Abstract
Investigators have long been interested in the natural phenomenon of fetal and placental cell trafficking into the maternal circulation. The scarcity of these circulating cells makes their detection and isolation technically challenging. However, as a DNA source of fetal origin not mixed with maternal DNA, they have the potential of considerable benefit over circulating cell-free DNA-based noninvasive prenatal genetic testing (NIPT). Endocervical trophoblasts, which are less rare but more challenging to recover are also being investigated as an approach for cell-based NIPT. We review published studies from around the world describing both forms of cell-based NIPT and highlight the different approaches’ advantages and drawbacks. We also offer guidance for developing a sound cell-based NIPT protocol.
INTRODUCTION
Cell-based noninvasive prenatal testing (cbNIPT) using fetal and placental cells offers unique advantages over both diagnostic invasive testing and cell-free DNA (cfDNA)-based NIPT. Chorionic villus sampling (CVS) and amniocentesis are the gold standard for prenatal diagnostic genetic testing, but they have a 0.3% to 0.1% procedure-associated risk of miscarriage.1 Although this risk is very low, it has driven high patient and provider demand for, and rapid clinical implementation of, noninvasive prenatal screening (NIPS) analyzing circulating cfDNA in maternal plasma. However, the resolution and positive predictive value of cfDNA-based NIPS are limited, primarily because circulating cfDNA is mostly maternal in origin; only 5-20%, referred to as the fetal fraction, derives from the placenta. In contrast, if circulating fetal nucleated red cells and trophoblasts, or trophoblasts from the endocervical canal, can be consistently recovered, they provide a pure source of fetal DNA. We review here recent progress in using fetal DNA for cbNIPT and predict future developments in this field.
FETAL AND PLACENTAL CELLS IN THE MATERNAL CIRCULATION
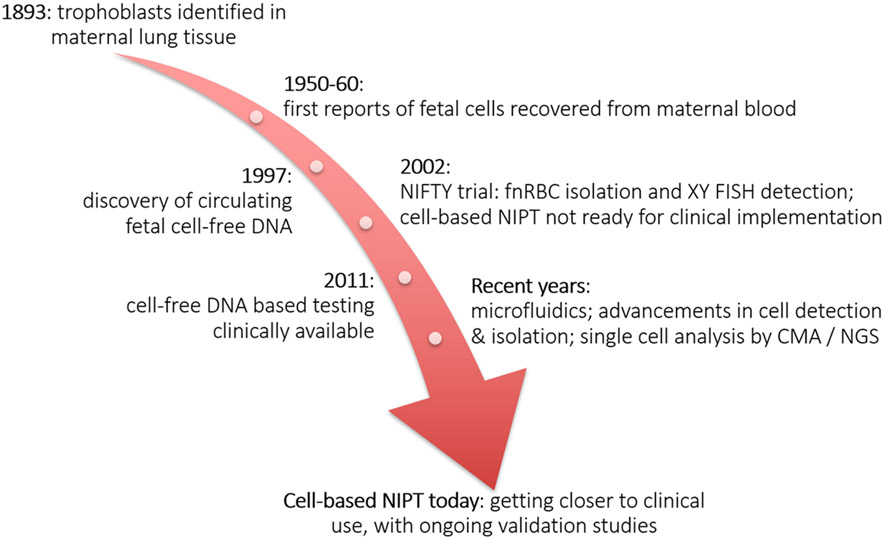
Schmorl first identified placental cells in the maternal circulation in 1893, when he found clusters of trophoblasts in the pulmonary capillaries of women who had died of eclampsia.2,3 Various studies conducted in the 1950’s and 60’s reported fetal cells in maternal blood samples collected mostly after delivery.4-8 In 1969, Walknowska and colleagues suggested using these circulating fetal cells for NIPT, after they found male cells in lymphocyte cultures prepared from maternal samples collected between 14 and 37 weeks gestation.9 Later, fetal cells in maternal blood were detected as early as 4 weeks gestation.10
Four different types of circulating fetal cells have been described: [1] trophoblasts, [2] fetal nucleated red blood cells (fnRBC), [3] lymphocytes, and [4] stem/progenitor cells. The latter two may build up maternal tolerance of the fetus or in the development of maternal autoimmune diseases,11 and they can persist for many years after delivery.12 This persistence makes fetal circulating lymphocytes and stem/progenitor cells unsuitable for non-invasive prenatal testing, as their presence in subsequent pregnancies could confound test results. In contrast, fnRBC and trophoblasts are rapidly cleared from the maternal circulation after delivery, with none detectable ≥8 weeks post-partum.13 Primitive erythroblasts from the yolk sac are the first fnRBC to appear at 7-12 weeks gestation, but disappear after 12w,14 when definitive erythroblasts, derived from the fetal liver, appear. The trophoblasts that enter the maternal circulation can originate from different trophoblast cell types. A detailed review of placental development and trophoblast lineage differentiation is beyond the scope of this text and can be found elsewhere.15 Briefly, upon implantation, the polar trophectoderm of the blastocyst invades the decidual stroma and differentiates into the earliest trophoblast lineages, the primitive mononuclear cytotrophoblast (CTB) and the multinucleated primitive syncytium (PS). Early proliferating cytotrophoblasts (CTB) then breach the uterine capillaries and form primary villi covered by CTB. Secondary and tertiary villi acquire mesoderm-derived stroma and blood vessels. These floating chorionic villi are covered by multinucleated syncytiotrophoblasts, formed through asymmetric division and fusion of CTB15,16. Villi at the periphery invade deeper into the uterine wall and become anchoring villi covered with mononuclear extravillous trophoblasts (EVT). EVT further differentiate into deeply invading interstitial EVT and endovascular EVT that replace the endothelium of maternal spiral arteries.15 These different stages of trophoblast development are marked by expression of different genes, and expression data support that primarily (endovascular) EVT are isolated from maternal blood samples.17-19 Whether more primitive trophoblast cell types circulate at earlier gestational ages needs further study.
The relative scarcity of fetal cells makes it challenging to isolate them from maternal blood. Without selecting for a specific cell type, the number of fetal cells is estimated to vary between 1-2 to 2-6 cells/ml of maternal blood.20,21 However, isolating cells without prior selection is labor-intensive, slow, and does not avoid cell types persisting from prior pregnancies. Thus, a cell type-specific enrichment procedure is recommended, even though it usually results in a lower fetal cell yield. The overall yield from maternal blood depends on multiple variables including gestational age10,22-27, pregnancy complications and pathologies28,29, fetal abnormalities20,30,31, maternal body mass index (BMI),27,32 physical activity before blood collection33, and various other factors,34 including individual biological variation. The optimal period for circulating trophoblast recovery is between 10-14 weeks gestation, coinciding with the high placental vascularization rate at the end of the first trimester,35 with a significantly lower yield after 15 weeks when there are fewer invading trophoblasts.23,25-27 The same downward trend has been reported for fnRBC,22,23 although one study found no correlation between the number of recovered fnRBC and gestational age.36 In contrast, in two studies with no cell selection procedure the number of recovered cells increased throughout pregnancy,10,23 indicating that although cell recovery varies and is methodology-dependent, circulating fetal cells can be isolated until advanced gestational age. As with fetal cfDNA,37,38 higher maternal BMI also reduces trophoblast yield, although with circulating cells, this trend is not always statistically significant.25,27,32
Some factors are associated with an increased number of fetal cells in the maternal circulation (e.g. with fetal trisomy 21 and other aneuploidies)20,30,31 including sex chromosome abnormalities, triploidy or +invdup(15).20,31 In contrast, fetal trisomy 18, a few cases of trisomy 13, and (in one study) 47,XXY were not associated with increased numbers of fetal cells.20,30,31 Trisomy 18 pregnancies usually have smaller placentas, which might release fewer trophoblasts into the maternal circulation. More circulating fetal cells have also been found with fetal growth restriction or other pregnancy complications such as eclampsia,29 hypertensive disorders of pregnancy (unpublished data, Crovetti et al.), or other adverse pregnancy outcomes, supporting that circulating cells have potential as a biomarker for abnormal placentation.28,39 One group reported that 30 minutes of moderate physical activity before blood draw led to an increased trophoblast yield.33 The same investigators also found that pregnancies with male fetuses were associated with a higher median number of circulating trophoblasts compared to female fetuses, and that the level of a few cytokines (RANTES, IL-2 and IL-5) correlates with trophoblast count.34
It is not known whether amniocentesis, CVS, or another prior procedure increases the numbers of circulating trophoblasts or fetal cells. However, since data from cffDNA analysis show a small but non-significant increase in the amount of cffDNA or fetal fraction immediately after CVS,40-42 we recommend that research on the yield of circulating trophoblast recovery be conducted with samples obtained before a prenatal diagnostic procedure or drawn at least one week afterward.
