Abstract
Nicotinamide adenine dinucleotide (NAD+) is an enzyme cofactor, co-substrate, and redox factor in all living cells and is necessary for maintaining cell metabolism. It has been shown that appropriate supplementation of NAD+ precursors or inhibition of NAD+-depleting enzymes can promote mitochondrial oxidative phosphorylation and improve host energy utilization efficiency. In addition, increasing evidence indicates that the gut microbiota plays a pivotal role in host metabolism. Theoretically, there should be a close correlation among NAD+, gut microbiota, and host metabolism; however, the information is limited. In this review, we summarize the metabolic process of NAD+ and its impact on host metabolism, the link between gut microbiota and host metabolism, as well as the potential effects of NAD+ on microbial metabolism, providing a new perspective on the interaction between gut microbiota and host metabolism.
Keywords: NAD+, Gut microbiota, Host metabolism, Metabolic diseases
1. Introduction
The importance of nicotinamide adenine dinucleotide (NAD+) in metabolism was first examined in a study on pellagra (Sydenstricker, 1958). Accumulating evidence has proven that NAD+ is an indispensable enzyme cofactor, co-substrate, and redox factor in all living cells, and it is important for maintaining many types of intracellular biological metabolisms (Ruszkiewicz et al., 2020). NAD+ is reduced to NADH by accepting electrons and then provides reducing equivalents for the mitochondrial respiratory chain, thereby promoting oxidative phosphorylation to produce ATP (Cantó et al., 2015). NAD+ is thought to bind more than 500 proteins and participate in the regulation of almost all major biological processes (Hopp et al., 2019; Ansari and Raghava, 2010). NAD+ can also directly combine with RNA to protect it from cleavage by ribonucleases (Cahová et al., 2015). Moreover, NAD+ can be phosphorylated to form nicotinamide adenine dinucleotide phosphate (NADP+). NADP+ is a cofactor in anabolic metabolism, and its reduced form, NADPH, is a powerful reducing agent in anabolic reactions, such as fatty acid and nucleic acid synthesis, as well as in the maintenance of cell redox homeostasis (Ying, 2008; Xiao et al., 2018). As an essential cofactor for glutathione reductase and thioredoxin reductase, NADPH is necessary for the production of glutathione peroxidase and peroxidase-mediated peroxide removal (Handy and Loscalzo, 2017). Decreased NAD+ anabolism (or increased catabolism) causes a decrease in NADPH levels, thereby resulting in impaired cell redox balance, which interferes with mitochondrial functioning and genomic signaling, which in turn increases cell sensitivity to necrosis and apoptosis pathways (Braidy et al., 2019). Additionally, NAD+ can be used as a co-substrate for NAD+-dependent enzymes, and thus plays an important role in mediating protein deacetylation, ADP-ribosyl modification, and the intracellular calcium signaling pathway (Chang and Guarente, 2014; Gupte et al., 2017; Chini et al., 2018). Increased intracellular NAD+ levels have been found to have a beneficial effect on pellagra, aging, metabolic disorders, muscular dystrophy, heart disease, kidney dysfunction, and neurodegenerative diseases (Katsyuba and Auwerx, 2017).
In recent years, an increasing number of studies have focused on the gut microbiota, considering it as a ‘new virtual metabolic organ’ (O'Hara and Shanahan, 2006; Milosevic et al., 2019). Microbial flora ferment undigested food to produce nutrients and various metabolites that affect host homeostasis. Short-chain fatty acids (SCFA) are the main final product of the fermentation of indigestible carbohydrates by intestinal microorganisms, and they have been shown to play an important role in host glucose and lipid metabolism (Morrison and Preston, 2016). Intestinal bacteria also affect the portal circulation of amino acids entering the body (Lin et al., 2017). It has been proven that the intestinal flora not only have the ability to promote the de novo synthesis of essential amino acids, but also use some of the proteins that escape digestion in the small intestine as a substrate for colonic microbial fermentation. Thus, they may produce a variety of amino acid metabolites; such as indole-3-propionic acid and phenylethylamine, which are involved to amino acid homeostasis in the host (Lin et al., 2017; Liu et al., 2020). The intestinal microbial metabolism of choline or choline-containing compounds in the diet leads to the formation of trimethylamine, which is then converted into trimethylamine-N-oxide (TMAO) by the host liver flavin monooxygenase (Canyelles et al., 2018). TMAO can effectively reduce host bile acid pools by reducing bile acid synthesis and the activity of liver bile acid transporters (Canyelles et al., 2018). A meta-analysis has shown that there is a positive correlation between TMAO and the risk of death in adult populations (Farhangi, 2020). In addition, intestinal microbes can convert primary bile acids into secondary bile acids, which are essential for improving the efficiency of fat emulsification and enterohepatic circulation of bile acids (Winston and Theriot, 2020). In summary, existing studies have shown that gut microbial metabolism can significantly affect metabolic homeostasis in the host.
Although there are many studies on NAD+ and gut microbiota independently, few has focused on the effects of NAD+ on the gut microbiota. It remains unclear how NAD+ acts on the structure and metabolism of gut microbes. This article reviews the metabolic process of NAD+ and its impact on host metabolism, as well as the link between gut microbiota and host metabolism. Based on the gathered information, this article speculates how NAD+ affects gut microbiota, providing a new understanding of the interaction between host metabolism and gut microbiota.
2. Metabolic process of NAD+
In mammals, the total ratio of NAD+/NADH is usually estimated to be between 3 and 10, depending on the metabolic state of the cell (Palzer et al., 2018). NAD+ exists in the cell in either a free or bound state. The concentration of free NAD+ in various subcellular compartments has been determined to be 92 to 122 μM in the cytoplasm, 87 to 136 μM in the nucleus, and 191 to 275 μM in the mitochondria (Cohen, 2020).
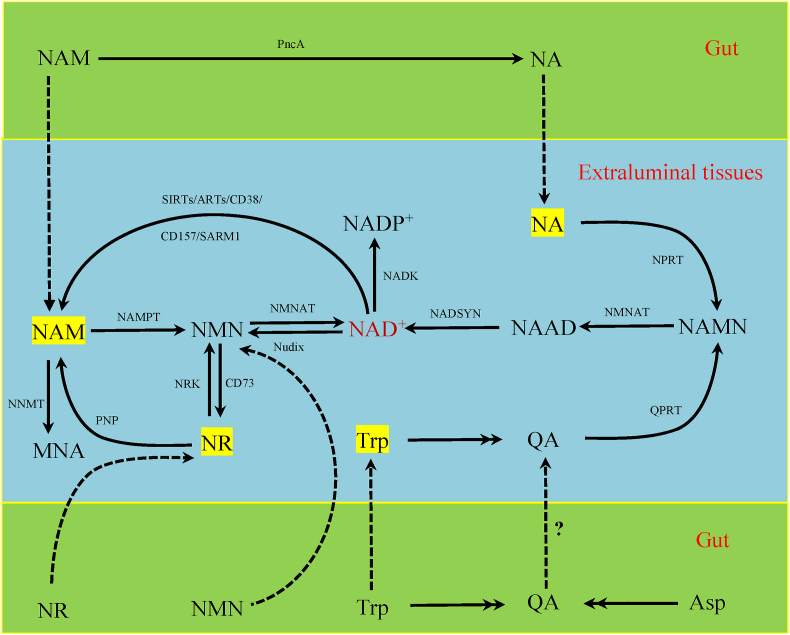
NAD+ is synthesized in four main ways in mammals: the de novo synthesis pathway, the salvage pathway, the nicotinamide ribose (NR) kinase pathway, and the Preiss-Handler pathway (Fig. 1). The NAD+ de novo synthesis pathway starts with tryptophan, which mainly occurs in the liver (Grant et al., 1999). The de novo synthesis pathway is regulated by a variety of enzymes, of which the combination of quinolinate phosphoribosyltransferase and indoleamine 2,3-dioxygenase or tryptophan 2,3-dioxygenase function to catalyze key reactions (Oxenkrug, 2013; Youn et al., 2016). Indoleamine 2,3-dioxygenase can be activated by pro-inflammatory mediators such as interferon, tumor necrosis factor, and lipopolysaccharide, while tryptophan 2,3-dioxygenase can be induced by stress hormones, including cortisol, estrogen, prolactin, and tryptophan (Oxenkrug, 2013). Quinolinate phosphoribosyltransferase catalyzes the conversion of quinolinic acid from tryptophan to produce nicotinic acid mononucleotide, which is subsequently converted to NAD+ (Youn et al., 2016). Most tissues use nicotinamide (NAM) to generate NAD+ through the salvage pathway in two steps; NAM is first converted to nicotinamide mononucleotide (NMN) under the catalysis of the key rate-limiting enzyme nicotinamide phosphoribosyltransferase (NAMPT), and then NMN is converted into NAD+ by nicotinamide mononucleotide adenylyltransferase (NMNAT) (Yang and Sauve, 2016). NAMPT exists in intracellular and extracellular subtypes, termed iNAMPT and eNAMPT, respectively (Hopp et al., 2019). There are three subtypes of NMNAT: NMNAT1 is located in the nucleus, NMNAT2 is located in the Golgi apparatus and neuronal axons, and NMNAT3 is located in the mitochondria (Hopp et al., 2019). NAD+ can also be generated from NR via the NR kinase pathway, with NMN as an intermediate (Bieganowski and Brenner, 2004). In the cell, NR can also be converted to NAM by purine nucleoside phosphorylase, which can then be converted into NAD+ via the salvage pathway (Mehmel et al., 2020). In addition, nicotinic acid (NA) synthesizes NAD+ through the Preiss-Handler pathway, and nicotinic acid mononucleotide is an important intermediate product in this synthesis (Preiss and Handler, 1958). The amount of NAD+ produced by 1 mg of NA requires the production of from 60 to 70 mg of tryptophan (Palzer et al., 2018), which suggests that using tryptophan to boost NAD+ production is inefficient.
Fig. 1.
NAD+ metabolic pathway in mammalian cells and gut microbes. ARTs = adenosine diphosphate-ribose transferases; NA = nicotinic acid; NAD = nicotinamide adenine dinucleotide; NADK = NAD kinase; NADP = nicotinamide adenine dinucleotide phosphate; NAM = nicotinamide; NAMN = nicotinic acid mononucleotide; NAMPT = nicotinamide phosphoribosyltransferase; NMN = nicotinamide mononucleotide; NMNAT = nicotinamide mononucleotide adenylyltransferase; NNMT = nicotinamide N-methyltransferase; NRK = nicotinamide ribose kinases; NR = nicotinamide ribose; NPRT = nicotinic acid phosphoribosyltransferase; MNA = 1-methylnicotinamide; PncA = nicotinamidase; PNP = purine nucleoside phosphorylase; QA = quinolinic acid; QPRT = quinolinic acid phosphoribosyltransferase; SIRTs = sirtuins; SARM1 = sterile alpha and Toll/interleukin-1 receptor motif-containing 1.
Microbial NAD+ metabolism has a number of differences compared to mammals. Some bacteria, such as Haemophilus influenzae, do not carry genes for the de novo pathway; whereas others, such as Cytophagahutchinsonii, have genes for the de novo pathway that are sourced from tryptophan (Gazzaniga et al., 2009). Some fungi, such as Candida glabrata, may also not have genes for the de novo pathway (Domergue et al., 2005). Both Escherichia coli and Saccharomyces cerevisiae encode homologous nicotinamidase, which can convert NAM to NA for use in Preiss-Handler rescue (Ghislain et al., 2002). Certain bacteria can also produce NAD+ from L-aspartate (Begley et al., 2001). These special reactions occurring in the gut are shown in Fig. 1.
