Abstract
Erwinia chrysanthemi 3937 secretes several pectinolytic enzymes, among which eight isoenzymes of pectate lyases with an endo-cleaving mode (PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ) have been identified. Two exo-cleaving enzymes, the exopolygalacturonate lyase, PelX, and an exo-poly-α-d-galacturonosidase, PehX, have been previously identified in other E. chrysanthemi strains. Using a genomic bank of a 3937 mutant with the major pel genes deleted, we cloned a pectinase gene identified as pelX, encoding the exopolygalacturonate lyase. The deduced amino acid sequence of the 3937 PelX is very similar to the PelX of another E. chrysanthemi strain, EC16, except in the 43 C-terminal amino acids. PelX also has homology to the endo-pectate lyase PelL of E. chrysanthemi but has a N-terminal extension of 324 residues. The transcription of pelX, analyzed by gene fusions, is dependent on several environmental conditions. It is induced by pectic catabolic products and affected by growth phase, oxygen limitation, nitrogen starvation, and catabolite repression. Regulation of pelX expression is dependent on the KdgR repressor, which controls almost all the steps of pectin catabolism, and on the global activator of sugar catabolism, cyclic AMP receptor protein. In contrast, PecS and PecT, two repressors of the transcription of most pectate lyase genes, are not involved in pelX expression. The pelX mutant displayed reduced pathogenicity on chicory leaves, but its virulence on potato tubers or Saintpaulia ionantha plants did not appear to be affected. The purified PelX protein has no maceration activity on plant tissues. Tetragalacturonate is the best substrate of PelX, but PelX also has good activity on longer oligomers. Therefore, the estimated number of binding subsites for PelX is 4, extending from subsites −2 to +2. PelX and PehX were shown to be localized in the periplasm of E. chrysanthemi 3937. PelX catalyzed the formation of unsaturated digalacturonates by attack from the reducing end of the substrate, while PehX released digalacturonates by attack from the nonreducing end of the substrate. Thus, the two types of exo-degrading enzymes appeared complementary in the degradation of pectic polymers, since they act on both extremities of the polymeric chain.
The enterobacterium Erwinia chrysanthemi causes soft rot disease of various plants. The maceration process involves the depolymerization of the pectin of plant cell walls and the middle lamella. Pectin is a heteropolysaccharide with a backbone consisting of partially esterified galacturonic acid. Pectin degradation is accomplished by a variety of pectinases, which differ mainly in the reaction mechanism (β-elimination or hydrolysis) and in the random or terminal mode of attack of the polymer (endo or exo).
Erwinia chrysanthemi is characterized by its ability to secrete multiple isoenzymes of endo-pectate lyases (endo-Pels), which play an important role in the soft rot disease (3). These enzymes randomly cleave, by β-elimination, internal glycosidic linkages in pectic polymers, preferentially polygalacturonate or low-methoxylated pectin (up to 30%) (43). They generate a series of oligogalacturonates with a 4,5-unsaturated residue at the nonreducing end. Eight E. chrysanthemi 3937 endo-Pels have been characterized, PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ (16, 29, 39). The corresponding genes are organized in four clusters on the bacterial chromosome (pelA-pelE-pelD, pelB-pelC-pelZ, pelI, and pelL). Pectate lyases are classified in five different families according to their primary amino acid sequences (4a). PelA, PelB, PelC, PelD, and PelE belong to family 1, which contains several bacterial pectate lyases, fungal pectin lyases, and plant proteins (14, 15). PelI belongs to family 3 (39), which includes enzymes of Erwinia carotovora and of the phytopathogenic fungus Nectria haematococca. Family 4 contains PelL and PelX of E. chrysanthemi (1, 23). PelZ of E. chrysanthemi is the sole characterized component of family 5 (29). Two exo-cleaving depolymerases have also been identified in E. chrysanthemi. A cell-bound exopolygalacturonate lyase and an extracellular exo-poly-α-d-galacturonosidase were found in E. chrysanthemi CUCPB1237 (7, 8). The corresponding genes, pelX and pehX, respectively, were isolated from E. chrysanthemi EC16 (6, 12). PehX and PelX contribute to pectin catabolism but they have little direct involvement in plant maceration.
Transcription of the E. chrysanthemi pectinase genes is dependent on several environmental conditions (16). The genes are all induced by pectin catabolic products and by the late exponential growth phase. Most of them are repressed under conditions of catabolite repression, nitrogen starvation, and high temperature. Other conditions affect the transcription of a limited set of genes: pelA, pelD, and pelE expression is increased by oxygen limitation, while elevation of osmolarity activates only pelE expression (17). The regulation of pectinase gene transcription involves several regulatory systems. The KdgR repressor mediates the induction of the pectinolytic genes in the presence of pectin catabolites (28, 33). The PecS and PecT proteins, which are members of the MarR and LysR families of regulators, respectively, repress the transcription of most of the pectinase genes (34, 41). The signals triggering PecS and PecT controls have not been identified. The cyclic AMP receptor protein (CRP), which modulates the expression of many catabolite operons, is essential for activating the transcription of the pectinase genes (32).
In this paper, we present the identification of the pelX gene of E. chrysanthemi 3937 and the characterization of its product, the exopolygalacturonate lyase, PelX. pelX expression was monitored by using an uidA transcriptional fusion. We assayed the fusion under various environmental conditions and tested the involvement of KdgR, PecS, PecT, and CRP in pelX regulation. PelX was purified to analyze its biochemical properties. To clarify the role of PelX in pectin degradation, we determined its best substrate by using purified oligogalacturonates and its cellular localization in E. chrysanthemi 3937.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| E. chrysanthemi 3937 derivatives | ||
| A350 | lacZ2 | 17 |
| A576 | lacZ2 arg-10 kdgK | 23 |
| A837 | lacZ2 kdgR | Laboratory collection |
| A1524 | lacZ2 pecS::Mu Cm | 34 |
| A2174 | lacZ2 pecT::Cm | 41 |
| A2507 | lacZ2 crp::Cm | 32 |
| A2524 | lacZ2 pelX::uidA | This work |
| A2699 | lacZ2 pelX::Cm | This work |
| A2737 | lacZ2 kdgR pelX::uidA | This work |
| A2738 | lacZ2 pecS::Mu Cm pelX::uidA | This work |
| A2739 | lacZ2 pecT::Cm pelX::uidA | This work |
| A2740 | lacZ2 kdgK pelX::uidA | This work |
| A2762 | lacZ2 crp::Cm pelX::uidA | This work |
| A3004 | pelX::uidA | This work |
| A3497 | lacZ2 kdgR ΔpehV-pehW-pehX::Km | This work |
| A3498 | lacZ2 kdgR ΔpehV-pehW-pehX::uidA pelX::Cm | This work |
| A849 | met-10 leu-1 xyl-1 | 18 |
| A2708 | met-10 arg-10 leu-1 kdgK | 18 |
| PMV4116 | ΔpelA-pelE-pelD-paeY ΔpelB-pelC-pelZ | 4 |
| E. coli | ||
| NM522 | Δ(lac-proAB) Δ(mcrB-hsdSM)5 supE thi (F′ proAB lacIqlacZΔM15) | Laboratory collection |
| BL21(DE3) | E. coli B, F−dcm ompT hsdS gal λ(DE3), T7 polymerase gene under the lacUV5 promoter | 40 |
| Plasmids | ||
| pUC18 | Apr | Laboratory collection |
| pBSAp | Bluescript KS(+), Apr | Stratagene |
| pBSCm | Bluescript KS(+), Cmr | Stratagene |
| pT7-5 | T7Φ10, Apr | 42 |
| pPN1 | pUC18 derivative with a 3.3-kb Sau3A fragment in the BamHI site, pelX+ | This work |
| pPN15 | pUC18 derivative with a 4.4-kb Sau3A fragment in the BamHI site, pelX+ | This work |
| pPN9 | pUC18 derivative with a 4.5-kb Sau3A fragment in the BamHI site, pelX+ | This work |
| pN496 | pBSCm with a 3.8-kb uidA-Km cassette | 17 |
| pPNa18 | pPN1 derivative with a uidA-Km insertion from pN496 in the NruI site, Apr, Kmr, pelX::uidA+ | This work |
| pTaX | pT7-5 derivative with a 2.9-kb MunI-XbaI fragment from pPN1, AprpelX+ | This work |
| pTaXD1 | pTaX derivative with a 0.9-kb NruI-SgrAI deletion | This work |
| pTaXD2 | pTaX derivative with a 0.6-kb MamI deletion | This work |
Media and growth conditions.
