Abstract
Pseudomonas aeruginosa is a ubiquitous environmental bacterium and an important human pathogen. The production of several virulence factors by P. aeruginosa is controlled through two quorum-sensing systems, las and rhl. We have obtained evidence that both the las and rhl quorum-sensing systems are also required for type 4 pilus-dependent twitching motility and infection by the pilus-specific phage D3112cts. Mutants which lack the ability to synthesize PAI-1, PAI-2, or both autoinducers were significantly or greatly impaired in twitching motility and in susceptibility to D3112cts. Twitching motility and phage susceptibility in the autoinducer-deficient mutants were partially restored by exposure to exogenous PAI-1 and PAI-2. Both twitching motility and infection by pilus-specific phage are believed to be dependent on the extension and retraction of polar type 4 pili. Western blot analysis of whole-cell lysates and enzyme-linked immunosorbent assays of intact cells were used to measure the amounts of pilin on the cell surfaces of las and rhl mutants relative to that of the wild type. It appears that PAI-2 plays a crucial role in twitching motility and phage infection by affecting the export and assembly of surface type 4 pili. The ability of P. aeruginosa cells to adhere to human bronchial epithelial cells was also found to be dependent on the rhl quorum-sensing system. Microscopic analysis of twitching motility indicated that mutants which were unable to synthesize PAI-1 were defective in the maintenance of cellular monolayers and migrating packs of cells. Thus, PAI-1 appears to have an essential role in maintaining cell-cell spacing and associations required for effective twitching motility.
Pseudomonas aeruginosa is ubiquitous and highly successful in a diversity of environments, including soil, rhizosphere, freshwater, and marine environments. It is an opportunistic pathogen of humans, causing infections in immunocompromised patients such as those with cancer or AIDS, as well as those suffering from cystic fibrosis and burns (16, 36, 42). In addition, P. aeruginosa has also been shown to infect plants and insects (21, 37). The success of P. aeruginosa in these diverse environments is attributed to its impressive arsenal of virulence factors, which include multiple cell-associated factors such as alginate, pili, and lipopolysaccharide (12, 21) and secreted virulence factors, including toxins (exotoxin A and exoenzyme S), proteases (elastase, LasA protease, and alkaline protease), and hemolysins (phospholipase and rhamnolipid) (16, 36).
In recent years, P. aeruginosa has become a model for studying quorum-sensing control of virulence factors. P. aeruginosa has two complete, semiindependent, quorum-sensing systems, designated las and rhl. Each quorum-sensing system consists of a transcriptional activator, LasR or RhlR, and an autoinducer synthase, LasI or RhlI. LasI directs the synthesis of the autoinducer PAI-1, N-(3-oxododecanoyl)-l-homoserine lactone, while RhlI directs the synthesis of the autoinducer PAI-2, N-butyryl-l-homoserine lactone (35). The las system has been shown to regulate several virulence factors, including the LasA and LasB proteases, alkaline protease, exotoxin A, and RhlR, as well as the protein secretory genes xcpP and xcpR (11, 19, 44, 46). To date, the rhl quorum-sensing system has been shown to control the expression of the genes coding for LasB protease, alkaline protease, pyocyanin, and rhamnolipid, as well as critical genes such as rpoS and the xcpP and xcpR genes (3, 4, 20, 30, 32, 32a).
Over the past few years, P. aeruginosa has also become the preferred model system for studies of type 4 pili and twitching motility (1, 5, 6). The polar, type 4 pili of P. aeruginosa are considered virulence factors and are multifunctional structures which play a role in (i) adherence to, and colonization of, mucosal surfaces (9, 12, 47), (ii) the initial stages of infection by bacteriophages such as the transposable Mu-like phages D3112 and B3 (7, 38, 39) and the temperate transducing phage F116 (25, 33), and (iii) twitching motility (2, 6).
Twitching motility is a flagellum-independent mode of surface translocation which requires functional type 4 pili (2, 5, 6, 14). Flagellum-dependent motility and chemotaxis in P. aeruginosa is reasonably well understood and the subject of recent research (18, 24). In contrast, the cellular and molecular bases of twitching motility are presently unknown but are thought to involve extension and retraction of the type 4 pili, the same mechanism that has been proposed for infection by pilus-specific bacteriophages (25). The biogenesis and function of type 4 pili are quite complex. Presently, over 30 genes, comprising 15 loci in three separate regions of the chromosome, appear to be involved in the biosynthesis, regulation of expression, and function of type 4 pili in P. aeruginosa (1, 15). In addition, an unlinked set of pil genes, pilGHIJKL chpAB, that are required for normal pilus production or twitching motility but whose exact roles remain unclear was recently discovered (5, 6, 26). Interestingly, each of these pil and chp genes has significant regions of homology to one of the che genes required for flagellar chemotaxis in enteric bacteria (6, 13) and to frz genes of Myxococcus xanthus (27).
