Abstract
Improving the thermal stability of biologics, including vaccines, is critical to reduce the economic costs and health risks associated with the cold chain. Here, we designed a versatile, safe, and easy-to-use reversible PEG-based hydrogel platform formed via dynamic covalent boronic ester cross-linking for the encapsulation, stabilization, and on-demand release of biologics. Using these reversible hydrogels, we thermally stabilized a wide range of biologics up to 65°C, including model enzymes, heat-sensitive clinical diagnostic enzymes (DNA gyrase and topoisomerase I), protein-based vaccines (H5N1 hemagglutinin), and whole viruses (adenovirus type 5). Our data support a generalized protection mechanism for the thermal stabilization of diverse biologics using direct encapsulation in reversible hydrogels. Furthermore, preliminary toxicology data suggest that the components of our hydrogel are safe for in vivo use. Our reversible hydrogel platform offers a simple material solution to mitigate the costs and risks associated with reliance on a continuous cold chain for biologic transport and storage.
A wide range of biologics, including viruses and vaccines, was thermally stabilized using reversible hydrogels.
INTRODUCTION
Biologics—defined here as biologically derived complex macromolecules used as pharmaceutical products—comprise most new drugs entering the market, including 8 of the top 10 grossing drug products globally in 2019 (1). Biologics include peptides, recombinant therapeutic proteins, enzymes, monoclonal antibodies, antibody-drug conjugates, vaccines, and viral vectors. The specificity and potency of biologics arise from their macromolecular composition, which provides chemical diversity and structural complexity (2). This complexity, however, means that biologics are often susceptible to physical and chemical deterioration, making them challenging to formulate, store, and even transport (3). In particular, the bioactivity of biologics is commonly reduced by thermal fluctuations during storage and transit, as aggregation, denaturation, and oxidative degradation render them ineffective (4). To ensure stable bioactivity of biologics, substantial resources are dedicated to maintaining a continuous cold chain throughout their manufacture and distribution; in 2020, the overall market for cold chain services was $17.2 billion and forecasted to rise to $21.3 billion by 2024 (5, 6). In addition to the economic burden, failures in the cold chain can lead to devastating consequences for public health, as in the case for ineffective vaccines (7, 8). Therefore, enabling ambient-temperature transportation and storage is an attractive strategy to reduce cold chain reliance and to minimize the associated economic costs and health risks.
Several approaches have been explored to improve the thermal stability of biomacromolecules (5). Biologics have been engineered during drug development (9–12) or conjugated to other molecules (13, 14) specifically to improve thermal stability. While extensive protection from thermal stresses has been achieved in some cases, directly altering biologics carries the risk of diminished drug efficacy and it is often difficult to generalize these successes to other biologics. Furthermore, additional regulatory approval is required for each new formulation. Another method to increase thermal stability is the removal of the solvent (usually water) from the formulation by lyophilization (freeze-drying), spray drying, or vacuum foam drying (15). This creates a solid powder that can be suitable for shipping without refrigeration (4). However, these drying approaches often expose the biologics to harsh processing conditions and, in some cases, the biologics remain susceptible to thermal stresses (16). Because of this, most freeze-dried formulations require stabilizers—for example, sugars, such as sucrose or trehalose, and polymers, such as poly(ethylene glycol) (PEG)—during drying. These excipients slow the loss of bioactivity by interacting with the biologics to maintain their preferred hydration state and/or to prevent aggregation (17). Most commonly used excipients, however, were developed for small-molecule drugs and were not optimized to stabilize biologics. Recently, advanced excipients have been developed to increase the thermal stability of biologics, including through the synthesis of trehalose glycopolymers (18), the design of random heteropolymers (19), and the use of anionic gold nanoparticles (20). Despite these advances, there are few available solutions that can effectively stabilize a broad selection of biologics with little formulation work required. Hence, there is a pressing need to develop next-generation excipients specifically designed to enhance the thermal stability of a diverse range of biologics.
A complementary and promising strategy to impart thermal stabilization is to immobilize biologics directly within a protective matrix that holds the molecules in place to resist conformational changes (5, 21). The biologics can be encapsulated within rigid crystalline materials, such as metal-organic frameworks (22–24), silica (25, 26), or sugar glasses (27, 28), or they can be immobilized within flexible polymeric matrices, such as hypromellose (29), silk fibroin–based biomaterials (30, 31), photodegradable hydrogels (32), or nanoparticle-based supramolecular hydrogels (33). Furthermore, the polymers comprising the network can be modified with functional groups that directly interact with regions of the protein, imparting additional stabilizing effects. Examples include hydrogels functionalized with trehalose (34, 35) and protein-adsorbing charged nanogels (36). One of the major challenges for matrix-based approaches, however, is to achieve on-demand release of the stabilized biologic from the protective matrix. Network degradation has been triggered by external stimuli such as pH (37) or light (32), but often these systems are limited by cost and the stimuli themselves can be harmful to the biologics. Another significant challenge for matrix-based approaches is to encapsulate biologics under gentle conditions. To successfully use hydrogels as stabilizing excipients for biologics, we need to design materials that allow mild encapsulation, effective thermal stabilization, and facile, on-demand recovery of the biologics.
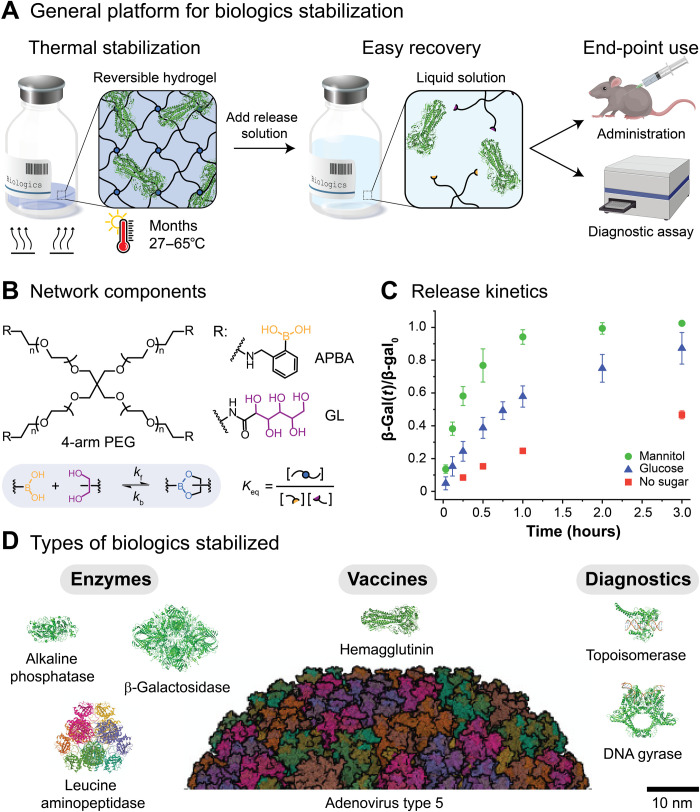
Therefore, to provide a user-friendly solution for the encapsulation, stabilization, and on-demand release of biologics, we designed a safe and easy-to-use reversible hydrogel for versatile stabilization of a broad range of biologics against thermal stress (Fig. 1A). Our approach was based on the use of dynamic covalent boronic ester–based cross-links, which readily form under physiological conditions (Fig. 1B). Boronic ester–based hydrogels have been used as responsive drug delivery systems, as dynamic cell culture scaffolds, and as sugar-sensing materials (37). Dynamic covalent hydrogels integrate properties of both chemically and physically cross-linked networks, enabling the formation of robust yet processable biomaterials that are self-healing and stimuli responsive (38, 39). Furthermore, the properties of dynamic covalent networks, such as the gel point, the plateau shear modulus, and the mechanical relaxation time, are dictated by the molecular behavior of the reversible cross-links (40). In the case of boronic ester–based materials, the stability of the cross-links is determined by the local environment, e.g., T, pH, and the concentration of competitive binders. Thus, on-demand recovery of the biologics is enabled by the addition of sugars that could displace the diols from the boronic ester cross-links, leading to erosion-mediated dissolution of the networks (Fig. 1C) (37). As a whole, we hypothesized that dynamic covalent networks would enable robust network formation to prevent translation and aggregation of the biologics while providing a handle for triggered dissolution, release, and recovery of the biologics.
Fig. 1. Thermal stabilization of biologics using reversible hydrogels.
(A) The thermal stabilization platform is based on dynamic covalent boronic ester–based hydrogels, where boronic acids (yellow) and diols (purple) form reversible boronic ester cross-links (blue). After encapsulation within the reversible hydrogels, the stored biologics are protected from thermal stresses. On-demand recovery and subsequent use of the biologics are enabled by the addition of sugars, which triggers network dissolution. (B) The reversible boronic ester–based hydrogels were formed from four-arm PEG macromers (Mn ≈ 10,000 g mol−1) end-functionalized with a phenylboronic acid derivative (APBA) or a cis-1,2-diol containing moiety [gluconolactone (GL)]. (C) The fractional release of β-galactosidase over time, β-gal(t)/β-gal0, from the PEG-APBA/GL gels [7.5% (w/w)] is modulated by the addition of cis-diol containing sugars such as mannitol and glucose (100 mg ml−1, pH 7.5). The release rate depends on the magnitude of the equilibrium binding constant, Keq,D, between PEG-APBA and the added sugars (Keq,mannitol = 20 and Keq,glucose = 3) (37). (D) Reversible hydrogels are suitable for the thermal stabilization of multiple classes of biologics, including model enzymes, protein-based vaccines, whole viruses, and clinical diagnostic enzymes with sizes ranging from 10 to 100 nm. For comparison, the mesh size of the reversible hydrogels, ξ, is estimated to be 7.5 nm.