Cells were grown in either Luria-Bertani or M63 medium (25). When required, the media were solidified with agar (15 g · liter−1). E. chrysanthemi and Escherichia coli cells were usually incubated at 30 and 37°C, respectively. Carbon sources were added at 2 g · liter−1 except for polygalacturonate (grade II; Sigma Chemical Co.), which was used at 4 g · liter−1. Pectate agar medium was used to detect pectinolytic activity, which is visualized, after addition of 1 M CaCl2, by pitting of the colony in the medium due to polygalacturonate cleavage (20). The media used to test the different physiological conditions have been described previously (17). When required, antibiotics were added at the following concentrations: kanamycin, 20 μg · ml−1; ampicillin, 50 μg · ml−1; chloramphenicol, 20 μg · ml−1; streptomycin, 100 μg · ml−1.
Matings and transductions.
Plasmid pULB110, a kanamycin-sensitive RP4::mini Mu derivative (44), was used for chromosome mobilization as previously described (18). The pelX-Km insertion was transduced into various strains by using the phi-EC2 generalized transducing phage (31).
Cellular fractionation and analytical procedures.
Periplasmic proteins were released from E. coli cells by osmotic shock (9) and from E. chrysanthemi cells by spheroplast formation (37). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on slab gels (4% stacking gel and 12% separating gel) with the mini-protean II system (Bio-Rad). Proteins were stained with Coomassie blue G-250. Isoelectrofocusing was performed in a 3.5 to 10 pH gradient with Pharmalytes. To perform the differential detection of pectin depolymerases after electrofocusing, the gel was incubated in 50 mM Tris-HCl (pH 8.5)–5 g of polygalacturonate liter−1 either with 1 mM CaCl2 (to detect the major pectate lyases) or with 1 mM MnCl2 (to detect PelX and PehX). After 10 to 60 min of incubation at 25°C, the gel was rinsed with water and stained with 0.05% ruthenium red.
Overproduction and purification of PelX.
The pelX gene was overexpressed by using the T7 promoter-T7 RNA polymerase system (42). The 2.9-kb MunI-XbaI fragment from plasmid pPN1 was inserted into the pT7-5 expression vector. The resulting plasmid, pTaX, was introduced into E. coli K38/pGP1.2, which contains the T7 RNA polymerase gene under the control of the cI857 thermosensitive repressor. The plasmid-encoded proteins were labelled with [35S]cysteine-[35S]methionine after thermal induction of the T7 polymerase (42).
For PelX purification, plasmid pTaX was introduced into E. coli BL21(DE3), which contains a chromosomal copy of the T7 RNA polymerase gene under the control of the lacUV5 promoter (40). The BL21(DE3)/pTaX cells were grown at 30°C in Luria-Bertani medium supplemented with ampicillin (150 μg · ml−1). At an optical density at 600 nm of 0.8 to 1, the synthesis of T7 RNA polymerase was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and the cells were grown for an additional 2 to 3 h.
Cells were harvested by centrifugation for 10 min at 5,000 × g at 4°C and then frozen at −80°C. The periplasmic fraction was extracted from cells by three cycles of freezing-thawing (19). Proteins were concentrated by 85% ammonium sulfate precipitation. The pellet was solubilized in 50 mM sodium phosphate buffer (pH 7)–5 mM EDTA–1.5 M ammonium sulfate and loaded onto a phenyl-TSK-Gel column that had been equilibrated and extensively washed with the same buffer. Upon application of a 1.5 to 0.5 M ammonium sulfate linear gradient, the PelX protein was eluted at about 0.7 M ammonium sulfate. The fractions containing the pure PelX protein were pooled and concentrated with Centricon 10 (Amicon).
Enzyme assays.
Pectate lyase activity was determined by spectrophotometrically monitoring the formation of unsaturated products from polygalacturonate at 230 nm. Unless otherwise specified, the standard assay mixture consisted of 50 mM Tris-HCl (pH 8.5), 0.1 mM CaCl2, and 0.5 g of polygalacturonate per liter in a total volume of 1 ml. The appearance of products was monitored at 37°C for 2 min. The molar extinction coefficient of unsaturated oligogalacturonates was assumed to be 5,200 (26). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of unsaturated product per min. The influence of Ca2+ was investigated by addition of CaCl2 concentrations ranging from 0 to 1 mM. EDTA (0.5 and 1 mM) was added to verify the cation requirement. The optimum pH was determined by using Tris-HCl (pH 7 to 9), N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-NaOH (pH 5.5 to 7.5), and sodium phosphate (pH 6 to 8), each at 50 mM. The activity on substrates with various degrees of methylation was determined by substituting 7, 22, 45, 60, 75, and 90% esterified citrus pectins (Copenhagen Pectin) for polygalacturonate. Oligogalacturonates of defined length (Gn, with n = 2 to 7) were prepared as previously described (21).
The β-glucuronidase activity was measured by monitoring the degradation of p-nitrophenyl-β-d-glucuronide into p-nitrophenol, which absorbs at 405 nm (2). Specific activities are expressed as nanomoles of products liberated per minute per milligram of bacterial dry weight.
Analysis of the depolymerase reaction products.
Thin-layer chromatography was used to identify the products obtained after polygalacturonate degradation (23). The chromatogram was developed with a 5:3:2 mixture of n-butanol, water, and acetic acid, and the products were visualized by treatment with phosphomolybdic acid (23).
Reaction products resulting from cleavage of hexagalacturonates were analyzed by high-performance anion-exchange chromatography with a Dionex BioLC system (5). Detection was carried out by pulsed amperometry and UV detection at 235 nm. Hexagalacturonate (G6) and reduced hexagalacturonate (rG6) were used to investigate the mode of action of PelX and PehX. rG6 was obtained after NaBH4 reduction of G6. The PehX reaction mixtures (0.5 ml) contained 0.5 mM substrate in 0.02 M Tris-HCl (pH 7) and 25 or 500 ng of enzyme for G6 and rG6, respectively. The PelX reaction mixtures (0.5 ml) contained 0.5 mM substrate in 0.02 M Tris-HCl (pH 8), 1 mM CaCl2, and 10 or 200 ng of enzyme for G6 and rG6, respectively. Reactions were performed at 30°C for 30 min. The enzyme reactions were stopped by the addition of 0.1 volume of 1% acetic acid, which lowered the pH to 4.
Recombinant DNA techniques.
Preparation of plasmid or chromosomal DNA, restriction digestions, ligations, DNA electrophoresis, and transformations were carried out as described by Sambrook et al. (37). For the construction of the genomic library, E. chrysanthemi PMV4116 chromosomal DNA was partially digested with Sau3A and 3- to 6-kb fragments were isolated by electroelution. The sized fragments were ligated into pUC18 digested with BamHI and treated with alkaline phosphatase (Pharmacia). The deleted proteins PelXD1 and PelXD2 were obtained by in-frame deletions of the pelX gene present in plasmid pTaX. pTaXD1, encoding PelXD1, was obtained by deletion of a 0.9-kb NruI-SgrAI fragment (the SgrAI protruding end was filled in with Klenow enzyme before ligation). pTaXD2, encoding PelXD2, was obtained by deletion of a 0.6-kb MamI fragment.
For nucleotide sequence analysis, deletions were generated with restriction endonucleases. The chain termination method was performed with double-stranded DNA templates, M13 primer or M13 reverse primer, [35S]dATP, and T7 DNA polymerase (Pharmacia sequencing kit). Some sequences were performed by Genome Express SA (Grenoble). The resulting data were analyzed with the Mac Molly program (SoftGene, Berlin, Germany).
Construction of the pelX::uidA fusion.
Insertion of a uidA-Km cassette in the correct orientation generates transcriptional fusion (2). The uidA-Km cassette was liberated by SmaI digestion of pN416 and inserted into the NruI site of plasmid pPN1. In one of the recombinant plasmids, pPNa18, uidA is oriented in the same transcriptional direction as pelX, giving rise to a pelX::uidA fusion. The pPNa18 plasmid was introduced into E. chrysanthemi cells by electroporation. The pelX mutation was then integrated into the E. chrysanthemi chromosome by marker exchange recombination after successive cultures in low-phosphate medium in the presence of the appropriate antibiotic (36).
