Abstract
Comparing the genomes of mammals which evolved to have poor vision identifies an important gene for eyesight.
Research organism: Mouse, Zebrafish
Related research article Indrischek H, Hammer J, Machate A, Hecker N, Kirilenko B, Roscito J, Hans S, Norden C, Brand M, Hiller M. 2022. Vision-related convergent gene losses reveal SERPINE3’s unknown role in the eye. eLife 11:e77999. doi: 10.7554/eLife.77999.
With hundreds of cell types smoothly working together to form clear images of the world, the vertebrate eye can put the most sophisticated digital cameras to shame. Yet many of the genes which establish and maintain this delicate machine remain unknown.
Most mammals have good vision, yet some species have naturally evolved poor eyesight: mice and rats, for instance, have very poor eyesight, while species like the naked mole rat have lost their vision entirely. One way to identify the genetic sequences important for vision is to compare the genomes of species with contrasting visual capacities. Now, in eLife, Michael Hiller (Senckenberg Research Institute), Michael Brand (TU Dresden) and colleagues – including Henrike Indrischek (Max Planck Institute for Molecular Cell Biology and Genetics) as first author – report that a largely uncharacterized gene called Serpine3 is inactivated in many animals with poor or compromised vision, suggesting it may play an important role in the eye (Indrischek et al., 2022).
First, the team screened the genomes of 49 mammalian species for mutations associated with a severe loss in eye function. This sample included ten species which had poor visual capacity, such as rodents, moles and echolocating bats. A gene was classified as playing a role in eyesight if mutations stopped it from working in more than three species with poor vision. This led to the identification of 29 genes, 15 of which had not been linked to eye development or function before. However, poor vision is mostly restricted to mammals living in low-light habitats which are often limited in nutrients and biodiversity (Olsen et al., 2021). As such, the loss-of-function mutations detected by Indrischek et al. may be unrelated to vision and instead be the result of animals adapting to these challenging environments.
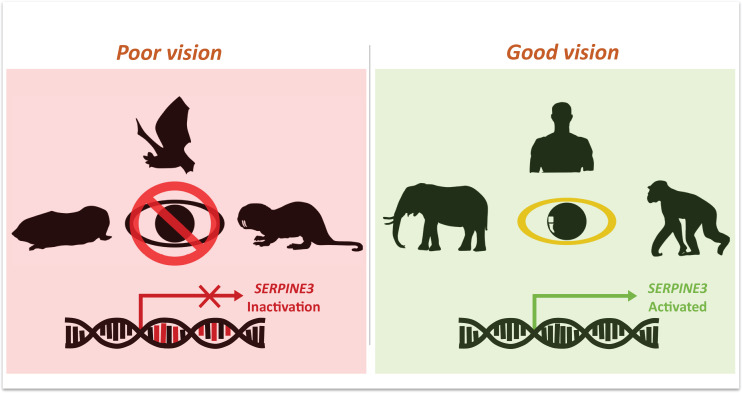
Indrischek et al. then focused on one gene, Serpine3, which was predicted to be inactive in seven out of the ten low-vision species (Figure 1). Conversely, animals with excellent vision, such as elephants and chimpanzees, have intact Serpine3 coding regions (Figure 1). To strengthen their hypothesis, Indrischek et al. added 381 other species with varying visual capabilities to their analysis. Out of the 430 species studied, 70 with poor eyesight had inactivated Serpine3.
Figure 1. Mutations in Serpine3 are associated with vision loss.
To identify genes that shape the eyes of vertebrates, Indrischek et al. screened the genome of mammals with poor (left, red) and good (right, green) vision. Most animals with poor eyesight – such as cape-golden moles, bats and naked mole rats – had mutations in the gene Serpine3 which led to its inactivation. However, in mammals with better vision – such as elephants, humans and chimpanzees – the coding region for Serpine3 was intact and the gene was active. Further experiments confirmed that the product of the Serpine3 gene is important for good vision.
