Summary
Directly ex vivo, peptide-specific CD8+ T cells are present at relatively low frequency and are typically in a resting state. This protocol details the expansion of memory peptide-specific CD8+ T cells by in vitro stimulation, which can be subsequently characterized using a range of assays including tetramer staining and intracellular cytokine staining.
For complete details on the use and execution of this protocol, please refer to Lineburg et al. (2021).
Subject areas: Cell Biology, Immunology
Graphical abstract
Highlights
-
•
Peptide-specific CD8+ T cells can be difficult to detect directly ex vivo
-
•
Protocol to expand memory peptide-specific CD8+ T cells in vitro
-
•
Protocol to increase the frequency of memory peptide-specific CD8+ T cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Directly ex vivo, peptide-specific CD8+ T cells are present at relatively low frequency and are typically in a resting state. This protocol details the expansion of memory peptide-specific CD8+ T cells by in vitro stimulation, which can be subsequently characterized using a range of assays including tetramer staining and intracellular cytokine staining.
Before you begin
Timing: 3 h
This protocol outlines the expansion of CD8+ T cells from HLA-B∗07:01+ individuals, against the HLA-B∗07:02 restricted SPRWYFYYL (SPR)-peptide as described in Lineburg et al. (2021) (https://doi.org/10.1016/j.immuni.2021.04.006). However, we and others have successfully used this protocol for the expansion CD8+ T cells against individual peptides or pools of peptides derived from a range of viruses.
This protocol, or minor variations of, can be utilized to expand CD8+ T cells for a variety of downstream applications, including but not limited to:
The identification of peptide(s) that can induce a functional CD8+ T cell response, and are therefore considered immunogenic peptides (Quinones-Parra et al., 2014; Hensen et al., 2021; Grant et al., 2018).
The quantification or characterization of CD8+ T cell responses toward known immunogenic peptides (Quinones-Parra et al., 2014; Grant et al., 2016, 2018).
The identification of previously unknown epitopes within whole proteins by utilizing overlapping peptides followed by screening with truncated peptides to identify the minimal, or core, peptide that induces a CD8+ T cell response (Grant et al., 2013; Wu et al., 2011).
Institutional permissions
Samples and experiments performed for this manuscript were undertaken in accordance with the Declaration of Helsinki, with Ethics approval from the La Trobe Human Ethics Committee (#HEC21097).
Note: Tissue culture needs to be undertaken in sterile conditions. All work should be undertaken within a Class II Biohazard cabinet using aseptic technique.
-
1.Preparation of Supplementum Completum (SC).
-
a.Prepare SC media by combining all reagents as outlined in the materials and equipment section, filter using a 0.22 μM filter, prepare 30 mL aliquots and store at −20°C.
-
a.
-
2.Heat inactivate fetal calf serum (FCS).
-
a.Thaw FCS (if frozen) at 37°C in a water-bath and heat inactivate as per manufacturers suggestions. Prepare 50 mL aliquots and store at −20°C.
-
a.
-
3.Preparation of R0 and RF10 media.
-
a.Prepare R0 and RF10 media as outlines in the materials and equipment section of this protocol.
-
b.Pre-warm these to 37°C using a water-bath or incubator before use.
-
a.
-
4.Reconstitute and make working stocks of your peptide.
-
a.Peptides are purchased lyophilized and we recommend >90% purity for tissue culture.
-
b.Reconstitute peptides at a concentration of 10 mM in 100% Dimethyl sulfoxide (DMSO) and prepare a tube of 1 mM working stock by diluting an aliquot 1 in 10 in R0 media. Both 10 mM and 1 mM stocks should be stored at −20°C.
-
a.
-
5.Reconstitute recombinant human IL-2 (rIL2).
-
a.Recombinant human IL-2 (rIL2) is purchased lyophilized. Reconstitute rIL2 as per manufacturers recommendations to a concentration of 105 IU/mL. Prepare small aliquots of 20–50 μL to prevent the need for many freeze/thaw cycles, and store at −20°C.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Donor-derived PBMCs | Australian Red Cross Lifeblood | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 | Thermo Fisher Scientific | Cat# 21870092 |
| 100× MEM non-essential amino acid solution | Gibco | 11140-050 |
| HEPES | Sigma | H4034 |
| L-glutamine | Sigma | G3126 |
| β-mercaptoethanol | Sigma | M3148 |
| 100× penicillin/streptomycin/Gentamycin (PSG) | Gibco | 10378-016 |
| Fetal calf serum (FCS) | Scientifix | FBSFR-18444 |
| Recombinant human IL-2 (rIL2) | PeproTech | 200-02 |
| Dimethyl Sulfoxide (DMSO) | Sigma | D2650 |
| SARS-CoV-2 Nucleocapsid protein-derived peptide N105–113 SPRWYFYYL | GenScript | N/A |
| Other | ||
| CO2 incubator with humidity | N/A | N/A |
| Waterbath | N/A | N/A |
| Class II Biohazard Cabinet | N/A | N/A |
| Benchtop Centrifuge | N/A | N/A |
| Tissue culture grade plasticware, 24 or 48 well plates | N/A | N/A |
| Sterile plasticware including 10 mL and 50 mL tubes, 5 mL, 10 mL and 25 mL Stripettes etc | N/A | N/A |
| Inverted Microscope | N/A | N/A |
Materials and equipment
CRITICAL: Please consult the relevant Material and Safety Data Sheet (MSDS) and follow all safety procedures before handling any chemicals.