The role of autoinducer-mediated signaling, or quorum sensing, in the control and expression of twitching motility has not been previously explored. We were intrigued by the possibility of a connection between quorum sensing and twitching motility because twitching motility is so clearly associated with high cell densities and cell-cell communication, which are the hallmarks of quorum-sensing systems in bacteria. Recently, quorum sensing has been shown to be involved in initiating and controlling swarming motility and the multicellular behavior of Serratia liquifaciens (10). We have been examining the roles that the las and rhl quorum-sensing systems play in the production of functional type 4 pili and the regulation of twitching motility in P. aeruginosa PAO1. In this report, we present evidence that both quorum-sensing systems of P. aeruginosa, las and rhl, are required for normal twitching motility and that the rhl system, but not the las system, is required for infection by pilus-specific phages and adherence to human bronchial epithelial cells.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) was routinely used to propagate P. aeruginosa strains. Broth was solidified with agar at the appropriate concentrations for the individual assays described below. Wild-type and mutant strains of P. aeruginosa were maintained as glycerol stocks and stored at −70°C. Freshly isolated colonies on LB agar plates were used for each experiment. Medium was supplemented with tetracycline at 50 μg/ml and mercuric chloride at 15 μg/ml, when needed.
TABLE 1.
Bacterial strains used
| P. aeruginosa strain | Relevant characteristic | Source |
|---|---|---|
| PAO1 | Wild-type prototroph | B. Iglewski |
| PDO100 | ΔrhlI::Tn501-2 derivative of PAO1 | D. Ohman |
| PAO-JP1 | ΔlasI::Tet derivative of PAO1 | B. Iglewski |
| PAO-JP2 | ΔlasI ΔrhlI::Tn501 | B. Iglewski |
Twitching-motility assays.
Twitching motility was assayed by two methods. Plates with LB agar (1%) poured to an average depth of 3 mm were prepared and dried briefly, and the strains to be tested were stab inoculated with a needle to the bottom of the petri dish. Plates were incubated at 37°C for 48 h, except where noted otherwise below. After the incubation period, the zone between the agar and the bottom of the petri dish, the twitch zone, was measured (5). A slide culture assay was used to assess twitching motility at the microscopic level (5). Strains were point inoculated onto the surface of a slab of LB agar (1%) placed on a microscope slide. The inoculated LB agar was covered with a coverslip, and the slide cultures were incubated for 3 to 5 h at 37°C in a humid petri dish. Twitching motility was visualized under an Olympus BH/2 microscope by using Nomarski optics and a 60× lens objective. The wild-type strain of PAO1 was included as a positive control (twt+) in both assays.
Phage sensitivity and adsorption assays.
Phage sensitivity was assayed by the plaque assay. Ten-microliter aliquots of diluted D3112cts phage lysates (103 to 109 PFU/ml) were spotted onto LB top agar (0.3%) seeded with 0.1 ml of a stationary-phase culture of the strain being tested (39).
Western blot analysis.
A polyclonal antibody prepared against the type 4 pili of P. aeruginosa, kindly provided by Randall Irvin (University of Alberta, Edmonton, Alberta, Canada) was used to determine the relative amounts of pilin protein produced by wild-type and mutant cells. Bacterial strains were grown from frozen stocks on LB agar (2%) plates for 7 to 8 h at 37°C, resuspended in double-distilled water, and adjusted to equivalent protein concentrations. Cells were solubilized by the addition of Laemmli sample buffer and boiled for 10 min (Bio-Rad, Cambridge, Mass.). Whole-cell lysates were separated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (12% [wt/vol] acrylamide). Prestained molecular weight standards (Bio-Rad) were used to assess efficiency of protein transfer to nitrocellulose membranes. Western immunoblotting was performed with polyclonal PAO1 anti-pilin antibodies, and detection was accomplished with the ProtoBlot II AP system (Bio-Rad).
Enzyme-linked immunosorbent assays (ELISAs).
