Abstract
Typing phages for Salmonella and the prophages of their typical propagation strains were analyzed at the DNA level. Most of them belong to the P22 branch of the lambdoid phages. Acquisition of new plating properties of the typing phages by propagation in particular strains can be due to different host specific modifications of the DNA or to recombination events with residing prophages which are reflected by changes in the respective DNA restriction patterns. It is concluded that the actually available set of typing phages is a historically unique combination of strains.
Salmonella spp., in particular Salmonella typhimurium, play a dominant role in food poisoning. Outbreaks of salmonellosis are observed at an increasing rate. In order to trace infections to the manifold possible sources, strain identification is of great importance. Many systems have been developed that are based on the electromobility of enzymes, on the immunological properties of surface antigens, etc. The method of phage typing has the longest tradition. The first system (designated “Scheme 1”) was established in 1943 (8); later it was extended (“Scheme 2”) (5). Anderson et al. (1) refined it to a form which still is one of the main identification systems used worldwide and which is known as the Anderson phage typing scheme.
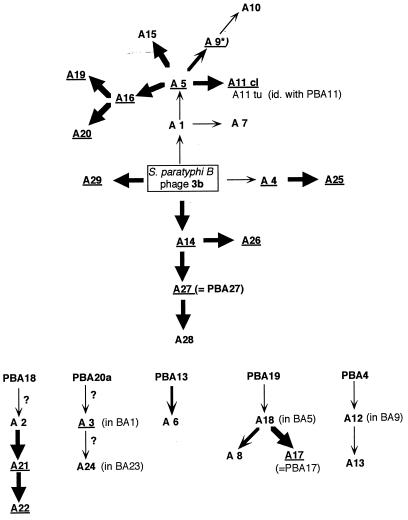
The basis of this typing system is a collection of Salmonella phages which were propagated on particular hosts of the species S. typhimurium. All Anderson typing phages used worldwide today are aliquots of phage stocks prepared decades ago. The Salmonella strains used for phage propagation are also used as reference typing strains. By propagation on these hosts, the phages gained new plating properties. This process has been called “modification,” a term not necessarily describing what is understood in the sense of “modification-restriction.” The genealogy of various typing phages, i.e., which phage results from the infection of a particular propagating strain with a defined “starting phage,” is described elsewhere (5). A graphical presentation of the pedigrees for most phages is shown in Fig. 1. All starting phages of the different lineages trace back to supernatants of Salmonella cultures, in most cases propagating strains (all of the phages with the prefix PBA in Fig. 1, followed by the number of the propagating strain according to Callow [5]), resulting thus from the spontaneous induction of residing prophages and the subsequent release of the temperate phages. Callow has noted that “the great majority of S. typhimurium are lysogenic.” This has been confirmed by our own observations (12, 13); moreover, we have shown that most of the phages released by various Salmonella isolates are closely related to the well-known phage P22 and therefore belong to the P22 branch of the lambdoid phages. The genomes of this phage group represent similar arrangements of modules, i.e., DNA segments coding for characteristic functions, which can be exchanged among each other, leading thus to high genetic variability (15).
FIG. 1.
Genealogy of Anderson typing phages. ∗, Phage A9 not available. Since A10 is unlike A5, although BA10 has no P22 sequences, A9 should already have undergone the change and should be identical to A10. ?, Propagating strain or infecting phage not available, (therefore no information about homologous sequences). Underlined terms indicate the phage has an EcoRI restriction pattern that is different from the infecting parental phage; nonunderlined terms indicate no change in the restriction pattern compared to the immediate ancestor; e.g., A8 (not underlined) has the same restriction pattern as A18, but A17 (underlined), which was propagated in BA17 with prophage sequences strongly homologous to P22/ES18, has a pattern different from A18. Thin, medium, and thick arrows indicate no homology, weak homology, or strong homology to P22/ES18, respectively.
Consequently, it was of interest to study the Anderson typing phages and their propagating strains in view of the prophage content of the latter and of the relationships among the Anderson phages and the possible prophages residing in their propagating strains. This study should elucidate the nature of the “modifications” altering the plating properties of the various typing phages.
MATERIALS AND METHODS
Bacterial strains.
