Abstract
Di (2-ethyl-hexyl) phthalate (DEHP) is a wildly used plasticizer. Maternal exposure to DEHP during pregnancy blocks the placental cell cycle at the G2/M phase by reducing the efficiency of the DNA repair pathways and affects the health of offsprings. However, the mechanism by which DEHP inhibits the repair of DNA damage remains unclear. In this study, we demonstrated that DEHP inhibits DNA damage repair by reducing the activity of the DNA repair factor recruitment molecule PARP1. NAD+ and ATP are two substrates necessary for PARP1 activity. DEHP abated NAD+ in the nucleus by reducing the level of NAD+ synthase NMNAT1 and elevated NAD+ in the mitochondrial by promoting synthesis. Furthermore, DEHP destroyed the mitochondrial respiratory chain, affected the structure and quantity of mitochondria, and decreased ATP production. Therefore, DEHP inhibits PARP1 activity by reducing the amount of NAD+ and ATP, which hinders the DNA damage repair pathways. The supplement of NAD+ precursor NAM can partially rescue the DNA and mitochondria damage. It provides a new idea for the prevention of health problems of offsprings caused by DEHP injury to the placenta.
Keywords: Di (2-ethyl-hexyl) phthalate, Nicotinamide adenine dinucleotide, ATP, DNA damage, Placental development
Graphical abstract
DEHP decreases the overall NAD+ level of the placenta and reduces the NAD+ level in the placental cell nucleus by participating in the regulation of NAD+ regionalization. By destroying the placental cell mitochondrial structure, reducing the number of mitochondria, hindering the way of electron transfer in the respiratory chain, DEHP reduces ATP production. Under the combined action of NAD+ and ATP depletion, inhibition of PARP1 activity leads to the accumulation of DNA damage and ultimately to the reduction of the placental trophoblast area. The partially damaging effects of DEHP on DNA and mitochondrial structures can be alleviated by moderate supplementation of NAD+ precursor NAM.
Highlights
-
•
DEHP reduces placental NAD + by compartmental regulation.
-
•
DEHP inhibits ATP production by disrupting mitochondrial structure quantity and function.
-
•
Limited NAD+ and ATP together inhibit DNA damage repair activity of PARP1.
-
•
Appropriate NAM can alleviate DNA and mitochondria damage to some extent.
1. Introduction
Fetal growth restriction (FGR) is a worldwide problem [1]. Based on the Developmental Origins of Health and Disease (DOHaD) hypothesis, it is related to the susceptibility to some diseases in later life [[1], [2], [3]]. FGR was inversely correlated with placental weight in an epidemiological, population-based study [4]. The placenta is a temporary yet essential organ that connects mother and fetus, which has barrier, transport, and endocrine functions [5] and the development of which is sensitive to some outer environment factors, such as Di (2-ethyl-hexyl) phthalate (DEHP).
DEHP is a wildly used plasticizer and potential carcinogen. It is a known environmental endocrine disruptor (EED) [6]. Since DEHP is not covalently attached to PVC products, it is easy to release into various environmental media [7]. It enters the body through the skin, digestive tract, and respiratory tract [8]. DEHP is hydrolyzed rapidly in vivo by esterase into mono (2-ethylhexyl) phthalate (MEHP), a biologically active monoester metabolite which is a reproductive poison [9]. DEHP has toxic effects on the occurrence of sperm and ovum and affects the development of fertilized eggs and embryos [[10], [11], [12]]. Epidemiological evidence showed that DEHP can affect semen parameters and testosterone levels in adult men [13] and increase the odds of premature delivery in women [14]. Under the toxicity of DEHP and MEHP, key processes related to placental development, such as implantation, differentiation, invasion, and angiogenesis, were negatively affected, which can further induce increased placental apoptosis [15,16]. Our previous work has shown that DEHP disrupts placental growth in a dual blocking mode [17]. It has been reported that DEHP causes DNA damage [18,19] and our further analyses suggested the DNA damage repair pathways were affected [17]. However, the detailed mechanism remains unelucidated.
DNA repair is essential for cells to maintain genome stability and cell survival [20]. PARP1, a nuclear enzyme sensing DNA damage [21], set up the damage repair platform through Poly-ADP-ribosylation (PARylation) itself and other repair factors in the repair pathways [[22], [23], [24]] under the assistance of nicotinamide adenine dinucleotide (NAD+) and ATP. NAD+ is a coenzyme involved in cellular energy metabolism and homeostasis, adaptive response, chromatin stability, gene expression [25,26] and DNA repair [27]. ATP is formed by glycolysis in the cytoplasm and oxidative phosphorylation in mitochondria. On one hand, DEHP can down-regulate the level of ATP in mouse oocytes [28] and testis [29]. On the other hand, our previous study found that NAD+ concentration decreased in the presence of DEHP [17]. Therefore, we hypothesized that DEHP interferes with DNA damage repair by inhibiting NAD+/ATP- PARP1 signal axis in placental cells.
In this study, we examined the mechanism of DNA damage in placental cells exposed to DEHP. We found that DEHP has a compartmentalized regulatory effect on NAD+ in placental cells, which reduced the concentration of NAD+ in nuclear and cytoplasmic through inhibiting NAD+ synthase NMNAT1 and NMNAT2, respectively. Meanwhile, NAD+ hydrolase CD38 and CD157 were observed to increase, which further reduced the intracellular NAD+ level. On the contrary, DEHP increased the concentration of NAD+ in mitochondria by up-regulating NMNAT3 synthetase. Although a large amount of NAD+ accumulates in mitochondria, DEHP inhibits the rate of ATP production by blocking the electron transport chain, reducing the number of mitochondria, and damaging the mitochondrial structure. Under the nuclear NAD+ and ATP deficiency, the activity of PARP1 is inhibited, resulting in the accumulation of DNA damage, reducing the proliferation of placental cells, and ultimately thinning the layer of placental trophoblast. Precursor nicotinamide (NAM) pre-supplementation alleviated the DNA damage and the structural damage of placental mitochondria caused by DEHP.
Therefore, DEHP inhibited the activity of PARP1 by regulating the levels of NAD+ and ATP, thus hindering the repair of DNA damage and blocking the proliferation of placental cells. Appropriate NAM can partially rescue these damages.
2. Materials and methods
2.1. Reagents
DEHP (Sigma-Aldrich, USA, D201154, purity ≥99.5%). MEHP (Sigma-Aldrich, USA, 796832, purity ≥97%). Corn oil (COFCO, 370131751–1). Tween 80 (Sigma-Aldrich, P1754). Nicotinic acid (NA) (Topscience, T0879). Nicotinamide (Topscience, T0934). β-Nicotinamide mononucleotide (NMN) (Topscience, T4721). Nicotinamide riboside (NR) (Topscience, T13795). DMSO (Sigma-Aldrich, RNBG2257). RPMI Medium Modified 1640 (Hyclone, SH30809.01). Fetal bovine serum (Sangon Biotech, Shanghai, E600001). HE Staining Kit (Solarbio, G1120). NAD+/NADH Assay Kit (Beyotime Biotechnology, S0175). BCA Protein Assay Kit (Thermo Scientific, 23225). Comet assay kit (R&D Systems, 4250-050-K).
2.2. Animal feeding and treatment
SPF-grade ICR mice (8 weeks old) were obtained from Beijing Vital River (license number: SCXK (Beijing, China) 2016–0011). The animals were first adaptively housed in a specific-pathogen-free (SPF) environment for one week and given access to sterile food and water (packed in a glass kettle). The room environment was maintained at 22 ± 2 °C and 50 ± 5% humidity under an automatically controlled cycle of 12 h light/dark. On the 7th night, one male and two female mice were mated in a new cage at 8:00 p.m. Female mice were checked by 8:00 a.m. the next morning. If a vaginal plug was found, it was considered as gestational (GD 0). Then pregnant mice were randomized and placed into different groups based on weight before drug treatment. Repeat the above operation until enough pregnant mice were obtained. In DEHP-treated groups, pregnant mice were administered with 0 mg/kg/d (Tween 80% and 1% corn oil emulsion), 5 mg/kg/d, 50 mg/kg/d, and 200 mg/kg/d DEHP by gavage (1% of body weight) during GD 0–15. The dose of DEHP (0, 5, 50, 200 mg/kg/d) was selected according to a previous study [17]. In intervention groups, pregnant mice were administered with 0 mg/kg/d (water), 500 mg/kg/d NAM, 50 mg/kg/d DEHP, and 500 mg/kg/d NAM + 50 mg/kg/d DEHP by gavage (1% of body weight) during GD 0–15. Pregnant mice were sacrificed at GD 15. The use of animals and the protocols are licensed by the Anhui Medical University Animal Care and Use Committee (20170394, LLSC20200694).
2.3. Cell culture
HTR-8/SVneo cell line, one of the first-trimester extravillous trophoblast cell lines, was purchased from the Shanghai ATCC cell bank. This cell line has been universally accepted to possess all the characteristics of first trimester human primary placental cells. Cells were grown in RPMI 1640 medium supplemented with 5% FBS and 1% penicillin/streptomycin (Sangon Biotech) in a maintained chamber at 37 °C with 5% CO2. The dose of MEHP (0, 20, 200, 500 μM) was selected according to a previous study [17]. MEHP was prepared in DMSO at a final concentration of 0.1% by volume. NA (0.5 mM), NAM (0.5 or 5 mM), NMN (0.5 mM), and NR (0.5 mM) were dissolved in distilled water at a final concentration of 0.1% by volume.
2.4. Placental histopathology
The placenta was removed and soaked in 4% paraformaldehyde (PFA) on a horizontal shaker overnight and embedded with paraffin [30]. Following this step, the placental section was dyed using hematoxylin and eosin (H&E). According to the structure, the placenta was divided into four zones: decidua (dec), junctional zone (jz), labyrinth (lab), and chorionic plate (cp) [30].
2.5. Mitochondrial DNA copy number quantification
Genomic DNA from the placenta was isolated using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer's instructions. The relative copy number of mitochondrial DNA per nuclear DNA ratio was measured by qPCR. The sequences of primers for the mitochondrial segment were as follows [31]: (F) CTAGAAACCCCGAAACCAAA and (R) CCAGCTATCACCAAGCTCGT. The sequences of primers for the single-copy nuclear control were as follows: (F) ATGGGAAGCCGAACATACTG and (R) CAGTCTCAGTGGGGGTGAAT.
