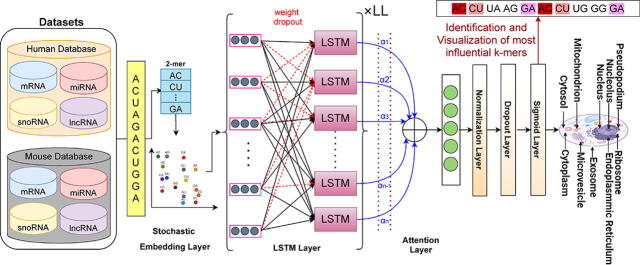
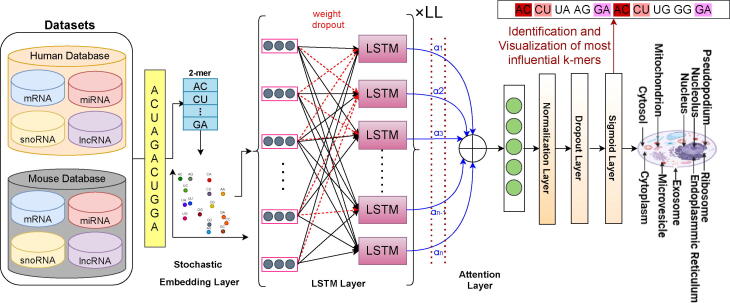
Graphical abstract
Keywords: RNA subcellular localization prediction, Single or multi compartment, Multi-class, Multi-label, GeneticSeq2Vec, Attention mechanism, LSTM, Neural tricks, Deep learning, Explainable, Human, Mouse
Highlights
-
•
A graph based novel approach for transforming RNA sequences into statistical vectors.
-
•
Development of a robust and precise deep learning classifier.
-
•
Utilization of attention mechanism capable of generating comprehensive feature space.
-
•
Incorporation of explainability suitable for highlighting distributions of nucleotides.
-
•
Development of a public web server use-able for predicting subcellular localizations.
Abstract
Subcellular localization of Ribonucleic Acid (RNA) molecules provide significant insights into the functionality of RNAs and helps to explore their association with various diseases. Predominantly developed single-compartment localization predictors (SCLPs) lack to demystify RNA association with diverse biochemical and pathological processes mainly happen through RNA co-localization in multiple compartments. Limited multi-compartment localization predictors (MCLPs) manage to produce decent performance only for target RNA class of particular sub-type. Further, existing computational approaches have limited practical significance and potential to optimize therapeutics due to the poor degree of model explainability. The paper in hand presents an explainable Long Short-Term Memory (LSTM) network “EL-RMLocNet”, predictive performance and interpretability of which are optimized using a novel GeneticSeq2Vec statistical representation learning scheme and attention mechanism for accurate multi-compartment localization prediction of different RNAs solely using raw RNA sequences. GeneticSeq2Vec generates optimized statistical vectors of raw RNA sequences by capturing short and long range relations of nucleotide k-mers. Using sequence vectors generated by GeneticSeq2Vec scheme, Long Short Term Memory layers extract most informative features, weighting of which on the basis of discriminative potential for accurate multi-compartment localization prediction is performed using attention layer. Through reverse engineering, weights of statistical feature space are mapped to nucleotide k-mers patterns to make multi-compartment localization prediction decision making transparent and explainable for different RNA classes and species. Empirical evaluation indicates that EL-RMLocNet outperforms state-of-the-art predictor for subcellular localization prediction of 4 different RNA classes by an average accuracy figure of 8% for Homo Sapiens species and 6% for Mus Musculus species. EL-RMLocNet is freely available as a web server at (https://sds_genetic_analysis.opendfki.de/subcellular_loc/).
1. Introduction
Biological functions of a variety of Ribonucleic Acid (RNAs) such as messenger RNA (mRNAs) [1], [2], microRNA (miRNAs) [3], [4], small nucleolar RNA (snoRNAs), long non-coding RNAs [5], [6] rely on their localization in various subcellular compartments such as nucleus, cytoplasm, cytosol [7], [8]. mRNAs localization in nucleus regulate gene expression by eliminating defective RNAs from the cell and tweaking the expression levels of various non protein coding RNAs[9], [10]. It provides quantitative as well as spatial control over the production of proteins by localizing in cytoplasm [2]. mRNA localization in cytosol helps to maintain cell membrane and control the use of nutrients for metabolism [11], [12]. miRNAs localization in nucleus play key role in cell division where each cell divides into identical daughter cells with an objective to promote organism growth and well being by replacing worn out cells [13]. Further, miRNAs localization in cytoplasm causes gene silencing by binding to mRNA molecules [13]. Small nucleolar RNAs (snRNAs) play a key role in post-transcriptional regulation by guiding RNA modifications of ribosomal RNAs (rRNA), transfer RNAs (tRNAs), and small nuclear ribonucleic acid RNAs (snRNAs) molecules by localizing in the nucleus [14]. Long non-coding RNAs (lncRNAs) control gene expression through chromatin remodelling by localizing in nucleus [15]. In cytoplasm, lncRNAs avoid mRNAs degradation as well as repress miRNAs to reduce their regulatory effects on mRNAs [16].
The subcellular localization of RNAs is an efficient and a widespread strategy to target the gene products to a particular region of various cells. Localization of various RNA molecules controls the translation of mRNAs into proteins in a temporal and spatial manner. It influences which type and number of proteins will be produced within certain cell by regulating the production of mRNA molecules and the amount of time they reside in the cytoplasm. Likewise, the spatial distribution of the RNA molecules mainly influences cellular concentration as well as location of its corresponding proteins which impact the cell function and its aptitude to interact with neighbouring cells or response to environmental changes. Further, it has the potential to avoid toxicity of various protein products, generate fast cellular responses, and determine molecular interactions [17], [18], [19]. It provides the basis for spatial differences in shape, structure, and function of a variety of cells in order to ensure that each cell exhibits a unique form of polarization [20], [21]. Characterizing RNA subcellular localization is essential for thorough categorization of different cell types and cell states [22]. In addition to facilitating a deep understanding of molecular and cellular biology, knowledge of RNA subcellular localization is also beneficial for the development of heterogeneous biomedical applications [22]. Like subcellular localization of messenger RNAs (mRNAs) assists to identify and treat Huntington’s disease by eliminating active mRNAs of disease specific gene in nucleus and cytoplasm [23]. Also, mRNAs guide protein synthesis by localizing in cytoplasm [2], paving way for the production of most effective recombinant proteins [24]. Furthermore, considering the association between RNA expression levels in different subcellular compartments with a variety of diseases such as Cancer [23], accurately determining RNA subcellular localization can largely assist to demystify their roles in various disease as well as to optimize therapeutics responsible to increase or decrease various RNAs expression levels in the target subcellular compartment.
A number of wet-lab experimental techniques based on single-molecule fluorescent in situ hybridization [25], epifluorescent [26], and confocal microscopic approaches [26] have been used to determine the subcellular localization of RNAs. However, such approaches are expensive, time consuming, and are not suitable for large scale characterisation of subcellular localization of diverse RNA classes across multiple species [22]. Considering the efficiency and robustness of computational approaches shown in various fields such as Natural Language Processing [27] and Bioinformatics [28], to date, a number of Artificial Intelligence based RNA associated subcellular localization predictors have been developed which are summarized in Table 1. The paradigms of existing approaches can be broadly classified into 2 categories, single compartment localization prediction (SCLP) [29], [30], [31], [32], [33], and multi-compartment localization prediction (MCLP) [7]. To better illustrate SCLP and MCLP paradigms, consider a hypothetical collection X of 5 RNA sequences represented as and a collection of 5 subcellular compartments L represented as . In SCLP, each RNA sequence belongs to exactly one subcellular compartment , such as belongs to Nucleus, belongs to Cytoplasm, and so on. Whereas, in MCLP, each RNA sequence belongs to more than one subcellular compartment at the same time, such as belongs to {Nucleus, Exosome}, belongs to {Mitochondria, Cytosol, Cytoplasm}, and so on.
Table 1.
A summary of existing computational subcellular localization predictors for miRNA, lncRNA, and mRNA molecules.
| Approach | Subcellular Localization Cardinality | Nucleotide Encoding | Classifier |
|---|---|---|---|
| RNA Type: miRNA | |||
| L2S-MirLoc [34] | Multi-Label | Electronion Interaction PseudoPotentials (EIIP) | Random Forest (RF) |
| miRNALoc [13] | pseudo dinucleotide compositions | Support Vector | |
| and di-nucleotide properties | Machine (SVM) | ||
| MirLocPredictor [35] | positional and semantic | Convolutional Neural | |
| information of k-mers (kmerPR2Vec) | Network (CNN) | ||
| MirGOFS [36] | functional similarity based encoding matrix | microRNA-based | |
| similarity inference model | |||
| MiRLocator [37] | K-mer embeddings using Word2vec (RNA2Vec) | BiLSTM | |
| encoder-decoder model | |||
| RNA Type: LncRNA | |||
| iLoc-LncRNA 2.0 7 | Multi-Class | fusing mutual information algorithm | SVM |
| and incremental feature selection strategy | |||
| lncLocation [38] | k-mer frequency, physicochemical properties, | SVM, RF, Logistic | |
| and secondary structure features Autoencoder | regression, XGBoost, | ||
| and binomial distribution based feature selection | lightGBM, DNN and CNN | ||
| Locate-R [39] | K-mer composition and Pearson based filtering | Deep SVM | |
| lncLocator 2.0 [5] | Glove embeddings | CNN, BiLSTM, MLP | |
| lncLocator [40] | k-mer frequency and stacked autoencoder | stacked ensemble classifier | |
| (SVM, RF) | |||
| iLoc-lncRNA[41] | binomial distribution-based feature selection, | SVM | |
| Pseudo K-tuple Nucleotide Composition | |||
| DeepLncLoc [16] | subsequence embeddings | CNN | |
| lncLocPred | k-mer, triplet, and PseDNC VarianceThreshold, | Logistic Regression | |
| binomial distribution, and F-score based feature selection | |||
| Yang et al. | kmer nucleotide composition, Analysis Of | SVM | |
| LncRNAPred [42] | Variance (ANOVA) based feature selection | ||
| DeepLncRNA [43] | k-mer, RNA binding motifs Genomic loci | feed-forward multi-layer | |
| deep neural network | |||
| KD-KLNMF [44] | k-mer and dinucleotide based | SVM | |
| spatial autocorrelation, KLD non-negative | |||
| matrix factorization based feature selection | |||
| RNA Type: mRNA | |||
| mLoc-mRNA [11] | Multi-Label | k-mer frequency and elastic-net based feature selection | RF |
| DM3Loc [45] | One-hot encoding | Attention based CNN | |
| Zhang mRNALoc [12] | Multi-Class | 9-mer, binomial distribution and one-way | SVM |
| analysis of variance based features | |||
| RNATracker [46] | One-hot encoding | Hybrid (CNN + LSTM + Attention) | |
| mRNAloc [9] | psuedo k-tuple nucleotide composition | SVM | |
| mRNALocater [10] | psuedo k-tuple nucleotide composition electron–ion | ||
| interaction pseudopotential, correlation coefficient filtering | Ensemble(CatBoost+ | ||
| LightGBM + XGBoost) | |||
| SubLocEP [47] | Nucleotide physicochemical properties | Weighted LightGBM | |
| NN-RNALoc [48] | k-mer frequency, distance-based sub-sequence | Multi-Layer DNN | |
| profiling and PCA for dimensionality reduction | |||
A critical analysis of existing computational approaches (Table 1) reveals that, to our knowledge, up to date, 5 multi-compartment localization predictors have been developed for miRNA molecules and 2 predictors have been developed for the mRNA molecule. A total of 6 single compartment localization predictors have been developed for mRNA and 11 have been developed for lncRNA biomolecule. It is evident from Table 1 that predominant RNA associated subcellular localization predictors [29], [30], [31], [32], [33] handle the problem of SCLP. However, these approaches are not effective to decode RNA association with various biochemical and pathological processes mainly happen through RNA concurrent presence in multiple compartments [7].
