Abstract
Pantothenic acid deficiency (PAD) in animals causes growth depression, fasting hypoglycemia and impaired lipid and glucose metabolism. However, a systematic multi-omics analysis of effects of PAD on hepatic function has apparently not been reported. We investigated liver proteome and metabolome changes induced by PAD to explain its effects on growth and liver metabolic disorders. Pekin ducks (1-d-old, n = 128) were allocated into 2 groups, with 8 replicates and 8 birds per replicate. For 16 d, all ducks had ad libitum access to either a PAD or a pantothenic acid adequate (control, CON) diet, formulated by supplementing a basal diet with 0 or 8 mg pantothenic acid/kg of diet, respectively. Liver enlargement, elevated liver glycogen concentrations and decreased liver concentrations of triglyceride and unsaturated fatty acids were present in the PAD group compared to the CON group. Based on integrated liver proteomics and metabolomics, PAD mainly affected glycogen synthesis and degradation, glycolysis and gluconeogenesis, tricarboxylic acid (TCA) cycle, peroxisome proliferator-activated receptor (PPAR) signaling pathway, fatty acid beta oxidation, and oxidative phosphorylation. Selected proteins were confirmed by Western blotting. Downregulation of proteins and metabolites involved in glycogen synthesis and degradation, glycolysis and gluconeogenesis implied that these processes were impaired in PAD ducks, which could have contributed to fasting hypoglycemia, liver glycogen storage, insufficient ATP production, and growth retardation. In contrast, PAD also upregulated proteins and metabolites involved in fatty acid beta oxidation, the TCA cycle, and oxidative phosphorylation processes in the liver; presumably compensatory responses to produce ATP. We inferred that PAD decreased liver triglyceride and unsaturated fatty acids by activating fatty acid beta oxidation and impairing unsaturated fatty acid synthesis. These findings contributed to our understanding of the mechanisms of PAD-induced changes in hepatic metabolism.
Keywords: Pantothenic acid, Hepatic metabolism, Proteomics, Metabolomics, Hypoglycemia
1. Introduction
Pantothenic acid, an essential water-soluble vitamin, is a component of coenzyme A (CoA) and acyl carrier protein. The coenzyme form of this vitamin is involved in various metabolic reactions including carbohydrates, lipids, and proteins (Miller and Rucker, 2012; Smith and Song, 1996). Due to its involvement in primary metabolic pathways, pantothenic acid deficiency (PAD) causes growth depression, skin lesions, and diarrhea in mammals such as rats, cats, and pigs (Gershoff and Gottlieb, 1964; Nelson, 1968; Smith and Song, 1996; Youssef et al., 1997; Zucker, 1958). Furthermore, in chicks, turkeys, and geese, PAD causes growth retardation, dermatosis, rough feathers, and high mortality (Bauerrnfeind et al., 1942; Hegsted and Riggs, 1949; Jukes, 1939; Kratzer and Williams, 1948; Lepkovsky et al., 1945; Wang et al., 2016). Similarly, in Pekin ducks, PAD causes growth depression, excessive eye secretions, poor feathers, and high mortality (Hegsted and Perry, 1948; Tang et al., 2020a, 2020b).
It is clear that PAD alters carbohydrate metabolism, with low fasting blood glucose concentrations and increased sensitivity to insulin in rats and dogs (Arnrich et al., 1956a, 1956b; Hurley and Morgan, 1952; Schultz et al., 1952; Winters et al., 1952), as well as fasting hypoglycemia in ducks (Tang et al., 2021). It has been speculated that pantothenic acid is part of a glucose carrier system (Huan and Hung, 1972). Furthermore, pantothenic acid is particularly important in fatty acid metabolism. In fatty acid synthesis and degradation, pantothenic acid coenzymes carry the acids (as acyl groups) through repetitive synthetic or degradative cycles. Fatty acids must also be “activated” by CoA before they can be synthesized into triglycerides. A series of previous studies demonstrated that dietary PAD disrupted lipid metabolism (Lin et al., 2012; Qian et al., 2015; Shiau and Hsu, 1999; Shibata et al., 2013; Wang et al., 2016; Wen et al., 2009; Wittwer et al., 1990), caused fat accumulation (Shibata et al., 2013), and elevated serum concentrations of triglycerides and free fatty acids in rats (Wittwer et al., 1990). Similarly, PAD elevated liver lipid content of fish (Lin et al., 2012; Wen et al., 2009) and shrimp (Shiau and Hsu, 1999), and decreased expression of various genes involved in liver fatty acid synthesis (Qian et al., 2015). In geese, PAD elevated serum concentrations of total cholesterol and triglyceride, but decreased serum high density lipoprotein cholesterol (Wang et al., 2016).
Effects of dietary PAD on carbohydrate and lipid metabolism in ducks, as well as underlying molecular mechanisms, have not been well characterized. Although it is clear that PAD affects carbohydrate and lipid metabolism, detailed characterization of underlying processes and the extent of changes induced by PAD have apparently not been reported. Here, we used an integrated proteomic and metabolomic approach in starter Pekin ducks, a well-established model of PAD, to investigate the effects of PAD on liver protein and metabolites in relation to changes in hepatic lipid profiles.
2. Materials and methods
2.1. Animal ethics
All experimental procedures were approved by the Animal Welfare Committee of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, and performed according to the guidelines for animal experiments established by the National Institute of Animal Health.
2.2. Animals and housing
Male white Pekin ducks (Anas platyrhynchos; 1-d-old, n = 128) were obtained from the Pekin duck breeding center (Chinese Academy of Agricultural Sciences) and randomly allocated to 16 raised plastic-floor pens with 8 birds per pen. All ducks were assigned into 2 experimental groups, each containing 8 replicates with 8 birds per replicate. From hatch to 16 d of age, the ducks had ad libitum access to water and either a PAD or a control diet (CON). During this interval, there was continuous light. The temperature was kept at 33 °C from 1 to 3 d of age, then gradually reduced to approximately 25 °C at 16 d of age.
2.3. Diet
The basal diet was pantothenic acid-deficient, containing only 4.65 mg pantothenic acid/kg of diet (Table 1). The PAD and control diets were produced by supplementing this basal diet with 0 and 8 mg/kg diet, respectively, of crystalline calcium pantothenate (purity, 99%; Xinfu Technology Co. Ltd, Hangzhou, China). The pantothenic acid concentration of the control diet met NRC (1994) requirements for starter ducks.
Table 1.
Composition of pantothenic acid-deficient basal diet from hatch to 16 d of age (as-fed basis).
| Item | Content |
|---|---|
| Ingredients, % | |
| Corn | 79.7 |
| Soy isolate protein | 16.0 |
| Limestone | 1.0 |
| Dicalcium phosphate | 1.6 |
| Vitamin and trace mineral premix1 | 1.0 |
| Sodium chloride | 0.3 |
| DL-Methionine | 0.3 |
| L-Lysine·HCl | 0.1 |
| Total | 100.0 |
| Calculated composition | |
| Metabolizable energy2, MJ/kg | 13.35 |
| Crude protein | 20.39 |
| Calcium | 0.93 |
| Nonphytate phosphorus | 0.43 |
| Lysine | 1.17 |
| Methionine | 0.57 |
| Methionine + cysteine | 0.80 |
| Threonine | 0.77 |
| Tryptophan | 0.19 |
| Arginine | 1.38 |
| Pantothenic acid3, mg/kg | 4.65 |
Supplied per kilogram of total diet: Cu (CuSO4•5H2O), 10 mg; Fe (FeSO4•7H2O), 60 mg; Zn (ZnO), 60 mg; Mn (MnSO4•H2O), 80 mg; Se (NaSeO3), 0.3 mg; I (KI), 0.2 mg; choline chloride, 1,000 mg; vitamin A (retinyl acetate), 10,000 IU; vitamin D3 (cholcalciferol), 3,000 IU; vitamin E (DL-α-tocopheryl acetate), 20 IU; vitamin K3 (menadione sodium bisulfate), 2 mg; thiamin (thiamin mononitrate), 2 mg; riboflavin, 10 mg; pyridoxine hydrochloride, 4 mg; cobalamin, 0.02 mg; nicotinic acid, 50 mg; folic acid, 1 mg; biotin, 0.2 mg.
The value is calculated according to the apparent metabolizable energy (AME) of ducks (Ministry of Agriculture of China, 2012).
The value was based on high performance liquid chromatography coupled with triple quadrupole mass spectrometry.
2.4. Data and sample collection
At 16 d of age, after overnight fasting, the ducks and residual diet from each pen were weighed to determine the final body weight and cumulative feed intake during the experimental period.
Two ducks from each pen were randomly selected and blood collected from a wing vein into heparin sodium-containing tubes, centrifuged at 1,500 × g for 10 min, and plasma stored at −20 °C. Thereafter, these ducks were euthanized by CO2 inhalation, and livers were immediately obtained, snap frozen in liquid nitrogen, and stored at −80 °C.
2.5. Pantothenic acid content
Pantothenic acid concentrations in feed and liver were determined by high performance liquid chromatography (HPLC) coupled with triple quadrupole mass spectrometry (Agilent 6470), as previously described (Lu et al., 2008). An agilent 1290 HPLC system consisting of a ZORBAX Eclipase Plus C18 column (3.0 mm × 150 mm, 1.8 μm) was used for pantothenic acid separation. The column oven was maintained at 35 °C and the flow rate of the mobile phase was 0.2 mL/min. The binary mobile phase consisted of acetonitrile and water containing 0.1% formic acid. Before LC/MS analysis, feed samples were prepared as previously described (Woollard et al., 2000), and liver samples were also prepared as previously described (Tang et al., 2020b). The peak was identified and quantified using pure authentic standards (Sigma-Aldrich, St. Louis, MO, USA).
2.6. Plasma parameters
Plasma parameters, including glucose, pyruvate, lactate, uric acid, alanine transaminase (ALT), aspartate transaminase (AST), cholesterol (CHO), triglyceride (TG), high density lipoprotein cholesterol (HDL-C), and low density lipoprotein cholesterol (LDL-C) were determined using commercial kits according to manufacturer's instructions (BioSino Bio-technology and Science Inc., Beijing, China).