CELL ISOLATION FROM MATERNAL BLOOD
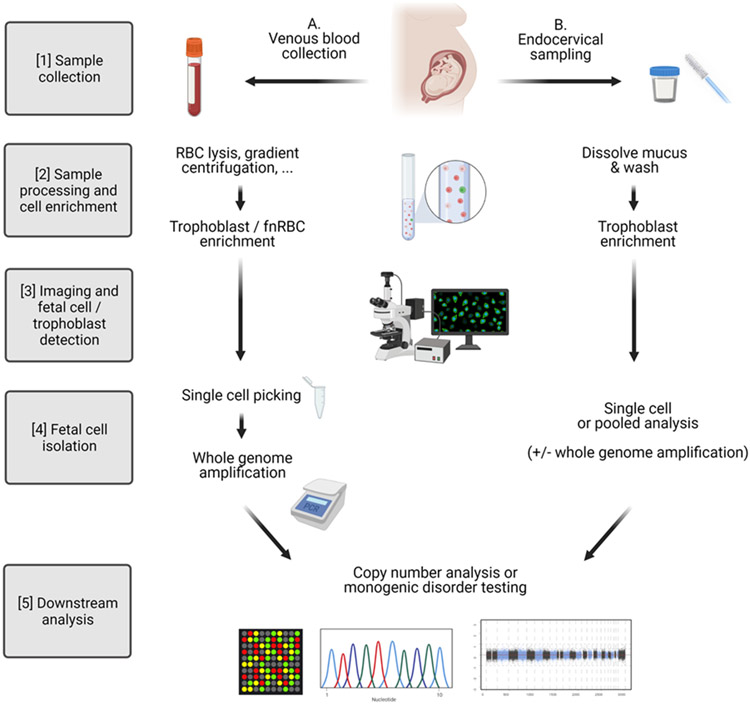
The general workflow of a cbNIPT protocol includes: [1] maternal sample collection, [2] initial sample processing and fetal cell enrichment (based on specific markers or size) with or without maternal cell depletion, [3] fetal cell identification, [4] fetal cell isolation, and [5] downstream analysis (Figure 1). Many different fetal cell isolation and detection techniques have been investigated over the years (overview in Supplementary tables). Earlier studies mainly utilized Y-chromosome detection (e.g., by fluorescence in situ hybridization (FISH)), which allows for broader capture of different cell types but precludes testing pregnancies with female fetuses. Hence, investigators have also explored specific isolation protocols independent of fetal sex.
Figure 1. Cell-based NIPT workflow.
(Created with BioRender.com)
Fetal nucleated red blood cells
Maternal red blood cells can be removed by bulk separation or density gradient centrifugation using Ficoll43-46 or Percoll47-50 in different gradient combinations, to separate the nucleated cell fraction (including fetal cells) from the maternal RBC and plasma fraction. That said, using Ficoll could result in 60-80% fetal cell loss,51 making the use of less aggressive methods more desirable. Lowering the pH of diluted blood samples by adding ACD (sodium citrate/citric acid/dextrose) improves retention of fnRBC during density gradient separation by increasing the mean corpuscular volume and lowering the density of the RBC, allowing use of a density gradient cut-off that better retains fnRBC.52 Franchi and colleagues refined this low pH protocol and conducted a trial on 46 women:53 using FISH for chromosomes 21 and 18, they noninvasively diagnosed 7 fetal aneuploidies. Alternatively, selective maternal RBC lysis based on differential erythrocyte carbonic anhydrase I and II activity can be used.54,55 Carbonic anhydrase I is active in maternal RBC but not in fetal RBC. Using isotonic bicarbonate and ammonium chloride solutions in combination with a specific anhydrase II inhibitor, maternal cells will produce ammonium bicarbonate, which attracts water into the cells, causing them to burst. Additional depletion of maternal white blood cells (WBC) can be achieved by specifically targeting surface markers as CD45 and CD14 with magnetic-activated cell sorting (MACS).
Herzenberg et al. first reported in 1979 a fetal cell selection step that consisted of fluorescence-activated cell sorting (FACS) specific for paternal HLA antigens expressed on fetal cells but not maternal cells. They confirmed by Y chromosome quinacrine staining that they isolated fetal cells from the blood of women carrying male fetuses.56,57 However, HLA-antigens require prior knowledge of the paternal HLA-genotype and optimization for each individual sample. Furthermore, as indicated above, Y-chromosome based confirmation is only applicable to male pregnancies. Hence, considerable efforts were made from the 1980’s to early 2000’s to find adequate, general markers for specific fnRBC selection. The surface marker CD71 (transferrin receptor) combined with CD36 and/or glycophorin A (GPA) are the most widely used targets,48,58-60and Bianchi and colleagues showed that adding anti-GPA antibodies to anti-CD71 or anti-CD36 substantially improves fnRBC selection and accuracy of fetal sex prediction.61,62 Choolani et al. confirmed that primitive erythroblasts were the predominant cell type up to 12 weeks gestation, declining rapidly thereafter.63 Both primitive and definitive erythroblasts were GPA+, CD47+, CD45− and CD35−, but CD71 and CD36 staining was very weak for primitive erythroblasts. Other strategies included targeting surface antigen i (anti-i),44 CD34,64 galactose-specific lectin65 or using the HAE9 antibody44, or focusing on physical fnRBC characteristics, including the use of charge flow separation based on cell-specific surface charge densities.66,67 These latter methods were less successful and/or have not been further examined.
Investigators have attempted to incrementally improve the fnRBC yield by using embryonic (epsilon, ε and zeta, ζ) and fetal (gamma, γ, F) hemoglobin (Hb) for cell selection and/or identification. HbF expression is significantly higher in fnRBC compared to adult nRBC,68 but under certain conditions (e.g., for beta-thalassemia carriers), maternal HbF expression is upregulated.60 Choolani and colleagues described decreasing ε-globin expression with increasing gestation from 7 to 14 weeks, when the number of ε-positive cells became almost negligible,49 and similarly, Christensen et al. detected only one ε-positive cell in 18 samples collected between 8-12 weeks.69 Similarly, zeta Hb, which decreases even earlier than ε-globin,70 is an inadequate fetal cell marker.
One of the largest early cell-based NIPT trials so far, the NICHD (National Institute of Child Health and Human Development) Fetal Cell Isolation Study (NIFTY), included 2,700 samples collected between 1995 and 1999.71 Samples were first subjected to density gradient centrifugation, with or without MACS or FACS depletion based on CD14, CD15 and/or CD45. Fetal nRBC were then enriched by positive MACS-based selection (CD71+) or by FACS sorting for HbF+/CD45− cells. Confirmation of fetal origin was done with X and Y− FISH in male pregnancies, and FISH for chromosomes 13, 18 and 21, to detect possible fetal aneuploidies. The results indicated that prenatal diagnosis based on fnRBC analysis was not yet ready for clinical implementation (Figure 2), mainly due to low fetal cell yield. (XY cells were found in only 41% of all male pregnancies.)
Figure 2. Timeline depicting milestones in the field of noninvasive prenatal testing.
Circulating fnRBC nevertheless remain an attractive target because they avoid confined placental mosaicism (CPM), which is an issue for trophoblasts (discussed below). However, none of the many published methods have yet been validated for clinical use, in part due to lack of specific antibodies. By screening 690 different hybridomas, Zimmerman et al. found two antibody clones with the desired specificity for fetal erythroblast cell membranes and that target antigens other than known markers CD71, CD36, GPA, antigen i and galactose.72 A recent study included the use of RBC aggregation followed by RBC lysis and maternal WBC depletion with anti-CD45 and -CD66b antibodies,73 yielding two fnRBC from 1 ml samples collected at 12 and 16 weeks gestation, and on average 48.8 cells/ml in 16 samples at 23-27 weeks. Staining included antibodies for GPA and CD45. Y-PCR was positive for pools of isolated cells, although it was unclear whether these pools also contained maternal cells. Huang et al. reported the recovery of both trophoblasts and fnRBC in five samples using a novel silicon immune-affinity method.74 Trisomy was convincingly documented for three of those cases by using both array comparative genomic hybridization (aCGH) and next generation sequencing (NGS) analysis of 2-, 3-, or 10-cell pools, and STR evidence showed that fnRBC were recovered in all samples. However, this study used a complex enrichment platform not easily replicated or commercially available. The same group recently reported 14 additional cases for which 2-71 confirmed fnRBC were collected from 2 ml.75 Zhang et al.76 used a microfluidic chip coated with an anti-CD71 antibody, and identified 5–35 fnRBC per 2 ml of maternal blood starting at 7 weeks gestation, and fetal origin was demonstrated by SRY-PCR.