Maceration of plant tissue.
Small cubes (3-mm sides) were cut from commercial Bindge potato tubers. They were placed into 0.1 M Tris-HCl (pH 8)–0.5 mM CaCl2, and 0.1 U of PelX was added per ml. Samples were incubated at 30°C and examined after 1 to 24 h. The degree of tissue maceration was estimated by determining the ease with which the tissue could be pulled apart with a spatula. The PelD macerating enzyme was used as the positive control (23).
Pathogenicity test.
Saintpaulia ionantha potted plants were infected as previously described (10). Plants were inoculated, after wounding of a leaf, with 50 μl of a bacterial suspension (108 bacteria). Results of infections were scored daily for 10 days. For inoculation of potato tubers, sterile pipette tips containing 5 μl of bacterial suspension (107 bacteria) were inserted into the tuber parenchyma (22). The tubers were inoculated and incubated in a dew chamber. After 1, 2, or 3 days, the tubers were sliced vertically through the inoculation point and the weight of decayed tissue was taken as the characteristic of disease severity. Chicory leaves were infected with 50 μl of a diluted bacterial suspension (106 bacteria). After incubation in a dew chamber for 24 h, the length of rotted tissue was measured to estimate the disease severity. Pathogenicity tests were repeated in three independent experiments. In each experiment, both the wild-type strain and the pelX mutant were used and 30 plants were infected for each strain.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. Y16797.
RESULTS
Isolation of the pelX gene and determination of its nucleotide sequence.
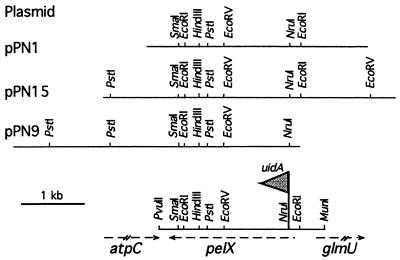
A genomic library containing inserts of 3 to 6 kb was constructed in pUC18 by using DNA extracted from strain PMV4116, a 3937 derivative with the pelA-pelE-pelD and pelB-pelC-pelZ gene clusters deleted (4). About 3,500 clones were screened for pectinolytic activity on various media. Using a pectate semisolid agar medium, we could identify six transformants presenting a weak pectinolytic activity. The assay of enzymatic activity revealed that three clones encode a pectate lyase activity. Restriction analysis of the plasmids present in these three clones showed that pPN1, pPN15, and pPN9 contained overlapping fragments of 3.3, 4.4, and 4.5 kb, respectively (Fig. 1). Deletion analysis and subcloning experiments enabled us to reduce the size of the DNA fragment exhibiting the pectate lyase activity to a 2.8-kb MunI-PvuII fragment (Fig. 1).
FIG. 1.
Physical map of the E. chrysanthemi 3937 pelX gene. The restriction map of the three PelX-encoding plasmids is indicated. The arrows below the MunI-PvuII fragment indicate the position and the transcription direction of the pelX gene and of adjacent genes. The flag shows the site of insertion of the uidA-Km cassette.
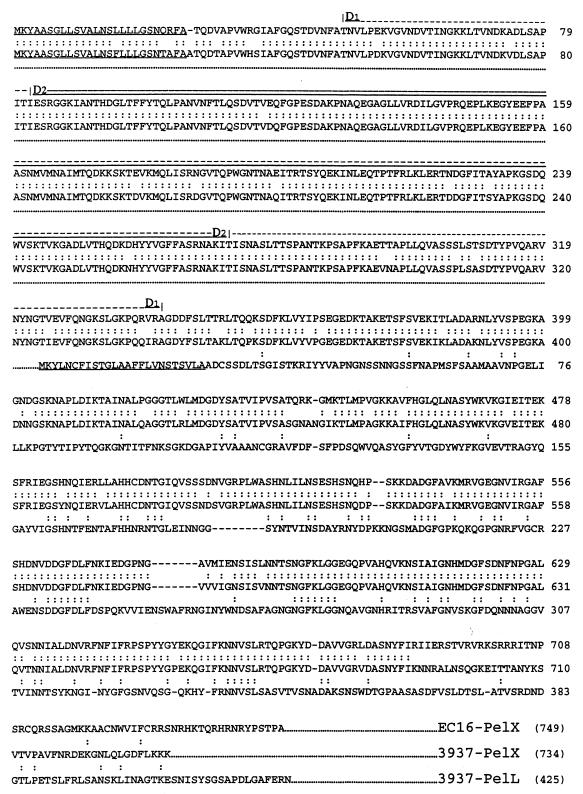
The nucleotide sequence of the 2,789-nucleotide (nt) MunI-PvuII DNA fragment was determined (GenBank accession no. Y16797). Sequence analysis revealed the presence of a unique complete open reading frame (ORF) which began with an ATG codon at position 503 and ended with TAA at position 2705. Insertion in the NruI site present in this ORF abolished pectate lyase production, demonstrating that this ORF codes for the pectate lyase activity. The orientation of the uidA gene in the insertion giving rise to a gene fusion is in accordance with the transcription direction of the identified ORF. The deduced amino acid sequence shows 91% identity to PelX of strain EC16 (6) between residues 1 and 690 (strain 3937 PelX numbering), with only two deletion-insertion mismatches (positions 27 and 446) (Fig. 2). However, no homology between EC16-PelX and 3937-PelX was detected after residue 691 (Fig. 2). This discrepancy involving the C-terminal part of the proteins is surprising when considering the high homology in the upstream sequence of the proteins. We noticed high homology at the DNA level all along the pelX genes of the two strains. The difference in the amino acid sequences is caused by the presence of a +1 frameshift at position 2574 of the 3937 pelX sequence. Since this difference may result from an error in one of the nucleotide sequences, we carefully verified the 3937 pelX sequence and analyzed the coding probability of each frame of the 3937 and EC16 sequences. In the 3937 sequence, the coding probability is high in frame 2 from positions 526 to 2704 while for EC16, the coding probability declines at the position corresponding to 2578, just downstream from the frameshift observed between the two strains. From these observations, we suppose that one error in the EC16-pelX sequence altered the correct reading frame. Alternatively, the EC16 strain may present a mutation leading to a change in the reading frame of the pelX gene without inactivating the enzymatic activity of its product.
FIG. 2.
Comparison of the amino acid sequences of the PelX and PelL proteins. The PelX sequence of E. chrysanthemi EC16 is from reference 6, and PelL of strain 3937 was described by Lojkowska et al. (23). Colons indicates identical residues. The signal sequence of the proteins are underlined. D1 and D2 indicate the ends of the two deletions on 3937 PelX.
The pelX ATG start is preceded by the potential ribosome-binding site GGGGAA (3 nt upstream) and by a potential promoter (16 nt upstream), with the following homology to the classical ς70 consensus: 6 of 6 nt (TTGACA) for the −35 region and 3 of 6 nt (ACAAAT) for the −10 region, with a spacing of 17 nt (residues matching the consensus are underlined) (Fig. 3). Since most of the pectinase genes are controlled by the KdgR repressor, we looked for a potential KdgR-binding site in the promoter region. The KdgR box corresponds to two imperfect inverted repeats of 9 nt. A potential KdgR box, with 17 of 18 nt conserved, AAAGAAACANTGTTTCATT (where N indicates 1 nt) is centered 12 nt upstream from the putative pelX promoter. A potential CRP-binding site (TGTGAN6CAAAA) partially overlaps the KdgR-binding site. Alignment with the 3937 and EC16 pelX regulatory regions indicates that these sites potentially involved in pelX transcription (−35 and −10 regions, CRP-binding site, and KdgR box) are totally conserved between the two strains (Fig. 3).
FIG. 3.
Alignment of the pelX promoters of E. chrysanthemi 3937 and EC16. The PelX sequence of E. chrysanthemi EC16 is from reference 6. Vertical lines indicate identical bases. The putative sequences corresponding to the translation start, Shine-Dalgarno (S.D.), −10 and −35 promoter, KdgR, and CRP-binding sites are underlined.
