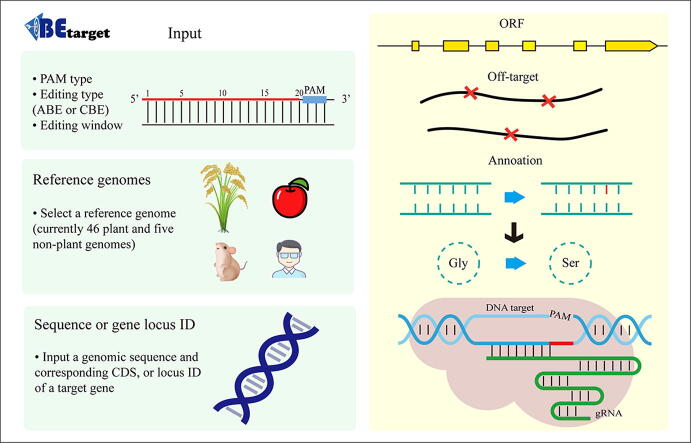
Graphical abstract
Abbreviations: ABE, adenine base editor; CBE, cytosine base editor; CDS, coding sequence; CRISPR, clustered regularly interspaced short palindromic repeats; DSB, double-strand break; ORF, open reading frame; NHEJ, non-homologous end joining; PAM, protospacer adjacent motif; gRNA, guide RNA
Keywords: CRISPR, Base editing, gRNA design, Genome editing
Highlights
-
•
BEtarget supports the gRNA design of base editing with different types of PAM.
-
•
BEtarget provides an interactive and customized visualization interface.
-
•
BEtarget can automatically detect the coordinates of coding regions (exons) in the genomic sequence of the target gene.
Abstract
CRISPR-dependent base editors enable direct nucleotide conversion without the introduction of double-strand DNA break or donor DNA template, thus expanding the CRISPR toolbox for genetic manipulation. However, designing guide RNAs (gRNAs) for base editors to enable gene correction or inactivation is more complicated than using the CRISPR system for gene disruption. Here, we present a user-friendly web tool named BEtarget dedicated to the design of gRNA for base editing. It is currently supported by 46 plant reference genomes and 5 genomes of non-plant model organisms. BEtarget supports the design of gRNAs with different types of protospacer adjacent motifs (PAM) and integrates various functions, including automatic identification of open reading frame, prediction of potential off-target sites, annotation of codon change, and assessment of gRNA quality. Moreover, the program provides an interactive interface for users to selectively display information about the desired target sites. In brief, we have developed a flexible and versatile web-based tool to simplify complications associated with the design of base editing technology. BEtarget is freely accessible at https://skl.scau.edu.cn/betarget/.
1. Introduction
Clustered regularly interspaced short palindromic repeats associated protein (CRISPR/Cas) systems are adaptive defense systems that protect bacteria and archaea from invading viruses or plasmids [1], [2], [3]. They act through at least three general steps, which are as follows. In the adaptive stage, organisms respond to viral or plasmid challenges by integrating short fragments of foreign sequence into the CRISPR locus. In the expression and interference stages, the CRISPR array is transcribed and processed into short crRNAs, which guide Cas proteins to cleave the invading foreign sequence [4]. Currently, CRISPR/Cas systems are engineered into versatile tools for genome editing [5], [6], [7]. Cas endonuclease targets a specific sequence through base pairing with the help of a guide RNA (gRNA) and creates a double-strand break (DSB) at the cleavage site [8]. In most eukaryotes, DSB is predominantly repaired by the error-prone non-homologous end joining (NHEJ) pathway, generally resulting in random nucleotide insertion or deletion mutations [9], [10]. Although the NHEJ-dependent gene disruption is efficient, it is frequently hard to achieve the expected accuracy in gene correction [11], [12], [13]. In the presence of a donor template, the homology-directed repair (HDR) pathway at the DSB site enables a symmetric sequence correction or insertion at the target site [5]. However, the low efficiency of HDR in plant cells and the lack of an efficient donor DNA delivery method limit its application in plants [14], [15].