Propagating strains of the typing phages are designated by numbers corresponding to the propagating strain number given previously (Table 4, column 3, of reference 5) with the prefix “BA.” These strains were kindly provided by L. R. Ward, Colindale, United Kingdom, via W. Rabsch, Wernigerode, Germany. Other strains were as follows: DB21, a wild-type strain of S. typhimurium LT2 positive for all three known modification-restriction systems (LT, SA, and SB [7]); DB21(sie1), DB21 which is lysogenic for P22 sieA sieB ts12.1 ts2.1, a P22 mutant defective in both superinfection exclusion systems (19); DB21(ES18), DB21 lysogenic for phage ES18; and LB5000 and LB5010, derivatives of LT2, defective in all three restriction functions but active in the corresponding modifications (4). LB5010 is a rough mutant. HisHB22(P22), harboring a deletion in the histidine operon, and Trp-8(P22), both of which are P22 lysogenic auxotrophic mutants of S. typhimurium LT2, were used as recipients in transduction experiments.
Phages.
Anderson typing phages were designated with the number according to column 1 of Table 4 in reference 5 with the prefix “A”; S. paratyphi phage 3b, being the direct or indirect ancestor of many typing phages, was also included in the study. All phages were kindly provided by L. R. Ward via W. Rabsch. Original lysates used in the typing assays were plated in suitable dilutions on DB21, and lysates were prepared on the same strain by a single plaque infection. Salmonella phages P22H5, a c2-mutant, and ES18h1, which is related to P22 (14), were also used. ES18 also infects, in contrast to P22, rough strains (11). Phages released from the Anderson propagating strains (i.e., the BA strains) are designated by the name of their host strain with a preceding “P”; e.g., PBA22 is the phage released from strain BA22, the propagating strain for Anderson phage A22.
Media.
Luria-Bertani (LB) medium was used for bacterial growth and phage propagation. M9 agar (16) was used for selection of the prototrophic transductants.
Transductions.
Transductions were performed as described elsewhere (10).
DNA preparation and hybridization.
Phage DNA was isolated from liquid phage lysates as described previously (2). DNA was digested with the restriction enzyme EcoRI (Boehringer Mannheim) as recommended by the supplier. DNA fragments were separated by agarose gel electrophoresis at 10 V/cm−1 in 0.8% (wt/vol) agarose gels (UltraPure; Gibco BRL) prepared with TBE buffer (89 mM Tris base, 89 mM boric acid). The DNA fragments were transferred to Biodyne A transfer membranes (Pall) by Southern blotting (17) and cross-linked to the membrane by exposure to UV light (254 nm, 120 mJ, 30 s) in a Stratalinker 1800 (Stratagene). DNAs from phages P22 and ES18, respectively, were used as probes and labelled with digoxigenin-11-dUTP by using the Random Primed Labelling Kit (Boehringer Mannheim). Prehybridizations, washes, and detection of hybridization signals were performed as recommended by Boehringer Mannheim. The hybridization temperature was 42°C.
RESULTS AND DISCUSSION
Growth of typing phages on DB21.
The original lysates of the Anderson phages used for typing assays and the lysate of phage 3b as obtained from L. Ward were prepared on specific propagating strains as described previously (5). In order to keep host influences as low as possible, they were tested for their ability to plate on our standard phage indicator DB21. All phages formed plaques; however, the efficiencies of plating varied to a great extent, possibly due to the restriction systems active in the LT2 strain DB21. This is supported by the observation that especially phage A8 yielded very few plaques on DB21. However, after isolation of a plaque from DB21, high yields of A8 could be obtained on this strain. Most of the phages formed plaques of similar size to those produced by P22, only phage A8 produced very small plaques. Although the plaques of several phage strains appeared rather clear, they undoubtedly were plaques of temperate phages because all of them could be traced back to prophages. Phage lysate A11 was a mixture of two different phages distinguishable by plaque morphology (clear, turbid). The different types were propagated separately (Table 1). Study of the DNA restriction patterns confirmed that we were dealing with different phages (as described below).
TABLE 1.