Using these reversible hydrogels, we thermally stabilized a wide range of biologics, starting with model enzymes [β-galactosidase (β-gal), leucine aminopeptidase (LAP), and alkaline phosphatase (ALP)]. Protection was attributed to the formation of polymer networks around the biologics, which restricted their mobility and physically prevented them from interacting with each other, reducing aggregation and bioactivity loss after exposure to heat. Encouraged by these results, we extended our approach to the thermal stabilization of heat-sensitive clinical diagnostic enzymes [DNA gyrase and human topoisomerase I (Top1)] and a protein-based vaccine [recombinant influenza A H5N1 hemagglutinin (HA)]. Last, we demonstrated that gel encapsulation could be used to thermally stabilize a particularly challenging class of biologics, whole viruses [adenovirus type 5 (Ad5)]. In total, our work demonstrates that reversible hydrogels are suitable for the thermal stabilization of multiple classes of biologics, with little formulation work required to stabilize each type of biologic (Fig. 1D). Furthermore, preliminary toxicology data suggest that the components of our hydrogel are safe for in vivo use. The ability to protect a wide range of biologics, including vaccines, from thermal stress with a simple material solution will help to mitigate the risks and costs associated with reliance on a continuous cold chain during transport and storage.
RESULTS
Fabrication of reversible hydrogels via boronic ester formation
As a strategy to stabilize biologics that allows on-demand release, we designed a versatile and low-cost reversible hydrogel platform based on dynamic covalent bonds (Fig. 1A). The reversible hydrogels were formed from four-arm PEG macromers (Mn ≈ 10,000 g mol−1) end-functionalized with a phenylboronic acid derivative (APBA) or a cis-1,2-diol containing moiety [gluconolactone (GL)] (Fig. 1B). Upon mixing, robust hydrogels formed quickly (~1 to 10 s) via boronic ester–based cross-links, with a final shear storage modulus in the range of G′ ≈ 5 kPa [7.5% (w/w) PEG]. The mechanical properties of the gels were not significantly affected by the biologic loading (up to 40 mg ml−1; fig. S1, A to C). Furthermore, as the dynamic covalent cross-links could break and reform under ambient conditions, the formed hydrogels (both biologic loaded and unloaded) were moldable and self-healing, which facilitated processing (fig. S1, D and E).
Hydrogel dissolution and release of biologics
Critically, the reversible boronic ester cross-links also enabled facile and on-demand recovery of the biologics, as subtle shifts in the environmental conditions can transform the material from a stable network to a soluble melt and back again (40). Biologic release from boronic ester–based dynamic covalent hydrogels is modulated by the addition of free competitor diols (such as simple sugars), as competitive displacement triggers network dissolution and release of cargo (37). The dissolution (and thus release) rate depends on the concentration of sugar added and on the magnitude of the equilibrium binding constant, Keq,D, between PEG-APBA and the added sugar. Here, dissolution of the gels upon addition of glucose (100 mg ml−1; Keq,D = 3) required much more time (>3 hours) when compared with mannitol (100 mg ml−1; Keq,D = 20), which required <1 hour (Fig. 1C). Dissolution of the gels without a sugar required >24 hours (fig. S2A). The dissolution rate also depended on the geometry of the gels. The dissolution time was reduced from ~1 hour to <10 min by flattening the moldable gels into disks with an increased surface area–to–volume ratio (40 μl of gel, 100 mg ml−1 of mannitol; fig. S2B and movies S1 and S2). Thus, biologic release could be tuned by selecting different releasing sugars and gel geometries. On the other hand, biologic release rates were not significantly affected by the gel volume, the biologic loading, or the dilution factor after release (fig. S2, C and D). For instance, the time required to recover a β-gal concentration of 10 mg ml−1 (4:1 release dilution from a 0.5-ml gel loaded with a β-gal concentration of 40 mg ml−1) or a β-gal concentration of 0.2 mg ml−1 (50:1 release dilution from a 40-μl gel loaded with a β-gal concentration of 5 mg ml−1) was similar (≈1 hour), given the same releasing sugar and gel geometry (fig. S2C).
Thermal stabilization requires network formation
After establishing the utility of the reversible hydrogels for the encapsulation and on-demand release of biologics, we investigated whether the gels could be used for the stabilization of temperature-sensitive biologics. We encapsulated β-gal, a commonly used model enzyme that catalyzes the hydrolysis of β-galactosides into monosaccharides and is industrially used for the removal of lactose from milk (41). We hypothesized that network formation in the presence of β-gal would restrict the movement of the proteins and physically prevent them from interacting with each other, reducing aggregation, and, hence, bioactivity loss after heat exposure (Fig. 2A). After 3 days at 50°C, β-gal activity was reduced to less than 50% activity in unencapsulated (free) samples, whereas gel-encapsulated enzyme retained greater than 85% activity (Fig. 2B). Full protection occurred only when the enzyme was encapsulated within the cross-linked gel. Limited protection was observed when β-gal was formulated alone with the individual gel components (PEG-APBA or PEG-GL; Fig. 2B). Furthermore, protection was not limited by gel volume or biologic loading. Indeed, 0.5 ml gels loaded with a β-gal concentration of 40 mg ml−1 performed as well as the smaller 40 μl gels loaded with a β-gal concentration of 5 mg ml−1 (fig. S3).
Fig. 2. Reversible hydrogels protect biologics from thermal stress.
(A) When unencapsulated (free) biologics are exposed to heat, irreversible aggregation occurs, leading to permanent bioactivity loss. Conversely, gel-encapsulated biologics are protected from the heat because of the formation of polymer networks around the biologics, which restricts their mobility and physically prevents them from interacting with each other, reducing aggregation and thus retaining bioactivity upon recovery after release from the gels. (B) β-Gal samples were stored for 3 days at 50°C under the following conditions: with both of the gel components (PEG-APBA and PEG-GL), with only one of the gel components (PEG-APBA or PEG-GL), and without any of the gel components. Following storage, β-gal was released from the samples using mannitol (100 mg ml−1, pH 7.5; t = 2 hours) and β-gal activity was measured and normalized to freshly prepared β-gal. Each plotted data point corresponds to an independent experiment. For each time point, the mean ± SEM is shown. (C) Free β-gal samples (dried and nondried) were stored at 50°C for 1 week. Following release, the extent of β-gal aggregation in the samples was quantified using size exclusion chromatography–multiangle light scattering (SEC-MALS). (D) The diffusion coefficient within the gels, Dgel, was determined by monitoring the fluorescence recovery after photobleaching (FRAP) of gel-encapsulated fluorescein isothiocyanate (FITC), fluorescently labeled bovine serum albumin (FITC-BSA), and fluorescently labeled dextran (FITC-dextran). (E) The protein-unfolding transition or the melting temperature, Tm, of gel-encapsulated and free β-gal was determined using a thermal shift assay involving a fluorescent dye that binds to hydrophobic surfaces and is quenched by water. By plotting the first derivative of the fluorescence emission as a function of temperature, dF/dT, Tm was determined.
Network prevents aggregation by restricting the mobility of the biologics
Network formation was crucial for the thermal stabilization of β-gal. While the extent of β-gal aggregation in the unencapsulated (free) samples increased significantly after 1 week at 50°C (Fig. 2C and fig. S5), no significant difference in the extent of aggregation was observed in the gel-encapsulated samples after 1 week at 50°C (fig. S6). The cross-linked polymer network constrained the mobility of encapsulated biologics, regardless of their size (Fig. 2D). The diffusion coefficient of fluorescently labeled bovine serum albumin (FITC-BSA; 66 kDa) decreased by two orders of magnitude, from 60 μm2 s−1 in the free state to 0.29 μm2 s−1 in the gel-encapsulated state (42, 43). A similar decrease was observed for fluorescently labeled dextran (FITC-dextran; 500 kDa), from 23.2 to 0.15 μm2 s−1 (44). Although the movement of the biologics was reduced, it was not stopped completely, owing to the reversible nature of the cross-links. Therefore, in most formulations, the samples were dried after encapsulation, which completely prevented translation of encapsulated biologics. After drying, the residual moisture content of the samples was ≈2.5% (fig. S4). We observed that the gel did not strongly affect the melting temperature of the proteins (Fig. 2E). In total, these data show that encapsulated biologics can explore conformational space at elevated temperatures but that the formed network limits aggregation of partially denatured proteins and irreversible loss of bioactivity.
Long-term thermal stabilization of enzymes
After establishing that gel encapsulation of β-gal reduced bioactivity loss upon exposure to thermal stress, we explored the utility of the reversible hydrogels as excipients for the long-term thermal stabilization of biologics. Accelerated thermal stress tests were performed at 50°C, which showed that gel-encapsulated β-gal retained more than 90% activity after 2 weeks and ≥75% activity after 4 weeks, while free β-gal was reduced to ~30% activity after 2 weeks and 0% after 4 weeks (Fig. 3, A and B). In addition, when compared to gel-encapsulated β-gal, the sample variability in free β-gal was much higher, with two populations appearing—one with nearly full activity and the other one with almost no activity—until 4 weeks when all samples lost activity. Thus, gel encapsulation not only prevented bioactivity loss over time but also reduced sample variability, enabling the use of reversible hydrogels as excipients for the long-term thermal stabilization of biologics.
Fig. 3. Thermal stabilization of β-gal, ALP, and LAP.
(A) The activity of the enzymes in the samples was quantified using colorimetric assays. After releasing the enzymes from the samples using mannitol (100 mg ml−1, pH 7.5; t = 2 hours), test solutions were taken from the released supernatant and mixed on a 96-well plate with a chromogenic substrate [ortho-nitrophenyl-β-galactopyranoside (oNPG) for β-gal, para-nitrophenyl phosphate disodium salt (pNPP) for ALP, and l-leucine-para-nitroanilide (LLpN) for LAP]. The absorbance change over time was measured using a plate reader (420 nm for β-gal and 405 nm for ALP and LAP). To calculate the activity of the enzymes, the mean slope of the enzyme test solution was normalized to that of freshly prepared enzyme solution. (B) Gel-encapsulated and unencapsulated (free) β-gal samples were stored at 50°C for up to 4 weeks. (C) Gel-encapsulated and free ALP samples were stored at 65°C for up to 8 days. (D) Gel-encapsulated and free LAP samples were stored at 50°C for up to 4 weeks. Each plotted data point corresponds to an independent experiment. For each time point, the mean ± SEM is shown.