Cultures were grown as described above for Western blot analysis. Growth was resuspended to an A600 of approximately 1.0 in LB medium. Cell suspensions were adjusted to contain the same number of cells as determined by direct microscopic counts. Aliquots of the cell suspensions were transferred to microtiter plate wells (Corning flat bottom, tissue culture treated), and incubated (16 h) at 4°C. All washes and antibody incubations of the microtiter plate wells were carried out following the attachment of cells. The wells were washed three times with phosphate-buffered saline (PBS) containing 0.05% Tween 20. Blocking was accomplished with 1% bovine serum albumin in PBS for 1 h at 37°C. The wells were washed three times with PBS. A 1/100 dilution of the primary anti-pilin antibody in 0.5% bovine serum albumin was added to the wells, and the plates were incubated for 1 h at 37°C. The developing antibody, an alkaline phosphatase conjugate (Bio-Rad), and the detection substrate (Sigma Fast p-Nitrophenyl Phosphate Tablet Sets) were used according to the manufacturer’s directions. Absorbance was measured with a Dynatech MR500 plate reader. Multiple trials of five replicates of each strain were tested, and mean values were calculated.
Cell adherence assays.
Bacterial strains were grown from frozen stocks on LB plates supplemented with an appropriate antibiotic(s). Bacteria were swabbed from the surfaces of the plates and suspended in LB medium, with or without a 10 μM concentration of one or 10 μM concentrations of both autoinducers, and grown at 37°C to an A660 of approximately 1.0. A 100-μl volume of cell suspension was plated onto LB plates, with or without autoinducers, and incubated overnight at 37°C. Bacteria were resuspended in 1 ml of LB medium to an A660 of approximately 0.4. Bacteria were labeled with [35S]methionine for 10 min, washed four times with Hanks balanced salt solution, and resuspended in a 1-ml volume. The concentration of bacterial cells was determined by plate counts. Human bronchial epithelial (16HBE) cells were grown to confluency in collagen-coated 24-well plates with minimum essential medium plus 10% fetal calf serum. 35S-labeled bacterial cells were added to the wells containing the 16HBE cells and incubated at 37°C for 1 h with slow rocking. Following this incubation period, the wells were washed four times with Hanks balanced salt solution to remove nonadherent bacterial cells. Cells and adherent bacteria were lysed with 2% SDS and counted in a scintillation counter. Each strain was tested in quadruplicate in three separate trials and standardized to 5 × 108 CFU of added bacterial cells.
RESULTS
Twitching motility of las and rhl quorum-sensing mutants.
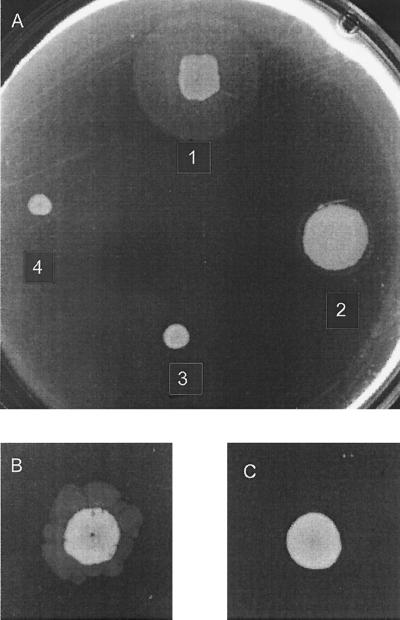
Macroscopic twitching-motility assays revealed that pilus-mediated motility was aberrant in all the mutant strains (Fig. 1A). However, flagellar motility was found to be unchanged (data not shown). The wild-type strain, PAO1, formed a twitch zone between the agar and the bottom of the petri dish and a normal surface colony (Fig. 1A and Table 2) as previously reported (5). In contrast, PDO100 (rhlI) was capable of producing only highly irregular twitch zones after extended periods and PAO-JP2 (lasI rhlI) was unable to form detectable twitch zones in this assay. PAO-JP1 (lasI) was found to produce a reduced twitch zone; however, the surface colony exhibited enhanced spreading. Growth rates of the individual mutant strains were not substantially different from that of the wild type.
FIG. 1.
