Revill P. A., Wieland S. F., Menne S., Hu J., Characterization and application of precore/core-related antigens in animal models of hepatitis B virus infection. Hepatology 74, 99–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J. M., Sorzano C. O. S., Vargas J., DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure Validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4