Next, we evaluated whether the reversible hydrogels could be used to thermally stabilize other biologics, because the proposed protection mechanism is not specific to β-gal. To this end, we investigated additional model enzymes, such as ALP. ALP is a commonly used research enzyme in molecular biology that catalyzes the removal of 5′ phosphate groups from nucleic acids (45). After 8 days at 65°C, gel-encapsulated ALP retained ≥60% activity, while free ALP was reduced to less than 10% activity, supporting that gel encapsulation protected ALP from thermal stresses even at high temperatures (Fig. 3C). Next, we assessed LAP, a metallopeptidase enzyme involved in cellular maintenance that cleaves the N-terminal residues from proteins and peptides (46). Initially, after 1 day at 50°C, the activity levels of gel-encapsulated and free LAP were similar (≥80%), but after 4 weeks at 50°C, the activity of free LAP decreased to ~30%, while the activity of gel-encapsulated LAP remained at ~60% (Fig. 3D). In total, robust long-term thermal stabilization of β-gal, ALP, and LAP was achieved, demonstrating that gel encapsulation is a broadly effective strategy to prevent bioactivity loss in enzymes.
Shelf-stable formulations of heat-sensitive clinical diagnostic enzymes
Encouraged by the ability of the gels to thermally stabilize model enzymes, we evaluated whether the gels could be used to protect heat-sensitive clinical diagnostic enzymes, such as Escherichia coli DNA gyrase (gyrase) and Top1. Gyrase and Top1 are DNA binding enzymes that are used in biomedical research and as clinical diagnostics; gyrase is used for the discovery of antibacterial drugs (47), and Top1 is used to identify the molecular mechanisms responsible for cancer (48). Both enzymes are highly temperature sensitive and labile, requiring temperatures at or below −20°C for storage. This limits their use, because the distribution of cryopreserved biologics is challenging and costly, requiring failure-prone cold chain solutions, such as shipping on dry ice and electronic monitoring of each package. In addition, improper handling and customs delays, with an average waiting time of 7 days (49), greatly increase the risk of spoilage during shipping (50).
Using our gels, gyrase and Top1 could be stored for months at room temperature. Gel-encapsulated gyrase retained greater than 70% activity for 4 weeks at 27°C and ≥50% activity after 8 weeks, outperforming free gyrase, which fell to less than 20% activity over the same period of time (Fig. 4, A and B). Furthermore, gel-encapsulated Top1 exhibited ≥85% activity for at least 6 months at 27°C, whereas free Top1 lost all activity after 2 months (Fig. 4, C and D). In this way, gel encapsulation could eliminate the need for dry ice shipping of gyrase and Top1, paving the way for the development of shelf-stable formulations for these, and other, heat-sensitive clinical diagnostic enzymes.
Fig. 4. Thermal stabilization of gyrase and Top1.
After releasing the enzymes from the samples using mannitol (100 mg ml−1, pH 7.5; t = 2 hours), the activity of gyrase and Top1 was quantified using gel-based assays. (A) Gyrase activity was determined by measuring the intensity of the supercoiled pHOT-1 bands by densitometry [1% tris-borate EDTA (TBE)/agarose gel]. (B) Gel-encapsulated and unencapsulated (free) gyrase samples were stored at 27°C for up to 8 weeks. (C) Top1 activity was determined by measuring the intensity of the relaxed pHOT-1 bands by densitometry (1% TBE/agarose gel). (D) Gel-encapsulated and free Top1 samples were stored at 27°C for up to 6 months. Each plotted data point corresponds to an independent experiment. For each time point, the mean ± SEM is shown.
Thermal stabilization of a protein-based vaccine
After demonstrating the effectiveness of gel encapsulation for the thermal stabilization of enzymes, we explored the platform for the stabilization of vaccines. Vaccine delivery requires a robust cold chain infrastructure, as most vaccines are sensitive to heat and freezing (51). This creates a large barrier for global immunization campaigns, because vaccine distribution and administration costs often exceed the production costs (29). There is a pressing need to increase the thermal stability of vaccines to keep them outside of the traditional +2° to +8°C range cold chain. Ensuring higher thermal stability of vaccines would greatly improve global vaccine access, as was the case for the meningococcal A conjugate vaccine MenAfriVac, which was granted approval for use in a controlled temperature chain at temperatures of 40°C for up to 4 days (52).
Therefore, we investigated whether gel encapsulation could increase the thermal stability of a protein-based vaccine. HA is a glycoprotein found on the surface of influenza viruses, which is integral to their infectivity. Recombinant HA-based influenza vaccines prevent infection by induction of HA inhibition antibodies (53, 54). The stability of HA was assessed for a dried formulation (with removal of solvent after gel encapsulation) and for a liquid formulation (without removal of solvent). After drying, gel-encapsulated HA retained greater than 90% functionality after 1 day at 60°C and ≥30% functionality after 3 days, whereas the functionality of dried free HA was ~45% after 1 day and <10% after 3 days (Fig. 5, A and B). These results indicate that the dried gel-encapsulated HA can withstand temperature spikes of 60°C for up to 1 day without losing functionality, which could help mitigate the harmful effects of abnormal temperature exposure during the distribution of vaccines (55). In the liquid formulation, gel-encapsulated HA retained ≥70% functionality after 7-day storage at 37°C, while the functionality of free HA in the liquid formulation dropped to <10% over the same period of time (Fig. 5C). Thus, thermal stabilization of HA was achieved without the need for solvent removal and drying. Gel encapsulation (without drying) decreased the diffusion coefficient of several biologics, including FITC-BSA (66 kDa) and FITC-dextran (500 kDa), by several orders of magnitude (Fig. 2D). Therefore, similarly to the dried samples, this decrease in biologic mobility in nondried samples likely leads to less aggregation and reduced bioactivity loss.
Fig. 5. Thermal stabilization of recombinant influenza A H5N1 HA.
(A) The functionality of HA in the samples was quantified with an enzyme-linked immunosorbent assay (ELISA). After releasing HA from the samples using mannitol (100 mg ml−1, pH 7.5; t = 2 hours), HA test solutions were taken from the released supernatant and added to a 96-well plate that was previously coated with a capture antibody. Next, a biotinylated detection antibody was bound to the immobilized HA. Last, streptavidin–horseradish peroxidase (HRP) was bound to the immobilized biotin and a substrate (TMB) was added to catalyze a color reaction. To quantify the functionality of HA, the optical density (450 nm) of the HA test solution was normalized to that of freshly prepared HA. (B) Gel-encapsulated and unencapsulated (free) ΗΑ samples (dried) were stored at 60°C for up to 3 days. (C) Gel-encapsulated and free HA samples (nondried) were stored at 37°C for up to 7 days. Each plotted data point corresponds to an independent experiment. For each time point, the mean ± SEM is shown.
Thermal stabilization of an adenovirus
Last, we stabilized Ad5 using gel encapsulation. Viral vectors, such as Ad5, are central to gene therapy and applications using genome editing techniques (i.e., CRISPR-Cas9) (56). In addition, recombinant adenovirus vectors are becoming increasingly important for use in vaccine development, especially for the recently developed mRNA vaccines. For example, adenoviruses are used in the production of several viral vector coronavirus disease 2019 (COVID-19) vaccines, including a replication-deficient chimpanzee adenovirus vaccine vector (ChAdOx1) for the Oxford-AstraZeneca COVID-19 vaccine (57) and a modified recombinant Ad26 for the Johnson & Johnson COVID-19 vaccine (58). Viruses such as Ad5 present a particular challenge for thermal stabilization because both protein conformation and nucleic acid integrity must be preserved to retain viral infectivity and immunogenicity; even simple drying is a challenge (28).
Within the gels, Ad5 was protected from desiccation and rehydration stresses. Moreover, after storage for 24 hours, gel-encapsulated Ad5 outperformed free Ad5 at all tested temperatures (4° to 65°C; Fig. 6, A and B). Furthermore, the infectivity of gel-encapsulated Ad5 at 27°C remained stable over a 4-week period, within 2 logs of infectivity, whereas free Ad5 lost >4 logs over the same time period (Fig. 6C). When gel-encapsulated Ad5 was formulated in the liquid state, the infectivity loss was reduced even further. After 4 weeks in the liquid state at 27°C, gel-encapsulated Ad5 lost almost no infectivity, while the infectivity of free Ad5 decreased by ~3 logs (Fig. 6D). As a whole, these results suggest that gel encapsulation of viruses could effectively prevent the loss of viral infectivity during storage and transport.
Fig. 6. Thermal stabilization of Ad5 expressing enhanced green fluorescent protein (GFP).
(A) The viral titers of Ad5 in the samples were quantified with a standard flow cytometric assay. After releasing Ad5 from the samples using mannitol (100 mg ml−1, pH 7.5; t = 2 hours), Ad5 test solutions were taken from the released supernatant and incubated with human embryonic kidney (HEK)–293T cells for 48 hours (multiplicity of infection = 10). The amount of GFP+ cells was determined using a flow cytometer and expressed as geometric mean fluorescence intensity per unit volume (gMFI ml−1). The log (gMFI) change of the Ad5 test solutions was compared to that of freshly prepared Ad5. (B) Gel-encapsulated and unencapsulated (free) Ad5 samples (dried) were stored at a range of temperatures (4° to 65°C) for 24 hours. (C) Gel-encapsulated and free Ad5 samples (dried) were stored at 27°C for up to 4 weeks. (D) Gel-encapsulated and free Ad5 samples (nondried) were stored at 27°C for up to 4 weeks. Each plotted data point corresponds to an independent experiment. For each time point, the mean ± SEM is shown.
Safety of the gel components
Encouraged by the high levels of protection conferred to the biologics by the reversible hydrogels, we investigated their suitability for clinical use. After releasing the biologics using a simple sugar-containing solution, we envision that a liquid formulation could be administered to the patients, containing both the released biologic and the dissolved hydrogel components (PEG-APBA and PEG-GL macromers). Therefore, the design of our material from safe and nontoxic components is a key feature for future clinical use.