Intracellular NAD+ is mainly consumed by 3 classes of enzymes: sirtuins (SIRTs), ADP-ribose transferases, and cyclic ADP-ribose (cADPR) synthases. SIRTs use NAD+ as a co-substrate to remove acetyl moieties from lysine on proteins, releasing NAM and O-acetyl-adenosine diphosphate-ribose (Chang and Guarente, 2014). Increasing evidence demonstrates that SIRTs not only are an important energy state sensor, but also protect cells from metabolic stress (Chang and Guarente, 2014). ADP-ribose (ADPR) transferases, along with NAD+ as a co-substrate, are able to transfer one (mono ADPR polymerases) or multiple (poly ADPR polymerases, PARP) ADPR moieties to the specific amino acid receptor site of the target protein or ribonucleotide (Gupte et al., 2017). PARPs contain 17 enzymes in humans; 16 of which catalyze the transfer of ADPR from NAD+ to macromolecular targets (proteins, DNA, and RNA) (Cohen, 2020). CD38 and CD157, the two main cADPR synthetases, can hydrolyze NAD+ to generate cADPR and NAM (Quarona et al., 2013). CD38 is a multifunctional protein that catalyzes the cleavage of the high-energy β-glycosidic bond between NAM and ribose (Chini et al., 2018). It can also act as an intracellular NAD hydrolase, which is involved in the production of the second messenger ADPR and cADPR of intracellular calcium signal transduction (Chini et al., 2018). CD157 is a homologous enzyme of CD38 and its ADP-ribosyl cyclase activity is much weaker than that of CD38 (Higashida et al., 2019).
Other enzymes consume NAD+. In neuronal axons, sterile alpha and Toll/interleukin-1 receptor motif-containing 1 is a negative regulator of the transcription program activated by Toll-like receptors, which have NAD+ glycohydrolase activity and can cleave NAD+ into ADPR, cADPR, and NAM (Essuman et al., 2017). Moreover, NAD+ can also be hydrolyzed into adenine ribonucleotides (AMP) and NMN by the Nudix hydrolase family (Bessman, 2019). In addition, NAD+ can be converted into NADH, which can then be catalyzed by NAD kinase to generate NADP+, which can be converted into NADPH (Ying, 2008).
Since mammalian cells are inefficient in directly absorbing NAD+ from dietary or microbial sources, it is necessary to convert NAD+ into NR, NMN, NAM, or NA in the gut. NAD+ that enters the intestines of animals through their diet can be converted into NAM and ADPR by CD38 (Durnin et al., 2020). ADPR is then degraded to AMP by extracellular pyrophosphatases, and AMP is finally cleaved into adenosine by 5′-nucleotidase (Durnin et al., 2020). Extracellular pyrophosphatases can also directly hydrolyze NAD+ into NMN and AMP. The extracellular nucleotidase activity of 5′-nucleotidase allows the hydrolysis of AMP and NMN, leading to the accumulation of adenosine and NR, respectively (Wilk et al., 2020). Moreover, NAM can be converted to NA by nicotinamidase in gut bacteria, thus rescuing dietary NAMPT inhibitor-induced toxicity (Shats et al., 2020). After these transformations, NAD+ is broken down into a variety of precursors that help the host adapt to complex environments.
In short, the intracellular NAD+ level is affected by its anabolism and catabolism, and NAMPT is a key factor affecting intracellular NAD+ level (Revollo et al., 2004; Yang et al., 2007). NAD+ precursors other than NAM can avoid NAMPT in the synthesis of NAD+, but the synthesized NAD+ will be converted into NAM after being metabolized. These endogenous NAM must re-generate NAD+ through the salvage pathway under the action of NAMPT, otherwise may accumulate to cause toxicity or deplete methyl groups to be permanently excreted, leading to various side effects.
3. Strategies to boost NAD+ and its effect on host metabolism
3.1. Nicotinamide ribose
As a precursor of NAD+, NR can strongly influence host metabolism by synthesizing it. Chronic NR supplementation increases NAD+ levels in some tissues, including the liver and muscle, but not in other tissue such as the brain or white adipose tissue, suggesting that NR metabolism is tissue specific (Cantó et al., 2012). It is generally believed that the optimal oral dose of NR for mice is 400 mg/kg per day (Trammell et al., 2016; Gariani et al., 2016; Crisol et al., 2018, 2020; Wang et al., 2018; Pham et al., 2019). Supplementing NR in mammalian cells and mouse tissues increases NAD+ levels and activates SIRT1 and SIRT3, which ultimately leads to enhanced oxidative metabolism and prevents metabolic abnormalities caused by a high-fat diet (HFD) (Cantó et al., 2012). Moreover, 5 w of NR supplementation increased NMNAT1 and NMNAT3 protein content in brown adipose tissue, promoted thermogenesis, and reduced fat mass accumulation, which may be linked to the increase in the expression of peroxisome proliferator receptor gamma coactivator alpha (PGC1α) mRNA and uncoupling protein 1 (Crisol et al., 2018). However, in another study, NR supplementation had no significant effect on NMNAT expression in mouse quadricep muscles (Cantó et al., 2012). In pre-diabetic and diabetic mouse models, supplementation with NR was accompanied by substantial resistance to weight gain and improvement in dyslipidemia, liver function, and glycemic control; however, NR did not normalize any of these metabolic parameters (Trammell et al., 2016).
NR supplementation is closely related to the promotion of mitochondrial function and is therefore linked to improved glucose and lipid metabolism. Oral administration of NR has been reported to increase the level of intramuscular NAD+ in C57BL/6J mice (Crisol et al., 2020). When combined with aerobic training, this increased level of NAD+ could promote the expression of proteins that make up the mitochondrial complex, such as mitochondrial cytochrome c oxidase subunit 1 (COX1) and ATP synthase subunit 5α and promote a slight increase in the number of type I fibers (Crisol et al., 2020). NR inclusion to a high-fat and high-sucrose diet enhanced mitochondrial COX and succinate dehydrogenase activities in the mouse liver, enhanced mitochondrial function, and mediated the reduction of hepatic steatosis by increasing β-oxidation and oxidative phosphorylation capacity (Gariani et al., 2016). Similarly, NR supplementation can promote the expression of mitochondrial transcription factor A, carnitine palmitoyltransferase 1β (CPT1B), uncoupling protein 2, and citrate synthase, thereby promoting mitochondrial biological functions and improving energetic efficiency (Pham et al., 2019). Furthermore, electron microscopy has shown that the mitochondria in the brown adipose tissue of NR-fed mice have more abundant ridges (Cantó et al., 2012). NR can also activate SIRT1/PGC1α/mitochondrial biosynthesis, which protects against ethanol-induced liver lipid accumulation and mitochondrial dysfunction, playing an important role in alcoholic fatty liver disease (Wang et al., 2018). In addition, NR supplements can reduce the level of reactive oxygen species in aging oocytes, reduce spindle abnormalities, increase mitochondrial membrane potential (ΔΨm), and decrease mitochondrial clustering (Yang et al., 2020).
Appropriate NR supplementation has beneficial effects; however, excessive NR may impair the normal metabolism of the host. Compared with mice fed a normal diet, mice fed with 9,000 mg of NR per kg of diet showed reduced metabolic flexibility, lower glucose clearance, and increased systemic insulin resistance (Shi et al., 2019). Moreover, excessive NR supplementation interferes with the energy and redox metabolism of healthy rats and impairs their physical performance, which might be attributed to the pleiotropic metabolism and redox properties of NAD+ and NADP+ (Kourtzidis et al., 2016, 2018).
3.2. Nicotinamide mononucleotide
As nicotinamide mononucleotide (NMN) can be converted into NAD+ in one step, it is an effective supplement. A recent study showed that NMN can be directly transported into cells by solute carrier family 12 member 8 (Grozio et al., 2019). Orally administered NMN can be quickly absorbed, effectively transported to the blood circulation, and immediately converted into NAD+ in the main metabolic tissues, causing liver NAD+ levels to increase steadily from 15 to 30 min after administration (Mills et al., 2016). In addition to being a precursor for NAD+ synthesis, NMN also inhibits the activity of PARP1 and CD38, which act as NAD+ glycohydrolases (Klimova and Kristian, 2019). The optimal oral dosage of NMN for mice is generally 300–500 mg/kg per day (Spinnler et al., 2013; Mills et al., 2016; Uddin et al., 2016, 2017; Xie et al., 2020a).
NMN plays an important role in maintaining host glucose and insulin homeostasis. In the offspring of obese female mice fed a HFD, supplementation with NMN enhanced the clearance of glucose from the circulation by increasing plasma insulin levels, and the levels of NAD+ and NADH in the liver were increased (Uddin et al., 2017). Spinnler et al. reported that NMN did not affect the viability of pancreatic islet β cells or cell apoptosis but could enhance insulin secretion under glucose stimulation (Spinnler et al., 2013). A study conducted in 2016 observed the metabolic effects of supplementation of NMN on mice which were obese due to a HFD, and found that NMN significantly improved glucose tolerance of obese mice, increased NAD+ levels in the liver and muscle, and increased the activity of citrate synthase in the liver (Uddin et al., 2016). Moreover, NMN administration restored normal levels of glucose, pyruvate, and acetyl-CoA in depressed mice (Xie et al., 2020a). NMN administration also caused a significant increase in serine/threonine kinase phosphorylation, thus improving liver insulin sensitivity of age-induced type 2 diabetic mice (Yoshino et al., 2011). NMN is reported to play a major role in different target tissues between men and women; specifically, NMN improves impaired glucose tolerance in diabetic men, but its effect is less evident in women (Yoshino et al., 2011).
The increase in NAD+ induced by NMN is closely related to fatty acid metabolism. Supplementation with NMN significantly increases NAD+ levels and mainly affects fatty acid metabolism in the liver of mice fed HFD through three aspects: increased fat catabolism by improving mitochondrial activity and β-oxidation, decreased fat anabolism by decreasing genes involved in fat synthesis and storage, and reduced fat import into the liver by decreasing the CD36 value (Uddin et al., 2017). Due to NMN treatment, chow- and HFD-fed offspring of obese female mouse had reduced liver lipid accumulation by 50% and 23%, respectively, accompanied by reduced liver genes involved in fat synthesis, transport, and uptake, as well as increased liver genes involved in fatty acid oxidation (Uddin et al., 2020). Transcriptome and metabolome analyses have shown that in depressed mice, NMN reduces the mRNA expression of genes involved in fatty acid synthesis, stimulates β-oxidation and glycolysis, and increases acetyl-CoA in the tricarboxylic acid (TCA) cycle (Xie et al., 2020a). Thus, NMN could improve mitochondrial energy metabolism in the hippocampus and liver of mice treated with corticosterone (Xie et al., 2020a).