Macroscopic stab assay for twitching motility. LB agar (1%) plates were stab inoculated with a needle to the bottom of the plate and incubated for 24 h at 37°C. The diffuse zone, the twitch zone, between the agar and the petri dish is a measure of twitching motility. The smaller, denser zone represents surface colony growth. (A) PAO1 wild type and “I” mutants. Colony 1, PAO1; colony 2, PAO-JP1 (ΔlasI); colony 3, PDO100 (ΔrhlI); colony 4, PAO-JP2 (ΔlasI rhlI). PAO-JP2 (ΔlasI rhlI), with PAI-1 and PAI-2 added to the agar at 10 μM each (B) and without PAI-1 and PAI-2 (C) was incubated for 48 h at 37°C.
TABLE 2.
Phage sensitivity, twitching-motility phenotype, and piliation of wild-type and mutant strains
| P. aeruginosa strain | Sensitivity to phage D3112a | Area of twitching motilityb | Surface piliationc |
|---|---|---|---|
| PAO1 | 103 | 20 | 0.407 ± 0.110 |
| PDO100 (rhlI) | 107d | 0e | 0.028 ± 0.009 |
| PAO-JP1 (lasI) | 103 | 12 | 0.351 ± 0.033 |
| PAO-JP2 (lasI rhlI) | 107d | 0 | 0.045 ± 0.014 |
Lowest titer of phage stock (dilutions of 103 to 109 PFU/ml were tested) resulting in plaque formation when 10 μl was spotted onto agar overlays containing the strain tested.
Diameters (in millimeters) of twitch zones in macroscopic twitch stab assay plates as shown in Fig. 1.
Mean A410 values ± standard deviations from ELISAs with PAO1 anti-pilin antibodies against whole cells and alkaline phosphatase-conjugated goat anti-rabbit developing antibodies.
Turbid plaques were observed.
Very small twitch zones were observed after extended incubation periods (>72 h) in some trials.
Twitching motility was partially restored in the mutant strain PAO-JP2 (lasI rhlI) by the addition of 10 μM concentrations (each) of PAI-1 and PAI-2 to twitch agar plates (Fig. 1B). The addition of neither PAI-1 nor PAI-2 alone was sufficient to restore twitching motility in PAOJP2. However, the addition of pure PAI-2 to twitch plates inoculated with PDO100 (rhlI) resulted in a partial restoration of twitching motility, but the concentration required varied from trial to trial (data not shown). In all instances, exogenous additions of autoinducers to the twitching-motility assay plates resulted in irregular and atypical twitch zones.
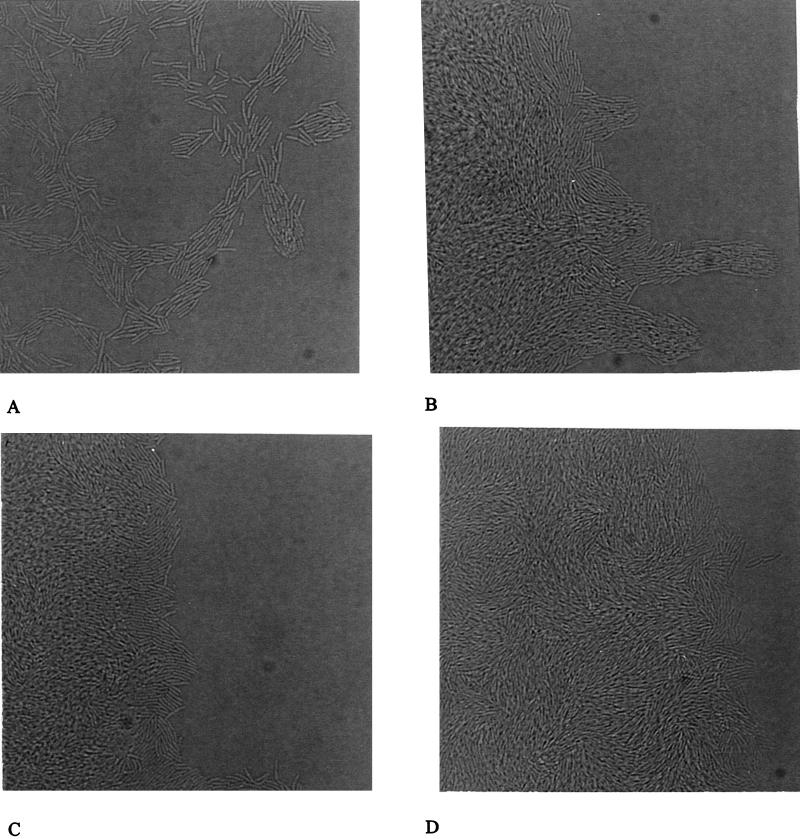
Microscopic analysis of twitching motility by a slide culture assay was used to examine twitching motility defects at the cellular level. As shown in Fig. 2A, the wild-type (PAO1) slide culture revealed the presence of a monolayer of motile “rafts” of cells at the leading edge of the colony periphery, which are characteristic of twitching motility in this bacterium (5). PAO-JP1 (lasI) cells were motile and formed rafts; however, the rafts contained dramatically larger numbers of cells and were often multilayered (Fig. 2B). In contrast, the edges of the microcolonies formed by the strains PDO100 (rhlI) and PAO-JP2 (lasI rhlI) were similar and characterized by multiple layers of cells and very few rafts (Fig. 2C to D). While PAO-JP2 was nonmotile in this assay, PDO100 (rhlI) was observed to be capable of movement, albeit at severely reduced rates and frequencies. In contrast, in the plate assay (Table 2) PDO100 was nonmotile after 24 h incubation.