Regardless of whether an existing approach addresses the problem of SCLP or MCLP, all existing approaches are not well generalized as they are designed to predict subcellular localization of one particular RNA type. Due to sub-optimal feature extraction and localization prediction paradigms, existing approaches are not powerful enough to handle different kinds of RNAs which vary in terms of sequence length, nucleotide k-mers composition, chemical structures and molecular interactions. Further, majority of the approaches are based on deep neural networks which are black boxes as they do not explain which features are important for the accurate identification of which subcellular compartment of particular RNA and species. The poor degree of model explainability hinders the researchers to accurately estimate the effects of diverse trade-offs in a model. To best of our knowledge, there is only one generic multi-compartment localization predictor [7] for multiple RNA types (mRNAs, snoRNAs, miRNAs, lncRNAs) and species (Homo sapiens, Mus musculus). However, this approach is computationally expensive and relies on manually curated features which is why it lacks to produce promising performance for the subcellular localization prediction of different types of RNAs across multiple species. Building on the need of a robust and explainable sequence based computational approach, the paper in hand presents an end-to-end deep learning approach “EL-RMLocNet” for multi-compartment localization prediction of 4 different RNAs (mRNAs, snoRNAs, miRNAs, lncRNAs) across 2 distinct species (Homo sapiens, Mus musculus). This paper presents novel approaches to optimize multi-compartment subcellular localization predictive pipeline at different levels:
-
•
It presents a novel GeneticSeq2vec approach that transforms raw RNA sequences into graphical space where it captures comprehensive relations of nucleotides to generate effective statistical vectors of nucleotides.
-
•
It develops a robust and precise classifier “EL-RMLocNet” which makes use of Long Short Term Memory layers to extract informative features that are further optimized through attention layer.
-
•
It presents the very first deep learning based predictor that highlights distribution patterns of nucleotides associated to particular subcellular locations for 4 different RNA classes (mRNA, snoRNA, miRNA, lncRNA) and 2 distinct species (Homo Sapiens, Mus Musculus).
-
•
A comprehensive performance comparison of proposed EL-RMLocNet approach with state-of-the-art RNA subcellular localization predictor reveals that EL-RMLocNet achieves an average accuracy increment of 8% for Homo Sapiens and 6% for Mus Musculus species.
-
•
To enable the scientific community to infer RNA subcellular localization on the go, it presents an interactive and user-friendly web server which is publicly available at https://sds_genetic_analysis.opendfki.de/subcellular_loc/.
2. Materials and Methods
This section describes the proposed RNA associated multi-compartment subcellular localization prediction approach, benchmark datasets, and evaluation measures used to assess the performance of the proposed approach.
2.1. A K-hop Neighbourhood Relation based Statistical Representation Scheme for RNA Sequences (GeneticSeq2Vec)
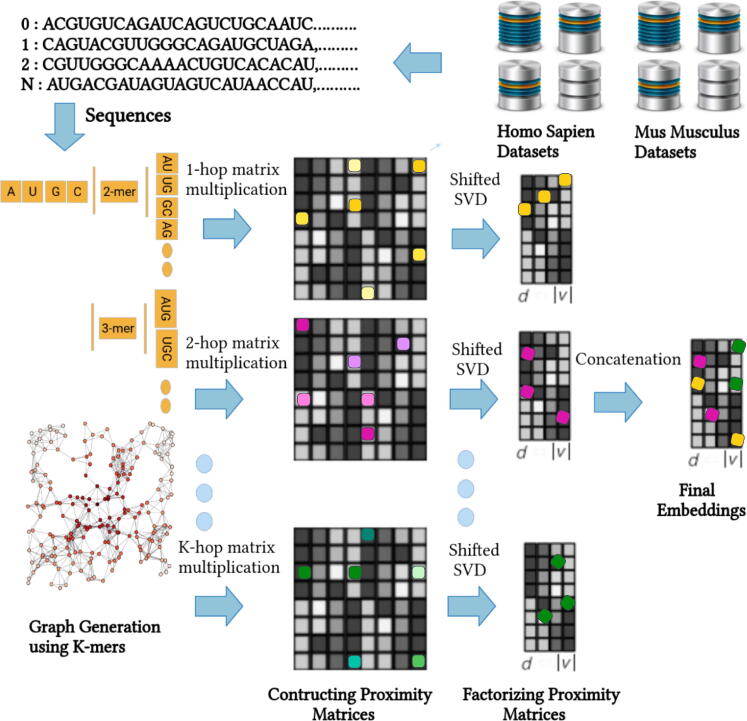
Machine and deep learning approaches require statistical representation of RNA sequences to extract useful nucleotide k-mers patterns for accurate target RNA multi-compartment subcellular localization prediction. Evidently, the better the statistical representation is, the better features are extracted which eventually help the model to accurately predict various subcellular compartments of target RNA type. Pre-dominantly, existing RNA subcellular localization predictors generate statistical representation by dividing the RNA sequences into nucleotide k-mers and capturing different characteristics of nucleotide k-mers such as order, frequency, and physicochemical properties [49]. However, these statistical representation learning approaches lack to capture a comprehensive context of nucleotide k-mers at different granularities, which negatively impacts the predictive performance and generalizeability of existing RNA subcellular localization predictors. Considering the effectiveness of graph based representation learning approaches for a variety of Natural Language Processing [50] and Bioinformatics tasks [28] mainly due to their ability to capture comprehensive semantic information and translational invariance of words. We present a novel graph based approach GeneticSeq2Vec to generate a rich statistical representation of RNA sequences, complete working paradigm of which is illustrated in Fig. 2 and summarized by the pseudo-code 1.
Fig. 2.
Workflow of Novel K-hop Neighbourhood Relation based Statistical Representation Learning Scheme for RNA Sequences.
Generation of statistical representation of raw RNA sequences using the proposed GeneticSeq2Vec approach is mainly comprised of four steps: 1) an un-directed k-mer graph generation, 2) k-hop proximity matrices construction, 3) k-hop proximity matrices factorization, 4) k-hop representation concatenation. In the step, sequences of particular RNA class (mRNA, snoRNA, miRNA, lncRNA) and species (homo sapien, mus musculus) are divided into nucleotide k-mers. Then, nucleotide k-mers of all the RNA sequences are concatenated to generate a nucleotide k-mers list. Using nucleotide k-mers list, unique nucleotide k-mer pairs are generated by rotating a window of 2 with the stride size of 1. To effectively model the correlations of nucleotide k-mers at different granularity, an un-directed graph is generated where the set of nucleotide k-mers are represented as vertices and their interaction as edges primarily treating nucleotide k-mer pairs collection as connection reference. To perform computational analysis of sized un-directed graph G, a numerical representation of the graph G is generated through an adjacency matrix where = 1 as well as = 1 if there is an edge between vertex and vertex . On the other hand, if there is no edge between vertex and vertex then = 0 and = 0, revealing each entry in adjacency matrix indicates whether the pair of vertices have any association.
With an aim to capture proximity which measures diverse relational information and semantic closeness of one vertex to another vertex, in step, it transforms adjacency matrix into proximity matrix by performing multiple operations. Firstly, by computing the summation of every row of adjacency matrix S, a normalized adjacency matrix is generated. To match the size of adjacency matrix S, normalized adjacency matrix X is extended to the size by repeating its only row. Afterward, using Eq. 1, transition probability of each vertex to its immediate neighbouring vertex is computed to produce proximity matrix A, where is the transition probability from vertex to its immediate neighbouring vertex .
| (1) |
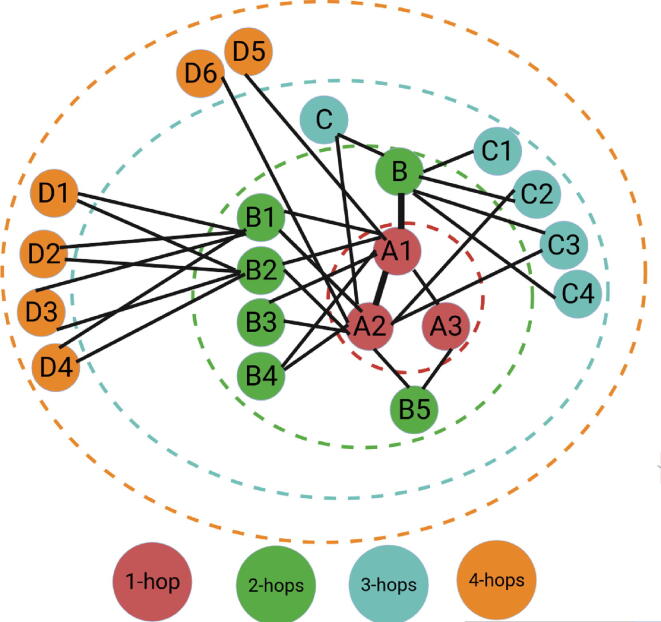
The proximity matrix A is multiplied by an identity matrix to generate a first-order (1-hop) proximity matrix . The first-order (1-hop) proximity matrix models whether there exists a direct connection between vertices by modeling the pairwise closeness between vertices. In Fig. 1, analysis of the edges connecting different vertices within the boundary of red dotted circle reveals that first-order proximity (1-hop) captures two kind of information: 1) vertex A1 is directly connected to vertex A2 as well as vertex A3, 2) vertex A1 and vertex A2 has strong relation represented with thick line, and vertex A1 and vertex A3 has weak connection represented with thin line.
Fig. 1.
Illustration of K-order (K-hop) Proximity Information, Red Dotted Circle Represents First-Order proximity(), Green Dotted Circle Indicates Second-Order Proximity (), Aqua Dotted Circle Represents Third-Order () Proximity, and Orange Dotted Circle Indicates Fourth-Order Proximity ().
By extending this paradigm to all vertices pairs present in the vocabulary, first-order (1-hop) proximity matrix captures the most fundamental relation between vertices. Considering, the extraction of information regarding whether two vertices are directly connected to each other (1-hop) is not sufficient to capture heterogeneous relations of k-mer vertices. Hence, it is important to capture higher-order (k-hop) proximity which can effectively model the complex relationships of vertices. More specifically, the second-order (2-hops) proximity information captures the common neighbours among two vertices, the more neighbours are shared among vertices, the stronger the connection is. In Fig. 1, analysis of the vertices connection within the boundary of green dotted circle indicates that, vertex A1 and A2 has 4 common neighbours (B1, B2, B3, B4), hence these vertices have a far more stronger connection as compared to A2 and A3 vertices which have only common neighbour (B5). This paradigm is extended to all vertices pairs to generate second-order (2-hops) proximity matrix. Clearly, second-order (2-hops) proximity information is important to determine the strength of vertices connection on the basis of number of common neighbours, extracting key nucleotide k-mers information such as most frequently co-occurring nucleotide k-mers as well as common contexts.