2.7. Liver proteomics
Six liver samples (3 biological replicates per group) were used to conduct the isobaric tags for relative and absolute quantification (iTRAQ) assays. Proteins were extracted and digested as previously described (Tang et al., 2019). Each digested sample was labelled with iTRAQ 8-plex reagents (AB Sciex, Foster City, CA, USA) according to the manufacturer's instructions. The PAD samples were labelled with iTRAQ tags 113, 114, and 115, and the CON samples were labelled with tags 116, 117, and 118. Labelled samples were mixed and fractionated into 20 fractions by HPLC (DINOEX Ultimate 3000 BioRS, Thermo Fisher, Waltham, MA, USA) using a Durashell C18 column (5 μm, 100 Å, 4.6 mm × 250 mm). The LC-electrospray ionization-MS/MS analysis was conducted with a Triple TOF 5600 plus system (AB SCIEX, Framingham, MA, USA). The original MS/MS file data for identification and quantitation were analyzed against the database UniProt_Mallard_8839 using ProteinPilot Software version 4.0 (AB SCIEX). To minimize the false discovery rate (FDR), a threshold for protein identification was applied. Only unique peptides with confidences >95% were contained in the iTRAQ labelling quantification and used for further analysis.
For analysis of proteomic results, the relative expressions of identified proteins were based on the ratio of the reporter ions of the peptides between the 2 groups (PAD vs CON). A protein was considered differentially expressed when it had both a fold change (FC) > 1.5 and P-value < 0.05.
To enrich the differentially expressed proteins with respect to specific functional terms, the protein lists were analyzed using ClueGo software (http://www.ici.upmc.fr/cluego/) with the Gene Ontology (GO) database (release date: February 2020). Pathway enrichment analysis of differentially expressed proteins (Ashburner et al., 2000) was performed using ClueGo software and applying the database from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (release date: February 2020).
2.8. Liver metabolomics
A total of 24 liver samples (12 biological replicates per group) were used to conduct metabolome assays. Each liver sample was ground in liquid nitrogen and 60 mg of the resulting powder was lysed in 800 μL of cold acetonitrile/methanol (1:1, vol/vol) and 200 μL water. The lysate was homogenized, and then sonicated at low temperature (twice, 30 min each). The mixture was centrifuged for 15 min (13,000 × g, 4 °C) and the supernatant dried in a vacuum centrifuge. For LC-MS analysis, samples were re-dissolved in 100 μL acetonitrile/water (1:1, vol/vol) solvent. Analyses were performed using an UHPLC (1290 Infinity LC, Agilent Technologies) coupled to a quadrupole time-of-flight (AB Sciex TripleTOF 6600). Raw MS data were converted to MzXML files using ProteoWizard MSConvert and the converted data imported into XCMS software.
After being normalized to total peak intensity, the processed data were imported into SIMCA-P (Version 14.1, Umetrics, Umea, Sweden), and subjected to multivariate data analysis, including pareto-scaled principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA). The variable importance in the projection (VIP) value of each variable in the PLS-DA model was calculated to determine its contribution to the classification. Metabolites with VIP value > 1 and P value < 0.05 were considered significant. For bioinformatic analysis, metabolites were subsequently mapped to pathways in KEGG.
2.9. Interaction analysis of proteomics and metabolomics
Integrated Molecular Pathway Level Analysis (IMPaLA, http://impala.molgen.mpg.de/) was used to analyze the significantly altered canonical pathways and molecular interaction networks of differentially expressed proteins and metabolites affected by PAD. Multiple testing corrections were performed with the FDR method (Benjamini-Hochberg) and significance set to q < 0.01.
2.10. Western blot analyses
Western blot analysis of for proteins, phosphoglucomutase 1 (PGM1), malic enzyme 1 (ME1), isocitrate dehydrogenase (IDH1), and malate dehydrogenase 2 (MDH2), were performed following the method as described previously (Zhang et al., 2021). Primary antibodies (1 μg/mL) against PGM1 (A6303; ABclonal), ME1 (A3956; ABclonal), IDH1 (A13245; ABclonal), and MDH2 (A13516; ABclonal) were used. Vinculin (A2752; ABclonal) served as a loading control.
2.11. Statistical analyses
All data were analyzed by Student's t-test, P < 0.05 was considered significant and variability was reported as the standard deviation. These statistical analyses were conducted using SAS software (SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. Growth performance
Compared to the CON group, the final body weight and cumulative feed intake of ducks were decreased in the PAD group (P < 0.001; Table 2).
Table 2.
Growth performance on d 16 of ducks in the pantothenic acid-deficient (PAD) and control (CON) group (g/bird).1
| Variable | PAD | CON | P-value |
|---|---|---|---|
| Initial body weight | 52.77 ± 0.51 | 52.70 ± 0.42 | 0.795 |
| Final body weight | 221.4 ± 43.8b | 553.1 ± 42.8a | <0.001 |
| Cumulative feed intake | 268.5 ± 28.0b | 640.8 ± 53.0a | <0.001 |
a,b Within a row, means without a common superscript differed (P < 0.05).
Each value represents the mean ± SD of 8 replicates (n = 8).
3.2. Plasma parameters
Compared to the CON group, plasma lactate, HDL-C, and LDL-C content were decreased in the PAD group (P < 0.01; Table 3). Furthermore, plasma uric acid, AST, and TG were elevated in the PAD group (P < 0.001; Table 3). However, there were no significant differences between groups for plasma pyruvate, ALT, or CHO (P > 0.05; Table 3).
Table 3.
Plasma parameters in pantothenic acid-deficient (PAD) and control (CON) ducks.1
| Variable | PAD | CON | P-value |
|---|---|---|---|
| Pyruvate, mmol/L | 0.23 ± 0.07 | 0.24 ± 0.03 | 0.424 |
| Lactate, mmol/L | 3.29 ± 0.90b | 5.79 ± 1.49a | <0.001 |
| Uric acid, μmol/L | 542.4 ± 187.9a | 331.6 ± 134.1b | 0.002 |
| ALT, U/L | 65.64 ± 29.30 | 64.25 ± 12.74 | 0.880 |
| AST, U/L | 36.93 ± 22.39a | 20.42 ± 13.57b | 0.036 |
| CHO, mmol/L | 7.38 ± 2.48 | 7.06 ± 0.76 | 0.599 |
| TG, mmol/L | 1.93 ± 1.57a | 0.56 ± 0.12b | <0.001 |
| HDL-C, mmol/L | 2.30 ± 0.59b | 4.00 ± 0.25a | <0.001 |
| LDL-C, mmol/L | 1.10 ± 0.41b | 2.61 ± 0.46a | <0.001 |
ALT = alanine transaminase; AST = aspartate transaminase; CHO = cholesterol; TG = triglyceride; HDL-C = high density lipoprotein cholesterol; LDL-C = low density lipoprotein cholesterol.
a,b Within a row, means without a common superscript differed (P < 0.05).
Each value represents the mean ± SD of 8 replicates (n = 8).
3.3. Liver parameters
Compared to the CON group, PAD decreased liver pantothenic acid, triglyceride, and free fatty acids (P < 0.001; Table 4), whereas PAD increased relative liver weight and liver glycogen content (P < 0.001; Table 4).
Table 4.
Liver parameters in pantothenic acid-deficient (PAD) and control (CON) ducks.1
| Variable | PAD | CON | P-value |
|---|---|---|---|
| Relative liver weight, g/g | 4.15 ± 0.36a | 3.73 ± 0.24b | 0.001 |
| Pantothenic acid, μg/g | 24.13 ± 4.75b | 40.61 ± 7.47a | <0.001 |
| Total lipid, % of fresh liver | 4.96 ± 0.32 | 5.16 ± 0.71 | 0.374 |
| Triglyceride, mg/g fresh liver | 3.08 ± 0.81b | 4.47 ± 2.02a | 0.039 |
| Cholesterol, mg/g fresh liver | 12.95 ± 2.43 | 13.49 ± 1.99 | 0.572 |
| Free fatty acids, μmol/g | 268.3 ± 14.63b | 294.0 ± 14.27a | <0.001 |
| Glycogen, mg/g | 92.14 ± 4.59a | 77.44 ± 5.82b | <0.001 |
a,b Within a row, means without a common superscript differed (P < 0.05).
Each value represents the mean ± SD of 8 replicates (n = 8).
There were no differences in liver saturated fatty acid (SFA), polyunsaturated fatty acid (PUFA), C16:0, C18:0, C18:2n6c, C18:3n6, C18:3n3, C20:0, C20:4n6, C22:0 between the PAD and CON groups. However, compared to the CON group, PAD decreased liver total fatty acid (TFA), unsaturated fatty acid (UFA), monounsaturated fatty acid (MUFA), C14:0, C16:1, C18:1n9c, C20:1, C20:2, C20:3n6, C20:5n3, C22:1, C22:2, and C22:6n3 (P < 0.05; Table 5). Whereas, it increased C24:0 and C24:1 (P < 0.05; Table 5).
Table 5.
Liver fatty acid composition in pantothenic acid-deficient (PAD) and control (CON) ducks (mg/g).1
| Fatty acid | PAD | CON | P-value |
|---|---|---|---|
| C14:0 | 0.19 ± 0.11b | 0.37 ± 0.12a | 0.001 |
| C16:0 | 21.37 ± 7.25 | 21.85 ± 5.29 | 0.857 |
| C16:1 | 0.64 ± 0.55b | 1.13 ± 0.35a | 0.021 |
| C18:0 | 18.12 ± 4.53 | 17.79 ± 4.10 | 0.855 |
| C18:1n9c | 38.51 ± 26.79b | 73.35 ± 28.10a | 0.006 |
| C18:2n6c | 11.95 ± 4.79 | 10.05 ± 2.15 | 0.243 |
| C18:3n6 | 0.23 ± 0.13 | 0.20 ± 0.034 | 0.493 |
| C18:3n3 | 0.11 ± 0.09 | 0.12 ± 0.05 | 0.717 |
| C20:0 | 0.17 ± 0.04 | 0.18 ± 0.05 | 0.761 |
| C20:1 | 0.30 ± 0.14b | 0.53 ± 0.15a | 0.001 |
| C20:2 | 0.93 ± 0.23b | 2.60 ± 0.33a | <0.001 |
| C20:3n6 | 1.57 ± 0.72b | 2.87 ± 0.64a | <0.001 |
| C20:4n6 | 24.36 ± 4.09 | 25.25 ± 2.25 | 0.533 |
| C20:5n3 | 0.10 ± 0.03b | 0.23 ± 0.07a | <0.001 |
| C22:0 | 0.34 ± 0.05 | 0.36 ± 0.05 | 0.300 |
| C22:1 | 0.071 ± 0.011b | 0.091 ± 0.013a | <0.001 |
| C22:2 | 0.10 ± 0.07b | 0.28 ± 0.06a | <0.001 |
| C24:0 | 0.69 ± 0.12a | 0.57 ± 0.06b | 0.006 |
| C24:1 | 2.96 ± 0.85a | 1.50 ± 0.32b | <0.001 |
| C22:6n3 | 0.34 ± 0.05b | 0.45 ± 0.07a | <0.001 |
| TFA | 123.1 ± 43.73b | 159.8 ± 40.03a | 0.049 |
| SFA | 40.88 ± 10.97 | 41.12 ± 9.40 | 0.957 |
| UFA | 82.18 ± 33.27b | 118.65 ± 30.92a | 0.013 |
| MUFA | 42.49 ± 27.22b | 76.59 ± 28.70a | 0.008 |
| PUFA | 39.69 ± 7.65 | 42.05 ± 4.25 | 0.378 |
TFA = total fatty acid; SFA = saturated fatty acid; UFA = unsaturated fatty acid; MUFA = monounsaturated fatty acid; PUFA = polyunsaturated fatty acid.
a,b Within a row, means without a common superscript differed (P < 0.05).
