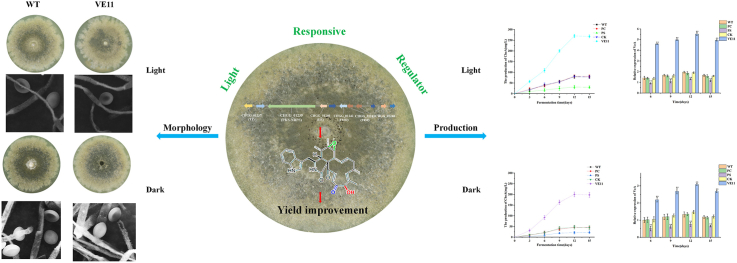
Abstract
Cytochalasans, with diverse structures and pharmacological activities, are a class of compounds containing isoindolinone moieties fused to the tricyclic or tetracyclic ring system. Chaetoglobosin A (cheA), mainly produced by Chaetomium globosum, is the most abundant cytochalasan. However, limited understanding of transcriptional regulation of morphological development and cheA biosynthesis in C. globosum has hindered cheA application in agriculture and biomedical field. This study examined the regulatory role of CgVeA gene in C. globosum. CgVeA had significant effect on secondary metabolites production in C. globosum, similar to that reported in other filamentous fungi. Inactivation of CgVeA caused an obvious decrease in cheA production from 51.32 to 19.76 mg/L under dark conditions. In contrast, CgVeA overexpression resulted in a dramatic increase in cheA production, reaching 206.59 mg/L under light conditions, which was higher than that noted under dark condition. The RT-qPCR results confirmed that CgVeA, as a light responsive regulator, positively regulated cheA biosynthesis by controlling the expression of core genes of the cheA biosynthetic gene cluster and other relevant regulators. Electrophoretic mobility shift assays proved that CgVeA directly regulated LaeA, cheR, and p450, and indirectly regulated PKS. Moreover, CgVeA had a significant effect on the regulation of asexual spores production. When compared with wild-type C. globosum, CgVeA-silenced and CgVeA overexpression mutants presented remarkable differences in sporulation, irrespective of light or dark condition. Besides, CgVeA expression was speculated to negatively regulate spore formation. These findings illustrated the regulatory mechanism of a hypothetical global regulator, CgVeA, in C. globosum, suggesting its potential application in industrial-scale cheA biosynthesis.
Keywords: Chaetoglobosin A, CgVeA, Chaetomium globosum
Graphical abstract
1. Introduction
Chaetomium spp. are globally ubiquitous fungi found in various habitats, such as soil, plants, wastes, and other distinct environments [[1], [2], [3], [4]]. They are well-known prolific producers of bioactive secondary metabolites, including azaphilones, terpenoids, and cytochalasans [2,5,6]. C. globosum is an important biocontrol species with significant capacity to produce these bioactive secondary metabolites. Cytochalasans, the major secondary metabolites produced by C. globosum, are a class of compounds consisting of tricyclic or tetracyclic ring systems fused to isoindolinone moieties. The macrocyclic ring of these products can undergo different modifications to form novel skeletons, exhibiting a broad range of biological activities, such as phytotoxic, antitumor, antifouling, and immunomodulatory activities [2,[7], [8], [9]], and have significant importance in pharmacological, agricultural, and industrial applications. CheA, one of the most abundant cytochalasans, is catalyzed and synthesized by polyketide-non-ribosomal peptide synthetase (PKS-NRPS) [10]. Owing to its significant research and commercial value, many studies have successively analyzed cheA. Previous works have clearly elucidated the function of the cheA biosynthetic gene cluster. By using an RNA-mediated gene silencing strategy, the gene cluster involved in cheA biosynthesis, which is most similar to the aspyridone gene cluster in Aspergillus nidulans, has been identified in Penicillium expansum [11]. Bioinformatics analysis has revealed that the cheA biosynthetic gene cluster consists of seven elements, such as the core region PKS-NRPS, a trans enoyl-reductase (ER), an FAD-dependent monooxygenase (FMO), two putative p450 monooxygenases, and two putative C6 transcription factors [12,13]. PKS-NRPS plays a major role in the condensation of acetate/malonate to form the carbon skeleton in the biosynthesis of chaetoglobosins [14]. ER forms a reductive carbon skeleton in the hybrid synthetase [15]. The polyketide portion of cheA is a non-nucleotide composed of three methyl groups from S-adenosyl methionine [16]. Once the non-peptides are assembled, NRPS is linked to an activated tryptophan via condensation reaction. When the cyclization intermediate is formed, the putative p450 would react and form the final product [12].
The global transcription factors have been confirmed to be a large family of pleiotropic regulators, which have been found to exert profound influence on fungi in many aspects, such as enhancing or reducing secondary metabolites production [17,18], activating silent or cryptic gene clusters [19,20], morphology development changes [20,21], etc. LaeA, the nuclear localized histone methyltransferase and a well-studied and major protein, was first identified in the model fungus A. nidulans and noted to be extensively distributed in fungi [22,23]. Ochratoxin A (OTA), which was first reported in 1965 in Aspergillus spp., is a potent nephrotoxin exerting carcinogenic, teratogenic, and immunotoxic effects on animals and possibly humans [24]. Several studies have been conducted to determine the function of LaeA gene in OTA biosynthesis [25,26]. It has been reported that LaeA is a positive regulator of OTA biosynthesis, and contributes to Aspergillus carbonarius pathogenicity by regulating the expression level of Acgox. Moreover, LaeA has been found to function by forming heterotrimeric velvet complexes with VeA and VelB [27]. VeA, a light-dependent transcription factor, is another well-known genetic link associated with morphogenesis and biosynthesis of natural products. Disruption of Apc.laeA and Apc.veA in Aspergillus pachycristatus NRRL 11440 has been observed to significantly reduce, but not eliminate, the production of echinocandin B. In addition, the two mutants exhibited aberrant phenotype, including development of aerial hyphae, conidiophores, ascospore, and pigmentation when grown under light or dark conditions, when compared with the parent strain [28]. Inactivation of VeA in Alternaria alternata by a homologous recombination strategy has been noted to significantly reduce sporulation and strongly compromise mycotoxin production, both in vitro and during pathogenesis of tomato fruits [29]. Overexpression of the major conserved regulatory VeA in Aspergillus fumigatus has been reported to cause a dramatic decrease in conidia development and an obvious increase in gliotoxin production, when compared with those in the wild-type control and inactivation mutants [30]. Moreover, VeA is also well known for its ability to adjust to environmental stress, such as salinity, osmotic pressure, temperature, and pH [31,32].
Over the past 5 years, our research team has attempted to identify and study the function of regulators that play an essential role in morphological development and cheA biosynthesis in C. globosum. The pathway-specific regulator CgcheR, which has an essential role in modulating sporulation and cheA production, was identified via bioinformatics analysis in C. globosum [13]. Disruption of CgcheR caused a significant decrease in the expression of genes associated with cheA biosynthetic gene cluster, and the CgcheR disruptant lost the ability to produce spores. In contrast, constitutive overexpression of CgcheR evidently improved the cheA titer from 52 to 260 mg/L, and the yield variation was mainly owing to the efficient activation of the biosynthetic gene cluster. Similarly, CgLaeA, a ubiquitous regulator in filamentous fungi, was also identified in C. globosum [33]. In the CgLaeA overexpression mutant, the cheA production significantly increased, the expression of transcription factor CgcheR was positively regulated, and the expression of another global regulator CgVelB was negatively regulated. CgLaeA usually functions by forming a complex with the VeA gene. The VeA gene, which is involved in the regulation of asexual and sexual sporulation and biosynthesis of various secondary metabolites, is highly conserved in filamentous fungi. However, little is known about the transcriptional regulation of cheA biosynthesis and morphological development in C. globosum. In the present study, the CgVeA gene of C. globosum, which encodes a global regulator, was characterized. A verification process based on CgVeA-silencing and CgVeA-overexpression strategies was performed to investigate the extent of CgVeA regulon in detail. Subsequently, the influence of CgVeA on cheA biosynthesis and sporulation was analyzed under light and dark conditions, respectively, with wild-type C. globosum W7 used as the control. The findings of the present study help to better understand the regulatory mechanism of CgVeA, and suggest that CgVeA could be a potential target for developing control strategies for cheA biosynthesis in C. globosum.