In 2017 and 2018, three publications with overlapping authors described similar technologies with positive selection for CD147, also known as extracellular matrix metalloproteinase inducer, which had not previously been used to recover fnRBC. Using biocompatible hydroxyapatite/chitosan nanoparticles microchips,.77, biotin-doped polypyrrole-based microchips,.78 , or bio-conjugated SiO2 gel microbeads,79all three reported up to 50-75 fnRBC/ml (peaking at 16-20 weeks gestation), using “anti-ε-HbF” staining as evidence for the cells’ fetal origin. But it is not clear which antibody was used, meaning some cells could have been maternal nRBC.
Trophoblasts
Other studies focused on isolating circulating trophoblasts, by selecting cells that express HLA-G, a nonclassical major histocompatibility complex class I antigen specific to EVT. In an initial study, cells were enriched with a Percoll density gradient and stained on slides with anti-HLA-G antibody, followed by FISH with probes for chromosomes X, Y and 21.80 One or more XY HLA-G+ cells were identified for 7/10 male pregnancies, but there was also non-specific staining of maternal cells attributed to secondary and tertiary antibodies.
Investigators in Denmark conducted microarray gene expression experiments on trophoblastic cell pools to find better markers for enrichment.17-19 They developed a proprietary protocol using the surface markers CD105 (endoglin) and CD141 (thrombomodulin) for MACS-based trophoblast selection, and cytokeratin (CK) staining for identification, as CK expression shows a typical vesicular pattern unique to trophoblasts compared to maternal WBC (Figure 3). In collaboration with our group, 100% recovery was achieved in a series of 111 samples, with on average 12.8 cells obtained from 30 ml of maternal blood.81 Fetal origin was confirmed by XY FISH for all included male pregnancies and demonstrated that the observed CK staining pattern was sufficiently specific for circulating trophoblasts. Downstream analysis by chromosomal microarray analysis (CMA) and/or NGS in this study and follow-up reports correctly identified fetal (mosaic) autosomal trisomies, sex chromosome abnormalities, and several large copy number changes..82,83
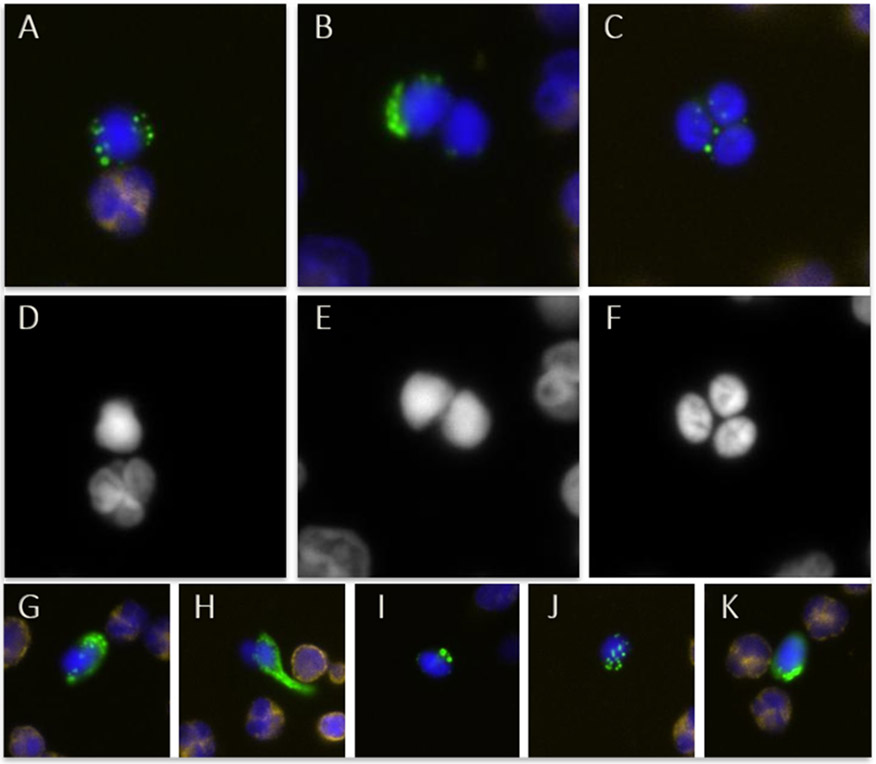
Figure 3. Immunostaining images of circulating trophoblasts.
The staining antibody cocktail includes anti-cytokeratin (CK; green) and anti-CD45 (yellow) antibodies, and DAPI nuclear stain. Panel A depicts a single trophoblast with a maternal cell, panel B a trophoblast doublet and panel C a cluster of three trophoblasts. Panels D, E, and F illustrate the respective DAPI staining and nuclear morphology. Panels G through K demonstrate the diversity in CK staining patterns that can be seen.
Our group developed a novel maternal cell depletion protocol in collaboration with RareCyte, by using a gradient of different high-density fluids in combination with a rubber float to lift and separate the nucleated cell fraction containing the trophoblasts above the float.84 We selected candidate trophoblasts based on nuclear morphology and CK staining pattern, and also used an anti-CD45 antibody to identify and exclude maternal WBC.84 After further protocol adjustments, we recovered on average 0.18 trophoblasts/ml (range: 0-1.58 cells/ml) from 30-40 ml blood samples from 125 women.85 DNA from individual trophoblasts was analyzed by low-coverage NGS for genome-wide copy number analysis, with correct identification of fetal aneuploidy in 11 cases and a known CNV in four cases (ranging from 1.1 Mb to 19 Mb in size). A new genotyping method confirmed the fetal origin of all analyzed cells by comparing fetal single nucleotide polymorphisms (SNPs) to the maternal (and when available, paternal) SNP profile (based on 50-60 SNPs to genome-wide analysis, described in 86 and unpublished data). Our most recent method includes an enrichment strategy based on a combination of anti-HLA-G, anti-Trop2, and anti-EpCAM antibodies resulting in an average yield of 0.20 trophoblasts/ml from 95 blood samples.25
Moving away from targeting specific antigens, Vona et al. explored trophoblast selection based on size using their “Isolation by Size of Epithelial Tumor/Trophoblastic cells” (ISET) protocol.87 The blood sample is filtered through an 8 μm-pore polycarbonate filter to retain a population of larger cells, which includes trophoblasts. After staining, trophoblasts are isolated by laser capture microdissection for downstream analysis. The authors reported recovery of 1-7 trophoblasts per 2 ml of maternal blood from six pregnant women carrying a male fetus.
More recently, several commercial and academic groups have explored microfluidics platforms (originally developed for CTC isolation) for cbNIPT applications, with variable success. These rare cell isolation platforms capture trophoblasts by immunomagnetic enrichment,88,89 size-based selection90,91 or specific dielectrophoretic separation,92 although for some only proof-of-principle data is available. Compared to manual processing, these platforms require lower sample volumes and less cell manipulation, and their approach for cell enrichment is often less harsh, lowering the risk for cell loss or inadequate cell quality.
Promising results were obtained with the anti-EpCAM NanoVelcro microfluidic chip enrichment after immunostaining for CK, HLA-G, and CD45,88 after which putative fetal cells are isolated by laser capture microdissection and analyzed by FISH, CMA, and short tandem repeat (STR) analysis. In a study of 15 women, up to 3 trophoblasts/ml were recovered for healthy pregnancies, and 7.5 cells/ml if there was a fetal genetic abnormality.88 Zhang et al. claimed recovery in 31 samples of 2-20 circulating fetal cells from only 2 ml of blood using a novel stimuli-responsive microfluidic interface with EpCAM-based capture and HLA-G/CK staining, but they also found stained cells in control and postpartum groups, suggesting this approach is less specific.93 Winter and colleagues used spike-in experiments to optimize their size-selection inertial microfluidics system, and subsequently recovered six trophoblasts from only 7 ml of blood from a pregnant woman whose fetus had trisomy 21.90 Gur et al. spiked blood samples with JEG3 cells, a choriocarcinoma cell line which expresses EVT markers. Immunomagnetic selection based on EpCAM and HLA-G through a two-tiered microchip system, resulted in high purity JEG3 cell recovery.89 Vander Plaetsen and colleagues recently tested the VTX-1 Liquid Biopsy system, which uses laminar microscale vortices for size-based trophoblast isolation, and recovered 2-6 trophoblasts from 8 ml of blood in 7/10 women carrying male pregnancies.91
TROPHOBLASTS FROM ENDOCERVICAL COLLECTION
In addition to maternal blood, trophoblasts can also be collected from the endocervical canal. One group developed a protocol for trophoblast retrieval and isolation from the cervix (TRIC) by inserting 2 cm of a cytobrush into the endocervical canal, collecting cervical mucus, and performing an anti-HLA-G immunomagnetic trophoblast enrichment.94 They reported a recovery of 500–1,500 EVT per sampling with minimal maternal cell contamination, as confirmed by β-human chorionic gonadotropin (hCG) staining for 95-100% of the cells and XY FISH for male pregnancies.95,96 Based on protein expression analysis, they determined the TRIC protocol yields EVT. The larger number of trophoblasts obtained via endocervical sampling contrasts with the scarcity of fetal cells or trophoblasts isolated from maternal blood samples, and could therefore provide a more fruitful sample source. In addition, the degree of DNA fragmentation in these cells was comparable to that of control cells, indicating adequate quality for further genetic testing. Of note, endocervical cell collection requires trained personnel. Although it is somewhat invasive, studies reported to date94-99 support that it is safe; cervical cancer screening using a similar approach can be done safely during pregnancy.