Further, the third-order (3-hops) proximity information is essential to measure the impact of common neighbours on the strength of long range connection between vertices. In Fig. 1, analysis of the trajectory A1-B-C-A2 within the boundary of aqua color dotted circle reveals that despite the strong connection among vertex A1 and vertex B, the connection between vertex A1 and A2 can be significantly weakened because of two weaker connections between vertex B and vertex C as well as vertex C and vertex A2. On the contrary, the trajectory A1-B- indicates that the relationship between vertex A1 and A2 remains very strong primarily due to the decent number of common neighbors between vertex A2 and vertex B which greatly strengthens their relationship. Likewise, the fourth-order (4-hops) proximity information is also crucial to capture global relations of vertices. In Fig. 1, analysis of the vertices connection inside the boundary of orange dotted circle reveals that the relation between vertex A1 and vertex A2 remains very strong because their connection partners B1 and B2 have four common neighbours D1-to-D4 which strengthen the relation of vertex A1 and A2. On the other hand, vertex A1 and vertex A2 becomes totally unrelated if we only consider their relation with vertex D5 and vertex D6 respectively, mainly because no path is left which connects vertex A1 to vertex A2. By extending the paradigms of third-order (3-hops) and fourth-order (4-hops) proximity to all possible vertices trajectories, global relations of the vertices can be captured in 3-hops and 4-hops proximity matrices which corresponds to long range contextual information of nucleotide k-mers.
It is evident from a thorough analysis of high order (k-hops) proximity modelling that each higher order (k-hop) proximity matrix captures different kind of relations among k-mer vertices. Therefore, instead of mapping heterogeneous nucleotide k-mer relations in a common subspace, GeneticSeq2Vec generates k-hop proximity matrices to retain heterogeneous relational information in different subspaces. Considering sequences vary across different RNA subtypes in terms of sequence length, nucleotide k-mer distribution, the idea of generating different subspaces helps to find optimal value of k-hop proximity for each RNA subtype as it avoids the influence of higher order proximity modelling to lower order proximity modelling. Building on, first order (1-hop) proximity matrix is computed through the multiplication of proximity matrix A to an identity matrix. Higher order (k-hop) proximity matrices can be computed by multiplying the proximity matrix A k-times to itself.
| (2) |
Where the proximity from the vertex to is mainly an entry in i-th row and j-th column of k-order (k-hops) proximity matrix . The k-hop multiplications of proximity matrix A helps to capture diverse interactions and global relations of the vertices, indicating higher order proximity matrices encode translational invariance information of nucleotide k-mers by to generate heterogeneous context aware representations. More specifically, the step produces k-hop representation matrices for the input graph G where the i-th row of each Wi represents a continuous value vector of d dimension for the nucleotide k-mer vertex learned by modeling its proximal k-hop relations with respect to all nucleotide k-mer vertices present in the vocabulary.
| Algorithm 1: A K-hop Neighbourhood Relation based Statistical Representation Scheme for RNA Sequences |
|---|
 |
In step, proposed GeneticSeq2Vec factorizes proximity matrices produced by different k-hops using Singular Value Decomposition (SVD) approach in order to learn precise k-hops representation matrices . Using Eq. 3, SVD decomposes each k-hops proximity matrix into the product of three matrices, two of them U and V are orthogonal matrices and serves as a diagonal matrix which is comprised of an ordered set of singular values.
| (3) |
Finally, in step, it combines the precise representation produced by different k-values to generate k-order (k-hops) relations aware representations of all vertices, which can be expressed as follows:
| (4) |
To facilitate the readers, complete GeneticSeq2Vec algorithm working paradigm is described in terms of pseudo-code 1.
2.2. Explainable Deep Learning based RNA Associated Multi-Compartment Localization Predictor
To accurately predict subcellular localization patterns of different RNA classes in multiple species, we have developed an explainable deep learning classifier “EL-RMLocNet”. EL-RMLocNet leverages the stochastic embedding layer to optimize the embedding matrix generated by the novel GeneticSeq2Vec approach. It uses LSTM to find and retain most informative features as well their long range dependencies from statistical vectors of RNA sequences. Unlike a trivial recurrent neural network (RNN), LSTM does not face the problem of vanishing gradients because it utilizes a gating mechanism to regulate the flow of information. The length of sequences and distribution of nucleotide k-mers vary across different RNA classes, indicating accurate subcellular localization of target RNA class relies on certain set of nucleotide k-mers patterns. EL-RMLocNet captures potential nucleotide k-mers patterns using attention mechanism which weights the features on the basis of their potential to accurately predict subcellular localization of target RNA class. By revealing potential nucleotide k-mers patterns for different RNA classes and species, attention mechanism also makes the decision making of deep learning model quite transparent. To significantly reduce the classification error, predictive potential and generalizeability of proposed classifier are optimized using multiple neural strategies such as normalization, dropout, and learning rate decay. Considering, the performance of the deep learning model is largely influenced by different hyperparameters such as number of layers, learning rate, batch size, etc, we optimize hyperparameters using grid search and facilitate optimal values of different hyperparameters in Table 2. Architecture of proposed deep learning model EL-RMLocNet is given in Fig. 3, Fig. 4, and details of various inherent layers is provided in following subsections.
Table 2.
Optimal Parameter Values of Proposed EL-RMLocNet Approach for 8 Benchmark Datasets Belonging to 4 different RNA classes and 2 Species.
| Benchmark Dataset | K-mer | Stride Size (s) | Embedding Dimension (d) | Embedding Dropout (ed) | LSTM Layers (ll) | LSTM Hidden Units (ld) | Attention Dimension(ad) | Dropout (dp) | Learning Rate (lr) | Learning Rate Decay (ld) | Batch Size (b) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo Sapien species | |||||||||||||
| mRNA | 3 | 2 | 200 | 0.005 | 1 | 200 | 50 | 0.01 | 0.05 | 0.001 | 32 | ||
| miRNA | 1 | 1 | 32 | 0.0025 | 1 | 32 | 60 | 0.005 | 0.06 | 0.1 | 32 | ||
| snoRNA | 2 | 2 | 64 | 0.0025 | 1 | 64 | 50 | 0.005 | 0.06 | 0.01 | 32 | ||
| lncRNA | 2 | 2 | 200 | 0.005 | 1 | 200 | 50 | 0.1 | 0.05 | 0.1 | 64 | ||
| Mus Musculus species | |||||||||||||
| mRNA | 2 | 1 | 200 | 0.0025 | 4 | 64 | 90 | 0.05 | 0.06 | 0.1 | 32 | ||
| miRNA | 1 | 1 | 32 | 0.0025 | 1 | 32 | 60 | 0.005 | 0.06 | 0.1 | 32 | ||
| snoRNA | 2 | 2 | 16 | 0.0025 | 1 | 16 | 50 | 0.005 | 0.06 | 0.0001 | 32 | ||
| lncRNA | 3 | 2 | 200 | 0.0025 | 4 | 60 | 50 | 0.05 | 0.05 | 0.01 | 128 | ||
Fig. 3.
Workflow of an Explainable Deep Learning Model for RNA Associated Multi-Compartment SubCellular Localization Prediction.
Fig. 4.
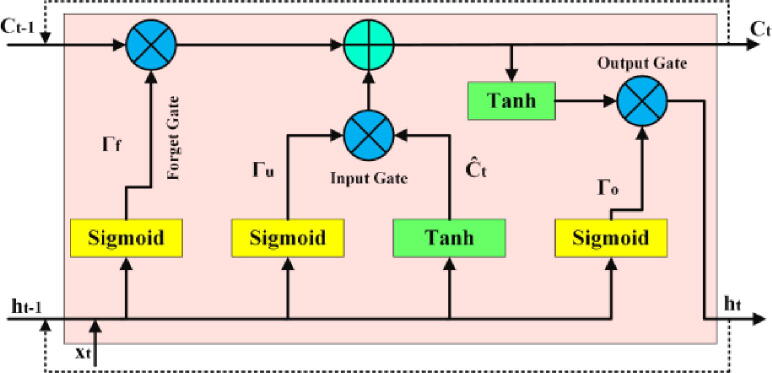
Information Flow in Standard LSTM Cell.
2.2.1. Stochastic Embedding Layer
The process of predicting RNA associated subcellular localization starts by dividing the RNA sequences into nucleotide k-mers by sliding a window of size w with the stride size of s. For every RNA sequence, statistical vector of each nucleotide k-mer is retrieved at the embedding layer mainly using embedding matrix of size vocabulary * vector dimensions produced by novel graph based representation learning module, discussed in Section 2.1. To optimize embedding matrix, 2 distinct embedding dropout tricks are utilized in order to avoid model over-fitting which happens due to over-specialization of only few features. In k-mer embedding dropout, entire k-mer has the dropout probability of dp whereas in k-mer vector dimension dropout, each k-mer vector dimension has the likelihood of dp to be replaced by zero. Optimized statistical vectors of RNA sequences are obtained by averaging the respective k-mer statistical vectors. The d-dimensional RNA sequence vectors are passed to LSTM network having ll layers, ld hidden units which finds and retain most informative features along with their dependencies.
2.2.2. Optimized Long Short Term Memory (LSTM) Layers
Contrary to the traditional recurrent neural network, LSTM controls the information flow by making use of 3 distinct gates. Update gate or Input gate or update gate, indicated as (Eq. 5) mainly regulates the flow of naive information in current time step. Forget gate, indicated as (Eq. 6) decides whether memory information of last time step shall be dropped to taken forward. Third gate known as output gate is indicated by (Eq. 7). It determines up to what extent information from previous time step will be transferred to next time step by taking currently available information into account. In these mathematical expressions, [] refer to weight matrices, indicate bias vectors, represents d-dimensional nucleotide k-mer vector fed at particular time-step and refer to next and previous time steps respectively, refers to current hidden state, indicates memory cell state, and represents element-wise product.
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
These 3 different gates mainly get activated or de-activated on the basis of corresponding weight matrices and behave on the basis of the corresponding activation function (e.g sigmoid (), tanh). In Eq. 6, weight matrix controls the working of forget gate. For example, if forget gate vector is completely zero, then content will not be considered at all, indicating all information provided by the will be discarded. Contrarily, if forget gate vector contains one, then the model preserve the information. These 3 different gates of LSTM perform a variety of operations to regulate nucleotide k-mers information represented as a floating point vector falling in range of 0-to-1. Each cell of LSTM is comprised of these three gates. To preserve long term information of nucleotide k-mers, hidden state h of every cell is saved at each time step.
To regularize LSTM ll layers, considering, dropping hidden state of LSTM layers can significantly hinder the aptitude of LSTM to retain long term dependencies. We optimize LSTM layers by applying weight dropout on recurrent weight matrices [] as well non-recurrent weight matrices [] of LSTM layers where we randomly drop subset of weights in the network instead of dropping subset of activations. While weight dropout on recurrent weights avoid overfitting on the recurrent connections of LSTM layers, weight dropout on non-recurrent weight matrices enhance the LSTM ability to extract important residue dependencies. In this manner, LSTM layers produce d-dimensional feature vectors for RNA sequences which are passed forward in the network.
2.2.3. Attention Layer
One of the most important feature of the human perception is its ability to focus on only most important parts of the input to make sense of the information present in outside world. Similarly, the significance of various nucleotide k-mers patterns for accurate RNA associated subcellular localization prediction varies across RNA classes and species, some nucleotide k-mers patterns are more discriminative while others are completely redundant. Considering, accurate multi-compartment subcellular localization prediction of various RNA classes and species mainly depends on the set of most relevant features. We utilize attention paradigm to optimize input d-dimensional RNA sequence vectors by weighting the features on the basis of their importance for hand on task.