2. Materials and methods
2.1. Fungal strain, plasmids, and culture conditions
The fungal strain and plasmids used in this study are summarized in Table 1. The sequences of primers employed in this study are shown in Table S1. C. globosum was cultivated on potato glucose agar (PDA) at 28 °C. pET28a, pBARGP-mcherry, and pSilent were stored at −80 °C. Escherichia coli DH5α and E. coli BL21(DE3) cells were cultured in LB medium (10 g of NaCl, 10 g of tryptone, and 5 g of yeast extract; pH of 7.2) with antibiotics. C. globosum was cultured under light and dark conditions, respectively, and the differences were compared.
Table 1.
Strains and plasmids used in this study.
| Strains/plasmids | Characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli DH5a | lacZΔM15 | Takara |
| E. coli BL21(DE3) | Protein expression of strain | Takara |
| C. globosum W7 | Wild-type strain | This study |
| PS | CgVeA silencing mutant | This study |
| PC | CgVeA scramble control mutant | This study |
| CK | CgVeA overexpression control mutant | This study |
| VE11 | CgVeA overexpression mutant | This study |
| Plasmids | ||
| pSlient-1 | Vector used for gene silencing | Takara |
| pET28a | Vector used for protein expression | Takara |
| pSL-VeA | A DNA fragment containing the upstream flanking sequence of VeA was inserted into pSlient-1 | This study |
| pS-VeA | A DNA fragment containing the downstream flanking sequence of VeA was inserted into pSL-VeA | This study |
| pSCL-VeA | A DNA fragment containing the upstream flanking sequence of SC-VeA was inserted into pSlient-1 | This study |
| pSC-VeA | A DNA fragment containing the downstream flanking sequence of SC-VeA was inserted into pSCL-VeA | This study |
| pBARGPE | Vector used for overexpression | Takara |
| pBG-VeA | A DNA fragment containing VeA, its putative promoter, and terminator was inserted into pBARGPE | This study |
| pBG-VeAC | Control overexpression plasmid without any fragment insertion | This study |
2.2. Plasmid construction
2.2.1. Construction of silencing and scramble control plasmid
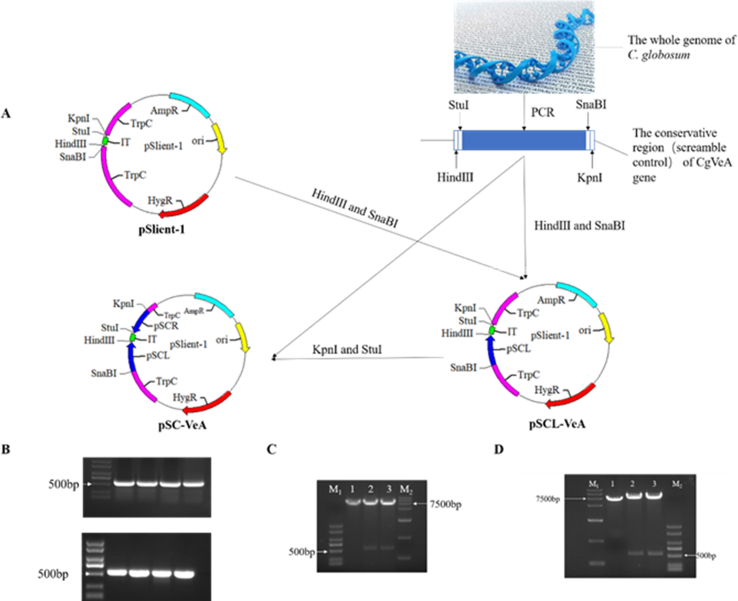
The conserved region of the CgVeA gene was analyzed by the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) and Conserved Domains (https://www.ncbi.nlm.nih.gov/cdd/). According to the results of conserved region analyses, primers VeA-pSLR and VeA-pSLL were designed to amplify the pSL (Table S2) of CgVeA. The plasmid pSlient-1 and pSL were digested with SnaBI and HindIII, respectively, ligated overnight with T4 DNA ligase, and transformed into E. coli DH5α competent cells. The pSR fragment was inserted via seamless cloning using the primers, VeA-pSRR and VeA-pSRL, to amplify the target fragment. The linearized vector pSL-VeA was digested with KpnI and StuI, and the fragment and plasmid were ligated according to the manufacturer's instructions (ClonExpress MultiS One Step Cloning Kit (Vazyme)) and transformed into E. coli DH5α competent cells to obtain plasmid pS-VeA. The scramble control plasmid was constructed in the same way as the silencing plasmid and labeled as pSC-VeA.
2.2.2. Construction of overexpression plasmid and overexpression control plasmid
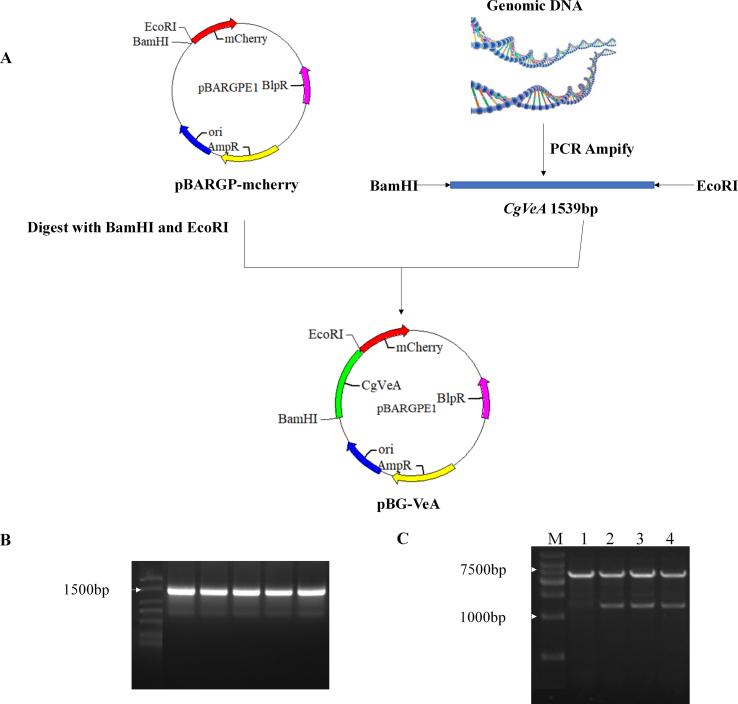
Based on the search of genomic information of C. globosum in NCBI (National Center for Biotechnology Information), along with the characteristics of VeA, it was found that CgVeA (GenBank: XP_001228297.1) has high homology with other filamentous fungi. The primers VeA-L and VeA-R were designed to amplify the sequence of the CgVeA gene, with BamHI and EcoRI sites. The CgVeA overexpression vector was constructed with pBARGP-mcherry. The target fragment after PCR amplification was 1539 bp. The pBARGP-mcherry plasmids and fragments were digested with BamHI and EcoRI, ligated using T4 DNA ligase, and transformed into E. coli DH5α competent cells to produce the overexpression plasmid pBG-VeA. Subsequently, overexpression control plasmid, comprising an empty vector pBARGP-mcherry without any inserted fragment, was constructed to verify the role of CgVeA. This plasmid was designated as pBG-VeAC and selected for transformation by protoplast transformation.