Properties of Anderson typing phages
| Phage | Plaque morphology (on DB21) | Transduction | Immunity to P22a | Anti-repressor | EcoRI patternb |
|---|---|---|---|---|---|
| 3b | Turbid | + | Hetero | ? | A |
| A1 | Clear | + | Hetero | ? | A |
| A2 | Turbid | + | Hetero | ? | |
| A3 | Clear | + | Hetero | ? | B |
| A4 | Turbid | + | Hetero | ? | |
| A5 | Clear | + | Hetero | ? | C |
| A6 | Turbid | + | Hetero | ? | C |
| A7 | Turbid | + | Hetero | ? | A |
| A8 | Clear | + | Homo | No | D |
| A10 | Turbid | + | Hetero | ? | E |
| A11tu | Turbid | + | Hetero | ? | |
| A11cl | Clear | NTc | Hetero | ? | F |
| A12 | Clear | + | Hetero | ? | G |
| A13 | Clear | + | Hetero | ? | G |
| A14 | Clear | + | Hetero | ? | |
| A15 | Turbid | + | Hetero | ? | C |
| A16 | Turbid | + | Hetero | ? | E |
| A17 | Clear | + | Homo | No | |
| A18 | Clear | + | Homo | No | D |
| A19 | Turbid | + | Hetero | ? | F |
| A20 | Clear | + | Hetero | ? | |
| A21 | Turbid | + | Hetero | ? | |
| A22 | Turbid | + | Hetero | ? | |
| A23 | Clear | + | Hetero | ? | E |
| A24 | Clear | + | Hetero | ? | B |
| A25 | Clear | + | Hetero | ? | F |
| A26 | Clear | + | Hetero | ? | |
| A27 | Clear | + | Hetero | ? | H |
| A28 | Turbid | + | Hetero | ? | H |
| A29 | Clear | + | Hetero | ? | |
| A32 | Clear | + | Hetero | ? | |
| A35 | Turbid | + | Hetero | ? |
Immunity type—heteroimmune (Hetero) or homoimmune (Homo)—is indicated.
Identical letters indicate identical EcoRI restriction patterns. Blank boxes indicate individual patterns which are represented only one time.
NT, not tested.
Transduction.
All phages were assayed for their capacity to encapsulate and to transfer host genetic material. The auxotrophic mutants HisB22(P22) and Trp-8(P22) were infected with phage particles of the various Anderson typing phages, and wild-type transductants were selected on minimal medium. All phages appeared to be transducers, although with different efficiencies, a result obviously due to different capacities for lysogenization (Table 1). Phages which formed “clear” plaques, produced considerably fewer wild-type transductants, and a poor background lawn of nontransduced recipient cells indicated extensive lytic reactions. Since both markers, his+ and trp+, could be efficiently transduced, although located at different positions on the Salmonella chromosome, these phages are considered to be generalized transducers (13).
Immunity.
As will be shown later, all of the typing phages and phage 3b are relatives of phage P22 with only two exceptions (A12 and A13). Therefore, it was of interest to determine their immunity. Phage P22 has a C-immunity system which is comparable in organization and function to the immunity region of phage lambda. In addition, P22 has a second system, immunity I. This system codes for an antirepressor which is able to inactivate the C repressor (18). Phage ES18, also related to P22, is homoimmune to P22, i.e., it has the same immunity as P22, but it has no antirepressor (14). When a new phage is plated on indicator strain DB21 which is lysogenic for P22 or ES18, three reactions are possible. (i) If the unknown phage lyses both lysogenic strains, it is heteroimmune to P22/ES18. (ii) If it lyses none of them, it is homoimmune to them, and it has no antirepressor (no imm-I region). (iii) If it lyses DB21(ES18) but not DB21(P22), it is homoimmune to P22/ES18 and—like P22—expresses an antirepressor. This biological assay is not suitable for testing heteroimmune phages for the presence of an antirepressor. A complication results from the existence of two superinfection exclusion systems (sieA, sieB) (19) in addition to the C immunity expressed by P22 but not by ES18: P22 prophage would exclude other phages even being heteroimmune. Therefore, this test can only be performed with cells lysogenic for a P22 mutant defective in both systems, i.e., P22 sie1. The results of this study are included in Table 1. They show that only A8, A17, and A18 are homoimmune to P22 and that these phages do not have the imm-I system. All other phages are heteroimmune to P22/ES18, and the existence of imm-I cannot be assayed biologically. At present we do not know to how many different immunity systems these P22-heteroimmune phages belong.
EcoRI restriction pattern and hybridization with P22 and ES18 DNA.
DNA from all typing phages was prepared, and EcoRI restriction patterns were monitored, followed by blotting for hybridization. Several typing phages show identical restriction patterns (Table 1). For example, phages 3b, A1, and A7 exhibit the same pattern. A closer look at Fig. 1 shows that these individual typing phages derive from each other and differ only in the bacterial hosts used for propagation. Therefore, phage 3b is the ancestor of A1, which was obtained by propagating phage 3b in strain BA1; phage A7 was obtained by propagating phage A1 in bacterial strain BA7. This demonstrates that these phages form a direct genealogic line and did not undergo visible changes in their DNA compositions by passage through different bacterial strains.