Wild-type Cas9 contains two conversed nuclease domains (RuvC and HNH-like domains) in which each cleaves one strand of the double-helix DNA. By mutating one of the two critical residues (D10A or H840A) in the nuclease domains, nickase Cas9 (nCas9) can be generated which cuts only one strand of the target sites [16], [17]; if two nuclease domains are mutated, the catalytically dead Cas9 (dCas9) shows inactive nuclease activity while retaining its ability to bind DNA [18]. Engineering of Cas9 variants with different deaminases has generated diverse base editing tools, including cytosine base editor (CBE) and adenine base editor (ABE) [19], [20], [21]. The most commonly used base editors are nCas9 or dCas9, which are fused with cytidine deaminase (CBE) or adenosine deaminase (ABE) to simulate cellular mismatch repair and subsequent suitable base substitution [22], [23]. Guided by sgRNA, fused protein complexes are enabled to produce site-specific C-to-T or A-to-G substitutions without producing DSBs or introducing donor DNA templates [22], [23].
Owing to the unique capability of fine-tuned mutagenesis, base editors are widely applied to both model and non-model organisms for genetic manipulation [24], [25], [26], [27]. For example, base substitutions in the codons of an open reading frame (ORF) potentially enable amino acid substitutions (missense mutation) or cause gene inactivation by introducing premature STOP codons (nonsense mutation) [11], [12], [28], [29], [30], [31]. Moreover, multiple base substitutions can occur in the editing window, thus ensuring the role of base editors in large-scale saturation mutation, regulatory element editing, therapeutic gene correction, and crop improvement [32], [33], [34], [35], [36]. However, the design of gRNAs for base editors is complicated, and several specific criteria are carefully considered [24], including the preferred editing window, bystander effect, potential codon change, and off-target effect. Thus a convenient tool for the rapid design of gRNAs for base editors is required and currently, there are a few programs available [37].
Based on the extensive analysis of genome-wide base editing outcomes in mammalian cells, several machine learning models with corresponding design programs, including BE-Hive, BE-DICT, DeepBaseEditor, and FORECasT-BE, were developed to predict the editing efficiency and the bystander effect [38], [39], [40], [41]. In other organisms, a few gRNA design tools were developed to assist researchers in the rapid choosing of appropriate target sites [42], [43], [44], [45], [46]. Due to the difficulties faced while performing a large-scale genetic transformation method, especially in plants, these tools majorly focus on the function of searching all possible target sites in the target gene and the annotation of the codon changes in the editing window. For example, CRISPR-BETS and CRISPR-CBEI focus on designing gRNAs for CBE-mediated nonsense mutation [42], [43], CRISPyweb 2.0 is a tool developed only for Streptomyces coelicolor [44], beditor is a specialized tool used to design genome-wide gRNA libraries in cell lines [45], and BE-Designer provides a comprehensive analysis of all possible candidate target sites with useful information, including potential off-target sites for base editors in various organisms [46].
Considering the wide and prospective applications of base editors in plants, we have developed a dedicated web-based tool named BEtarget to aid researchers in the design of gRNAs for genes of interest by visiting directly at http://skl.scau.edu.cn/betarget/, or from our previous CRISPR-GE website [47] (http://skl.scau.edu.cn/). The program provides a user-friendly submission interface by supporting fully customizable settings of input parameters and target genes. BEtarget lists all possible target sites in the ORF of a given gene as an interactive table and a graph along with comprehensive information, including amino acid change, potential off-target sites, and quality assessment.
2. Workflow and implementation
2.1. Workflow of BEtarget
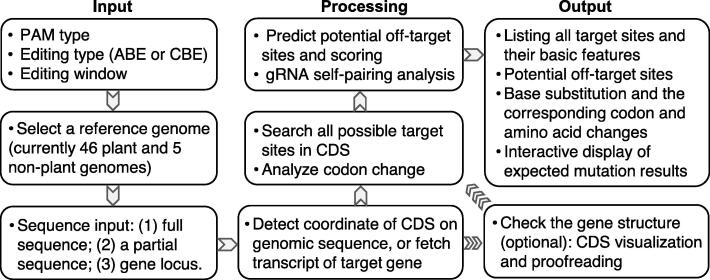
BEtarget is a web-based application in a “browser-server” mode. Its front web pages are implemented with HTML5, JavaScript, and Bootstrap, and the backend is constructed using the Django framework. Programs for processing the uploaded data and analyzing sequences are written with Python 3. The workflow of BEtarget is shown in Fig. 1. Overall, it is designed with three major steps: (i) the input page accepts user-defined parameters and a target sequence or a gene locus identity (ID) number; (ii) the backend processes the uploaded data, including automatic detection of the ORF, identification of all possible target sites in the ORF, and prediction of potential off-target sites; (iii) the output page retrieves the resulting data in the JSON format and displays results in an interactive table with useful information.