2.3. Protoplast transformation
After constructing the plasmids pS-VeA, pSC-VeA, pBG-VeA, and pBG-VeAC, PEG-mediated protoplast transformation was performed as described previously [34]. The wild-type C. globosum was cultured in PDA broth for 3 days, and the mycelia were filtered and digested using lywallzyme in an orbital shaker incubator at 30 °C and 110 rpm. After digestion, the mixture was filtered through a double lens paper and centrifuged at 4000 rpm for 5 min at 4 °C, the supernatant was discarded, and the sediments were resuspended in 0.06 mol/L STC (27.3 g of sorbitol, 0.75 g of Tris, 0.13 g of CaCl2, and 125 mL of water; pH of 7.5). This step was repeated two or three times, and the concentration of protoplasts was determined after resuspending the sediments in 200 μL of STC. Then, 10 μg of plasmids were added to the STC and the mixture was incubated at 0 °C for 20 min. Subsequently, 50% PEG 4000 (50 g of PEG 4000, 8.48 g of Tris, 0.11 g of CaCl2, and 100 mL of water; pH of 7.5) was added to the protoplast-plasmid mixture and incubated at 0 °C for 20 min. After that, the mixture was cultured in 10 mL of regeneration medium (0.1 g of yeast extract and peptone, 3.42 g of sucrose, 0.15 g of agar, and 10 mL of water; pH of 7.0) for 12 h at 28 °C, and then poured into the screening medium (0.1 g of agar, 10 mL of PDA, and appropriate concentrations of antibiotics). After 24 h of incubation, the growth of the transformants was observed and single colonies on the second layer were collected.
2.4. Morphological observation
2.4.1. Scanning electron microscopy
The spores grown on the PDA plate were collected by cutting off the culture medium (length of side: 0.5 cm) using a surgical knife, fixed in 50% glutaraldehyde solution, and dehydrated with 50%, 75%, 80%, 90%, 95%, and 100% ethanol, respectively. Subsequently, all the samples were passed through tert-butanol and subjected to critical-point drying by lyophilizer. The dried samples were placed onto a stub bearing adhesive and viewed under a scanning electron microscope.
2.4.2. Enumeration of C. globosum spores
The spores from the wild-type and mutant C. globosum colonies grown on PDA plates were collected and suspended in glycerinum (50%). Then, the spore suspension was added to the blood cell count plate and the spores were counted under a light microscope. The procedure was repeated thrice at each time point, and the average value was taken into consideration.
2.5. RNA isolation and quantitative real-time PCR
The mycelium pellets in the PDA broth were filtered and washed twice with sterile water. After absorbing the water with filter paper, the mycelium pellets were transferred to the mortar, mixed with liquid nitrogen, and ground to powder. The RNA was extracted by using the RNA prep pure plant kit (Tiangen, China) and translated into cDNA using a reverse transcription kit (Novoprotein Scientific Inc, China). The RNA quality was analyzed by agarose gel electrophoresis, and 10-fold diluted cDNA was employed as template to perform quantitative real-time PCR (RT-qPCR) using Trans Start Top Green qPCR SuperMix (TransGen Biotech, China) in a Real-time PCR System (Thermo Fisher Scientific, Carlsbad, CA, USA). The β-actin gene (GenBank Accession No. CH408033.1) was used as an internal control for C. globosum. The relative abundances of CgVeA, LaeA, cheR, PKS, and p450 genes were standardized by the expression levels of the β-actin gene, and the relative quantification of each transcript was achieved using the 2−ΔΔCT method [35]. The enumerations were performed in triplicate for each group of data, and the average was taken into consideration.
2.6. Determination of C. globosum biomass and cheA concentration
The wild-type and mutant C. globosum strains were cultivated in 50 mL of PDA medium under light or dark condition at 28 °C and 180 rpm. The biomass of all the incubated strains was determined at different time points by harvesting and weighing the cells by dry weight method [12]. The filtrate was extracted with an equal volume of ethyl acetate, and the extract was filtered and dried in a rotary evaporator at 35 °C. The crude extract was dissolved in 1.5 mL of methanol, filtered through a 0.22-μm filter, and assayed using high-performance liquid chromatography (HPLC). The HPLC conditions were as follows: TC-C18 column (Agilent, 4.6 mm × 250 mm, 5 μm) and 45% CH3CN (v/v) in H2O at a flow rate of 1.0 mL/min. The cheA (C32H36N2O5) standard (Sigma-Aldrich, Germany) was used as the control.
2.7. Detection of the role of CgVeA in the inhibitory activity of C. globosum against phytopathogenic fungi
The wild-type and mutant C. globosum strains were cultured in PDA broth for 7 days. After incubation, the fermentation broth was filtered, diluted to 20%, mixed with PDA solid medium, and poured onto plates. The control plate was not inoculated with C. globosum strains. After solidification of the medium, single mycelial plugs of various phytopathogenic fungi from 3-day-old PDA cultures were inoculated in the center of the PDA-C. globosum plates and incubated until phytopathogenic fungal colonies filled the control plate. Then, the diameter of the phytopathogenic fungal colony in the experimental plates was determined. The investigation was performed in triplicate, and the average value was taken into consideration.
2.8. Expression and purification of CgVeA protein
Based on the VeA gene in the C. globosum genomic DNA, the coding region for the production of CgVeA protein was generated by PCR. The DNA fragment was amplified using PF-CgVeA and PR-CgVeA primer pairs. After digesting the pET28a plasmid vector with NcoI and XhoI, the amplified fragment was ligated into this vector to obtain pET28a-CgVeA. Then, pET28a-CgVeA was introduced into E. coli BL21(DE3) cells for heterologous expression, and the cells were cultivated in 1.5 mL of LB medium with kanamycin in a shaking incubator (200 rpm) at 37 °C. When the optical density of the culture at 600 nm reached 0.6–0.8, protein expression was induced by adding 0.5 mM isopropyl thio-d-galactopyranoside to the culture and incubating the culture at 15 °C for 28 h. Subsequently, the culture was collected and mixed with 5 mL of 1 × PBS, and subjected to ultrasonication. Then, the mixture was centrifuged at 12,000 rpm for 1 min, and the supernatant obtained was the CgVeA protein. The protein size was assessed by SDS-PAGE, and the protein concentration was measured by BCA. Finally, the protein samples were stored at −80 °C or directly used in electrophoretic mobility shift assays (EMSA).
2.9. EMSA
Fragments comprising 500-bp (from −400 to +100) upstream of the four known secondary metabolite biosynthetic gene clusters, including PKS, LaeA, p450, and cheR, were PCR-amplified using the primers listed in Table S2. The CgVeA protein at different concentrations were incubated with DNA probe in 20 μL of the reaction mixture, including 20 mM Tris base, 2 mM dithiothreitol, 5 mM MgCl2, 0.5 μg of calf BSA, 5% (v/v) glycerol, and, if needed, 500 ng of poly[d(I–C)]. The DNA probe and DNA–protein complexes were separated by electrophoresis on non-denaturing 4% (wt/vol) polyacrylamide gel for 25 min, with 0.5 × TBE as the running buffer. After electrophoresis, the DNA was stained with SYBR Gold nucleic acid gel stain for 30 min in 1 × TBE and photographed under UV using Quantity One.