Supplimentum Completum (SC)
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | N/A | 400 mL |
| 100× MEM Non-essential Amino Acids | 20% (v/v) | 100 mL |
| L-Glutamine | 0.6% (w/v) | 3 g |
| HEPES | 2% (w/v) | 11.9 g |
| β-mercaptoethanol | 0.007% (v/v) | 35 μL |
| Total | N/A | 500 mL |
Prepare 30 mL aliquots and store at −20°C until use.
Alternatives: Many variations of this media supplement exist in the literature, including the use of variations of the core ingredients at varying final concentrations, and are all used for successful CD8+ T cell culture.
R0 media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | N/A | 500 mL |
| SC | 5% (v/v) | 30 mL |
| 100× Penicilin/Streptimycin/Gentamycin | 1% (v/v) | 5 mL |
| Total | N/A | 535 mL |
Store at 4°C ideally for no longer than a month. Discard if media turns bright pink due to alkalization caused by aeration. Media should be clear, discard if media becomes cloudy as it is suggestive of contamination.
RF10 media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | N/A | 500 mL |
| SC | 5% (v/v) | 30 mL |
| 100× Penicilin/Streptimycin/Gentamycin | 1% (v/v) | 6 mL |
| FCS | 10% (v/v) | 60 mL |
| Total | N/A | 596 mL |
Store at 4°C ideally for no longer than a month. Discard if media turns bright pink due to alkalization caused by aeration. Media should be clear, discard if media becomes cloudy as it is suggestive of contamination.
Step-by-step method details
This method describes the generation of a CD8+ T cell line against the HLA-B∗07:02 restricted SARS-CoV-2 SPR peptide using PBMCs from a healthy HLA-B∗07:02+ individual.
Peptide stimulation of PBMCs to establish CD8+ T cell lines
In this step, PBMCs are stimulated with the peptide of interest, in this case the HLA-B∗07:02-restricted SPR peptide, to begin the activation and proliferation of peptide-specific CD8+ T cell for the establishment of peptide-specific CD8+ T cell lines. All work should be undertaken in a Class II Biohazard cabinet to prevent contamination.
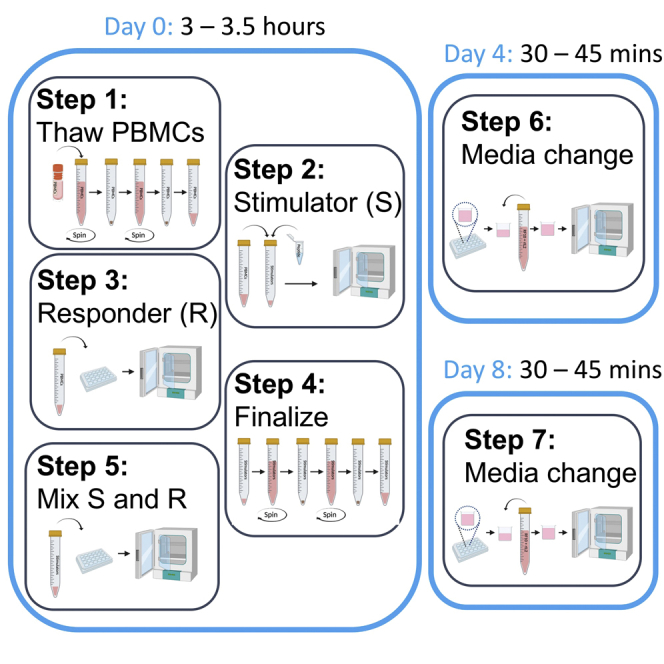
Day 0
Timing: 3–3.5 h
-
1.Thaw and prepare PBMCs derived from a healthy HLA-B∗07:02+ donor.
-
a.Thaw 3 × 106–1 × 107 cryogenically frozen PBMCs per CD8+ T cell line into 9 mL pre-warmed RF10 in a 10–15 mL tube.
-
i.Warm the cryovial in a 37°C water-bath until half of the cells are thawed.
-
ii.Using a transfer pipette, add a small amount of pre-warmed media from your tube into the cryovial to thaw the remaining cells, then transfer everything into your tube.Note: Aim to use between 3 × 106 and 1 × 107 PBMCs per CD8+ T cell line.Note: Cells are typically cryogenically frozen in media containing 10% DMSO, which prevents the formation of ice that would shear up the membrane of the cells during the freezing and storing process. Once thawed, DMSO can be toxic to cells, so it is important to work quickly to get cells into pre-warmed FCS containing media to prevent cell death.Alternatives: Freshly isolated PBMCs can also be used by harvesting into a 10 mL tube and topping to 10 mL with pre-warmed RF10 media.