2.6. NAD+/NADH assay
NAD+/NADH levels were measured with NAD+/NADH Assay Kit according to the manufacturer's instructions. Animal tissue was homogenized using a tissue homogenizer after adding the extracting solution. HTR-8 cells were seeded on six-well plates (1.0 × 106 cells/well) and 400 μL of extracting solution was added. Absorbance at 450 nm was measured using a multifunctional microplate reader (BioTek, SENERGX2 Synergy 4). Finally, BCA assay was done to determine the protein concentration.
2.7. Immunoblotting
Total placental and HTR8/SVneo cell lysates were heated for 10 min at 100 °C and then electrophoretically separated by 10–12.5% SDS-PAGE. After electrophoresis, the protein was transferred to the PVDF membrane and blocked in 5% skim milk formulated in TBST for 1 h. After blocking, the membranes were individually incubated with PCNA (Abcam, ab18197), γH2AX (Ser139, CST, 9718S), PARP1(CST, 46D11), Poly (ADP-ribose) (Trevigen, 4335-MC-100), NMNAT1 (GeneTex, GTX66480), NMNAT2 (Affbiotech, DF13581), NMNAT3 (Abcam, ab121030), ACPM (Omnimabs, OM633568), Total Oxphos (Abcam, ab110413), CD38 (Abcam, ab216343), CD157 (Affbiotech, DF3744), and NAMPT (Abcam, ab236874) antibodies for 1–4 h. Following the primary antibody incubation, the membranes were washed in TBST and subsequentially incubated with an appropriate secondary antibody. The membranes were washed again in TBST and detected with the enhanced chemiluminescence solution (Advansta, K-12043-D20). The protein bands were analyzed by Image J software for gray value.
2.8. Comet assay
The comet assay was performed with the Comet assay kit according to the manufacturer's instructions. HTR-8 cells were mixed into LMAgarose (at 37 °C) in a 1:10 vol ratio and immobilized onto a comet assay glass slide (50 μL each well). Then, the glass slides were placed in a wet box at 4 °C for 30 min. Following this step, slides were immersed in pre-chilled lysis solution in the dark at 4 °C for 1 h. The excess fluid was subsequentially removed and the slides were incubated in pre-chilled neutral electrophoresis buffer in the dark at 4 °C for 30 min. Later, the slides were placed in a new neutral buffer and then electrophoresed at 25 V for 1 h in the dark at 4 °C. With the excess electrophoresis solution aspirated, the slides were immersed in the DNA Precipitation Solution, and placed in the dark at RT for 30 min followed by incubation in 70% ethanol for 30 min in the dark at RT, and dried at 40 °C for 15 min. The nuclei were stained with 5 μg/mL of ethidium bromide (EB) at RT in the dark for 20 min. The images were observed by Olympus fluorescence microscope BX53 (Olympus Japan). DNA damage and migration were assessed (100 cells in each group) using the CASP software package [32].
2.9. Transmission electron microscopy (TEM)
The placental samples were post-fixed with 4% glutaraldehyde and washed in phosphate buffer four times for 1 h each. After that, the samples were post-fixed with 1% osmium tetroxide for 2 h at 4 °C. Then the samples were rinsed in ddH2O three times for 10 min each, followed by rinsing further in 2% aqueous uranyl acetate for 2 h at RT, protected from light. Sample dehydration: 50% ethanol for 15 min, 70% ethanol for 15 min, 90% ethanol for 15 min, 100% ethanol for 15 min, and 100% acetone two times for 20 min each. Following the dehydration step, the samples were treated with 100% acetone: embedding agent Epon 812 (1:1) at RT for 2 h. Later, the placental tissues were embedded, polymerized, cut into slices, stained using lead citrate, and aired. Finally, the sections were observed using a transmission electron microscope (Thermo scientific, Talos L120C G2).
2.10. Seahorse analysis
HTR-8 cells were seeded at 25,000 cells per well 24 h before the measurement and equilibrated in a CO2-free incubator at 37 °C for 1 h. Analyses were performed using Oligomycin (1 μM), and Rotenone (1 μM) plus Antimycin A (1 μM) as indicated. Seahorse measurements were performed on Seahorse XFp Extracellular Flux Analyzer (Agilent).
2.11. Mitochondrial OXPHOS complex enzyme activity assay
Placental mitochondria complex enzyme activity was detected following the manufacturer's instruction. The enzymatic activities of complex I (NADH dehydrogenase), complex II (succinate-coenzyme Q reductase), complex III (CoQ-cytochrome C reductase), complex IV (Cytochrome C oxidase) and complex V (F1F0-ATP synthase) were assayed using Mitochondrial Respiratory Complex I–V Activity Assay Kit (SolarBio, BC0515/BC3235/BC3245/BC0945/BC1445). Absorbance was measured using a multifunctional microplate reader (BioTek, SENERGX2 Synergy 4). The BCA method was then used to determine the protein concentration.
2.12. Live cell imaging
HTR-8 cells were inoculated with 6 × 105 per well in 35 mm confocal dishes (Biosharp, BS-20-GJM) before MEHP treatment. MitoTracker (Maokangbio, M7514) and Hoechst 33342 (Invitrogen, H3570) were used to label mitochondria and nuclei respectively. Live cells were visualized 24 h post the MEHP treatment by confocal microscopy (LEICA, THUNDER Imager 3D Live Cell).
2.13. Statistical analysis
The data were analyzed with SPSS, version 23.0 (SPSS, Chicago, IL, USA), and represented as means ± standard error (SEM). Student's t-test, Analysis of variance (ANOVA), Mann-Whitney U test, and Kruskal-Wallis H test were used as appropriate. Statistical graphs were produced in GraphPad Prism 7. P < 0.05 was considered a significant difference.
3. Results
3.1. Trophoblast area reduced by DEHP in placenta
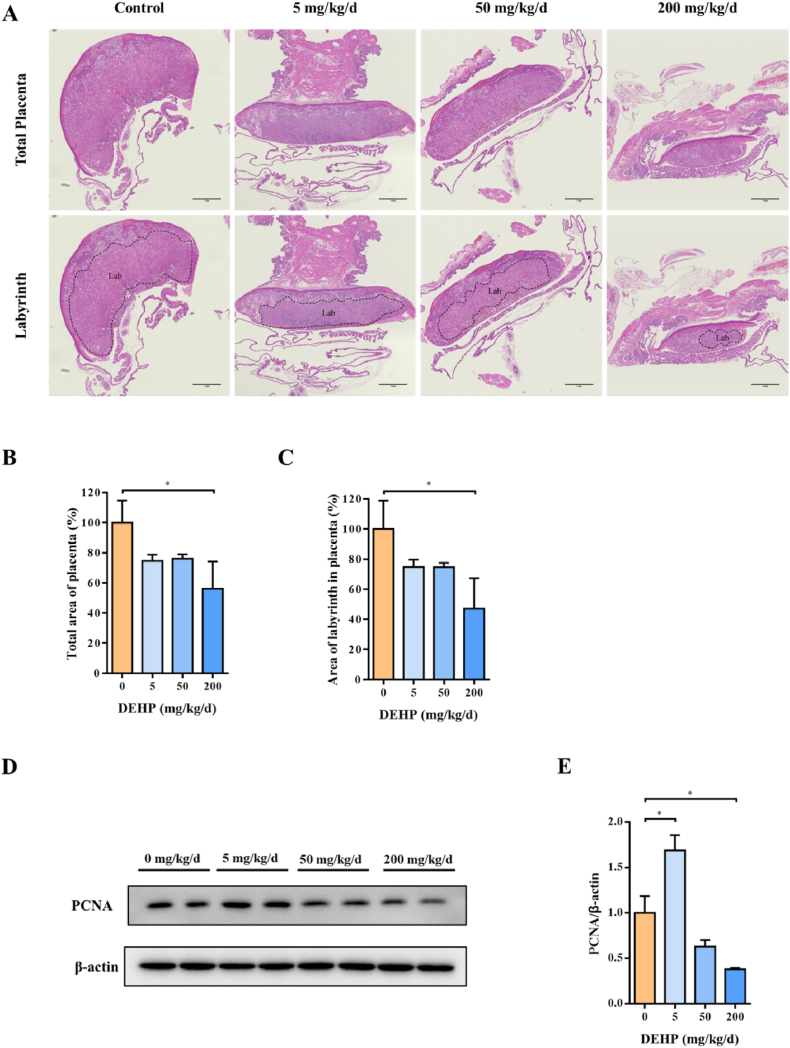
Pregnant mice were treated with DEHP during gestational day 0–15 (GD 0–15) (Supplementary Figs. 1A–G). Placental sections from GD 15 were stained with hematoxylin and eosin to explore the effect of DEHP on placenta development. Upon the treatment of DEHP, the total placental area decreased with the increase of drug dosage and showed a clear decrease at 200 mg/kg/d (Fig. 1A and B). The area ratio of the labyrinth trophoblast layer also decreased (Fig. 1C). PCNA protein is a marker of proliferation, and its expression was negatively correlated with DEHP concentration (Fig. 1D and E). These evidences suggest that maternal exposure to DEHP during pregnancy inhibits placental trophoblast development.
Fig. 1.
Comparison of the histopathology in normal and DEHP exposed placentas. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. (A) Representative images of the largest longitudinal section of GD 15 placenta detected by H&E staining. The ranges of the labyrinth were outlined in the lower part of the figures. Data are normalized by the control. (B) Total area of placenta. (C) Area of labyrinth in placenta. Lab: labyrinth. Scale bar: 1 mm. (D) Representative images of PCNA using Western blot, and (E) quantitative analysis of PCNA protein levels in mice placenta. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference compared to control (P < 0.05).