3. Results
3.1. Silencing and overexpression of CgVeA in C. globosum
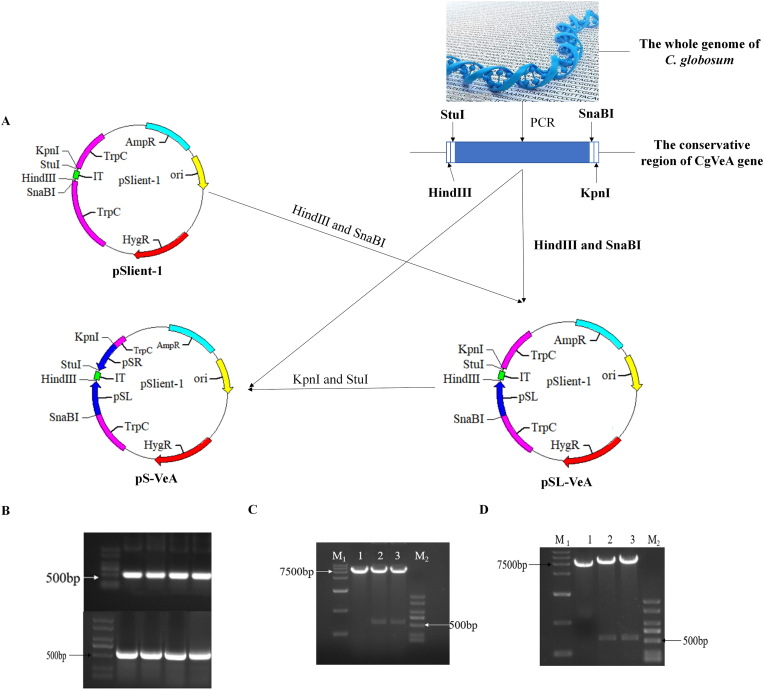
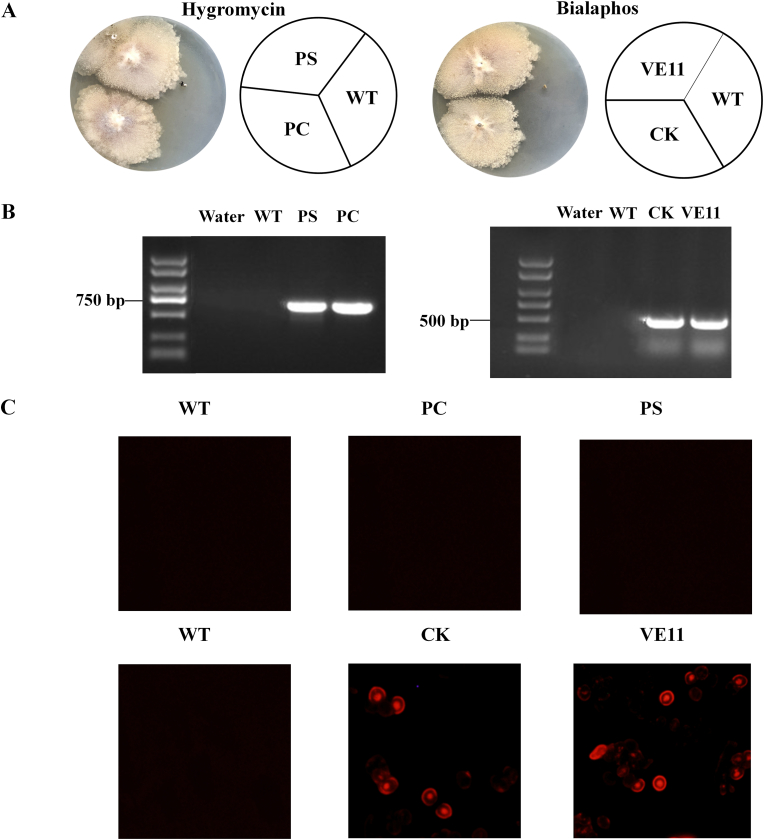
To determine the function of CgVeA in C. globosum, the target gene was inactivated by utilizing the RNAi strategy (Fig. 1A). The conserved region of the CgVeA gene was obtained by SMART and Conserved Domains. Based on the results of conserved region analysis, primers VeA-pSLL and VeA-pSLR were designed to amplify the pSL of CgVeA (Fig. 1B), ligated to pSilent plasmid through SnaBI and HindIII sites, and transformed into E. coli DH5α competent cells. The two selected transformants formed two bright bands, whereas the vector control did not exhibit any interference band, indicating that pSL was accurately ligated to the corresponding genomic locus of pSilent plasmid (Fig. 1C). The insertion of pSR fragment was carried out by seamless cloning (Fig. 1B), and the results showed that pSR was accurately inserted into the vector pSL-VeA plasmid (Fig. 1D). The pS-VeA and pSC-VeA plasmids construction strategy and validation results are displayed in Fig. 1 and Fig. S1. After transforming the plasmids pS-VeA and pSC-VeA into C. globosum, the corresponding CgVeA-silenced mutant (PS) and scramble control mutant (PC) obtained were subjected to inverted culture on PDA medium for five generations, and incubated on hygromycin (200 μg/mL)-resistant screening plates to determine the genetic stability of all derivatives. While all the mutants could grow on the screening plate, the parent cells could not grow (Fig. 2A), thus, confirming that the resistance genes had been successfully inserted into the C. globosum genome. Moreover, the two mutants were further verified by diagnostic PCR using primers Hyg-F and Hyg-R, and the findings showed that both the mutants formed a 752-bp band, whereas the water control and wild-type C. globosum did not produce any band (Fig. 2B), which were consistent with the results of hygromycin resistance screening.
Fig. 1.
Construction and validation of CgVeA silencing vector. (A) Strategy for the construction of CgVeA silencing vector. (B) Electrophoresis of PCR products of pSL and pSR fragments. (C) Verification results of double digestion by SnaBI and HindIII. 1: Linearized pSilent plasmid fragment (7056 bp); 2–3: Two bright bands obtained after digestion indicate the linearized pSL-VeA plasmid fragment and interference sequence (539 bp). (D) Validation results of double digestion by KpnI and StuI. 1: Linearized pSilent plasmid fragment (7056 bp); 2–3: Two bright bands obtained after digestion indicate the linearized pS-VeA plasmid fragment and interference sequence (539 bp).
Fig. 2.
Silencing and overexpression of CgVeA in C. globosum. (A) WT, PS, PC, CK, and VE11 strains were cultured on resistance plates for 7 days. (B) PCR results for Hyg and BlpR resistance genes. (C) Strains observed under fluorescence microscope. (WT: Wild-type strain; PC: Vector control; PS: CgVeA-silenced mutant, CK: Scramble control; VE11: CgVeA overexpression mutant).
The CgVeA overexpression mutant (VE11) was constructed using pBARGP-mcherry as the original plasmid, and the construction strategy and validation results are shown in Fig. S2. The plasmids pBG-VeA and pBG-VeAC were transformed into C. globosum by protoplast transformation, and the corresponding transformants VE11 and vector control (CK) were continuously cultured for five generations and then cultivated on bialaphos resistance screening plates for 7 days. While the wild-type C. globosum could not grow on the screening plate, VE11 and CK could grow well (Fig. 2A). Subsequently, the strains were verified by diagnostic PCR using primers BlpR-F and BlpR-R, and the results showed that only VE11 and CK formed a 496-bp band, proving that the BlpR resistance gene had been successfully inserted into the C. globosum genome (Fig. 2B). Furthermore, to detect the expression of the red fluorescence marker, the wild-type C. globosum, PS, PC, CK, and VE11 were observed under fluorescence microscope. The results showed that the red fluorescence protein was accurately expressed in CK and VE11, whereas no fluorescence was detected in the other strains (Fig. 2C).