The pelX translational stop is followed by a GC-rich inverted repeat located 26 nt downstream (GAGGGCGCCN7GGCGCCCGTC; calculated free energy of formation, −67 kJ mol−1). This sequence may be involved in the termination of pelX transcription.
The pelX gene is preceded and followed by partial potential ORFs transcribed on the other DNA strand. DNA homology searching revealed a high similarity between the ORF situated on the 3′ pelX side and the atpC gene of E. coli, encoding the ɛ subunit of ATP synthase (38), and between the ORF situated on the 5′ pelX side and the glmU gene of E. coli, encoding N-acetylglucosamine-1-phosphate uridyltransferase, involved in peptidoglycan and lipopolysaccharide biosynthesis (24). It is interesting that glmU and atpC are adjacent on the E. coli chromosome. Thus, acquisition of the pelX gene by E. chrysanthemi appeared to result from an insertion between the home genes glmU and atpC present on the ancestral enterobacterial genome.
Expression of the pelX gene.
A pelX::uidA transcriptional fusion was constructed by insertion of an uidA-Km cassette in the NruI site situated inside the pelX ORF (Fig. 1). After recombination into the E. chrysanthemi chromosome, the expression of the fusion was followed in various conditions (Table 2). Under noninducing conditions, pelX showed a low basal level of expression. In the presence of polygalacturonate, its transcription was stimulated about fourfold. By monitoring the expression of the fusion during the growth curve, we showed that pelX transcription increased about fourfold when the cells entered the late exponential growth phase and was coincident with production of the endo-Pels, as measured by pectate lyase activity (data not shown). In the presence of a readily utilizable carbon source, such as glucose, a fourfold decrease in pelX transcription was observed (Table 2). Oxygen limitation slightly increased pelX expression in the presence of polygalacturonate, while nitrogen starvation strongly inhibited pelX expression. Variations of the growth temperature (25, 30, or 37°C) or modification of the medium osmolarity had no significant effect on the pelX expression (Table 2).
TABLE 2.
Expression of the pelX::uidA fusion under various growth conditions
| Growth conditionsa
|
β-Glucuronidase sp actb (mean ± SD) | ||
|---|---|---|---|
| Carbon source | Inducer | Specific condition | |
| Glycerol | None | None | 62 ± 10 |
| Glycerol | Polygalacturonate | None | 232 ± 22 |
| Glucose | None | None | 16 ± 3 |
| Glycerol | None | Oxygen limitation | 86 ± 20 |
| Glycerol | Polygalacturonate | Oxygen limitation | 405 ± 31 |
| Glycerol | None | N starvation | 13 ± 3 |
| Glycerol | Polygalacturonate | N starvation | 11 ± 3 |
| Glycerol | None | 25°C | 54 ± 13 |
| Glycerol | None | 37°C | 57 ± 11 |
| Glycerol | None | + NaCl (0.3 M) | 57 ± 13 |
Cultures of strain A2524 were grown in M63 minimal medium to late log phase in the presence or absence of the inducing compound (polygalacturonate) under different physiological conditions and, unless stated, at 30°C.
β-Glucuronidase specific activity reflects the expression of the pelX::uidA fusion and is expressed as nanomoles of product liberated per minute per milligram of bacterial dry weight.
The pelX fusion was transduced into strains containing mutations affecting pectinase production (Table 3). Pectate lyase activity, corresponding to the endo-Pels, strongly increased in the kdgR mutant in the absence of polygalacturonate, due to inactivation of the KdgR repressor, and similarly in the kdgK mutant in the presence of polygalacturonate, due to the accumulation of the intracellular inducer 2-keto-3-deoxygluconate (KDG). The pelX transcription, as estimated by measurement of pelX::uidA expression, was clearly affected by the kdgR or kdgK mutations. An increase of about 33-fold was observed under noninducing conditions in the kdgR mutant, and a 3-fold increase was observed under inducing conditions in the kdgK mutant (Table 3). This result suggests that KDG is responsible for induction of pelX expression in the presence of polygalacturonate and that pelX expression is KdgR dependent. Like pectate lyase activity, expression of the pelX fusion is very low in the crp mutant, demonstrating the important role played by the global activator CRP in the pelX transcription. Mutation in pecS or pecT does not significantly affect the expression of the pelX fusion. The pecS and pecT genes are thus not involved in pelX regulation.
TABLE 3.
Expression of the pelX::uidA fusion in various mutants
| Strain (main genotype) | Carbon source | Inducera | Sp actb of:
|
|
|---|---|---|---|---|
| β-Glucuronidase | Pectate lyase | |||
| A2524 (pelX::uidA) | Glycerol | None | 62 | 0.06 |
| Glycerol | Polygalacturonate | 232 | 2.27 | |
| Glucose | None | 16 | 0.04 | |
| Glucose | Polygalacturonate | 209 | 1.78 | |
| A2737 (kdgR pelX::uidA) | Glycerol | None | 2,055 | 1.11 |
| Glycerol | Polygalacturonate | 1,551 | 4.32 | |
| A2740 (kdgK pelX::uidA) | Glycerol | None | 66 | 0.06 |
| Glycerol | Polygalacturonate | 642 | 18.23 | |
| A2738 (pecS pelX::uidA) | Glycerol | None | 54 | 0.36 |
| Glycerol | Polygalacturonate | 225 | 3.49 | |
| A2739 (pecT pelX::uidA) | Glycerol | None | 49 | 0.45 |
| Glycerol | Polygalacturonate | 186 | 3.66 | |
| A2762 (crp pelX::uidA) | Glucose | None | 4 | 0.01 |
| Glucose | Polygalacturonate | 3 | 0.16 | |
Cultures were grown at 30°C in M63 glycerol minimal medium in the presence or absence of polygalacturonate (0.25%) to late log phase.
β-Glucuronidase specific activity reflects the expression of the pelX::uidA fusion and is expressed as nanomoles of product liberated per minute per milligram of bacterial dry weight. Pectate lyase specific activity corresponds to the action of the endo-Pels and is expressed as micromoles of product liberated per minute per milligram of bacterial dry weight. The results reported are the mean of at least three independent experiments with standard deviations corresponding to less than 20%, except for the very low activities in the crp mutant, for which standard deviations reached about 30%.
Analysis of the pelX mutant.
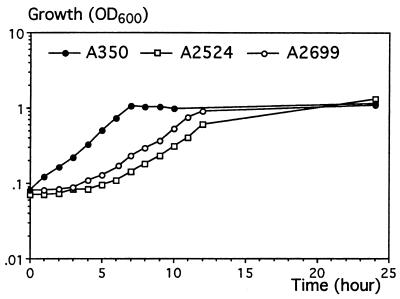
Growth of the pelX mutants A2524 and A2699 with polygalacturonate as the sole carbon source was compared with that of the parental strain, A350. The final growth yields were about 109 bacteria per mg of polygalacturonate for the two strains. However, a clear difference was noticed in the lag phase and doubling times during exponential growth (about 110 min for A350 and 160 min for the pelX mutants) (Fig. 4). Thus, the pelX mutation affects the initiation of polygalacturonate catabolism.
FIG. 4.
Growth of E. chrysanthemi pelX mutants on polygalacturonate. The sole carbon source was polygalacturonate (4 g · liter−1) in M63 medium. Growth was monitored by measuring the optical density of the culture at 600 nm (OD600). A350 is the parental strain of the two pelX mutants A2524 and A2699.
We also compared the maceration provoked by the pelX mutant A3004 and wild-type strain 3937 on chicory leaves and potato tubers. On chicory leaves, the length of rotted tissues measured 24 h after inoculation appeared to be reduced for the pelX mutant in comparison with the 3937 strain (29 ± 9 and 45 ± 11 mm, respectively). No significant reduction was observed on potato tubers 1, 2, or 3 days after infection (data not shown). We also compared the pathogenic behavior of the E. chrysanthemi pelX mutant on potted plants of Saintpaulia ionantha with that of the wild-type strain. After infecting 30 plants with strains 3937 and A3004, we monitored the appearance of symptoms for 10 days. We observed no significant difference in the progress of the disease between the pelX mutant and the wild type strain (data not shown).