3.2. DNA damage in placenta triggered by DEHP
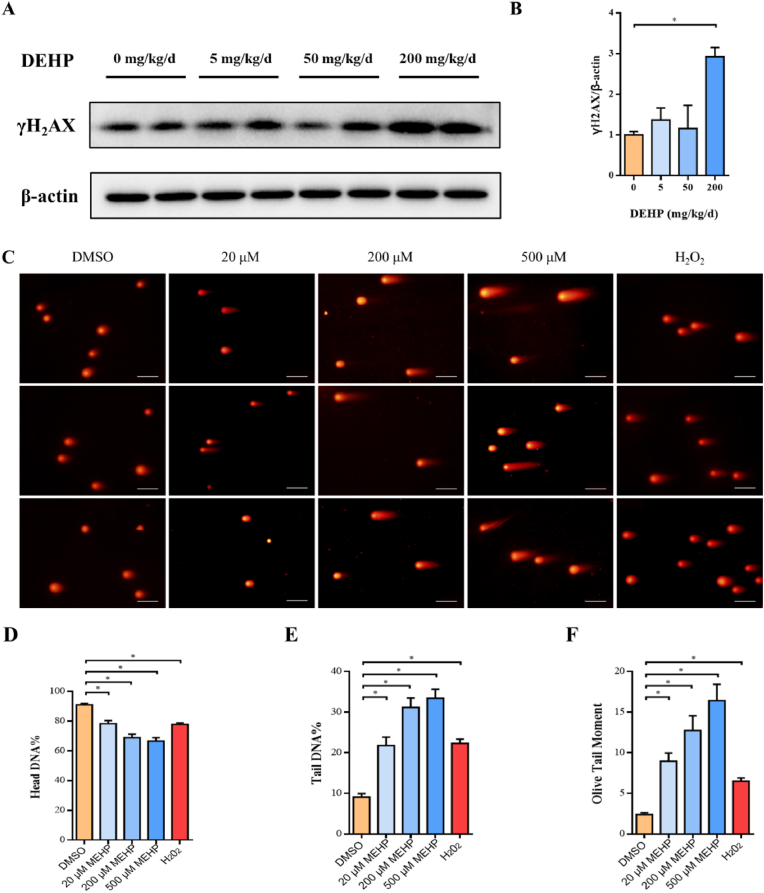
In order to determine whether trophoblast proliferation deficiency was related to DNA damage, we detected the level of γH2AX phosphorylation, the marker of DNA damage, in placenta by Western-Blotting assay (Fig. 2A). Compared with the control group, the phosphorylation levels of γH2AX increased in DEHP-treated groups when the DEHP concentration reaches 200 mg/kg/d (Fig. 2B). A comet assay was then carried out in HTR-8 cells to further confirm the formation of DNA damage (Fig. 2C). As cultured cells in vitro lack a complete metabolic system similar to the animal, we treated HTR-8 cells with Mono-(2-ethylhexy) phthalate (MEHP), the bioactive metabolite of DEHP. The head DNA percentage, tail DNA percentage, and olive tail moment were analyzed, and the results showed the head DNA percentage is negatively correlated with MEHP concentration, while the percentage of tail DNA and the comet tail moment increased in a MEHP dose-dependent manner (Fig. 2D–F). These results suggest that DEHP exposure leads to the accumulation of placental DNA damage.
Fig. 2.
DNA damage in placenta triggered by DEHP. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. HTR-8 cells were treated with MEHP (0, 20, 200, 500 μM) for 24 h or 500 μM H2O2 for 30 min. (A) Representative images of phosphorylated γ-H2AX using Western blot, and (B) quantitative analysis of phosphorylated γ-H2AX levels in mice placenta. (C) Representative images of the comet assays. (D) Head DNA percentage, (E) Tail DNA percentage, and (F) Olive tail moment was analyzed in HTR-8 cells, respectively. Scale bar: 100 μm. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference compared to control (P < 0.05).
3.3. Effect of DEHP exposure on PARP1 protein expression and activity
When DNA damage occurs, PARP1 will be activated and binds to the damaged site to build a repair factors platform [33]. To evaluate whether the DNA damage repair pathways were activated, we examined the protein level and activity of PARP1 in mouse placenta. In the DEHP-treated placenta, the protein concentration of PARP1 hardly changed, while the concentration of cleaved PARP1 was up-regulated along with the increase of DEHP dosage (Fig. 3A–C). Since DNA repair proteins and PARP1 itself renovate DNA damage through Poly-ADP-ribosylation (PARylation) [34], the PARylation levels were also evaluated. Compared with the control group, the PARylation level of PARP1 was down-regulated along with the increase of DEHP concentration (Fig. 3D–E). These results indicate that DEHP did not affect the level of PARP1 protein but restrained PARP1 activity and thus likely hindered DNA repair in mouse placenta.
Fig. 3.
Effect of DEHP exposure on PARP1 protein expression and activity. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. (A) PARP-1, Cleaved PARP-1, (D) Total protein PARylation representative images of Western blot. (B) PARP-1, (C) Cleaved PARP-1, (E) PAR protein levels of quantitative analysis in mice placenta. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference (P < 0.05).
3.4. The distribution of NAD+ in three pools in placenta treated with DEHP
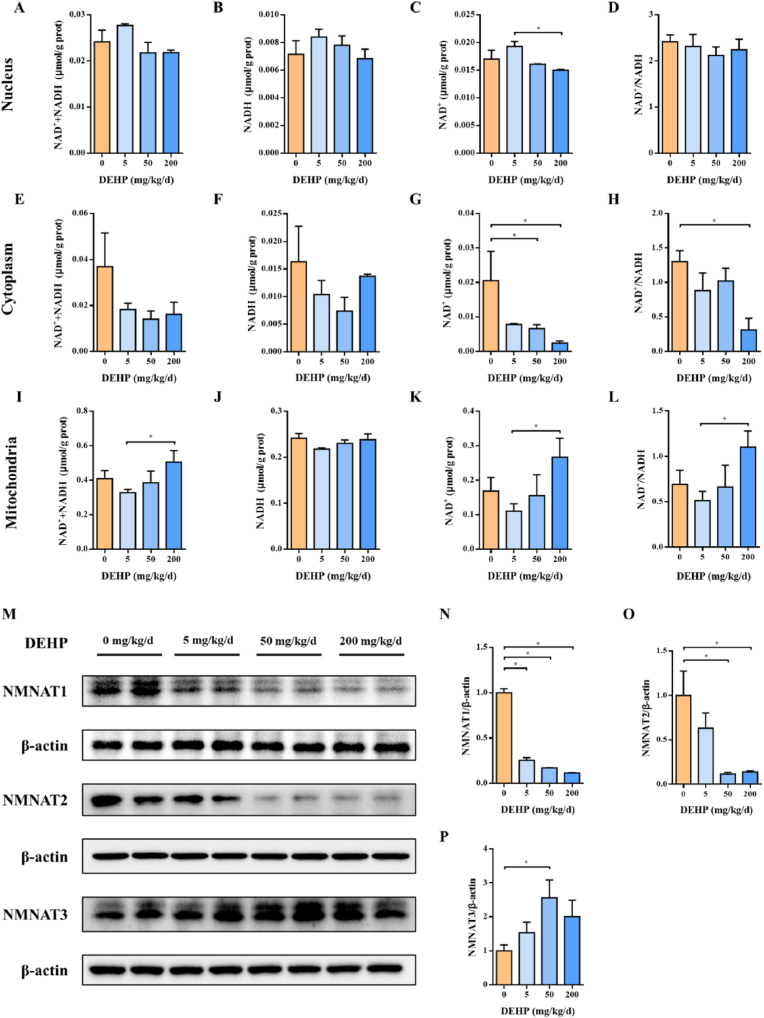
To execute the DNA repair function, PARP1 should be kept active in the presence of NAD+ in the nucleus and ATP [35,36]. Many biological functions of NAD+ are organized and coordinated through its compartmentalization within the cell [37]. Bio-synthesis of NAD+ in different pools requires NMNAT enzymes at different subcellular locations [38]: NMNAT1/2/3 is located at the nucleus, cytoplasm, and mitochondria, respectively. To assess the level of NAD+ located in different pools, we fractionated mouse placenta to isolate the nucleus, cytoplasm, and mitochondria. As shown in Fig. 4, NAD+ levels declined in the nucleus and cytoplasm, while ascended in the mitochondria in the DEHP-treated mouse placenta (Fig. 4A-L). To check whether the NAD+ level change is due to the change of their respective NMNATs, the NMNAT1/2/3 levels were examined. Interestingly, with the rising of DEHP concentration, the protein levels of NMNAT1 and NMNAT2 decreased, while the protein level of NMNAT3 increased (Fig. 4M–P), which is consistent with our hypothesis. These results showed that DEHP reduced nuclear and cytoplasmic NAD+ concentrations by reducing the protein level of NAD+ synthetase NMNAT1 and NMNAT2 while increasing mitochondrial NAD+ levels by elevating the protein level of synthetase NMNAT3.
Fig. 4.
NAD + concentration and bio-synthesis changed in different subcellular compartments after DEHP treatment. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. NAD+ content was measured in placental (A–D) nucleus, (E–H) cytoplasm, and (I–L) mitochondria, respectively. (M) NMNAT1, NMNAT2, and NMNAT3 representative images of Western blot. Quantifications of (N) NMNAT1, (O) NMNAT2, and (P) NMNAT3 expression in mice placenta based on band intensity were shown on the right side of the blots. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference (P < 0.05).
3.5. NAD + depletion in pregnant mice triggered by DEHP
From the experiments above, we have observed that DEHP regulated the compartmentalized distribution of NAD+ in placental cells. Furthermore, we observed that DEHP also reduced the concentration of NAD+ in serum and placenta during pregnancy (Fig. 5A–H). Similar results were also observed in HTR-8 cells (Fig. 5I-L). These results suggest that DEHP not only regulates the regional distribution of NAD+ in placental cells but also influences the source of NAD+ entering the placenta. To investigate whether the change of the NAD+ level was due to its metabolic processes, the NAD+ hydrolases and synthases levels were measured. The protein levels of NAD+ hydrolase CD38 and CD157 in DEHP-treated groups were higher than those in the control groups (Fig. 5M–O). However, the protein level of NAMPT, a synthetase that converts NAM to NMN, almost remained unchanged (Fig. 5P and Q). Overall, DEHP may induce an increase in the expression level of NAD+ hydrolases, leading to the depletion of NAD+ in serum and placenta during pregnancy.
Fig. 5.
NAD + depletion in pregnant mice triggered by DEHP. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. HTR-8 cells were treated with MEHP (0, 20, 200, 500 μM) for 24 h. NAD+ content was measured in (A–D) serum, (E–H) placenta, and (I–L) HTR-8 cells, respectively. (M) CD38, CD157, and (N) NAMPT representative images of Western blot. Quantifications of (O) CD38, (P) CD157, and (Q) NAMPT expression in mice placenta based on band intensity were shown below the blots. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference compared to control (P < 0.05).