Similar groups of related phages with identical EcoRI patterns, like phage A5 (ancestor)-A15 (progeny of A5), A27-A28, etc., can be seen in Fig. 1. Arrows show the various lineages; normal characters indicate that the respective phage shows the same EcoRI restriction pattern as its immediate predecessor: phage 3b→A4; A27→A28, etc.
Nevertheless, these obviously identical typing phages show different lytic activities on type strains (see Table 2 in reference 5). In fact, this different plating behavior is the basis for the typing scheme. These phages differ only in their bacterial history. Therefore, it is highly probable that at least these plating differences result from different host-controlled modification-restriction systems. This aspect will be demonstrated elsewhere (11a).
On the other hand, there are many examples showing that propagation of one Anderson typing phage in a particular propagation strain results in a phage exhibiting a different EcoRI DNA pattern (underlined terms in Fig. 1). Examples are phage A5 and its many descendants. A5 resulting from A1 by propagation in strain BA5 has an EcoRI pattern different from that of A1. Exception for A15, the descendents of A5 (A9, A11cl, and A16) are different from their ancestor.
This is either an indication that the infecting phage may recombine with a possibly related prophage residing in the propagation strain or that by superinfection a prophage may be activated and predominate in the resulting lysate. Therefore, the propagation strains were also studied in more detail (see below).
As Fig. 1 shows, the typing phages A8 and A18 also show the same restriction pattern; this is not surprising, because A18 is the ancestor of A8. The pattern is identical to that of phage ES18 (data not shown). This finding is in accordance with the notion of the discoverers of ES18 (11), who described this transducing phage as a single plaque isolate from Anderson typing phage 18 (i.e., A18).
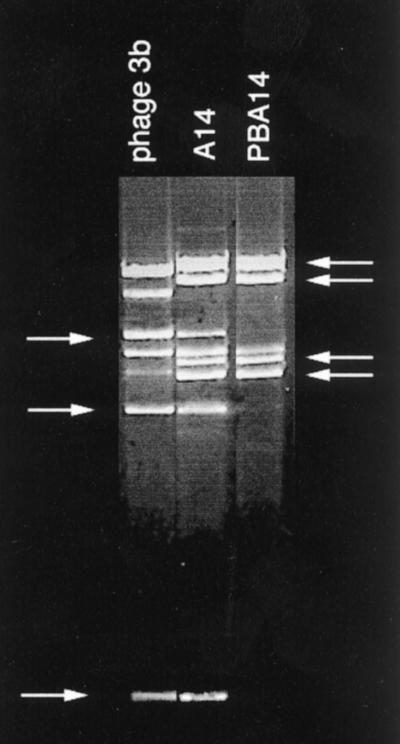
Furthermore, the study of restriction patterns has shown that the two plaque morphologically distinguishable phages in the typing lysates A11, A11cl, and A11tu, are indeed two different phages and not a mixture of wild-type phages and their clear-plaque mutants (Fig. 2).
FIG. 2.
EcoRI restriction patterns of phage DNAs as indicated.
Since we have demonstrated in a recent study (15) that most of the transducing Salmonella phages belong to the P22 branch of the lambdoid phages, it was worthwhile to test also the Anderson typing phages for a possible relationship to P22 and the related phage ES18. EcoRI-cleaved DNAs of the Anderson typing phages were blotted in duplicate after gel electrophoresis and subsequently hybridized against genomes of phages P22 and ES18, respectively. Only phages A12 and A13 gave no signals with these probes. All other phages hybridized with both of them, although not all bands hybridized with equal intensity. In most phages there were several bands which hybridized only with the ES18 probe, but not with P22, indicating that these genomes carry sequences which are typical of ES18.
This is remarkable for the following reason. As we have shown elsewhere (14), about half of the genome of phage ES18 represents sequences of phage P22, in particular the immediate-early genes; some other modules correspond to E. coli phage lambda, and the remainder, especially the head and base plate genes, is characteristic for ES18 (which recognizes a different receptor). In an extensive study on the modular genome composition of several collections of Salmonella phages, none were found to carry ES18-typical sequences (15).
Propagation strains for Anderson typing phages: prophages and their DNA.