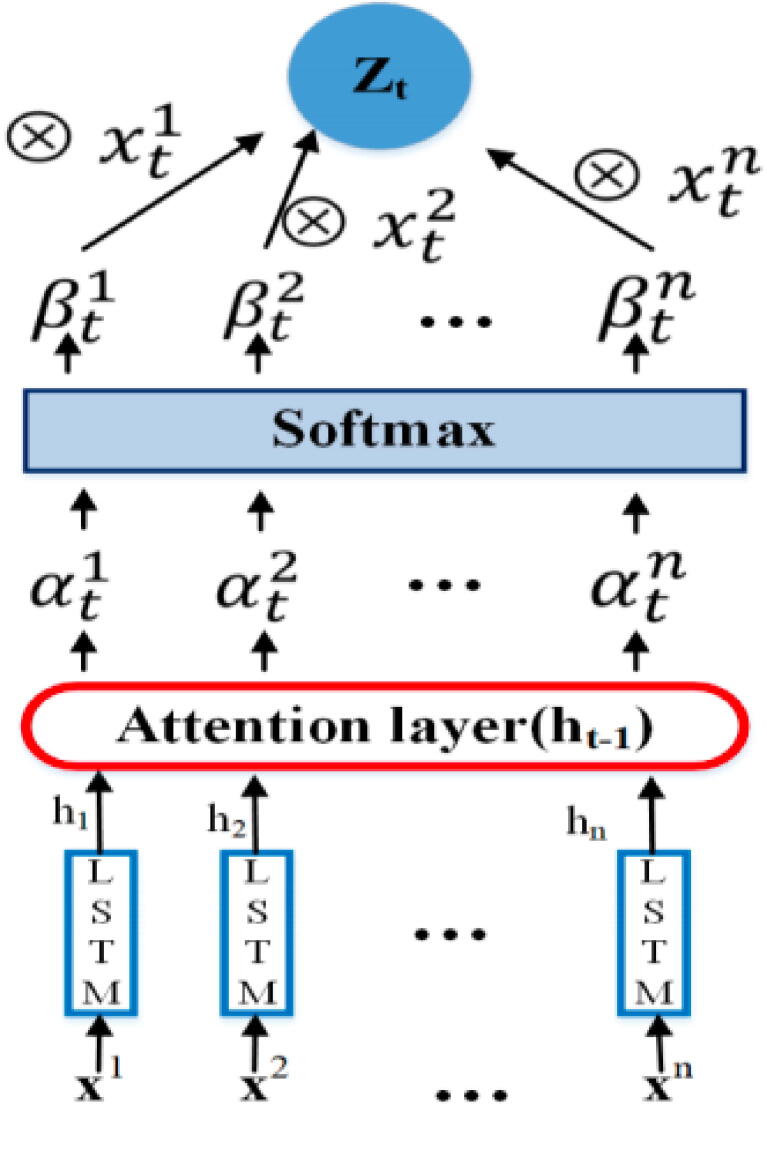
The workflow of attention paradigm which involves the generation of attention weights and optimizing input features using attention weights is summarized in the Fig. 5. First of all, we map the input d-dimensional LSTM feature vectors represented as to using Eq. 11, where refers to non-linear activation function, and represents hidden state at the time step t with size s.
| (11) |
Fig. 5.
Architecture of the Attention Model.
In order to avoid the issue of long-term dependencies which can significantly derail multi-compartment subcellular localization prediction performance, we utilize LSTM as non-linear activation function . Then attention mechanism is constructed using a deterministic attention based deep learning model. For a particular sequence , using previous hidden state represented as as well as cell state within LSTM cell, and can be defined using Eq. 12 and Eq. 13 respectively:
| (12) |
| (13) |
In these equations, matrices .and v are hyperparameters of the attention model that can be learned through backpropagation. The vector is of length m where value estimates the significance of given feature sequence for a particular time step t. These values are mainly normalized using softmax. Whereas represents attention weight that contains a value indicating the amount of attention should be placed on input feature sequences. Output produced by attention model can be obtained at a particular time step t where the weighted and optimized input feature sequence represented as will be equivalent to (Eq. 14):
| (14) |
By replacing the normal d-dimensional LSTM feature vector with and updating attention model, we manage to obtain optimized attention based feature vectors for RNA sequences. Unlike where all input features are treated equally, enables more attention to specific features in order to extract most potential features effectively by eliminating the impact of redundant features for target RNA associated subcellular localization prediction. Optimized ad dimensional attention based feature vectors are passed forward in the network.
2.2.4. Bag of tricks for Optimizing the Training and Prediction of EL-RMLocNet Approach
To optimize the training of deep learning model EL-RMLocNet, 3 distinct optimization tricks are utilized. The ad dimensional vectors produced by attention layer are passed to the normalization layer [51]. Normalization performs standardization of the input to hidden layer for every batch b mainly by calculating mean and variance of each batch b. This ensures that input-to-output mapping of proposed deep learning model does not overly generalize certain part of input distribution which results faster training, improve convergence, and generalizeability. By leveraging normalized input and hyperparameter, normalization layer alleviates internal co-variance shift mainly by stabilizing hidden distribution , mathematical expression of which is given in Eq. 19.
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
Further, we also apply traditional dropout to avoid model overfitting occurred due to neuron co-adaptation where neurons stop operating independently and rely on other neurons to make decisions. Through random sampling based on the Bernoulli distribution (Eq. 20), we apply traditional dropout on hidden neurons where each hidden neuron has the likelihood of dp to be dropped.
| (20) |
Considering choosing an optimal learning rate lr for deep learning model is not a straightforward task, another optimization trick used in proposed deep learning model EL-RMLocNet is learning rate decay. Learning rate decay trick smartly updates the learning rate in such a manner that global minima is computed and model converges to the best possible weights. By making use of adaptive moment estimation based on weight decay (ADAMW) optimizer, learning rate lr value is optimized using decay rate of ld during weight update, which can be mathematically expressed as:
| (21) |
Using one-hot encoded actual subcellular localization compartments, probability score for each subcellular localization compartment present in benchmark dataset is computed through the application of softmax before computing cross-entropy loss CE, which can be mathematical expressed as:
| (22) |
Using the batch size b, proposed EL-RMLocNet predictor accurately infers the multi-compartment subcellular localization of various RNAs across multiple species.
2.3. Benchmark RNA-Associated SubCellular Localization Prediction Datasets
RNAs are broadly segregated into two categories, coding RNA and non-coding RNA. Coding RNAs like messenger RNAs (mRNAs) play a vital role in transcription. Non-coding RNAs like long non-coding RNA (lncRNA), microRNA (miRNA), small nucleolar RNA (snoRNA) play a regulatory role in diverse biological processes ranging from epigenetic modifications to gene expression. These RNAs exist in multiple subcellular compartments illustrated in Fig. 6 in order to perform various functions. Considering biological functionalities of diverse RNAs are strongly associated to their subcellular localization patterns, we collect 8 different RNA subcellular localization datasets belonging to homo sapiens and mus musculus species from literature [7]. Wang et al. [7] utilized a public metathesaurus RNALocate [52] which contains subcellular localization of more than 65 organisms (e.g musculus, homo sapiens, saccharomyces cerevisiae), 9 RNA classes, and 42 subcellular compartments (e.g., nucleus, cytoplasm, ribosomes) to prepare RNA associated multi-compartment subcellular localization datasets of 4 different RNA classes (miRNAs, snoRNAs, lncRNAs, mRNAs) for 2 different species (homo sapiens, musculus). Using RNALocate database [52], after downloading subcellular localization sequences of 4 RNA classes, RNA classes which have sequences more than defined threshold . are under-sampled through CD-HIT tool using cut-of threshold of 80% to obtain miRNAs, snoRNAs, lncRNAs, and mRNAs datasets for homo sapiens and mus musculus species.
Fig. 6.
Schematic Illustration of RNA Associated Multi-Compartment Subcellular Localization in Cells.
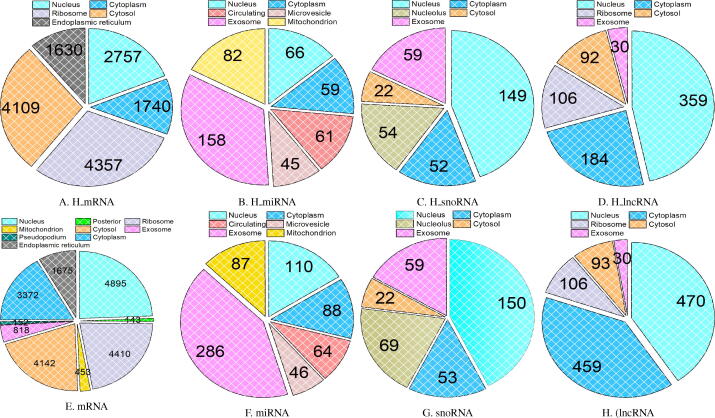
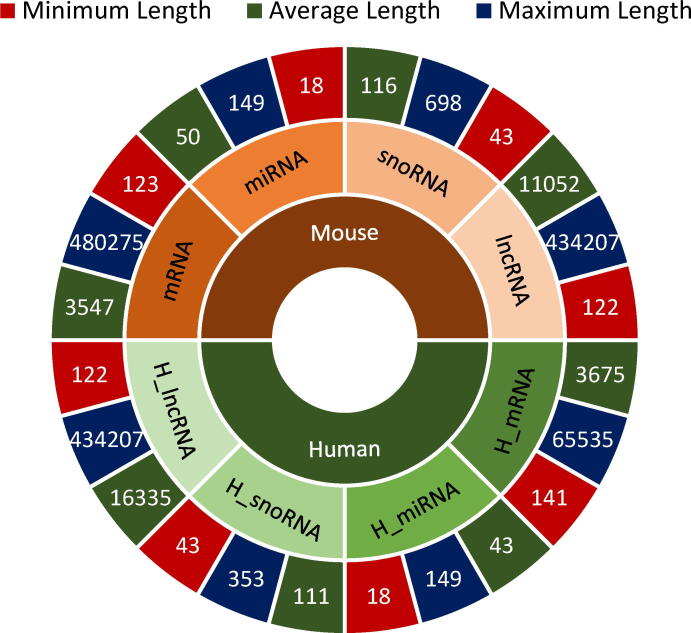
For 8 benchmark datasets, statistical distribution of 4 different RNAs in diverse subcellular compartments is provided in Fig. 7. More specifically, 4 pie graphs in first row of the Fig. 7 indicate the statistical distribution of mRNA, miRNA, snoRNA, and lncRNA sequences in multiple subcellular compartments for homo sapien species, whereas second row pie graphs reveal the statistical distribution of 4 different RNAs in diverse cellular compartments. Further, in order to analyze the variation in sequence length across all 8 benchmark datasets, donut chart in Fig. 8 reports the minimum, maximum, and average sequence length of 4 different RNA subtypes datasets for homo sapien species represented as and for mus musculus species represented as .
Fig. 7.
Statistical Distribution of Benchmark RNA Associated Multi-Compartment Localization Prediction Datasets Belonging to Homo Sapien species (A-D) and Mus Musculus species (E-H).
Fig. 8.
A Comparison of Variations in Sequence Length across 8 Benchmark RNA Associated Multi-Compartment Subcellular Localization Datasets.
For homo sapien species, dataset contains the most lengthier sequences whose average length falls around 16,335 nucleotides. The dataset contains the second most lengthier sequences followed by , and dataset with average length of 3,675, 111, and 43 nucleotides respectively. For mus musculus species, lncRNA dataset contains longer sequences followed by mRNA, snoRNA, and miRNA dataset with average sequence length of 11,052, 3,547, 116, and 50 nucleotides respectively. Comparing the variations in sequence length across all 8 benchmark datasets indicates that all 4 RNA subtypes datasets have slightly longer sequences in homo sapien species as compared to mus muscluss species.