3.6. DEHP exposure causes a decrease in ATP levels by blocking the mitochondrial electron transport chain
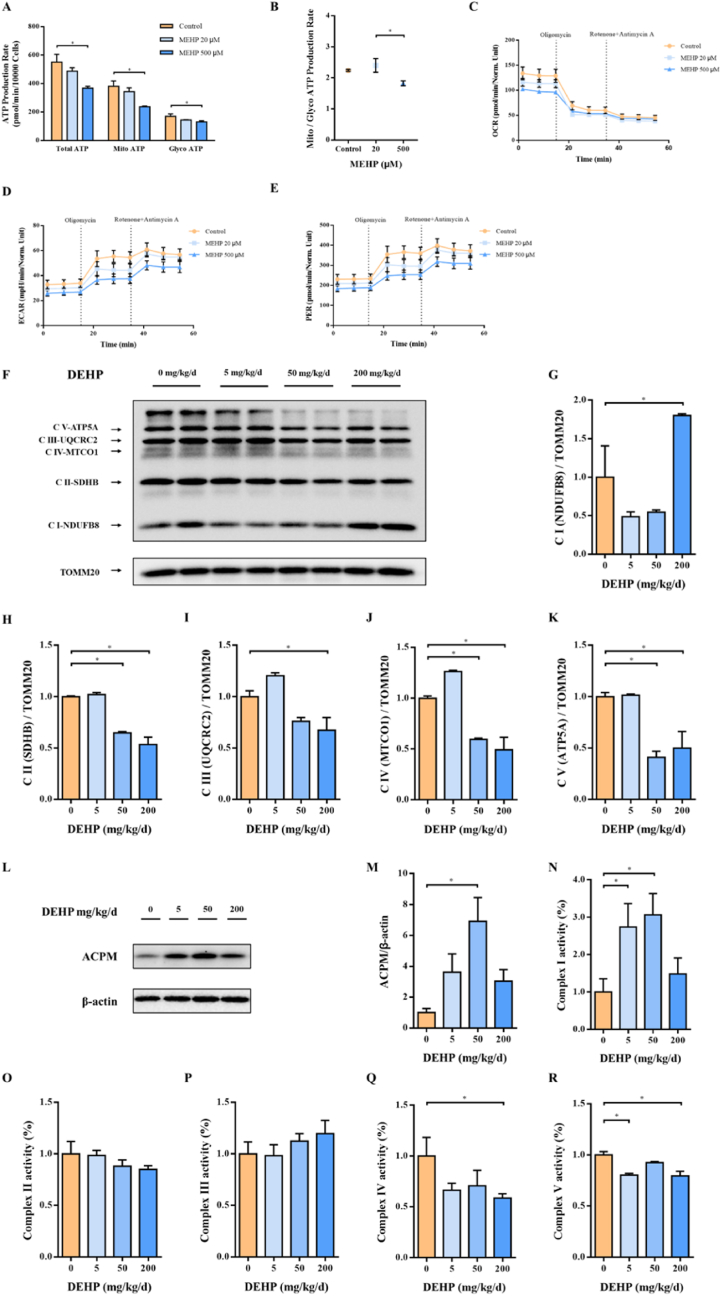
To evaluate the level of ATP, another donor's contribution to PARP1 activity upon DEHP exposure, we performed Seahorse XF Real-Time ATP Rate assay in HTR-8 cells exposed to MEHP, the bioactive metabolite of DEHP. ATP is primarily produced by oxidative phosphorylation and glycolysis in the cell. Oxygen consumption rate (OCR) is generated by mitochondrial electron transport and used to evaluate oxidative phosphorylation function. Extracellular acidification rate (ECAR) results from glycolytic acidification and mitochondrial acidification. After subtracting mitochondrial acidification, the resulting value is called the glycolytic proton efflux rate (PER). ECAR and PER can use together to evaluate glycolysis states. As shown in Fig. 6A and B, the ATP production rate was repressed by MEHP in both cytoplasm and mitochondria. OCR, ECAR, and PER had a downward trend (Fig. 6C–E). In order to figure out the reason for the decrease of ATP, we detected the protein levels and complex enzyme activity of the mitochondrial respiratory chain. The protein levels of complex II-V of the mitochondrial respiratory chain decreased significantly, while complex I increased (Fig. 6F–K). To confirm the change of complex I, we detected the protein level of ACPM, the accessory and non-catalytic subunit of NADH dehydrogenase complex I, and it was indeed elevated in the placenta of DEHP treated mouse (Fig. 6L and M). In terms of OXPHOS complex enzyme activity, complex I activity was elevated, while complex IV and complex V activities were reduced (Fig. 6N–R). In summary, these results indicate that DEHP inhibits placental mitochondrial respiration and electron transport.
Fig. 6.
DEHP causes mitochondrial function reduced and electron-transport chain arrested in the placenta. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. HTR-8 cells were treated with MEHP (0, 20, 500 μM) for 24 h. ATP production rates due to glycolysis or mitochondrial respiration as measured with the Seahorse XF Real-Time ATP Rate assay. (A) ATP production rate, (B) Mitochondria/Glycolysis ATP production rate. (C) Oxygen consumption rate, (D) Extracellular acidification rate, and (E) Proton efflux rate, were measured in HTR-8 cells. (F) OXPHOS representative images of Western blot. Quantifications of (G) Complex I, (H) Complex II, (I) Complex III, (J) Complex IV, and (K) Complex V expression in mice placental mitochondria based on band intensity were shown on the right side of the blots. (L) Representative images of ACPM using Western blot, and (M) quantitative analysis of ACPM protein levels in mice placenta. Activity of mitochondrial (N) complex I, (O) complex II, (P) complex III, (Q) complex IV, and (R) complex V. Enzyme activity data are normalized by the control. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference compared to control (P < 0.05).
3.7. DEHP exposure caused a decrease in ATP level by destroying mitochondrial structure and reducing mitochondrial number in placenta
To investigate whether the decrease of ATP is related to the structure and quantity of mitochondria, we observed morphological changes of mitochondria in placenta by TEM and used a mitochondrial fluorescent probe in living cell images to represent the mitochondrial area. Under DEHP treatment, the structure of mitochondria was disrupted, and both the number and area of mitochondria were reduced in mouse placenta (Fig. 7A–C). The mitochondrial DNA copy number also declined (Fig. 7D). In addition, along with the rise of the MEHP concentration, the ratio of the mitochondrial area also decreased (Fig. 7E and F).
Fig. 7.
Effect of the structure and number of placental mitochondria affected by DEHP. Pregnant mice were administrated by DEHP (0, 5, 50 and 200 mg/kg/d) during GD 0–15. HTR-8 cells were treated with MEHP (0, 20, 200, 500 μM) for 24 h. (A) Respective mitochondrial morphology analysis of trophoblast cells in mice placenta. (B) Number of mitochondria, and (C) mean mitochondrial area. 13500 × , Scale bar: 500 nm. 45000 × , Scale bar: 200 nm. (D) Mitochondrial DNA copy number was detected in mice placenta. (E) Mitochondrial immunofluorescence representative image in live HTR-8 cells in control or MEHP treatment (after 24 h). 40 × , Scale bar: 50 μm. 100 × , Scale bar: 20 μm. (F) The area of mitochondria was measured per live HTR-8 cells and normalize. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference compared to control (P < 0.05).
Together, under DEHP exposure, the mitochondrial structure was disrupted, the mitochondrial respiratory chain was blocked and the quantity and area of mitochondria decreased, resulting in the reduction of ATP production.
3.8. NAM can alleviate DNA and mitochondrial damage in placenta induced by DEHP
Since the deficiency of NAD+ and ATP reduces the activity of PARP1, hinders the repair of DNA damage, and thus inhibits the proliferation of placental cells, we want to know whether supplementation of the precursor of NAD+ can rescue the phenotypes induced by DEHP treatment. There are many precursors to supplement NAD+, such as NA, NAM, NMN, and NR. NAM was selected as the appropriate supplemental precursor by detecting the NAD+ concentration in the serum of mice exposed to different precursors (Supplemental Fig. 2A). Mice treated with 500 mg/kg/d NAM for 2 h were selected through time course and concentration gradient assays (Supplemental Figs. 2B and C). Compared with the DEHP group, NAM intervention significantly reduced the level of γH2AX phosphorylation (Fig. 8A and B). The ability of NMN, NR, and NAM to produce NAD+ in different exposure duration was tested in HTR-8 cells (Supplemental Figs. 3A–H). HTR-8 cells pre-treated with 5 mM NAM for 6 h were selected. Compared with the MEHP group in the comet assay, the comet head DNA% increased, tail DNA% and tail moment showed a significant decrease in the group of NAM + MEHP (Fig. 8C–F). Surprisingly, through TEM, we found that NAM can better alleviate the damage of mitochondrial structure caused by DEHP treatment (Fig. 8G). Under NAM supplementation, the number and area of placental mitochondria also recovered (Fig. 8H and I). These results indicated that NAM supplementation alleviated the DNA and mitochondrial damage in the mouse of maternal exposed to DEHP during pregnancy.
Fig. 8.
NAM alleviated DNA damage and mitochondrial damage in placenta induced by DEHP. Pregnant mice were administrated by water, NAM (500 mg/kg/d), DEHP (50 mg/kg/d) during GD 0–15. NAM + DEHP group was firstly treated with NAM (500 mg/kg/d), then add DEHP (50 mg/kg/d) after 2 h, during GD 0–15. HTR-8 cells were treated with DMSO, NAM (5 mM), MEHP (200 μM), H2O2 (500 μM) for 24 h, 30 h, 24 h, 0.5 h, respectively. NAM + MEHP group was firstly treated with NAM (5 mM) for 6 h, then add MEHP (200 μM) to the medium for 24 h. (A) Representative images of phosphorylated γ-H2AX using Western blot, and (B) quantitative analysis of phosphorylated γ-H2AX protein levels in mice placenta. (C) Representative images of the comet assays. (D) Head DNA percentage, (E) Tail DNA percentage, and (F) Olive tail moment was analyzed in HTR-8 cells, respectively. Scale bar: 100 μm. (G) Respective mitochondrial morphology analysis of trophoblast cells in mice placenta. (H) Number of mitochondria, and (I) Mean mitochondrial area. 13500 × , Scale bar: 500 nm. 45000 × , Scale bar: 200 nm. All data are expressed as mean ± standard errors. (N = 6). * indicate a statistically significant difference (P < 0.05).