-
i.
-
b.Centrifuge tubes to pellet cells, 600 × g, for 5 min at 20°C–25°C is sufficient to pellet cells.
-
c.Remove supernatant by decanting.
-
i.Decant by pouring in a single swift motion to prevent losing your cell pellet.
-
i.
-
d.Resuspend cells in 10 mL R0 and take a small aliquot (∼20 μL) into an Eppendorf tube or 96 well plate for cell counting.Note: FCS is thought to perhaps interfere with peptide presentation, therefore this wash in FCS free media is important to remove any remaining FCS from the thawing and first wash step.
-
e.Centrifuge tubes 600 × g for 5 min at 20°C–25°C to pellet cells.
-
f.While cells are spinning, perform a cell count using the trypan blue exclusion method and a Neubauer hemocytometer.Alternatives: Any manual or machine-assisted cell counting methods that can quantify the number of live cells can be used.
-
g.Remove supernatant by decanting.
-
h.Resuspend PBMCs in 300 μL R0.
-
i.Aiming to use 3 × 106 and 1 × 107 PBMCs per CD8+ T cell line, resuspend PBMCs in 300 μL R0 for each CD8+ T cell line, so 600 μL for two CD8+ T cell lines, 900 μL for three CD8+ T cell lines etc.
-
a.
-
2.Prepare the stimulators.Note: One third of your PBMCs become your stimulators (Figure 1). These are then peptide pulsed, and since all nucleated cells present HLA-I on their cell surface, they have the potential to present the peptide at their cell surface. Upon exposure to the responders, CD8+ T cells in the responder fraction become activated, the stimulators are likely to be killed off. Therefore, peptide pulsing and “sacrificing” just a fraction of your PBMCs as stimulators, rather than just adding peptide into all of the cells, can result in less overall cell death.
-
a.Remove 100 μL (1/3 of 300 μL) into a new 10 mL tube to become stimulators.
-
b.Add 1 μL of 1 mM SPR peptide stock to the tube, resulting in a final concentration of 10 μM of peptide in the tube.Note: Peptide pulse up to 3 × 106 cells in 100 μL (1/3 of a starting number of 1 × 107 PBMCs), if you are using more cells to generate larger CD8+ T cell lines, scale up the volume and amount of peptide added accordingly, keeping the same final concentration of peptide as 10 μM.Alternatives: We have set up CD8+ T cell lines with as little as 1 μM of peptide, but 10 μM for individual peptides results is our most common approach as it is well used in the literature and results in robust and reproducible results.Alternatives: You can stimulate PBMCs with pools of peptide rather than individual peptides. We have published concentrations of peptide from as low as 2 μM per peptide (Szeto et al., 2021), and up to 10 μM per peptide (Grant et al., 2018), with many other concentrations found in the literature. For optimized results it is best to test various concentrations for your specific work.
-
c.Mix well by vortexing for 5 s.
-
d.Incubate for 90 min at 37°C with 5% CO2, keeping lids loose (not sealed tightly but still closed to prevent contamination) to permit CO2 exchange.
-
e.Proceed immediately to step 3 once your incubation has begun.
-
a.
-
3.Prepare the responders.
-
a.During the 90 min incubation (step 2), plate out the remaining 200 μL of PBMCs, or 2/3, (from step 1) into a single well of a flat bottom tissue culture grade plate.
-
i.Use a 48 well plate for CD8+ T cell lines with a starting number of 3–5 × 106 PBMCs.
-
ii.Use a 24 well plate for CD8+ T cell lines with a starting number of 1 × 107 PBMCs.Note: If setting up CD8+ T cell lines with a starting number of more than 1 × 107 PBMCs, but less than 2 × 107 PBMCs, start with a 24 well plate and assess cell density for overcrowding daily (for more details, see step 6). If setting up CD8+ T cell lines with a starting number of more than 2 × 107 PBMCs, consider starting with a 12 well plate, and assess your cell density for overcrowding or sparsity (see step 6 for more details) daily.Note: If setting up multiple CD8+ T cell lines and therefore using multiple wells of the tissue culture plate, use wells towards the middle of the plate as these tend to be subject to less evaporation.Note: If setting up multiple CD8+ T cell lines and therefore using multiple wells of the tissue culture plate, consider spacing them out on the plate in case of any unexpected splashes which may cause media to enter an adjacent well.
-
i.
-
b.Top well with RF10 until the well is approximately half full.
-
c.Leave cells to rest until step 5, at 37°C with 5% CO2.Note: If using multiple plates, label both the base (on the side) and the lid of the plate, or draw a line down the plate in different colors or positions so they can be easily matched (Figure 2) in case several lids are off at once.