Each value represents the mean ± SD of 8 replicates (n = 8).
3.4. Changes in the liver proteomics of duck in response to PAD
Using iTRAQ analysis, a total of 18,985 peptide spectral matches were identified, from which 2,945 proteins were identified in the livers of the 2 groups. In comparisons of the relative abundance of proteins from liver of PAD versus CON ducks, a total of 275 proteins had an FC > 1.5, of which 170 proteins were upregulated and 105 proteins were downregulated. The selected proteins regulated by PAD are presented in Table 6, whereas a complete list is presented in Appendix Table 1.
Table 6.
Selected differentially expressed proteins in duck liver caused by pantothenic acid deficiency (PAD).
| UniProtKB ID | Protein name | Short name | Fold change1 | P-value |
|---|---|---|---|---|
| Glycogen synthesis and degradation | ||||
| U3J9R8 | Alpha-1,4 glucan phosphorylase | PYGL | −2.16 | 3.45E-06 |
| U3J383 | Phosphoglucomutase 1 | PGM1 | −2.30 | 1.06E-07 |
| R0LET4 | Glycogen synthase | GYS2 | −1.56 | 5.41E-04 |
| R0LKW3 | UTP--glucose-1-phosphate uridylyltransferase | UGP2 | −3.37 | 5.52E-11 |
| Glycolysis and gluconeogenesis | ||||
| R0LKR3 | Glucose-6-phosphate isomerase | GPI | −2.85 | 9.18E-06 |
| P13743 | L-Lactate dehydrogenase B chain | LDHB | −1.64 | 7.07E-06 |
| R0LQQ0 | Phosphoenolpyruvate carboxykinase 1 | PCK1 | −2.45 | 4.21E-05 |
| U3IEW2 | Pyruvate dehydrogenase E1 beta subunit | PDHB | −1.85 | 2.32E-04 |
| U3ILL1 | Pyruvate dehydrogenase E1 component subunit alpha | PDHA1 | −2.17 | 9.48E-05 |
| U3J1L1 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | −3.24 | 4.64E-05 |
| U3IVG9 | Hexokinase domain containing 1 | HKDC1 | −3.62 | 2.53E-07 |
| U3I9L2 | phosphofructokinase, liver type | PFKL | −4.05 | 3.87E-07 |
| U3IR48 | Dihydrolipoyl dehydrogenase | DLD | −1.76 | 2.46E-05 |
| U3IHG8 | Fructose-bisphosphate aldolase | ALDOB | −1.67 | 8.52E-03 |
| U3ILF5 | Phosphoglycerate kinase | PGK1 | −2.23 | 5.75E-08 |
| U3I8D8 | Triosephosphate isomerase | TPI1 | −2.04 | 3.58E-04 |
| U3ICQ7 | Acetyltransferase component of pyruvate dehydrogenase complex | DLAT | −2.83 | 4.17E-03 |
| U3IW82 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | PDHX | −2.23 | 3.71E-05 |
| U3J383 | Phosphoglucomutase 1 | PGM1 | −2.30 | 1.06E-07 |
| U3IM27 | Aldehyde dehydrogenase 2 family | ALDH2 | −1.50 | 6.32E-06 |
| U3IE74 | Lactate dehydrogenase A | LDHA | 3.33 | 5.50E-12 |
| U3IWS8 | Hexokinase 2 | HK2 | 2.56 | 2.22E-02 |
| PPAR signaling pathway | ||||
| A0A0H3U2H5 | Stearoyl-CoA desaturase1 | SCD1 | −3.31 | 4.65E-05 |
| R0LQQ0 | Phosphoenolpyruvate carboxykinase 1 | PCK1 | −2.45 | 4.21E-05 |
| U3I7T9 | Acyl-CoA synthetase family member 2 | ACSF2 | −1.55 | 1.07E-04 |
| U3I8S1 | Acyl-CoA synthetase bubblegum family member 2 | ACSBG2 | −5.91 | 7.92E-07 |
| U3IDQ1 | Acyl-coenzyme A oxidase | ACOX2 | −2.05 | 1.06E-06 |
| U3IIW2 | Fatty acid desaturase 2 | FADS2 | −3.32 | 2.90E-04 |
| U3IKU2 | Sterol carrier protein 2 | SCP2 | −2.51 | 9.84E-07 |
| U3IL38 | Malic enzyme 1 | ME1 | −4.04 | 5.86E-14 |
| U3INH3 | Sorbin and SH3 domain containing 2 | SORBS2 | −1.58 | 2.74E-02 |
| U3J4C8 | Fatty acid binding protein 7 | FABP7 | −2.56 | 6.74E-03 |
| Fatty acid oxidation | ||||
| U3J8S7 | Carnitine palmitoyltransferase 1A | CPT1A | 2.37 | 7.85E-05 |
| U3INM7 | Carnitine palmitoyltransferase 2 | CPT2 | 1.57 | 6.26E-03 |
| R0JG91 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS | 1.50 | 4.00E-03 |
| U3ITA9 | Acyl-CoA dehydrogenase medium chain | ACADM | 1.91 | 1.18E-02 |
| U3IAY7 | Acyl-CoA dehydrogenase long chain | ACADL | 1.88 | 4.37E-04 |
| U3J4J3 | Acetyl-CoA acetyltransferase 1 | ACAT1 | 3.50 | 8.53E-06 |
| U3IU30 | Acyl-CoA synthetase long chain family member 1 | ACSL1 | 1.91 | 1.29E-06 |
| U3J4Z9 | Acyl-CoA synthetase long chain family member 5 | ACSL5 | 3.13 | 3.36E-09 |
| U3I806 | Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha | HADHA | 2.00 | 1.49E-07 |
| U3I6S1 | Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta | HADHB | 2.23 | 7.26E-04 |
| U3J3G1 | Enoyl-CoA hydratase, short chain 1 | ECHS1 | 1.79 | 5.23E-03 |
| R0LHZ1 | 3-Ketoacyl-CoA thiolase, mitochondrial | ACAA2 | 1.78 | 1.44E-04 |
| U3IKG5 | Aldehyde dehydrogenase 9 family member A1 | ALDH9A1 | 1.51 | 5.09E-05 |
| U3IZY1 | Aldehyde dehydrogenase 7 family member A1 | ALDH7A1 | 1.68 | 4.77E-05 |
| U3IHS8 | Carnitine O-acetyltransferase | CRAT | 1.69 | 3.88E-03 |
| U3I624 | Glycerol kinase | GK | 1.60 | 9.37E-04 |
| Oxidative phosphorylation | ||||
| U3I998 | NADH:ubiquinone oxidoreductase core subunit S1 | NDUFS1 | 1.92 | 1.28E-05 |
| U3IVZ4 | NADH:ubiquinone oxidoreductase subunit B5 | NDUFB5 | 1.78 | 3.00E-02 |
| U3J3L1 | NADH:ubiquinone oxidoreductase core subunit V1 | NDUFV1 | 1.91 | 5.99E-03 |
| U3J9G0 | NDUFA4 mitochondrial complex associated | NDUFA4 | 1.99 | 1.90E-02 |
| U3IMS0 | NADH:ubiquinone oxidoreductase core subunit S3 | NDUFS3 | 1.69 | 2.49E-04 |
| R0JL39 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | 1.64 | 4.12E-03 |
| U3I2D1 | Ubiquinol-cytochrome c reductase core protein 1 | UQCRC1 | 1.60 | 1.13E-04 |
| U3I342 | Ubiquinol-cytochrome c reductase core protein 2 | UQCRC2 | 1.93 | 1.57E-03 |
| A6ZJ02 | Cytochrome c oxidase subunit 2 | COX2 | 1.80 | 2.33E-02 |
| R0KK84 | ATP synthase subunit beta, mitochondrial | ATP5F1B | 1.74 | 1.46E-02 |
| R0LYJ7 | ATP synthase subunit d, mitochondrial | ATP5H | 1.82 | 7.41E-03 |
| U3IK89 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | ATP5C1 | 2.28 | 2.95E-04 |
| U3IVL6 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | ATP5A1 | 2.30 | 1.96E-03 |
| U3J9J8 | ATPase H+ transporting V1 subunit B2 | ATP6V1B2 | 1.71 | 1.95E-03 |
| U3IXX4 | Solute carrier family 25 member 5 | SLC25A5 | 4.27 | 1.97E-02 |
| TCA cycle | ||||
| R0J775 | Aconitase 1 (Fragment) | ACO1 | 1.69 | 1.43E-04 |
| R0L7Q0 | Fumarate hydratase (Fragment) | FH | 2.57 | 3.79E-04 |
| U3J597 | Isocitrate dehydrogenase | IDH1 | 3.80 | 2.72E-13 |
| U3IA60 | Malate dehydrogenase 2 | MDH2 | 3.95 | 2.45E-06 |
TCA = tricarboxylic acid.
Fold change is expressed as the ratio of the pantothenic acid-deficient (PAD) to the control (CON) group. For downregulated proteins, the fold change was transformed to the corresponding negative value.
We performed GO categories of biological process, cellular component, and molecular function, and pathway analysis on the set of 275 differentially expressed proteins in livers from the PAD group compared to those in the CON group. As shown in Fig. 1, the top 15 enriched terms under biological process included: small molecule metabolic process, oxoacid metabolic process, oxidation-reduction process, small molecule catabolic process, single-organism catabolic process, monocarboxylic acid metabolic process, carboxylic acid catabolic process, organonitrogen compound metabolic process, cofactor metabolic process, small molecule biosynthetic process, coenzyme metabolic process, single-organism biosynthetic process, alpha-amino acid metabolic process, organic substance catabolic process, and cellular catabolic process.
Fig. 1.
Top 15 significantly enriched biological processes, cellular components, and molecular functions in ducks with pantothenic acid deficiency (PAD) compared to control ducks.