We found that the hydrogel components were nontoxic in vivo up to 200 mg kg−1 in Sprague-Dawley rats (fig. S7). Furthermore, clinical chemistry and hematology blood analysis showed that, at supratherapeutic levels of the hydrogel components, there were no changes to white cell number or platelet number (i.e., no acute inflammatory response; figs. S8 and S9). In addition, no changes in the organs were observed when grossly compared to vehicle controls. As expected with PEG molecules with Mn < 60,000 g mol−1, renal clearance of the gel components still occurred (fig. S7) (59). These results suggest that an average human male (average weight of 88.8 kg) could tolerate up to 17.8 g and an average human female (average weight of 76.4 kg) could tolerate up to 14.9 g of the hydrogel components with one shot. Therefore, because we anticipate that much less than 1 g of hydrogel would be required to stabilize most biologics, the reversible hydrogels could be used as safe and easy-to-use off-the-shelf excipients for many potential pharmaceutical applications, including the thermal stabilization of biologics.
DISCUSSION
There is a pressing need to improve the thermal stability of biologics, including vaccines, to reduce the economic costs and health risks associated to the cold chain. In this work, we demonstrated that reversible hydrogels are suitable for the long-term thermal stabilization of biologics, up to 65°C. Protection was broad, covering multiple classes of biologics, including model and clinical diagnostic enzymes, protein-based vaccines, and whole viruses, with sizes ranging from 10 to 100 nm. Our data point toward a generalized protection mechanism for the thermal stabilization of diverse biologics using direct encapsulation in reversible hydrogels. We attributed protection to the formation of polymer networks around the biologics, which restricted their mobility and physically prevented them from interacting with each other, reducing aggregation and bioactivity loss after exposure to heat.
The platform has the potential to be a very user-friendly solution for the thermal stabilization of biologics. The rapid release enabled by the reversible hydrogel design allows facile recovery of the bioactive cargo. Furthermore, the reversible cross-linking will facilitate processing, as the hydrogels are moldable and self-healing. Another advantage of our platform is that protection was achieved using both dried and nondried formulations, which would reduce the amount of formulation work required to translate the platform as an off-the-shelf excipient in the biotechnology industry for the thermal stabilization of biologics. The technology is not limited by the gel volume or the biologic loading; we successfully encapsulated, thermally stabilized, and recovered functional biologics at therapeutically relevant concentrations (gel loading of 40 mg ml−1; 10 mg ml−1 after release) and volumes (0.5 ml of gels; 2 ml after release).
Nevertheless, limitations remain for the platform. Preliminary toxicology data suggest that the gel components are safe for in vivo use; however, one of the gel components could be found to be unsuitable for clinical translation. As the stabilization mechanism is generalized, this limitation could be overcome by another reversible cross-linking chemistry (or by physical cross-linking) or a different polymer backbone. We envision that the platform will be used in combination with other approaches, as an extra layer of protection or as part of a toolbox to improve the thermal stability of biologics. We anticipate that the ability to protect biologics from thermal stress with reversible hydrogels offers a useful and complementary approach to mitigate the costs and risks associated with reliance on a continuous cold chain for biologic transport and storage.
MATERIALS AND METHODS
Materials
Four-arm PEG-NH2HCl (Mn ≈ 10,000 g mol−1) was purchased from JenKem Technology USA. Regenerated cellulose dialysis tubing [1 kDa molecular weight cutoff (MWCO)] was purchased from Spectrum Labs. Methanol (MeOH), triethylamine, 2-formylphenylboronic acid, sodium borohydride (NaBH4), d-(+)-gluconic acid δ-lactone (GL), 2,2-dihydroxyindane-1,3-dione (ninhydrin), deuterated water (D2O), sodium dihydrogen phosphate (monobasic; NaH2PO4), sodium hydrogen phosphate (dibasic; Na2HPO4) β-gal from Aspergillus oryzae (catalog no. G5160), ALP from bovine intestinal mucosa (catalog no. P7640), microsomal LAP from porcine kidney (catalog no. L5006), trehalose, mannitol, glucose, ortho-nitrophenyl-β-galactopyranoside (oNPG), l-leucine-para-nitroanilide (LLpN), polysorbate 20 (Tween 20), BSA, 3,3′,5,5′-tetramethylbenzidine (TMB), 30% (w/w) hydrogen peroxide solution in H2O (H2O2), citric acid, sulfuric acid (H2SO4), FITC, albumin-FITC conjugate (FITC-BSA), FITC-dextran (Mn ≈ 500 kDa), and SYPRO Orange Protein Gel Stain (SYPRO Orange) were purchased from Sigma-Aldrich. Para-nitrophenyl phosphate disodium salt (pNPP), phosphate-buffered saline (PBS), and tris-buffered saline (TBS) were purchased from Thermo Fisher Scientific. Gyrase (catalog no. TG2000G) and Top1 (catalog no. TG2005HRC) were received from TopoGEN Inc. Recombinant influenza A H5N1 HA (catalog no. FR-59) was received from the International Reagent Resource. Ad5 expressing enhanced green fluorescent protein (GFP) (catalog no. 1060) was purchased from Vector Biolabs. Human embryonic kidney (HEK)–293 cells were purchased from the American Type Culture Collection (HEK-293T; catalog no. CRL-3216) and from Thermo Fisher Scientific (HEK-293A; catalog no. R70507).
Analytical techniques
Nuclear magnetic resonance
1H 1D nuclear magnetic resonance (NMR) spectra were acquired on Bruker Avance III 400 (Bruker BioSpin GmbH). The residual undeuterated solvent peak (4.70 parts per million for D2O) was used for reference. The relative integration is reported as the number of protons (H), and the following abbreviations were used to denote multiplicities: s = singlet, d = doublet, t = triplet, m = multiplet, and br = broad.
Plate reader
Absorbance measurements were acquired with a Hidex Sense microplate reader (Hidex; Turku, Finland) using clear 96-well tissue culture plates (flat bottom).
Flow cytometer
The quantification of GFP+ cells was done using a CytoFLEX flow cytometer (Beckman Coulter Life Sciences, Indianapolis, IN, USA).
Size exclusion chromatography–multiangle light scattering
Size exclusion chromatography–multiangle light scattering (SEC-MALS) measurements were performed using a DAWN HELEOS II MALS detector (Wyatt Technology, Santa Barbara, CA, USA) coupled to an Infinity II 1260 high-performance liquid chromatography system (Agilent Technologies, USA). For separation, a Superdex 200 Inc. 10/300 column (GE Healthcare, UK) was used with PBS as the mobile phase. Flow rates of either 0.5 or 0.75 ml min−1 were applied. Data were collected and analyzed in ASTRA software (Wyatt Technology, Santa Barbara, CA, USA).
Confocal microscope
Fluorescence recovery after photobleaching (FRAP) experiments were performed on a Leica TCS SP8 STED confocal microscope (Leica Microsystems, Heerbrug, Switzerland) using a ×63 magnification oil immersion objective [numerical aperture (NA) = 1.40].
Real-time polymerase chain reaction
The thermal shift assay was performed using an Applied Biosystems StepOnePlus real-time polymerase chain reaction (PCR) system (Thermo Fisher Scientific AG, Basel, Switzerland).
Thermogravimetric analysis
Thermogravimetric analysis (TGA) was performed by using a TGA/DSC 3+ thermogravimetric analyzer (Mettler Toledo, Greifensee, Switzerland) to study the residual moisture content of the gels before and after vacuum drying. Samples were exposed first to dry N2 for 10 min at 25°C, and then heated to 120°C (10°C min−1) and held for 1 hour, before being cooled back to 25°C.
Rheometer
Rheometric characterization was performed using a strain-controlled shear rheometer (MCR 502; Anton-Paar, Zofingen, Switzerland) equipped with a Peltier stage to control the temperature. Silicone oil was applied to the samples to prevent drying, and the experiments were performed at 25°C unless stated otherwise. The measurements were performed using a 20-mm parallel plate geometry with a gap of 0.5 mm. Motor adjustments were performed before each experiment. The samples were prepared at least 1 hour before each experiment to allow complete gelation. The samples were loaded by placing them directly on the Peltier plate and lowering the geometry to the desired gap. After loading, the samples were then equilibrated to the set temperature for at least 15 min (γ = 0.1%; ω = 1 rad s−1). Strain sweep (SS) experiments were performed at ω = 1 rad s−1 for γ = 0.001 to 1000%. Frequency sweep (FS) experiments were performed at γ = 1% (within the linear viscoelastic regime as determined from SS experiments) for ω = 100 to 0.01 rad s−1. Stress relaxation experiments were performed by holding the samples at γ = 10% and measuring the relaxation modulus G(t) every 0.25 s for a total duration of 1 hour. Step strain recovery experiments were performed at ω = 1 rad s−1 by alternating periods of high strain (γ = 100%) and low strain (γ = 1%) for multiple cycles.
Experimental design
Synthesis of four-arm PEG-APBA
The synthesis of four-arm PEG-APBA was performed according to a published procedure with some modifications (40). Four-arm PEG-NH2HCl (2.0 g, 0.2 mmol; Mn ≈ 10,000 g mol−1) was dissolved in anhydrous MeOH (10 ml) in a round-bottom flask equipped with a stir bar. Subsequently, triethylamine (0.5 ml, 3.6 mmol) and 2-formylphenylboronic acid (180 mg, 1.2 mmol) were added to the reaction, which was left to proceed for 72 hours under argon gas at room temperature. Next, NaBH4 (90 mg, 2.4 mmol) was added portion wise after cooling the reaction on ice to 4°C and the reaction was left to proceed for 12 hours at room temperature. Last, MeOH was evaporated and the remaining product was dissolved in dH2O. The pH of the aqueous solution was balanced to 7 (using 1 M HCl), dialyzed against dH2O for 72 hours (1 kDa MWCO), filtered (0.2 μm), and then lyophilized to yield a white powder. 1H (400 MHz, D2O): δ 7.4 to 7.1 (m, 16 H), 4.0 (s, 8 H), 3.6 (m, 892 H), and 3.0 (t, 8 H). The degree of functionalization determined from 1H NMR was ≈85% (theoretical Mn = 10,480 g mol−1; fig. S10).