The product of the Serpine3 gene belongs to a family of proteins secreted into the extracellular space and implicated in blood clotting, neuroprotection and some human diseases (Law et al., 2006; Barnstable and Tombran-Tink, 2004). To better understand the role Serpine3 plays in vision, the team carried out functional experiments in zebrafish, as their retinas are organized into layers which are similar to those found in humans (Fadool, 2003). This showed that Serpine3 is highly expressed in the zebrafish eye, particularly in the inner nuclear layer of the retina. Next, Indrischek et al. deleted Serpine3 during zebrafish development, causing adult animals to have deformed eyes and disrupting the organization of the cell layers in the retina. This suggests that vertebrate eyes need the product of Serpine3 in order to function properly.
Finally, Indrischek et al. analyzed a human dataset of genomic sequences from patients with eye-related diseases. They found mutations near to the transcription start site for Serpine3 are associated with refractive errors in the eye and macular degeneration, suggesting that this gene may also play a role in human eye diseases.
Taken together, these findings suggest that Serpine3 is important for good vision. In the future, it would be interesting to explore whether the gene is primarily important during development, or to maintain retinal cells and layers during adulthood. This knowledge could help identify new therapies for debilitating eye diseases associated with Serpine3 or other related genes.
More importantly, Indrischek et al. elegantly demonstrate how studying natural variation in traits such as eyesight can identify the function of uncharacterized genes and the role they may play in disease. Nature is full of characteristics which converged between species over the course of evolution (Farhat et al., 2013; Bergmann and Morinaga, 2019). Applying a comparative genomic approach similar to the one used in this study could transform our understanding of a multitude of other biological processes (Liu et al., 2021; Valenzano et al., 2015; Rohner, 2018). We just have to look.
Biographies
Tathagata Biswas is in the Stowers Institute for Medical Research, Kansas City, United States
Jaya Krishnan is in the Stowers Institute for Medical Research, Kansas City, United States
Nicolas Rohner is in the Stowers Institute for Medical Research and Department of Molecular and Integrative Physiology, University of Kansas Medical Center, Kansas City, United States
Competing interests
No competing interests declared.
References
- Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of PEDF in the eye: molecular targets and therapeutic potential. Progress in Retinal and Eye Research. 2004;23:561–577. doi: 10.1016/j.preteyeres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Bergmann PJ, Morinaga G. The convergent evolution of snake-like forms by divergent evolutionary pathways in squamate reptiles. Evolution; International Journal of Organic Evolution. 2019;73:481–496. doi: 10.1111/evo.13651. [DOI] [PubMed] [Google Scholar]
- Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Developmental Biology. 2003;258:277–290. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PKC, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nature Genetics. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrischek H, Hammer J, Machate A, Hecker N, Kirilenko B, Roscito J, Hans S, Norden C, Brand M, Hiller M. Vision-related convergent gene losses reveal SERPINE3’s unknown role in the eye. eLife. 2022;11:e77999. doi: 10.7554/eLife.77999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RHP, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biology. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Gao J, Cui X, Li Z, Chen L, Yuan Y, Zhang Y, Mei L, Zhao L, Cai D, Hu M, Zhou B, Li Z, Qin T, Si H, Li G, Lin Z, Xu Y, Zhu C, Yin Y, Zhang C, Xu W, Li Q, Wang K, Gilbert MTP, Heller R, Wang W, Huang J, Qiu Q. A towering genome: experimentally validated adaptations to high blood pressure and extreme stature in the giraffe. Science Advances. 2021;7:eabe9459. doi: 10.1126/sciadv.abe9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L, Thum E, Rohner N. Lipid metabolism in adaptation to extreme nutritional challenges. Developmental Cell. 2021;56:1417–1429. doi: 10.1016/j.devcel.2021.02.024. [DOI] [PubMed] [Google Scholar]
- Rohner N. Cavefish as an evolutionary mutant model system for human disease. Developmental Biology. 2018;441:355–357. doi: 10.1016/j.ydbio.2018.04.013. [DOI] [PubMed] [Google Scholar]
- Valenzano DR, Benayoun BA, Singh PP, Zhang E, Etter PD, Hu CK, Clément-Ziza M, Willemsen D, Cui R, Harel I, Machado BE, Yee MC, Sharp SC, Bustamante CD, Beyer A, Johnson EA, Brunet A. The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell. 2015;163:1539–1554. doi: 10.1016/j.cell.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]