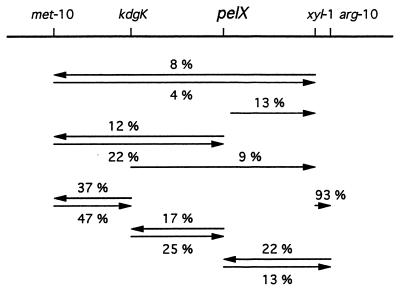
The pelX locus was identified by using the Kmr marker of the pelX::uidA fusion. Chromosomal mobilization mediated by plasmid pULB110 was used for conjugation with various polyauxotrophic recipients (18). The pelX-Km marker cotransferred with the met-10 and xyl-1 markers. A more precise mapping in this region indicated that pelX is located between kdgK and arg-10. The resulting gene order is shown in Fig. 5.
FIG. 5.
Location of the pelX gene on the genetic map of E. chrysanthemi 3937. Localization was performed by chromosomal mobilization with plasmid pULB110. The numbers indicate the percentage of cotransfer between two markers. Arrowheads point to the unselected marker.
Identification and purification of PelX.
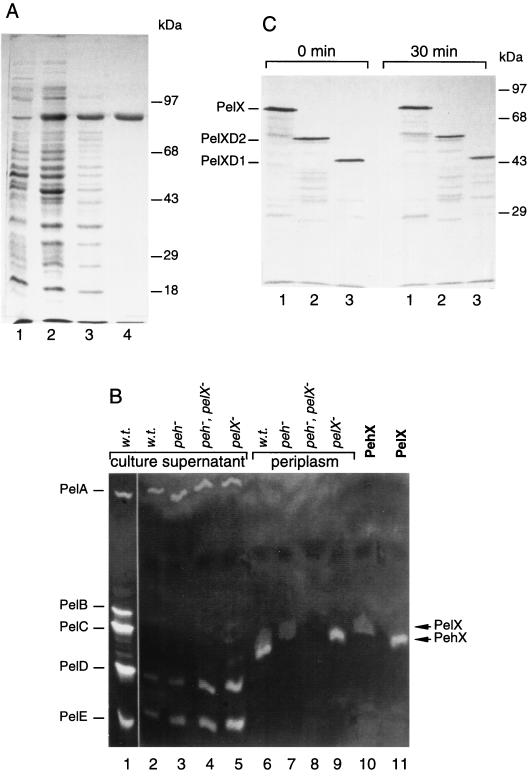
The PelX protein was overproduced in E. coli and purified to analyze its properties. Analysis by SDS-PAGE revealed the presence of a protein of about 76 kDa (Fig. 6A). Isoelectrofocusing followed by specific staining of pectinolytic activity indicated that the isoelectric point of PelX is about 9 (Fig. 6B). These data are in agreement with that deduced from the nucleotide sequence of the pelX gene. Indeed, the pelX gene of strain 3937 encodes a 734-amino-acid protein, including a typical amino-terminal signal sequence with a potential cleavage site between the two alanine residues at positions 26 and 27 (Fig. 2). A cleavage site was observed at the same position in EC16-PelX and confirmed by sequencing the N terminus of the mature protein (6). The mature 3937 PelX protein contains 708 amino acids and has a calculated molecular mass of 76,938 daltons and a calculated pI of 7.7.
FIG. 6.
Identification of the PelX protein. (A) SDS-PAGE separation was followed by staining with Coomassie blue. Lanes: Whole-cell extract of BL21(DE3)/pTaX before (lane 1) and after (lane 2) IPTG induction; periplasmic fraction of BL21(DE3)/pTaX after IPTG induction (lane 3); pure PelX protein (lane 4). Molecular mass markers are indicated. (B) Electrofocusing was performed either at 2,500 V-h (lane 1) or at 1,500 V-h (lanes 2 to 11) and followed by specific revelation of pectate lyase activity in the presence of either CaCl2 for 10 min (lane 1) or MnCl2 for 40 min (lanes 2 to 11). Culture supernatants (lanes 1 to 5) and periplasmic fractions (lanes 6 to 9) of the E. chrysanthemi strains are shown: A837 (parental strain) (lanes 1, 2, and 6), A3497 (Δpeh) (lanes 3 and 7), A3498 (Δpeh pelX::Cm) (lanes 4 and 8), and A2737 (pelX::Km) (lanes 5 and 9). Lanes 10 and 11 contain purified PehX and PelX, respectively. The positions of the five major pectate lyases (PelA to PelE), PelX, and PehX are indicated. w.t., wild type. (C) Stability of the PelX deletion derivatives. E. coli K38/pGP1.2 cells carrying pTaX (lane 1), pTaXD2 (lane 2), or pTaXD1 (lane 3) were labelled with [35S]cysteine-[35S]methionine. An excess of “cold” cysteine-methionine was added 10 min later, and the cells were collected immediately (0 min) or incubated for additional 30 min at 37°C (30 min).
PelX has homology to the endo-Pel PelL (1, 23) (Fig. 2). However, compared with PelL, PelX possesses an N-terminal extension of 324 residues, suggesting that it is organized in two domains. This observation led us to construct PelX derivatives presenting large deletions in their N-terminal part but retaining the signal sequence. PelXD1 and PelXD2 lack 294 and 191 residues, respectively (Fig. 2). These two proteins were overproduced in E. coli. Both are relatively stable (Fig. 6C) but present no pectinolytic capacity, demonstrating that the N-terminal region is important for PelX activity.
PelX has been suggested to be localized in the periplasm in E. chrysanthemi EC16 (6). However, “the localization of the enzyme has not been rigorously determined”. To verify the localization of PelX in E. chrysanthemi 3937, we developed a differential detection of pectinases after electrofocusing. The high activity of the major pectate lyases can mask the weak activity of PelX. Since the major E. chrysanthemi pectate lyases are inhibited in the presence of Mn2+ (43), Ca2+ cations were replaced by Mn2+ for the detection of PelX, which has a similar activity in the presence of the two cations (Fig. 6B). In addition, the E. chrysanthemi exo-poly-α-d-galacturonosidase PehX has almost the same pI as PelX and its activity is not affected by cations. To suppress PehX, we used a mutant with the pehX gene and adjacent regions deleted. After cellular fractionation, PelX was detected only in the periplasm of E. chrysanthemi (Fig. 6B). Surprisingly, the polygalacturonase activity was also detected in the periplasm of E. chrysanthemi 3937, while PehX was shown to be extracellular in E. chrysanthemi CUCPB1237 and EC16 (8, 13). The localization of PehX was confirmed by using a specific detection of polygalacturonase activity in the presence of 10 mM EDTA, which completely inhibits the pectate lyases (data not shown).
Biochemical characterization of PelX.
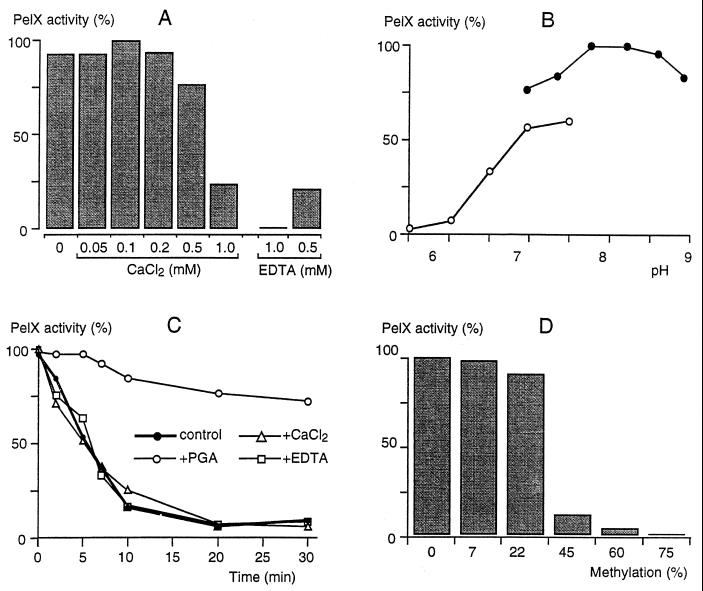
The addition of EDTA totally inhibits PelX activity, but the enzyme is weakly activated by the addition of Ca2+ (Fig. 7A). Moreover, in contrast to the major E. chrysanthemi pectate lyases, PelX activity is not inhibited but weakly activated by other bivalent cations, such as Mn2+, Co2+, Ni2+, and Cu2+ (data not shown). Increasing the Tris-HCl concentration activated PelX, with a maximal activity at 100 mM (data not shown). This effect is linked to the ionic strength of the medium, since NaCl also affected PelX activity, mainly at low Tris-HCl concentrations. With 20 mM Tris-HCl, the addition of 50 mM NaCl provoked a twofold activation of PelX. With 100 mM Tris-HCl, the addition of 50 mM NaCl did not affect PelX activity. The optimum pH for the reaction is about 8.5, but the enzyme is active within a large range of pH (Fig. 7B). PelX retains more than 50% of its activity at neutral pH. Even at pH 7, Tris-HCl increased PelX activity in comparison with other tested buffers (Fig. 7B and data not shown). The stability of PelX (Fig. 7C) was studied by its incubation at 45°C under different conditions. In contrast to the extracellular E. chrysanthemi pectate lyases, addition of Ca2+ or EDTA to the protein did not modify its stability, since a decrease of 50% in PelX activity was observed after 5 min, either in the presence of one of these compounds or in their absence. In the presence of polygalacturonate, a decrease of only 25% in activity was observed after 30 min, indicating that the interaction of PelX with its substrate increased its thermostability. Pectins with up to 22% methylation were as good as polygalacturonate as substrates (Fig. 7D). The ability of PelX to macerate plant tissues was analyzed by incubating serial dilutions of purified enzymes with potato cubes. PelX was totally inactive in tissue maceration, since the addition of 0.1 U · ml−1 caused no tissue softening even after 24 h (data not shown).