FIG. 2.
Slide culture assay for twitching motility. The P. aeruginosa PAO1 wild type and las and rhl quorum-sensing mutants were inoculated onto slide cultures and incubated for 3 to 5 h at 37°C. The areas in the photomicrographs represent the edges of cell growth (magnification, ca. ×570). (A) PAO1 (wild type); (B) PAO-JP1 (ΔlasI); (C) PDO100 (ΔrhlI); (D) PAO-JP2 (ΔlasI rhlI).
Piliation of las and rhl quorum-sensing mutants.
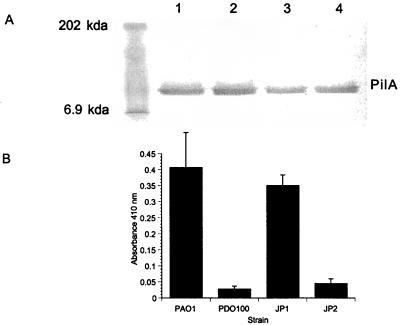
Western blot analysis and ELISAs were used to determine levels of pilin protein produced by the las and rhl quorum-sensing mutants and levels of surface piliation, respectively. Western blot analysis of whole-cell lysates showed that the total levels of pilin were approximately equivalent for wild-type and mutant strains (Fig. 3A). However, ELISAs of whole cells revealed distinct differences in the levels of pili on the surfaces of wild-type and mutant cells (Fig. 3B). Cells of PAO-JP1 (lasI) had levels of surface pili equivalent to that of wild-type cells. In contrast, the levels of pili detected on the surfaces of cells of the mutant strains PDO100 (rhlI) and PAO-JP2 (lasI rhlI) were significantly reduced (Fig. 3B).
FIG. 3.
Western blot analysis and ELISA of pilin. (A) Western blot analysis of pilin in whole-cell lysates. Whole-cell lysates were adjusted to contain equal amounts of protein, solubilized by boiling in SDS-polyacrylamide gel electrophoresis sample buffer, and separated by electrophoresis on an SDS–12% polyacrylamide gel by using a standard Tris-glycine buffer system. Western blots were developed with PAO1 anti-pilin polyclonal antiserum and an alkaline phosphatase-conjugated antibody as the developing antibody (Promega). (B) ELISA against whole cells. Bacterial cells were bound overnight to microtiter plate wells, followed by incubation with the PAO1 anti-pilin polyclonal antiserum. The developing antibody was an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin. The A410 was read with a Dynetech MR500 plate reader. Values are the means of results from three trials of five replicates each.
Phage sensitivity and adsorption.
Sensitivity to infection by the pilus-specific phage D3112cts was measured by titration. Aliquots (10 μl) of various dilutions of a phage stock were spotted onto agar overlays containing the wild-type strain or mutant strains. Wild-type PAO1 and the mutant PAO-JP1 (lasI) were both sensitive to the lowest titer (103 PFU/ml) of phage tested, resulting in total clearing in the area of the applied phage (Table 2). In contrast, PDO100 (rhlI) and PAO-JP2 (lasI rhlI) were highly resistant, forming only turbid plaques at the highest titers tested (107 and 109 PFU/ml).
Adherence to human bronchial epithelial cells.