4. Discussion
Our previous study has shown that maternal exposure to environmental pollutants DEHP during pregnancy induces fetal growth restriction by inhibiting the proliferation of placental cells through DNA damage [17]. Placental development defects are associated with fetal growth retardation, leading to adverse pregnancy outcomes and some diseases in adulthood of offsprings [39,40]. Therefore, it is very essential to understand the mechanism of DNA damage in placental cells treated by DEHP. From a toxicological point of view, we started with DEHP concentrations that produced embryotoxicity and then considered its mechanism. This study explored the mechanism of DNA damage of placental cells by DEHP. The concentration of NAD+ in the nucleus and the production rate of ATP were down-regulated by DEHP treatment in placental cells, thus inhibiting the activity of PARP1 and hampering the DNA repair pathways, and finally leading to placental development disorder (Fig. 9).
Fig. 9.
Summary diagram. DEHP decreases the overall NAD + level of the placenta and reduces the NAD+ level in the placental nucleus by participating in the regulation of NAD+ regionalization. By destroying the placental mitochondrial structure, reducing the number of mitochondria, hindering the way of electron transfer in the respiratory chain, DEHP reduces ATP production. Under the combined action of NAD+ and ATP depletion, inhibition of PARP1 activity leads to the accumulation of DNA damage and ultimately to the reduction of the placental trophoblast area. The partially damaging effects of DEHP on DNA and mitochondrial structures can be alleviated by moderate supplementation of NAD+ precursor NAM.
A fetal growth restriction model of DEHP-treated in whole pregnancy was constructed in our previous study in which exposure to DEHP at different stages of fetal development reduced placental weight and diameter, especially on days 6–12 of pregnancy [41]. In this study, the same obstacles to the development of placenta and offsprings were found during DEHP exposure in GD 0–15 (Supplementary Figs. 1A–G). Impaired trophoblast proliferation can lead to placental tissue maldevelopment [42,43]. Consistent with the results of other research groups [15], we found that both the total area of placental and the proportion of placental trophoblast area in the total area decreased in the DEHP-treated groups compared with the control groups (Fig. 1). For proliferation, we found that the expression of PCNA protein increased under 5 mg/kg/d DEHP, but this did not change the placental pathology. Low-dose DEHP can promote cell proliferation, while high-dose DEHP inhibits cell proliferation [44]. This result may be caused by cellular stress responses, dose effect [45] and hormesis [46,47].
Occupational DEHP exposure may lead to oxidative distress and DNA damage [19]. DEHP and its metabolites can break down DNA in the liver [48], ovary [49], oocytes [28], Leydig cells [50], and placental trophoblasts cells [51] through reactive oxygen species (ROS). We also observed DNA damage in the placenta of mouse and HTR-8 cells. Normally, the repair pathways should be launched once DNA damage occurs [52]. However, the repair approach did not work effectively after DEHP induced DNA damage. Thus, we decided to investigate the mechanism of DNA damage caused by DEHP from the perspective of DNA repair failure. PARP1 is a nuclear enzyme that is activated by DNA damage and plays a crucial role in repairing DNA breaks [53]. In zebrafish larvae and HEK293T cells treated with DEHP or MEHP, the mRNA level of PARP1 increased [54]. In MEHP-treated HTR-8 cells, however, PARP1 protein expression significantly decreased [17]. In this study, we discovered that the PARP1 protein level was almost unchanged in placenta, but its activity was down-regulated. This may be related to differences in biological samples, drug concentrations, and processing times. NAD+ and ATP are the essential substrates for PARP1 activity [23,24]. We attempted to clarify the reasons for the change of PARP1 activity by considering these two aspects.
NAD+ is a cofactor of key enzymes in glycolysis, TCA cycle, and oxidative phosphorylation, participating in a variety of redox reactions in cells [55]. NAD+ is distributed in the nucleus, cytoplasm, and mitochondria and is bio-synthesized by NAMAT1, 2 and 3, respectively [38]. PARP1 is the major consumer of the nuclear NAD+ pool [56]. NMNAT1 is recruited to the target gene promoters by PARP1 to generate NAD+ to support the catalytic activity of PARP1, and the reduction of NMNAT1 hinders DNA repair [57]. Over-activation of PARP1 consumes a large amount of NAD+ and reduces it [58,59] and reduction of NAD+ further inhibited the activity of PARP1 [[60], [61], [62]], which might suggest a negative feedback regulation. The study by Santhosh A. et al. showed that DEHP inhibits NAD+ bio-synthesis in erythrocytes, resulting in a decrease in NAD+ levels [63]. Similarly, we found that DEHP decreased the NAD+ concentration in the placenta and blood of mice. Moreover, DEHP also decreased the level of NMNAT1. Therefore, DEHP down-regulates the bio-synthesis of NAD+ by abating the protein level of NMNAT1, thereby interfering with the activity of PARP1.
We found an interesting phenomenon that the level of NAD+ decreased in both nucleus and cytoplasm but increased in mitochondria, which is challenging to explain. A major cause of cell death resulting from genotoxic stress is believed to be the depletion of NAD+ in the nucleus and cytoplasm. Even though the nucleus and cytoplasm pools of NAD+ are depleted, the higher NAD+ level in mitochondria can still maintain cell viability and protect cells from death [64]. There is evidence that the increase of mitochondrial NAD+ can promote cell survival during genotoxic stress, and the content of mitochondrial NAD+ is the main determinant of cell apoptosis [64]. Sirt3, also an NAD+-dependent enzyme, plays a key role in the maintenance of mitochondrial function [65]. Under DEHP treatment, the protein expression of Sirt3 increased in the mouse placenta (data not shown). High concentrations of NAD+ in mitochondria may provide the last chance for cell survival.
Liu H. et al. demonstrated that DEHP inhibits the Nrf2 pathway and mitochondrial biogenesis pathway in HepG2 cells, resulting in excessive autophagy and down-regulated ATP level [66]. In the testis microenvironment, DEHP causes lactic acid accumulation and insufficient ATP supply by disrupting the tricarboxylic acid cycle and gluconeogenesis [29]. Similarly, a decrease in ATP levels was also observed in cardiomyocytes [67] and mouse oocytes [28] treated with DEHP. In this study, we found that DEHP down-regulated ATP production by disrupting placental mitochondrial structure, reducing mitochondrial numbers, and inhibiting OXPHOS complex protein expression and enzymatic activity. These phenomena could be a direct toxic effect of DEHP/MEHP, or an indirect toxic effect caused by ROS. Firstly, MEHP can pass through the placenta [68]. It may have direct toxic effects on placental mitochondria. Next, mitochondria are major sources of reactive oxygen species [69], electrons in the mitochondrial electron transport chain combine with molecular oxygen to form ROS superoxide anion radical (O2•−) [70] which can directly damage mitochondrial and cellular molecules or is converted into other oxidants [71]. ROS mainly includes superoxide anion radical, hydrogen peroxide, hydroxyl radical, and others [72]. Studies have shown that DEHP [73] and MEHP [74] can cause oxidative distress in mitochondria and generate a large amount of ROS (hydrogen peroxide or superoxide anion radical). High level of oxidative challenge is associated with biomolecular damage [75]. This oxidative damage may further inhibit mitochondrial respiration and electron transport, creating a vicious circle. At the same time, the complex antioxidant network possessed by mitochondria may also be affected [76,77]. This may lead to the disruption of the vitagenes network including heme oxygenase, γ-glutamyl cysteine ligase, and sirtuins [78]. Eventually resulting in mitochondrial fragmentation, mitophagy and cell death [[79], [80], [81]]. But it is still unclear why the respiratory chain was inhibited in the context of elevated NAD+, which is the focus of our future studies.
Nicotinamide is a vitamin that serves as a precursor of NAD+ and has DNA repair and anti‐inflammatory properties in different studies [[82], [83], [84], [85]]. Li F. et al. shown that nicotinamide benefits both dams and pups [86]. Dietary nicotinamide improved the maternal condition, prolongs pregnancies, and prevents FGR in two contrasting mouse models of preeclampsia [86]. For DNA damage, NAM can suppress cisplatin-induced DNA damage in kidney [84], enhance DNA repair in keratinocytes [87] and decrease DNA fragmentation in neure [88,89] and microvascular endothelial cell [90]. Supplementing NAD+[91] or other NAD + precursors, such as NMN [92], NR [93], can also alleviate DNA damage. However, there are also study indicated that NAM can cause DNA damage in tumours/normal tissues in mice [94] and inhibit DNA repair [95]. In this study, we found NAM can alleviate DNA damage. induced by DEHP. The main reason is the supplementation of NAD+. NMNAT1 is the last enzyme of the synthesis of NAD+ in nuclear. DEHP decreased the level of NMNAT1. However, the supplementation of NAD+ precursors still elevated the concentration of NAD+. This is also part of our future research. Niacinamide supplementation might stimulate the activity or expression of NMNATs. In the presence of sufficient NAM, NAD+ is continuously supplied for DNA repair pathways. Moreover, NAM could prevent PARP1 degradation and permit DNA repair by directly inhibiting caspase 3-like activity [96]. Under the intervention of NAM, DNA damage in the DEHP groups was significantly alleviated in our study. DNA damage in the NAM + DEHP group was lower than that in the NAM and DEHP groups alone. The same results were obtained from in vitro experimental validation. We thus speculated that this may reflect the antagonistic effects between DEHP and NAM.
Fluctuations in NAD+ impact mitochondrial function and metabolism [97]. NAD+ and NAD+ precursors had been shown to be capable of boosting mitochondrial function [98]. The treatment of different species or cells with NAM or NR can improve mitochondrial biogenesis and function [98,99]. Through TEM, we also observed that NAM supplementation could partially ameliorate structural damage to mitochondria.
In the future, efforts will be made to understand the molecular mechanisms underlying this current study. Such as, whether there is a negative feedback relationship between NAD+ and ATP, whether NAM can promote the production of ATP, and whether NAM supplementation can regulate mitochondrial bio-synthesis, etc. We hope to provide insights into alleviating genotoxic stress induced by exogenous chemicals.
5. Conclusion
In conclusion, DEHP blocks the activity of PARP1 in DNA damage repair by down-regulating the bio-synthesis of nuclear NAD+ and mitochondrial ATP, resulting in the inhibition of placental cell proliferation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [82073593], the Scientific Research Projects in Colleges and Universities of Anhui Education Department [KJ2019A0281 and KJ2020A0666], Domestic Visiting and Training Project for Excellent Young Key Teachers of Colleges and Universities [gxgnfx2020115], Postgraduate Scientific Research Projects of Universities in Anhui Province [YJS20210289] and Research level improvement program of Anhui Medical University [2021xkjT010]. We thank Yang Cai and Xia Ning (Anhui Medical University) for material preparation. We thank Dr. Bo Wang (Anhui Medical University) for its generous donation of antibodies. We frankly thank all participants involved in this study, finally.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102414.