FIG. 7.
Enzymatic properties of PelX. (A) The influence of Ca2+ was tested by using various concentrations of CaCl2 in 50 mM Tris-HCl (pH 8.5)–0.5 g of polygalacturonate per liter. The cation requirement was confirmed by addition of 0.5 and 1 mM EDTA. (B) The effect of pH was tested with 0.5 g of polygalacturonate per liter as substrate, in the presence of 0.1 mM CaCl2 in 50 mM ACES-NaOH buffer (pH 5.5 to 7.5 [open symbols]) or in 50 mM Tris-HCl buffer (pH 7 to 9 [solid symbols]). (C) The thermostability of PelX was monitored at 45°C after various incubation times in 50 mM Tris-HCl (pH 8.5) without (control) or with the addition of CaCl2 (0.5 mM), EDTA (0.5 mM), or polygalacturonate (0.5 g · liter−1). The residual activity is given as a percentage of the initial activity. Activity toward pectins presenting various degrees of methylation (D) was determined in 50 mM Tris-HCl (pH 8.5)–0.1 mM CaCl2–0.5 g of substrate liter−1.
We analyzed PelX activity on oligogalacturonates of various lengths (G2 to G7), which are more likely to be present in the periplasmic space than is polygalacturonate (Table 4). PelX did not cleave digalacturonate. Trigalacturonate was a poor substrate of PelX, while higher oligomers (G4 to G7) were better substrates than polygalacturonate. Maximal activity was observed with tetragalacturonate. For longer chains, the cleavage rate slowly decreased when the degree of polymerization increased (Table 4). Determination of Km and Vmax values on some substrates indicated that PelX presents the highest affinity for G4. This suggests that the number of binding subsites for PelX is 4. The higher activity observed with G4 than with shorter oligogalacturonates suggested that the PelX subsites −2 and +2 have a high affinity for the substrate.
TABLE 4.
Effect of substrate length on PelX activity
| Substrate | Voa (U/mg) | Vmaxb (U/mg) | Kmb |
|---|---|---|---|
| PGA | 83 | 90 ± 10 | 0.25 ± 0.05 g · liter−1 |
| G2 | 0 | ||
| G3 | 31 | 40 ± 5 | 75 ± 10 μM |
| G4 | 253 | 520 ± 30 | 25 ± 5 μM |
| G5 | 148 | NDc | ND |
| G6 | 142 | ND | ND |
| G7 | 134 | ND | ND |
The initial enzymatic rate was measured in 100 mM Tris-HCl (pH 8.5)–0.1 mM CaCl2–0.5 g of polygalacturonate (PGA) per liter or 0.5 mM oligogalacturonates (Gn, n = 2 to 7).
For Km and Vmax determination, PelX was incubated with PGA concentrations ranging from 0.025 to 0.25 g · liter−1 and with Gn concentrations ranging from 10 to 100 μM.
ND, not determined.
Analysis of the PelX reaction products.
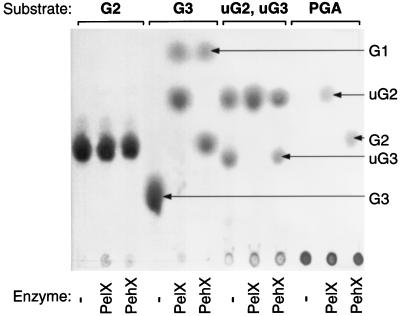
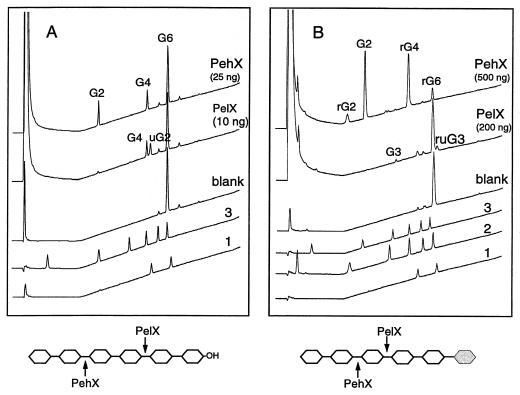
To precisely determine the mode of action of PelX and PehX, the reaction products obtained on various substrates were characterized by thin-layer chromatography and high-performance anion-exchange chromatography. As previously observed (6, 8), with polygalacturonate as the substrate, both enzymes catalyze the formation of only one product. PehX liberates digalacturonate, while PelX liberates unsaturated digalacturonate (Fig. 8). PelX and PehX have no activity on saturated or unsaturated dimers. Both enzymes cleaved saturated and unsaturated trimers, but PehX activity was lower on unsaturated trimers (Fig. 8), indicating that modification of the residue situated at the nonreducing end influences the action of PehX. It was previously shown that PehX releases dimers from the nonreducing end of polygalacturonate (8). To clarify from which end of the substrate PelX releases dimers, we analyzed the products obtained after degradation of hexamers (G6) and reduced hexamers (rG6) by PelX and PehX (Fig. 9). PehX hydrolyzed G6 into tetragalacturonate (G4) and digalacturonate (G2), while PelX cleaved G6 into G4 and unsaturated digalacturonate (uG2) (Fig. 9A). The action of PehX on rG6 demonstrated hydrolysis from the nonreducing end, since it led mainly to the appearance of G2 and reduced tetramers (rG4) (Fig. 9B). A small quantity of reduced dimers (rG2) also appeared, due to further hydrolysis of rG4 into G2 and rG2. PelX degraded rG6 with very low efficiency, since a large decrease in reaction rate was observed by comparison with the action on G6 (Fig. 9). Moreover, the weak cleavage of rG6 by PelX did not lead to dimer formation but resulted in the formation of reduced unsaturated trigalacturonate (ruG3) and trigalacturonate (G3). Thus, modification of the reducing end inhibited PelX activity and provoked a shift in the PelX cleavage site (Fig. 9). These results indicate that the exopolygalacturonate lyase PelX attacks the reducing end of the substrate while the exo-poly-α-d-galacturonosidase PehX attacks its nonreducing end (Fig. 10).
FIG. 8.
Separation of the PelX and PehX reaction products by thin-layer chromatography. The reaction mixtures contained 0.1 M Tris-HCl (pH 8), 0.1 mM CaCl2, 5 U of enzymatic activity per ml, and 1.5 mg of the following substrates per ml: digalacturonic acid (G2), trigalacturonic acid (G3), a mixture of unsaturated di- and trigalacturonic acids (uG2, uG3), or polygalacturonic acid (PGA). Incubations were performed at 30°C for 3 h after addition of PelX or PehX or without enzyme (−). A 5-μl sample from each reaction mixture was applied to chromatogram sheets. The positions of the individual compounds are indicated by arrows. It should be noted that the staining method used is not sensitive enough to detect the unsaturated monomer which appears by degradation of uG3 with PelX.
FIG. 9.