-
a.
-
4.Finish preparing your stimulators.Note: These wash steps remove excess unbound peptide so that it can’t be bound by your “responder” fraction when added together, ensuring only the stimulators are presenting peptide. Since FCS is not useful at this stage, using R0 is best to prevent wasting FCS.
-
a.Add 10 mL of R0 to your stimulators.
-
b.Centrifuge tube 600 × g for 5 min at 20°C–25°C to pellet cells.
-
c.Remove supernatant by decanting.
-
d.Resuspend cells in another 10 mL of R0.
-
e.Centrifuge tube 600 × g for 5 min at 20°C–25°C to pellet cells.
-
f.Remove supernatant by decanting.
-
g.Resuspend stimulators in 50 μL RF10.Alternatives: Although we typically resuspend our stimulators in 50 μL, you can resuspend them in any volume that will fit within the wells containing your responders, which are approximately half full as per step 3b. By the end of this protocol, all wells will be topped up with media (step 5d), therefore the exact volume doesn’t matter at this stage. If you resuspend your cells in more media, you will simply add less media in the top up step (step 5d).
-
a.
-
5.Mix your stimulators and responders together.
-
a.Add the 50 μL of stimulators into the tissue culture plate into the half-full wells (step 3b) containing your responders.
-
b.Mix gently by pipetting.
-
c.Top wells with RF10.Note: Do not fill the wells to the brim, leave 2–3 mm free at the top of the well to prevent splashing of media onto the lid of the plate when the plate is moved around.
-
d.Incubate plates at 37°C with 5% CO2 4 days.
-
a.
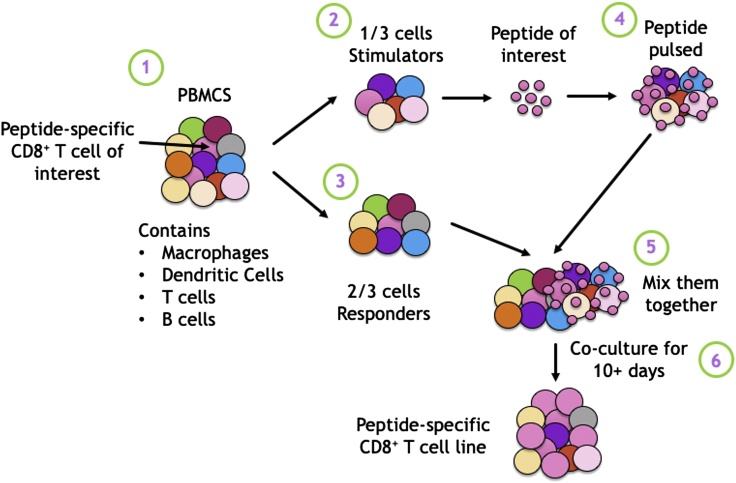
Figure 1.
Theory behind the expansion of peptide-specific CD8+ T cells from PBMCs
Peptide-specific CD8+ T cells are at low frequency within PBMCs, therefore to increase their frequency above the limit of detection and prime them for functional assays, they can be expanded in vitro. PBMCs, which contain a range of cells including but not limited to Macrophages, Dendritic cells, T cell and B cells, are split into 1/3 stimulators and 2/3 responders. The stimulators are peptide pulsed with the peptide of interest and subsequently co-cultured with the responder fraction. Over time, peptide-specific CD8+ T cells grow in size, and proliferate, while other cells dye off, thereby increasing the relative frequency of the peptide-specific cells of interest.
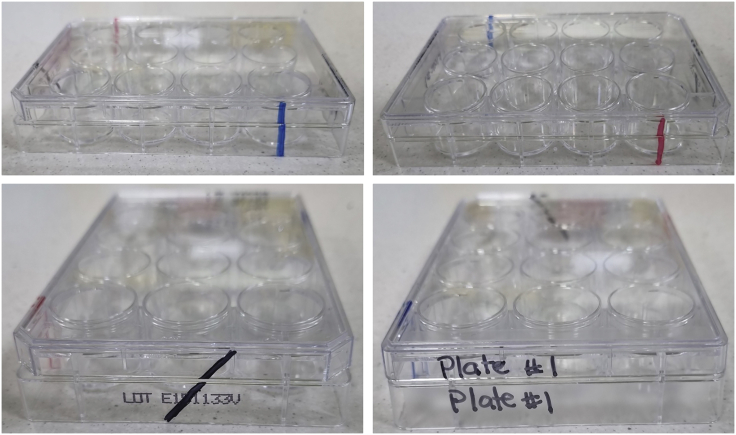
Figure 2.
Tips for labeling tissue culture plates so that lids can be matched to their base
In the instance you have multiple tissue culture plates in use, it is worth making sure that the lid and base of the plates can be matched in case several lids are off at once. A simple way to do this is to use a marker and draw a line down the plate. This can be done in different colors or in different positions, allowing the correct lid to be matched to the correct base quickly and easily. Alternatively, you can label both the lid and the side of the plates with an appropriate identifier.