NMN supplementation also affected mitochondrial function. In mice, NMN is quickly converted to NAD+. NMN administration in mice has been shown to increase the ratio of mitochondrial DNA (mtDNA)-encoded mitochondrial protein COX1 and mitochondrial protein ATP synthase subunit 5α or increase the nuclear DNA-encoded succinate dehydrogenase complex subunit B, suggesting that NMN induces two critically correlated mitochondrial alterations in skeletal muscle, mitochondrial protein imbalance, and mitochondrial oxidative metabolism (Mills et al., 2016). In a mouse model of forebrain ischemia, the administration of NMN not only prevented the depletion of mitochondrial NAD+ after ischemia, increased acetylation of mitochondrial proteins, and deficiency of blood-induced phosphorylation of dynamin-related protein 1, but it also inhibited the increase in superoxide dismutase 2 acetylation and reactive oxygen species production; thus reversing mitochondrial rupture after ischemia (Klimova et al., 2020). Davila et al. (2018) showed that isolated mitochondria synthesize NAD+ from NMN instead of NAM, and there is an undiscovered carrier in mammalian mitochondrial membranes that can directly import NAD+. Further research is required to elucidate the importance of NMN in mitochondria.
Importantly, without any obvious toxicity or harmful effects, the increase in NAD+ caused by NMN supplementation prevents age-related gene expression changes in key metabolic organs of mice, thereby inhibiting age-related weight gain, enhancing energy metabolism, promoting physical exercise, and improving insulin sensitivity and the plasma lipid profile (Mills et al., 2016). In contrast, inhibiting the synthesis of NAD+ by NMN would result in reduced mitochondrial oxidative phosphorylation transcripts, mtDNA content, and ATP levels (Gomes et al., 2013).
3.3. Nicotinamide
Nicotinamide supplementation has complex effects on the metabolism of animal cells. As a precursor of NAD+, it can produce a series of beneficial effects by promoting the biosynthesis of NAD+. After NAM was administered to rats fed a HFD at a rate of 100 mg/kg per day, NAMPT, NAD+, the NAD+/NADH ratio, SIRTs mRNA expression, and SIRT1 activity in rat liver cells increased in combination with an increase in peroxisome proliferator-activated receptor gamma, PGC1α, and mtDNA levels (Yang et al., 2014). Quantitative targeted metabolomics studies of the NAMPT-NAD-SIRT1 pathway have shown that NAM supplementation reduced liver NAMPT abundance and increased liver SIRT1 levels in normal diet-fed or HFD-fed mice, while hepatic NAD+ remained stable (Mitchell et al., 2018). Non-targeted metabolomics analysis has shown that NAM supplementation enhances liver glucose catabolism by promoting both glycolysis and glycogen storage (Mitchell et al., 2018). Additionally, NAM reduced the abundance of glucose 6 phosphate and glucose 1 phosphate while increasing the level of citric acid, an intermediate product of the TCA cycle, in mice fed a HFD (Mitchell et al., 2018). In diabetic mouse models, NAM administration did not affect body weight or blood sugar levels, but did rebalance protein anabolism and catabolism by inactivating the transforming growth factor beta 1/Smad2 signaling pathway, thereby improving muscle atrophy (Guo et al., 2019). Furthermore, NAM might regulate glucose metabolism in mice through SIRT1-PGC1α signaling pathways and regulate lipid metabolism through the SIRT1-liver X receptor alpha (Wan et al., 2019).
Alternatively, NAM is often used as an inhibitor of SIRTs, a class of NAD+ consumption enzymes that play an important role in the deacetylation of key proteins in cells (Zhang et al., 2015). Nicotinamide treatment has been reported to reduce the expression of SIRT1 mRNA and proteins in the skeletal muscles of mice, impairs skeletal muscle mitochondrial respiratory capacity and energy production, and induces glucose intolerance and skeletal muscle lipid accumulation (Qi et al., 2016). Furthermore, these changes were accompanied by reduced exogenous fatty acid oxidation and increased triglyceride esterification, coinciding with the production of ATP, the activity of mitochondrial complexes I and IV, and the decrease in mtDNA content (Qi et al., 2016). Accordingly, NAM treatment at a rate of 250 mg/kg per day for 28 days significantly reduced the expression of mitochondria and nuclear-encoded electron transfer chain mRNA expression in the mouse cerebral cortex (Naia et al., 2017). Moreover, NAM is a substrate of nicotinamide N-methyltransferase, which catalyzes the conversion of NAM and S-adenosylmethionine to 1-methylnicotinamide and S-adenosyl homocysteine (Komatsu et al., 2018). 1-Methylnicotinamide is subsequently irreversibly oxidized to N-methyl-2-pyridone-5-carboxamide or N-methyl-4-pyridone-5-carboxamide, which is excreted in urine (Li et al., 2013). Therefore, excessive NAM methylation affects the level of S-adenosylmethionine, which in turn leads to a decrease in the degree of liver DNA methylation, causing liver DNA damage and thereby impairing glucose tolerance and insulin sensitivity. As a result, excessive NAM has severe effects of liver toxicity.
Therefore, supplementation with NAM is beneficial when NAD+ is lacking due to certain pathological conditions, but excessive NAM supplementation is potentially toxic if NAD+ is sufficient to meet the needs of cells.
3.4. Nicotinic acid
NA has been widely used as a vitamin for many years. NA can boost NAD+ via the Preiss-Handler pathway. It has been shown that supplementing NA can increase NAD+ level in the body, thereby bringing about a series of beneficial effects (Shi et al., 2017; Pirinen et al., 2020). Currently, an increasing number of studies have focused on the therapeutic potential of NA.
NA is the first anti-dyslipidemic drug, as it has a significant impact on cell lipid metabolism. It has been reported that the levels of triglycerides, total cholesterol, and low-density lipoprotein cholesterol in mice are significantly reduced with feed supplemented with 0.5% NA, while the levels of high-density lipoprotein and high-density lipoprotein cholesterol are markedly increased (Yang et al., 2019). NA upregulates the expression of apolipoprotein M by increasing the expression of liver X receptor alpha, a new type of apolipoprotein related to high-density lipoprotein cholesterol, thereby improving cholesterol metabolism (Yang et al., 2019). NA has been shown to reduce hepatic triglyceride synthesis and subsequently cause very low-density lipoprotein/low-density lipoprotein secretion via direct and non-competitive inhibition of hepatocyte diacylglycerol acyltransferase 2 (Kamanna et al., 2013). In addition, NA-activated G protein-coupled receptor109A (GPR109A) reduces the absorption of sterols and fatty acids in the intestines of mice and reduces the de novo synthesis of liver fatty acids (Ye et al., 2019). A study in 2020 reported that NA inhibits de novo fat production to improve liver steatosis in C57BL/6 mice fed a HFD via the GPR109A-mediated protein kinase C-extracellular regulatory protein kinase 1/2-AMP-activated protein kinase (AMPK) signaling pathway (Ye et al., 2020). Moreover, 1 g of NA supplemented daily for 4 w dramatically increased the mRNA expression levels of mitochondrial fatty acid intake-related genes (CPT1B and solute carrier family 25 member 20), TCA cycle-related genes (succinate dehydrogenase), and mitochondrial respiratory chain-related genes (COX5A and COX6A1) in sheep skeletal muscle (Khan et al., 2013). A meta-analysis demonstrated that the administration of NA could dramatically improve the lipid metabolism in patients with type 2 diabetes mellitus, but it had little effect on glucose metabolism (Xiang et al., 2020). Furthermore, NA may induce miR-502-3p expression, which impairs insulin sensitivity in human adipocytes (Montastier et al., 2019). Studies have also shown that NA-activated GPR109A inhibits glucose-stimulated insulin secretion, which is associated with hyperglycemia (Chen et al., 2015a; Wang et al., 2016).
Supplementation with NA has beneficial effects in some metabolic diseases. In adult-onset mitochondrial myopathy, NA supplementation, which causes a significant increase in the concentration of NAD+ and its metabolites in the blood, can not only restore the plasma concentration of alanine (a biomarker of mitochondrial disease) and the branched-chain amino acids (valine and isoleucine) for mitochondrial catabolism, but also enhances mitochondrial biogenesis and respiratory chain activity by inhibiting the mechanistic target of the rapamycin signaling pathway and activating the peroxisome proliferator-activated receptor signaling pathway, thus improving the muscle strength of mitochondrial myopathy (Pirinen et al., 2020). A review article showed that NA might be effective in the treatment of non-alcoholic fatty liver disease (NAFLD), the mechanisms of which are oxidative stress reduction and diacylglycerol acyltransferase 2 inhibition (Kashyap et al., 2019). However, in B6129SF2/J mice with NAFLD, NA treatment reduced the expression of 4-hydroxyphenylpyruvate related to fatty acid oxidation and decreased the abundance of phosphocholine, which is essential for the production and secretion of very low-density lipoproteintriglycerides, thereby potentiating hepatic steatosis (Fang et al., 2020).
Specifically, the effects of NA on host metabolism can differ among hosts and be influenced by many factors. For example, there is a difference in the biosynthesis of unsaturated fatty acids in mice between acute and long-term NA treatment; specifically, n-6 polyunsaturated fatty acids prevail during acute NA treatment, while after prolonged NA treatment, n-3 polyunsaturated fatty acids prevail (Heemskerk et al., 2014a). According to a report, the gene encoding the cyclic AMP-degrading enzyme, phosphodiesterase 3B, was down-regulated in mice treated with 0.3% NA for 15 w, thus counteracting the inhibitory effect of short-term NA treatment on cyclic AMP production in adipocytes (Heemskerk et al., 2014b). Moreover, the effect of NA administration was related to genetic factors. It has been reported that NA had no impact on diet-induced NAFLD development in C57BL/6J mice but potentiated hepatic steatosis in HFD-fed B6129SF2/J mice (Fang et al., 2020). Additionally, NA usually causes liver toxicity at doses higher than 3 g/day, which may be caused by mitochondrial damage (Leung et al., 2018). The most obvious known side effect of NA administration is flushing, which is caused by the production of prostaglandins D2 and E2 by subcutaneous Langerhans cells through GPR109A (Kamanna and Kashyap, 2008).
Unlike NR, NMN, and NAM, NA can exert its effects through ways other than the NAD+ pathway, making it more difficult to attribute its effects. There is no doubt that NA can activate GPR109A and inhibit diacylglycerol acyltransferase 2. However, many studies have only focused on these factors and have ignored the beneficial effect of NA as an NAD+ booster, which was described in a 2019 review (Romani et al., 2019). Since boosting NAD+ can improve lipid metabolism and has a certain therapeutic effect on metabolic diseases, it is recommended that research conducted on NA should study both the role of NA itself and its impact on NAD+.
3.5. Others
When other precursors are insufficient to meet the body's NAD+ needs, the de novo synthesis pathway from tryptophan plays an important role in maintaining the level of NAD+ in the cell. The lack of key enzymes in the de novo synthesis pathway has been reported to cause congenital malformations and NAD+ deficiency in humans and mice (Shi et al., 2017). Genetic and pharmacological suppression of α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase — a master regulator located at the intersection of cetyl-CoA and NAD+ metabolism in the kynurenine pathway — enhanced de novo NAD+ synthesis and SIRT1 activity (Katsyuba et al., 2018). This ultimately enhances mitochondrial functions, as indicated by an increased mtDNA:nDNA ratio, citrate synthase activity, and oxidative phosphorylation gene expression at the transcript and protein levels (Palzer et al., 2018; Katsyuba et al., 2018). In contrast, the inhibition of key enzymes in the kynurenine-NAD+ pathway could lead to the transfer of excess kynurenine from the formation of NAD+ to the production of xanthurenic acid and other diabetic derivatives of kynurenine, ultimately inducing insulin resistance (Begley et al., 2001).