While trophoblast retrieval from maternal blood samples shows a downward trend with increasing gestational age,22,23,25-27 neither gestational age98 nor maternal BMI98 influences endocervical EVT recovery, though fewer EVTs were recovered from women with early pregnancy loss.97 EVTs from these pregnancies and those with fetal growth restriction and preeclampsia had altered expression of various markers such as AFP, ENG, and FLT-1, and less PAPP-A, LGALS14, and PGF compared to controls. Hence, endocervical trophoblasts have potential as biomarkers for pregnancy complications.
Investigators in South Korea recently adjusted the TRIC protocol to include a pre-enrichment fixation step, and noted a higher number of β-hCG-positive cells post-formalin-fixation.100 Also, the Paterlini group adjusted their size-based selection ISET protocol87 for trophoblasts collected with a cytobrush rotated at the external os, similar to a standard PAP smear.101 For a set of 21 samples (8-21 weeks gestation), they were able to recover 2–12 trophoblasts/sample.
CELL-BASED NIPT DATA ANALYSIS CONSIDERATIONS
Although cfDNA screening for fetal chromosomal abnormalities is well integrated into the clinic, its main disadvantage is that the fetal cfDNA (fetal fraction) is only 5-20% of all cfDNA in maternal blood.102 Furthermore, cfDNA-based NIPT results are influenced by maternal factors such as BMI, chromosomal mosaicism, fibroids, malignancies, and by CPM. Using circulating fetal cells or endocervical trophoblasts as a fetal DNA source avoids maternal DNA contamination if cells are carefully isolated and genotyped to be fetal. This eliminates the effect of most maternal confounding factors. With pure fetal DNA, smaller copy number changes down to 1-2 Mb in size can be detected by CMA or low-coverage NGS, which is in the range of those commonly identified by CMA on CVS or amniocentesis samples.82-85 Similar to CMA on invasively obtained samples, cbNIPT currently cannot detect copy-neutral rearrangements such as balanced translocations or inversions.
An essential step in single circulating fetal cell analysis is whole genome amplification (WGA), to ensure sufficient input DNA for the downstream analysis. Each method has advantages and drawbacks, but the goal is to use a method that performs adequately for both CNV analysis and monogenic disorder testing.e.g.103,104 A highly uniform genome coverage is required for CNV analysis, while low allelic-drop out (ADO) and false positive rate (FPR) are essential for point mutation detection. ADO (absence of amplification of one allele for a heterozygous locus) can occur due to unequal amplification (e.g., because of DNA degradation), and will lead to inaccurate NGS results. On the other hand, suboptimal polymerase fidelity and false positive results due to sequencing and, more importantly, amplification errors, can also pose a problem. One of the studies comparing three WGA technologies (five kits)105 showed that degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR106) had good uniformity and reproducibility but performed worst for other features (including ADO, FPR), while multiple displacement amplification (MDA107) had the lowest FPR, reasonable ADO, but low reproducibility. Multiple annealing and looping-based amplification cycles (MALBAC108) demonstrated high uniformity and reproducibility, and the lowest ADO rate.
The quality of the single cells will also determine the success of the WGA reaction. For trophoblasts isolated from maternal blood, there can be considerable cell-to-cell variability in whole-genome copy number profiles, making analysis of each single cell profile separately more advantageous than pooling cells or merging profiles of multiple cells from the same individual.25 Two main biological factors are responsible for this (Figure 4). Trophoblasts in an advanced stage of apoptosis109 give noisy whole genome profiles because the timing and degree of DNA degradation varies across the genome. Cells presumed to be in the S-phase of the cell cycle have noisy profiles because genomic regions replicate asynchronously, resulting in apparent random copy number gains and losses.110 Both patterns are recognizable with experience. Pooling a “good-quality” cell with a cell with the aforementioned patterns or low-quality profiles can compromise the overall result and is therefore not recommended. Yet, sometimes trophoblasts are isolated as clusters of two, three, or more cells that cannot be separated (Figure 3), and pooled analysis cannot be avoided. Of note, a higher proportion of lower quality cells might be seen for certain conditions or fetal abnormalities, such as a pregnancy affected with Turner syndrome.25
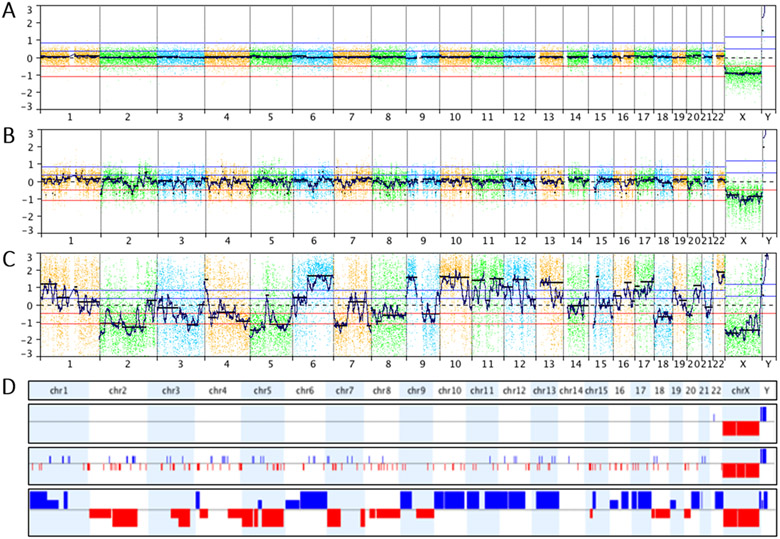
Figure 4. Whole genome profiles from circulating trophoblasts.
[A] High-quality whole genome profile of a single trophoblast obtained from a pregnancy with a male fetus. The profile was obtained by comparison to a female reference, hence the relative loss of one X chromosome and the relative gain of Y. [B] Whole genome profile of a male trophoblast presumed in S-phase. Random gains and losses can be seen throughout the genome. [C] Whole genome profile demonstrating genomic degradation of an apoptotic trophoblast. [D] Whole genome plots specifically depicting the copy number gains (blue) and losses (red) detected in all three cells above. All profiles were created in NxClinical (BioDiscovery).
Given the number of trophoblasts obtained from endocervical samples, analysis of a larger pool can potentially be done, and might be less sensitive to the effect of a few low-quality cells, provided most cells are of adequate quality.95
Separate analysis of each individual circulating cell enables the detection of distinct genetic signatures in case of non-identical twins (and theoretically, higher multiples), and confirms the presence of different karyotypes in CPM or other forms of mosaicism, provided enough cells can be tested. CPM for aneuploidy is found in 1-2% of all CVS samples111 and can lead to abnormal results with cfDNA-based aneuploidy screening, but mosaicism cannot be confirmed noninvasively with cfDNA. In contrast, with single trophoblast-based NIPT, single cell analysis will allow detection of aneuploid and euploid cells separately, as illustrated by five CPM cases from our group.25,84,85 For one mosaic trisomy 13 case, one case with 46,XX/45,X mosaicism, and one with two mosaic copy number losses on chromosome 13, both euploid and aneuploid cells were found. In two additional cases (trisomy 13 and trisomy 15), only two and one aneuploid cells in total were recovered, respectively, highlighting the importance of the number of recovered cells to detect CPM. The same applies to multiple pregnancies to ensure the recovery of cells from each fetus.25,85 Determining from which fetus an individual cell originates is not possible in same-sex twin pairs, and additional genotyping is needed to determine zygosity.