It is a characteristic feature of the typing phages that any that have been propagated on a particular host strain undergo specific “modifications.” At least in some cases these modifications included also changes in the DNA restriction patterns (Fig. 1). These changes are possibly due to recombination with residing prophages. We have shown that most, if not all, natural Salmonella isolates are lysogenic at least for one prophage (12) and that most of them are related to phage P22 (15). Therefore, it was interesting to examine whether this holds true also for the Anderson propagating strains. If the situation here were similar, at least some of the “modifications” of the typing phages could result from recombination with the resident prophages as indicated by the EcoRI patterns, thus producing new module combinations and consequently different biological properties. Through acquisition of new base plate modules a different host range might result (e.g., ES18 which infects rough strains), and exchange of immunity regions would also influence plating properties, depending on the prophage residing in the strain to be tested.
Supernatants of overnight cultures of the various propagating strains were spotted on three different indicator strains: DB21, LB5000, and LB5010, the latter one being a rough strain. Table 2 shows that only 6 of 25 strains did not release a detectable phage. Strain BA19 released two different phages: PBA19A and PBA19B. From some supernatants only very tiny plaques could be detected on one or both of the strains LB5000 and LB5010; from some of these phages we did not obtain enough particles for DNA preparation. The DNAs of all other phages were cleaved with EcoRI and separated by gel electrophoresis. Three restriction patterns were observed to occur several times: PBA13 and PBA14; PBA16 and PBA20; and PBA19A, PBA25, PBA26, and PBA29. Phage PBA19B showed the same restriction pattern as ES18.
TABLE 2.
Spontaneous phage release by Anderson propagation strains
| Strain | Spontaneous phage release
|
Comments | Hybridization of bacterial DNA with:
|
EcoRI patterna | |||
|---|---|---|---|---|---|---|---|
| DB21 | LB5000 | LB5010 | P22 | ES18 | |||
| BA1 | − | − | − | − | − | ||
| BA2 | − | ++ | +++ | Very small, not propagated | − | (+) | NT |
| BA3 | Strain not available | ||||||
| BA4 | − | +++ | − | Very small, not propagated | − | − | NT |
| BA5 | − | − | − | − | − | ||
| BA6 | − | − | − | − | (+) | ||
| BA7 | +++ | +++ | +++ | Very small, not propagated | − | − | NT |
| BA8 | − | +++ | − | Not related to P22 | − | (+) | + |
| BA9 | − | − | +++ | Very small, not propagated | − | (+) | NT |
| BA10 | − | ++ | +++ | Very small, not propagated | − | − | NT |
| BA11 | +++ | ++ | +++ | + | + | NT | |
| BA12 | Strain not available | ||||||
| BA13 | +++ | +++ | − | + | + | A | |
| BA14 | +++ | +++ | +++ | + | + | A | |
| BA15 | − | − | − | + | + | ||
| BA16 | +++ | +++ | − | + | + | B | |
| BA17 | − | ++ | +++ | Very small, not propagated | + | + | NT |
| BA18 | Strain not available | ||||||
| BA19 | +++ (two types) | +++ | +++ | A, turbid; B, clear | +, + | +, + | C, ES18 |
| BA20 | +++ | +++ | +++ | + | + | B | |
| BA21 | − | − | − | + | + | ||
| BA22 | +++ | +++ | − | + | + | + | |
| BA23 | − | − | − | − | (+) | ||
| BA24 | Strain not available | ||||||
| BA25 | +++ | +++ | − | + | + | C | |
| BA26 | +++ | +++ | ++ | + | + | C | |
| BA27 | +++ | +++ | − | + | + | + | |
| BA28 | − | ++ | ++ | Very small, not propagated | (+) | + | NT |
| BA29 | +++ | +++ | +++ | + | + | C | |
Restriction pattern of released phages: identical letters = identical patterns; +, = individual pattern. NT, not tested.
Hybridization experiments with P22- and ES18-DNA as probes showed that, except for PBA8, all testable phages are related to P22 and ES18 (Table 2). Both phages released by strain BA19, PBA19A and PBA19B, are P22-related phages. Hybridization of EcoRI-cleaved bacterial DNA of all propagating strains with these probes not only confirmed that the phages reside as prophages in the strains but also that at least strain BA15, which did not release detectable phages, contained P22/ES18-related material (e.g., BA15; Table 2). It cannot be determined whether phages were produced but could not be detected (plaques were too small or there was a lack of suitable indicator strains) or whether the hybridization signals resulted from defective prophage genomes. Nevertheless, this means that at least 15 of 25 available propagating strains harbor genomes or parts of genomes of phages related to P22 and, hence, to most of the Anderson typing phages. Therefore, in most propagation strains the precondition is fulfilled for general recombination between an infecting typing phage and the resident prophage, leading to a new typing phage with new plating properties (indicated by the thick arrows in Fig. 1).