Fig. 1.
Overall workflow chart of BEtarget. The program searches all possible target sites in a target gene based on the user’s defined parameters and performs a comprehensive analysis of the target sites, including their basic features, potential off-target sites, and changes in codons and amino acids within the editing widows.
2.2. Sequence input and setting of parameters
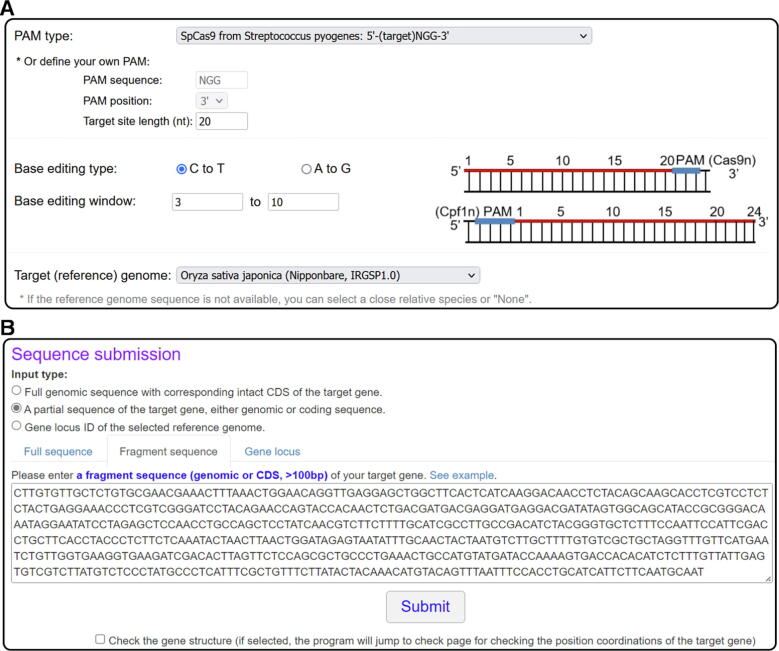
The submission interface consists of two parts, namely a panel for parameter settings, including the setting of a protospacer adjacent motif (PAM), editing type (CBE or ABE), editing window and a reference genome selection, and another panel for target sequence (or gene locus) input (Fig. 2). BEtarget presently supports the searching of targets sites with commonly used PAM (including NGG, NG, TTN, and TTTN) or any specific PAM defined by users. Based on the type and efficiency of the base editor used (either CBE or ABE), users can manually define the editing window accordingly. Currently, 46 plant reference genomes and 5 genomes of non-plant model organisms are provided as references to evaluate potential off-target sites. Users can also select “None” if no target reference genome is available for which the program will not predict potential off-target sites. Alternatively, users can request the developer to add more desired reference genome(s). In the sequence input panel, three types are supported as input: (i) the genomic sequence with the corresponding intact coding sequence (CDS) of a target gene; (ii) a partial sequence of the target gene; (iii) a gene locus ID of the selected reference genome. When a partial sequence of the target gene as input which can either be a genomic sequence or CDS, BEtarget will automatically extract the corresponding genomic sequence and CDS of the gene by invoking a subprogram called GeneCat for gRNA design. The GeneCat tool can also be accessed independently from the website (http://skl.scau.edu.cn/) to extract the genomic sequence and corresponding CDS of a gene (Supplementary Fig. 1). Below the “Submit” button, users can select to check for gene structure (default is not selected).
Fig. 2.
Submission page of BEtarget. (A) Settings panel for PAM type, editing type, editing window, and reference genome. (B) Input panel for target sequence (or gene). Three types are supported as input: (i) the genomic sequence with the corresponding CDS of a target gene; (ii) a partial sequence of target gene; (iii) a gene locus ID of the selected reference genome.