Since the massive activation of PARP will seriously deplete the level of NAD+ in the cell, inhibiting PARP can bring beneficial effects by increasing cellular NAD+ content. A study reported that an increase of NAD+ content and SIRT1 activity in brown adipose tissue and muscles of mice with PARP1 gene deletion resulted in higher mitochondrial content, increased energy consumption, and protection against metabolic diseases (Bai et al., 2011). PARP1 inhibition leads to increased levels of SIRT1 and PGC1α in the cardiac tissue of diabetic mice (Waldman et al., 2018). In alcoholic and non-alcoholic fatty liver mouse models, pharmacological and genetic inhibition of PARP1 can restore liver NAD+ content, reduce the decrease in SIRT1 activation, and beneficially affect liver mitochondrial damage, steatosis, and metabolic disorders (Mukhopadhyay et al., 2017).
As CD38 is one of the main NAD+-degrading enzymes in mammalian tissues, CD38 inhibition is also an effective way to increase NAD+ bioavailability, resulting in improved metabolism. The increase in CD38 expression in cells leads to mitochondrial metabolic dysfunction, while the mitochondria of knockout CD38 mice have higher NAD+ levels, increased mitochondrial membrane potential, and increased oxygen consumption (Camacho-Pereira et al., 2016). In line with this, another study found that CD38 inhibition increased NAD+ levels, activated SIRTs, AMPK, and PARPs, negatively regulated the mechanistic targets of rapamycin ribosomal protein S6 kinase and extracellular regulatory protein kinase, and attenuates telomere-related DNA damage (Tarragó et al., 2018).
Due to the low efficiency of the conversion of tryptophan to NAD+ and the certain physiological functions of PARPs and CD38, these three methods are not usually used as strategies to increase NAD+ content in the body. However, these treatment strategies may have unexpected performance when NAD+ deficiency is caused by certain conditions. In addition, the combined effect of various strategies for increasing NAD+ need to be studied.
4. Gut microbiota and host metabolism
4.1. Gut flora composition and structure
The gut microbiota is a collection of all microbes living in the intestines of animals, including; bacteria, archaea, viruses, and some single-celled eukaryotes (Salazar et al., 2017). The gut microbiota consists of 1013 to 1014 microorganisms whose genome (microbiome) has at least 100 times the genes of the human genome (Gill et al., 2006). The duodenum contains approximately 103 microbial cells per gram of content, while each gram of colon content contains about 1012 microbial cells (Gomes et al., 2018). Generally, the intestinal flora consists of four main phyla: Firmicutes, Bacteroides, Actinomyces, and Proteus (Qin et al., 2010; Jin et al., 2020). In adults, Bacteroides and Firmicutes constitute the majority of the intestinal flora, and the Bacteroides/Firmicutes ratio is regarded as a healthy indicator of the intestinal flora (Jin et al., 2020). Interestingly, it has been discovered that microbes also exist in the blood, likely due to their relationship with intestinal microbes, implying the complexity of the role of intestinal microbes (Castillo et al., 2019).
The composition and structure of tract microorganisms are affected by many factors, including genetics, host physiology, and environmental factors (Holscher, 2017). It has been shown that Christensenellaceae is a highly heritable bacterium, and environmental factors mainly affect the Bacteroides community (Goodrich et al., 2014). Compared to young adults, the elderly have lower levels of Firmicutes and a higher proportion of Proteobacteria (Salazar et al., 2017). The intestinal flora can respond quickly to changes in the diet. The colonic microbiota composition in mice fed a processed meat protein diet was significantly different from that in mice fed a casein or soy protein diet, and the former had a higher abundance of colonic microbiota (Xie et al., 2020b). Moreover, dietary fatty acids can regulate the structure of gut microbes by stimulating the secretion of bile acids from the host (Holscher et al., 2020).
There are two sequencing methods, 16S rDNA sequencing and metagenomic sequencing, which are commonly used to assess the composition of microorganisms and their relative abundances. Shotgun metagenomics can detect the entire genome of any organism, including known and unknown microorganisms, with higher sensitivity than either of the common sequencing methods (Jin et al., 2020). With the advancement of technology, the role of intestinal microbes has become clearer. Although there have been many studies on gut microbes, there is still no clear answer to the question of what composition of gut microbiota is the best.
4.2. Gut microbiota and nutrient metabolism
Dietary nutrients, including proteins, carbohydrates, and lipids, are digested in the digestive tract through complex interactions between the host and intestinal microbiota. Dietary nutrients provide the source of energy and amino acids for the host, as well as substrates for intestinal microbial fermentation; therefore, dietary nutrients are an important factor in gut microbiota composition and structure.
4.2.1. Gut microbiota and protein metabolism
Most of the dietary protein is digested in the small intestine but excessive intake of protein leads to microbial fermentation in the large intestines (He et al., 2015). Primary bacteria related to protein metabolism in the small intestine consist of Escherichia coli (E. coli), Klebsitella spp., Succinivibrio dextrinosolvens, Streptococcus spp., Mitsuokella spp., and Anaerovibrio lipolytica. These bacteria can secrete various proteases and peptidases and some of them can also directly metabolize amino acids (Dai et al., 2010). In the large intestine of monogastric animals, proteolytic activity has been mainly attributed to the genera of Bacteroides, Fusobacterium, Propionibacterium, Streptococcus, Lactobacillu, and Clostridium (Davila et al., 2013; Macfarlane et al., 1988). Bacteroides can secrete proteases near the brush border of absorptive cells. Furthermore, Butyrivibrio fibrisolvens, Prevotella ruminicola, Mitsuokella multiacidas, and Streptococcus bovis can secrete dipeptidyl peptidase for protein digestion and absorption (Ma et al., 2017). Excessive protein intake leads to an increase in the β-hemolytic enterotoxigenic strains of E. coli (Chen et al., 2015b). In contrast to high-protein diets, low-protein diets cause a shift in intestinal microbial composition toward higher counts of beneficial bacteria, which preferentially ferment carbohydrates (Opapeju et al., 2009).
4.2.2. Gut microbiota and energy metabolism
There is a complex interaction between gut microbiota and energy metabolism. The small intestines of most mammals cannot secret enzymes to digest complex polysaccharides; therefore, the degradation of fiber components, such as cellulose, depends mainly on the metabolic activities of microorganisms in the large intestine. In the colon, dietary fiber is fermented by gut microbiota, and the major metabolites of fiber are short-chain fatty acids (SCFAs) (Binder, 2011). High-carbohydrate and high-fiber diets increase the expression of butyrate-producing related genes in the gut microbiota, whereas HFD can reverse these positive effects at the transcriptional level (Turnbaugh et al., 2009; Wu et al., 2011; Qin et al., 2012; Koh et al., 2016; Zhao et al., 2018). The decrease of microbial SCFAs could promote the features of metabolic syndrome (Zhao et al., 2018). Gut microbial SCFAs modulate fat metabolism directly in host adipose tissue through the activation/inhibition of multiple receptor signaling pathways, but may also mediate host feeding behavior through G-protein coupled receptor 43 pathway; both in the nervous system and in the intestine (Xiong et al., 2004; Zaibi et al., 2010; Kimura et al., 2013; Chambers et al., 2015; Perry et al., 2016; Williams et al., 2016). High-fat diet-induced alterations of the gut microbiota is often accompanied by disruption of intestinal barrier function, which can lead to lipopolysaccharide infiltration (Peng et al., 2009). Lipopolysaccharide-induced activation of toll-like receptor 4 launches a signaling cascade to release many proinflammatory cytokines (Janssens and Beyaert, 2003; O'Neill et al., 2013). Previous studies have found that lipopolysaccharide affects host energy metabolism via endocannabinoid system, vagal nerve stimulation, and the circadian clock (Muccioli et al., 2010; Vaughn et al., 2017; Wang et al., 2017a).
4.3. Gut microbiota and host metabolic diseases
Obesity and its associated metabolic diseases are increasing in prevalence owing to decreases in physical activity levels and a shift to diets that include addictive and/or high-calorie foods. Currently, the hypothesis that the gut microbiota are one of the key modulators that improve metabolic diseases due to their close link to metabolism has been improved (Cani, 2017).
4.3.1. Gut flora and obesity
Obesity, a common metabolic disease, has been found to be closely related to intestinal microbes. Previous studies of animal and human models have shown that obesity is mainly related to changes in the relative abundances of Bacteroidetes and Firmicutes (Turnbaugh et al., 2006; Ley et al., 2006; Armougom et al., 2009). Other researchers have suggested that changes in the ratio of Firmicutes to Bacteroidetes should not be regarded as a general feature that distinguishes the intestinal flora of normal and obese individuals (Walters et al., 2014). Changes in the intestinal flora may lead to changes in the intestinal metabolites related to glycolysis, the TCA cycle, and homolactic fermentation regulation (Li et al., 2020).
Improving intestinal microbiota has a therapeutic effect on obesity. Allicin treatment significantly improved gut microbiota composition, induced the most significant alteration enrichment of Bifidobacterium and Lactobacillus, and reduced body weight gain and fat accumulation in HFD mice (Zhang et al., 2020). Importantly, transplantation of allicin-induced gut microbiota to HFD mice plays a remarkable role in decreasing adiposity, maintaining glucose homeostasis, and ameliorating hepatic steatosis (Zhang et al., 2020). This flora transplantation treatment also significantly suppressed the expression of lipogenesis-related genes (including fatty acid synthase, sterol regulatory factor binding protein 1, fatty acid binding protein 4, and acetyl-CoA carboxylase) in subcutaneous white adipose tissue and epididymal white adipose tissue, while significantly increasing the ratio of Bacteroidetes to Firmicutes, the relative abundance of the Bifidobacterium and SCFA-producing microbial Blautia, and the expression of genes related to fatty acid oxidation, including CPT, medium-chain acyl-CoA dehydrogenase, and peroxisome proliferator activated receptor alpha (Zhang et al., 2020). A HFD supplemented with resveratrol (200 mg/kg/day) decreased body fat and weight of Kunming mice as well as the relative abundance of Enterococcus faecalis (positively correlated with body weight) and increased Lactobacillus and Bifidobacterium (negatively correlated with body weight) (Zhao et al., 2017).
4.3.2. Gut flora and type 2 diabetes
Type 2 diabetes (T2D), which is caused by glucose metabolism disorders and insulin resistance, is also linked to intestinal microbes. Bifidobacterium, Bacteroides, Faecalibacterium, Akkermansia, and Roseburia are negatively correlated with T2D, while Ruminococcus, Fusobacterium, and Blautia are positively correlated with T2D (Gurung et al., 2020). In db/db mice (a T2D mouse model), the β diversity and relative abundance of gut bacteria were significantly different from those of healthy mice, and these differences were related to a significant increase in Verrucomicrobia and a significant decrease in Bacteroidaceae (Yu et al., 2019). It has also been shown that the level of Lactobacillus in patients with T2D is significantly higher than that of healthy individuals, whereas the incidence of Bifidobacterium is dramatically lower than that of healthy individuals (Sedighi et al., 2017). However, the abundance of Bifidobacterium spp. in the stool of patients with T2D is higher than that of healthy individuals (Adachi et al., 2019). This difference may be related to genetics and diet.