2.4. Evaluation Measures
Following the evaluation criterion used by existing RNA subcellular localization predictors [7], performance of proposed approach is evaluated using 6 different evaluation measures including Accuracy, Average Precision, Coverage, Ranking Loss, One-error, and area under receiver operating characteristic. Accuracy estimates performance of the model by dividing the number of sequences where the target subcellular compartments are correctly predicted with total number of sequences. Average precision indicates the ability of the model to correctly predict positive sequences and avoid miss-classification of negative sequences as positive sequences. Coverage estimates up to what average number of steps required by the model to cover all the true subcellular compartments of sequences. Ranking loss measures how many times the wrong subcellular compartment is ranked above the true subcellular compartment. One error monitors the performance of the model by measuring number of sequences in which top-ranked subcellular compartments given by the model are absent from the true subcellular compartments set associated with the sequences. In addition, to make sure that proposed deep learning model EL-RMLocNet performance is neither biased towards majority majority subcellular compartment nor minority subcellular compartment, we analyze the model performance using AU-ROC. AU-ROC deeply investigates the trade-off between true positive rate and false positive rate by giving equivalently importance to true positives and true negatives. The higher the accuracy, average precision, and AU-ROC are, and the lower the coverage, ranking loss, and one error values are, the better the model predictability is for the hand-on task.
| (23) |
In Eq. 23, denotes number of RNA sequences, refers to number of subcellular localization compartments, refers to the rank of subcellular localization compartment y in all compartments Y, f(y) denotes the score of y inferred by machine learning classifier, Y refers to actual subcellular localization compartment set, denotes the predicted subcellular localization compartments set which is the symmetric difference between actual and predicted subcellular localization compartment set.
3. Experimental Setup
Proposed methodology is implemented in Python using an open source deep learning framework Pytorch [53]. In order to perform a fair performance comparison of proposed approach with existing state-of-the-art RNA multi-compartment localization predictor, 10-fold cross validation is performed. We use GridSearch [54] to optimize a variety of hyperparameters. To capture hidden pattern of nucleotides, considering RNA sequences are comprised of only 4 unique bases, we perform experimentation with 5 different k-mers ranging from 1-to-5 generated using stride size of 1-to-3. To capture comprehensive relations and positional in-variances of nucleotide k-mers, novel k-hop neighbourhood based statistical representation learning scheme performs experimentation with 2 to 7 hop based proximity matrices to generate rich d-dimensional vectors for RNA sequences.
Proposed EL-RMLocNet classifier is trained by tweaking an embedding dropout from 0.004 to 0.005, LSTM neurons from 100-to-400, batch size from 32-to-128, adaptive moment estimation based on weight decay (ADAMW) as an optimizer, learning rate from 0.04-to-0.05, decay rate from 1e-05-to-1e-07, standard dropout from 0.1-to-0.05, and categorical cross entropy as a loss function. Model checkpoint which achieves lowest training error is saved to make prediction on test sequences for the task of RNA subcellular localization prediction. To ensure the reproduceability of reported results, optimal values of different hyperparameters are summarized in Table 2.
4. Results and Discussion
This section quantifies the impact of 6 different sequence fixed length generation approaches over the performance of the proposed EL-RMLocNet approach for RNA multi-compartment subcellular localization prediction. Further, it performs a comprehensive assessment of the predictive performance and generalizeability of proposed EL-RMLocNet approach for RNA associated multi-compartment subcellular localization prediction using a variety of evaluation metrics. It compares the performance of proposed EL-RMLocNet approach with state-of-the-art RNA associated multi-compartment subcellular localization predictor using 8 benchmark datasets. It also performs intrinsic analysis of the key nucleotide k-mers patterns found by proposed approach EL-RMLocNet to accurately predict the subcellular localization of different RNA classes in distinct species.
4.1. Performance Assessment of EL-RMLocNet for Multi-Compartment RNA Localization Prediction
It is evident from the donut chart 8 that in both homo sapien and mus musculus species, sequence length of all 4 RNA subtypes including mRNA, miRNA, snoRNA, and lncRNA significantly differ from each other. Considering machine and deep learning classifiers operate on fixed length genomic sequences, we perform experimentation with 6 different settings based on copy padding, sequence truncation and hybrid paradigms to fix the length of RNA sequences across all 8 benchmark datasets of 2 distinct species.
In copy padding paradigm, first of all, maximum possible sequence length is computed by comparing all the sequences of particular dataset. Afterward, all the sequences whose lengths are less than maximum threshold, are extended in order to justify maximum length by inserting a specific constant at starting or ending region of sequences. Another paradigm to fix the length of sequences is sequence truncation where first of all minimum possible sequence length is computed. Then, nucleotides from starting or ending region of all those sequences whose lengths are greater than minimum threshold are truncated in order to reduce the length up to minimum threshold. Considering copy padding paradigm may create an unnecessary bias to fade out discriminative sequence patterns and sequence truncation paradigm is vulnerable to loose important nucleotide distribution information. Hybrid paradigm first finds average sequence length and then utilize copy padding trick to fix the length of those sequences whose lengths are shorter than average length threshold and leverage sequence truncation trick for sequences whose lengths are greater than average length threshold.
Considering accurate RNA subcellular localization prediction relies on certain distributional patterns of nucleotides which can be present in any region of the sequences. We perform experimentation with all 3 sequence fixed length generation paradigms using 6 different settings. Table 3 quantifies the impact of 6 different sequence fixed length generation settings over the performance of proposed EL-RMLocNet approach in terms of average precision, whereas the performance figures in terms of other most widely used evaluation metrics are given in supplementary file-1. In Table 3, 2 settings related to copy padding are represented as , sequence truncation settings are shown as , and hybrid paradigm settings are shown as , where the setting names reveal the region of the sequences targeted for extension or truncation along with length threshold criteria. As is evident from the Table 3, for homo sapien species, from both copy padding settings, EL-RMLocNet approach achieves better average precision with setting across all RNA subtypes except where setting performs better. A similar performance trend can be seen with sequence truncation settings where EL-RMLocNet attains better average precision with as compared to across most RNA subtypes. Unlike copy padding and sequence truncation settings, from 2 hybrid paradigm settings, EL-RMLocNet approach produces better average precision with across all RNA subtypes except lncRNA where its counterpart setting performs better. Overall, EL-RMLocNet achieves peak performance with setting for miRNA and snoRNA biomolecules, with for mRNA biomolecule, and with for lncRNA biomolecule, obtaining the average precision of 86%, 83%, 77%, and 85% respectively. This indicates that all 3 sequence fixed length generation paradigms (copy padding, sequence truncation, and hybrid) manage to achieve good performance for one or the other RNA multi-compartment subcellular localization prediction.
Table 3.
Comparing the Impact of 6 Different Sequence Fixed Length Generation Approaches over the Performance of Proposed EL-RMLocNet Approach Produced for 8 Benchmark Datasets of 2 different Species in terms of Average Precision
| RNA Subtype |
Sequence Length Variation |
|||||
|---|---|---|---|---|---|---|
| Start_Max | End_Max | Start_Average | End_Average | Start_Min | End_Min | |
| Homo Sapien | ||||||
| mRNA | 0.72 | 0.70 | 0.77 | 0.72 | 0.73 | 0.71 |
| miRNA | 0.85 | 0.86 | 0.85 | 0.84 | 0.77 | 0.77 |
| lncRNA | 0.83 | 0.84 | 0.83 | 0.84 | 0.82 | 0.85 |
| snoRNA | 0.77 | 0.83 | 0.80 | 0.78 | 0.80 | 0.80 |
| Mus Musculus | ||||||
| mRNA | 0.66 | 0.65 | 0.71 | 0.68 | 0.60 | 0.63 |
| miRNA | 0.86 | 0.87 | 0.86 | 0.86 | 0.84 | 0.83 |
| lncRNA | 0.73 | 0.70 | 0.77 | 0.73 | 0.72 | 0.69 |
| snoRNA | 0.82 | 0.81 | 0.82 | 0.81 | 0.80 | 0.81 |
Analyzing the performance trends for mus musculus species (Table 3) indicates that from 2 copy padding settings, EL-RMLocNet approach achieves superior average precision using for 3 RNA subtypes including mRNA, lncRNA, and snoRNA and attains better performance using for miRNA biomolecule. Whereas from 2 sequence truncation settings, EL-RMLocNet approach produces good performance with setting for miRNA and lncRNA bimolecules, and with for mRNA and snoRNA bimolecules. Contrarily, from 2 hybrid paradigm settings, EL-RMLocNet approach produces better average precision with setting as compared to setting across all 4 different RNA subtypes. Overall, EL-RMLocNet approach achieves peak performance with setting for mRNA, lncRNA biomolecules, with for miRNA biomolecule, and with for snoRNA biomolecule, obtaining highest average precision of 71%, 77%, 87%, and 82% respectively.
Further, in order to describe the effectiveness of proposed EL-RMLocNet approach for accurate multi-compartment subcellular localization prediction of diverse RNA classes without being biased towards the distribution of subcellular compartments, performance of proposed EL-RMLocNet approach is analyzed in terms of AU-ROC. Using one-versus-rest strategy, one particular subcellular compartment of target RNA class is treated as positive and all other subcellular compartments are treated as negative to compute AU-ROC with respect to particular subcellular compartment. By iteratively selecting one subcellular compartment as positive and remaining ones as negative, multiple AU-ROCs are computed. By averaging multiple AU-ROCs, AU-ROC for multi-compartment localization prediction of target RNA class is computed.
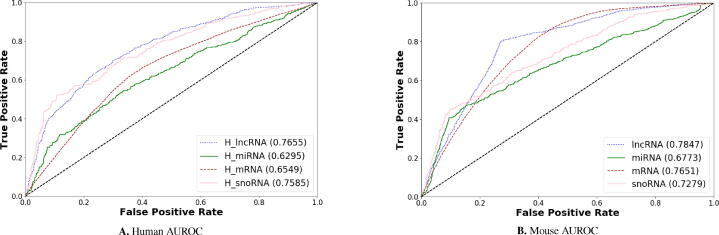
ROC probability curves and AU-ROC scores produced by EL-RMLocNet approach for mRNA, miRNA, snoRNA, and lncRNA multi-compartment subcellular localization across 2 distinct species including homo sapien and mus musculus are provided in Fig. 9. As is shown by the Fig. 9, for homo sapien species, from all 4 different RNAs, EL-RMLocNet approach achieves higher degree of seperability for lncRNA multi-compartment subcellular localization prediction, achieving the peak AU-ROC score of 77%. This is primarily due to the characteristics shown in pie graphs 7 and dount chart 8, lncRNA dataset contains most lengthier sequences having average length around 16,335 nucleotides which expose a greater probability to extract comprehensive nucleotide k-mers patterns for proposed novel K-hop neighbourhood based representation learning scheme and precise deep neural network. For homo sapien species, EL-RMLocNet achieves second best degree of separability around 76% for snoRNAs followed by 65% of mRNA, and 62% of miRNA biomolecules. Among all biomolecules datasets belonging to homo sapien species, miRNA dataset has highest subcellular compartment cardinality. Also, each subcellular compartment has limited number of sequences which are also the shortest among all datasets with average length around 43 nucleotides. These charateristics eventually lead to slightly lower the performance of EL-RMLocNet approach for miRNA biomolecule. Whereas, EL-RMLocNet approach manages to achieve decent performance for snoRNA and mRNA biomolecules having sufficient length sequences as well as sequence to label distribution.