Cell-based testing for single gene disorders has also been explored. The Paterlini group published two sets of samples in which they correctly identified unaffected and affected fetuses with cystic fibrosis or spinal muscular atrophy using their ISET protocol.112,113 Chang et al. reported success with cbNIPT for one pregnancy at risk for congenital deafness and one for epidermolytic ichthyosis.114 Fetal cell analysis for each known familial mutation agreed with amniocentesis results.
In a recent report on a woman carrying monozygotic twins at risk for cystic fibrosis, from which 7 circulating trophoblasts were isolated, the twins were correctly identified as unaffected carriers, but ADO was noted for 1/3 STRs included in the genotyping analysis.115 Because of ADO, SNP-based genotyping may be better after WGA than STR-analysis, as more SNPs can be included.103,116-119 Chen and colleagues developed a double-negative selection protocol,120 and subjected the recovered fetal cells first to low-coverage NGS (and subsequent high-coverage whole genome sequencing for the best quality cell) for point mutation detection. Although they noted good performance overall, they observed an ADO rate of 24.9%, and (not unexpectedly) less uniform coverage for WGA products compared to unamplified genomic DNA. Cayrefourcq et al. investigated trophoblast analysis for monogenic diseases caused by point mutations or triplet repeat expansion.92 They only obtained a conclusive fetal result for one out of 7 families at risk of Huntington disease, and obtained a similar result when attempting mini-exome sequencing for 9 couples at risk for a specific monogenic disorder. They concluded that due to technical issues (including cell loss throughout sample processing) and ADO (3-fold higher in trophoblasts compared to maternal single cells), further improvements would be necessary for cbNIPT to be applicable for both single gene disorders and aneuploidies/CNV.
Huang and colleagues tweaked the TRIC protocol to replace intracellular immunostaining with endoplasmic reticulum tracker dye, which is nondestructive and potentially ensures a better DNA quality for endocervically isolated trophoblasts.121 They accurately identified the fetal genotypes in a series of 11 pregnancies at risk for alpha- or beta-thalassemia, and noted adequate SNV detection when performing whole exome sequencing in an additional five samples. Platforms avoiding WGA could also be helpful, including a sensitive single cell digital droplet PCR method recently described by Sato and colleagues.122
WORKING TOWARDS A CLINICAL TEST AND FUTURE OUTLOOKS
Because it is unhindered by maternal DNA contamination, cbNIPT could potentially be offered as a noninvasive diagnostic test rather than a screening assay (Table 1). In addition, cbNIPT can be provided early on, offering flexibility for further decision-making and management of the pregnancy. Endocervical cbNIPT has been performed at 5 weeks gestation and blood-based cbNIPT can also potentially be performed before 10 weeks.95,96 cfDNA-based NIPS is already in widespread use for detection of common fetal aneuploidies. To a more limited extent, it is used for subchromosomal copy number abnormalities associated with common deletion syndromes (in some cases across the entire genome), but the resolution and positive predictive values remain low. Since cbNIPT is based on pure fetal DNA from intact cells, it could be used for testing smaller copy number variants, currently at an approximate 1 megabase resolution.85 For the same reason, cbNIPT may also offer more flexibility and resolution for combined analysis for copy number alterations and monogenic disorder testing on the same sample. Further studies are needed to assess the analytical and clinical utility of these approaches on both circulating fetal cells and endocervical trophoblasts.
Table 1. Overview of different sample sources and their characteristics.
β-hCG: beta human choriogonadotropin, CPM: confined placental mosaicism, (f)nRBC: (fetal) nucleated red blood cell, GA: gestational age, HbF: fetal hemoglobin
| Cell type | fnRBC from blood | Trophoblasts from blood |
Endocervically obtained trophoblasts |
|---|---|---|---|
| Sampling | Maternal peripheral blood sample | Maternal peripheral blood sample | Endocervical collection with a cytobrush |
| Workflow |
|
|
|
| Major markers for enrichment and/or detection | CD71, GPA, HbF | HLA-G, EpCAM, CK | HLA-G, β-hCG |
|
Cell recovery
(normal pregnancies) |
Range: 0.04-10/ml | Range: up to 500 nRBC per ml reported (not all confirmed fetal) |
Range: 2-1,500/sample |
| Advantages |
|
|
|
| Drawbacks |
|
|
|
One of the main challenges in cbNIPT on circulating trophoblasts or fnRBC is consistent recovery of a sufficient number of cells from each patient to reduce the % test failures. Current studies indicate higher cell numbers for endocervical trophoblasts, but here the challenge ispurification from the endocervical mucus and surrounding maternal cells. Significant improvements have been made for both approaches. Other aspects to consider when evaluating a test’s potential for clinical implementation are turn-around-time (TAT), scalability, throughput, and cost. The TAT of library preparation, sequencing, and data analysis would be similar to cfDNA-based NIPT, but extra time is required for sample processing, including maternal cell depletion, fetal cell enrichment, and cell isolation.
Cell-based NIPT can potentially reach a large pregnant population, including those lacking access to a larger center offering diagnostic amniocentesis or CVS. More work is needed, however, to increase the throughput of current protocols and guarantee consistent test performance. The available data on (automated) microfluidics platforms are very promising, and these assays could boost throughput while also lowering TAT for cbNIPT from circulating cells. The current cost (a few thousand dollars per sample) is prohibitive for wider commercialization, but a more affordable test can be developed through further optimization of the processing protocol, bulk production, and decreasing sequencing costs.
Supplementary Material
Bulleted statements.
What is already known about this topic?
Several different protocols have been published to date for circulating fetal cell enrichment, detection, isolation, and analysis for the purpose of cell-based noninvasive prenatal testing (NIPT).
What does this review add?
This review offers a comprehensive and up-to-date critical analysis of published studies on cell-based NIPT, focusing on more recent developments.
Acknowledgements
The authors wish to acknowledge the leadership and contributions of Dr. Arthur L. Beaudet in developing and validating cell-based NIPT studies at Baylor College of Medicine, resulting in a number of publications included in this review. The authors thank Ms. Bettina Siegel for expert scientific editing support.
Funding statement
Our cell-based NIPT research is funded by Baylor College of Medicine and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD055651-12S1. The authors are solely responsible for this content, which does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
No ethical approval was required for this study, as this is a review article.
Data availability statement
Data sharing is not applicable to this review article, as no datasets were generated during the current study.