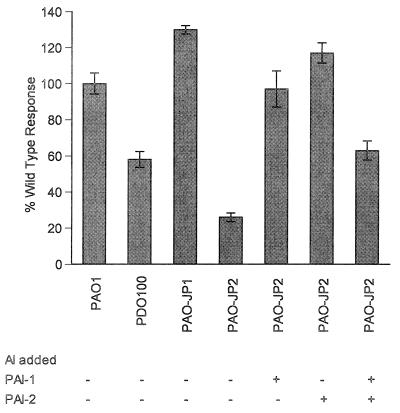
Wild-type PAO1 and PAO-JP1 (lasI) cells bound to human bronchial epithelial (16HBE) cells equally well (Fig. 4). Interestingly, the PDO100 (rhlI) and PAO-JP2 (lasI rhlI) strains exhibited a two- to fivefold decrease in the level of adherence to 16HBE cells compared to that of wild-type bacteria. When a 10 μM concentration of either PAI-1 or PAI-2 was added back to PAO-JP2 cells, the level of binding was restored to that of wild-type bacteria (Fig. 4). Interestingly, the addition of both PAI-1 and PAI-2 at 10 μM each resulted in a significant reduction in binding ability.
FIG. 4.
Epithelial cell binding assay. P. aeruginosa strains labeled with [35S]methionine were incubated with confluent monolayers of 16HBE cells. After 1 h cells were washed to remove nonadherent bacteria. Cells were lysed, and numbers of CFU of bacteria were determined. Data was standardized to 5 × 108 bacteria added and was done with quadruplicate 16HBE cultures. PAO1 (wild type), PAO-JP2 (ΔlasI rhlI), PDO100 (ΔrhlI), and PAO-JP1 (ΔlasI) were used + indicates the addition of the appropriate autoinducer (10 μM PAI-1 or PAI-2) to PAO-JP2. Values are representative of results of three separate experiments and are expressed as percentages of the wild-type response.
DISCUSSION
P. aeruginosa has two quorum-sensing systems, las and rhl, each of which has been shown to activate numerous genes in a hierarchial manner (20, 41). Some genes (e.g., lasB) can be activated by either system (20, 41). In addition, the las system regulates the rhl system in several ways. First, LasR–PAI-1 activates transcription of rhlR (20, 34). Second, in the absence of PAI-2, PAI-1 can activate the RhlR protein, albeit not as well as the RhlR cognate autoinducer PAI-2 (32). Third, PAI-1 very efficiently competes with PAI-2 for binding to RhlR and results in the inhibition of PAI-2 activation of RhlR when both autoinducers are present (32, 34). These complex interrelationships between the las and rhl systems are exemplified by the findings of the present study.
The results described above show that both autoinducer compounds, PAI-1 and PAI-2, play important roles in maintaining the normal twitching motility and pilus function of P. aeruginosa. Our results indicate that the rhl quorum-sensing system, but not the las system, affects the export and/or assembly of pilin into functional pili on the cell surface as shown by ELISAs with anti-pilin antibodies against intact cells. It has been previously shown that pilin mRNA levels in the lasI and rhlI mutants are equivalent to those of the wild-type strain (32), and our Western blot analysis likewise indicated that pilin synthesis is unaffected by mutations in autoinducer synthases. Thus, it seems clear that PAI-2 synthesis somehow affects the formation of functional pili on the cell surface at the level of pilin export and/or assembly, not at the level of pilin synthesis. Such export and assembly of pilin is a crucial aspect of twitching motility since previous studies (1, 2) suggest that twitching motility operates by the repeated extension and retraction of polar type 4 pili, possibly involving assembly and disassembly. The rhlI mutant strain used in this study has been shown to have a partial deletion in the pheC gene immediately downstream of rhlI (3). However, the addition of phenylalanine had no effect on the restoration of twitching motility. Additionally, we have found that the lasR rhlR double mutant, which contains a normal pheC gene, is also deficient in twitching motility (data not shown).
Our results indicate that the las quorum-sensing system is also required for normal twitching motility. The lasI mutant was not deficient in surface pili and was therefore expected to retain wild-type twitching motility and susceptibility to infection by D3112cts. The lasI mutant was indeed found to retain wild-type susceptibility to phage D3112; thus, the pili of this mutant are at least partially functional. However, the lasI mutant was significantly altered in twitching motility, as shown in both macroscopic and microscopic assays. These differences in cell-cell associations suggest that the role(s) of PAI-1 may be to keep the cells at a certain cell density, promote optimal cell-cell associations, and direct cellular movement.
A role for the las system in maintaining normal twitching motility is further supported by the requirement of both PAI-1 and PAI-2 for restoration of twitching motility in PAO-JP2 (lasI rhlI). Alternatively, this may simply reflect the fact that LasR–PAI-1 is required for normal expression of the rhlR gene. Pesci et al. (34) showed that PAI-2 alone has no effect on the autoinduction of a rhlR′-lacZ fusion in PAO-JP2. Thus, PAI-1 is required to activate the rhl system.