Fig. 9.
AU-ROC Produced by Proposed EL-RMLocNet Approach for Multi-Compartment Subcellular Localization of mRNA, miRNA, snoRNA, and lncRNA across 2 Species A. Homo Sapien and B. Mus Musculus.
On the other hand, for mus musculus species, EL-RMLocNet approach produces best degree of separability for lncRNA biomolecules, achieving AU-ROC figure of 78%. As discussed for homo sapien species, this is mainly due to the highest average sequence length shown in donut chart 8 which allows novel representation learning based deep learning classifier to extract more discrminative set of features for subcellular localization prediction. Further, EL-RMLocNet approach achieves second best degree of separability around 77% for mRNA followed by 73% for snoRNA, and 68% for miRNA multi-compartment subcellular localization prediction. Despite very high subcellular compartment cardinality in mRNA dataset, EL-RMLocNet approach manages to achieve promising performance due to good length and sufficient number of sequences against every subcellular compartment. Among all biomolecules datasets belonging to mus musculs species, miRNA dataset contains the shortest sequences with average length of just 50 nucleotides which leads to slightly lower the performance of EL-RMLocNet approach. With the increase of average sequence length up to 116 nucleotides for snoRNA biomolecule, performance of EL-RMLocNet approach gets increased, a performance trend which is quite evident and discussed for most RNA subtypes in homo sapien species as well.
To summarize, proposed EL-RMLocNet approach achieves slightly better degree of seperability for mus musculus species where it attains better peak performance for most RNA classes. Further, unlike most predictive approaches whose performances significantly plunge on account of different sized dataset or species, proposed EL-RMLocNet approach shows quite consistent performance and robustness across multiple datasets and species mainly due to its aptitude to capture comprehensive relations of nucleotide k-mers as well as to select most relevant features for target RNA class and species.
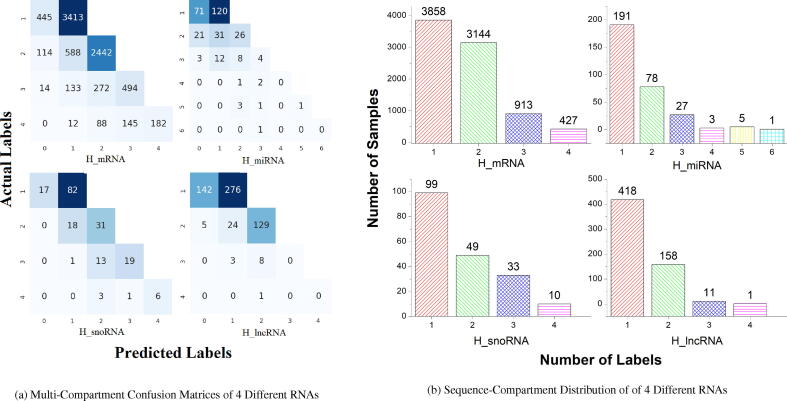
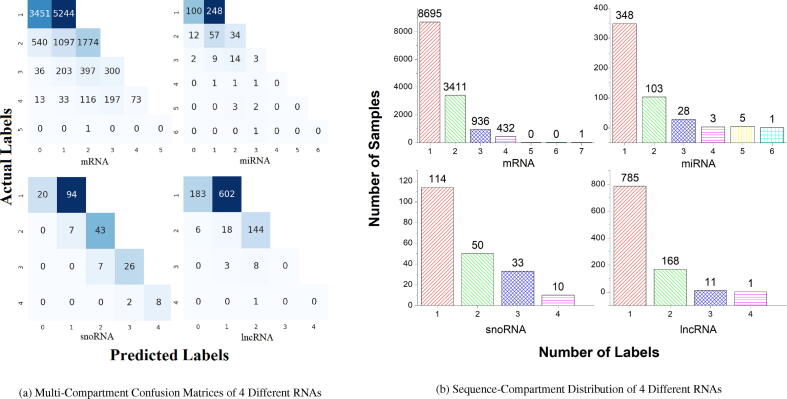
Further, to analyze up to what extent EL-RMLocNet approach manages to correctly predict various combinations of subcellular compartmens on account of heterogeneous subcellular compartment cardinality across 8 different benchmark datasets, multi-compartment confusion matrices along with sequence-to-compartment distributions bar graphs for homo sapien species and mus musculus species are given in Fig. 10 and Fig. 11 respectively. We leverage one-versus-rest strategy in order to generate confusion matrices across all 8 benchmark datasets where false negatives (fn), false positives (fp), true negatives (tn), and true positives (tp) are computed by considering one subcellular compartment as positive and other subcellular compartments as negative. By averaging fn, fp, tn,and tp using total number of available subcellular compartments, confusion matrix for target RNA associated subcellular localization dataset is computed. This is primarily to assess the robustness of EL-RMLocNet when positive subcellular compartment has few number of RNA sequences and negative subcellular compartment has large number of RNA sequences.
Fig. 10.
Multi-Compartment Localization Prediction Performance Produced by EL-RMLocNet on 4 Benchmark Homo Spaien Datasets of mRNA, miRNA, snoRNA, and lncRNA Corresponding to Unique Sequence-Compartment Distribution.
Fig. 11.
Multi-Compartment Localization Prediction Performance Produced by EL-RMLocNet on 4 Benchmark Mus Musculus Datasets of mRNA, miRNA, snoRNA, and lncRNA Corresponding to Unique Sequence-Compartment Distribution.
From accuracy confusion matrices (Fig. 10) produced by proposed EL-RMLocNet approach for homo sapien species, performance analysis for mRNA multi-compartment localization prediction indicates that, from 3,858 uni-compartment RNA sequences, subcellular localization of 3,413 sequences are correctly predicted by proposed EL-RMLocNet approach, indicating over 88% uni-compartment RNA sequences are correctly predicted. From 3,144 bi-compartments RNA sequences, 2,442 RNA sequences are correctly classified into 2 cellular compartments, making it to 78% of total bi-compartment sequences. For tri-compartment and tetra-compartment cardinalities, almost 54% and 43% RNA sequences of respective cardinalities are correctly classified in appropriate subcellular compartments. For homo sapien miRNA subcellular localization, 63% of total uni-compartment, 33% of total bi-compartment, and 15% of total tri-compartment RNA sequences are accurately categorized in respective subcellular localization compartments by EL-RMLocNet approach. For homo sapien snoRNA subcellular localization prediction, EL-RMLocNet approach accurately categorizes 83% of uni-compartment 63% of bi-compartment, 58% of tri-compartment, and 60% of tetra-compartment RNA sequences. Further, for lncRNA multi-compartment subcellular localization prediction, 66% of uni-compartment, and 82% of bi-compartment RNA sequences are correctly predicted. Whereas, no tri-compartment or tetra-compartment RNA sequence is correctly classified in respective subcellular compartment by EL-RMLocNet approach.
It is evident that a significant number of genomic sequences having different subcellular compartment cardinalities are accurately predicted by EL-RMLocNet approach across different RNA classes. Overall, for homo sapien species, EL-RMLocNet achieves better performance on mRNA followed by snoRNA, lncRNA, and miRNA biomolecules. It manages to correctly predict 88% of mRNA uni-compartment, 82% of lncRNA bi-compartment, 58% of snoRNA tri-compartment, and 60% of snoRNA tetra-compartment RNA sequences. Unlike existing RNA associated multi-compartment localization predictors whose performance significantly drops on account of different sized dataset as well as with the increase of subcellular compartment cardinality, proposed EL-RMLocNet approach shows promising performance across muliple datasets and shows robustness for different subcellular compartment cardinalities.
Turning towards the accuracy confusion matrices produced by proposed EL-RMLocNet approach for 4 different RNAs belonging to mus musculus species, performance analysis of mRNA multi-compartment localization prediction indicates that from 8,695 uni-compartment RNA sequences, 5,244 are correctly predicted which makes up to 60% of uni-compartment sequences. Further, 28% of bi-compartment, 32% of tri-compartment, and 17% of tetra-compartment RNA sequences are accurately inferred in respective cellular compartments. For miRNA subcellular localization, decent percentages of uni-compartment, bi-compartment, and tri-compartment RNA sequences are accurately predicted which falls around 71%, 33%, and 11% respectively. For snoRNA subcellular localization prediction, 82% of uni-compartment, 86% of bi-compartment, 79% of tri-compartment, and 80% of tetra-compartment RNA sequences are corrected predicted by EL-RMLocNet approach. Similarly for lncRNA subcellular localization prediction, 77% of uni-compartment and 86% of bi-compartment RNA sequences are accurately predicted into respective localization compartments. To summarize accuracy confusion matrices performance across both species, it is easy to understand that unlike existing computational approaches whose performance decline on account of different species, proposed EL-RMLocNet achieves promising performance across all 4 different RNA classes. Contrary to homo sapien species, for mus musculus species, EL-RMLocNet achieves better performance on snoRNA followed by lncRNA, mRNA, and miRNA biomolecules. It manages to accurately predict 82% of snoRNA uni-compartment, 86% of snoRNA and lncRNA bi-compartment, 79% of snoRNA tri-compartment, and 88% of snoRNA tetra-compartment RNA sequences, revealing once again a promising robustness towards different subcellular compartment cadinalities.
In a nutshell, a comprehensive and multi-dimensional assessment indicates that proposed EL-RMLocNet approach marks promising performance for multi-compartment subcellular localization of 4 different RNAs across 2 different species. It achieves higher performance figures for mus musculus species for most RNA classes. While the novel approach based on the idea of using RNA-As-Graphs assists to capture comprehensive semantic and structural information of nucleotide k-mers. The gating mechanism of LSTM helps to find and retain long range dependencies of the features, and attention mechanism assists to find most relevant features for target RNA class and species. By optimizing feature extraction and target specific subcellular localization prediction, proposed EL-RMLocNet manages to achieve promising performance over multiple different sized benchmark datasets for RNA associated multi-compartment subcellular localization prediction.
4.2. Comparison of EL-RMLocNet with Existing Multi-Compartment RNA Localization Predictors
Considering the significance of determining co-localization of biomolecules in multiple subcellular compartments for deep understanding of cellular biology and to develop diverse biochemical applications [55], Wang et al. [7] developed the state-of-the-art multi-compartment localization predictor for 4 different RNA classes of 2 distinct species. They utilized 6 different nucleotide composition and statistics based sequence encoding schemes including nucleotide property composition, nucleotide k-mers composition, reverse compliment k-mer, nucleic acid composition, di-nucleotide composition, tri-nucleotide composition, and composition of k-spaced nucleic acid pairs to adequately represent the nucleotide information present in RNA sequences. By fusing multivariate information using Hilbert–Schmidt independence criterion based multiple kernel learning, they found an optimal combined kernel for SVM classifier for multi-compartment localization prediction of mRNAs, miRNAs, snoRNAs, and lncRNAs for home sapiens and mus musculus species.