Based on analysis of the KEGG pathway, differentially expressed proteins were enriched in amino acid metabolism, glycolysis and gluconeogenesis, fatty acid beta oxidation, peroxisome proliferator-activated receptor (PPAR) signaling pathway, tricarboxylic acid (TCA) cycle, tryptophan metabolism, folate metabolism, oxidative stress, cori cycle, and oxidative phosphorylation (Fig. 2).
Fig. 2.
The pathway analysis by the Kyoto Encyclopedia of Genes and Genomes (KEGG) on differentially expressed proteins in ducks with pantothenic acid deficiency (PAD) compared to control ducks.
To be specific, PAD downregulated 2 proteins involved in glycogenolysis (alpha-1,4 glucan phosphorylase [PYGL] and PGM1) and 2 proteins involved in glycogenesis (glycogen synthase [GYS2] and UTP-glucose-1-phosphate uridylyltransferase [UGP2]). Eighteen proteins in glycolysis and gluconeogenesis pathways were differentially expressed after PAD. Of these, 2 proteins were enhanced (lactate dehydrogenase A [LDHA] and hexokinase 2 [HK2]) and 16 proteins were diminished (glucose-6-phosphate isomerase [GPI], L-lactate dehydrogenase B chain [LDHB], phosphoenolpyruvate carboxykinase 1 [PCK1], pyruvate dehydrogenase E1 beta subunit [PDHB], pyruvate dehydrogenase E1 component subunit alpha [PDHA1], glyceraldehyde-3-phosphate dehydrogenase [GAPDH], hexokinase domain containing 1 [HKDC1], phosphofructokinase, liver type [PFKL], dihydrolipoyl dehydrogenase [DLD], fructose-bisphosphate aldolase [ALDOB], phosphoglycerate kinase [PGK1], triosephosphate isomerase [TPI1], acetyltransferase component of pyruvate dehydrogenase complex [DLAT], dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex [PDHX], PGM1, and aldehyde dehydrogenase 2 family [ALDH2]). In PAD ducks, there was upregulation of 4 proteins involved in the TCA cycle, including aconitase 1 (ACO1), fumarate hydratase (FH), IDH1, and MDH2. Dietary PAD downregulated 10 proteins involved in the PPAR signaling pathway, including stearoyl-CoA desaturase1 (SCD1), PCK1, acyl-CoA synthetase family member 2 (ACSF2), acyl-CoA synthetase bubblegum family member 2 (ACSBG2), acyl-coenzyme A oxidase (ACOX2), fatty acid desaturase 2 (FADS2), sterol carrier protein 2 (SCP2), ME1, sorbin and SH3 domain containing 2 (SORBS2), and fatty acid binding protein 7 (FABP7). Dietary PAD upregulated 16 proteins involved in fatty acid beta oxidation, including carnitine palmitoyltransferase 1A (CPT1A), carnitine palmitoyltransferase 2 (CPT2), short-chain specific acyl-CoA dehydrogenase, mitochondrial (ACADS), acyl-CoA dehydrogenase medium chain (ACADM), acyl-CoA dehydrogenase long chain (ACADL), acetyl-CoA acetyltransferase 1 (ACAT1), acyl-CoA synthetase long chain family member 1 (ACSL1), acyl-CoA synthetase long chain family member 5 (ACSL5), hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha (HADHA), hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta (HADHB), enoyl-CoA hydratase, short chain 1 (ECHS1), 3-ketoacyl-CoA thiolase, mitochondrial (ACAA2), aldehyde dehydrogenase 9 family member A1 (ALDH9A1), aldehyde dehydrogenase 7 family member A1 (ALDH7A1), carnitine O-acetyltransferase (CRAT), and glycerol kinase (GK). Dietary PAD upregulated 15 proteins involved in oxidative phosphorylation, including NADH:ubiquinone oxidoreductase core subunit S1 (NDUFS1), NADH:ubiquinone oxidoreductase subunit B5 (NDUFB5), NADH:ubiquinone oxidoreductase core subunit V1 (NDUFV1), NDUFA4 mitochondrial complex associated (NDUFA4), NADH:ubiquinone oxidoreductase core subunit S3 (NDUFS3), cytochrome b-c1 complex subunit Rieske, mitochondrial (UQCRFS1), ubiquinol-cytochrome c reductase core protein 1 (UQCRC1), ubiquinol-cytochrome c reductase core protein 2 (UQCRC2), cytochrome c oxidase subunit 2 (COX2), ATP synthase subunit beta, mitochondrial (ATP5F1B), ATP synthase subunit d, mitochondrial (ATP5H), ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (ATP5C1), ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle (ATP5A1), ATPase H+ transporting V1 subunit B2 (ATP6V1B2), and solute carrier family 25 member 5 (SLC25A5).
3.5. Changes in the liver metabolomics of duck in response to PAD
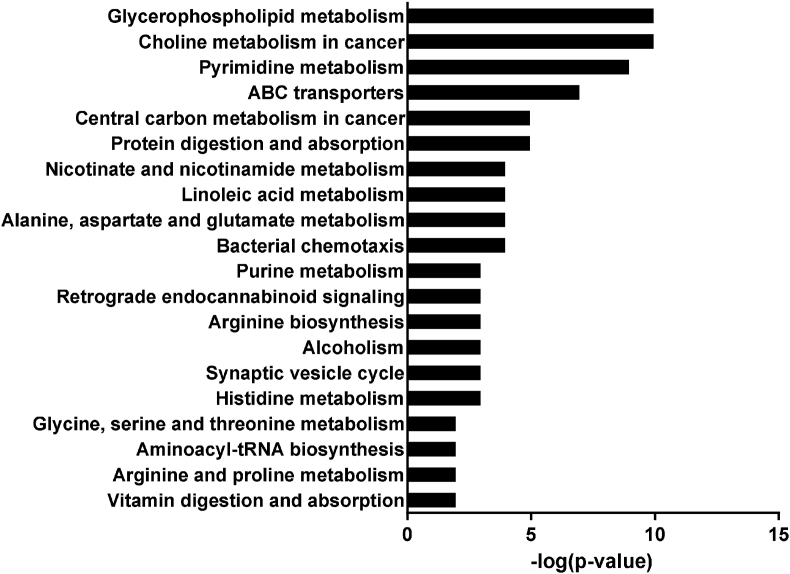
Metabolic profiles of differences between PAD and CON group liver samples were detected using untargeted metabolomics analysis by UPLC-Q-TOF/MS (Appendix Fig. 1). A total of 99 significant variations in metabolites were selected with the criteria of a PLS-DA model VIP >1 and P < 0.05 (Table 7). Compared to CON, 53 metabolites of differential metabolites were up-regulated by PAD, whereas 46 metabolites were down-regulated. Based on the analysis of KEGG pathways, differential metabolites were enriched in the pathways of glycerophospholipid metabolism, choline metabolism in cancer, pyrimidine metabolism, ABC transporters, central carbon metabolism in cancer, protein digestion and absorption, nicotinate and nicotinamide metabolism, alanine, aspartate and glutamate metabolism, linoleic acid metabolism, and bacterial chemotaxis (Fig. 3). Compared to the CON group, the levels of alpha-D-glucose 1-phosphate, L-palmitoylcarnitine, L-carnitine, stearoylcarnitine, succinate, L-malic acid, and argininosuccinic acid in liver were markedly increased in the PAD ducks, whereas the concentrations of maltotriose, maltopentaose, cellobiose, and 3-alpha-mannobiose were significantly decreased.
Table 7.
Liver metabolites in ducks significantly affected by pantothenic acid deficiency (PAD).