Synthesis of four-arm PEG-GL
The synthesis of four-arm PEG-GL was performed according to a published procedure with some modifications (40). Four-arm PEG-NH2HCl (2.0 g, 0.2 mmol; Mn ≈ 10,000 g mol−1) was dissolved in anhydrous MeOH (50 ml) in a round-bottom flask equipped with a stir bar. Next, triethylamine (1.0 ml, 7.2 mmol) and d-(+)-gluconic acid δ-lactone (285 mg, 1.6 mmol) were added to the PEG mixture. After allowing the reaction to proceed for 72 hours at room temperature, the extent of the reaction was monitored by performing a ninhydrin test to detect the presence of free amines (blue, unreacted amines still present; yellow/colorless, amines successfully coupled). Additional triethylamine (1.0 ml, 7.2 mmol) and d-(+)-gluconic acid δ-lactone (285 mg, 1.6 mmol) were added if free amines persisted, and the reaction was left to proceed for another 24 to 72 hours at room temperature. Last, MeOH was evaporated and the remaining product was dissolved in dH2O. The pH of the aqueous solution was balanced to 7 (using 1 M NaOH), dialyzed against dH2O for 72 hours (1 kDa MWCO), filtered (0.2 mm), and then lyophilized to yield a white powder. 1H (400 MHz, D2O): δ 4.3 (d, 4 H), 4.0 (t, 4 H), and 3.6 (m, 924 H). The degree of functionalization determined from 1H NMR was ≈85% (theoretical Mn = 10,713 g mol−1; fig. S11).
Formation of reversible networks and gel encapsulation of biologics
The following biologic stock solutions were used for gel encapsulation: β-gal [40 mg ml−1 in 0.1 M phosphate buffer (pH 7.5)], ALP [160 mg ml−1 in TBS (pH 7.5)], LAP (4.6 mg ml−1, as received from Sigma-Aldrich), gyrase (~80 U μl−1, as received from TopoGEN Inc.), Top1 (~200 U μl−1, as received from TopoGEN Inc.), recombinant influenza A H5N1 HA (1 mg ml−1, as received from the International Reagent Resource), and Ad5 expressing enhanced GFP [1.2 × 109 plaque-forming units (PFU) ml−1, as received from Vector Biolabs]. In addition, stock solutions of trehalose, PEG-APBA, and PEG-GL were prepared for each biologic formulation in their respective buffers (0.1 M phosphate buffer for β-gal, LAP, and HA; TBS for ALP; molecular biology–grade dH2O for gyrase and Top1; and PBS for Ad5). Hydrogel networks were formed by mixing equivalent volumes of PEG-APBA and PEG-GL stock solutions and replacing the balance volume in each gel formulation with the respective volumes of biologic stock solution (β-gal, ALP, LAP, gyrase, Top1, HA, or Ad5), trehalose stock solution (such that the mass of trehalose = 2.5× mass of biologic in the final gel), and the corresponding buffer. After mixing, gelation occurred within 10 s. The final polymer content in the gels was 5% (w/w) for ALP, 7% (w/w) for Ad5, 7.5% (w/w) for β-gal, 10% (w/w) for LAP and HA, and 11.7% (w/w) for gyrase and Top1. The final volume of the gels was 4 μl for gyrase and Top1, 20 μl for ALP and HA, and 40 μl for β-gal, LAP, and Ad5. The final biologic concentrations in the gels were 5 mg ml−1 for β-gal, 27 mg ml−1 for ALP, 0.2875 mg ml−1 for LAP, 20 U μl−1 for gyrase, 50 U μl−1 for Top1, 0.05 mg ml−1 for HA, and 3.0 × 108 PFU ml−1 for Ad5. Unless otherwise stated, samples (both gel-encapsulated and unencapsulated) were subsequently dehydrated under vacuum (25 mbar) for 2 hours at room temperature to remove residual moisture (fig. S3A). In the case of gyrase and Top1, samples were dehydrated under vacuum (250 mbar) overnight at room temperature. Unless otherwise stated, samples were prepared in 2 ml of poly(propylene) reaction tubes (art. no. 623201, Greiner Bio-One). Samples were then stored at the test temperature for the indicated period of time before release and analysis. In the case of β-gal (50°C), ALP (65°C), LAP (50°C), HA (37 and 60°C), and Ad5 (40 and 65°C), samples were stored in a universal convection oven (UN 30, Memmert GmbH). In the case of gyrase (27°C), Top1 (27°C), and Ad5 (27°C), samples were stored at room temperature on a bench. In the case of Ad5 (4°C), samples were stored in a fridge.
Triggered network dissolution and release of encapsulated biologics
After storage with (or without) gel encapsulation, the biologics were released (solubilized) by immersing the samples in a release buffer [100 mg ml−1 of mannitol (pH 7.5)] under agitation at room temperature (2 hours for β-gal, ALP, LAP, and HA; 45 min for gyrase and Top1; and 15 min for Ad5). The final concentrations of the biologics in the supernatant were 100 μg ml−1 for β-gal, 270 μg ml−1 for ALP, 5.75 μg ml−1 for LAP, 2 U μl−1 for gyrase, 12.5 U μl−1 for Top1, 0.5 μg ml−1 for HA, and 3.0 × 107 PFU ml−1 for Ad5. The bioactivity of the biologics in each sample was quantified and normalized to a freshly prepared solution at the same concentration (storage: −20° or −80°C) to compare the control (free) and treatment groups (gel-encapsulated). The equilibrium binding constant, Keq,D, between PEG-APBA and the releasing sugars (mannitol, fructose, and glucose) was taken from a previous study (37).
β-Gal activity assay
β-Gal activity was measured using oNPG according to the manufacturer’s protocol (Sigma-Aldrich). Briefly, room temperature oNPG [16 mM in 0.1 M phosphate buffer (pH 7.5)] was mixed 2:1 with the β-gal test solution in a 96-well plate (oNPG:β-gal test solution; 100 μl:50 μl). β-Gal catalyzes the hydrolysis of oNPG to release ortho-nitrophenol, a chromogenic substrate with maximal absorbance at 420 nm. Absorbance at 420 nm was measured every 60 s for 10 min, and the mean slope of the resultant curve was recorded. To calculate the activity of β-gal, the mean slope of the β-gal test solution was normalized to that of freshly prepared β-gal solution (100 μg ml−1).
ALP activity assay
ALP activity was measured using pNPP according to the manufacturer’s protocol (Thermo Fisher Scientific). Briefly, room temperature pNPP was mixed 1:1 with the ALP test solution in a 96-well plate (pNPP:ALP test solution; 50 μl:50 μl). ALP catalyzes the hydrolysis of pNPP to release para-nitrophenol, a chromogenic substrate with maximal absorbance at 405 nm. Absorbance at 405 nm was measured every 60 s for 10 min, and the mean slope of the resultant curve was recorded. To calculate the activity of ALP, the mean slope of the ALP test solution was normalized to that of freshly prepared ALP solution (270 μg ml−1).
LAP activity assay
LAP activity was measured using LLpN according to the manufacturer’s protocol (Sigma-Aldrich). Briefly, room temperature LLpN [1.6 mM in 0.1 M phosphate buffer with 6.6% (v/v) MeOH (pH 7.5)] was mixed 1:1 with the LAP test solution in a 96-well plate (LLpN:LAP test solution; 100 μl:100 μl). LAP catalyzes the hydrolysis of LLpN to release para-nitroanilide, a chromogenic substrate with maximal absorbance at 405 nm. Absorbance at 405 nm was measured every 60 s for 10 min, and the mean slope of the resultant curve was recorded. To calculate the activity of LAP, the mean slope of the LAP test solution was normalized to that of freshly prepared LAP solution (5.75 μg ml−1).
Gyrase activity assay
Gyrase activity (TopoGEN Inc.) was measured by supercoiling of relaxed pHOT-1 DNA, following the manufacturer’s protocol. Briefly, 1 μl of gyrase test solution, 4 μl of 5× gyrase “assay” buffer (TopoGEN Inc.), and 250 ng of relaxed pHOT-1 DNA (TopoGEN Inc.) were combined in molecular biology–grade dH2O at a final assay volume of 20 μl and incubated at 37°C for 60 min. After incubation, 4 μl of 5× loading “stop” buffer (TopoGEN Inc.) was added and mixed by vortex. The reaction products were separated on a 1% tris-borate EDTA (TBE)/agarose gel [no ethidium bromide (EtBr)] and stained with EtBr, and the bands were imaged. Enzyme activity was quantified by measuring the intensity of the supercoiled pHOT-1 bands by densitometry using ImageJ (fig. S12).
Top1 activity assay
Top1 activity (TopoGEN Inc.) was measured by relaxation of supercoiled pHOT-1 DNA, following the manufacturer’s protocol. Briefly, 1 μl of test solution, 2 μl of 10× Top1 “reaction” buffer (TopoGEN Inc.), and 125 ng of supercoiled pHOT-1 DNA (TopoGEN Inc.) were combined in molecular biology–grade dH2O at a final assay volume of 20 μl and incubated at 37°C for 30 min. After incubation, 4 μl of 5× loading stop buffer (TopoGEN Inc.) was added and mixed by vortex. The reaction products were separated on a 1% TBE/agarose gel (no EtBr) and stained with EtBr, and the bands were imaged. Enzyme activity was quantified by measuring the intensity of the relaxed pHOT-1 bands by densitometry using ImageJ (fig. S13).
HA quantification assay
The functionality of recombinant influenza A H5N1 HA was quantified with a solid-phase sandwich H5N1 HA enzyme-linked immunosorbent assay kit (Sino Biological Inc., catalog no. SEK002) according to the manufacturer’s protocol (Fig. 5A). Briefly, 100 μl of capture antibody (diluted to 0.5 μg ml−1 with PBS) was added to each well in a 96-well plate and incubated overnight at 4°C after sealing the plate. After washing the plate three times with 300 μl of wash buffer [0.05% Tween 20 in TBS (pH 7.2 to 7.4)], 300 μl of blocking buffer (2% BSA in wash buffer) was added to each well and incubated for 1 hour at room temperature under agitation. After a washing step, 100 μl of the HA test solution (diluted 1:50 with 0.1% BSA in wash buffer) was added to each well and the sealed plate was incubated for 2 hours at room temperature under agitation. After another washing step, 100 μl of the biotinylated detection antibody (diluted to 1.5 μg ml−1 with 0.5% BSA in wash buffer) was added to each well and the sealed plate was incubated for 1 hour at room temperature under agitation. After another wash, 100 μl of horseradish peroxidase (HRP)–conjugated streptavidin (diluted 1:2000 with 0.5% BSA in wash buffer) was added to each well and the sealed plate was incubated for 1 hour at room temperature under agitation. After a final washing step, 200 μl of substrate solution [TMB (0.1 mg ml−1) and 0.0024% H2O2 in 0.05 M Na2HPO4 and 0.025 M citric acid buffer (pH 5.5)] was added to each well and the sealed plate was covered in foil and incubated for 20 min at room temperature under agitation. Last, 50 μl of stop solution (2 M H2SO4) was added to each well and mixed thoroughly with the pipette. The optical density at 450 nm of each well was immediately determined using a microplate reader. For all experiments, a seven-point standard curve using twofold serial dilutions with a high standard of 5 ng ml−1 was made (fig. S14).