Analysis of PelX and PehX reaction products on hexagalacturonate (A) and reduced hexagalacturonate (B) by high-pressure liquid chromatography. For PelX, the reaction mixtures (0.5 ml) contained 0.5 mM substrate in 20 mM Tris-HCl (pH 8)–1 mM CaCl2. A 10- or 200-ng sample of PelX was used in a 30-min reaction for G6 (A) and rG6 (B), respectively. For PehX, the reaction mixtures (0.5 ml) contained 0.5 mM substrate in 20 mM Tris-HCl pH 7. 25 ng or 500 ng of PehX were used in a 5 min reaction for G6 (A) and rG6 (B), respectively. Reactions were carried out at 30°C. The blanks contained the substrate prior to enzyme addition. The reaction products were identified by using standard mixtures of uG2 and uG3 (curve 1), oligogalacturonates G1 to G6 (curve 3), and reduced oligogalacturonates rG1 to rG6 (curve 2).
FIG. 10.
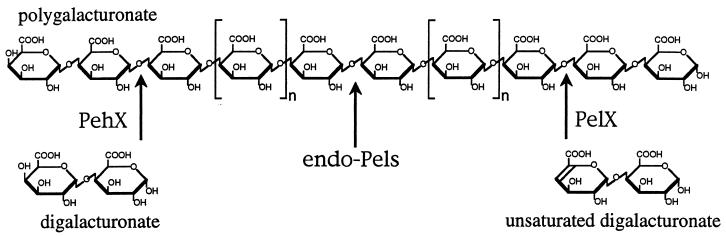
Sites of attack of the E. chrysanthemi exo-enzymes PelX and PehX. The arrows indicate the site of cleavage of the polymer, polygalacturonate, for each type of enzyme produced by E. chrysanthemi, PehX, PelX, and the endo-Pels (PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ). The sole product resulting from the action of the exo-polygalacturonase, PehX, or the exopolygalacturonate lyase, PelX, is indicated below the corresponding arrow.
DISCUSSION
This paper reports the characterization of the pelX gene of E. chrysanthemi 3937 and of its product, the exopolygalacturonate lyase, PelX, which cleaves polygalacturonate by β-elimination. E. chrysanthemi produces another type of exo-cleaving enzyme, the exo-poly-α-d-galacturonosidase, PehX, which hydrolyzes polygalacturonate (12). We showed that the two exo-cleaving pectinases, PelX and PehX, are located in the periplasm of E. chrysanthemi 3937. The exopolygalacturonate lyase activity was previously reported to be cell bound in E. chrysanthemi CUCPB1237 (7) and was supposed to be periplasmic in E. chrysanthemi EC16 (6). In contrast, PehX was found extracellularly in E. chrysanthemi CUCPB1237 and EC16 (8, 13). Analysis of various strains indicated that the polygalacturonase activity is detected in the culture supernatant of strains isolated from dicot but not from monocot hosts (35). These last strains could produce a periplasmic exo-poly-α-d-galacturonosidase, as shown for strain 3937.
PelX of strain 3937 was purified from recombinant E. coli cells. The activity of PelX on polygalacturonate and oligogalacturonates is inhibited by EDTA and requires low concentrations of Ca2+ or other bivalent cations, such as Mn2+, Co2+, Ni2+, or Cu2+. In contrast to the E. chrysanthemi endo-Pels (43), PelX is not highly dependent on pH, since good activity is observed within a wide range of pH, from 7 to 9.5. PelX can utilize polygalacturonate and partially methylated pectins as substrates. However, in contrast to EC16 PelX (6), 3937 PelX is not active on highly methylated pectins and its activity is only slightly stimulated in the presence of NaCl. The optimal conditions for 3937 PelX activity are also different from those observed for 3937 PelL, an endo-Pel having homology to the C-terminal amino acid sequence of PelX (23). This region contains a series of four to six identical residues that could be involved in the active site of these enzymes (Fig. 2). The E. chrysanthemi pectate lyases PelL and PelX belong to pectate lyase family 4 and have no homology to other described pectic enzymes. However, compared with PelL, PelX possesses an N-terminal extension of 324 residues, suggesting an organization of PelX in two domains. Exo-cleaving polysaccharidases, such as cellulases or xylanases, may present an N-terminal domain important for substrate recognition (11). To assess the role of the N-terminal extension of PelX, we constructed two derivatives containing important deletions in this region. The lack of pectinolytic activity of these deleted proteins indicated that the N-terminal region of PelX is necessary for its enzymatic activity.
Analysis of a pelX::uidA transcriptional fusion indicated that pelX expression increased in the presence of polygalacturonate, in the late exponential growth phase, and during oxygen limitation. In contrast, pelX expression was repressed in the presence of glucose and during nitrogen starvation. Therefore, like the other pel genes, the expression of pelX is controlled by several environmental stimuli. Inducibility of pectinases by pectic derivatives is mediated by the KdgR repressor, which binds to a specific DNA sequence present in the vicinity of the promoters of the controlled genes (28). The formation of pectin catabolic products, mainly KDG, provokes the dissociation of KdgR from its operators. The fact that expression of the pelX::uidA fusion is affected by kdgR or kdgK mutations indicates that pelX induction in the presence of polygalacturonate is dependent on the KdgR-KDG couple. Since pelX expression is very low in the crp mutant, CRP is necessary for activation of its expression. The presence of sequences highly homologous to the consensus determined for the KdgR-binding site, and for the CRP-binding site in the pelX regulatory region (Fig. 3), suggests that both KdgR and CRP probably control pelX transcription by direct binding. In contrast, pelX expression is not controlled by PecS or PecT, two other proteins involved in pectate lyase regulation in E. chrysanthemi (16).
While endo-enzymes catalyze the formation of multiple products of various sizes (30), the action of exo-cleaving enzymes on polymeric substrates generates a single product. The reaction products of PehX and PelX on polygalacturonate are in accordance with their exo-activity, since they generate only digalacturonate and unsaturated digalacturonate, respectively. To determine whether PehX and PelX attack from the nonreducing or the reducing end of the substrate, we used HPAEC to analyze the products obtained after degradation of hexamers, modified or not modified on the reducing end. The exopolygalacturonate lyase, PelX, degraded the hexagalacturonate to an unsaturated dimer and tetramer. Reduced hexagalacturonate was a very poor substrate of PelX, and its degradation resulted in a shift of the bond cleaved, with formation of a trimer and an unsaturated reduced trimer. The fact that modification of the reducing extremity interfered with PelX activity demonstrated that PelX attacks the oligomer from the reducing end. The exo-poly-α-d-galacturonosidase, PehX, degraded the hexagalacturonate into a dimer and a tetramer. The rate of hydrolysis of the reduced hexagalacturonate was similar (data not shown) and led to dimer and reduced-tetramer formation. As expected (8), the action of PehX is typical of an enzyme that attacks from the nonreducing end. It is remarkable that E. chrysanthemi, in addition to a set of at least eight endo-Pels (39), produces two exo-enzymes able to act on both ends of the substrate (Fig. 10). These two types of activity are complementary and may be necessary when one extremity of the polymer is blocked by modification of the terminal residue (by glycosylation with a neutral sugar or by esterification).
PelX plays some role in pectin catabolism since, during growth on polygalacturonate, the pelX mutant showed a longer lag phase and doubling time than did the wild-type strain. The virulence of the pelX mutant appeared not to be affected on Saintpaulia plants or potato tubers, but the maceration observed on chicory leaves slightly decreased, indicating that PelX action may be necessary for the total effect of E. chrysanthemi on some host plants. The periplasmic enzyme PelX is probably involved in the degradation of pectic oligomers that could enter the periplasm. Analysis of the PelX activity on oligomers of various length (2 to 7 residues) demonstrated that tetragalacturonate is its best substrate. While longer oligomers remain good substrates, trigalacturonate is a poor substrate of PelX. The data obtained with oligomers suggest that PelX recognizes only 4 residues at the reducing end of the polysaccharide. In E. coli, oligosaccharides as large as maltoheptaose can enter the periplasm by specific porines (45). The high PelX activity on tetra- to heptagalacturonates suggests that these pectic oligomers could enter the E. chrysanthemi periplasm. In the periplasm, the action of PelX enables further degradation of oligomers up to dimers and trimers that can probably enter the cytoplasm. Oligogalacturonates longer than trimers are poor substrates for the first enzyme of the cytoplasmic catabolic pathway, oligogalacturonate lyase (27). The role of the periplasmic exo-enzymes PelX and PehX is thus probably to be intermediate elements in pectin degradation, acting between the extracellular endo-Pels and the cytoplasmic oligogalacturonate lyase.