| No. | Name | rt(s) | m/z | VIP | Metabolite | Fold change1 | P-value |
|---|---|---|---|---|---|---|---|
| 1 | M522T869 | 869 | 522.20 | 6.64 | Maltotriose | −19.89 | 2.14E-03 |
| 2 | M846T989 | 989 | 846.31 | 1.22 | Maltopentaose | −11.07 | 2.86E-03 |
| 3 | M360T752 | 752 | 360.15 | 3.30 | Cellobiose | −10.37 | 1.66E-03 |
| 4 | M325T753 | 753 | 325.11 | 2.68 | 3alpha-Mannobiose | −8.89 | 1.63E-03 |
| 5 | M269T405 | 405 | 269.09 | 3.10 | Inosine | −8.84 | 2.66E-06 |
| 6 | M383T72 | 72 | 383.33 | 1.80 | 25-hydroxyvitamin D3 | −4.76 | 8.61E-07 |
| 7 | M468T357 | 357 | 468.31 | 3.18 | LysoPC(14:0) | −3.48 | 3.31E-04 |
| 8 | M216T754 | 754 | 216.06 | 5.63 | sn-Glycerol 3-phosphoethanolamine | −3.47 | 2.17E-07 |
| 9 | M385T59 | 59 | 385.35 | 1.94 | Desmosterol | −3.28 | 1.91E-06 |
| 10 | M284T870 | 870 | 284.05 | 1.01 | N-Acetyl-D-Glucosamine 6-Phosphate | −3.20 | 8.57E-08 |
| 11 | M489T857 | 857 | 489.11 | 1.05 | Cytidine 5′-diphosphocholine (CDP-choline) | −3.11 | 3.39E-02 |
| 12 | M744T962 | 962 | 744.08 | 1.48 | Nicotinamide adenine dinucleotide phosphate | −3.09 | 3.46E-09 |
| 13 | M326T60 | 60 | 326.30 | 1.81 | N-Oleoylethanolamine | −2.95 | 8.28E-04 |
| 14 | M302T868 | 868 | 302.06 | 1.14 | N-Acetylglucosamine 1-phosphate | −2.93 | 7.21E-05 |
| 15 | M420T61 | 61 | 420.38 | 1.65 | 22beta-Hydroxycholesterol | −2.57 | 6.72E-04 |
| 16 | M113T298_2 | 298 | 113.03 | 5.62 | Uracil | −2.44 | 9.55E-07 |
| 17 | M613T964 | 964 | 613.16 | 4.44 | Glutathione disulfide | −2.37 | 1.92E-03 |
| 18 | M295T888 | 888 | 295.15 | 1.58 | Glycyl-Arginine | −2.35 | 1.12E-02 |
| 19 | M809T130 | 130 | 808.58 | 6.17 | PC(18:1(9Z)/18:1(9Z)) | −2.32 | 3.68E-02 |
| 20 | M262T298 | 298 | 262.10 | 4.01 | Uridine | −2.31 | 1.36E-04 |
| 21 | M779T108 | 108 | 778.54 | 1.72 | PC(16:0/16:0) | −2.30 | 3.42E-02 |
| 22 | M249T809 | 809 | 249.11 | 1.40 | Threoninyl-Glutamate | −2.29 | 8.45E-03 |
| 23 | M142T879 | 879 | 142.03 | 1.36 | O-Phosphoethanolamine | −2.27 | 1.14E-04 |
| 24 | M112T273 | 273 | 112.09 | 1.20 | Histamine | −2.13 | 4.09E-03 |
| 25 | M517T237 | 237 | 517.33 | 9.06 | Taurodeoxycholic acid | −2.06 | 4.75E-02 |
| 26 | M338T56_2 | 56 | 338.34 | 1.92 | Erucamide | −1.93 | 2.21E-03 |
| 27 | M118T665 | 665 | 118.06 | 4.86 | Guanidoacetic acid | −1.88 | 8.09E-06 |
| 28 | M121T209 | 209 | 121.05 | 1.05 | Purine | −1.88 | 2.16E-02 |
| 29 | M335T885 | 885 | 335.06 | 1.41 | Beta-nicotinamide D-ribonucleotide | −1.83 | 3.57E-02 |
| 30 | M339T82 | 82 | 339.25 | 1.50 | Cis-(6,9,12)-linolenic acid | −1.77 | 1.43E-02 |
| 31 | M337T888 | 888 | 337.17 | 1.26 | Arginyl-Tyrosine | −1.76 | 1.46E-02 |
| 32 | M664T841 | 841 | 664.12 | 1.56 | Nicotinamide adenine dinucleotide (NAD) | −1.75 | 7.93E-05 |
| 33 | M123T196 | 196 | 123.05 | 2.87 | Nicotinamide | −1.66 | 6.11E-03 |
| 34 | M235T834 | 834 | 235.09 | 1.29 | Glutamyl-Serine | −1.61 | 8.77E-04 |
| 35 | M500T288 | 288 | 500.30 | 2.78 | Tauroursodeoxycholic acid | −1.55 | 2.42E-02 |
| 36 | M522T341 | 341 | 522.35 | 5.17 | LysoPC(18:1(9Z)) | −1.53 | 2.84E-02 |
| 37 | M156T716_2 | 716 | 156.08 | 4.44 | L-Histidine | −1.47 | 3.06E-04 |
| 38 | M321T80 | 80 | 321.24 | 1.13 | 20-Hydroxyeicosatetraenoic acid | −1.47 | 3.10E-02 |
| 39 | M147T998 | 998 | 147.11 | 3.12 | L-Pipecolic acid | −1.35 | 2.57E-04 |
| 40 | M322T70 | 70 | 322.27 | 3.32 | Arachidonic acid (peroxide free) | −1.29 | 1.29E-02 |
| 41 | M222T490 | 490 | 222.10 | 1.36 | N-Acetyl-D-glucosamine | −1.26 | 2.41E-02 |
| 42 | M165T560_2 | 560 | 165.05 | 1.90 | Trans-2-hydroxycinnamic acid | −1.21 | 1.39E-03 |
| 43 | M136T560 | 560 | 136.07 | 1.45 | Dopamine | −1.21 | 7.62E-04 |
| 44 | M203T754 | 754 | 203.05 | 1.10 | Myo-inositol | −1.20 | 7.02E-04 |
| 45 | M182T560_2 | 560 | 182.08 | 2.43 | L-Tyrosine | −1.15 | 2.13E-02 |
| 46 | M120T502 | 502 | 120.08 | 1.12 | Tyramine | −1.14 | 1.09E-02 |
| 47 | M70T586 | 586 | 70.06 | 1.61 | Diethanolamine | 1.14 | 1.45E-02 |
| 48 | M116T586 | 586 | 116.07 | 4.47 | D-Proline | 1.16 | 1.99E-03 |
| 49 | M90T655 | 655 | 90.05 | 1.96 | L-Alanine | 1.23 | 5.66E-04 |
| 50 | M377T398 | 398 | 377.14 | 1.44 | Riboflavin | 1.33 | 6.44E-03 |
| 51 | M134T776 | 776 | 134.04 | 4.31 | L-Aspartate | 1.49 | 1.29E-04 |
| 52 | M291T907 | 907 | 291.13 | 1.10 | Argininosuccinic acid | 1.53 | 8.50E-03 |
| 53 | M204T194_1 | 194 | 204.09 | 1.22 | N-Acetylmannosamine | 1.55 | 2.07E-02 |
| 54 | M126T557_3 | 557 | 126.02 | 10.39 | Taurine | 1.62 | 8.20E-08 |
| 55 | M159T337 | 337 | 159.05 | 1.05 | Allantoin | 1.69 | 2.37E-02 |
| 56 | M162T746 | 746 | 162.11 | 1.67 | L-Carnitine | 1.74 | 1.27E-04 |
| 57 | M811T133 | 133 | 810.60 | 3.07 | 1-Stearoyl-2-oleoyl-sn-glycerol 3-phosphocholine (SOPC) | 1.80 | 4.10E-02 |
| 58 | M168T191 | 191 | 168.06 | 2.26 | Pyridoxal (vitamin B6) | 1.85 | 1.79E-02 |
| 59 | M285T411 | 411 | 285.08 | 1.86 | Xanthosine | 1.89 | 1.16E-05 |
| 60 | M243T775 | 775 | 243.03 | 1.09 | Alpha-D-glucose 1-phosphate | 1.90 | 8.96E-06 |
| 61 | M136T294 | 294 | 136.06 | 1.87 | Adenine | 2.09 | 5.87E-04 |
| 62 | M229T205 | 205 | 229.08 | 2.19 | 2′-Deoxyuridine | 2.12 | 2.56E-03 |
| 63 | M160T732 | 732 | 160.13 | 2.59 | Cyclohexylamine | 2.15 | 1.13E-05 |
| 64 | M169T629_2 | 629 | 169.04 | 5.10 | Uric acid | 2.22 | 1.20E-06 |
| 65 | M146T676_2 | 676 | 146.09 | 4.94 | 4-Guanidinobutyric acid | 2.23 | 3.15E-05 |
| 66 | M568T318 | 318 | 568.34 | 1.81 | 1-Stearoyl-sn-glycerol 3-phosphocholine | 2.35 | 7.61E-03 |
| 67 | M298T145 | 145 | 298.10 | 3.15 | S-Methyl-5′-thioadenosine | 2.43 | 4.88E-06 |
| 68 | M204T576 | 576 | 204.12 | 3.38 | Acetylcarnitine | 2.44 | 1.19E-03 |
| 69 | M209T478 | 478 | 209.09 | 2.67 | L-Kynurenine | 2.45 | 5.20E-05 |
| 70 | M114T78 | 78 | 114.09 | 2.38 | Triethanolamine | 2.45 | 5.53E-08 |
| 71 | M192T478_1 | 478 | 192.06 | 2.23 | 5-Hydroxyindoleacetate | 2.49 | 2.13E-05 |
| 72 | M127T179 | 179 | 127.05 | 3.26 | Thymine | 2.55 | 2.65E-03 |
| 73 | M130T581 | 581 | 130.09 | 3.52 | D-Pipecolic acid | 2.66 | 7.27E-04 |
| 74 | M112T385 | 385 | 112.05 | 1.25 | Cytosine | 2.66 | 2.18E-04 |
| 75 | M127T615 | 615 | 127.05 | 3.42 | Imidazoleacetic acid | 2.68 | 6.25E-07 |
| 76 | M243T180 | 180 | 243.10 | 4.04 | Thymidine | 2.74 | 3.38E-03 |
| 77 | M160T784 | 784 | 160.06 | 1.89 | DL-2-Aminoadipic acid | 2.75 | 3.51E-02 |
| 78 | M184T68 | 68 | 184.06 | 1.61 | 4-Pyridoxic acid | 2.95 | 1.88E-02 |
| 79 | M130T119 | 119 | 130.09 | 3.91 | 2-Pyrrolidineacetic acid | 2.99 | 2.70E-04 |
| 80 | M133T778 | 778 | 133.01 | 3.60 | L-Malic acid | 3.21 | 3.40E-02 |
| 81 | M428T286 | 286 | 428.37 | 2.50 | Stearoylcarnitine | 3.41 | 1.93E-05 |
| 82 | M241T772 | 772 | 241.01 | 1.08 | Alpha-D-galactose 1-phosphate | 3.48 | 8.04E-03 |
| 83 | M170T681 | 681 | 170.09 | 2.90 | 1-Methylhistidine | 3.78 | 1.60E-05 |
| 84 | M151T377 | 377 | 151.04 | 2.36 | p-Hydroxyphenylacetic acid | 4.09 | 5.36E-03 |
| 85 | M241T212_2 | 212 | 241.09 | 1.33 | Acadesine (Drug) | 4.10 | 5.67E-03 |
| 86 | M61T181 | 181 | 61.04 | 1.96 | Urea | 4.56 | 1.84E-05 |
| 87 | M209T552 | 552 | 209.07 | 1.33 | D-Ribose | 4.77 | 1.30E-02 |
| 88 | M211T637 | 637 | 211.08 | 1.72 | Ribitol | 4.79 | 9.37E-03 |
| 89 | M277T873 | 873 | 277.14 | 3.03 | L-Saccharopine | 4.96 | 1.35E-04 |
| 90 | M400T294 | 294 | 400.34 | 3.20 | L-Palmitoylcarnitine | 5.05 | 3.55E-06 |
| 91 | M131T745 | 745 | 131.04 | 1.22 | Glutaric acid | 5.09 | 6.56E-03 |
| 92 | M129T106_2 | 106 | 129.06 | 12.59 | ketoisocaproic acid | 5.20 | 2.75E-03 |
| 93 | M181T200_2 | 200 | 181.07 | 1.66 | Methoxyacetic acid | 5.94 | 8.58E-05 |
| 94 | M254T364 | 364 | 254.09 | 1.74 | Succinate | 6.20 | 8.40E-06 |
| 95 | M163T74_2 | 74 | 163.04 | 9.12 | Phenylpyruvate | 6.21 | 1.69E-03 |
| 96 | M114T312 | 312 | 114.06 | 3.54 | Creatinine | 6.22 | 1.84E-04 |
| 97 | M455T385 | 385 | 455.19 | 1.04 | Deoxycytidine | 8.59 | 1.86E-03 |
| 98 | M136T173 | 173 | 136.04 | 1.32 | Anthranilic acid (Vitamin L1) | 12.42 | 5.45E-04 |
| 99 | M132T659_2 | 659 | 132.08 | 12.68 | Creatine | 13.01 | 5.81E-06 |
m/z = mass-to-charge ratio; VIP = variable importance in the projection.