3.2. Role of CgVeA in C. globosum development
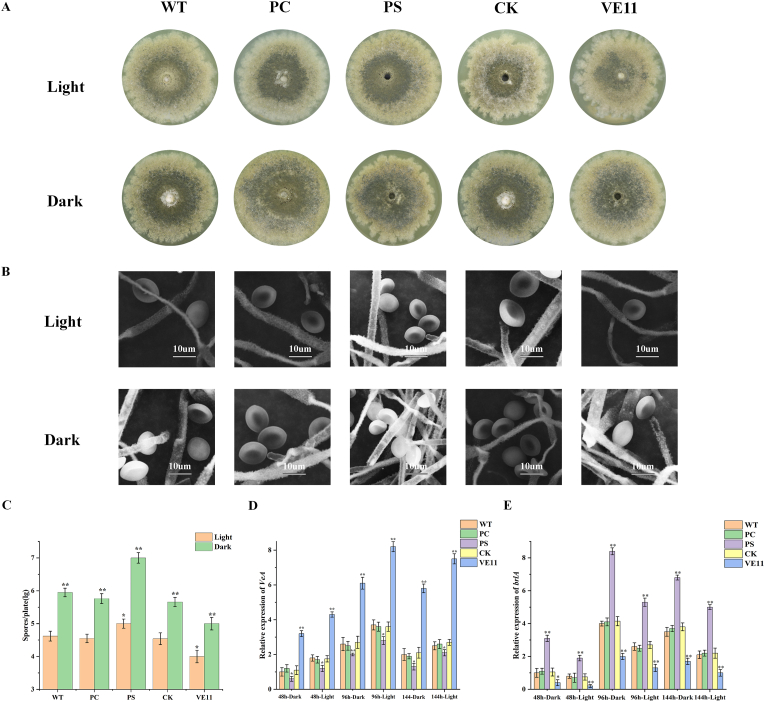
To explore the role of CgVeA on the development of C. globosum, the wild-type C. globosum, VE11, CK, PS, and PC strains were cultured on PDA plates for 7 days at 28 °C under light and dark conditions, respectively. To reduce the influence of other factors, the spores concentration for all the strains was adjusted to 1 × 107 CFU/mL (oxford cup was placed on each plate and 50 μL of spore suspension were added). The results showed reduced spores production in VE11, whereas increased spores production in PS, when compared with that in the wild-type C. globosum, under light condition (Fig. 3A). In contrast, the number of spores in VE11 obviously enhanced from 1.23 × 104 to 1.02 × 105 CFU/mL under dark condition, when compared with that under illumination (Fig. 3C). Similar trend in spores production was also observed in the other strains. Thus, these findings indicated that the number of spores obviously decreased under light condition, when compared with that under dark condition. Furthermore, scanning electron microscopy analysis revealed that CgVeA did not affect the morphology of conidia and the diameter of mycelia under light or dark conditions (Fig. 3B).
Fig. 3.
Effect of CgVeA on C. globosum colony, morphology, and conidiation under dark and light conditions. (A) Sporulation of WT, PC, PS, CK, and VE11 strains on PDA plates. (B) Morphological characteristics of all the tested strains observed under scanning electron microscope. (C) Number of spores produced by WT, PC, PS, CK, and VE11 strains on PDA medium. (D) Transcription level of CgVeA in WT, PC, PS, CK, and VE11 strains under dark and light conditions. (E) Expression level of brlA in WT, PC, PS, CK, and VE11 strains under dark and light conditions. The relative abundance of mRNAs was standardized against the levels of the β-actin gene. Error bars represent standard deviations from three independent experiments. Differences were analyzed by Student's t-test, and p < 0.05 was considered statistically significant. The levels of significance are **p < 0.01, *p < 0.05. (WT: Wild-type strain; PC: Vector control; PS: CgVeA-silenced mutant, CK: Scramble control; VE11: CgVeA overexpression mutant).
Subsequently, RT-qPCR was performed to further examine the influence of CgVeA on the development of C. globosum, and the findings demonstrated that CgVeA expression under light condition was higher than that under dark condition, which proved that CgVeA negatively regulated sporulation (Fig. 3D). It has been reported that the brlA gene (GenBank Accession No: XP_001223891.1) has a key role in regulating conidiation in Penicillium decumbens [36]. In the present study, the expression of brlA in PS was 2.01-fold higher than that in the wild-type C. globosum under dark condition (Fig. 3E), suggesting that brlA was negatively regulated by CgVeA.
3.3. Role of CgVeA in C. globosum biomass and cheA production
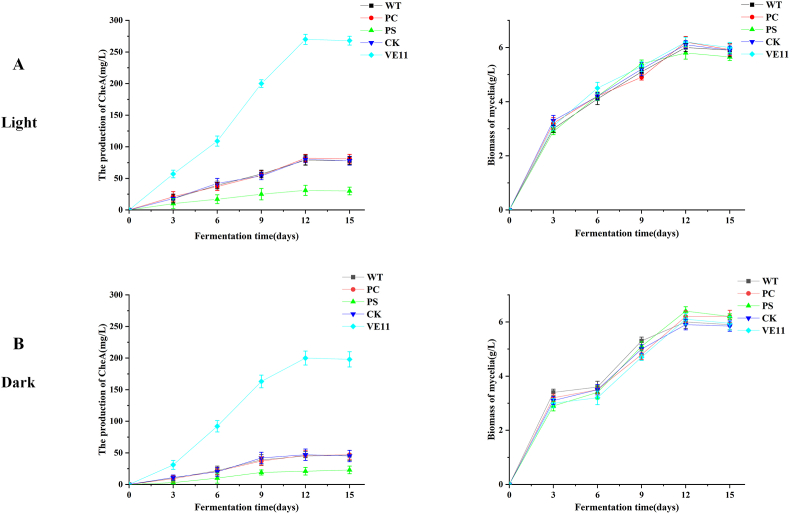
To determine the regulatory effect of CgVeA on secondary metabolites production in C. globosum, the wild-type and mutant C. globosum strains were cultured in PDA broth under light and dark conditions, respectively, and sampled at various time points in an interval of 3 days. The cheA production reached an equilibrium in 12 days of incubation. The cheA yield of VE11 reached 270.48 and 200.71 mg/mL under light and dark conditions, respectively, which were markedly higher than those of the wild-type C. globosum (Fig. 4A and B). This result indicated that CgVeA expression is essential for cheA production, and that CgVeA functions as a light-responsive positive regulator of the biosynthesis of target compounds in C. globosum. The biomass results obtained under both light and dark conditions revealed that the biomass of all the tested strains linearly increased from 3 to 9 days and presented no significant changes at the equilibrium stage (Fig. 4A and B).
Fig. 4.
Biomass and cheA yield of WT, PC, PS, CK, and VE11 strains under light and dark conditions. Biomass and cheA production of WT, PC, PS, CK, and VE11 strains incubated in PDA medium for different durations under (A) light condition and (B) dark condition. Data were averaged using triplicate measurements. (WT: Wild-type strain; PC: Vector control; PS: CgVeA-silenced mutant, CK: Scramble control; VE11: CgVeA overexpression mutant).