First half media change of CD8+ T cell lines
This step is to allow for fresh media replacement, as well as adding IL-2 to assist in the growth of the CD8+ T cell lines. All work should be undertaken in a Class II Biohazard cabinet to prevent contamination.
Day 4
Timing: 30–45 min
Alternatives: Although we typically do this on day 4, if media in the CD8+ T cell line is already starting to change color (compared to the RF10 used when setting up the CD8+ T cell lines), this step can be performed on day 3.
-
6.Half media change of your cultures.
-
a.Prepare some pre-warmed RF10 containing 20 IU/mL recombinant human IL2 (rIL2).
-
b.Remove plate carefully from the incubator to prevent splashing.
-
c.Look at your cells under a microscope, typically a 10× objective is sufficient, making note if your cells look particularly sparse (Figure 3Ai) or overcrowded (Figure 3Aii).
 CRITICAL: Cell density is very important in the first few days of establishing a CD8+ T cell line to ensure that responders are interacting with the stimulators, but that they are not so overcrowded that there is nowhere to proliferate. If your cells are too sparse (step 6c, Figure 3Ai) or overcrowded (step 6c, Figure 3Aii) see the alternate protocol below.Note: You would not typically expect to need to condense or split your cells this early on in the protocol if the recommended cell number is used in the recommended well size.
CRITICAL: Cell density is very important in the first few days of establishing a CD8+ T cell line to ensure that responders are interacting with the stimulators, but that they are not so overcrowded that there is nowhere to proliferate. If your cells are too sparse (step 6c, Figure 3Ai) or overcrowded (step 6c, Figure 3Aii) see the alternate protocol below.Note: You would not typically expect to need to condense or split your cells this early on in the protocol if the recommended cell number is used in the recommended well size. -
d.Using a pipette, remove approximately half of the media from the top of the well.
-
i.Take care not to disturb your cells which have settled on the bottom of the well.
-
i.
-
e.Top the wells (leaving 2–3 mm gap) with fresh pre-warmed RF10 media.
-
f.Gently mix the media into the well, resuspending the cells at the same time.Note: Cells like to sometimes gather and settle around the edges of wells, while leaving the middle sparser (Figure 3B). Mixing helps to redistribute the cells within the well if this has occurred.
-
g.Gently measure the volume of media in your well by pipette.
-
h.Add rIL2 directly into the well for a final concentration of 10 IU/mL and gently mix.
-
i.Incubate plates at 37°C with 5% CO2.
-
j.Check your cells under a microscope every few days, and more frequently if the cells are looking overcrowded or sparse, typically a 10× objective is sufficient.
-
a.
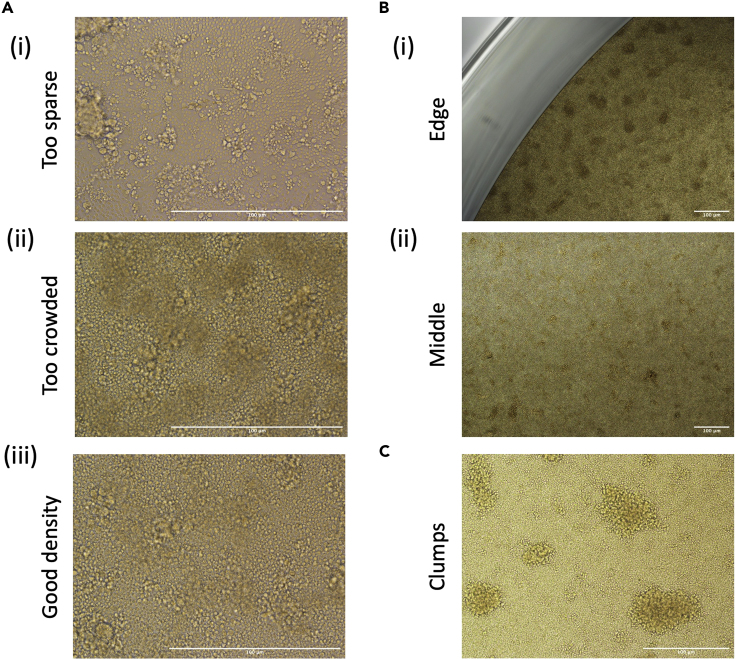
Figure 3.
Microscope images of CD8+ T cell lines
HLA-A∗02:01 restricted M158-specific CD8+ T cells were expanded from a HLA-A∗02:01+ individual as per the protocol described in this manuscript. Images were taken on a Nikon Eclipse Ti microscope on a ×4,×10 and ×20 objective daily from Day 1 to Day 10.
(A) Representative photo depicting cells that are too sparse, which can limit interactions between cells which can prevent adequate activation.
(B) Images demonstrating how cells sometimes preferentially clump around the edges of tissue culture wells, therefore it is important to resuspend and redistribute cells during media changes.