2.3. Data processing
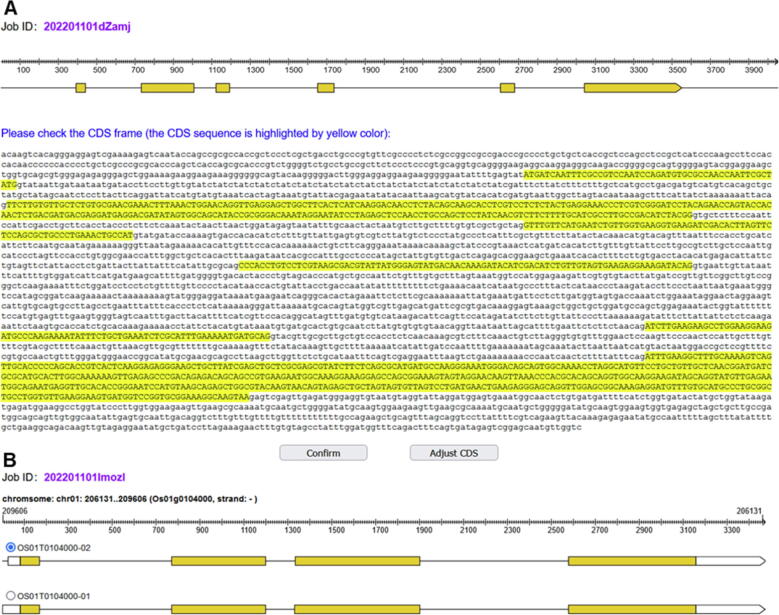
After submitting a task, BEtarget first preprocesses the uploaded target sequence or gene locus. It is important to exactly define ORF in the given genomic sequence to identify the locations of target sites. After uploading the genomic sequence and intact CDS, the program aligns CDS to the genomic sequence and automatically detects the boundaries of each exon. If gene locus ID is used as input, the program fetches the genomic coordinates of all transcripts of the input gene from the reference genome database. If users choose to check gene structure at the submission page, detailed coordinate information and sequence are returned to the front check page for the users to confirm the exon/intron structure or select a preferred transcript from possible multiple alternative transcripts (Fig. 3). Minor modifications in the exon positions are allowed when necessary, in case the position offset happens in the program. Otherwise, in general, the program uses genomic coordinates judged by the program. Once the coordinates of ORF or the transcript are confirmed, all possible target sites are extracted, and only those located in or overlapping with the coding regions are kept for further analysis. Potential off-target sites and their scores are then predicted by invoking the offTarget program in the CRISPR-GE toolkit [41]. Possible secondary structures of the targets that pair with the gRNA scaffold sequence are also analyzed. Finally, possible base substitutions within the editing windows are subjected to the annotation of codon change and corresponding amino acid change. The resulting data, including the target sites and their correlated information, are returned to the results page in the JSON format.
Fig. 3.
Proofreading of CDS and transcript selection of the target gene. (A) When the genomic sequence with corresponding CDS is used as input, the program automatically detects the coordinates of CDS on the genomic sequence. If users select to check the gene structure at the submission page, BEtarget jumps to the check page, which displays the exon/intron structure of the target gene sequence in which the exons are highlighted in yellow. Coordinates can be modified by clicking the “Adjust CDS” button. (B) When gene locus is used as input, BEtarget displays all possible alternative transcripts of the target gene, and users can select the preferred transcript for target design. The yellow boxes indicate ORFs of the transcript. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Output and result visualization
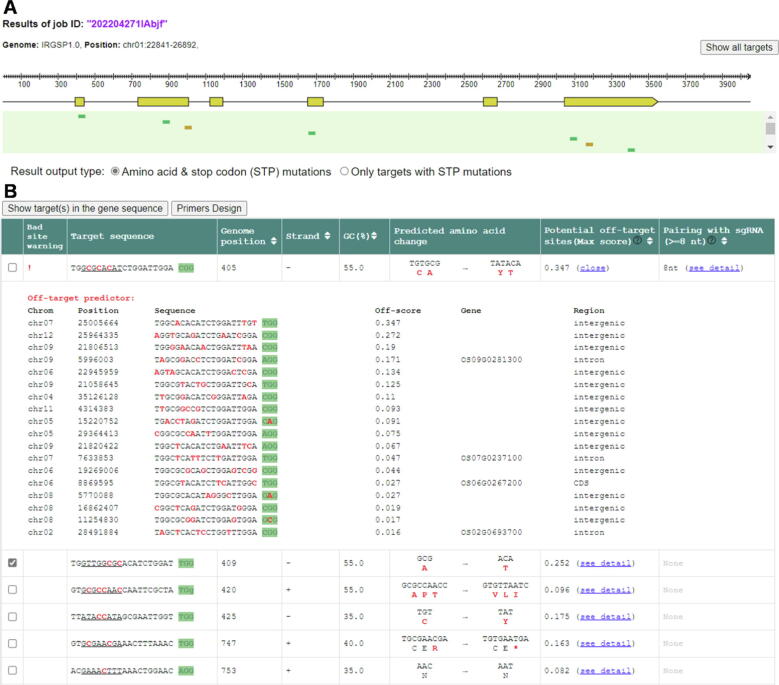
BEtarget produces an interactive graph and table for displaying the results (Fig. 4). The graph shows the structure of a target gene in which the exons are indicated as yellow-colored boxes (Fig. 4A). When the mouse pointer is moved on an exon, the table lists only candidate target sites in the exon, and when clicked on “show all target”, it displays all candidate target sites in the gene. The initial results table lists all candidate target sites and their positions, strands, GC content, potential off-target sites, and corresponding codon changes (Fig. 4B). Possible base substitutions and corresponding amino acid changes, including stop codon mutations within the editing windows are highlighted in red. Users can select to display target sites with the expected mutation type, either as all candidate sites or only those with stop codon mutations (nonsense mutations). By clicking the “see detail” link, users can examine detailed potential off-target sites with their scores.