Ad5 quantification assays
Two different assays were used to quantify the viral titers of Ad5 expressing enhanced GFP. For the dried samples, a standard flow cytometric assay was used to determine the viral titers by transduction efficiency. The assay was performed using 80 to 90% confluent HEK-293T cells. Briefly, 60 μl of the released Ad5 test solution was mixed with 940 μl of growth medium [Gibco DMEM (Dulbecco’s modified Eagle’s medium) High Glucose, containing penicillin (100 U/ml), streptomycin (100 μg ml−1), l-glutamine (292 μg ml−1), and 10% fetal bovine serum (FBS); Thermo Fisher Scientific] and added dropwise to a single well of HEK-293T cells that had been seeded in 12-well plates the previous day (20 to 50% confluent), resulting in a multiplicity of infection (MOI) of 10 when Ad5 (3.0 × 108 PFU ml−1) was encapsulated in the samples. Fresh positive controls were prepared on the day of infection by diluting the master virus stock (1.2 × 109 PFU ml−1) with growth medium to a final concentration of 1.8 × 106 PFU ml−1, resulting in MOI = 10 when 1 ml was added to a single well. Dilutions were prepared in growth medium for MOI = 3 and MOI = 1 controls. In addition, several wells were left uninfected as negative controls. After incubating at 37°C with 5% CO2 for 48 hours, GFP expression was determined by flow cytometry. For this, the cells were washed with sterile PBS, trypsinized, and resuspended into a 96-well plate with growth medium. After performing a live/dead stain using the Zombie NIR dye (BioLegend), the cells were washed with a fluorescence-activated cell sorting buffer and analyzed in a flow cytometer. Data analysis was performed using FlowJo 10.7.2 software. For the gating strategy, single and live cells were preselected, followed by the selection of a GFP+ gate using the uninfected wells (fig. S15). Using this gating strategy, the amount of GFP+ cells was determined and expressed as geometric mean fluorescence intensity per unit volume (gMFI ml−1) (fig. S16). For the nondried samples, a standard plaque-forming assay was used to determine the viral titers. The assay was performed using 90 to 100% confluent HEK-293A cells seeded in six-well plates. Briefly, the Ad5 test solution was serially diluted from 10−2 to 10−6, and 100 μl from each dilution was mixed directly with the cells in the wells. On the next day, 5% SeaPlaque agarose gel (Lonza Group) was mixed 1:3 with complete medium [Gibco DMEM, containing 10% FBS, penicillin (200 U ml−1), streptomycin (200 μg ml−1), and 2 mM l-glutamine; Thermo Fisher Scientific]. The cell monolayers were overlaid with 3 ml of the agarose gel mixture and kept at 25°C for 15 min for the gel to solidify. After incubating at 37°C with 5% CO2 for 7 days, the plaques were counted, and the titers were calculated and expressed as plaque-forming units per unit volume (PFU ml−1) (fig. S17).
Aggregation of β-gal
The aggregation of β-gal was assessed using SEC-MALS, which combines MALS with SEC. The SEC separates polymer/protein molecules according to their hydrodynamic radius. Basic physical equations connect the molar mass with scattered light intensity (measured by MALS) and concentration [measured by ultraviolet and differential refractive index (dRI)]. Thus, SEC-MALS can determine both the molecular weight and the size of the polymer/protein molecules. In this work, the following samples were prepared and analyzed using SEC-MALS: the unencapsulated (free) β-gal samples (nondried and dried, unstressed and stressed for 1 week at 50°C; fig. S5), the released gel components (PEG-APBA and PEG-GL without β-gal; fig. S6A), and the released gel-encapsulated β-gal samples (nondried and dried, unstressed and stressed for 1 week at 50°C; fig. S6, B to D). Unless stated otherwise, the concentration of β-gal in the samples was 5 mg ml−1, the total PEG content in the gels was 7.5% (w/w), the samples were dried under vacuum (25 mbar, 2 hours), and the gel-encapsulated β-gal was released using mannitol (100 mg ml−1, 2 hours). In the free β-gal samples (nondried and unstressed; fig. S5A), the β-gal main peak was identified at 12.2 ml, corresponding to the β-gal monomer (Mn = 121 kDa). The β-gal main peak could no longer be identified in the nondried, free β-gal samples that had been stressed for 1 week at 50°C, as the sample was heavily degraded. Instead, aggregate peaks were identified at 7, 11.3, 13, and 14.6 ml (Mn > 2000 kDa) (fig. S5B). Therefore, to quantify the extent of aggregation in the free β-gal samples, the overall amount of aggregates in the samples was compared to the overall calculated sample mass. In the sample containing the gel components alone (PEG-APBA and PEG-GL; fig. S6A), the PEG monomer and dimer peaks were identified at 13.8 ml (Mn = 9.6 kDa) and at 11.8 ml (Mn = 19.8 kDa), respectively. In the mixed samples (fig. S6, B to D), the PEG gel components and the β-gal monomer coelute because they have a similar hydrodynamic radius (although their Mn values are different by a factor of 6). Therefore, it was not possible to quantify the extent of aggregation in the gel-encapsulated β-gal samples. However, because the peak corresponding to the β-gal aggregates was still identified at 7 ml (Mn > 2000 kDa), a qualitative assessment of β-gal aggregation could still be made for the gel-encapsulated samples.
Fluorescence recovery after photobleaching
PEG-APBA/GL gels [10% (w/w); 0.1 M phosphate buffer (pH 7.5)] were loaded with either FITC, FITC-BSA, or FITC-dextran (1 mg ml−1) (Mn ≈ 500 kDa). The loaded gels were transferred to the imaging chambers (Lab-Tek II four-well chamber slides with #1.5 German borosilicate cover glass; Thermo Fisher Scientific) and equilibrated overnight at room temperature. Recovery curves were taken with a Leica TCS SP8 STED confocal microscope using the FRAP booster option, a ×63 magnification oil immersion objective (NA = 1.40), and a 488-nm argon laser for excitation. Three different regions of interest (ROIs) were selected for each sample: a bleached area (ROI 1; 20 μm diameter), a fluorescent background area (ROI 2), and a nonfluorescent background area (ROI 3). All areas were monitored by 30 prebleach image scans (1-s interval; 1% laser power). ROI 1 was bleached by 10 image scans (1-s interval; 100% laser power). The fluorescence recovery in all areas was monitored by 450 postbleach image scans (5-s interval; 1% laser power) (fig. S18). ImageJ and the EasyFRAP-web tool were used for quantitative analysis of the FRAP measurements (60). Briefly, fluorescence intensity was normalized using the full-scale method, and last curve fitting was performed using the double term exponential equation to determine the mobile fraction, Mf, and the half-maximal recovery time, t1/2. Last, the diffusion coefficient for the molecules within the gels, Dgel, was calculated according to Dgel = 0.224 r2/t1/2, where r is the radius of the bleached spot (10 μm) (table S1) (61).
Thermal shift assay
The fluorescent dye SYPRO Orange was used to monitor the protein-unfolding transition of β-gal (62). SYPRO Orange binds nonspecifically to hydrophobic surfaces, and water strongly quenches its fluorescence. When a protein unfolds, the exposed hydrophobic surfaces bind the dye, resulting in an increase in fluorescence by excluding water. The thermal shift assay was conducted using a real-time PCR instrument (Applied Biosystems, StepOnePlus), which provides accurate temperature control and contains a charge-coupled device detector to monitor the fluorescence in the wells. To perform the assay, gels were formed [20 μl, 7.5% (w/w)] directly inside the wells of a 96-well PCR plate by adding 7.5 μl of PEG-APBA [10% (w/w) in PBS], 7.5 μl of PEG-GL [10% (w/w) in PBS], 2.5 μl of β-gal (40 mg ml−1), and 2.5 μl of SYPRO Orange (8× dilution from 1000× stock). In the case of the nonencapsulated samples, the volume of PEG-GL and PEG-APBA was replaced with PBS. In both gel-encapsulated and unencapsulated samples, the final concentration of β-gal was ≈10 μM (5 mg ml−1; Mn ≈ 465,400 g mol−1). After sealing, the plate was heated from 25° to 97°C with a heating rate of 0.3°C min−1 and the fluorescence intensity of the samples was determined using the ROX dye filter (excitation/emission maxima were at 586 and 621 nm, respectively). The melting temperature, Tm, which occurs at the midpoint of the unfolding transition, was determined by plotting the first derivative of the fluorescence emission as a function of temperature (dF/dT) (Fig. 2E).