Table 4 compares the performance produced by proposed EL-RMLocNet approach with stat-of-the-art approach [7] for the subcellular localization of 4 different RNAs (mRNAs, miRNAs, snoRNAs, and lncRNAs) for home sapien species. As is indicated by the Table 4, proposed approach EL-RMLocNet outperforms state-of-the-art approach [7] across all 4 benchmark datasets belonging to different RNAs in terms of 5 different evaluation measures. EL-RMLocNet achieves the average precision increment of 7%, 1%, 1%, and 10% as compared to state-of-the-art [7] performance for miRNA, mRNA, snoRNA, and lncRNA multi-compartment localization prediction. EL-RMLocNet improves state-of-the-art accuracy by 11%, 5%, 1%, and 13% for miRNA, mRNA, snoRNA, and lncRNA multi-compartment localization prediction. Performance analysis in terms of coverage, ranking loss, and one-error where lower value indicates better predictive performance, EL-RMLocNet surpasses the previous best performance by a decent margin for all 4 RNAs across all evaluation metrics.
Table 4.
Performance Comparison of Proposed EL-RMLocNet Approach with State-of-the-art Approach for Multi-Compartment Localization Prediction of miRNA, mRNA, snoRNA, and lncRNA using 8 Benchmark Datasets of Homo Sapiens (Human) and Mus Musculus (Mouse) Species.
| Average Precision |
Accuracy |
Coverage |
Ranking Loss |
One error |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Datasets | State-of-the-art [7] | EL-RMLocNet | State-of-the-art [7] | EL-RMLocNet | State-of-the-art [7] | EL-RMLocNet | State-of-the-art [7] | EL-RMLocNet | State-of-the-art [7] | EL-RMLocNet | |
| Human | miRNA | 0.79 | 0.86 | 0.52 | 0.63 | 1.46 | 0.70 | 0.17 | 0.11 | 0.29 | 0.26 | |
| mRNA | 0.76 | 0.77 | 0.41 | 0.46 | 1.69 | 0.68 | 0.24 | 0.23 | 0.37 | 0.35 | ||
| snoRNA | 0.82 | 0.83 | 0.54 | 0.55 | 1.54 | 0.45 | 0.18 | 0.17 | 0.24 | 0.20 | ||
| lncRNA | 0.75 | 0.85 | 0.42 | 0.55 | 1.18 | 0.45 | 0.22 | 0.17 | 0.37 | 0.20 | ||
| Mouse | miRNA | 0.79 | 0.87 | 0.58 | 0.69 | 1.31 | 0.50 | 0.18 | 0.10 | 0.31 | 0.28 | |
| mRNA | 0.70 | 0.71 | 0.34 | 0.37 | 1.71 | 0.87 | 0.14 | 0.13 | 0.44 | 0.40 | ||
| snoRNA | 0.80 | 0.82 | 0.52 | 0.56 | 1.59 | 0.29 | 0.21 | 0.20 | 0.25 | 0.20 | ||
| lncRNA | 0.76 | 0.77 | 0.43 | 0.47 | 0.95 | 0.60 | 0.19 | 0.18 | 0.40 | 0.36 | ||
Furthermore, performance comparison of proposed EL-RMLocNet approach with stat-of-the-art approach [7] for the subcellular localization of 4 different RNAs (mRNAs, miRNAs, snoRNAs, and lncRNAs) for Mus Musculus species (Table 4) indicates that proposed EL-RMLocNet approach once again outperforms previous best performance across all 4 benchmark datasets in terms of five different evaluation metrics. EL-RMLocNet outperforms state-of-the-art average precision by 8%, 1%, 2%, and 1% for miRNA, mRNA, snoRNA, and lncRNA multi-compartment subcellular localization. In terms of accuracy, EL-RMLocNet outperforms previous best performance by 11%, 3%, 4%, and 4% for all 4 miRNA, mRNA, snoRNA, lncRNA multi-compartment localization prediction. Similarly, performance analysis in terms of coverage, ranking loss, and one-error reveals that EL-RMLocNet achieves lower error values across most evaluation metrics for all 4 different RNA classes.
To sum up, proposed EL-RMLocNet approach achieves better performance across most datasets from 8 benchmark datasets belonging to 4 different RNAs and 2 species. Overall EL-RMLocNet achieves higher performance increment for homo sapiens species as compared to Mus musculus. It outperforms stat-of-the-art approach [7] by an average accuracy figure of 8% for homo sapiens species and 6& for Mus musculus species. Unlike traditional nucleotide frequency and physico-chemical properties based sequence encoding schemes used by stat-of-the-art approach [7] which lack to capture comprehensive relations of nucleotides. EL-RMLocNet uses a novel weighted graph based statistical representation learning schemes which treats nucleotide k-mers as nodes and their interactions as edges to better characterize nucleotide k-mers relations. Further, unlike machine learning based stat-of-the-art approach [7], proposed EL-RMLocNet makes use of a precisely deep neural network which utilizes gating mechanism to retain informative features and their dependencies, and attention mechanism to find RNA class and species specific discriminative distribution of features to accurately predict target species RNA subcellular localization.
4.3. Visualization of Most Informative Nucleotide k-mers Patterns
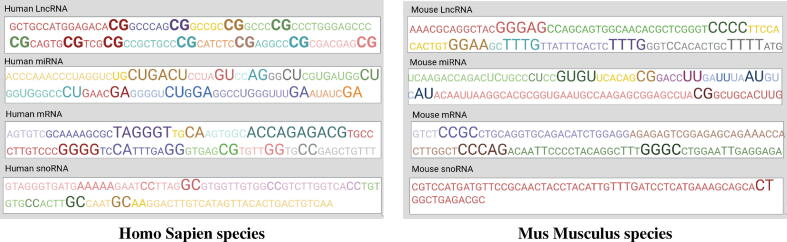
Proposed EL-RMLocNet approach effectively predicts the subcellular localization of various RNAs mainly by finding the most discriminative features with the help of attention mechanism. The mapping of statistical feature space having certain attention weights to their corresponding nucleotides k-mers is essential to elaborate which nucleotide k-mer distribution is most informative to accurately predict various subcellular compartments of target RNA subtype of particular species. The acquisition and interactive visualization of such information effectively interpret and explain the decision making of deep learning model, actualize the generalizeability and practical significance of the model to facilitate biomedical researchers and practitioners. Considering, sequence length largely fluctuates across different RNA subtypes and species ranging from few hundreds nucleotides to thousands of nucleotides. We visualize the importance given by attention mechanism of proposed EL-RMLocNet approach to nucleotide k-mer distribution within the range of 100 nucleotides across all 8 benchmark datasets of 4 different RNAs (mRNA, miRNA, snoRNA, and lncRNA) and 2 species (Homo Sapien, Mus Musculus) to avoid repetition of information and improve readability.
Considering, attention mechanism can even assign different weights to same nucleotide k-mer and same weight to different nucleotide k-mers depending on the short and long range contextual information. Fig. 12 highlights nucleotide k-mer distribution of 4 different RNAs across 2 species on a gradient scale from light to darker shade of a specific color, and size scale from shorter to larger fonts, indicating more darker and standout nucleotide k-mers are most informative for target RNA subtype. For instance, for homo sapien lncRNA multi-compartment subcellular localization prediction, nucleotide bi-mer “CG” is most informative across different distributions of nucleotides. For Mus Musculus miRNA multi-compartment subcellular localization prediction, nucleotide bi-mers CC,GC, AG, GG are most informative followed by AA, and TT within certain nucleotide distributions. Similarly for other RNA subtypes across both species, most informative and least informative nucleotide k-mers and their different nucleotide distributions (unique color shaded) are evident in the Fig. 12. We believe that an interactive intrinsic analysis of various RNAs helps to identify the most appropriate degree of nucleotide k-mer (e.g bi-mer, tri-mer), identify region containing most useful nucleotide k-mer distribution, providing a direction to optimize the performance and generalizeability of various other RNA sequence analysis tasks.
Fig. 12.
Most Informative and Least Informative Nucleotide K-mers Patterns for 4 different RNAs belonging to Homo Sapien and Musculus Species Identified by Attention Layer of Proposed EL-RMLocNet Approach.
5. An Interactive and User-Friendly EL-RMLocNet Web Server
In order to facilitate scientific community, proposed EL-RMLocNet approach is deployed as a public RNA subcellular localization prediction platform. This interactive and user-friendly web server can be used to infer subcellular localization and to validate experimentally identified subcellular localization of distinct RNA classes for multiple species. Contrary to other web server, this web server allows the researchers to train and optimize deep neural network from scratch, analyze the impact of different hyperparameters over the quality of sequence vectors generated by graph based representation and generalizeability of deep neural network. It can also be used to perform subcellular localization prediction over novel RNA sequences of various classes on the go for Homo Sapiens, Mus Musculus, and many other species.
6. Conclusion
In this study, we establish an effective multi-compartment localization prediction landscape for 4 different RNA classes and 2 distinct species to better understand the functional dynamics of RNAs. Unlike existing computational approaches which lack to capture context of residues at different granularity while generating statistical representation of RNA sequences as well as potential residue patterns important for accurate multi-compartment localization prediction. Our proposed approach EL-RMLocNet generates a comprehensive local and global residue contextual information aware statistical vectors of RNA sequences by treating RNA-As-Graph captures. It makes use of LSTM network to extract features, short and long range dependencies, and attention mechanism to assign the weights to the features on the basis of their importance for accurate multi-compartment localization of target RNA class. Visualization of important higher order residue patterns using X technique can assist researchers to draw important insights while comparing sequences of homogeneous or heterogeneous RNA classes. A comprehensive comparison of proposed EL-RMLocNet approach with state-of-the-art approach using 8 benchmark datasets of 4 different RNA classes and 2 distinct species proves that EL-RMLocNet is the first most effective generic yet explainable model for RNA multi-compartment localization prediction. We expect public availability of EL-RMLocNet will prove a valuable asset for subcellular localization prediction of various RNAs across multiple species, as well as an additional tool for the classification and localization prediction of other biomolecules.
Funding
This work is supported by the Sartorius Artificial Intelligence Lab.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Yan Z., Lécuyer E., Blanchette M. Prediction of mrna subcellular localization using deep recurrent neural networks. Bioinformatics. 2019;35(14):i333–i342. doi: 10.1093/bioinformatics/btz337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. Li, L. Zhang, S. He, F. Guo, Q. Zou, Sublocep: a novel ensemble predictor of subcellular localization of eukaryotic mrna based on machine learning, Briefings in Bioinformatics. [DOI] [PubMed]
- 3.Asim M.N., Malik M.I., Zehe C., Trygg J., Dengel A., Ahmed S. Mirlocpredictor: A convnet-based multi-label microrna subcellular localization predictor by incorporating k-mer positional information. Genes. 2020;11(12):1475. doi: 10.3390/genes11121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M.N. Asim, M.A. Ibrahim, C. Zehe, O. Cloarec, R. Sjogren, J. Trygg, A. Dengel, S. Ahmed, L2s-mirloc: A lightweight two stage mirna sub-cellular localization prediction framework (2021) 1–8.