Fig. 4.
Visualization of the results. (A) The graph shows the structure of a target gene. When the mouse pointer is moved on an exon, the results table lists the target sites present in the exon. When the “Show all target” is clicked on, it displays all candidate target sites in the gene. Users can selectively display target sites with the expected mutation type, either as all candidate sites or only those with stop codon mutations. (B) The table lists candidate target sites and their useful information, including basic features, predicted amino acid changes, potential off-target sites, and scores. Users can select target site(s) (on the left-most side) and click “Show target(s) in the gene sequence” or “Primer design” for these purposes.
Although there is a lack of large-scale analysis of base editing outcomes for predicting the efficiency because of the low-efficiency of delivery method in plants, it is possible to anticipate low-efficiency target sites, including those with very low or high GC content (≤25 % or ≥ 80 %), poly-T site(s), contiguous base-pairing with the sgRNA sequence, or potential off-target sites of high score value (≥0.7). These candidate low-quality target sites are marked with warning indicators (!, !!, or !!!). Finally, users can select target site(s) (on the left-most side) and click on “Show target(s) in the gene sequence” or “Primer design” (below the candidate targets) for the purposes indicated.
4. Conclusion
BEtarget is a user-friendly tool to select optimal gRNAs in a given gene for base editing. The program extracts comprehensive information of all candidate target sites in the coding regions, including their basic features, predicted codon changes, potential off-target sites, and annotations. The results are displayed using an interactive graph and table. In this study, BEtarget was carefully compared with other similar design tools that foucus on the function of searching target sites for base editing, including CRISPR-BETS [42], CRISPR-CBEI [43], PnB Designer [48], and BE-Designer [46], as summarized in Supplementary Table 1. The comparison showed that BEtarget has the following advantages. Firstly, BEtarget supports gRNA design for current base editors, including ABE and CBE, with no limitation of PAM variants. By modifying the parameters of the PAM setting, users can define their PAM type. Secondly, BEtarget is flexible for sequence input, which allows users input a partial sequence of target gene, and the program can automatically fetch the genomic sequence and corresponding CDS. Thirdly, BEtarget provides an interactive and customized visualization interface to display information about target sites. In summary, BEtarget is an innovative tool for researchers to rapidly choose appropriate target sites for base editing.
Author contributions
YL and XX drafted the project. XX, FL, and XT wrote the programs for BEtarget. YL and QZ provided critical comments on the design of BEtarget. WL and DZ tested the programs. XX and FL wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31991223) and the Major Program of Guangdong Basic and Applied Basic Research (2019B030302006).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.07.046.
Contributor Information
Xianrong Xie, Email: xiexianrong@scau.edu.cn.