ACKNOWLEDGMENTS
Appreciation is expressed to Valerie James for reading the manuscript. We thank our colleagues, Sylvie Reverchon, Guy Condemine, and William Nasser, for valuable discussions. We thank Copenhagen Pectin for the gift of well-characterized pectins.
This work was supported by grants from the Centre National de la Recherche Scientifique (UMR 5577), from the Ministère de l’Education Nationale de la Recherche et de la Technologie and from the Commission of the European Communities (AIR 2-CT-941345).
REFERENCES
- 1.Alfano J R, Ham J H, Collmer A. Use of Tn5tac1 to clone a pel gene encoding a highly alkaline, asparagine-rich pectate lyase isozyme from an Erwinia chrysanthemi EC16 mutant with deletions affecting the major pectate lyase isozymes. J Bacteriol. 1995;177:4553–4556. doi: 10.1128/jb.177.15.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardonnet N, Blanco C. uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusion and genetic constructions. FEMS Microbiol Lett. 1992;93:243–248. doi: 10.1016/0378-1097(92)90469-5. [DOI] [PubMed] [Google Scholar]
- 3.Barras F, Van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 4.Beaulieu C, Boccara M, Van Gijsegem F. Pathogenic behaviour of pectinase-defective Erwinia chrysanthemi mutants on different plants. Mol Plant-Microbe Interact. 1993;6:197–202. [Google Scholar]
- 4a.Beer, S. Personal communication.
- 5.Benen J, Kester H, Parenicova L, Visser J. Kinetics and mode of action of Aspergillus niger polygalacturonases. Prog Biotechnol. 1996;14:221–230. [Google Scholar]
- 6.Brooks A D, He S Y, Gold S, Keen N T, Collmer A, Hutcheson S W. Molecular cloning of the structural gene for exopolygalacturonate lyase from Erwinia chrysanthemi EC16 and characterization of the enzyme product. J Bacteriol. 1990;172:6950–6958. doi: 10.1128/jb.172.12.6950-6958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collmer A, Bateman D F. Impaired induction and self-catabolite repression of extracellular pectate lyase in Erwinia chrysanthemi mutants deficient in oligogalacturonide lyase. Proc Natl Acad Sci USA. 1981;78:3920–3924. doi: 10.1073/pnas.78.6.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collmer A, Whalen C H, Beer S V, Bateman D F. An exo-poly-alpha-d-galacturonosidase implicated in the regulation of extracellular pectate lyase production in Erwinia chrysanthemi. J Bacteriol. 1982;149:626–634. doi: 10.1128/jb.149.2.626-634.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland B R, Richter R J, Furlong C E. Renaturation and identification of periplasmic proteins in two-dimensional gels of Escherichia coli. J Biol Chem. 1982;257:15065–15071. [PubMed] [Google Scholar]
- 10.Expert D, Toussaint A. Bacteriocin-resistant mutants of Erwinia chrysanthemi: possible involvement of iron acquisition in phytopathogenicity. J Bacteriol. 1985;163:221–227. doi: 10.1128/jb.163.1.221-227.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilkes N, Henrissat B, Kilburn D, Miller J R, Warren R. Domains in microbial β-1-4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He S Y, Collmer A. Molecular cloning, nucleotide sequence, and marker exchange mutagenesis of the exo-poly-alpha-d-galacturonosidase-encoding pehX gene of Erwinia chrysanthemi EC16. J Bacteriol. 1990;172:4988–4995. doi: 10.1128/jb.172.9.4988-4995.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffron S, Henrissat B, Yoder M D, Lietzke S, Jurnak F. Structure-based multiple alignment of extracellular pectate lyase sequences. Mol Plant-Microbe Interact. 1995;8:331–334. doi: 10.1094/mpmi-8-0331. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B, Heffron S E, Yoder M D, Lietzke S E, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 17.Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect the transcription of the pectinase genes of Erwinia chrysanthemi 3937. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugouvieux-Cotte-Pattat N, Reverchon S, Robert-Baudouy J. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol Microbiol. 1989;3:573–581. doi: 10.1111/j.1365-2958.1989.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 19.Johnson B H, Hecht M H. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Bio/Technology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- 20.Keen N T, Dahlbeck D, Staskawicz B, Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984;159:825–831. doi: 10.1128/jb.159.3.825-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kester H, Visser J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem. 1990;12:150–160. [PubMed] [Google Scholar]
- 22.Lojkowska E, Dorel C, Reignault P, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Use of GUS fusions to study the expression of Erwinia chrysanthemi pectinase genes during infection of potato tubers. Mol Plant-Microbe Interact. 1993;6:488–494. [Google Scholar]
- 23.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 24.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 26.Moran F, Nasuno S, Starr M P. Extracellular and intracellular polygalacturonic acid trans eliminase of Erwinia carotovora. Arch Biochem Biophys. 1968;123:298–306. doi: 10.1016/0003-9861(68)90138-0. [DOI] [PubMed] [Google Scholar]
- 27.Moran F, Nasuno S, Starr M P. Oligogalacturonide trans-eliminase of Erwinia carotovora. Arch Biochem Biophys. 1968;125:734–741. doi: 10.1016/0003-9861(68)90508-0. [DOI] [PubMed] [Google Scholar]
- 28.Nasser W, Reverchon S, Condemine G, Robert-Baudouy J. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J Mol Biol. 1994;236:427–440. doi: 10.1006/jmbi.1994.1155. [DOI] [PubMed] [Google Scholar]
- 29.Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. REgulation of pelZ, a gene of the pelBC cluster encoding a new pectate lyase in Erwinia chrysanthemi 3937. J Bacteriol. 1996;178:7187–7196. doi: 10.1128/jb.178.24.7187-7196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston J, Rice J, Ingram L, Keen N T. Differential depolymerization mechanisms of pectate lyase secreted by Erwinia chrysanthemi EC16. J Bacteriol. 1992;174:2039–2042. doi: 10.1128/jb.174.6.2039-2042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resibois A, Colet M, Faelen M, Schoonejans E, Toussaint A. PhiEC2, a new generalised transducing phage of Erwinia chrysanthemi. Virology. 1984;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 32.Reverchon S, Expert D, Robert-Baudouy J, Nasser W. The cyclic AMP receptor protein is the main activator of the pectinolysis genes in Erwinia chrysanthemi. J Bacteriol. 1997;179:3500–3508. doi: 10.1128/jb.179.11.3500-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reverchon S, Nasser W, Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 34.Reverchon S, Nasser W, Robert-Baudouy J. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol. 1994;11:1127–1139. doi: 10.1111/j.1365-2958.1994.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 35.Ried J L, Collmer A. Comparison of pectic enzymes produced by Erwinia chrysanthemi, Erwinia carotovora subsp. carotovora, and Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 1986;52:305–310. doi: 10.1128/aem.52.2.305-310.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeder D L, Collmer A. Marker-exchange mutagenesis of pectate lyase isozyme gene in Erwinia chrysanthemi. J Bacteriol. 1985;164:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Saraste M, Gay N, Runswik M, Walker J. The atp operon: nucleotide sequence of the genes for the gamma, beta, and epsilon subunits of Escherichia coli ATP synthase. Nucleic Acids Res. 1981;9:5287–5296. doi: 10.1093/nar/9.20.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevchik V E, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. The pectate lyase PelI of Erwinia chrysanthemi belongs to a new family. J Bacteriol. 1997;179:7321–7330. doi: 10.1128/jb.179.23.7321-7330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier W F, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Surgey N, Robert-Baudouy J, Condemine G. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J Bacteriol. 1996;178:1593–1599. doi: 10.1128/jb.178.6.1593-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabor S, Richardson C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Gijsegem F, Toussaint A, Schoonejans E. In vivo cloning of pectate lyase and cellulase genes of Erwinia chrysanthemi. EMBO J. 1985;4:787–795. doi: 10.1002/j.1460-2075.1985.tb03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wandersman C, Schwartz M, Ferenci T. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 1979;140:1–13. doi: 10.1128/jb.140.1.1-13.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]