Fold change is expressed as the ratio of the PAD to the control (CON) group. For downregulated metabolites, the fold change was transformed to the corresponding negative value.
Fig. 3.
The pathway analysis by the Kyoto Encyclopedia of Genes and Genomes (KEGG) on differentially expressed metabolites in ducks with pantothenic acid deficiency (PAD) compared to control ducks.
3.6. Interaction network and pathway analysis
A total of 275 differential proteins and 99 differential metabolites affected by PAD were entered into the IMPaLA for canonical pathway analysis. There was significant enrichment of the interaction network of liver proteins and metabolites affected by PAD. The top 10 altered canonical pathways are listed in Fig. 4, whereas selected altered canonical pathways are listed in Table 8. Liver alterations induced by PAD were related to amino acid metabolism, glycolysis and gluconeogenesis, fatty acid beta oxidation, trans-sulfuration pathway, PPAR signaling pathway, TCA cycle, alanine and aspartate metabolism, glycogen synthesis and degradation, selenium micronutrient network, and oxidative phosphorylation.
Fig. 4.
Integrative analysis of differentially expressed proteins and metabolites in ducks with pantothenic acid deficiency (PAD) compared to control ducks.
Table 8.
Integrated pathway analysis of liver proteins and metabolites in ducks significantly affected by pantothenic acid deficiency (PAD).
| Pathway | Protein | Metabolite1 | q-value |
|---|---|---|---|
| Glycogen synthesis and degradation | PYGL (↓), PGM1 (↓), GYS2 (↓), UGP2 (↓) | Alpha-D-glucose 1-phosphate (↑), maltotriose (↓), maltopentaose (↓), cellobiose (↓), 3alpha-mannobiose (↓) | 2.45E-05 |
| Fatty acid beta oxidation | CPT1A (↑), CPT2 (↑), ACADS (↑), ACADM (↑), ACADL (↑), ACAT1 (↑), ACSL1 (↑), ACSL5 (↑), HADHA(↑), HADHB (↑), ECHS1 (↑), ACAA2 (↑), ALDH9A1 (↑), ALDH7A1 (↑), CRAT (↑), GK (↑) | L-palmitoylcarnitine (↑), L-carnitine (↑), stearoylcarnitine (↑) | 3.25E-15 |
| Oxidative phosphorylation | NDUFS1 (↑), NDUFB5 (↑), NDUFV1 (↑), NDUFA4 (↑), NDUFS3 (↑), UQCRFS1 (↑), UQCRC1 (↑), UQCRC2 (↑), COX2 (↑), ATP5F1B (↑), ATP5H (↑), ATP5C1 (↑), ATP5A1 (↑), ATP6V1B2 (↑), SLC25A5 (↑) | Succinate (↑) | 1.82E-04 |
| TCA cycle | ACO1 (↑), FH (↑), IDH1 (↑), MDH2 (↑) | Succinate (↑), L-malic acid (↑), argininosuccinic acid (↑) |
1.25E-09 |
PYGL = alpha-1,4 glucan phosphorylase; PGM1 = phosphoglucomutase 1; GYS2 = glycogen synthase; UGP2 = UTP-glucose-1-phosphate uridylyltransferase; CPT1A = carnitine palmitoyltransferase 1A; CPT2 = carnitine palmitoyltransferase 2; ACADS = acyl-CoA dehydrogenase medium chain; ACADM = acyl-CoA dehydrogenase medium chain; ACADL = acyl-CoA dehydrogenase long chain; ACAT1 = acetyl-CoA acetyltransferase 1; ACSL1 = acyl-CoA synthetase long chain family member 1; ACSL5 = acyl-CoA synthetase long chain family member 5; HADHA = hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha; HADHB = hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta; ECHS1 = enoyl-CoA hydratase, short chain 1; ACAA2 = 3-ketoacyl-CoA thiolase, mitochondrial; ALDH9A1 = aldehyde dehydrogenase 9 family member A1; ALDH7A1 = aldehyde dehydrogenase 7 family member A1; CRAT = carnitine O-acetyltransferase; GK = glycerol kinase; NDUFS1 = NADH:ubiquinone oxidoreductase core subunit S1; NDUFB5 = NADH:ubiquinone oxidoreductase subunit B5; NDUFV1 = NADH:ubiquinone oxidoreductase core subunit V1; NDUFA4 = NDUFA4 mitochondrial complex associated; NDUFS3 = NADH:ubiquinone oxidoreductase core subunit S3; UQCRFS1 = cytochrome b-c1 complex subunit Rieske, mitochondrial; UQCRC1 = ubiquinol-cytochrome c reductase core protein 1; UQCRC2 = ubiquinol-cytochrome c reductase core protein 2; COX2 = cytochrome c oxidase subunit 2; ATP5F1B = ATP synthase subunit beta, mitochondrial; ATP5H = ATP synthase subunit d, mitochondrial; ATP5C1 = ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; ATP5A1 = ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle; ATP6V1B2 = ATPase H+ transporting V1 subunit B2; SLC25A5 = solute carrier family 25 member 5; ACO1 = aconitase 1; FH = fumarate hydratase; IDH1 = isocitrate dehydrogenase; MDH2 = malate dehydrogenase 2.
Arrows indicates protein or metabolite down- (↓) or up-regulation (↑).
3.7. Western blot analyses
To validate the iTRAQ results, the abundance of PGM1, ME1, IDH1, and MDH2 were analyzed by Western blot. Compared to the CON group, the protein expressions of PGM1 and ME1 were decreased in the PAD group (Fig. 5A and B), while the protein expressions of IDH1 and MDH2 were increased (Fig. 5C and D), which were consistent with the iTRAQ results.
Fig. 5.
Western blot analysis of phosphoglucomutase 1 (PGM1; A), malic enzyme 1 (ME1; B), isocitrate dehydrogenase (IDH1; C), and malate dehydrogenase 2 (MDH2; D) protein expression of liver tissue of ducks in the pantothenic acid deficient (PAD) and Control (CON) groups. Loading control, vinculin, was used to normalize the levels of PGM1, ME1, IDH1, and MDH2. Representative Western blots are shown. Values are means with their standard errors. a, b Mean values with unlike letters were significantly different (P < 0.05). Data were analyzed by the Student's t-test.
4. Discussion
Retarded growth, high mortality, and fasting hypoglycemia occurred in PAD ducks in the present study (Tang et al., 2021), consistent with previous reports (Arnrich et al., 1956a, 1956b; Hurley and Morgan, 1952; Schultz et al., 1952; Winters et al., 1952). Furthermore, poor pantothenic acid status in PAD ducks was confirmed by a marked reduction of liver pantothenic acid contents, as tissue pantothenic acid is a useful biomarker for pantothenic acid status (Lin et al., 2012; Qian et al., 2015; Shiau and Hsu, 1999). A severe PAD animal model was successfully established. Compared to the CON group, average daily feed intake was decreased by 60% in PAD. The effects in the present study may be directly affected by PAD, or attributed to the reduced feed intake caused by PAD, which requires further investigation.
In the present study, PAD caused liver damage, as indicated by elevated plasma AST activity and relative liver weight. Liver damage in the PAD group may have been due to abnormal lipid metabolism indicated by reduced liver TG and free fatty acids content, as well as impaired glucose metabolism indicated by increased glycogen. The present study was apparently the first to use a proteomic and metabolomic approach to investigate liver damage and growth depression induced by PAD. Based on liver proteomics and metabolomics profiles, PAD mainly affected glycogen synthesis and degradation, glycolysis and gluconeogenesis, TCA cycle, PPAR signaling pathway, fatty acid beta oxidation, and oxidative phosphorylation.
Pantothenic acid is involved in carbohydrate metabolism, and it has been speculated that pantothenic acid is part of a glucose carrier system (Huan and Hung, 1972). Deficiency of this vitamin results in abnormal glucose metabolism, with low fasting blood glucose concentrations and increased sensitivity to insulin in rats and dogs (Arnrich et al., 1956a, 1956b; Hurley and Morgan, 1952; Schultz et al., 1952; Winters et al., 1952). In this study, PAD caused fasting hypoglycemia (Tang et al., 2021) and elevated liver glycogen in ducks, which is in agreement with previous studies (Arnrich et al., 1956a, 1956b; Hurley and Morgan, 1952; Schultz et al., 1952; Winters et al., 1952). In the PAD ducks, there was downregulation of 2 proteins involved in glycogenolysis and 2 others involved in glycogenesis. Whereas, PYGL catalyzes the cleavage of alpha-1,4-glucosidic bonds to release glucose-1-phosphate from liver glycogen stores, and PGM1 catalyzes the conversion of glucose 1-phosphate and glucose 6-phosphate. The reduction of PGM1 expression in PAD ducks was consistent with increased alpha-D-glucose 1-phosphate. Decreased protein expression of PYGL and PGM1 implied that glycogenolysis was impaired in response to PAD (Fig. 6), accounting for hypoglycemia. Similarly, in glycogen storage disease type VI (also known as Hers disease) with mutations in PYGL that inhibited conversion of glycogen to glucose, there was moderate hypoglycemia, liver damage and inflammation (Luo et al., 2020; Wilson et al., 2019), whereas mutations in PGM1 cause glycogen storage disease type XIV (Voermans et al., 2017). Furthermore, markedly decreased concentrations of maltotriose, maltopentaose, cellobiose, and 3-alpha-mannobiose in the liver of PAD ducks in the present study also supported this implication.
Fig. 6.
Disturbed metabolic pathways in liver tissue caused by pantothenic acid deficiency (PAD) based on proteomics and metabolomics profiles. Regulation is color and arrow coded in which red arrow ( ) stands for upregulated, blue arrow (
) stands for upregulated, blue arrow ( ) for downregulated in PAD group compared to control group. PYGL = alpha-1,4 glucan phosphorylase; PGM1 = phosphoglucomutase 1; FADH2 = Flavin adenine dinucleotide; TCA = tricarboxylic acid.
) for downregulated in PAD group compared to control group. PYGL = alpha-1,4 glucan phosphorylase; PGM1 = phosphoglucomutase 1; FADH2 = Flavin adenine dinucleotide; TCA = tricarboxylic acid.
In addition, PAD downregulated 2 proteins involved in glycogenesis, GYS2 and UGP2. As a key enzyme in glycogenesis, GYS2 converts glucose into glycogen. Mutations in the GYS2 gene in children are associated with hypoglycemia due to glycogen storage disease type 0 (Orho et al., 1998). In addition, UGP2 is also involved in glycogenesis, specifically synthesis of UDP-glucose from glucose-1-phosphate and UTP (Hu et al., 2020). Therefore, downregulation of these proteins would have impaired glycogenesis in response to PAD.