The intestinal flora can affect the metabolism of patients with T2D by producing a variety of metabolites, such as SCFA, bile acids, branched-chain amino acids, and trimethylamine (Vrieze et al., 2012; Pedersen et al., 2016; Adachi et al., 2019; Leylabadlo et al., 2020). Prevotellacopri and Bacteroides vulgatus is considered as the main species driving the link between branched-chain amino acids and insulin resistance (Pedersen et al., 2016). The number of extracellular vesicles derived from Pseudomonas panacis in HFD-fed mice increased, which blocked the insulin signaling pathway in skeletal muscle and adipose tissue (Choi et al., 2015). In addition, previous studies have shown that probiotic treatment can positively affect T2D by affecting gut microbes. The addition of Lactobacillus acidophilus can reshape the intestinal flora, increase the levels of SCFA-producing bacteria (Blautia, Roseburia, and Anaerotruncus), and reduce the relative abundance of gram-negative bacteria (such as Desulfovibrio and Alipites) (Yan et al., 2019). Furthermore, addition of L. acidophilus has been shown to downregulate the expression of glycogen synthase kinase 3β, fatty acid synthase, and sterol regulatory factor binding protein 1C, and upregulate expression of serine/threonine kinase, which is related to glucose and lipid metabolism (Yan et al., 2019). Similarly, treatment with 109 CFU of Lactobacillus casei can also reduce several symptoms of diabetes, including fasting blood sugar, postprandial blood sugar, glucose intolerance, and insulin resistance (Wang et al., 2017b).
4.3.3. Gut flora and nonalcoholic fatty liver disease
NAFLD is a disease characterized by excessive accumulation of fat in liver cells, is often accompanied by portal vein and lobular inflammation and liver cell damage and is closely related to changes in the intestinal flora (Brunt et al., 2015). Compared with subjects without NAFLD, those with NAFLD had reduced bacterial alpha diversity, showed higher Firmicutes to Bacteroidetes ratios, and had lower abundances of Bacteroidetes, Prevotella, Gemmiger, and Oscillospira and increased proportions of Bradyrhizobium, Anaerococcus, Peptoniphilus, Propionibacterium acnes, and Dorea (Del Chierico et al., 2017; Monga Monga Kravetz et al., 2020).
Previous studies have confirmed the key role of gut microbes in NAFLD progression. Germ-free mice do not develop NAFLD even if they are fed a HFD (Fei et al., 2020). However, NAFLD develops in germ-free mice fed with HFD when associated with Enterobacter cloacae B29, Escherichia coli PY102, and Klebsiella pneumoniae A7, three endotoxin-producing strains that exhibited overgrowth in the gut of morbidly obese volunteers with severe fatty liver disease (Fei et al., 2020). Gut microbiota may affect the progression of NAFLD in two ways. Intestinal endothelial barrier dysfunction caused by pathogenic bacteria causes the translocation of bacterial components and triggers innate immunity, which in turn leads to liver inflammation (Safari and Gérard, 2019). Furthermore, various metabolites produced by intestinal flora may affect the liver, thereby regulating NAFLD sensitivity (Safari and Gérard, 2019).
In addition, probiotic treatment can alleviate NAFLD by improving the composition of the intestinal microbes. Lactobacillus rhamnosus treatment may reduce the abundance of Clostridium and Streptococcus and increase the proportion of Bifidobacterium in the gut of mice with NAFLD (Wang et al., 2020). The levels of Bifidobacterium, Parabacteroides, Bacteroides, and Akkermansia are significantly increased in the intestines of NAFLD mice treated with Bifidobacterium adolescentis, which counteracts the increases of Proteobacteria and Clostridium, which are induced by high-fat and high-cholesterol diets (Wang et al., 2020). Probiotics induced a significant reduction in liver fat and induced changes in gut microbiota composition and a beneficial clinical response (i.e. the increase in Agathobaculum, Dorea, Blautia, and Ruminococcus was associated with steatosis reduction) (Ahn et al., 1999). Probiotics could improve liver pathology in NAFLD models, as indicated by reduced steatosis and inflammatory cell infiltration, as well as improved liver enzymes, hepatocyte ballooning, and liver fibrosis (Xue et al., 2017; Duseja et al., 2019). Compared with the NAFLD model group, probiotic-treated mice had reduced activities of alanine aminotransferase, aspartate aminotransferase, γ-glutamyl aminotransferase, and alkaline phosphatase (Xue et al., 2017).
5. The potential effects of NAD+ on gut microbiota
Since both promoting NAD+ and improving the intestinal microflora have certain therapeutic effects on host metabolic disorders, there may be a potential connection between NAD+ and the gut microbiota. Intestinal microbes can promote host NAD+ pools through the bacteria-enabled deamidated pathway and the de novo synthesis pathway (Magnúsdóttir et al., 2015; Shats et al., 2020). However, there is a lack of reports on the influence of NAD+ on gut microbiota. We suppose that NAD+ mainly affects intestinal microbes in two ways as follows.
As an enzyme cofactor, substrate, or redox factor necessary for microbial metabolism, NAD+ can directly affect the metabolism of intestinal microbes. NAD+ is an important component in the energy metabolism of microorganisms. Systematic genome evaluation has shown that most gut microbes have the ability to synthesize NAD+ (Magnúsdóttir et al., 2015). NAD+ is a cofactor for many key enzymes involved in energy metabolism, such as glyceraldehyde phosphate dehydrogenase, pyruvate dehydrogenase, 3-hydroxyacyl-CoA dehydrogenase, malate dehydrogenase, α-ketoglutarate dehydrogenase, and lactate dehydrogenase. As these enzymes are widely involved in the production of cellular ATP, it is speculated that boosting NAD+ can promote microbial energy production (Katsyuba et al., 2020). Furthermore, enzyme molecules with NAD+ as the substrate are essential for the survival and reproduction of microorganisms. Some bacteria have NAD+-dependent DNA ligases, which play key roles in DNA replication, repair, and recombination (Luo and Barany, 1996). In animal intestines, Bifidobacterium longum and Lactobacillus acidophilus can catalyze protein/histone deacetylation by using SIRT2, an NAD+-dependent enzyme, thus responding to oxidative stress (Guo et al., 2017). It has been reported that SIRT2 is a key component of yeast longevity (Lamming et al., 2004). Moreover, multiple thiamine genes in Saccharomyces cerevisiae are regulated by intracellular NAD+ concentration through NAD+-dependent histone deacetylases, Hst1 and SIRT2 (Li et al., 2010).
In addition, boosting NAD+ can indirectly affect gut microbiota by regulating the host's immune and metabolic statuses. NAD+ can activate SIRT1, which mediates the resistance of mice to Mycobacterium tuberculosis infection (Cheng et al., 2017). It has been shown that CD38-deficient mice are more susceptible to bacterial infections (Partida-Sánchez et al., 2001). Moreover, supplementation with NAM, which is the precursor of NAD+, has been shown to increase the lethality of the host against Staphylococcus aureus, thus restricting the infection of Staphylococcus aureus in mice (Kyme et al., 2012). Therefore, it is speculated that boosting NAD+ can prevent the colonization of pathogenic bacteria in the intestinal tract. As a key inhibitory neurotransmitter in the colon, β-NAD can also regulate the movement of the colon, which is important since the colon is a critical cite for intestinal microbial fermentation (Durnin et al., 2020). Furthermore, NAD+ can act on the gut microbiota by affecting the synthesis of host bile acids. The synthesis of cholesterol in the endoplasmic reticulum requires multiple steps of electronic input, and NADH and NADPH are used as electron sources (Porter, 2015). In the liver, cholesterol is converted into bile acids, which have long been recognized to promote the absorption, transportation, and metabolism of dietary fat and fat-soluble nutrients (Jin et al., 2019). Moreover, the bile acids secreted into the intestine can also be antimicrobial when they destroy the bacterial cell membrane, thus inhibiting the proliferation of bacteria (Kurdi et al., 2006; Inagaki et al., 2006; Jin et al., 2019). Conversely, obstruction of bile flow can lead to the proliferation of harmful bacteria in the intestines (Inagaki et al., 2006).
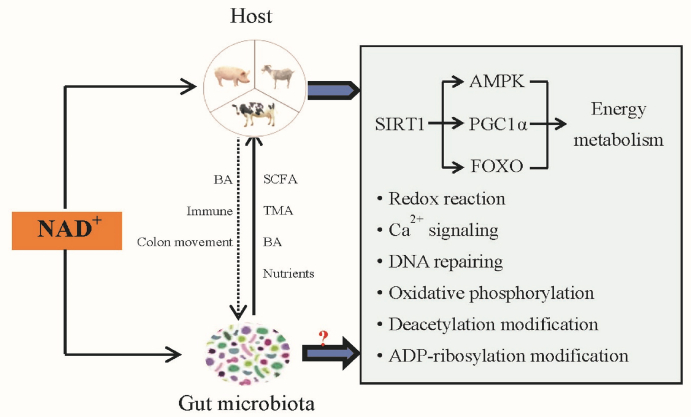
In general, boosting microbial NAD+ metabolism may help maintain the normal metabolism of microbes and boosting host NAD+ may promote beneficial intestinal bacteria and inhibit harmful bacteria. The effects of boosting NAD+ on host metabolism and its possible impact on gut microbiota are shown in Fig. 2.
Fig. 2.
The roles of NAD+ on the host metabolism and its speculated regulatory effects on gut microbiota. AMPK = AMP-activated protein kinase; BA = bile acids; FOXO = forkhead box O; NAD+ = nicotinamide adenine dinucleotide; PGC1α = peroxisome proliferator receptor gamma coactivator alpha; SIRT1 = sirtuin1; SCFA = short-chain fatty acids; TMA = trimethylamine.
6. Conclusions and perspectives
In conclusion, NAD+ participates in almost all cellular metabolic processes, and it is an essential factor for host metabolism and has a potentially regulatory function on gut microbiota, which is important for improving host nutrient metabolism and metabolic disorders. Further research is needed to prove the metabolic function of NAD+ in intestinal microbes and its related mechanisms. Additionally, the optimal selection of NAD+-boosting strategies and the optimal combination of various methods under different metabolic states of the host are topics that remain to be explored.
Author contributions
Zhongxiang Ren drafted the manuscript. Yetong Xu, Tiejun Li, Weizhong Sun, Zhiru Tang, Yongsheng Wang, Kaifeng Zhou, Jigang Li, Qi Ding, Kaiyang Liang, and Liuting Wu reviewed the manuscript. Yulong Yin and Zhihong Sun conceptualized and proofread the manuscript.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (31872370), the Fundamental Research Funds for the Central Universities (XDJK2019B014), and the Natural Science Foundation Project of CQ CSTC (cstc2018jcyjAX0025).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Yulong Yin, Email: yinyulong@isa.ac.cn.
Zhihong Sun, Email: sunzh2002cn@aliyun.com.