Contributor Information
Tao Xu, Email: xutao@hfuu.edu.cn.
Ling-li Zhao, Email: zhaolingli@ahmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Wang H., Zhang J.M., Xu Z.J., Yang J.X., Xu Y., Liu Y., Li B.H., Xie J.S., Li J. Circular RNA hsa_circ_0000848 promotes trophoblast cell migration and invasion and inhibits cell apoptosis by sponging hsa-miR-6768-5p. Front. Cell Dev. Biol. 2020;8:278. doi: 10.3389/fcell.2020.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maitre L., Fthenou E., Athersuch T., Coen M., Toledano M.B., Holmes E., Kogevinas M., Chatzi L., Keun H.C. Urinary metabolic profiles in early pregnancy are associated with preterm birth and fetal growth restriction in the Rhea mother-child cohort study. BMC Med. 2014;12:110. doi: 10.1186/1741-7015-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du T.T., Fernandez C., Barshop R., Guo Y.J., Krousel-Wood M., Chen W., Qi L., Harville E., Mauvais-Jarvis F., Fonseca V., Bazzano L. Sex differences in cardiovascular risk profile from childhood to midlife between individuals who did and did not develop diabetes at follow-up: the bogalusa heart study. Diabetes Care. 2019;42:635–643. doi: 10.2337/dc18-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pásztor N., Sikovanyecz J., Keresztúri A., Kozinszky Z., Németh G. Evaluation of the relation between placental weight and placental weight to foetal weight ratio and the causes of stillbirth: a retrospective comparative study. J. Obstet. Gynaecol. 2018;38:74–80. doi: 10.1080/01443615.2017.1349084. [DOI] [PubMed] [Google Scholar]
- 5.Maltepe E., Fisher S.J. Placenta: the forgotten organ. Annu. Rev. Cell Dev. Biol. 2015;31:523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- 6.Latorre I., Hwang S., Sevillano M., Montalvo-Rodriguez R. PVC biodeterioration and DEHP leaching by DEHP-degrading bacteria. Int. Biodeterior. Biodegrad. 2012;69:73–81. doi: 10.1016/j.ibiod.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raeisi A., Faghihi K., Shabanian M. Designed biocompatible nano-inhibitor based on poly(β-cyclodextrin-ester) for reduction of the DEHP migration from plasticized PVC. Carbohydr. Polym. 2017;174:858–868. doi: 10.1016/j.carbpol.2017.06.105. [DOI] [PubMed] [Google Scholar]
- 8.Zou Q.Y., Hong S.L., Kang H.Y., Ke X., Wang X.Q., Li J., Shen Y. Effect of di-(2-ethylhexyl) phthalate (DEHP) on allergic rhinitis. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grady R., Sathyanarayana S. An update on phthalates and male reproductive development and function. Curr. Urol. Rep. 2012;13:307–310. doi: 10.1007/s11934-012-0261-1. [DOI] [PubMed] [Google Scholar]
- 10.Al-Saleh I., Coskun S., Al-Doush I., Abduljabbar M., Al-Rouqi R., Al-Rajudi T., Al-Hassan S. Couples exposure to phthalates and its influence on in vitro fertilization outcomes. Chemosphere. 2019;226:597–606. doi: 10.1016/j.chemosphere.2019.03.146. [DOI] [PubMed] [Google Scholar]
- 11.Khasin L.G., Rosa J.D., Petersen N., Moeller J., Kriegsfeld L.J., Lishko P.V. The impact of di-2-ethylhexyl phthalate on sperm fertility. Front. Cell Dev. Biol. 2020;8:426. doi: 10.3389/fcell.2020.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parra-Forero L.Y., Veloz-Contreras A., Vargas-Marín S., Mojica-Villegas M.A., Alfaro-Pedraza E., Urióstegui-Acosta M., Hernández-Ochoa I. Alterations in oocytes and early zygotes following oral exposure to di(2-ethylhexyl) phthalate in young adult female mice. Reprod. Toxicol. 2019;90:53–61. doi: 10.1016/j.reprotox.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Radke E.G., Braun J.M., Meeker J.D., Cooper G.S. Phthalate exposure and male reproductive outcomes: a systematic review of the human epidemiological evidence. Environ. Int. 2018;121:764–793. doi: 10.1016/j.envint.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radke E.G., Glenn B.S., Braun J.M., Cooper G.S. Phthalate exposure and female reproductive and developmental outcomes: a systematic review of the human epidemiological evidence. Environ. Int. 2019;130 doi: 10.1016/j.envint.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong T., Lai L., Hu J., Guo M.J., Li M., Zhang L., Zhong C.X., Yang B., Wu L., Zhang D.L., Tang M., Kuang H.B. Maternal exposure to di-(2-ethylhexyl) phthalate disrupts placental growth and development in pregnant mice. J. Hazard Mater. 2015;297:25–33. doi: 10.1016/j.jhazmat.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Razo L.D., Martínez-Ibarra A., Vázquez-Martínez E.R., Cerbón M. The impact of Di-(2-ethylhexyl) Phthalate and Mono(2-ethylhexyl) Phthalate in placental development, function, and pathophysiology. Environ. Int. 2021;146 doi: 10.1016/j.envint.2020.106228. [DOI] [PubMed] [Google Scholar]
- 17.Sun C.C., Zhao S., Chu L.L., Zhang S.Y., Li Y.L., Sun M.F., Wang Q.N., Huang Y.C., Zhang J., Wang H., Gao L., Xu D.X., Zhang S.C., Xu T., Zhao L.L. Di (2-ethyl-hexyl) phthalate disrupts placental growth in a dual blocking mode. J. Hazard Mater. 2022;421 doi: 10.1016/j.jhazmat.2021.126815. [DOI] [PubMed] [Google Scholar]
- 18.Yin J.C., Liu R., Jian Z.H., Yang D., Pu Y.P., Yin L.H., Wang D.Y. Di (2-ethylhexyl) phthalate-induced reproductive toxicity involved in dna damage-dependent oocyte apoptosis and oxidative stress in Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 2018;163:298–306. doi: 10.1016/j.ecoenv.2018.07.066. [DOI] [PubMed] [Google Scholar]
- 19.Gurdemir G., Erkekoglu P., Balci A., Sur U., Ozkemahli G., Tutkun E., Yilmaz H., Asci A., Kocer-Gumusel B. Oxidative stress parameters, selenium levels, DNA damage, and phthalate levels in plastic workers. J. Environ. Pathol. Toxicol. Oncol. 2019;38:253–270. doi: 10.1615/JEnvironPatholToxicolOncol.2019026470. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch S., Marshall L.V., Lechon F.C., Pearson A.D.J., Moreno L. Targeted approaches to childhood cancer: progress in drug discovery and development. Expet Opin. Drug Discov. 2015;10:483–495. doi: 10.1517/17460441.2015.1025745. [DOI] [PubMed] [Google Scholar]
- 21.Kim H., Park J., Kang H., Yun S.P., Lee Y.S., Lee Y.I., Lee Y.J. Activation of the Akt1-CREB pathway promotes expression to inhibit PARP1-mediated neuronal death. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aax7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri A.R., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alemasova E.E., Lavrik O.I. Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019;47:3811–3827. doi: 10.1093/nar/gkz120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D.L., Hu X., Li J., Liu J., Bulte L.B.T., Wiersma M., Malik N.U.A., Marion D.M.S., Tolouee M., Hoogstra-Berends F., Lanters E.A.H., Roon A.M., Vries A.A.F., Pijnappels D.A., Groot N.M.S., Henning R.H., Brundel B.J.J.M. DNA damage-induced PARP1 activation confers cardiomyocyte dysfunction through NAD(+) depletion in experimental atrial fibrillation. Nat. Commun. 2019;10:1307. doi: 10.1038/s41467-019-09014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Z.F., Zhang X.N., Nasertorabi F., Cheng Q.Q., Pei H., Louie S.G., Stevens R.C., Zhang Y. Facile chemoenzymatic synthesis of a novel stable mimic of NAD. Chem. Sci. 2018;9:8337–8342. doi: 10.1039/c8sc03899f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang N.C., Cho Y.H., Lee I. The lifespan extension ability of nicotinic acid depends on whether the intracellular NAD(+) level is lower than the sirtuin-saturating concentrations. Int. J. Mol. Sci. 2019;21:142. doi: 10.3390/ijms21010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Geng A., Zhang W.N., Qian Z., Wan X.P., Jiang Y., Mao Z.Y. Fight to the bitter end: DNA repair and aging. Ageing Res. Rev. 2020;64 doi: 10.1016/j.arr.2020.101154. [DOI] [PubMed] [Google Scholar]
- 28.Lu Z.Z., Zhang C.T., Han C.Q., An Q.L., Cheng Y.Y., Chen Y.Z., Meng R., Zhang Y., Su J.M. Plasticizer bis(2-ethylhexyl) phthalate causes meiosis defects and decreases fertilization ability of mouse oocytes in vivo. J. Agric. Food Chem. 2019;67:3459–3468. doi: 10.1021/acs.jafc.9b00121. [DOI] [PubMed] [Google Scholar]
- 29.Shen G.L., Zhou L.L., Liu W., Cui Y., Xie W.P., Chen H.M., Yu W.L., Li W.T., Li H.S. Di(2-ethylhexyl)phthalate alters the synthesis and β-oxidation of fatty acids and hinders ATP supply in mouse testes via UPLC-Q-exactive orbitrap MS-based metabonomics study. J. Agric. Food Chem. 2017;65:5056–5063. doi: 10.1021/acs.jafc.7b01015. [DOI] [PubMed] [Google Scholar]
- 30.Isaac S.M., Langford M.B., Simmons D.G., Adamson S.L. 4 - anatomy of the mouse placenta throughout gestation. Guide to Investig Mouse Pregn. 2014:69–73. doi: 10.1016/B978-0-12-394445-0.00004-7. [DOI] [Google Scholar]
- 31.Malik A.N., Czajka A., Cunningham P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion. 2016;29:59–64. doi: 10.1016/j.mito.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Glei M., Schneider T., Schlörmann W. Comet assay: an essential tool in toxicological research. Arch. Toxicol. 2016;90:2315–2336. doi: 10.1007/s00204-016-1767-y. [DOI] [PubMed] [Google Scholar]