3.4. Role of CgVeA in the expression of cheA biosynthetic genes and other regulator genes
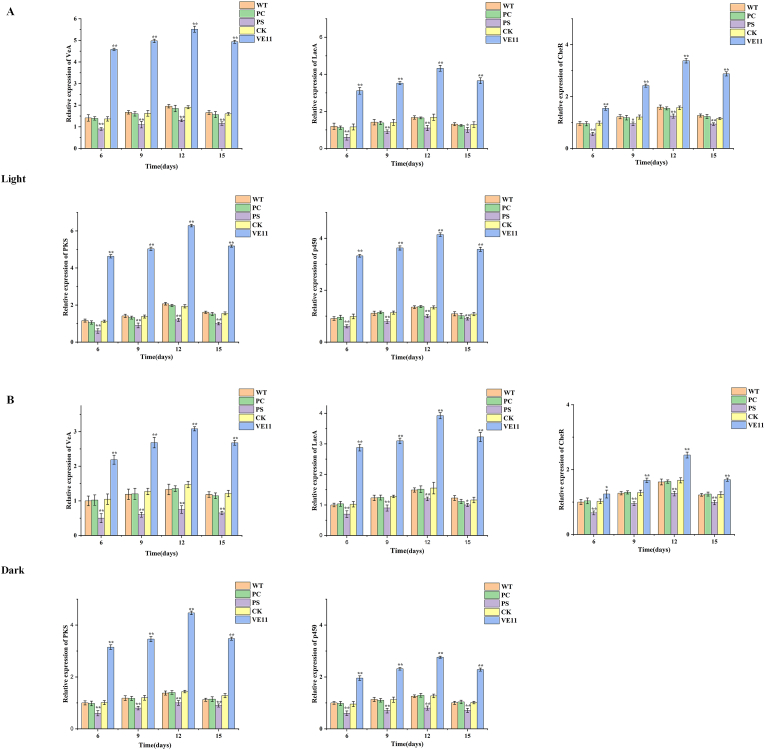
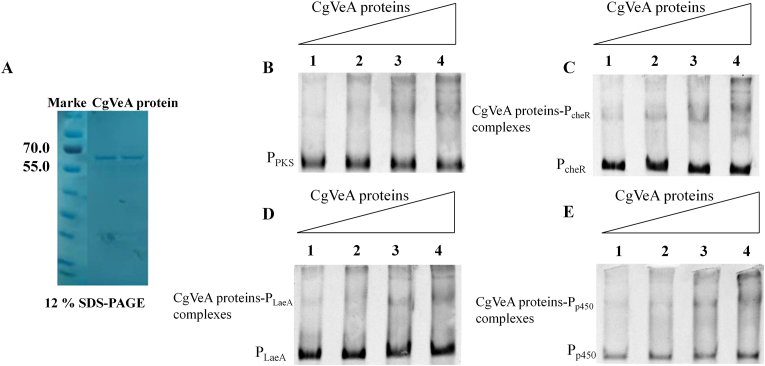
To examine the effects of CgVeA on the transcription of cheA biosynthetic genes, development-related genes, and other regulator genes, including PKS, p450, brlA, LaeA, and cheR genes, RT-qPCR was performed. The cells were harvested and extracted after 6, 9, 12, and 15 days of incubation, and quantitatively analyzed under light and dark conditions, respectively. The PKS gene, which is responsible for polymerizing monomer structure to form carbon skeleton, has an extremely important role in cheA biosynthesis. The p450 gene is another crucial gene in the cheA biosynthetic gene cluster, and is involved in converting the nascent substrate into terminal product cheA by redox reaction. The expression levels of both these genes were obviously enhanced in VE11 (containing the CgVeA overexpression cassette), but were dramatically decreased in the CgVeA-silenced mutant, PS (Fig. 5). These findings, combined with the results of cheA production (Fig. 4), indicated that CgVeA played an important regulatory role in the biosynthesis of cheA by controlling the transcription levels of the core enzymes in the gene cluster. Furthermore, the results showed that effectively enhanced expression of CgVeA had a positive impact on the expression levels of LaeA and cheR, which were identified to be positively correlated with cheA biosynthesis [13], especially under light condition. In contrast, both LaeA and cheR were obviously reduced in PS (Fig. 5). These results suggested that CgVeA might form a regulatory network with LaeA and cheR to interact with cheA biosynthetic gene cluster to enhance cheA production. EMSA was performed to directly verify the role of CgVeA in the regulation of cheA biosynthesis, and the CgVeA protein was assessed by SDS-PAGE (Fig. 6A). The EMSA results showed clear bands corresponding to p450, cheR, and LaeA; however, no band corresponding to PKS was detected (Fig. 6B–D). These findings indicated that CgVeA regulated the biosynthesis of cheA through direct interaction with the promoter region of p450. The p450 gene has an important role in oxidation reactions for the biosynthesis of cheA [12]. Similarly, CgVeA directly regulated LaeA, a global regulator, and cheR, a pathway-specific regulator, by binding to the promotor regions of these two genes (Fig. 6C and D). On the contrary, CgVeA indirectly regulated PKS (Fig. 6E).
Fig. 5.
Gene expression in WT, PC, PS, CK, and VE11 strains. Expressions of CgVeA, LaeA, CheR, PKS, and p450 in WT, PC, PS, CK, and VE11 strains at 6, 9, 12, and 15 days of incubation in PDA medium at 28 °C under (A) light condition and (B) dark condition. Data were averaged using triplicate measurements. Differences were analyzed by Student's t-test, and p < 0.05 was considered statistically significant. The levels of significance are **p < 0.01, *p < 0.05. (WT: Wild-type strain; PC: Vector control; PS: CgVeA-silenced mutant, CK: Scramble control; VE11: CgVeA overexpression mutant).
Fig. 6.
Binding ability of CgVeA protein to PKS, LaeA, cheR, and p450. (A) SDS-PAGE of the purified CgVeA protein. EMSA of CgVeA protein with (B) PKS, (C) cheR, (D) LaeA, (E) p450. Channels 1–4: 0, 2.5, 5, and 10 μg of CgVeA, respectively.
3.5. Role of CgVeA in the inhibitory activity of C. globosum against phytopathogenic fungi
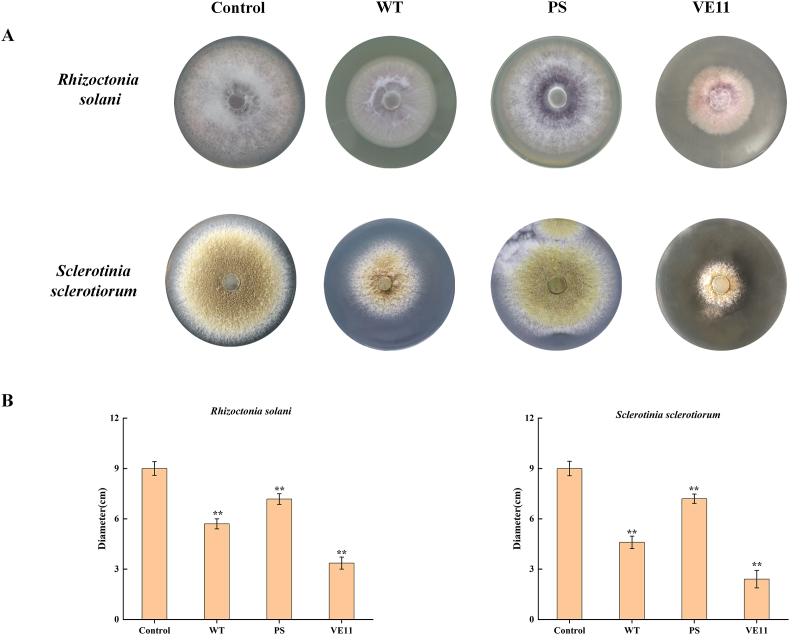
Fig. 7 shows the results of the effect of CgVeA on the inhibitory activity of the culture filtrates of wild-type C. globosum, PS, and VE11 strains against phytopathogenic fungi. The colony diameters of Rhizoctonia solani on wild-type C. globosum, VE11, and PS culture plates were 5.72, 3.36, and 7.18 cm, respectively, indicating that the culture filtrate of CgVeA overexpression mutant showed a dramatic increase in the inhibitory activity against phytopathogenic fungal cells. Furthermore, the colony diameters of Sclerotinia sclerotiorum on wild-type C. globosum and PS culture plate were 4.62 and 7.21 cm, respectively, demonstrating lower inhibitory activity of the filtrate of the CgVeA-silenced mutant against phytopathogenic fungi, when compared with that of the wild-type strain. These results suggested that CgVeA plays an important role in the inhibitory activity of C. globosum against phytopathogenic fungi.