- 5.Y. Lin, X. Pan, H.-B. Shen, lnclocator 2.0: a cell-line-specific subcellular localization predictor for long non-coding rnas with interpretable deep learning, Bioinformatics. [DOI] [PubMed]
- 6.Fan Y., Chen M., Zhu Q. lnclocpred: predicting lncrna subcellular localization using multiple sequence feature information. IEEE Access. 2020;8:124702–124711. [Google Scholar]
- 7.Wang H., Ding Y., Tang J., Zou Q., Guo F. Identify rna-associated subcellular localizations based on multi-label learning using chou’s 5-steps rule. BMC genomics. 2021;22(1):1–14. doi: 10.1186/s12864-020-07347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., et al. Rna maps reveal new rna classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 9.Garg A., Singhal N., Kumar R., Kumar M. mrnaloc: a novel machine-learning based in-silico tool to predict mrna subcellular localization. Nucl Acids Res. 2020;48(W1):W239–W243. doi: 10.1093/nar/gkaa385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Q. Tang, F. Nie, J. Kang, W. Chen, mrnalocater: Enhance the prediction accuracy of eukaryotic mrna subcellular localization by using model fusion strategy, Molecular Therapy. [DOI] [PMC free article] [PubMed]
- 11.Meher P.K., Rai A., Rao A.R. mloc-mrna: predicting multiple sub-cellular localization of mrnas using random forest algorithm coupled with feature selection via elastic net. BMC Bioinformatics. 2021;22(1):1–24. doi: 10.1186/s12859-021-04264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z.-Y., Yang Y.-H., Ding H., Wang D., Chen W., Lin H. Design powerful predictor for mrna subcellular location prediction in homo sapiens. Briefings Bioinform. 2021;22(1):526–535. doi: 10.1093/bib/bbz177. [DOI] [PubMed] [Google Scholar]
- 13.Meher P.K., Satpathy S., Rao A.R. mirnaloc: predicting mirna subcellular localizations based on principal component scores of physico-chemical properties and pseudo compositions of di-nucleotides. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-71381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Streit D., Shanmugam T., Garbelyanski A., Simm S., Schleiff E. The existence and localization of nuclear snornas in arabidopsis thaliana revisited. Plants. 2020;9(8):1016. doi: 10.3390/plants9081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges M.C., Daulagala A.C., Kourtidis A. Lnccation: lncrna localization and function. J Cell Biology. 2021;220(2) doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Zeng, Y. Wu, C. Lu, F. Zhang, F.-X. Wu, M. Li, Deeplncloc: a deep learning framework for long non-coding rna subcellular localization prediction based on subsequence embedding, bioRxiv. [DOI] [PubMed]
- 17.Savulescu A.F., Brackin R., Bouilhol E., Dartigues B., Warrell J.H., Pimentel M.R., Beaume N., Fortunato I.C., Dallongeville S., Boulle M., et al. Interrogating rna and protein spatial subcellular distribution in smfish data with dypfish. Cell Reports Methods. 2021;1(5) doi: 10.1016/j.crmeth.2021.100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahbabian K., Chartrand P. Control of cytoplasmic mrna localization. Cellular Mol Life Sci. 2012;69(4):535–552. doi: 10.1007/s00018-011-0814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zappulo A., Van Den Bruck D., Mattioli C.C., Franke V., Imami K., McShane E., Moreno-Estelles M., Calviello L., Filipchyk A., Peguero-Sanchez E., et al. Rna localization is a key determinant of neurite-enriched proteome. Nature Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilbertz J.H., Voigt F., Horvathova I., Roth G., Zhan Y., Chao J.A. Single-molecule imaging of mrna localization and regulation during the integrated stress response. Mol Cell. 2019;73(5):946–958. doi: 10.1016/j.molcel.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Padròn A., Iwasaki S., Ingolia N.T. Proximity rna labeling by apex-seq reveals the organization of translation initiation complexes and repressive rna granules. Mol Cell. 2019;75(4):875–887. doi: 10.1016/j.molcel.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savulescu A.F., Bouilhol E., Beaume N., Nikolski M. Prediction of rna subcellular localization: learning from heterogeneous data sources. Iscience. 2021 doi: 10.1016/j.isci.2021.103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didiot M.-C., Ferguson C.M., Ly S., Coles A.H., Smith A.O., Bicknell A.A., Hall L.M., Sapp E., Echeverria D., Pai A.A., et al. Nuclear localization of huntingtin mrna is specific to cells of neuronal origin. Cell reports. 2018;24(10):2553–2560. doi: 10.1016/j.celrep.2018.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T.B. Kallehauge, S. Kol, M. Rørdam Andersen, C. Kroun Damgaard, G.M. Lee, H. Faustrup Kildegaard, Endoplasmic reticulum-directed recombinant mrna displays subcellular localization equal to endogenous mrna during transient expression in cho cells, Biotechnology journal 11 (10) (2016) 1362–1367. [DOI] [PubMed]
- 25.Arora A., Goering R., Velez P.T., Taliaferro J.M. Visualization and quantification of subcellular rna localization using single-molecule rna fluorescence in situ hybridization. Methods Mol Biol. 2022:247–266. doi: 10.1007/978-1-0716-1851-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deprey K., Batistatou N., Kritzer J.A. A critical analysis of methods used to investigate the cellular uptake and subcellular localization of rna therapeutics. Nucl Acids Res. 2020;48(14):7623–7639. doi: 10.1093/nar/gkaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D.W. Otter, J.R. Medina, J.K. Kalita, A survey of the usages of deep learning for natural language processing, IEEE Transactions on Neural Networks and Learning Systems. [DOI] [PubMed]
- 28.H.-C. Yi, Z.-H. You, D.-S. Huang, C.K. Kwoh, Graph representation learning in bioinformatics: trends, methods and applications, Briefings in Bioinformatics. [DOI] [PubMed]
- 29.Cheng L., Leung K.-S. Quantification of non-coding rna target localization diversity and its application in cancers. J Molecular Cell Biol. 2018;10(2):130–138. doi: 10.1093/jmcb/mjy006. [DOI] [PubMed] [Google Scholar]
- 30.Feng P., Zhang J., Tang H., Chen W., Lin H. Predicting the organelle location of noncoding rnas using pseudo nucleotide compositions. Interdisciplinary Sci: Comput Life Sci. 2017;9(4):540–544. doi: 10.1007/s12539-016-0193-4. [DOI] [PubMed] [Google Scholar]
- 31.Cao Z., Pan X., Yang Y., Huang Y., Shen H.-B. The lnclocator: a subcellular localization predictor for long non-coding rnas based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y., Fu X., Qu W., Xiao Y., Shen H.-B. Mirgofs: a go-based functional similarity measurement for mirnas, with applications to the prediction of mirna subcellular localization and mirna–disease association. Bioinformatics. 2018;34(20):3547–3556. doi: 10.1093/bioinformatics/bty343. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z.-Y., Yang Y.-H., Ding H., Wang D., Chen W., Lin H. Design powerful predictor for mrna subcellular location prediction in homo sapiens. Briefings Bioinform. 2021;22(1):526–535. doi: 10.1093/bib/bbz177. [DOI] [PubMed] [Google Scholar]
- 34.M.N. Asim, M.A. Ibrahim, C. Zehe, O. Cloarec, R. Sjogren, J. Trygg, A. Dengel, S. Ahmed, L2s-mirloc: A lightweight two stage mirna sub-cellular localization prediction framework (2021) 1–8.
- 35.Asim M.N., Malik M.I., Zehe C., Trygg J., Dengel A., Ahmed S. Mirlocpredictor: A convnet-based multi-label microrna subcellular localization predictor by incorporating k-mer positional information. Genes. 2020;11(12):1475. doi: 10.3390/genes11121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Fu X., Qu W., Xiao Y., Shen H.-B. Mirgofs: a go-based functional similarity measurement for mirnas, with applications to the prediction of mirna subcellular localization and mirna–disease association. Bioinformatics. 2018;34(20):3547–3556. doi: 10.1093/bioinformatics/bty343. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Y., Cai J., Yang Y., Zhao H., Shen H. 2018 IEEE International Conference on Data Mining (ICDM) 2018. Prediction of microrna subcellular localization by using a sequence-to-sequence model; pp. 1332–1337. [Google Scholar]
- 38.Feng S., Liang Y., Du W., Lv W., Li Y. Lnclocation: efficient subcellular location prediction of long non-coding rna-based multi-source heterogeneous feature fusion. Int J Mol Sci. 2020;21(19):7271. doi: 10.3390/ijms21197271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad A., Lin H., Shatabda S. Locate-r: Subcellular localization of long non-coding rnas using nucleotide compositions. Genomics. 2020;112(3):2583–2589. doi: 10.1016/j.ygeno.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Cao Z., Pan X., Yang Y., Huang Y., Shen H.-B. The lnclocator: a subcellular localization predictor for long non-coding rnas based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 41.Su Z.-D., Huang Y., Zhang Z.-Y., Zhao Y.-W., Wang D., Chen W., Chou K.-C., Lin H. iloc-lncrna: predict the subcellular location of lncrnas by incorporating octamer composition into general pseknc. Bioinformatics. 2018;34(24):4196–4204. doi: 10.1093/bioinformatics/bty508. [DOI] [PubMed] [Google Scholar]
- 42.Yang X.-F., Zhou Y.-K., Zhang L., Gao Y., Du P.-F. Predicting lncrna subcellular localization using unbalanced pseudo-k nucleotide compositions. Current Bioinform. 2020;15(6):554–562. [Google Scholar]
- 43.Gudenas B.L., Wang L. Prediction of lncrna subcellular localization with deep learning from sequence features. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-34708-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Qiao H. Kd-klnmf: Identification of lncrnas subcellular localization with multiple features and nonnegative matrix factorization. Anal Biochem. 2020;610 doi: 10.1016/j.ab.2020.113995. [DOI] [PubMed] [Google Scholar]
- 45.Wang D., Zhang Z., Jiang Y., Mao Z., Wang D., Lin H., Xu D. Dm3loc: multi-label mrna subcellular localization prediction and analysis based on multi-head self-attention mechanism. Nucleic Acids Res. 2021;49(8):e46. doi: 10.1093/nar/gkab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Z., Lécuyer E., Blanchette M. Prediction of mrna subcellular localization using deep recurrent neural networks. Bioinformatics. 2019;35(14):i333–i342. doi: 10.1093/bioinformatics/btz337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Li, L. Zhang, S. He, F. Guo, Q. Zou, Sublocep: a novel ensemble predictor of subcellular localization of eukaryotic mrna based on machine learning, Briefings in Bioinformatics. [DOI] [PubMed]
- 48.N.S. Babaiha, R. Aghdam, C. Eslahchi, Nn-rnaloc: neural network-based model for prediction of mrna sub-cellular localization using distance-based sub-sequence profiles, bioRxiv. [DOI] [PMC free article] [PubMed]
- 49.Chen Z., Zhao P., Li F., Marquez-Lago T.T., Leier A., Revote J., Zhu Y., Powell D.R., Akutsu T., Webb G.I., et al. ilearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of dna, rna and protein sequence data. Briefings Bioinform. 2020;21(3):1047–1057. doi: 10.1093/bib/bbz041. [DOI] [PubMed] [Google Scholar]
- 50.S. Vashishth, Neural graph embedding methods for natural language processing, arXiv preprint arXiv:1911.03042.
- 51.Ioffe S., Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Mach Learn. 2015:448–456. [Google Scholar]
- 52.Zhang T., Tan P., Wang L., Jin N., Li Y., Zhang L., Yang H., Hu Z., Zhang L., Hu C., et al. Rnalocate: a resource for rna subcellular localizations. Nucleic Acids Res. 2017;45(D1):D135–D138. doi: 10.1093/nar/gkw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L., et al. Pytorch: An imperative style, high-performance deep learning library. Adv Neural Inform Processing Syst. 2019;32:8026–8037. [Google Scholar]
- 54.P. Liashchynskyi, P. Liashchynskyi, Grid search, random search, genetic algorithm: A big comparison for nas, arXiv preprint arXiv:1912.06059.
- 55.Asim M.N., Ibrahim M.A., Imran Malik M., Dengel A., Ahmed S. Advances in computational methodologies for classification and sub-cellular locality prediction of non-coding rnas. Int J Mol Sci. 2021;22(16):8719. doi: 10.3390/ijms22168719. [DOI] [PMC free article] [PubMed] [Google Scholar]