Eighteen proteins in glycolysis and gluconeogenesis pathways were differentially expressed after PAD, making it the largest category of identified proteins. Of these, 2 proteins were enhanced and 16 proteins were diminished. The fact that 16 of 18 proteins were downregulated in the PAD group implied hepatic glycolysis was impaired (Fig. 6), probably followed by decreased glycogenolysis. This implication was supported by reduced liver lactate level.
Pantothenic acid is involved in fatty acid synthesis and degradation. Pantothenic acid coenzymes carry the acids as acyl groups through repetitive synthetic or degradative cycles. Dietary PAD disrupts lipid metabolism (Lin et al., 2012; Qian et al., 2015; Shiau and Hsu, 1999; Shibata et al., 2013; Wang et al., 2016; Wen et al., 2009; Wittwer et al., 1990), including increased serum triglyceride in rats (Wittwer et al., 1990) and geese (Wang et al., 2016) and decreased serum HDL-C in geese (Wang et al., 2016). In the present study, PAD elevated plasma TG concentrations but decreased HDL-C and LDL-C in ducks, which was consistent with previous findings (Wang et al., 2016; Wittwer et al., 1990). In this study, PAD reduced liver TG, free fatty acids, total fatty acid, and unsaturated fatty acid of ducks; whereas, it caused liver fat accumulation in rats (Shibata et al., 2013), fish (Lin et al., 2012; Wen et al., 2009) and shrimp (Shiau and Hsu, 1999), and decreased the expression of various genes involved in liver fatty acid synthesis (Qian et al., 2015). Perhaps these discrepancies were due to different species and PAD status. According to the proteomics analysis, PAD upregulated 16 proteins involved in fatty acid beta oxidation in the present study. In accordance with these findings, PAD increased hepatic concentrations of L-palmitoylcarnitine, L-carnitine, and stearoylcarnitine. Based on these changes, we inferred that fatty acid beta oxidation was activated by PAD, consistent with decreased liver TG, free fatty acids, and TFA content (Fig. 6).
Simultaneously, dietary PAD downregulated 10 proteins involved in the PPAR signaling pathway, such as SCD1 and FADS2, implying suppression of this signaling pathway. Of these, SCD1 is a rate-limiting enzyme to produce monounsaturated fatty acid oleic acid from the saturated fatty acid stearic acid, which is responsible for forming a double bond in stearoyl-CoA (Paton and Ntambi, 2009). In addition, FADS2 is involved in biosynthesis of highly unsaturated fatty acids from the essential PUFA linoleic acid (18:2n-6) and alpha-linolenic acid (18:3n-3) precursors, acting as a fatty acyl-CoA desaturase that introduces a cis double bond at carbon 6 of the fatty acyl chain (Stoffel et al., 2008). Decreased protein expressions of SCD1 and FADS2 due to PAD may provide a possible explanation for the observed reductions in C18:1n9c, C20:1, C20:2, C20:3n6, UFA, and MUFA.
Based on proteome and metabolome analyses, dietary PAD upregulated 4 proteins involved in the TCA cycle, as well as hepatic concentrations of succinate, L-malic acid, and argininosuccinic acid. Based on enhanced expression of all these proteins and elevated metabolites, we inferred that the TCA cycle was activated by PAD (Fig. 6). Simultaneously, dietary PAD upregulated 15 proteins involved in oxidative phosphorylation. The 5 proteins, NDUFS1, NDUFB5, NDUFV1, NDUFA4, NDUFS3, are subunits of complex I, with a direct role in complex I assembly (Lazarou et al., 2007; Stroud et al., 2016). In addition, ATP5F1B, ATP5H, ATP5C1, ATP5A1, ATP6V1B2 are subunits of complex V, with a direct role in complex V assembly (Brüggemann et al., 2017; Chinopoulos, 2017; Rönn et al., 2009). Upregulated expression of proteins involved in the oxidative phosphorylation process, including complex I and complex V, indicated that mitochondrial oxidative phosphorylation was promoted by PAD (Fig. 6), which was supported by the markedly enhanced liver succinate concentration.
Together, our liver proteomic and metabolomic analyses revealed glycogen synthesis and degradation, glycogenolysis and glycogenesis processes were impaired in PAD ducks, which probably resulted in fasting hypoglycemia, insufficient ATP production in the liver, and subsequent growth retardation. In contrast, PAD upregulated fatty acid oxidation, the TCA cycle, and oxidative phosphorylation processes in the liver to produce ATP, presumably as compensatory mechanisms. The previous observation that hypoglycemia induced activation of fatty acid oxidation in endothelial cells supported this inference (Kajihara et al., 2017; Yoshinaga et al., 2021). Furthermore, this explanation was supported by findings in fasted rats that glucose utilization was depressed, whereas liver fatty acid beta oxidation was stimulated to meet energy needs (Berry et al., 1993; Hue et al., 2009). During starvation, the glucose-fatty acid cycle integrates responses of muscle, white adipose tissue, and liver to promote a shift from carbohydrate to fat oxidation and maintain glucose homeostasis (Perry et al., 2018). The association of cycling with fatty acid oxidation from glucose provides a potential mechanism for hepatic thermogenesis, and may account for the oxidation of fatty acid by liver in the absence of overt ATP demands (Berry et al., 1983; Debeer et al., 1974). These findings add to our understanding of mechanisms underlying PAD-induced metabolic disorders.
5. Conclusions
Dietary PAD caused fasting hypoglycemia, liver damage, elevated liver glycogen, and decreased liver TG or TFA in ducks. Based on proteomic and metabolomic analyses, PAD downregulated proteins and metabolites involved in glycogen synthesis and degradation, glycolysis and gluconeogenesis, indicating these processes were impaired, which probably lead to fasting hypoglycemia, insufficient hepatic ATP production, and growth retardation. In contrast, PAD upregulated proteins and metabolites involved in fatty acid oxidation, the TCA cycle, and oxidative phosphorylation processes in liver, presumably as compensatory mechanisms to produce ATP.
Author contributions
Jing Tang: data curation, writing–original draft preparation. Yongbao Wu: formal analysis. Bo Zhang: investigation. Suyun Liang: conceptualization. Zhanbao Guo: investigation. Jian Hu: methodology. Zhengkui Zhou: methodology. Ming Xie: resources. Shuisheng Hou: supervision, writing–review & editing.
Data availability
All data are shown in supplementary materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/archive/) partner repository with the data set identifier PXD026607.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This work was supported by the earmarked fund for China Agricultural Research System (CARS-42), the science and technology innovation project of Chinese Academy of Agricultural Sciences (CXGC-IAS-09), and Taishan Industry Leadership Talent Project of Shandong province in China (TSCY20190108). We thank Prof. John Kastelic for his valuable suggestions on the manuscript. We thank the members of our laboratory for their assistance during the animal experiment and sample collection. We thank Wuhan GeneCreate Biological Engineering Co., Ltd. For performing the mass spectrometric analysis.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2022.03.008.
Appendix. Supplementary data
The following is the Supplementary data to this article:
References
- Arnrich L., Hurley L.S., Forker B.R., Morgan A.F. Response to stress by riboflavin-deficient and pantothenic acid-deficient dogs. Am J Physiol. 1956;184:515–520. doi: 10.1152/ajplegacy.1956.184.3.515. [DOI] [PubMed] [Google Scholar]
- Arnrich L., Nelson M.R., Gram M.R., Morgan A.F. Effect of adrenal hormones on carbohydrate metabolism in riboflavin and pantothenic acid-deficient dogs. Am J Physiol. 1956;186:427–434. doi: 10.1152/ajplegacy.1956.186.3.427. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerrnfeind J.C., Norris L.C., Heuser G.F. The pantothenic acid requirement of chicks. Poult Sci. 1942;21:142–146. [Google Scholar]
- Berry M.N., Clark D.G., Grivell A.R., Wallace P.G. The calorigenic nature of hepatic ketogenesis: an explanation for the stimulation of respiration induced by fatty acid substrates. Eur J Biochem. 1983;131:205–214. doi: 10.1111/j.1432-1033.1983.tb07251.x. [DOI] [PubMed] [Google Scholar]
- Berry M.N., Phillips J.W., Henly D.C., Clark D.G. Effects of fatty acid oxidation on glucose utilization by isolated hepatocytes. FEBS. 1993;319:26–30. doi: 10.1016/0014-5793(93)80030-x. [DOI] [PubMed] [Google Scholar]
- Brüggemann M., Gromes A., Poss M., Schmidt D., Klümper N., Tolkach Y., Dietrich D., Kristiansen G., Müller S.C., Ellinger J. Systematic analysis of the expression of the mitochondrial ATP synthase (complex V) subunits in clear cell renal cell carcinoma. Transl Oncol. 2017;10:661–668. doi: 10.1016/j.tranon.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinopoulos C. ATP synthase complex and the mitochondrial permeability transition pore: poles of attraction. EMBO Rep. 2017;18:1041–1042. doi: 10.15252/embr.201744412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeer L.J., Mannaerts G., De Schepper P.J. Effects of octanoate and oleate on energy metabolism in the perfused rat liver. Eur J Biochem. 1974;47:591–600. doi: 10.1111/j.1432-1033.1974.tb03730.x. [DOI] [PubMed] [Google Scholar]
- Gershoff S.N., Gottlieb L.S. Pantothenic acid deficiency in cats. J Nutr. 1964;82:135–138. doi: 10.1093/jn/82.1.135. [DOI] [PubMed] [Google Scholar]
- Hegsted D.M., Perry R.L. Nutritional studies with the duck Ⅴ. Riboflavin and pantothenic acid requirements. J Nutr. 1948;35:411–417. doi: 10.1093/jn/35.4.411. [DOI] [PubMed] [Google Scholar]
- Hegsted D.M., Riggs T.R. The pantothenic acid requirements of chicks receiving a purified diet. J Nutr. 1949;37:361–367. doi: 10.1093/jn/37.3.361. [DOI] [PubMed] [Google Scholar]