In vivo acute toxicity study
A non-GMP maximum tolerated dose study of the gel components (following Animal Research: Reporting of In Vivo Experiments guidelines) was performed by CARE Research LLC (Fort Collins, CO, USA). The purpose of the study was to determine the acute maximum tolerated dose for a single intraperitoneal injection of the gel components (PEG-APBA, PEG-GL, and degraded gel) into Sprague-Dawley rats (Charles River Laboratories). All materials were handled under aseptic conditions, and buffers used for reconstitution were either purchased as sterile or autoclaved before use. The gel components were not subjected to additional sterilization procedures. The study design and animal usage were reviewed and approved by the CARE Research Institutional Animal Care and Use Committee (IACUC) for compliance with regulations before study initiation (IACUC no. 1744). Animal welfare for this study was in compliance with the U.S. Department of Agriculture’s Animal Welfare Act (9 CFR Parts 1, 2, and 3), the Guide for the Care and Use of Laboratory Animals (63), and CARE Research standard operating procedures. Fifty female Sprague-Dawley rats were divided into 10 groups of 5 rats each and were separated on the basis of table S2. In addition, two animals were retained for replacement as deemed necessary by the study director. Animals were placed randomly into cages upon arrival and assigned groups in consecutive order. Predose body weights ranged from 250.1 to 302.1 g. Potential toxicity was evaluated by observing clinical signs and body weight change (fig. S7A). Detailed clinical observations were conducted after dose on day 1, then daily, and whenever an abnormal observation was found. Body weights were recorded before dose on day 1 and on days 2 and 6 (day of scheduled euthanasia) (fig. S7B). Additional cage-side morbidity/mortality/general health observations were conducted at least once daily. To obtain a more accurate evaluation of potential toxicity, a gross necropsy was performed on day 6 to collect organ weight data (fig. S7C) and terminal blood for clinical chemistry (fig. S8) and hematology (fig. S9). Terminal blood (abdominal vessel, maximum volume) was collected under anesthesia (isoflurane inhalation) from all study animals and analyzed for hematology (EDTA-containing tube) and chemistry (red-top tube). Samples were sent to Colorado Histo-Prep and the Colorado State University Diagnostic Laboratory on ice packs for analyses. Following blood collection and exsanguination, animals from the vehicle control group (group 1) and the high-dose groups for PEG-GL (group 4), PEG-APBA (group 7), and degraded gel (group 10) were subjected to a gross necropsy and organ weights were collected (adrenals, kidneys, thymus, spleen, heart, and liver). Organs were not saved. During the course of the study, one animal was found dead, roughly 10 min after dose. Necropsy-confirmed death was due to a dosing error. The animal was replaced with a replacement animal at the discretion of the study director.
Statistical analysis
Statistical analysis was performed using Origin 2021 (OriginLab Corporation). Unless otherwise noted, statistical significance for single comparisons was determined by Student’s t test; analysis of variance (ANOVA) with a Bonferroni posttest was used for multiple comparisons. For the in vivo acute toxicity study, the sample size was determined using G*Power. Statistical power analysis suggested that five rats per group were required to achieve statistical significance in the proposed study using the desired end points. The selected sample size achieves a statistical power (1 − β) of 0.8 with an α (type 1 error) of 5%. Body weights were subjected to two-way ANOVA, and all hematology and clinical chemistry results were subjected to one-way ANOVA.
Acknowledgments
We thank J. Kusch and the Scientific Center for Optical and Electron Microscopy ScopeM (ETH Zurich) for support in FRAP acquisition, D. Roessner and R. Mildner (Wyatt Technology Corporation) for assistance with the SEC-MALS measurements, F. Donat and C. Müller (ETH Zurich) for assistance with the TGA measurements, and D. Busha for assistance with designing the virus stabilization experiments. Aspects of the figures were prepared using BioRender.com. The images of the biologics in Fig. 1 were created with the PyMOL Molecular Graphics System (version 2.4.1, Schrödinger LLC) using structures obtained from the Protein Data Bank (rcsb.org) with the following identification codes: 6X1Q for β-gal (64), 1GYT for LAP (65), 1ANJ for ALP (66), 1RRJ for Top1 (67), 6RKU for E. coli DNA gyrase (68), 2VIU for HA (69), and 6B1T for Ad5 (70).
Funding: This work was supported by start-up funds from ETH Zurich, the Claude & Giuliana Foundation, and Nanoly Bioscience Inc. Research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH under award number 1R43GM128466-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: B.M.-D., B.V.S., and M.W.T. conceived the ideas and designed the experiments. J.R.J. designed the experiments for DNA gyrase and Top1 stabilization. J.R.J. and T.H. designed the experiments for Ad5 stabilization. B.M.-D., J.R.J., T.H., M.L., F.G., M.W., A.W., and S.T. executed the experiments and analyzed the data. B.M.-D. and M.W.T. wrote the manuscript with assistance from all authors. All authors have approved the final version of the manuscript.
Competing interests: B.V.S., J.R.J., T.H., and A.W. are/were employees of Nanoly Bioscience Inc., which develops polymer-based thermal stabilization technologies. B.V.S., J.R.J., and M.W.T. own stock in Nanoly Bioscience Inc. B.M.-D., B.V.S., J.R.J., and M.W.T. are inventors on a patent related to this work: PCT phase, filed by Nanoly Bioscience Inc. (U.S. Patent application no. 62/814,547, filed on 6 March 2019). All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Urquhart L., Top companies and drugs by sales in 2019. Nat. Rev. Drug Discov. 19, 228 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Mitragotri S., Burke P. A., Langer R., Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 13, 655–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 289, 1–30 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Wang W., Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 203, 1–60 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Welch R. P., Lee H., Luzuriaga M. A., Brohlin O. R., Gassensmith J. J., Protein–polymer delivery: Chemistry from the cold chain to the clinic. Bioconjug. Chem. 29, 2867–2883 (2018). [DOI] [PubMed] [Google Scholar]
- 6.N. Basta, M. Lipowicz, 2020 Biopharma Cold Chain Sourcebook (Pharmaceutical Commerce, ed. 11, 2020);www.pharmaceuticalcommerce.com/view/sourcebook.
- 7.Ashok A., Brison M., LeTallec Y., Improving cold chain systems: Challenges and solutions. Vaccine 35, 2217–2223 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Mihigo R., Okeibunor J., Cernuschi T., Petu A., Satoulou A., Zawaira F., Improving access to affordable vaccines for middle-income countries in the african region. Vaccine 37, 2838–2842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H., Liu L., Li J., Chen J., Du G., Rational design to improve protein thermostability: Recent advances and prospects. ChemBioEng Rev. 2, 87–94 (2015). [Google Scholar]
- 10.Hida K., Hanes J., Ostermeier M., Directed evolution for drug and nucleic acid delivery. Adv. Drug Deliv. Rev. 59, 1562–1578 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Gribenko A. V., Patel M. M., Liu J., McCallum S. A., Wang C., Makhatadze G. I., Rational stabilization of enzymes by computational redesign of surface charge–charge interactions. Proc. Natl. Acad. Sci. U.S.A. 106, 2601–2606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding H., Gao F., Liu D., Li Z., Xu X., Wu M., Zhao Y., Significant improvement of thermal stability of glucose 1-dehydrogenase by introducing disulfide bonds at the tetramer interface. Enzyme Microb. Technol. 53, 365–372 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Lee J., Mansfield K. M., Ko J. H., Sallam S., Wesdemiotis C., Maynard H. D., Trehalose glycopolymer enhances both solution stability and pharmacokinetics of a therapeutic protein. Bioconjug. Chem. 28, 836–845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alconcel S. N. S., Baas A. S., Maynard H. D., FDA-approved poly(ethylene glycol)-protein conjugate drugs. Polym. Chem. 2, 1442–1448 (2011). [Google Scholar]
- 15.Leclair D. A., Cranston E. D., Lichty B. D., Xing Z., Thompson M. R., Consecutive spray drying to produce coated dry powder vaccines suitable for oral administration. ACS Biomater Sci. Eng. 4, 1669–1678 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kasper J. C., Friess W., The freezing step in lyophilization: Physico-chemical fundamentals, freezing methods and consequences on process performance and quality attributes of biopharmaceuticals. Eur. J. Pharm. Biopharm. 78, 248–263 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Crowe J. H., Trehalose as a “chemical chaperone”: Fact and fantasy. Adv. Exp. Med. Biol. 594, 143–158 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Lee J., Lin E.-W., Lau U. Y., Hedrick J. L., Bat E., Maynard H. D., Trehalose glycopolymers as excipients for protein stabilization. Biomacromolecules 14, 2561–2569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panganiban B., Qiao B., Jiang T., DelRe C., Obadia M. M., Nguyen T. D., Smith A. A. A., Hall A., Sit I., Crosby M. G., Dennis P. B., Drockenmuller E., Olvera de la Cruz M., Xu T., Random heteropolymers preserve protein function in foreign environments. Science 359, 1239–1243 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Pelliccia M., Andreozzi P., Paulose J., D’Alicarnasso M., Cagno V., Donalisio M., Civra A., Broeckel R. M., Haese N., Jacob Silva P., Carney R. P., Marjomäki V., Streblow D. N., Lembo D., Stellacci F., Vitelli V., Krol S., Additives for vaccine storage to improve thermal stability of adenoviruses from hours to months. Nat. Commun. 7, 13520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senske M., Smith A. E., Pielak G. J., Protein stability in reverse micelles. Angew. Chem. Int. Ed. Engl. 55, 3586–3589 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Liang K., Ricco R., Doherty C. M., Styles M. J., Bell S., Kirby N., Mudie S., Haylock D., Hill A. J., Doonan C. J., Falcaro P., Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun. 6, 7240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbert F. C., Abeyrathna S. S., Abeyrathna N. S., Wijesundara Y. H., Brohlin O. R., Carraro F., Amenitsch H., Falcaro P., Luzuriaga M. A., Durand-Silva A., Diwakara S. D., Smaldone R. A., Meloni G., Gassensmith J. J., Stabilization of supramolecular membrane protein–lipid bilayer assemblies through immobilization in a crystalline exoskeleton. Nat. Commun. 12, 2202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luzuriaga M. A., Welch R. P., Dharmarwardana M., Benjamin C. E., Li S., Shahrivarkevishahi A., Popal S., Tuong L. H., Creswell C. T., Gassensmith J. J., Enhanced stability and controlled delivery of MOF-encapsulated vaccines and their immunogenic response in vivo. ACS Appl. Mater. Interfaces 11, 9740–9746 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ravindra R., Zhao S., Gies H., Winter R., Protein encapsulation in mesoporous silicate: The effects of confinement on protein stability, hydration, and volumetric properties. J. Am. Chem. Soc. 126, 12224–12225 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Chen Y.