References
- Adachi K., Sugiyama T., Yamaguchi Y., Tamura Y., Izawa S., Hijikata Y., et al. Gut microbiota disorders cause type 2 diabetes mellitus and homeostatic disturbances in gut-related metabolism in Japanese subjects. J Clin Biochem Nutr. 2019;64:231–238. doi: 10.3164/jcbn.18-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S.B., Jun D.W., Kang P.K., Lim J.H., Lim S., Chung M.J. Multispecies probiotic mixture in nonalcoholic fatty liver disease. Sci Rep. 1999;9:5688. doi: 10.1038/s41598-019-42059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari H.R., Raghava G.P. Identification of NAD interacting residues in proteins. BMC Bioinf. 2010;11:160. doi: 10.1186/1471-2105-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom F., Henry M., Vialettes B., Raccah D., Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P., Cantó C., Oudart H., Brunyánszki A., Cen Y., Thomas C., et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metabol. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T.P., Kinsland C., Mehl R.A., Osterman A., Dorrestein P. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam Horm. 2001;61:103–119. doi: 10.1016/s0083-6729(01)61003-3. [DOI] [PubMed] [Google Scholar]
- Bessman M.J. A cryptic activity in the Nudix hydrolase superfamily. Protein Sci. 2019;28:1494–1500. doi: 10.1002/pro.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieganowski P., Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Binder H.J. Role of colonic short-chain fatty acid transport in diarrhea. Annu Rev Physiol. 2011;72:297–313. doi: 10.1146/annurev-physiol-021909-135817. [DOI] [PubMed] [Google Scholar]
- Braidy N., Berg J., Clement J., Khorshidi F., Poljak A., Jayasena T., et al. Role of nicotinamide adenine dinucleotide and related precursors as therapeutic targets for age-related degenerative diseases: rationale, biochemistry, pharmacokinetics, and outcomes. Antioxidants Redox Signal. 2019;30:251–294. doi: 10.1089/ars.2017.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt E.M., Wong V.W., Nobili V., Day C.P., Sookoian S., Maher J.J., et al. Nonalcoholic fatty liver disease. Nat Rev Dis Prim. 2015;1 doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- Cahová H., Winz M.L., Höfer K., Nübel G., Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- Camacho-Pereira J., Tarragó M.G., Chini C.C.S., Nin V., Escande C., Warner G.M., et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metabol. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P.D. Gut microbiota-At the intersection of everything? Nat Rev Gastroenterol Hepatol. 2017;14:321–322. doi: 10.1038/nrgastro.2017.54. [DOI] [PubMed] [Google Scholar]
- Cantó C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y., et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metabol. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Menzies K.J., Auwerx J. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metabol. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canyelles M., Tondo M., Cedó L., Farràs M., Escolà-Gil J.C., Blanco-Vaca F. Trimethylamine N-oxide: a link among diet, gut microbiota, gene regulation of liver and intestine cholesterol homeostasis and hdl function. Int J Mol Sci. 2018;19:3228. doi: 10.3390/ijms19103228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D.J., Rifkin R.F., Cowan D.A., Potgieter M. The healthy human blood microbiome: fact or fiction? Front Cell Infect Microbiol. 2019;9:148. doi: 10.3389/fcimb.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers E.S., Viardot A., Psichas A., Morrison D.J., Murphy K.G., Zac-Varghese S.E., et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metabol. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., So W.Y., Li S.Y., Cheng Q., Boucher B.J., Boucher B.J., et al. Niacin-induced hyperglycemia is partially mediated via niacin receptor GPR109a in pancreatic islets. Mol Cell Endocrinol. 2015;404:56–66. doi: 10.1016/j.mce.2015.01.029. [DOI] [PubMed] [Google Scholar]
- Chen J., Li Y., Tian Y., Huang C., Li D., Zhong Q., et al. Interaction between microbes and host intestinal health: modulation by dietary nutrients and gut-brain-endocrine-immune Axis. Curr Protein Pept Sci. 2015;16:592–603. doi: 10.2174/1389203716666150630135720. [DOI] [PubMed] [Google Scholar]
- Cheng C.Y., Gutierrez N.M., Marzuki M.B., Lu X., Foreman T.W., Paleja B., et al. Host sirtuin 1 regulates mycobacterial immunopathogenesis and represents a therapeutic target against tuberculosis. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aaj1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini E.N., Chini C.C.S., EspindolaNetto J.M., de Oliveira G.C., van Schooten W. The pharmacology of CD38/NADase: an emerging target in cancer and diseases of aging. Trends Pharmacol Sci. 2018;39:424–436. doi: 10.1016/j.tips.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Kwon Y., Kim D.K., Jeon J., Jang S.C., Wang T., et al. Gut microbe-derived extracellular vesicles induce insulin resistance, thereby impairing glucose metabolism in skeletal muscle. Sci Rep. 2015;5 doi: 10.1038/srep15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.S. Interplay between compartmentalized NAD+ synthesis and consumption: a focus on the PARP family. Genes Dev. 2020;34:254–262. doi: 10.1101/gad.335109.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisol B.M., Veiga C.B., Lenhare L., Braga R.R., Silva V.R.R., da Silva A.S.R., et al. Nicotinamide riboside induces a thermogenic response in lean mice. Life Sci. 2018;211:1–7. doi: 10.1016/j.lfs.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Crisol B.M., Veiga C.B., Braga R.R., Lenhare L., Baptista I.L., Gaspar R.C., et al. NAD+ precursor increases aerobic performance in mice. Eur J Nutr. 2020;59:2427–2437. doi: 10.1007/s00394-019-02089-z. [DOI] [PubMed] [Google Scholar]
- Dai Z., Zhang J., Wu G., Zhu W. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids. 2010;39:1201–1215. doi: 10.1007/s00726-010-0556-9. [DOI] [PubMed] [Google Scholar]
- Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y., et al. Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol Res. 2013;68:95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Davila A., Liu L., Chellappa K., Redpath P., Nakamaru-Ogiso E., Paolella L.M., et al. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife. 2018;7 doi: 10.7554/eLife.33246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F., Nobili V., Vernocchi P., Russo A., De Stefanis C., Gnani D., et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451–464. doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- Domergue R., Castaño I., De Las Peñas A., Zupancic M., Lockatell V., Hebel J.R., et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- Durnin L., Kurahashi M., Sanders K.M., Mutafova-Yambolieva V.N. Extracellular metabolism of the enteric inhibitory neurotransmitter β-nicotinamide adenine dinucleotide (β-NAD) in the murine colon. J Physiol. 2020;598:4509–4521. doi: 10.1113/JP280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duseja A., Acharya S.K., Mehta M., Chhabra S., Shalimar Rana S., et al. High potency multistrain probiotic improves liver histology in non-alcoholic fatty liver disease (NAFLD): a randomised, double-blind, proof of concept study. BMJ Open Gastroenterol. 2019;6 doi: 10.1136/bmjgast-2019-000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K., Summers D.W., Sasaki Y., Mao X., DiAntonio A., Milbrandt J. The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron. 2017;93:1334–1343. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Li Z., Graff E.C., McCafferty K.J., Judd R.L. Niacin increases diet-induced hepatic steatosis in B6129 mice. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158731. [DOI] [PubMed] [Google Scholar]
- Farhangi M.A. Gut microbiota-dependent trimethylamine N-oxide and all-cause mortality: findings from an updated systematic review and meta-analysis. Nutrition. 2020;78 doi: 10.1016/j.nut.2020.110856. [DOI] [PubMed] [Google Scholar]
- Fei N., Bruneau A., Zhang X., Wang R., Wang J., Rabot S., et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio. 2020;11 doi: 10.1128/mBio.03263-19. e03263–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariani K., Menzies K.J., Ryu D., Wegner C.J., Wang X., Ropelle E.R., et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2016;63:1190–1204. doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaniga F., Stebbins R., Chang S.Z., McPeek M.A., Brenner C. Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev. 2009;73:529–541. doi: 10.1128/MMBR.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Talla E., François J.M. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene. PNC1. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.P., Price N.L., Ling A.J., Moslehi J.J., Montgomery M.K., Rajman L., et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.C., Hoffmann C., Mota J.F. The human gut microbiota: metabolism and perspective in obesity. Gut Microb. 2018;9:308–325. doi: 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.S., Passey R., Matanovic G., Smythe G., Kapoor V. Evidence for increased de novo synthesis of NAD in immune-activated RAW264.7 macrophages: a self-protective mechanism? Arch Biochem Biophys. 1999;372:1–7. doi: 10.1006/abbi.1999.1381. [DOI] [PubMed] [Google Scholar]
- Grozio A., Mills K.F., Yoshino J., Bruzzone S., Sociali G., Tokizane K., et al. Slc12a8 is a nicotinamide mononucleotide transporter. Nat Metab. 2019;1:47–57. doi: 10.1038/s42255-018-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Li S., Xie Y., Zhang Q., Liu M., Xu Z., et al. The NAD+-dependent deacetylase, Bifidobacterium longum Sir2 in response to oxidative stress by deacetylating SigH (σH) and FOXO3a in Bifidobacterium longum and HEK293T cell respectively. Free Radic Biol Med. 2017;108:929–939. doi: 10.1016/j.freeradbiomed.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Guo S., Chen Q., Sun Y., Chen J. Nicotinamide protects against skeletal muscle atrophy in streptozotocin-induced diabetic mice. Arch Physiol Biochem. 2019;125:470–477. doi: 10.1080/13813455.2019.1638414. [DOI] [PubMed] [Google Scholar]
- Gupte R., Liu Z., Kraus W.L. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M., Li Z., You H., Rodrigues R., Jump D.B., Morgun A., et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handy D.E., Loscalzo J. Responses to reductive stress in the cardiovascular system. Free Radic Biol Med. 2017;109:114–124. doi: 10.1016/j.freeradbiomed.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Han M., Qiao S., He P., Li D., Li N., et al. Soybean antigen proteins and their intestinal sensitization activities. Curr Protein Pept Sci. 2015;16:613–621. doi: 10.2174/1389203716666150630134602. [DOI] [PubMed] [Google Scholar]
- Heemskerk M.M., Dharuri H.K., van den Berg S.A., Jónasdóttir H.S., Kloos D.P., Giera M., et al. Prolonged niacin treatment leads to increased adipose tissue PUFA synthesis and anti-inflammatory lipid and oxylipin plasma profile. J Lipid Res. 2014;55:2532–2540. doi: 10.1194/jlr.M051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk M.M., van den Berg S.A., Pronk A.C., van Klinken J.B., Boon M.R., Havekes L.M., et al. Long-term niacin treatment induces insulin resistance and adrenergic responsiveness in adipocytes by adaptive downregulation of phosphodiesterase 3B. Am J Physiol Endocrinol Metab. 2014;306:E808–E813. doi: 10.1152/ajpendo.00641.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida H., Hashii M., Tanaka Y., Matsukawa S., Higuchi Y., Gabata R., et al. CD38, CD157, and RAGE as molecular determinants for social behavior. Cells. 2019;9:62. doi: 10.3390/cells9010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holscher H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microb. 2017;8:172–184. doi: 10.1080/19490976.2017.1290756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holscher H.D. Gut microbes: nuts about fatty acids. J Nutr. 2020;150:652–653. doi: 10.1093/jn/nxaa045. [DOI] [PubMed] [Google Scholar]