Recently, it was reported that both the las and rhl quorum-sensing systems regulate the xcpP and xcpR genes, which are components of the type II or general secretory pathway (GSP) of P. aeruginosa (4, 45). The GSP is responsible for the secretion of P. aeruginosa exoenzymes such as elastase, exotoxin A, phospholipase C, lipase, and alkaline phosphatase (30, 32, 40, 45). The assembly of type 4 pili is the function of a second branch of the GSP, termed the pilus biogenesis pathway (22). There are two sets of genes specifying the Xcp system and the pilus biogenesis pathway, and although the two branches operate independently, there is significant overlap as both appear to require the products of the pilD (xcpA) and pilA genes (17, 22, 23, 43, 45). It is not surprising, therefore, that assembly of pili is regulated by the quorum-sensing systems as well. Presently, the target of the rhl quorum-sensing system that controls twitching motility is unknown, but the genes involved in the pilus biogenesis pathway are likely candidates and are presently being studied in our laboratory.
The observation that the PDO100 (rhlI) and PAO-JP2 (lasI rhlI) mutants, which do not produce significant pili on their cell surfaces, have reduced levels of binding to human bronchial epithelial cell surfaces was not surprising. Reports have indicated that the type 4 pilus of P. aeruginosa accounts for approximately 90% of its adherence capability to a variety of cell types, with the pilin subunit itself carrying the adherence function (12, 47). Additionally, O’Toole and Kolter (31) have recently shown that the type 4 pili of P. aeruginosa are also necessary for attachment to surfaces in biofilm formation. On the surface, our results showing that the addition of PAI-1 or PAI-2 alone restores cell binding to wild-type levels but that the addition of both autoinducers results in a reduction of cell binding appear confusing. However, it has been shown that PAI-1 can partially activate RhlR, albeit much less effectively than PAI-2 (32). Furthermore, PAI-1 very effectively inhibits binding of PAI-2 to RhlR (34). Therefore, in the presence of both PAI-1 and PAI-2 there may be competition, which may lead to a reduction in the activity of RhlR.
The absolute levels, or dose, of autoinducers has been found to be critical to the expression levels and functions of quorum-sensing-regulated genes in P. aeruginosa (32). Recently, Nassar et al. (29) have shown that, depending on the concentration of the cognate autoinducer N-(3-oxohexanoyl)-homoserine lactone (OHHL), the transcriptional activator ExpR binds to DNA in different ways. Clarification of this finding awaits purification of the LasR and RhlR proteins. Twitching motility, adherence to cells and surfaces, and phage infection are likely to require or represent different levels of pilus functionality. Normal twitching motility certainly requires fully functional pili, while adherence and infection by phage may have different requirements.
The finding that pilus assembly and twitching motility is controlled by quorum sensing in P. aeruginosa has not been reported previously. Until now the focus has been on quorum-sensing control of virulence factors that are exoenzymes; however, pili are also considered an important virulence factor. The coordinated timing of the expression of multiple virulence factors at high cell densities is likely to confer a significant advantage to a pathogen in allowing it to overcome host defenses. Indeed, gliding motility of the opportunistic pathogen S. liquifaciens has been shown to be controlled by a quorum-sensing system (10). Additionally, autoinducers have recently been shown to play a role in the development of both naturally occurring and laboratory-created biofilms with P. aeruginosa (8, 28). These studies provide evidence that autoinducers and quorum-sensing systems may, in fact, be important in natural environments and complex microbial communities. Determination of exactly how the rhl and las quorum-sensing systems control twitching motility and cell-cell communication on cell surfaces awaits further study.
ACKNOWLEDGMENTS
This work was supported by NIH grant A133713 to B.H.I. R.S.S. was supported by NIH training grant HL07216.
We thank Randall Irvin for the generous gift of anti-pilin antibodies.