Interactions between infecting typing phages and prophages.
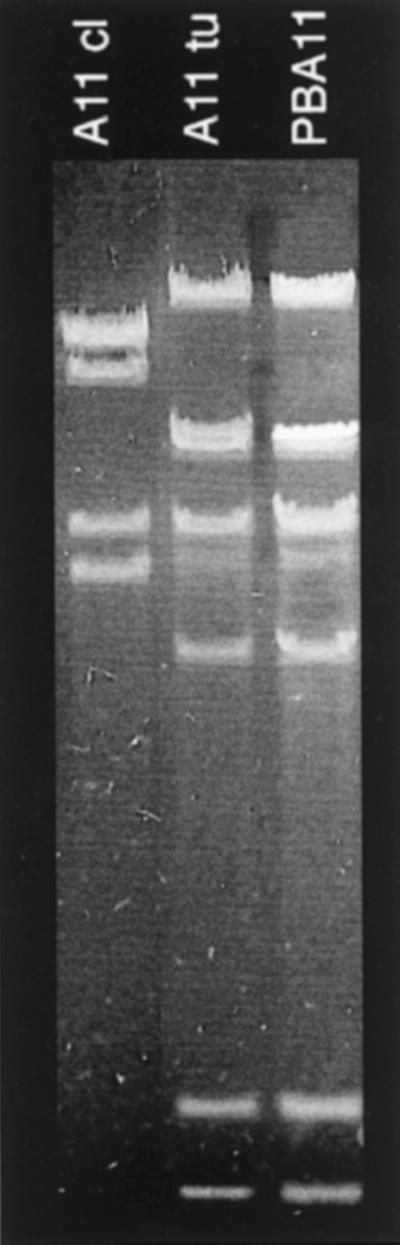
To test this hypothesis, the EcoRI restriction patterns of infecting typing phages, of the phages released by the respective propagation strains, and of the new typing phages resulting from these infections were compared. Figure 3 shows a very clear example of an altered DNA pattern due to recombination. Phage A14 results from propagation of phage 3b in strain BA14. This propagation strain harbors a P22-related prophage: PBA14. “Progeny” phage A14 shows three bands from “parent” 3b (arrows pointing to 3b) and four bands from the other “parent” PBA14 (arrows pointing to PBA14). One of the bands and the underrepresented band resulting from DNA packaging (9) have all three phages in common. Not all lineages are as clear as this one. Figure 4 shows the next step leading from typing phage A14 to the new typing phage A26 by propagation in strain BA26. Once more the DNA of newly arising phage shows changes, although most bands of “parent” A14 are conserved. However, three bands are missing, but no bands from the other “parent” PBA26 appear. In many cases the recombination processes may be even more complicated, possibly due to repeated recombination events during a propagation cycle. Also, the changes of the restriction patterns from phage 3b to A4 and from A1 to A5 cannot be interpreted in a simple way because propagating strains BA4 and BA5 did not release a detectable phage and because hybridization with P22 and ES18 probes did not indicate homology with these genomes. It cannot be excluded that the Salmonella typing strains which, according to a previous study (5), are also the propagation strains and which came to us recently have been replaced since the time of propagation of the typing phage stocks.
FIG. 3.
EcoRI restriction patterns of phage DNAs as indicated. DNA of phage A14 in the center is flanked by the DNA of its ancestor phage (phage 3b) and the DNA of the phage (PBA14) residing in the propagation strain (BA14). The arrows indicate bands retained from the respective “parent.”
FIG. 4.