- Hopp A.K., Grüter P., Hottiger M.O. Regulation of glucose metabolism by NAD+ and ADP-ribosylation. Cells. 2019;8:890. doi: 10.3390/cells8080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Moschetta A., Lee Y.K., Peng L., Zhao G., Downes M., et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S., Beyaert R. Role of toll-like receptors in pathogen recognition. Clin Microbiol Rev. 2003;16(4):637–646. doi: 10.1128/CMR.16.4.637-646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.H., Fang Z.P., Fan M.J., Huang W.D. Bile-ology: from bench to bedside. J Zhejiang Univ - Sci B. 2019;20:414–427. doi: 10.1631/jzus.B1900158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Shi X., Yang J., Zhao Y., Xue L., Xu L., et al. Gut microbes in cardiovascular diseases and their potential therapeutic applications. Protein & Cell. 2020;12:346–359. doi: 10.1007/s13238-020-00785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanna V.S., Kashyap M.L. Mechanism of action of niacin. Am J Cardiol. 2008;101:20B–26B. doi: 10.1016/j.amjcard.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Kamanna V.S., Ganji S.H., Kashyap M.L. Recent advances in niacin and lipid metabolism. Curr Opin Lipidol. 2013;24:239–245. doi: 10.1097/MOL.0b013e3283613a68. [DOI] [PubMed] [Google Scholar]
- Kashyap M.L., Ganji S., Nakra N.K., Kamanna V.S. Niacin for treatment of nonalcoholic fatty liver disease (NAFLD): novel use for an old drug? J Clin Lipidol. 2019;13:873–879. doi: 10.1016/j.jacl.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Katsyuba E., Auwerx J. Modulating NAD+ metabolism, from bench to bedside. EMBO J. 2017;36:2670–2683. doi: 10.15252/embj.201797135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyuba E., Mottis A., Zietak M., De Franco F., van der Velpen V., Gariani K., et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature. 2018;563:354–359. doi: 10.1038/s41586-018-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyuba E., Romani M., Hofer D., Auwerx J. NAD+ homeostasis in health and disease. Nat Metab. 2020;2:9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- Khan M., Couturier A., Kubens J.F., Most E., Mooren F.C., Krüger K., et al. Niacin supplementation induces type II to type I muscle fiber transition in skeletal muscle of sheep. Acta Vet Scand. 2013;55:85. doi: 10.1186/1751-0147-55-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T., et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimova N., Kristian T. Multi-targeted effect of nicotinamide mononucleotide on brain bioenergetic metabolism. Neurochem Res. 2019;44:2280–2287. doi: 10.1007/s11064-019-02729-0. [DOI] [PubMed] [Google Scholar]
- Klimova N., Fearnow A., Long A., Kristian T. NAD+ precursor modulates post-ischemic mitochondrial fragmentation and reactive oxygen species generation via SIRT3 dependent mechanisms. Exp Neurol. 2020;325 doi: 10.1016/j.expneurol.2019.113144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kanda T., Urai H., Kurokochi A., Kitahama R., Shigaki S., et al. NNMT activation can contribute to the development of fatty liver disease by modulating the NAD+ metabolism. Sci Rep. 2018;8:8637. doi: 10.1038/s41598-018-26882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtzidis I.A., Stoupas A.T., Gioris I.S., Veskoukis A.S., Margaritelis N.V., Tokizane K., et al. The NAD(+) precursor nicotinamide riboside decreases exercise performance in rats. J Int Soc Sports Nutr. 2016;13:32. doi: 10.1186/s12970-016-0143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtzidis I.A., Dolopikou C.F., Tsiftsis A.N., Margaritelis N.V., Theodorou A.A., Zervos I.A., et al. Nicotinamide riboside supplementation dysregulates redox and energy metabolism in rats: implications for exercise performance. Exp Physiol. 2018;103:1357–1366. doi: 10.1113/EP086964. [DOI] [PubMed] [Google Scholar]
- Kurdi P., Kawanishi K., Mizutani K., Yokota A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol. 2006;188:1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyme P., Thoennissen N.H., Tseng C.W., Thoennissen G.B., Wolf A.J., Shimada K., et al. C/EBPε mediates nicotinamide-enhanced clearance of Staphylococcus aureus in mice. J Clin Invest. 2012;122:3316–3329. doi: 10.1172/JCI62070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming D.W., Wood J.G., Sinclair D.A. Small molecules that regulate lifespan: evidence for xenohormesis. Mol Microbiol. 2004;53:1003–1009. doi: 10.1111/j.1365-2958.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- Leung K., Quezada M., Chen Z., Kanel G., Kaplowitz N. Niacin-induced anicteric microvesicularsteatotic acute liver failure. Hepatol Commun. 2018;2:1293–1298. doi: 10.1002/hep4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Leylabadlo H.E., Sanaie S., SadeghpourHeravi F., Ahmadian Z., Ghotaslou R. From role of gut microbiota to microbial-based therapies in type 2-diabetes. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104268. [DOI] [PubMed] [Google Scholar]
- Li M., Petteys B.J., McClure J.M., Valsakumar V., Bekiranov S., Frank E.L., et al. Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol Cell Biol. 2010:3329–3341. doi: 10.1128/MCB.01590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Tian Y.J., Guo J., Sun W.P., Lun Y.Z., Guo M., et al. Nicotinamide supplementation induces detrimental metabolic and epigenetic changes in developing rats. Br J Nutr. 2013;110:2156–2164. doi: 10.1017/S0007114513001815. [DOI] [PubMed] [Google Scholar]
- Li R., Huang X., Liang X., Su M., Lai K.P., Chen J., et al. Integrated omics analysis reveals the alteration of gut microbe-metabolites in obese adults. Briefings Bioinf. 2020;7:bbaa165. doi: 10.1093/bib/bbaa165. [DOI] [PubMed] [Google Scholar]
- Lin R., Liu W., Piao M., Zhu H. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. 2017;49:2083–2090. doi: 10.1007/s00726-017-2493-3. [DOI] [PubMed] [Google Scholar]
- Liu Y., Hou Y., Wang G., Zheng X., Hao H. Gut microbial metabolites of aromatic amino acids as signals in host-microbe interplay. Trends Endocrinol Metabol. 2020;31:818–834. doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- Luo J., Barany F. Identification of essential residues in Thermus thermophilus DNA ligase. Nucleic Acids Res. 1996;24:3079–3085. doi: 10.1093/nar/24.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Tian Y., Wu Y., Ma X. Contributions of the interaction between dietary protein and gut microbiota to intestinal health. Curr Protein Pept Sci. 2017;18(8):795–808. doi: 10.2174/1389203718666170216153505. [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Allison C., Gibson S.A., Cummings J.H. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64:37–46. doi: 10.1111/j.1365-2672.1988.tb02427.x. [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir S., Ravcheev D., de Crécy-Lagard V., Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmel M., Jovanović N., Spitz U. Nicotinamide riboside-the current state of research and therapeutic uses. Nutrients. 2020;12:1616. doi: 10.3390/nu12061616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.F., Yoshida S., Stein L.R., Grozio A., Kubota S., Sasaki Y., et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metabol. 2016;24:795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic I., Vujovic A., Barac A., Djelic M., Korac M., Radovanovic Spurnic A., et al. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: a review of the literature. Int J Mol Sci. 2019;20:395. doi: 10.3390/ijms20020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.J., Bernier M., Aon M.A., Cortassa S., Kim E.Y., Fang E.F., et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metabol. 2018;27:667–676. doi: 10.1016/j.cmet.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monga Kravetz A., Testerman T., Galuppo B., Graf J., Pierpont B., Siebel S., et al. Effect of gut microbiota and PNPLA3 rs738409 variant on nonalcoholic fatty liver disease (NAFLD) in obese youth. J Clin Endocrinol Metab. 2020;105:dgaa382. doi: 10.1210/clinem/dgaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montastier E., Beuzelin D., Martins F., Mir L., Marqués M.A., Thalamas C., et al. Niacin induces miR-502-3p expression which impairs insulin sensitivity in human adipocytes. Int J Obes (Lond) 2019;43:1485–1490. doi: 10.1038/s41366-018-0260-5. [DOI] [PubMed] [Google Scholar]
- Morrison D.J., Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microb. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccioli G.G., Naslain D., Bäckhed F., Reigstad C.S., Lambert D.M., Delzenne N.M., et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:392. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P., Horváth B., Rajesh M., Varga Z.V., Gariani K., Ryu D., et al. PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J Hepatol. 2017;66:589–600. doi: 10.1016/j.jhep.2016.10.023. [DOI] [PubMed] [Google Scholar]
- Naia L., Rosenstock T.R., Oliveira A.M., Oliveira-Sousa S.I., Caldeira G.L., Carmo C., et al. Comparative mitochondrial-based protective effects of resveratrol and nicotinamide in huntington's disease models. Mol Neurobiol. 2017;54:5385–5399. doi: 10.1007/s12035-016-0048-3. [DOI] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Golenbock D., Bowie A.G. The history of toll-like receptors- redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- Opapeju F.O., Krause D.O., Payne R.L., Rademacher M., Nyachoti C.M. Effect of dietary protein level on growth performance, indicators of enteric health, and gastrointestinal microbial ecology of weaned pigs induced with postweaning colibacillosis. J Anim Sci. 2009;87:2635–2643. doi: 10.2527/jas.2008-1310. [DOI] [PubMed] [Google Scholar]
- Oxenkrug G. Insulin resistance and dysregulation of tryptophan-kynurenine and kynurenine-nicotinamide adenine dinucleotide metabolic pathways. Mol Neurobiol. 2013;48:294–301. doi: 10.1007/s12035-013-8497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzer L., Bader J.J., Angel F., Witzel M., Blaser S., McNeil A., et al. Alpha-amino-beta-carboxy-muconate-semialdehyde decarboxylase controls dietary niacin requirements for NAD+ synthesis. Cell Rep. 2018;25:1359–1370. doi: 10.1016/j.celrep.2018.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Sánchez S., Cockayne D.A., Monard S., Jacobson E.L., Oppenheimer N., Garvy B., et al. Cyclic ADP-ribose production by CD38 regulates intracellular calcium release, extracellular calcium influx and chemotaxis in neutrophils and is required for bacterial clearance in vivo. Nat Med. 2001;7:1209–1216. doi: 10.1038/nm1101-1209. [DOI] [PubMed] [Google Scholar]
- Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- Peng L., Li Z.R., Green R.S., Holzman I.R., Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L., et al. Acetate mediates a microbiome-brain-β cell axis promoting metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.X., Bae M., Kim M.B., Lee Y., Hu S., Kang H., et al. Nicotinamide riboside, an NAD+ precursor, attenuates the development of liver fibrosis in a diet-induced mouse model of liver fibrosis. Biochim Biophys Acta, Mol Basis Dis. 2019;1865:2451–2463. doi: 10.1016/j.bbadis.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E., Auranen M., Khan N.A., Brilhante V., Urho N., Pessia A., et al. Niacin cures systemic NAD+ deficiency and improves muscle performance in adult-onset mitochondrial myopathy. Cell Metabol. 2020;31:1078–1090. doi: 10.1016/j.cmet.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Porter T.D. Electron transfer pathways in cholesterol synthesis. Lipids. 2015;50:927–936. doi: 10.1007/s11745-015-4065-1. [DOI] [PubMed] [Google Scholar]