References
- 1.Salomon LJ, Sotiriadis A, Wulff CB, et al. Risk of miscarriage following amniocentesis or chorionic villus sampling: systematic review of literature and updated meta-analysis [Internet]. Vol. 54, Ultrasound in Obstetrics and Gynecology. John Wiley and Sons Ltd; 2019; p. 442–51. [DOI] [PubMed] [Google Scholar]
- 2.Schmorl G Pathologisch-anatomische Untersuchungen über Puerperal-Eklampsie. Verlag FCW Vogel; 1893; [Google Scholar]
- 3.Lapaire O, Holzgreve W, Oosterwijk JC, et al. Georg Schmorl on Trophoblasts in the Maternal Circulation. Placenta 2007;28(1):1–5. [DOI] [PubMed] [Google Scholar]
- 4.Chown B Anaemia from bleeding of the fetus into the mother’s circulation. Lancet (London, England) 1954;266(6824):1213–5. [DOI] [PubMed] [Google Scholar]
- 5.Zipursky A, Hull A, White FD, Israels LG. Foetal erythrocytes in the maternal circulation. Lancet (London, England) 1959;1(7070):451–2. [DOI] [PubMed] [Google Scholar]
- 6.Gordon H, Bhoyroo SK. A study of foetal erythrocytes in the maternal circulation during the antenatal period. J Obstet Gynaecol Br Commonw 1966;73(4):571–4. [DOI] [PubMed] [Google Scholar]
- 7.Weiner W, Child RM, Garvie JM, Peek WH. Foetal cells in the maternal circulation during pregnancy. Br Med J 1958;2(5099):770–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas GW, Thomas L, Carr M, et al. Trophoblast in the circulating blood during pregnancy. Am J Obstet Gynecol 1959;78:960–73. [DOI] [PubMed] [Google Scholar]
- 9.Walknowska J, Conte F, Grumbach M. Practical and theoretical implications of fetal/maternal lymphocyte transfer. Lancet 1969;293(7606):1119–22. [DOI] [PubMed] [Google Scholar]
- 10.Ariga H, Ohto H, Busch MP, et al. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion 2001;41(12):1524–30. [DOI] [PubMed] [Google Scholar]
- 11.Bianchi D, Khosrotehrani K, Way S, et al. Forever Connected: The Lifelong Biological Consequences of Fetomaternal and Maternofetal Microchimerism. Clin Chem 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianchi DW, Zickwolf GK, Weil GJ, et al. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A 1996;93(2):705–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Looij A, Singh R, Hatt L, et al. Do fetal extravillous trophoblasts circulate in maternal blood postpartum? Acta Obstet Gynecol Scand 2020;99(6):751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z-W, Fong C-Y, Gauthaman K, et al. Biology of human primitive erythroblasts for application in noninvasive prenatal diagnosis. Prenat Diagn 2018;38(9):673–84. [DOI] [PubMed] [Google Scholar]
- 15.Knöfler M, Haider S, Saleh L, et al. Human placenta and trophoblast development: key molecular mechanisms and model systems [Internet]. Vol. 76, Cellular and Molecular Life Sciences. Birkhauser Verlag AG; 2019; p. 3479–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 2012;33(5):327–34. [DOI] [PubMed] [Google Scholar]
- 17.Brinch M, Hatt L, Singh R, et al. Identification of circulating fetal cell markers by microarray analysis. Prenat Diagn 2012;32(8):742–51. [DOI] [PubMed] [Google Scholar]
- 18.Hatt L, Brinch M, Singh R, et al. Characterization of fetal cells from the maternal circulation by microarray gene expression analysis--could the extravillous trophoblasts be a target for future cell-based non-invasive prenatal diagnosis? Fetal Diagn Ther 2014;35(3):218–27. [DOI] [PubMed] [Google Scholar]
- 19.Hatt L, Brinch M, Singh R, et al. A new marker set that identifies fetal cells in maternal circulation with high specificity. Prenat Diagn 2014;34(11):1066–72. [DOI] [PubMed] [Google Scholar]
- 20.Bianchi DW, Williams JM, Sullivan LM, et al. PCR Quantitation of Fetal Cells in Maternal Blood in Normal and Aneuploid Pregnancies. Am J Hum Genet 1997;61(4):822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krabchi K, Gros-Louis F, Yan J, et al. Quantification of all fetal nucleated cells in maternal blood between the 18th and 22nd weeks of pregnancy using molecular cytogenetic techniques. Clin Genet 2001;60(2):145–50. [DOI] [PubMed] [Google Scholar]
- 22.Bianchi DW, Stewart JE, Garber MF, et al. Possible effect of gestational age on the detection of fetal nucleated erythrocytes in maternal blood. Prenat Diagn 1991;11(8):523–8. [DOI] [PubMed] [Google Scholar]
- 23.Hamada H, Arinami T, Kubo T, et al. Fetal nucleated cells in maternal peripheral blood: frequency and relationship to gestational age. Hum Genet 1993;91(5):427–32. [DOI] [PubMed] [Google Scholar]
- 24.Lim TH, Tan AS, Goh VH. Relationship between gestational age and frequency of fetal trophoblasts and nucleated erythrocytes in maternal peripheral blood. Prenat Diagn 2001;21(1):14–21. [PubMed] [Google Scholar]
- 25.Vossaert L, Wang Q, Salman R, et al. Validation Studies for Single Circulating Trophoblast Genetic Testing as a Form of Noninvasive Prenatal Diagnosis. Am J Hum Genet 2019;105(6):1262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravn K, Singh R, Hatt L, et al. The Number of Circulating Fetal Extravillous Trophoblasts Varies from Gestational Week 6 to 20. Reprod Sci 2020;27(12):2170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panchalee T, Vossaert L, Wang Q, et al. The effect of maternal body mass index and gestational age on circulating trophoblast yield in cell-based noninvasive prenatal testing. Prenat Diagn 2020;40(11):1383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn S, Huppertz B, Holzgreve W. Fetal Cells and Cell Free Fetal Nucleic Acids in Maternal Blood: New Tools to Study Abnormal Placentation? Placenta 2005;26(7):515–26. [DOI] [PubMed] [Google Scholar]
- 29.Holzgreve W, Ghezzi F, Naro E Di, et al. Disturbed feto-maternal cell traffic in preeclampsia. Obstet Gynecol 1998;91(5 Pt 1):669–72. [DOI] [PubMed] [Google Scholar]
- 30.Krabchi K, Gadji M, Samassekou O, et al. Quantification of fetal nucleated cells in maternal blood of pregnant women with a male trisomy 21 fetus using molecular cytogenetic techniques. Prenat Diagn 2006;26(1):28–34. [DOI] [PubMed] [Google Scholar]
- 31.Krabchi K, Gadji M, Forest J-C, Drouin R. Quantification of all fetal nucleated cells in maternal blood in different cases of aneuploidies. Clin Genet 2006;69(2):145–54. [DOI] [PubMed] [Google Scholar]
- 32.Kruckow S, Schelde P, Hatt L, et al. Does Maternal Body Mass Index Affect the Quantity of Circulating Fetal Cells Available to Use for Cell-Based Noninvasive Prenatal Test in High-Risk Pregnancies? [Internet]. Fetal Diagnosis and Therapy S. Karger AG; May 1, 2018. p. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlütter JM, Kirkegaard I, Ferreira AS, et al. The Number of Endovascular Trophoblasts in Maternal Blood Increases Overnight and after Physical Activity: An Experimental Study. Fetal Diagn Ther 2015;40(1):54–8. [DOI] [PubMed] [Google Scholar]
- 34.Schlütter JM, Kirkegaard I, Petersen OB, et al. Fetal gender and several cytokines are associated with the number of fetal cells in maternal blood--an observational study. PLoS One 2014;9(9):e106934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turco MY, Moffett A. Development of the human placenta. Vol. 146, Development (Cambridge). Company of Biologists Ltd; 2019; [DOI] [PubMed] [Google Scholar]
- 36.Shulman LP, Phillips OP, Tolley E, et al. Frequency of nucleated red blood cells in maternal blood during the different gestational ages. Hum Genet 1998;103(6):723–6. [DOI] [PubMed] [Google Scholar]
- 37.Scott FP, Menezes M, Palma-Dias R, et al. Factors affecting cell-free DNA fetal fraction and the consequences for test accuracy. J Matern Fetal Neonatal Med 2018;31(14):1865–72. [DOI] [PubMed] [Google Scholar]
- 38.Hou Y, Yang J, Qi Y, et al. Factors affecting cell-free DNA fetal fraction: statistical analysis of 13,661 maternal plasmas for non-invasive prenatal screening. Hum Genomics 2019;13(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont RF, Richardson LS, Boniface JJ, et al. Commentary on a combined approach to the problem of developing biomarkers for the prediction of spontaneous preterm labor that leads to preterm birth. Vol. 98, Placenta. W.B. Saunders Ltd; 2020; p. 13–23. [DOI] [PubMed] [Google Scholar]
- 40.Vora NL, Johnson KL, Peter I, et al. Circulating cell-free DNA levels increase variably following chorionic villus sampling. Prenat Diagn 2010;30(4):325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Tommaso M, Seravalli V, Salvianti F, et al. Cell-free fetal DNA in maternal circulation after chorionic villous sampling. Prenat Diagn 2013;33(7):695–9. [DOI] [PubMed] [Google Scholar]
- 42.Samuel AR, Son M, Ananth CV, Wapner RJ. The effect of chorionic villus sampling on the fraction of cell-free fetal DNA in maternal plasma. J Matern Neonatal Med 2016;29(16):2654–7. [DOI] [PubMed] [Google Scholar]