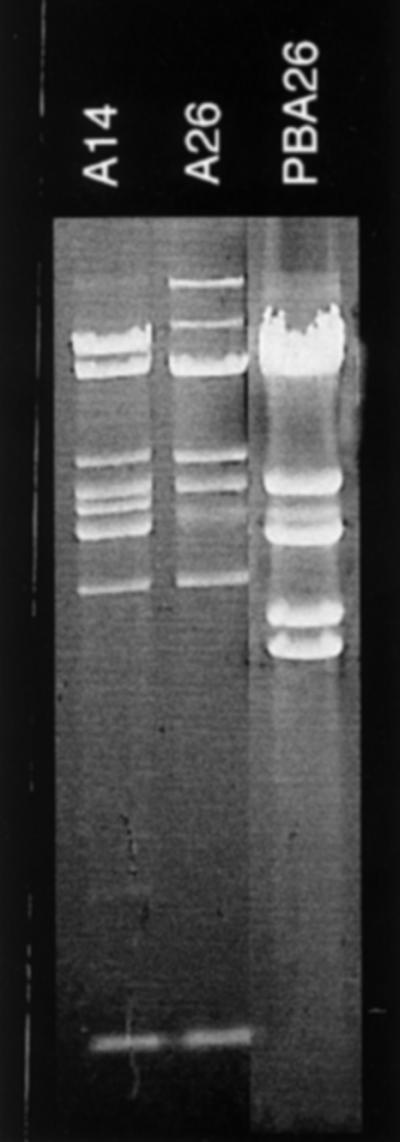
EcoRI restriction patterns of phage DNAs as indicated. DNA of phage A26 in the center is flanked by the DNA of its ancestor phage (phage A14) and the DNA of the phage (PBA26) residing in the propagation strain (BA26).
Figure 1 compiles these data, illustrating whether a propagating strain carries P22/ES18 related sequences suitable for recombination (thick arrows leading to the new phage) and whether the resulting new phage has a restriction pattern different than that of the parental phage (underlined phage name), thus indicating a possible recombination event during phage development in the lysogenic propagating strain.
For phage ES18 an interesting lineage can be observed (Fig. 1). According to Callow (5), typing strain A18 was obtained by propagating the phages released by strain BA19 in strain BA5. BA19, however, releases two different temperate phages, PBA19A and PBA19B, both hybridizing with P22/ES18-DNA. Obviously, PBA19B was more successful in BA5 because the resulting phage A18 (i.e., ES18) shows the same restriction pattern as PBA19B. From A18 a new typing phage, A8, was obtained by infecting strain BA8. A8 has retained the restriction pattern of its parental phage A18. A18 is also the ancestor of A17, which was obtained by passing A18 through propagating strain BA17. A17, however, shows a restriction pattern different from A18 but identical to the prophage residing in BA17, as identified by hybridization. Thus, it may be concluded that infection of BA17 by phage A18 has activated the resident prophage in some way which subsequently asserted itself over A18 and dominated in the resulting lysate.
Another interesting example of prophage activation after phage infection is typing phage lysate A11. As already mentioned A11 is a mixture of A11cl and A11tu, both of which show different EcoRI restriction patterns. The propagation strain for A11, BA11, released a P22-related phage, PBA11 (Table 2). Its EcoRI restriction pattern is exactly the same as that of A11tu (Fig. 3, lane 3).
These were only a few examples of the widely connected genealogy of the typing phages which indicated that propagation of phages in strains harboring related prophages may have all imaginable consequences: (i) the outcoming phage may be identical to the infecting phage (e.g., A5→A15); (ii) the outcoming phage may be identical to the resident prophage (e.g., A17 = PAB17); (iii) the resulting lysate may be a mixture of the infecting or a recombinant phage and the activated prophage of the propagating strain (e.g., phage A11); and (iv) the outcoming phage may be a recombinant between the infecting phage and the resident (in some cases rudimentary) prophage (e.g., A5→A16).
Another interesting aspect is the fact that there seem to be preferred points of recombination. Several phages belonging to different lineages and stemming from different ancestors end up with identical restriction patterns after several genomic changes. Examples are the phages A11cl, A19 and A25, or A6 and A5 (A15).
The Anderson collection of typing phages thus contains nice examples for the appearance of new phages due to recombination between infecting phages and residing prophages. These observations are in accordance with the predictions of the theory of modular evolution of bacteriophages (3, 6). It is true that this evolution took place during phage propagations under laboratory conditions. But all players in this game, the lysogenic Salmonella strains as well as the phages released by some of them, were natural isolates, unmodified and not selected for particular properties during a long laboratory history. Therefore, we may assume that similar events are taking place at any time in nature, thus creating permanently new phages with specific properties such as different host ranges.
In conclusion, phage typing mainly relies on three effects which result from propagation of particular phages in different host strains: (i) host-controlled modification (11a); (ii) adsorption properties, depending on the particular base plate; and (iii) susceptibility of the typing phage to repressors (immunity type) and to various superinfection exclusion systems controlled by residing prophages. Whereas item i depends only on the properties of the host strain, items ii and iii can be influenced by recombination with residing prophages.