Fig. 7.
Inhibitory effects of CgVeA mutation against phytopathogenic fungi. (A) Antifungal activity of culture filtrates against R. solani. Control: no culture filtrate; WT: culture filtrate of the wild-type C. globosum; PS, culture filtrate of CgVeA-silenced mutant; VE11: culture filtrate of CgVeA overexpression mutant. (B) Colony diameter of S. sclerotiorum. Line bars in each column denote standard errors of triplicate experiments. Differences were analyzed by Student's t-test, and p < 0.05 was considered statistically significant. The levels of significance are **p < 0.01, *p < 0.05.
4. Discussion
C. globosum is the most prevalent member of Chaetomiaceae, which has been well-studied owing to its various beneficial products such as cytochalasan [6], azaphilones [37], and terpenoids [38]. CheA, the major secondary metabolite of C. globosum, has a typical structure of cytochalasan. It exhibits antitumor activity and excellent broad-spectrum antimicrobial ability against phytopathogenic fungi such as Fusarium sporotrichioides, Setosphaeria turcica, and Rhizopus stolonifer [39,40]. Several well-designed studies have characterized the metabolites from “silent” or “repressed” secondary metabolic biosynthetic gene clusters in fungi via activation of cluster-specific or global transcription regulator. For instance, rsmA, a recently discovered YAP-like bZIP protein in Aspergillus nidulans, significantly activated the production of carcinogenic and anti-predation sterigmatocystin by binding to the two sites of a C6 transcription factor in AflR promoter region [41]. Deletion of rsdA, identified in Pestalotiopsis fici through genome-wide analysis, not only resulted in a significant reduction in secondary metabolites production, but also caused abnormal morphological development [42]. However, limited information about the transcriptional regulation of morphological development and cheA biosynthesis in C. globosum has hindered the application and popularization of cheA in agriculture and industries.
Therefore, in the present study, the CgVeA gene was silenced and overexpressed in C. globosum, respectively, to illustrate its functional mechanism in regulating cheA biosynthesis and asexual spores production. The RT-qPCR results and morphological culture characteristics demonstrated that CgVeA has a negative regulatory effect on spore formation, especially in the presence of light. The number of asexual spores decreased with the increasing expression level of CgVeA; however, CgVeA had no influence on the morphology of spores and hyphae. Similarly, negative regulation of asexual spores production by VeA gene has been reported in Aspergillus sp [43,44]. In the past 5 years, our research group has progressively explored the factors influencing cheA production. CgLaeA and CgcheR, the Zn(II)2Cys6 structural and pathway-specific transcription factors, respectively, have been identified in C. globosum, and have been found to exert positive effects (especially, CgcheR) on target compounds synthesis [13,33]. Furthermore, EMSA results have revealed that CgcheR shared a relatively strong shift signal to PKS gene, which has been reported to be associated with polymerization of monomeric substance to form the carbon backbone [12]. Similarly, in the present study, CgVeA was found to positively regulate cheA biosynthesis. Moreover, CgVeA was observed to directly regulate the LaeA gene, implicitly confirming interactions between CgVeA and LaeA. However, as a global regulator, CgVeA did not bind to the promoter region of the PKS gene, thus, necessitating further research on the interaction between CgVeA and PKS.
The cheA production effectively increased when a strong promoter gpdA was used to enhance the expression level of CgVeA. On the contrary, the cheA yield significantly decreased after CgVeA silencing. Furthermore, RT-qPCR results showed that CgVeA, as a light responsive regulator, could promote the expression of CgLaeA and CgcheR. The transcription levels of genes related to cheA biosynthetic gene cluster, including PKS and p450 genes, were further promoted, which might be the result of the interaction of regulatory network. Similar to these findings, it has been reported that biosynthesis of aflatoxins, which are cytotoxic and carcinogenic polyketide compounds, in Aspergillus flavus is VeA-dependent [45]. Moreover, VeA has been determined to regulate the expression of multiple genes related to secondary metabolites production in A. fumigatus [46]. In the present study, the expression of VeA was positively correlated with the cheA yield, which was achieved by modulating the expression of the key gene, PKS. When compared with the wild-type C. globosum, the inhibitory effects of the culture filtrate of CgVeA overexpression mutant against phytopathogenic fungi were significantly improved, which might be related to the enhanced cheA production or activation of the expression of other silent or cryptic secondary metabolite gene cluster.
In summary, the functional mechanism of CgVeA in C. globosum was determined in the present study, and CgVeA was found to have a positive effect on cheA biosynthesis and negative effect on asexual spore formation. These influences might be the consequence of the ability of CgVeA to regulate the core enzymes involved in cheA biosynthesis or simulate the expression of other regulators. The mass production of cheA not only yields high economic benefits, but also offers substantial environmental protection by providing a novel alternative to chemical pesticides.
CRediT authorship contribution statement
Conceptualization, Zhengran Wang; Data curation, Shanshan Zhao; Formal analysis, Kai Zhang; Funding acquisition, Qian Yang; Methodology, Congyu Lin; Project administration, Yang; Software, Shanshan Zhao; EMSA experiments, Xin Ru.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
We would like to thank Associate Professor Xiujie Gong at the Institute of Farming and Cultivation, Heilongjiang Academy of Agricultural Sciences, People's Republic of China, for providing us with the pathogenic fungal strains, Rhizoctonia solani and Sclerotinia sclerotiorum. This work was supported in part by grant from the Harbin Science and Technology Project (No. 2016AB3AP042). We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.07.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Guo Q.F., Yin Z.H., Zhang J.J., Kang W.Y., Wang X.W., Ding G., et al. Chaetomadrasins A and B, two new cytotoxic cytochalasans from desert soil-derived fungus Chaetomium madrasense 375. Molecules. 2019; Sep 5;24(18):3240. doi: 10.3390/molecules24183240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X.Y., Tan X.M., Yu M., Yang J., Sun B.D., Qin J.C., et al. Bioactive metabolites from the desert plant-associated endophytic fungus Chaetomium globosum (Chaetomiaceae) Phytochemistry. 2021;185 doi: 10.1016/j.phytochem.2021.112701. [DOI] [PubMed] [Google Scholar]
- 3.Salo J.M., Kedves O., Mikkola R., Kredics L., Andersson M.A., Kurnitski J., et al. Detection of Chaetomium globosum, Ch. Cochliodes and Ch. Rectangulare during the diversity tracking of mycotoxin-producing Chaetomium-like isolates obtained in Buildings in Finland. Toxins. 2020;12(7):443. doi: 10.3390/toxins12070443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., Yang J., Liao Y.Y., Cheng G., Chen J., Cheng X.D., et al. Cytotoxic nitrogenated azaphilones from the deep-sea-derived fungus Chaetomium globosum MP4-S01-7. J Nat Prod. 2020;83(4):1157–1166. doi: 10.1021/acs.jnatprod.9b01165. [DOI] [PubMed] [Google Scholar]
- 5.Kumar R., Kundu A., Dutta A., Saha S., Das A. Profiling of volatile secondary metabolites of Chaetomium globosum for potential antifungal activity against soil borne fungi. J Pharmacogn Phytochem. 2020;9:922–927. [Google Scholar]