- Hu Q.Y., Shen S., Li J.H., Liu L.W., Liu X., Zhang Y.Y., Zhou Y.J., Zhu W.W., Yu Y., Cui G.Y. Low UGP2 expression is associated with tumour progression and predicts poor prognosis in hepatocellular carcinoma. Dis Markers. 2020:3231273. doi: 10.1155/2020/3231273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan P.H., Hung L.V. Use of the double thiry-vella loop in the study of the effects of pantothenic acid on intestinal absorption of glucose. Br J Nutr. 1972;28:405–408. doi: 10.1079/bjn19720049. [DOI] [PubMed] [Google Scholar]
- Hue L., Taegtmeyer H. The randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297:E578–E591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley L.S., Morgan A.F. Carbohydrate metabolism and adrenal cortical function in the pantothenic acid-deficient rat. J Biol Chem. 1952;195:583–590. [PubMed] [Google Scholar]
- Jukes T.H. The pantothenic acid requirement of the chick. J Biol Chem. 1939;129:225–231. [Google Scholar]
- Kajihara N., Kukidome D., Sada K., Motoshima H., Furukawa N., Matsumura T., Nishikawa T., Araki E. Low glucose induces mitochondrial reactive oxygen species via fatty acid oxidation in bovine aortic endothelial cells. J Diabetes Invest. 2017;8:750–761. doi: 10.1111/jdi.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzer F.H., Williams D. The pantothenic acid requirement of poults for early growth. Poult Sci. 1948;27:518–523. [Google Scholar]
- Lazarou M., McKenzie M., Ohtake A., Thorburn D.R., Ryan M.T. Analysis of the assembly profiles for mitochondrial-and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepkovsky S., Bird F.H., Kratzer F.H., Asmundson V.S. The comparative requirements of chicks and Turkey poults for pantothenic acid. Poult Sci. 1945;24:335–339. [Google Scholar]
- Lin Y.H., Lin H.Y., Shiau S.Y. Estimation of dietary pantothenic acid requirement of grouper, Epinephelus malabaricus according to physiological and biochemical parameters. Aquaculture. 2012;324:92–96. [Google Scholar]
- Lu B., Ren Y., Huang B., Liao W., Cai Z., Tie X. Simultaneous determination of four water-soluble vitamins in fortified infant foods by ultra-performance liquid chromatography coupled with triple quadrupole mass spectrometry. J Chromatogr Sci. 2008;46:225–232. doi: 10.1093/chromsci/46.3.225. [DOI] [PubMed] [Google Scholar]
- Luo X.M., Hu J.C., Gao X.R., Fan Y.J., Sun Y., Gu X.F., Qiu W.J. Novel PYGL mutations in Chinese children leading to glycogen storage disease type VI: two case reports. BMC Med Genet. 2020;21:74. doi: 10.1186/s12881-020-01010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.W., Rucker R.B. In: Present Knowledge in Nutrition. 10th ed. Erdman J.W., Macdonald I.A., Zeisel S.H., editors. Wiley-Blackwell; Washington, DC, USA: 2012. Pantothenic acid; pp. 375–390. [Google Scholar]
- NRC . 9th ed. National Academies Press; Washington, DC: 1994. Nutrient Requirements of Poultry. [Google Scholar]
- Ministry of Agriculture of China . Nutrient Requirements of Meat-type Ducks of China. China Agriculture Press; Beijing, China: 2012. [Google Scholar]
- Nelson R.A. Intestinal transport, coenzyme A, and colitisin pantothenic acid deficiency. Am J Clin Nutr. 1968;21:495–501. doi: 10.1093/ajcn/21.5.495. [DOI] [PubMed] [Google Scholar]
- Orho M., Bosshard N.U., Buist N.R.M., Gitzelmann R., Aynsley-Green A., Blümel P., Gannon M.C., Nuttall F.Q., Groop L.C. Mutations in the liver glycogen synthase gene in children with hypoglycemia due to glycogen storage disease type 0. J Clin Invest. 1998;102:507–515. doi: 10.1172/JCI2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton C.M., Ntambi J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009;297:E28–E37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Wang Y., Cline G.W., Rabin-Court A., Song J.D., Dufour S., Zhang X.M., Petersen K.F., Shulman G.I. Leptin mediates a glucose-fatty acid cycle to maintain glucose homeostasis in starvation. Cell. 2018;172:234–248. doi: 10.1016/j.cell.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Li X.F., Zhang D.D., Cai D.S., Tian H.Y., Liu W.B. Effects of dietary pantothenic acid on growth, intestinal function, anti-oxidative status and fatty acids synthesis of juvenile blunt snout bream Megalobrama amblycephala. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönn T., Poulsen P., Tuomi T., Isomaa B., Groop L., Vaag A., Ling C. Genetic variation in ATP5O is associated with skeletal muscle ATP50 mRNA expression and glucose uptake in young twins. PLoS One. 2009;4:e4793. doi: 10.1371/journal.pone.0004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R.B., Winters R.W., Krehl W.A. The adrenal cortex of the pantothenic acid deficient rat: modification of the lesion by ACTH and cortisone treatment. Endocrinology. 1952;51:336–343. doi: 10.1210/endo-51-4-336. [DOI] [PubMed] [Google Scholar]
- Shiau S.Y., Hsu C.W. Dietary pantothenic acid requirement of juvenile grass shrimp, Penaeus monodon. J Nutr. 1999;129:718–721. doi: 10.1093/jn/129.3.718. [DOI] [PubMed] [Google Scholar]
- Shibata K., Fukuwatari T., Higashiyama S., Sugita C., Azumano I., Masaaki O. Pantothenic acid refeeding diminishes the liver, perinephrical fats, and plasma fats accumulated by pantothenic acid deficiency and/or ethanol consumption. Nutrition. 2013;29:796–801. doi: 10.1016/j.nut.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Smith C.M., Song W.O. Comparative nutrition of pantothenic acid. J Nutr Biochem. 1996;7:312–321. [Google Scholar]
- Stoffel W., Holz B., Jenke B., Binczek E., Gunter R.H., Kiss C., Karakesisoglou I., Thevis M., Weber A.A., Arnhold S., Addicks K. Delta 6-desaturase (FADS2) deficiency unveils the role of omega 3- and omega 6-polyunsaturated fatty acids. EMBO J. 2008;27:2281–2292. doi: 10.1038/emboj.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., Salim A., Ryan M.T. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- Tang J., Feng Y.L., Zhang B., Wu Y.B., Guo Z.B., Liang S.Y., Zhou Z.K., Xie M., Hou S.S. Severe pantothenic acid deficiency induces alterations in the intestinal mucosal proteome of starter Pekin ducks. BMC Genom. 2021;22:491. doi: 10.1186/s12864-021-07820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Zhang B., Liang S.Y., Wu Y.B., Feng Y.L., Guo Z.B., Xing G.N., Jiao J.L., Zhou Z.K., Xie M., Hou S.S. Effects of pantothenic acid on growth performance and antioxidant status of growing male white Pekin ducks. Poult Sci. 2020;99:4436–4441. doi: 10.1016/j.psj.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Zhang B., Xue M., Shi W.B., Wu Y.B., Feng Y.L., Huang W., Zhou Z.K., Xie M., Hou S.S. Pantothenic acid requirement of male White Pekin ducks from hatch to 21 days of age. Anim Feed Sci Tech. 2020;269:114637. [Google Scholar]
- Tang J., Hu J., Xue M., Guo Z., Xie M., Zhang B., Zhou Z., Huang W., Hou S. Maternal diet deficient in riboflavin induces embryonic death associated with alterations in the hepatic proteome of duck embryos. Nutr Metab. 2019;16:19. doi: 10.1186/s12986-019-0345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voermans N., Preisler N., Madsen K.L., Janssen M.C.H., Kusters B., Abu Bakar N., Conte F., Lamberti V.M.L., Nusman F., van Engelen B.G., van Scherpenzeel M., Vissing J., Lefeber D.J. PGM1 deficiency: substrate use during exercise and effect of treatment with galactose. Neuromuscul. Disord. 2017;27:370–376. doi: 10.1016/j.nmd.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Wang B., Zhang X., Yue B., Ge W., Zhang M., Ma C., Kong M. Effects of pantothenic acid on growth performance, slaughter performance, lipid metabolism, and antioxidant function of Wulong geese aged one to four weeks. Anim Nutr. 2016;2:312–317. doi: 10.1016/j.aninu.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z.P., Zhou X.Q., Feng L., Jiang J., Liu Y. Effect of dietary pantothenic acid supplement on growth, body composition and intestinal enzyme activities of juvenile Jian carp (Cyprinus carpio var. Jian) Aquacult Nutr. 2009;15:470–476. [Google Scholar]
- Wilson L.H., Cho J.H., Estrella A., Smyth J.A., Wu R., Chengsupanimit T., Brown L.M., Weinstein D.A., Lee Y.M. Liver Glycogen phosphorylase deficiency leads to profibrogenic phenotype in a murine model of glycogen storage disease type VI. Hepatol Commun. 2019;3:1544–1555. doi: 10.1002/hep4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters R.W., Schultz R.B., Krehl W.A. The adrenal cortex of the pantothenic acid-deficient rat: carbohydrate metabolism. Endocrinol. 1952;50:388–398. doi: 10.1210/endo-50-4-388. [DOI] [PubMed] [Google Scholar]
- Wittwer C.T., Beck S., Peterson M., Davidson R., Wilson D.E., Hansen R.G. Mild pantothenate deficiency in rats elevates serum triglyceride and free fatty acid levels. J Nutr. 1990;120:719–725. doi: 10.1093/jn/120.7.719. [DOI] [PubMed] [Google Scholar]
- Woollard D.C., Indyk H.E., Christiansen S.K. The analysis of pantothenic acid in milk and infant formulas by HPLC. Food Chem. 2000;69:201–208. [Google Scholar]
- Yoshinaga A., Kajihara N., Kukidome D., Motoshima H., Matsumura T., Nishikawa T., Araki E. Hypoglycemia induces mitochondrial reactive oxygen species production through increased fatty acid oxidation and promotes retinal vascular permeability in diabetic mice. Antioxid Redox Sign. 2021;34:1245–1259. doi: 10.1089/ars.2019.8008. [DOI] [PubMed] [Google Scholar]
- Youssef J.A., Song W.O., Badr M.Z. Mitochondrial, but not peroxisomal, β-oxidation of fatty acids is conserved in coenzyme A-deficient rat liver. Mol Cell Biochem. 1997;175:37–42. doi: 10.1023/a:1006877021617. [DOI] [PubMed] [Google Scholar]
- Zhang B., Tang J., Wu Y.B., Cao J.T., Xing G.N., Sun P.X., Huang W., Xie M., Hou S.S. Effects of riboflavin deficiency on the lipid metabolism of duck breeders and duck embryos. Poult Sci. 2021;100:101342. doi: 10.1016/j.psj.2021.101342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker T.F. Pantothenic acid deficiency and its effect on the integrity and functions of the intestines. Am J Clin Nutr. 1958;6:65–74. doi: 10.1093/ajcn/6.1.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are shown in supplementary materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/archive/) partner repository with the data set identifier PXD026607.