These results suggest that it may be very difficult to repropagate the set of typing phages in the same way as it exists because the passage of a particular phage through a host harboring a related prophage allows the production of various different recombinants which may have different plating properties. The actual collection may be a historically unique combination of these strains, and repeating exactly the procedure of Anderson et al. may lead to test phages with quite different characteristics. In conclusion, once the stocks of the original Anderson typing phages are exhausted, this may be the end of phage typing with the Anderson system.
ACKNOWLEDGMENTS
I am indebted to B. Zavari and U. Bergmann for skillful technical assistance; L. Ward, Colindale, United Kingdom, and W. Rabsch, Wernigerode, Germany, for providing phage and bacterial strains; and W. Rabsch and P. Schicklmaier, Munich, Germany, for many fruitful discussions.
This work was supported by a grant from the German Ministry for Education, Science, Research, and Technology (BEO 21/0311230).
REFERENCES
- 1.Anderson E S, Ward L R, de Saxe M J, de Sa J D. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 3.Botstein D. A theory of modular evolution for bacteriophages. Annu N Y Acad Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 4.Bullas L R, Ryu J-I. Salmonella typhimurium LT2 strains which are r−m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callow B R. A new phage-typing scheme for Salmonella typhimurium. J Hyg. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 7.Colson C, van Pel A. DNA restriction and modification systems in Salmonella. I. SA and SB, two Salmonella typhimurium systems determined by genes with a chromosomal location comparable to that of the Escherichia coli hsd genes. Mol Gen Genet. 1974;129:325–337. doi: 10.1007/BF00265696. [DOI] [PubMed] [Google Scholar]
- 8.Felix A, Callow B R. Typing of paratyphoid B bacilli by means of Vi bacteriophage. Br Med J. 1943;ii:127. doi: 10.1136/bmj.2.4308.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson E N, Miller H I, Adams M A. EcoRI restriction endonuclease cleavage site map of bacteriophage P22 DNA. J Mol Biol. 1978;118:347–363. doi: 10.1016/0022-2836(78)90233-4. [DOI] [PubMed] [Google Scholar]
- 10.Kufer B, Backhaus H, Schmieger H. The packaging initiation site of phage P22. Analysis of packaging events by transduction. Mol Gen Genet. 1982;187:510–515. doi: 10.1007/BF00332636. [DOI] [PubMed] [Google Scholar]
- 11.Kuo T-T, Stocker B A D. ES18, a general transducing phage for smooth and nonsmooth Salmonella typhimurium. Virology. 1970;42:621–632. doi: 10.1016/0042-6822(70)90308-9. [DOI] [PubMed] [Google Scholar]
- 11a.Rabsch, W. Unpublished data.
- 12.Schicklmaier P, Moser E, Wieland T, Rabsch W, Schmieger H. A comparative study on the frequency of prophages among natural isolates of Salmonella and Escherichia coli with emphasis on generalized transducers. Antonie Leeuwenhoek. 1998;73:49–54. doi: 10.1023/a:1000748505550. [DOI] [PubMed] [Google Scholar]
- 13.Schicklmaier P, Schmieger H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl Environ Microbiol. 1995;61:1637–1640. doi: 10.1128/aem.61.4.1637-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schicklmaier P, Schmieger H. Sequence comparison of the genes for immunity, DNA replication, and cell lysis of the P22-related Salmonella phages ES18 and L. Gene. 1997;195:93–100. doi: 10.1016/s0378-1119(97)00182-0. [DOI] [PubMed] [Google Scholar]
- 15.Schicklmaier, P., T. Wieland, and H. Schmieger. Molecular characterization and module composition of P22-related Salmonella phage genomes. J. Biotechnol., in press. [DOI] [PubMed]
- 16.Smith H O, Levine M. Two sequential repressions of DNA synthesis in the establishment of lysogeny by phage P22 and its mutants. Proc Natl Acad Sci USA. 1964;52:356–363. doi: 10.1073/pnas.52.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–514. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Susskind M M. A new gene of bacteriophage P22 which regulates synthesis of antirepressor. J Mol Biol. 1980;138:685–713. doi: 10.1016/0022-2836(80)90060-1. [DOI] [PubMed] [Google Scholar]
- 19.Susskind M M, Wright A, Botstein D. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. II. Genetic evidence for two exclusion systems. Virology. 1971;45:638–652. doi: 10.1016/0042-6822(71)90178-4. [DOI] [PubMed] [Google Scholar]