-C., Smith T., Hicks R. H., Doekhie A., Koumanov F., Wells S. A., Edler K. J., van den Elsen J., Holman G. D., Marchbank K. J., Sartbaeva A., Thermal stability, storage and release of proteins with tailored fit in silica. Sci. Rep. 7, 46568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung V., Mapletoft J., Zhang A., Lee A., Vahedi F., Chew M., Szewczyk A., Jahanshahi-Anbuhi S., Ang J., Cowbrough B., Miller M. S., Ashkar A., Filipe C. D. M., Thermal stabilization of viral vaccines in low-cost sugar films. Sci. Rep. 9, 7631 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcock R., Cottingham M. G., Rollier C. S., Furze J., De Costa S. D., Hanlon M., Spencer A. J., Honeycutt J. D., Wyllie D. H., Gilbert S. C., Bregu M., Hill A. V. S., Long-term thermostabilization of live poxviral and adenoviral vaccine vectors at supraphysiological temperatures in carbohydrate glass. Sci. Transl. Med. 2, 19ra12 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Bajrovic I., Schafer S. C., Romanovicz D. K., Croyle M. A., Novel technology for storage and distribution of live vaccines and other biological medicines at ambient temperature. Sci. Adv. 6, eaau4819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S., Wang X., Lu Q., Hu X., Uppal N., Omenetto F. G., Kaplan D. L., Stabilization of enzymes in silk films. Biomacromolecules 10, 1032–1042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinson J. A., Palmer C. R., Miller D. P., Li A. B., Lightner K., Jost H., Weldon W. C., Oberste M. S., Kluge J. A., Kosuda K. M., Thin silk fibroin films as a dried format for temperature stabilization of inactivated polio vaccine. Vaccine 38, 1652–1660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sridhar B. V., Janczy J. R., Hatlevik Ø., Wolfson G., Anseth K. S., Tibbitt M. W., Thermal stabilization of biologics with photoresponsive hydrogels. Biomacromolecules 19, 740–747 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Meis C. M., Salzman E. E., Maikawa C. L., Smith A. A. A., Mann J. L., Grosskopf A. K., Appel E. A., Self-assembled, dilution-responsive hydrogels for enhanced thermal stability of insulin biopharmaceuticals. ACS Biomater Sci. Eng. 7, 4221–4229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Ko J. H., Lin E.-W., Wallace P., Ruch F., Maynard H. D., Trehalose hydrogels for stabilization of enzymes to heat. Polym. Chem. 6, 3443–3448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Shea T. M., Webber M. J., Aimetti A. A., Langer R., Covalent incorporation of trehalose within hydrogels for enhanced long-term functional stability and controlled release of biomacromolecules. Adv. Healthc. Mater. 4, 1802–1812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macdougall L. J., Wechsler M. E., Culver H. R., Benke E. H., Broerman A., Bowman C. N., Anseth K. S., Charged poly( N -isopropylacrylamide) nanogels for the stabilization of high isoelectric point proteins. ACS Biomater. Sci. Eng. 7, 4282–4292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marco-Dufort B., Willi J., Vielba-Gomez F., Gatti F., Tibbitt M. W., Environment controls biomolecule release from dynamic covalent hydrogels. Biomacromolecules 22, 146–157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marco-Dufort B., Tibbitt M. W., Design of moldable hydrogels for biomedical applications using dynamic covalent boronic esters. Mater. Today Chem. 12, 16–33 (2019). [Google Scholar]
- 39.Webber M. J., Tibbitt M. W., Dynamic and reconfigurable materials from reversible network interactions. Nat. Rev. Mater. 7, 541–556 (2022). [Google Scholar]
- 40.Marco-Dufort B., Iten R., Tibbitt M. W., Linking molecular behavior to macroscopic properties in ideal dynamic covalent networks. J. Am. Chem. Soc. 142, 15371–15385 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Husain Q., β Galactosidases and their potential applications: A review. Crit. Rev. Biotechnol. 30, 41–62 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Rani S. A., Pitts B., Stewart P. S., Rapid diffusion of fluorescent tracers into staphylococcus epidermidis biofilms visualized by time lapse microscopy. Antimicrob. Agents Chemother. 49, 728–732 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaigalas A. K., Hubbard J. B., McCurley M., Woo S., Diffusion of bovine serum albumin in aqueous solutions. J. Phys. Chem. 96, 2355–2359 (1992). [Google Scholar]
- 44.Schnell E. A., Eikenes L., Tufto I., Erikson A., Juthajan A., Lindgren M., de L. Davies C., Diffusion measured by fluorescence recovery after photobleaching based on multiphoton excitation laser scanning microscopy. J. Biomed. Opt. 13, 064037 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Rittié L., Perbal B., Enzymes used in molecular biology: A useful guide. J. Cell Commun. Signal. 2, 25–45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsui M., Fowler J. H., Walling L. L., Leucine aminopeptidases: Diversity in structure and function. Biol. Chem. 387, 1535–1544 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Phillips J. W., Goetz M. A., Smith S. K., Zink D. L., Polishook J., Onishi R., Salowe S., Wiltsie J., Allocco J., Sigmund J., Dorso K., Lee S., Skwish S., De La Cruz M., Martín J., Vicente F., Genilloud O., Lu J., Painter R. E., Young K., Overbye K., Donald R. G. K., Singh S. B., Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chem. Biol. 18, 955–965 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Zhang L., Sullivan P., Suyama J., Marchetti D., Epidermal growth factor-induced heparanase nucleolar localization augments DNA topoisomerase I activity in brain metastatic breast cancer. Mol. Cancer Res. 8, 278–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Bank, Average time to clear exports through customs (days) | Data;https://data.worldbank.org/indicator/IC.CUS.DURS.EX.
- 50.Muller R., Betsou F., Barnes M. G., Harding K., Bonnet J., Kofanova O., Crowe J. H., Benson E., Bulla A., Chuaqui R., Clements J. A., Coppola D., De Souza Y., De Wilde A., Eliason J., Glazer B., Grizzle W., Gunter E., Hodgkinson V., Hubel A., Kokkat T., Koppandi I., Mackenzie-Dodds J., Pugh R., Rajasekhar V., Shea K., Skubitz A., Sobel M., Somiari S., Tybring G., Valyi-Nagy K., Preservation of biospecimens at ambient temperature: Special focus on nucleic acids and opportunities for the biobanking community. Biopreserv. Biobank 14, 89–98 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Lloyd J., Cheyne J., The origins of the vaccine cold chain and a glimpse of the future. Vaccine 35, 2115–2120 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zipursky S., Djingarey M. H., Lodjo J.-C., Olodo L., Tiendrebeogo S., Ronveaux O., Benefits of using vaccines out of the cold chain: Delivering meningitis A vaccine in a controlled temperature chain during the mass immunization campaign in Benin. Vaccine 32, 1431–1435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox M. M. J., Izikson R., Post P., Dunkle L., Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vaccines 3, 97–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richards K. A., Moritzky S., Shannon I., Fitzgerald T., Yang H., Branche A., Topham D. J., Treanor J. J., Nayak J., Sant A. J., Recombinant HA-based vaccine outperforms split and subunit vaccines in elicitation of influenza-specific CD4 T cells and CD4 T cell-dependent antibody responses in humans. npj Vaccines 5, 77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yakum M. N., Ateudjieu J., Pélagie F. R., Walter E. A., Watcho P., Factors associated with the exposure of vaccines to adverse temperature conditions: The case of North West region, Cameroon. BMC Res. Notes 8, 277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee C. S., Bishop E. S., Zhang R., Yu X., Farina E. M., Yan S., Zhao C., Zeng Z., Shu Y., Wu X., Lei J., Li Y., Zhang W., Yang C., Wu K., Wu Y., Ho S., Athiviraham A., Lee M. J., Wolf J. M., Reid R. R., He T.-C., Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 4, 43–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E. A., Dold C., Faust S. N., Finn A., Flaxman A. L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A. M., Pollock K. M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J.; Oxford COVID Vaccine Trial Group , Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A. M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., Berghmans P.-J., Kimmel M., Van Damme P., de Hoon J., Smith W., Stephenson K. E., De Rosa S. C., Cohen K. W., McElrath M. J., Cormier E., Scheper G., Barouch D. H., Hendriks J., Struyf F., Douoguih M., Van Hoof J., Schuitemaker H., Interim results of a phase 1–2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. 384, 1824–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang B.-B., Lum P. K., Hayashi M. M., Roskos L. K., Polyethylene glycol modification of filgrastim results in decreased renal clearance of the protein in rats. J. Pharm. Sci. 93, 1367–1373 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Koulouras G., Panagopoulos A., Rapsomaniki M. A., Giakoumakis N. N., Taraviras S., Lygerou Z., EasyFRAP-web: A web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 46, W467–W472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang M., Day C. A., Kenworthy A. K., DiBenedetto E., Simplified equation to extract diffusion coefficients from confocal FRAP data. Traffic 13, 1589–1600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huynh K., Partch C. L., Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1–28.9.14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Research Council, Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). [PubMed] [Google Scholar]
- 64.Merk A., Fukumura T., Zhu X., Darling J. E., Grisshammer R., Ognjenovic J., Subramaniam S., 1.8 Å resolution structure of β-galactosidase with a 200 kV CRYO ARM electron microscope. IUCrJ 7, 639–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strater N., X-ray structure of aminopeptidase A from Escherichia coli and a model for the nucleoprotein complex in Xer site-specific recombination. EMBO J. 18, 4513–4522 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy J. E., Tibbitts T. T., Kantrowitz E. R., Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases. J. Mol. Biol. 253, 604–617 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Chrencik J. E., Staker B. L., Burgin A. B., Pourquier P., Pommier Y., Stewart L., Redinbo M. R., Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 339, 773–784 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Broeck A. V., Lotz C., Ortiz J., Lamour V., Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex. Nat. Commun. 10, 4935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fleury D., Wharton S. A., Skehel J. J., Knossow M., Bizebard T., Antigen distortion allows influenza virus to escape neutralization. Nat. Struct. Biol. 5, 119–123 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Dai X., Wu L., Sun R., Zhou Z. H., Atomic structures of minor proteins VI and VII in human adenovirus. J. Virol. 91, e00850-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Tables S1 and S2
Movies S1 and S2