(C) Representative image of round clumps which can be indicative of peptide-specific proliferation. Scale bars depict 100 μm at the various magnifications.
Alternatives: If your cells are too sparse (steps 6c/6j, Figure 3Ai) consider placing them in a smaller tissue culture grade plate. Remove half of the media as per step 6d. Then, resuspend your cells in the existing media, taking care to resuspend all of your cells from the bottom of the plate thoroughly, and transfer to a smaller well (i.e., if using a 24 well plate, move to a 48 well plate), making note of the volume of your culture. Top the well with RF10, making note of the volume you have added. Then add rIL2 directly at a final concentration of 10 IU/mL.
Alternatives: If your cells are too overcrowded (steps 6c/6j, Figure 3Aii), consider moving the cells into a larger well in a tissue culture plate. Remove half of the media as per step 6d and replace with RF10 as per step 6e. Then, resuspend your cells, taking care to resuspend all of your cells from the bottom of the plate thoroughly, and either transfer to a larger well, or distribute into two wells. Top the well(s) with RF10 media. Gently measure the volume in each well by pipette, and add rIL2 directly into the well for a final concentration of 10 IU/mL.
Second half media change of CD8+ T cell lines
In this step, we will remove some of the existing media, replacing it with fresh media. All work should be undertaken in a Class II Biohazard cabinet to prevent contamination.
Day 8 and then twice weekly until used or discarded
Timing: 30–45 min
Alternatives: Although this is typically done on day 8, if media in the CD8+ T cell line is starting to change color (compared to the color after the last media change), this step can be performed on day 7.
CRITICAL: Cell density is very important for the continual proliferation and growth of CD8+ T cell lines. If your cells are too sparse (step 7c, Figure 3Ai) or overcrowded (step 7c, Figure 3Aii) see the alternate protocol below.
Note: You would not typically expect to need to condense or split your cells this at this stage of the protocol if the recommended cell number is used in the recommended well size. However, if the CD8+ T cells have expanded significantly, or there has been a large amount of cell death, this may be required.
-
7.Half media change of your cultures.
-
a.Repeat steps 6a to 6f, keeping note of how much fresh RF10 media is added into each well.
-
b.Add rIL2 for a final concentration of 10 IU/mL of the fresh rIL2 added into your well.Note: Your existing media now contains the remaining rIL2, and your new media contains 10 IU/mL rIL2, ensuring that the final concentration of rIL2 within the well remains at approximately 10 IU/mL IL2. If you are concerned about variability in rIL2 concentrations between CD8+ T cell lines, please see the alternative approach below.Alternative: If you are concerned about minor differences in the concentration of rIL2 between wells, harvest your CD8+ T cell lines into a 10 mL tube and centrifuge tube at 600 × g for 5 min at 20°C–25°C to pellet cells. Then resuspend cells in RF10 containing 10 IU/mL rIL2 and place them back into their original well.
-
c.Gently mix the media into the well, resuspending the cells at the same time.Note: Cells like to gather and settle around the edges of wells, while leaving the middle sparser (Figure 3). Mixing helps to redistribute the cells within the well.
-
d.Incubate plates at 37°C with 5% CO2 for an additional 3–4 days.
-
a.
Alternatives: If your cells are too sparse or crowded (step 6c, Figures 3Ai and 3Aii) follow the advice as per “Alternatives” in step 6. Make a note of any fresh media added into your wells and add rIL2 so that the concentration with the fresh media is 10 IU/mL rIL2.
Note: CD8+ T cell lines can be assayed from day 9 or 10, or continue to be grown, if required, by doing a half media change twice weekly. We have grown CD8+ T cell lines until day 21, and in theory they can be grown for longer, however, since these are not immortalized cell lines, they do start dying if subject to extended culture. Therefore, we typically do our first assays from day 10, and try to complete any subsequent assays by day 16 or day 17 where possible.
Expected outcomes
Over time, most commonly from day 5, onward (although sometime earlier), round clumps may be visible under the microscope (Figure 3C), and may be indicative that CD8+ T cell proliferation has occurred, although this cannot be confirmed without any CD8+ T cell specific assays such as tetramer staining or intracellular cytokine staining.
Peptide-specific CD8+ T cells have typically expanded enough by day 9 or 10 to be visible above the limit of detection in several assays, including by tetramer staining or intracellular cytokine staining assays. As standard, we typically assay CD8+ T cell lines from day 10 onwards.
Peptide-specific CD8+ T cell lines can range from 0%-specific to greater than 60% specific (Grant et al., 2018) (Quinones-Parra et al., 2014) (Grant et al., 2016) (Grant et al., 2013) and this varies significantly by peptide, and by donor. Many factors will influence the specificity of your CD8+ T cell line, some of which include how many peptide-specific CD8+ T cells were present in the original PBMCs an how much they proliferate within the CD8+ T cell line over the course of time.