Yao-Guang Liu, Email: ygliu@scau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Deveau H., Garneau J.E., Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 5.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., et al. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172(6):1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Cong L., Ran F.A., Cox D., Lin S., Barretto R., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 12.Kuscu C., Parlak M., Tufan T., Yang J., Szlachta K., et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14(7):710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 13.Voytas D.F., Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12(6):e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M., Rehman S., Tang X., Gu K., Fan Q., et al. Methodologies for improving HDR efficiency. Front Genet. 2019;9:691. doi: 10.3389/fgene.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao Y., Botella J.R., Liu Y., Zhu J.K. Gene editing in plants: progress and challenges. Natl Sci Rev. 2019;6(3):421–437. doi: 10.1093/nsr/nwz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Zeng D., Zheng Z., Lin Z., Xue Y., et al. The ScCas9(++) variant expands the CRISPR toolbox for genome editing in plants. J Integr Plant Biol. 2021;63(9):1611–1619. doi: 10.1111/jipb.13164. [DOI] [PubMed] [Google Scholar]
- 20.Tan J., Zeng D., Zhao Y., Wang Y., Liu T., et al. PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol J. 2022 doi: 10.1111/pbi.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19(12):770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molla K.A., Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37(10):1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol Cell. 2017;68(1):26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng D., Liu T., Tan J., Zhang Y., Zheng Z., et al. PhieCBEs: plant high-efficiency cytidine base editors with expanded target range. Mol Plant. 2020;13(12):1666–1669. doi: 10.1016/j.molp.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R., Liu J., Chai Z., Chen S., Bai Y., et al. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants. 2019;5(5):480–485. doi: 10.1038/s41477-019-0405-0. [DOI] [PubMed] [Google Scholar]
- 28.Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell. 2017;67(6):1068–1079. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua K., Tao X., Zhu J.K. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J. 2019;17(2):499–504. doi: 10.1111/pbi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., Li Y., Yang S., Huang S., Yan M., et al. CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat Cell Biol. 2018;20(11):1315–1325. doi: 10.1038/s41556-018-0202-4. [DOI] [PubMed] [Google Scholar]
- 32.Yan D., Ren B., Liu L., Yan F., Li S., et al. High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol Plant. 2021;14(5):722–731. doi: 10.1016/j.molp.2021.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Y., Li J., Li G., Huang S., Yu W., et al. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol Ther. 2018;26(11):2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24(10):1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 35.Li C., Zong Y., Wang Y., Jin S., Zhang D., et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19(1):59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharat S.S., Li S., Li J., Yan L., Xia L. Base editing in plants: Current status and challenges. The Crop J. 2020;8(3):384–395. [Google Scholar]
- 37.McDaniel S., Komor A., Goren A. The use of base editing technology to characterize single nucleotide variants. Comput Struct Biotechnol J. 2022;20:1670–1680. doi: 10.1016/j.csbj.2022.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arbab M., Shen M.W., Mok B., Wilson C., Matuszek Ż., et al. Determinants of base editing outcomes from target library analysis and machine learning. Cell. 2020;182(2):463–480. doi: 10.1016/j.cell.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquart K.F., Allam A., Janjuha S., Sintsova A., Villiger L., et al. Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nat Commun. 2021;12(1):5114. doi: 10.1038/s41467-021-25375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song M., Kim H.K., Lee S., Kim Y., Seo S.Y., et al. Sequence-specific prediction of the efficiencies of adenine and cytosine base editors. Nat Biotechnol. 2020;38(9):1037–1043. doi: 10.1038/s41587-020-0573-5. [DOI] [PubMed] [Google Scholar]
- 41.Pallaseni A., Peets E.M., Koeppel J., Weller J., Vanderstichele T., et al. Predicting base editing outcomes using position-specific sequence determinants. Nucleic Acids Res. 2022;50(6):3551–3564. doi: 10.1093/nar/gkac161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y., He Y., Sretenovic S., Liu S., Cheng Y., et al. CRISPR-BETS: a base-editing design tool for generating stop codons. Plant Biotechnol J. 2021;20(3):499–510. doi: 10.1111/pbi.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H., Wu Z., Chen X., Ji Q., Tao S. CRISPR-CBEI: a designing and analyzing tool kit for cytosine base editor-mediated gene inactivation. mSystems. 2020;5(5):e00350–e420. doi: 10.1128/mSystems.00350-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blin K., Shaw S., Tong Y., Weber T. Designing sgRNAs for CRISPR-BEST base editing applications with CRISPy-web 2.0. Synth Syst. Biotechnol. 2020;5(2):99–102. doi: 10.1016/j.synbio.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dandage R., Despres P.C., Yachie N., Landry C.R. beditor: a computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing. Genetics. 2019;212(2):377–385. doi: 10.1534/genetics.119.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang G.H., Park J., Lim K., Kim S., Yu J., et al. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018;19(1):542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X., Ma X., Zhu Q., Zeng D., Li G., et al. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant. 2017;10(9):1246–1249. doi: 10.1016/j.molp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Siegner S.M., Karasu M.E., Schroder M.S., Kontarakis Z., Corn J.E. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinf. 2021;22(1):101. doi: 10.1186/s12859-021-04034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.