- Preiss J., Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958;233:488–492. [PubMed] [Google Scholar]
- Qi Z., Xia J., Xue X., He Q., Ji L., Ding S. Long-term treatment with nicotinamide induces glucose intolerance and skeletal muscle lipotoxicity in normal chow-fed mice: compared to diet-induced obesity. J Nutr Biochem. 2016;36:31–41. doi: 10.1016/j.jnutbio.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Quarona V., Zaccarello G., Chillemi A., Brunetti E., Singh V.K., Ferrero E., et al. CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytometry B Clin Cytom. 2013;84:207–217. doi: 10.1002/cyto.b.21092. [DOI] [PubMed] [Google Scholar]
- Revollo J.R., Grimm A.A., Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Romani M., Hofer D.C., Katsyuba E., Auwerx J. Niacin: an old lipid drug in a new NAD+ dress. J Lipid Res. 2019;60:741–746. doi: 10.1194/jlr.S092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz J.A., Bürkle A., Mangerich A. NAD+ in sulfur mustard toxicity. Toxicol Lett. 2020;324:95–103. doi: 10.1016/j.toxlet.2020.01.024. [DOI] [PubMed] [Google Scholar]
- Safari Z., Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell Mol Life Sci. 2019;76:1541–1558. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar N., Valdés-Varela L., González S., Gueimonde M., de Los Reyes-Gavilán C.G. Nutrition and the gut microbiome in the elderly. Gut Microb. 2017;8:82–97. doi: 10.1080/19490976.2016.1256525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi M., Razavi S., Navab-Moghadam F., Khamseh M.E., Alaei-Shahmiri F., Mehrtash A. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- Shats I., Williams J.G., Liu J., Makarov M.V., Wu X., Lih F.B., et al. Bacteria boost mammalian host NAD metabolism by engaging the deamidated biosynthesis pathway. Cell Metabol. 2020;31:564–579. doi: 10.1016/j.cmet.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Enriquez A., Rapadas M., Martin E.M.M.A., Wang R., Moreau J., et al. NAD deficiency, congenital malformations, and niacin supplementation. N Engl J Med. 2017;377:544–552. doi: 10.1056/NEJMoa1616361. [DOI] [PubMed] [Google Scholar]
- Shi W., Hegeman M.A., Doncheva A., Bekkenkamp-Grovenstein M., de Boer V.C.J., Keijer J. High dose of dietary nicotinamide riboside induces glucose intolerance and white adipose tissue dysfunction in mice fed a mildly obesogenic diet. Nutrients. 2019;11:2439. doi: 10.3390/nu11102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinnler R., Gorski T., Stolz K., Schuster S., Garten A., Beck-Sickinger A.G., et al. The adipocytokine Nampt and its product NMN have no effect on beta-cell survival but potentiate glucose stimulated insulin secretion. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydenstricker V.P. The history of pellagra, its recognition as a disorder of nutrition and its conquest. Am J Clin Nutr. 1958;6:409–414. doi: 10.1093/ajcn/6.4.409. [DOI] [PubMed] [Google Scholar]
- Tarragó M.G., Chini C.C.S., Kanamori K.S., Warner G.M., Caride A., de Oliveira G.C., et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metabol. 2018;27:1081–1095. doi: 10.1016/j.cmet.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell S.A., Weidemann B.J., Chadda A., Yorek M.S., Holmes A., Coppey L.J., et al. Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci Rep. 2016;6 doi: 10.1038/srep26933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin G.M., Youngson N.A., Sinclair D.A., Morris M.J. Head to head comparison of short-term treatment with the NAD(+) precursor nicotinamide mononucleotide (NMN) and 6 weeks of exercise in obese female mice. Front Pharmacol. 2016;7:258. doi: 10.3389/fphar.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin G.M., Youngson N.A., Doyle B.M., Sinclair D.A., Morris M.J. Nicotinamide mononucleotide (NMN) supplementation ameliorates the impact of maternal obesity in mice: comparison with exercise. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin G.M., Youngson N.A., Chowdhury S.S., Hagan C., Sinclair D.A., Morris M.J. Administration of nicotinamide mononucleotide (NMN) reduces metabolic impairment in male mouse offspring from obese mothers. Cells. 2020;9:791. doi: 10.3390/cells9040791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn A.C., Cooper E.M., DiLorenzo P.M., O'Loughlin L.J., Konkel M.E., Peters J.H., et al. Energy-dense diet triggers changes in gut microbiota, reorganization of gutbrain vagal communication and increases body fat accumulation. Acta Neurobiol Exp. 2017;77(1):18–30. doi: 10.21307/ane-2017-033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R.S., Bartelsman J.F., et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- Waldman M., Nudelman V., Shainberg A., Abraham N.G., Kornwoski R., Aravot D., et al. PARP-1 inhibition protects the diabetic heart through activation of SIRT1-PGC-1α axis. Exp Cell Res. 2018;373:112–118. doi: 10.1016/j.yexcr.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Walters W.A., Xu Z., Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H.F., Li J.X., Liao H.T., Liao M.H., Luo L., Xu L., et al. Nicotinamide induces liver regeneration and improves liver function by activating SIRT1. Mol Med Rep. 2019;19:555–562. doi: 10.3892/mmr.2018.9688. [DOI] [PubMed] [Google Scholar]
- Wang N., Guo D.Y., Tian X., Lin H.P., Li Y.P., Chen S.J., et al. Niacin receptor GPR109A inhibits insulin secretion and is down-regulated in type 2 diabetic islet beta-cells. Gen Comp Endocrinol. 2016;237:98–108. doi: 10.1016/j.ygcen.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kuang Z., Yu X., Ruhn K.A., Kubo M., Hooper L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X., Zhao J., Zhang H. Lactobacillus casei CCFM419 attenuates type 2 diabetes via a gut microbiota dependent mechanism. Food Funct. 2017;8:3155–3164. doi: 10.1039/c7fo00593h. [DOI] [PubMed] [Google Scholar]
- Wang S., Wan T., Ye M., Qiu Y., Pei L., Jiang R., et al. Nicotinamide riboside attenuates alcohol induced liver injuries via activation of SirT1/PGC-1α/mitochondrial biosynthesis pathway. Redox Biol. 2018;17:89–98. doi: 10.1016/j.redox.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Jiao T., Xu Y., Li D., Si Q., Hao J., et al. Bifidobacterium adolescentis and Lactobacillus rhamnosus alleviate non-alcoholic fatty liver disease induced by a high-fat, high-cholesterol diet through modulation of different gut microbiota-dependent pathways. Food Funct. 2020;11:6115–6127. doi: 10.1039/c9fo02905b. [DOI] [PubMed] [Google Scholar]
- Wilk A., Hayat F., Cunningham R., Li J., Garavaglia S., Zamani L., et al. Extracellular NAD+ enhances PARP-dependent DNA repair capacity independently of CD73 activity. Sci Rep. 2020;10:651. doi: 10.1038/s41598-020-57506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.K., Chang R.B., Strochlic D.E., Umans B.D., Lowell B.B., Liberles S.D. Sensory neurons that detect stretch and nutrients in the digestive system. Cell. 2016;166:209–221. doi: 10.1016/j.cell.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston J.A., Theriot C.M. Diversification of host bile acids by members of the gut microbiota. Gut Microb. 2020;11:158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D., Zhang Q., Wang Y.T. Effectiveness of niacin supplementation for patients with type 2 diabetes: a meta-analysis of randomized controlled trials. Medicine (Baltim) 2020;99 doi: 10.1097/MD.0000000000021235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Wang R.S., Handy D.E., Loscalzo J. NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxidants Redox Signal. 2018;28:251–272. doi: 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Yu C., Zhou J., Xiao Q., Shen Q., Xiong Z., et al. Nicotinamide mononucleotide ameliorates the depression-like behaviors and is associated with attenuating the disruption of mitochondrial bioenergetics in depressed mice. J Affect Disord. 2020;263:166–174. doi: 10.1016/j.jad.2019.11.147. [DOI] [PubMed] [Google Scholar]
- Xie Y., Wang C., Zhao D., Wang C., Li C. Dietary proteins regulate serotonin biosynthesis and catabolism by specific gut microbes. J Agric Food Chem. 2020;68:5880–5890. doi: 10.1021/acs.jafc.0c00832. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Miyamoto N., Shibata K., Valasek M.A., Motoike T., Kedzierski R.M., et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., He J., Gao N., Lu X., Li M., Wu X., et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep. 2017;7 doi: 10.1038/srep45176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Li N., Shi J., Li H., Yue Y., Jiao W., et al. Lactobacillus acidophilus alleviates type 2 diabetes by regulating hepatic glucose, lipid metabolism and gut microbiota in mice. Food Funct. 2019;10:5804–5815. doi: 10.1039/c9fo01062a. [DOI] [PubMed] [Google Scholar]
- Yang Y., Sauve A.A. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta. 2016;1864:1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang T., Baur J.A., Baur J.A., Perez E., Matsui T., et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.J., Choi J.M., Kim L., Park S.E., Rhee E.J., Lee W.Y., et al. Nicotinamide improves glucose metabolism and affects the hepatic NAD-sirtuin pathway in a rodent model of obesity and type 2 diabetes. J Nutr Biochem. 2014;25:66–72. doi: 10.1016/j.jnutbio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Yang L., Li T., Zhao S., Zhang S. Niacin regulates apolipoprotein M expression via liver X receptor-α. Mol Med Rep. 2019;20:3285–3291. doi: 10.3892/mmr.2019.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Cong L., Wang Y., Luo X., Li H., Wang H., et al. Increasing ovarian NAD+ levels improve mitochondrial functions and reverse ovarian aging. Free Radic Biol Med. 2020;156:1–10. doi: 10.1016/j.freeradbiomed.2020.05.003. [DOI] [PubMed] [Google Scholar]
- Ye L., Cao Z., Lai X., Wang W., Guo Z., Yan L., et al. Niacin fine-tunes energy homeostasis through canonical GPR109A signaling. Faseb J. 2019;33:4765–4779. doi: 10.1096/fj.201801951R. [DOI] [PubMed] [Google Scholar]
- Ye L., Cao Z., Lai X., Shi Y., Zhou N. Niacin ameliorates hepatic steatosis by inhibiting de novo lipogenesis via a GPR109A-mediated PKC-ERK1/2-AMPK signaling pathway in C57BL/6 mice fed a high-fat diet. J Nutr. 2020;150:672–684. doi: 10.1093/jn/nxz303. [DOI] [PubMed] [Google Scholar]
- Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metabol. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn H.S., Kim T.G., Kim M.K., Kang G.B., Kang J.Y., Lee J.G., et al. Structural insights into the quaternary catalytic mechanism of hexameric human quinolinate phosphoribosyltransferase, a key enzyme in de novo NAD biosynthesis. Sci Rep. 2016;6 doi: 10.1038/srep19681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Han W., Zhan G., Li S., Jiang X., Wang L., et al. Abnormal gut microbiota composition contributes to the development of type 2 diabetes mellitus in db/db mice. Aging (Albany NY) 2019;11:10454–10467. doi: 10.18632/aging.102469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaibi M.S., Stocker C.J., O'Dowd J., Davies A., Bellahcene M., Cawthorne M.A., et al. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett. 2010;584:2381–2386. doi: 10.1016/j.febslet.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Zhang L., Ma R., Hu J., Ding X., Xu Y. Sirtuin inhibition adversely affects porcine oocyte meiosis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., He X., Sheng Y., Yang C., Xu J., Zheng S., et al. Allicin-induced host-gut microbe interactions improves energy homeostasis. Faseb J. 2020;34:10682–10698. doi: 10.1096/fj.202001007R. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang Q., Ma W., Tian F., Shen H., Zhou M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017;8:4644–4656. doi: 10.1039/c7fo01383c. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X., et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]