- 43.Gänshirt D, Smeets FW, Dohr A, et al. Enrichment of fetal nucleated red blood cells from the maternal circulation for prenatal diagnosis: experiences with triple density gradient and MACS based on more than 600 cases. Fetal Diagn Ther 1998;13(5):276–86. [DOI] [PubMed] [Google Scholar]
- 44.Troeger C, Holzgreve W, Hahn S. A comparison of different density gradients and antibodies for enrichment of fetal erythroblasts by MACS. Prenat Diagn 1999;19(6):521–6. [PubMed] [Google Scholar]
- 45.Samura O, Sekizawa A, Zhen DK, et al. Comparison of fetal cell recovery from maternal blood using a high density gradient for the initial separation step: 1.090 versus 1.119 g/ml. Prenat Diagn 2000;20(4):281–6. [DOI] [PubMed] [Google Scholar]
- 46.Prieto B, Alonso R, Paz A, et al. Optimization of nucleated red blood cell (NRBC) recovery from maternal blood collected using both layers of a double density gradient. Prenat Diagn 2001;21(3):187–93. [DOI] [PubMed] [Google Scholar]
- 47.Takabayashi H, Kuwabara S, Ukita T, et al. Development of non-invasive fetal DNA diagnosis from maternal blood. Prenat Diagn 1995;15(1):74–7. [DOI] [PubMed] [Google Scholar]
- 48.Smits G, Holzgreve W, Hahn S. An examination of different Percoll density gradients and magnetic activated cell sorting (MACS) for the enrichment of fetal erythroblasts from maternal blood. Arch Gynecol Obstet 2000;263(4):160–3. [DOI] [PubMed] [Google Scholar]
- 49.Choolani M, O’Donnell H, Campagnoli C, et al. Simultaneous fetal cell identification and diagnosis by epsilon-globin chain immunophenotyping and chromosomal fluorescence in situ hybridization. Blood 2001;98(3):554–7. [DOI] [PubMed] [Google Scholar]
- 50.D’Souza E, Ghosh K, Colah R. A comparison of the choice of monoclonal antibodies for recovery of fetal cells from maternal blood using FACS for noninvasive prenatal diagnosis of hemoglobinopathies. Cytometry B Clin Cytom 2009;76(3):175–80. [DOI] [PubMed] [Google Scholar]
- 51.Emad A, Drouin R. Evaluation of the impact of density gradient centrifugation on fetal cell loss during enrichment from maternal peripheral blood. Prenat Diagn 2014;34(9):878–85. [DOI] [PubMed] [Google Scholar]
- 52.Sitar G, Brambati B, Baldi M, et al. The use of non-physiological conditions to isolate fetal cells from maternal blood. Exp Cell Res 2005;302(2):153–61. [DOI] [PubMed] [Google Scholar]
- 53.Franchi PG, Palka C, Morizio E, et al. Sequential combined test, second trimester maternal serum markers, and circulating fetal cells to select women for invasive prenatal diagnosis. Schmidt EE, editor. PLoS One 2017;12(12):e0189235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyer SH, Noyes AN, Boyer ML. Enrichment of erythrocytes of fetal origin from adult-fetal blood mixtures via selective hemolysis of adult blood cells: an aid to antenatal diagnosis of hemoglobinopathies. Blood 1976;47(6):883–97. [PubMed] [Google Scholar]
- 55.Furbetta M, Angius A, Ximenes A, et al. Prenatal Diagnosis of β Thalassaemia by Fetal Red Cell Enrichment with NH4Cl-NH4HCO3 Differential Lysis of Maternal Cells. Br J Haematol 1980;44(3):441–50. [DOI] [PubMed] [Google Scholar]
- 56.Herzenberg LA, Bianchi DW, Schröder J, et al. Fetal cells in the blood of pregnant women: detection and enrichment by fluorescence-activated cell sorting. Proc Natl Acad Sci U S A 1979;76(3):1453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iverson GM, Bianchi DW, Cann HM, Herzenberg LA. Detection and isolation of fetal cells from maternal blood using the fluorescence-activated cell sorter (FACS). Prenat Diagn 1981;1(1):61–73. [DOI] [PubMed] [Google Scholar]
- 58.Price JO, Elias S, Wachtel SS, et al. Prenatal diagnosis with fetal cells isolated from maternal blood by multiparameter flow cytometry. Am J Obstet Gynecol 1991;165(6 Pt 1):1731–7. [DOI] [PubMed] [Google Scholar]
- 59.Reading JP, Huffman JL, Wu JC, et al. Nucleated erythrocytes in maternal blood: quantity and quality of fetal cells in enriched populations. Hum Reprod 1995;10(9):2510–5. [DOI] [PubMed] [Google Scholar]
- 60.Cheung MC, Goldberg JD, Kan YW. Prenatal diagnosis of sickle cell anaemia and thalassaemia by analysis of fetal cells in maternal blood. Nat Genet 1996;14(3):264–8. [DOI] [PubMed] [Google Scholar]
- 61.Bianchi DW, Flint AF, Pizzimenti MF, et al. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci U S A 1990;87(9):3279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bianchi DW, Zickwolf GK, Yih MC, et al. Erythroid-specific antibodies enhance detection of fetal nucleated erythrocytes in maternal blood. Prenat Diagn 1993;13(4):293–300. [DOI] [PubMed] [Google Scholar]
- 63.Choolani M, O’Donoghue K, Talbert D, et al. Characterization of first trimester fetal erythroblasts for non-invasive prenatal diagnosis. Mol Hum Reprod 2003;9(4):227–35. [DOI] [PubMed] [Google Scholar]
- 64.Little MT, Langlois S, Wilson RD, Lansdorp PM. Frequency of fetal cells in sorted subpopulations of nucleated erythroid and CD34+ hematopoietic progenitor cells from maternal peripheral blood. Blood 1997;89(7):2347–58. [PubMed] [Google Scholar]
- 65.Sekizawa A, Purwosunu Y, Farina A, et al. Development of noninvasive fetal DNA diagnosis from nucleated erythrocytes circulating in maternal blood. Prenat Diagn 2007;27(9):846–8. [DOI] [PubMed] [Google Scholar]
- 66.Wachtel SS, Sammons D, Manley M, et al. Fetal cells in maternal blood: recovery by charge flow separation. Hum Genet 1996;98(2):162–6. [DOI] [PubMed] [Google Scholar]
- 67.Wachtel SS, Sammons D, Twitty G, et al. Charge flow separation: quantification of nucleated red blood cells in maternal blood during pregnancy. Prenat Diagn 1998;18(5):455–63. [PubMed] [Google Scholar]
- 68.Zheng YL, Demaria M, Zhen D, et al. Flow sorting of fetal erythroblasts using intracytoplasmic anti-fetal haemoglobin: preliminary observations on maternal samples. Prenat Diagn 1995;15(10):897–905. [DOI] [PubMed] [Google Scholar]
- 69.Christensen B, Kølvraa S, Lykke-Hansen L, et al. Studies on the isolation and identification of fetal nucleated red blood cells in the circulation of pregnant women before and after chorion villus sampling. Fetal Diagn Ther 2003;18(5):376–84. [DOI] [PubMed] [Google Scholar]
- 70.Jackson L Fetal cells and DNA in maternal blood. Prenat Diagn 2003;23(10):837–46. [DOI] [PubMed] [Google Scholar]
- 71.Bianchi DW, Simpson JL, Jackson LG, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. Prenat Diagn 2002;22(7):609–15. [DOI] [PubMed] [Google Scholar]
- 72.Zimmermann S, Hollmann C, Stachelhaus SA. Unique monoclonal antibodies specifically bind surface structures on human fetal erythroid blood cells. Exp Cell Res 2013;319(17):2700–7. [DOI] [PubMed] [Google Scholar]
- 73.Byeon Y, Ki C-S, Han K-H. Isolation of nucleated red blood cells in maternal blood for Non-invasive prenatal diagnosis. Biomed Microdevices 2015;17(6):118. [DOI] [PubMed] [Google Scholar]
- 74.Huang C-E, Ma G-C, Jou H-J, et al. Noninvasive prenatal diagnosis of fetal aneuploidy by circulating fetal nucleated red blood cells and extravillous trophoblasts using silicon-based nanostructured microfluidics. Mol Cytogenet 2017;10(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma GC, Lin WH, Huang CE, et al. A silicon-based coral-like nanostructured microfluidics to isolate rare cells in human circulation: Validation by SK-BR-3 cancer cell line and its utility in circulating fetal nucleated red blood cells. Micromachines 2019;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Yang Y, Li X, et al. Frequency-enhanced transferrin receptor antibody-labelled microfluidic chip (FETAL-Chip) enables efficient enrichment of circulating nucleated red blood cells for non-invasive prenatal diagnosis. Lab Chip 2018;18(18):2749–56. [DOI] [PubMed] [Google Scholar]
- 77.He Z, Guo F, Feng C, et al. Fetal nucleated red blood cell analysis for non-invasive prenatal diagnostics using a nanostructure microchip. J Mater Chem B 2017;5(2):226–35. [DOI] [PubMed] [Google Scholar]
- 78.Feng C, He Z, Cai B, et al. Non-invasive Prenatal Diagnosis of Chromosomal Aneuploidies and Microdeletion Syndrome Using Fetal Nucleated Red Blood Cells Isolated by Nanostructure Microchips. Theranostics 2018;8(5):1301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei X, Ao Z, Cheng L, et al. Highly sensitive and rapid isolation of fetal nucleated red blood cells with microbead-based selective sedimentation for non-invasive prenatal diagnostics. Nanotechnology 2018;29(43):434001. [DOI] [PubMed] [Google Scholar]
- 80.van Wijk IJ, Griffioen S, Tjoa ML, et al. HLA-G expression in trophoblast cells circulating in maternal peripheral blood during early pregnancy. Am J Obstet Gynecol 2001;184(5):991–7. [DOI] [PubMed] [Google Scholar]
- 81.Kølvraa S, Singh R, Normand EA, et al. Genome-wide copy number analysis on DNA from fetal cells isolated from the blood of pregnant women. Prenat Diagn 2016;36(12):1127–34. [DOI] [PubMed] [Google Scholar]