- 33.Naumann M., Pal A., Goswami A., Lojewski X., Japtok J., Vehlow A., Naujock M., Günther R., Jin M.M., Stanslowsky N., Reinhardt P., Sterneckert J., Frickenhaus M., Pan-Montojo F., Storkebaum E., Poser I., Freischmidt A., Weishaupt J.H., Holzmann K., Troost D., Ludolph A.C., Boeckers T.M., Liebau S., Petri S., Cordes N., Hyman A.A., Wegner F., Grill S.W., Weis J., Storch A., Hermann A. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 2018;9:335. doi: 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bianchi A., Lopez S., Altwerger G., Bellone S., Bonazzoli E., Zammataro L., Manzano A., Manara P., Perrone E., Zeybek B., Han C., Menderes G., Ratner E., Silasi D.A., Huang G.S., Azodi M., Newberg J.Y., Pavlick D.C., Elvin J., Frampton G.M., Schwartz P.E., Santin A.D. PARP-1 activity (PAR) determines the sensitivity of cervical cancer to olaparib. Gynecol. Oncol. 2019;155:144–150. doi: 10.1016/j.ygyno.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannan S., Fang W., Song G.C., Mullighan C.G., Hammitt R., McMurray J., Zweidler-McKay P.A. Notch/HES1-mediated PARP1 activation: a cell type-specific mechanism for tumor suppression. Blood. 2011;117:2891–2900. doi: 10.1182/blood-2009-12-253419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai P., Houten S.M., Huber A., Schreiber V., Watanabe M., Kiss B., Murcia G., Auwerx J., Murcia J.M. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J. Biol. Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 37.Cambronne X.A., Kraus W.L. Location, location, location: compartmentalization of NAD(+) synthesis and functions in mammalian cells. Trends Biochem. Sci. 2020;45:858–873. doi: 10.1016/j.tibs.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdin E. NAD⁺ in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 39.Meyer N., Schumacher A., Coenen U., Woidacki K., Schmidt H., Lindquist J.A., Mertens P.R., Zenclussen A.C. Y-box binding protein 1 expression in trophoblast cells promotes fetal and placental development. Cells. 2020;9:1942. doi: 10.3390/cells9091942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrecht C., Chamley L., Charnock-Jones D.S., Collins S., Fujiwara H., Golos T., Grayo S., Hannan N., Harris L., Ichizuka K., Illsley N.P., Iwashita M., Kallol S., Al-Khan A., Lash G., Nagamatsu T., Nakashima A., Niimi K., Nomoto M., Redman C., Saito S., Tanimura K., Tomi M., Usui H., Vatish M., Wolfe B., Yamamoto E., O'Tierney-Ginn P. IFPA meeting 2018 workshop report II: abnormally invasive placenta; inflammation and infection; preeclampsia; gestational trophoblastic disease and drug delivery. Placenta. 2019;84:9–13. doi: 10.1016/j.placenta.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Shen R., Zhao L.L., Yu Z., Zhang C., Chen Y.H., Wang H., Zhang Z.H., Xu D.X. Maternal di-(2-ethylhexyl) phthalate exposure during pregnancy causes fetal growth restriction in a stage-specific but gender-independent manner. Reprod. Toxicol. 2017;67:117–124. doi: 10.1016/j.reprotox.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Zong S.S., Li C.Q., Luo C.F., Zhao X., Liu C.H., Wang K., Jia W.W., Bai M.L., Yin M.H., Bao S.H., Guo J., Kang J.H., Duan T., Zhou Q. Dysregulated expression of IDO may cause unexplained recurrent spontaneous abortion through suppression of trophoblast cell proliferation and migration. Sci. Rep. 2016;6 doi: 10.1038/srep19916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian F.J., Qin C.M., Li X.C., Wu F., Liu X.R., Xu W.M., Lin Y. Decreased stathmin-1 expression inhibits trophoblast proliferation and invasion and is associated with recurrent miscarriage. Am. J. Pathol. 2015;185:2709–2721. doi: 10.1016/j.ajpath.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Chen L.J., Ye A.F., Liu X.C., Lu J.C., Xie Q.X., Guo Y.X., Sun W.J. Combined effect of co-exposure to di (2-ethylhexyl) phthalates and 50-Hz magnetic-fields on promoting human amniotic cells proliferation. Ecotoxicol. Environ. Saf. 2021;224 doi: 10.1016/j.ecoenv.2021.112704. [DOI] [PubMed] [Google Scholar]
- 45.Mancuso C., Pani G., Bilirubin V. Calabrese. An endogenous scavenger of nitric oxide and reactive nitrogen species. Redox Rep. 2006;11:207–213. doi: 10.1179/135100006X154978. [DOI] [PubMed] [Google Scholar]
- 46.Calabrese V., Cornelius C., Dinkova-Kostova A.T., Calabrese E.J., Mattson M.P. Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxidants Redox Signal. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calabrese V., Mancuso C., Calvani M., Rizzarelli E., Butterfield D.A., Stella A.M.G. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat. Rev. Neurosci. 2007;8(10):766–775. doi: 10.1089/ars.2009.3074. [DOI] [PubMed] [Google Scholar]
- 48.Ha M., Wei L., Guan X., Li L.B., Liu C.J. p53-dependent apoptosis contributes to di-(2-ethylhexyl) phthalate-induced hepatotoxicity. Environ Pollut. 2016;208:416–425. doi: 10.1016/j.envpol.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Li L., Liu J.C., Lai F.N., Liu H.Q., Zhang X.F., Dyce P.W., Shen W., Chen H. Di (2-ethylhexyl) phthalate exposure impairs growth of antral follicle in mice. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erkekoglu P., Rachidi W., Yuzugullu O.G., Giray B., Favier A., Ozturk M., Hincal F. Evaluation of cytotoxicity and oxidative DNA damaging effects of di(2-ethylhexyl)-phthalate (DEHP) and mono(2-ethylhexyl)-phthalate (MEHP) on MA-10 Leydig cells and protection by selenium. Toxicol. Appl. Pharmacol. 2010;248:52–62. doi: 10.1016/j.taap.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 51.Tetz L.M., Cheng A.A., Korte C.S., Giese R.W., Wang P.G., Harris C., Meeker J.D., Loch-Caruso R. Mono-2-ethylhexyl phthalate induces oxidative stress responses in human placental cells in vitro. Toxicol. Appl. Pharmacol. 2013;268:47–54. doi: 10.1016/j.taap.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia J.J., Zeng X.S., Zhou X.S., Li Y., Bai J. The induction of thioredoxin-1 by epinephrine withdraws stress via interaction with β-arrestin-1. Cell Cycle. 2014;13:3121–3131. doi: 10.4161/15384101.2014.949214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Dong D.D., Tang S.W., Chen X., Gao Q. PPE38 of Mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics. J. Proteome Res. 2013;12:2055–2066. doi: 10.1021/pr301017e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu C.J., Luo J.J., Liu Y., Yang X.J. The oxidative stress responses caused by phthalate acid esters increases mRNA abundance of base excision repair (BER) genes in vivo and in vitro. Ecotoxicol. Environ. Saf. 2021;208 doi: 10.1016/j.ecoenv.2020.111525. [DOI] [PubMed] [Google Scholar]
- 55.Tarragó M.G., Chini C.C.S., Kanamori K.S., Warner G.M., Caride A., Oliveira G.C., Rud M., Samani A., Hein K.Z., Huang R.Q., Jurk D., Cho D.S., Boslett J.J., Miller J.D., Zweier J.L., Passos J.F., Doles J.D., Becherer D.J., Chini E.N. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD(+) decline. Cell Metabol. 2018;27:1081–1095. doi: 10.1016/j.cmet.2018.03.016. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marjanović M.P., Hurtado-Bagès S., Lassi M., Valero V., Malinverni R., Delage H., Navarro M., Corujo D., Guberovic I., Douet J., Gama-Perez P., Garcia-Roves P.M., Ahel I., Ladurner A.G., Yanes O., Bouvet P., Suelves M., Teperino R., Pospisilik J.A., Buschbeck M. MacroH2A1.1 regulates mitochondrial respiration by limiting nuclear NAD(+) consumption. Nat. Struct. Mol. Biol. 2017;24:902–910. doi: 10.1038/nsmb.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T., Berrocal J.G., Yao J., DuMond M.E., Krishnakumar R., Ruhl D.D., Ryu K.W., Gamble M.J., Kraus W.L. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J. Biol. Chem. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agathanggelou A., Smith E., Davies N.J., Kwok M., Zlatanou A., Oldreive C.E., Mao J.W., Costa D.D., Yadollahi S., Perry T., Kearns P., Skowronska A., Yates E., Parry H., Hillmen P., Reverdy C., Delansorne R., Paneesha S., Pratt G., Moss P., Taylor A.M.R., Stewart G.S., Stankovic T. USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood. 2017;130:156–166. doi: 10.1182/blood-2016-12-758219. [DOI] [PubMed] [Google Scholar]
- 59.Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerner R.R., Klepsch V., Macheiner S., Arnhard K., Adolph T.E., Grander C., Wieser V., Pfister A., Moser P., Hermann-Kleiter N., Baier G., Oberacher H., Tilg H., Moschen A.R. NAD metabolism fuels human and mouse intestinal inflammation. Gut. 2018;67:1813–1823. doi: 10.1136/gutjnl-2017-314241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colmone A. Helping T cells feel at home in the liver. Science. 2017;355:1277–1278. doi: 10.1126/science.355.6331.1277-g. [DOI] [PubMed] [Google Scholar]
- 62.Moore Z., Chakrabarti G., Luo X., Ali A., Hu Z., Fattah F.J., Vemireddy R., DeBerardinis R.J., Brekken R.A., Boothman D.A. NAMPT inhibition sensitizes pancreatic adenocarcinoma cells to tumor-selective, PAR-independent metabolic catastrophe and cell death induced by β-lapachone. Cell Death Dis. 2015;6:e1599. doi: 10.1038/cddis.2014.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santhosh A., Lakshmi L.R., Arun P., Deepadevi K.V., Nair K.G., Manojkumar V., Kurup P.A. Synthesis of NAD+ in erythrocytes incubated with nicotinic acid and the effect of di-(2-ethyl hexyl) phthalate (DEHP) Indian J. Biochem. Biophys. 1998;35:236–240. [PubMed] [Google Scholar]