REFERENCES
- 1.Alm R S, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Bradley D E. A function of Pseudomonas aeruginosa PAO pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LasR-LasI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 5.Darzins A. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric single-domain response regulator CheY. J Bacteriol. 1993;175:5934–5944. doi: 10.1128/jb.175.18.5934-5944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 7.Darzins A, Casadaban M J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989;171:3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Doig P, Todd T, Sastry P A, Lee K K, Hodges R S, Paranchych W, Irvin R T. Role of pili in adhesion of Pseudomonas aeruginosa to human epithelial cells. Infect Immun. 1988;56:1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberl L, Winson M K, Sternberg C, Stewart G S, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behavior of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 11.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 13.Hazelbauer G L, Berg H C, Matsumura P. Bacterial motility and signal transduction. Cell. 1993;73:15–22. doi: 10.1016/0092-8674(93)90156-k. [DOI] [PubMed] [Google Scholar]
- 14.Henrichson J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs M, Collie E S, Free P D, Livingston S P, Mattick J S. PilS and PilR, a two component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 16.Joklik W K, Willett H P, Amos D B, Wilfert C M. Pseudomonas. In: Joklik W K, editor. Zinsser microbiology. 20th ed. Norwalk, Conn: Appleton and Lange; 1992. pp. 576–583. [Google Scholar]
- 17.Koomey M. Prepilin-like molecules in type-4 pilus biogenesis: minor subunits, chaperones or mediators of organelle translocation. Trends Microbiol. 1995;3:409–411. doi: 10.1016/s0966-842x(00)88989-1. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda A, Kumano T, Taguchi K, Nikata T, Kato J, Ohtake H. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J Bacteriol. 1995;177:7019–7025. doi: 10.1128/jb.177.24.7019-7025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 20.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 21.Lory S. Pseudomonas and other nonfermenting bacilli. In: Davis B D, Dulbecco R, Eisen H N, Ginsberg H S, editors. Microbiology. Philadelphia, Pa: Lippincott; 1990. pp. 595–600. [Google Scholar]
- 22.Lory S, Lu H-M, Pepe J. Common steps in extracellular protein secretion and biogenesis of pili in Pseudomonas aeruginosa. In: Nakazawa T, editor. Molecular biology of Pseudomonas. Washington, D.C: American Society for Microbiology; 1996. pp. 417–426. [Google Scholar]
- 23.Lu H-M, Motley S T, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 24.Masduki A, Nakamura J, Olga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:333–343. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattick J S, Whitchurch C B, Alm R A. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene. 1996;179:147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 26.Mattick J S, Whitchurch C B, Martin P R, Alm R A, Hobbs M. Regulatory pathways affecting the biogenesis and function of type 4 fimbriae in Pseudomonas aeruginosa, abstr. XXI. VI International Congress on Pseudomonas; Molecular Biology and Biotechnology. 1997. [Google Scholar]
- 27.McBride M J, Weinberg R A, Zusman D R. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean R J C, Whiteley M, Stickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 29.Nasser W, Bouillant M L, Salmond G, Reverchon S. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 30.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 32.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1997;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pemberton J M. F116: a DNA bacteriophage specific for the pili of Pseudomonas aeruginosa strain PAO. Virology. 1973;55:558–560. doi: 10.1016/0042-6822(73)90203-1. [DOI] [PubMed] [Google Scholar]
- 34.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pesci E C, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C: ASM Press; 1999. pp. 147–155. [Google Scholar]
- 36.Pollack M. Pseudomonas aeruginosa. In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practices of infectious diseases. Edinburgh, Scotland: Churchill Livingstone; 1990. pp. 1673–1691. [Google Scholar]
- 37.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 38.Rehmat S, Shapiro J A. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol Gen Genet. 1983;192:416–423. doi: 10.1007/BF00392184. [DOI] [PubMed] [Google Scholar]
- 39.Roncero C, Darzins A, Casadaban M J. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J Bacterial. 1990;172:1899–1904. doi: 10.1128/jb.172.4.1899-1904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rust L, Pesci E C, Iglewski B H. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by the LasR and Pseudomonas autoinducer PAI-1: an autoinducer regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J J, Travis S, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 43.Strom M S, Nunn D, Lory S. Multiple roles of the pili biogenesis protein PilD: involvement of PilD in secretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991;173:1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toder D S, Gambello M J, Iglewski B H. Pseudomonas aeruginosa LasA; a second elastase gene under transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 45.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunski Andree. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992;103:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 46.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong W Y, Campbell A P, McInnes C, Sykes B D, Paranchych W, Irvin R T, Hodges R S. Structure-function analysis of the adherence-binding domain on the pilin of Pseudomonas aeruginosa PAK and KB7. Biochemistry. 1995;34:12963–12972. doi: 10.1021/bi00040a006. [DOI] [PubMed] [Google Scholar]