- 82.Vestergaard EM, Singh R, Schelde P, et al. On the road to replacing invasive testing with cell-based NIPT: five clinical cases with aneuploidies, microduplication, unbalanced structural rearrangement or mosaicism. Prenat Diagn 2017;37(11):1120–4. [DOI] [PubMed] [Google Scholar]
- 83.Hatt L, Singh R, Christensen R, et al. Cell-based noninvasive prenatal testing (cbNIPT) detects pathogenic copy number variations. Clin Case Reports 2020;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Breman AM, Chow JC, U’Ren L, et al. Evidence for feasibility of fetal trophoblastic cell-based noninvasive prenatal testing. Prenat Diagn 2016;36(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vossaert L, Wang Q, Salman R, et al. Reliable detection of subchromosomal deletions and duplications using cell-based noninvasive prenatal testing. Prenat Diagn 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhuo X, Wang Q, Vossaert L, et al. Use of amplicon-based sequencing for testing fetal identity and monogenic traits with Single Circulating Trophoblast (SCT) as one form of cell-based NIPT. El-Maarri O, editor. PLoS One 2021;16(4):e0249695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vona G, Béroud C, Benachi A, et al. Enrichment, immunomorphological, and genetic characterization of fetal cells circulating in maternal blood. Am J Pathol 2002;160(1):51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hou S, Chen J-F, Song M, et al. Imprinted NanoVelcro Microchips for Isolation and Characterization of Circulating Fetal Trophoblasts: Toward Noninvasive Prenatal Diagnostics. ACS Nano 2017;11(8):8167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gur O, Chang CL, Jain R, et al. High-purity isolation of rare single cells from blood using a tiered microchip system. PLoS One 2020;15(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rezaei M, Winter M, Zander-Fox D, et al. A Reappraisal of Circulating Fetal Cell Noninvasive Prenatal Testing. Vol. 37, Trends in Biotechnology. Elsevier Ltd; 2019; p. 632–44. [DOI] [PubMed] [Google Scholar]
- 91.Vander Plaetsen A, Weymaere J, Tytgat O, et al. Enrichment of circulating trophoblasts from maternal blood using laminar microscale vortices. Prenat Diagn 2021;pd.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cayrefourcq L, Vincent MC, Pierredon S, et al. Single Circulating Fetal Trophoblastic Cells Eligible for Non Invasive Prenatal Diagnosis: the Exception Rather than the Rule. Sci Rep 2020;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang H, Yang Y, Liu Y, et al. Stimuli-Responsive Microfluidic Interface Enables Highly Efficient Capture and Release of Circulating Fetal Cells for Non-Invasive Prenatal Testing. Anal Chem 2020;92(13):9281–6. [DOI] [PubMed] [Google Scholar]
- 94.Imudia AN, Suzuki Y, Kilburn BA, et al. Retrieval of trophoblast cells from the cervical canal for prediction of abnormal pregnancy: a pilot study. Hum Reprod 2009;24(9):2086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolnick JM, Kilburn BA, Bajpayee S, et al. Trophoblast retrieval and isolation from the cervix (TRIC) for noninvasive prenatal screening at 5 to 20 weeks of gestation. Fertil Steril 2014;102(1):135–142.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jain CV, Kadam L, Dijk M Van, et al. Fetal genome profiling at 5 weeks of gestation after noninvasive isolation of trophoblast cells from the endocervical canal. Sci Transl Med 2016;8(November):363re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fritz R, Kohan-Ghadr HR, Bolnick JM, et al. Noninvasive detection of trophoblast protein signatures linked to early pregnancy loss using trophoblast retrieval and isolation from the cervix (TRIC). Fertil Steril 2015;104(2):339–346.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fritz R, Kohan-Ghadr HR, Sacher A, et al. Trophoblast retrieval and isolation from the cervix (TRIC) is unaffected by early gestational age or maternal obesity. Prenat Diagn 2015;35(12):1218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bolnick JM, Kohan-Ghadr HR, Fritz R, et al. Altered biomarkers in trophoblast cells obtained noninvasively prior to clinical manifestation of perinatal disease. Sci Rep 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee MJ, Kim SH, Shim SH, et al. Optimization protocol of fixation method for trophoblast retrieval from the cervix (TRIC): A preliminary study. Diagnostics 2020;10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfeifer I, Benachi A, Saker A, et al. Cervical trophoblasts for non-invasive single-cell genotyping and prenatal diagnosis. Placenta 2016;37:56–60. [DOI] [PubMed] [Google Scholar]
- 102.Rose NC, Kaimal AJ, Dugoff L, Norton ME. Screening for Fetal Chromosomal Abnormalities: ACOG Practice Bulletin, Number 226. Obstet Gynecol 2020;136(4):e48–69. [DOI] [PubMed] [Google Scholar]
- 103.Normand E, Qdaisat S, Bi W, et al. Comparison of three whole genome amplification methods for detection of genomic aberrations in single cells. Prenat Diagn 2016;36(9):823–30. [DOI] [PubMed] [Google Scholar]
- 104.Liu WQ, Zhang HM, Hu D, et al. The performance of MALBAC and MDA methods in the identification of concurrent mutations and aneuploidy screening to diagnose beta-thalassaemia disorders at the single- and multiple-cell levels. J Clin Lab Anal 2018;32(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang L, Ma F, Chapman A, et al. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu Rev Genomics Hum Genet 2015;16(1):79–102. [DOI] [PubMed] [Google Scholar]
- 106.Telenius H, Carter NP, Bebb CE, et al. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 1992;13(3):718–25. [DOI] [PubMed] [Google Scholar]
- 107.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res 2001;11(6):1095–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 2012;338(6114):1622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kolialexi A, Tsangaris GT, Antsaklis A, et al. Apoptosis in maternal peripheral blood during pregnancy. Fetal Diagn Ther 2001;16(1):32–7. [DOI] [PubMed] [Google Scholar]
- 110.Dimitriadou E, der Aa N Van, Cheng J, et al. Single cell segmental aneuploidy detection is compromised by S phase. Mol Cytogenet 2014;7(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grati FR, Malvestiti F, Branca L, et al. Chromosomal mosaicism in the fetoplacental unit. Best Pract Res Clin Obstet Gynaecol 2017;42:39–52. [DOI] [PubMed] [Google Scholar]
- 112.Béroud C, Karliova M, Bonnefont JP, et al. Prenatal diagnosis of spinal muscular atrophy by genetic analysis of circulating fetal cells. Lancet 2003;361(9362):1013–4. [DOI] [PubMed] [Google Scholar]
- 113.Mouawia H, Saker A, Jais J-P, et al. Circulating trophoblastic cells provide genetic diagnosis in 63 fetuses at risk for cystic fibrosis or spinal muscular atrophy. Reprod Biomed Online 2012;25(5):508–20. [DOI] [PubMed] [Google Scholar]
- 114.Chang L, Zhu X, Li R, et al. A novel method for noninvasive diagnosis of monogenic diseases from circulating fetal cells. Prenat Diagn 2020; [DOI] [PubMed] [Google Scholar]
- 115.Jeppesen LD, Hatt L, Singh R, et al. Cell-based non-invasive prenatal diagnosis in a pregnancy at risk of cystic fibrosis. Prenat Diagn 2020; [DOI] [PubMed] [Google Scholar]
- 116.Hou Y, Wu K, Shi X, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience 2015;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Babayan A, Alawi M, Gormley M, et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 2017;8(34):56066–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deleye L, Tilleman L, Plaetsen A-S Vander, et al. Performance of four modern whole genome amplification methods for copy number variant detection in single cells. Sci Rep 2017;7(1):3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weymaere J, Vander Plaetsen AS, Tilleman L, et al. Kinship analysis on single cells after whole genome amplification. Sci Rep 2020;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen F, Liu P, Gu Y, et al. Isolation and Whole Genome Sequencing of fetal cells from maternal blood towards the ultimate non-invasive prenatal testing. Prenat Diagn 2017; [DOI] [PubMed] [Google Scholar]
- 121.Huang Y, Situ B, Huang L, et al. Nondestructive Identification of Rare Trophoblastic Cells by Endoplasmic Reticulum Staining for Noninvasive Prenatal Testing of Monogenic Diseases. Adv Sci 2020;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sato T, Ito Y, Samura O, et al. Direct Assessment of Single-Cell DNA Using Crudely Purified Live Cells: A Proof of Concept for Noninvasive Prenatal Definitive Diagnosis. J Mol Diagnostics 2020;22(2):132–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this review article, as no datasets were generated during the current study.