Limitations
It is important to note that a limitation of this protocol is that it is thought to expand only memory CD8+ T cell populations, but not naïve CD8+ T cells. With this knowledge, if you wish to characterize CD8+ T cell responses, or assess whether novel pathogen-derived peptides are immunogenic, it would be best to use PBMCs derived from donors with a known exposure to the pathogen of interest.
The frequency of peptide-specific CD8+ T cells present in an individual is influenced by many donor-centric factors including precursor frequency of naïve CD8+ T cells and other HLA’s possessed by an individual, to name a few. With this knowledge it is important to consider expanding CD8+ T cells in multiple donors (we prefer to use at least n=3–5 where possible) with the correct HLA-I profile to ensure that results are not being biased by individuals.
Additionally, there are several factors that can influence the exact specificity of CD8+ T cell lines including but not limited to: the starting number of PBMCs, their density, how many peptide-specific CD8+ T cells are present, how quickly peptide-specific CD8+ T cells expand, the quality of your FCS, the temperature maintenance of your incubator, as well as several factors listed below. Indeed, PBMCs harvested at the same time point, but expanded against the same peptide at a different time can give some variability. This is most obvious when the proportion of peptide specific CD8+ T cells is smaller. As such, it is important to consider your results within this limitation. For this reason, we typically assay different donors on different days, and assess the presence or absence of a CD8+ T cell response, rather than a direct comparison in the proportion of specificity.
Troubleshooting
Problem 1
PBMCs won’t resuspend and appear as sticky clumps.
When thawing or resuspending PBMCs before or after stimulation (Day 1, step 1 or step 4), they can appear as sticky clumps.
Potential solution
Sticky clumped cells are a result of DNA being released as cells die. It is not uncommon for cells to die during the freeze thaw cycle, however excessive cell death can result from poor freezing or thawing. Make sure cells are appropriately cryogenically frozen and stored, and work quickly when thawing. Using a DNase reagent when thawing can help limit cell death when working with samples that have not be processed or stored under optimal conditions. To prevent additional cell death, remove sticky clumps carefully by pipette as you notice them, most commonly during thawing or after the peptide-pulse step.
Problem 2
Cell number is decreasing/cells are dying in culture.
It appears that the density of cells is decreasing over time, evidenced by visual density changes by microscopy or the presence of dead cells using the trypan blue exclusion method of cell counting.
Potential solution
There are several factors that can influence the success of cell culture.
It is important to check that all of your reagent and plasticware is in date, particularly reagents or plasticware stored in large volumes or have longer shelf life.
Check that the CO2 incubator is working optimally, holding its temperature and CO2 level, and has enough water in the tray or water jacket (depending on the model of your incubator) to ensure that humidity conditions are maintained.
It is worth trying PBMCs from different donors/batches/sources in case the PBMCs were not well processed or stored, or if the cells from that specific individual are not responding well to cell culture conditions, which can happen with cells that are exhausted, often derived from individuals suffering from chronic inflammatory conditions or infections such as human immunodeficiency virus (HIV) or hepatitis C virus (HCV).
The density of cells within culture is important, especially initially, to ensure that the responder fraction is in contact with the stimulator fraction to provide adequate stimulation. Ideally you would check your CD8+ T cell cultures every 2–3 days by looking at them under the microscope to ensure they are at an optimal density to promote activation and proliferation.
Problem 3
Cultures are contaminated.
Tissue culture can be contaminated with bacteria or fungus causing cell death.
Potential solution
Tissue culture can be particularly susceptible to contamination, more commonly bacterial but also fungal contamination. If you have bacteria or fungal contamination within your tissue culture, it is best to dispose of the cultures without opening them if possible. For example, if possible (please refer to the relevant waste stream disposal guideline for your workplace) do not open plates so as to add bleach or other disinfection reagent, as this can cause the aerosolization of the bacteria or fungal spores which can significantly increase the potential for contamination. Instead, contain the plate within in the Class II Biohazard cabinet in a biohazard bin and dispose of through your waste stream.
Bacteria can be introduced from several sources, as such, it is important to work within a Class II Biohazard cabinet under aseptic conditions to limit the potential for contamination. Additionally, make sure that all water-baths including their water trays (if relevant) and incubators are cleaned frequently, that media and reagents are only opened within a Class II Biohazard cabinet or are filter sterilized prior to use and all plasticware and tips are sterile before use.
Problem 4
Cells are not peptide specific in downstream assays.
CD8+ T cell lines can look healthy (good density of cells, appearance of proliferation) but not be peptide specific in downstream assays.
Potential solution
Assuming that your CD8+ T cell cultures look healthy and alive (see troubleshooting tips above) there are several reasons why peptide-specific CD8+ T cell have not expanded.
-
•
Check that your donors have the correct HLA-I restriction for the peptide of interest. Since CD8+ T cells recognize and respond to peptides presented by HLA-I, it is vital that the individuals have the HLA-I molecule of interest for your experiment.