- 64.Yang H.Y., Yang T.L., Baur J.A., Perez E., Matsui T., Carmona J.J., Lamming D.W., Souza-Pinto N.C., Bohr V.A., Rosenzweig A., Cabo R., Sauve A.A., Sinclair D.A. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X.K., Ji R.P., Liao X.H., Castillero E., Kennel P.J., Brunjes D.L., Franz M., Möbius-Winkler S., Drosatos K., George I., Chen E., Colombo P.C., Schulze P.C. MicroRNA-195 regulates metabolism in failing myocardium via alterations in sirtuin 3 expression and mitochondrial protein acetylation. Circulation. 2018;137:2052–2067. doi: 10.1161/CIRCULATIONAHA.117.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H., Han W.N., Zhu S.Y., Li Z.Y., Liu C.H. Effect of DEHP and DnOP on mitochondrial damage and related pathways of Nrf2 and SIRT1/PGC-1α in HepG2 cells. Food Chem. Toxicol. 2021;158 doi: 10.1016/j.fct.2021.112696. [DOI] [PubMed] [Google Scholar]
- 67.Cai J.Z., Shi G.L., Zhang Y., Zheng Y.Y., Yang J., Liu Q., Gong Y.F., Yu D.H., Zhang Z.W. Taxifolin ameliorates DEHP-induced cardiomyocyte hypertrophy via attenuating mitochondrial dysfunction and glycometabolism disorder in chicken. Environ Pollut. 2019;255 doi: 10.1016/j.envpol.2019.113155. [DOI] [PubMed] [Google Scholar]
- 68.Latini G., Felice C.D., Presta G., Vecchio A.D., Paris I., Ruggieri F., Mazzeo P. In utero exposure to di-(2-ethylhexyl)phthalate and duration of human pregnancy. Environ. Health Perspect. 2003;111:1783–1785. doi: 10.1289/ehp.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J., Guan Z.Q., Murphy A.N., Wiley S.E., Perkins G.A., Worby C.A., Engel J.L., Heacock P., Nguyen O.K., Wang J.H., Raetz C.R.H., Dowhan W., Dixon J.E. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metabol. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Botting K.J., Skeffington K.L., Niu Y., Allison B.J., Brain K.L., Itani N., Beck C., Logan A., Murray A.J., Murphy M.P., Giussani D.A. Translatable mitochondria-targeted protection against programmed cardiovascular dysfunction. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan Y., Schroeder E.A., Ocampo A., Barrientos A., Shadel G.S. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metabol. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joshi A., Mitophagy M. Kundu. In hematopoietic stem cells: the case for exploration. Autophagy. 2013;9:1737–1749. doi: 10.4161/auto.26681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y., Shi G.L., Cai J.Z., Yang J., Zheng Y.Y., Yu D.H., Liu Q., Gong Y.F., Zhang Z.W. Taxifolin alleviates apoptotic injury induced by DEHP exposure through cytochrome P450 homeostasis in chicken cardiomyocytes. Ecotoxicol. Environ. Saf. 2019;183 doi: 10.1016/j.ecoenv.2019.109582. [DOI] [PubMed] [Google Scholar]
- 74.Savchuk I., Söder O., Svechnikov K. Mono-2-ethylhexyl phthalate stimulates androgen production but suppresses mitochondrial function in mouse leydig cells with different steroidogenic potential. Toxicol. Sci. 2015;145:149–156. doi: 10.1093/toxsci/kfv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sies H., Belousov V.V., Chandel N.S., Davies M.J., Jones D.P., Mann G.E., Murphy M.P., Yamamoto M., Winterbourn C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022;23:499–515. doi: 10.1038/s41580-022-00456-z. [DOI] [PubMed] [Google Scholar]
- 76.Murphy E., Ardehali H., Balaban R.S., DiLisa F., Dorn G.W., Kitsis R.N., Otsu K., Ping P., Rizzuto R., Sack M.N., Wallace D., Youle R.J. Mitochondrial function, biology, and role in disease: a scientific statement from the American heart association. Circ. Res. 2016;118:1960–1991. doi: 10.1161/RES.0000000000000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasahara E., Sato E.F., Miyoshi M., Konaka R., Hiramoto K., Sasaki J., Tokuda M., Nakano Y., Inoue M. Role of oxidative stress in germ cell apoptosis induced by di(2-ethylhexyl)phthalate. Biochem. J. 2002;365:849–856. doi: 10.1042/BJ20020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miquel S., Champ C., Day J., Aarts E., Bahr B.A., Bakker M., Bánáti D., Calabrese V., Cederholm T., Cryan J., Dye L., Farrimond J.A., Korosi A., Layé S., Maudsley S., Milenkovic D., Mohajeri M.H., Sijben J., Solomon A., Spencer J.P.E., Thuret S., Berghe W.V., Vauzour D., Vellas B., Wesnes K., Willatts P., Wittenberg R., Geurts L. Poor cognitive ageing: vulnerabilities, mechanisms and the impact of nutritional interventions. Ageing Res. Rev. 2018;42:40–55. doi: 10.1016/j.arr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 79.Chen X., Wang J.S., Qin Q.Z., Jiang Y., Yang G.T., Rao K.M., Wang Q., Xiong W., Yuan J. Mono-2-ethylhexyl phthalate induced loss of mitochondrial membrane potential and activation of Caspase3 in HepG2 cells. Environ. Toxicol. Pharmacol. 2012;33:421–430. doi: 10.1016/j.etap.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu J., Wang L.M., Zhang L.H., Zheng F., Wang F., Leng J.H., Wang K.Y., Héroux P., Shen H.M., Wu Y.H., Xia D.J. Mono-2-ethylhexyl phthalate drives progression of PINK1-parkin-mediated mitophagy via increasing mitochondrial ROS to exacerbate cytotoxicity. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wurtele H., Tsao S., Lépine G., Mullick A., Tremblay J., Drogaris P., Lee E.H., Thibault P., Verreault A., Raymond M. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 2010;16:774–780. doi: 10.1038/nm.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ballotti R., Healy E., Bertolotto C. Nicotinamide as a chemopreventive therapy of skin cancers. Too much of good thing? Pigment. Cell Melanoma Res. 2019;32:601–602. doi: 10.1111/pcmr.12772. [DOI] [PubMed] [Google Scholar]
- 84.Wu W.W., Fu Y., Liu Z.W., Shu S.Q., Wang Y., Tang C.Y., Cai J., Dong Z. NAM protects against cisplatin-induced acute kidney injury by suppressing the PARP1/p53 pathway. Toxicol. Appl. Pharmacol. 2021;418 doi: 10.1016/j.taap.2021.115492. [DOI] [PubMed] [Google Scholar]
- 85.Wang R.H., Sengupta K., Li C.L., Kim H.S., Cao L., Xiao C.Y., Kim S., Xu X.L., Zheng Y., Chilton B., Jia R., Zheng Z.M., Appella E., Wang X.W., Ried T., Deng C.X. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li F., Fushima T., Oyanagi G., Townley-Tilson H.W.D., Sato E., Nakada H., Oe Y.J., Hagaman J.R., Wilder J., Li M.Y., Sekimoto A., Saigusa D., Sato H., Ito S., Jennette J.C., Maeda N., Karumanchi S.A., Smithies O., Takahashi N. Nicotinamide benefits both mothers and pups in two contrasting mouse models of preeclampsia. Proc. Natl. Acad. Sci. U.S.A. 2016;113:13450–13455. doi: 10.1073/pnas.1614947113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Surjana D., Halliday G.M., Damian D.L. Nicotinamide enhances repair of ultraviolet radiation-induced DNA damage in human keratinocytes and ex vivo skin. Carcinogenesis. 2013;34:1144–1149. doi: 10.1093/carcin/bgt017. [DOI] [PubMed] [Google Scholar]
- 88.Chong Z.Z., Lin S.H., Li F.Q., Maiese K. The sirtuin inhibitor nicotinamide enhances neuronal cell survival during acute anoxic injury through AKT, BAD, PARP, and mitochondrial associated "anti-apoptotic" pathways. Curr. Neurovascular Res. 2005;2:271–285. doi: 10.2174/156720205774322584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin S.H., Vincent A., Shaw T., Maynard K.I., Maiese K. Prevention of nitric oxide-induced neuronal injury through the modulation of independent pathways of programmed cell death. J. Cerebr. Blood Flow Metabol. 2000;20:1380–1391. doi: 10.1097/00004647-200009000-00013. [DOI] [PubMed] [Google Scholar]
- 90.Chong Z.Z., Lin S.H., Maiese K. Nicotinamide modulates mitochondrial membrane potential and cysteine protease activity during cerebral vascular endothelial cell injury. J. Vasc. Res. 2002;39:131–147. doi: 10.1159/000057762. [DOI] [PubMed] [Google Scholar]
- 91.McReynolds M.R., Chellappa K., Baur J.A. Age-related NAD(+) decline. Exp. Gerontol. 2020;134 doi: 10.1016/j.exger.2020.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia Y., Kang X., Tan L.S., Ren Y.F., Qu L., Tang J.W., Liu G., Wang S.X., Xiong Z.Y., Yang L. Nicotinamide mononucleotide attenuates renal interstitial fibrosis after AKI by suppressing tubular DNA damage and senescence. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.649547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hou Y.J., Lautrup S., Cordonnier S., Wang Y., Croteau D.L., Zavala E., Zhang Y.Q., Moritoh K., O'Connell J.F., Baptiste B.A., Stevnsner T.V., Mattson M.P., Bohr V.A. NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E1876–E1885. doi: 10.1073/pnas.1718819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olsson A.R., Sheng Y., Pero R.W., Chaplin D.J., Horsman M.R. DNA damage and repair in tumour and non-tumour tissues of mice induced by nicotinamide. Br. J. Cancer. 1996;74:368–373. doi: 10.1038/bjc.1996.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim J.Y., Lee H., Woo J.M., Wang Y., Kim K., Choi S., Jang J.J., Kim Y., Park I.A., Han D., Ryu H.S. Reconstruction of pathway modification induced by nicotinamide using multi-omic network analyses in triple negative breast cancer. Sci. Rep. 2017;7:3466. doi: 10.1038/s41598-017-03322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li F.Q., Chong Z.Z., Maiese K. Cell Life versus cell longevity: the mysteries surrounding the NAD+ precursor nicotinamide. Curr. Med. Chem. 2006;13:883–895. doi: 10.2174/092986706776361058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajman L., Chwalek K., Sinclair D.A. Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metabol. 2018;27:529–547. doi: 10.1016/j.cmet.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., Guarente L., Auwerx J. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.