-
•
Your peptide may not be immunogenic. Not all peptides capable of being presented by HLA-I molecules are immunogenic (Hensen et al., 2021; Koutsakos et al., 2019), and not all known immunogenic peptides are immunogenic or highly immunogenic in all donors (Quinones-Parra et al., 2014; Sant et al., 2020; Lineburg et al., 2021). Therefore, it is important to screen several donors with the correct HLA-I profile to ensure that your results are representative.
-
•
Your peptide may not be pure enough. Although many people generate successful CD8+ T cell lines using crude peptides, we prefer a purity of >90% for our CD8+ T cell line to ensure that the effect of any impurities on our CD8+ T cell cultures is limited.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof Stephanie Gras, S.Gras@latrobe.edu.au.
Materials availability
This study did not generate new unique regents.
Acknowledgments
We would like to thank the La Trobe Institute of Molecule Sciences (LIMS) Bioimaging platform for their assistance in taking and processing the microscopy images within this manuscript. This work was funded by the Australian National Health and Medical Research Council (NHMRC) and an MRFF grant; E.J.G. is supported by an Australian Research Council DECRA Fellowship (DE210101479) and an AINSE Early Career Research Grant, and S.G. is supported by an NHMRC Senior Research Fellowship (#1159272).
Author contributions
Conceptualization, S.G. and E.J.G.; methodology, E.J.G.; investigation, E.J.G.; writing – original draft, E.J.G.; writing – review & editing, E.J.G. and S.G.; funding acquisition, S.G. and E.J.G.; resources, S.G. and E.J.G.; supervision, S.G.
Declaration of interests
The authors have no interests to declare.
Contributor Information
Emma J. Grant, Email: e.grant@latrobe.edu.au.
Stephanie Gras, Email: s.gras@latrobe.edu.au.
Data and code availability
The datasets/code supporting this protocol publication has not been deposited in a public repository because they are stand-alone experiments and do not form a part of a research publication, but are available from the corresponding author on request.
References
- Grant E., Wu C., Chan K.F., Eckle S., Bharadwaj M., Zou Q.M., Kedzierska K., Chen W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T-cell responses. Immunol. Cell Biol. 2013;91:184–194. doi: 10.1038/icb.2012.78. [DOI] [PubMed] [Google Scholar]
- Grant E.J., Josephs T.M., Loh L., Clemens E.B., Sant S., Bharadwaj M., Chen W., Rossjohn J., Gras S., Kedzierska K. Broad CD8(+) T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 2018;9:5427. doi: 10.1038/s41467-018-07815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant E.J., Josephs T.M., Valkenburg S.A., Wooldridge L., Hellard M., Rossjohn J., Bharadwaj M., Kedzierska K., Gras S. Lack of heterologous cross-reactivity toward HLA-A∗02:01 restricted viral epitopes is underpinned by distinct alphabetaT cell receptor signatures. J. Biol. Chem. 2016;291:24335–24351. doi: 10.1074/jbc.M116.753988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensen L., Illing P.T., Bridie Clemens E., Nguyen T.H.O., Koutsakos M., Van De Sandt C.E., Mifsud N.A., Nguyen A.T., Szeto C., Chua B.Y., et al. CD8(+) T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A∗24:02 allomorph. Nat. Commun. 2021;12:2931. doi: 10.1038/s41467-021-23212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., et al. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S., et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065.e5. doi: 10.1016/j.immuni.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Parra S., Grant E., Loh L., Nguyen T.H.O., Campbell K.A., Tong S.Y.C., Miller A., Doherty P.C., Vijaykrishna D., Rossjohn J., et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. USA. 2014;111:1049–1054. doi: 10.1073/pnas.1322229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant S., Quinones-Parra S.M., Koutsakos M., Grant E.J., Loudovaris T., Mannering S.I., Crowe J., Van De Sandt C.E., Rimmelzwaan G.F., Rossjohn J., et al. HLA-B∗27:05 alters immunodominance hierarchy of universal influenza-specific CD8+ T cells. PLoS Pathog. 2020;16:e1008714. doi: 10.1371/journal.ppat.1008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto C., Chatzileontiadou D.S.M., Nguyen A.T., Sloane H., Lobos C.A., Jayasinghe D., Halim H., Smith C., Riboldi-Tunnicliffe A., Grant E.J., Gras S. The presentation of SARS-CoV-2 peptides by the common HLA-A(∗)02:01 molecule. iScience. 2021;24:102096. doi: 10.1016/j.isci.2021.102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Zanker D., Valkenburg S., Tan B., Kedzierska K., Zou Q.M., Doherty P.C., Chen W. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc. Natl. Acad. Sci. USA. 2011;108:9178–9183. doi: 10.1073/pnas.1105624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets/code supporting this protocol publication has not been deposited in a public repository because they are stand-alone experiments and do not form a part of a research publication, but are available from the corresponding author on request.