- 6.Li T.T., Wang Y., Li L., Tang M.Y., Meng Q.H., Zhang C., et al. New cytotoxic cytochalasans from a plant-associated fungus Chaetomium globosum kz-19. Mar Drugs. 2021;19:438. doi: 10.3390/md19080438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng X.G., Liu J., Gao Y., Cheng F., Chang J.L., Chen J., et al. Pchaeglobolactone A, spiropchaeglobosin A, and pchaeglobosals A and B: four rearranged cytochalasans from Chaetomium globosum P2-2-2. Org Lett. 2020;22(24):9665–9669. doi: 10.1021/acs.orglett.0c03623. [DOI] [PubMed] [Google Scholar]
- 8.Zheng C.J., Shao C.L., Wu L.Y., Chen M., Wang K.L., Zhao D.L., et al. Bioactive phenylalanine derivatives and cytochalasins from the soft coral-derived fungus, Aspergillus elegans. Mar Drugs. 2013;11:2054–2068. doi: 10.3390/md11062054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou H., Song Y.X., Liu X.Q., Gong W., Li E.G., Tan R.X., et al. Chaetoglobosin Fex from the marine-derived endophytic fungus inhibits induction of inflammatory mediators via Toll-like receptor 4 signaling in macrophages. Biol Pharm Bull. 2011;34:1864–1873. doi: 10.1248/bpb.34.1864. [DOI] [PubMed] [Google Scholar]
- 10.Skellam E. The biosynthesis of cytochalasans. Nat Prod Rep. 2017;34:1252–1263. doi: 10.1016/B978-0-12-670650-5.50014-5. [DOI] [PubMed] [Google Scholar]
- 11.Schümann J., Hertweck C. Molecular basis of cytochalasan biosynthesis in fungi: gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J Am Chem Soc. 2007;129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 12.Ishiuchi K., Nakazawa T., Yagishita F., Mino T., Noguchi H., Hotta K., et al. Combinatorial generation of complexity by redox enzymes in the Chaetoglobosin A biosynthesis. J Am Chem Soc. 2013;135:7371–7377. doi: 10.1021/ja402828w. [DOI] [PubMed] [Google Scholar]
- 13.Cheng M., Zhao S.S., Liu H., Liu Y.T., Lin C.Y., Song J.Z., et al. Functional analysis of a chaetoglobosin A biosynthetic regulator in Chaetomium globosum. Fungal Biol. 2021;125:201–210. doi: 10.1016/j.funbio.2020.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Hong E.J., Kim N.K., Lee D., Kim W.G., Lee I. Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 2015;19:973–983. doi: 10.1016/j.funbio.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Halo L.M., Marshall J.W., Yakasai A.A., Song Z., Butts C.P., Crump M.P., et al. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. Chembiochem. 2008;9:585–594. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- 16.Sekita S., Yoshihira K., Natori S., et al. Chaetoglobosins, cytotoxic 10-(indol-3-yl)-[13]cytochalasans from Chaetomium spp. I. Production, isolation and some cytological effects of chaetoglobosins A-J[J] Chem Pharm Bull. 1982;30:1609–1617. doi: 10.1248/cpb.30.1609. [DOI] [PubMed] [Google Scholar]
- 17.Hou B., Zhu X., Kang Y., Wang R., Wu H., Ye J., et al. LmbU, a cluster-situated regulator for Lincomycin, consists of a DNA-binding domain, an auto-inhibitory domain, and forms homodimer. Front Microbiol. 2019;10:989. doi: 10.3389/fmicb.2019.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chettri P., Bradshaw R.E. LaeA negatively regulates dothistromin production in the pine needle pathogen Dothistroma septosporum. Fungal Genet Biol. 2016;97:24–32. doi: 10.1016/j.fgb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Wiemann P., Keller N.P. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41(2):301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 20.Derntl C., Kluger B., Bueschl C., Schuhmacher R., Mach R.L., Mach-Aigner A.R. Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc Natl Acad Sci USA. 2017;114(4):560–569. doi: 10.1073/pnas.1609348114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhi Q.Q., He L., Li J.Y., Li J., Wang Z.L., He G.Y., et al. The kinetochore protein spc105, a novel interaction partner of LaeA, regulates development and secondary metabolism in Aspergillus flavus. Front Microbiol. 2019;10:1881. doi: 10.3389/fmicb.2019.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bok J.W., Keller N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3(2):527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain S., Keller N. Insights to fungal biology through LaeA sleuthing. Fungal Biol Rev. 2013;27:51–59. doi: 10.1016/j.fbr.2013.05.004. [DOI] [Google Scholar]
- 24.van der Merwe, Steyn P.S., Fourie L., Scott D.B., Theron J.J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature. 1965;205(976):1112–1113. doi: 10.1038/2051112a0. [DOI] [PubMed] [Google Scholar]
- 25.Maor U., Barda O., Sadhasivam S., Bi Y., Levin E., Zakin V., et al. Functional roles of LaeA, polyketide synthase, and glucose oxidase in the regulation of ochratoxin A biosynthesis and virulence in Aspergillus carbonarius. Mol Plant Pathol. 2021;22(1):117–129. doi: 10.1111/mpp.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G., Zhang H., Wang Y., Liu F., Li E., Ma J., et al. Requirement of c, VeA, and VelB on asexual development, Ochratoxin A biosynthesis, and fungal virulence in Aspergillus ochraceus. Front Microbiol. 2019;10:2759. doi: 10.3389/fmicb.2019.02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayram O., Krappmann S., Ni M., Bok J.W., Helmstaedt K., Valerius O., et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320(5882):1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 28.Lan N., Yue Q., An Z., Bills G.F. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J Ind Microbiol Biotechnol. 2020;47(1):155–168. doi: 10.1007/s10295-019-02250-x. [DOI] [PubMed] [Google Scholar]
- 29.Estiarte N., Lawrence C.B., Sanchis V., Ramos A.J., Crespo-Sempere A. LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int J Food Microbiol. 2016;238:153–164. doi: 10.1016/j.ijfoodmicro.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Dhingra S., Andes D., Calvo A.M. VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2012;11(12):1531–1543. doi: 10.1128/EC.00222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan Y., Wang H., Wang Y., Ge Y., Ren X., Ren C., et al. The role of the veA gene in adjusting developmental balance and environmental stress response in Aspergillus cristatus. Fungal Biol. 2018;122(10):952–964. doi: 10.1016/j.funbio.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Chen H., Sumarah M.W., Gao Q., Wang D., Zhang Y. veA gene acts as a positive regulator of conidia production, ochratoxin A biosynthesis, and oxidative stress tolerance in Aspergillus niger. J Agric Food Chem. 2018;66(50):13199–13208. doi: 10.1021/acs.jafc.8b04523. [DOI] [PubMed] [Google Scholar]
- 33.Cheng M., Zhao S., Lin C., Song J., Yang Q. Requirement of LaeA for sporulation, pigmentation and secondary metabolism in Chaetomium globosum. Fungal Biol. 2021;125(4):305–315. doi: 10.1016/j.funbio.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Tamano K., Satoh Y., Ishii T., Terabayashi Y., Ohtaki S., et al. The beta-1,3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci Biotechnol Biochem. 2007;71(4):926–934. doi: 10.1271/bbb.60591. [DOI] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, CA, U S) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Long L.K., Wang Y., Yang J., Xu X., Liu G. A septation related gene AcsepH in Acremonium chrysogenum is involved in the cellular differentiation and cephalosporin production. Fungal Genet Biol. 2013;50:11–20. doi: 10.1016/j.fgb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Youn U.J., Sripisut T., Park E.J., Kondratyuk T.P., Fatima N., Simmons C.J., et al. Determination of the absolute configuration of chaetoviridins and other bioactive azaphilones from the endophytic fungus Chaetomium globosum. Bioorg Med Chem Lett. 2015;25(21):4719–4723. doi: 10.1016/j.bmcl.2015.08.063. [DOI] [PubMed] [Google Scholar]
- 38.Ruan B.H., Yu Z.F., Yang X.Q., Yang Y.B., Hu M., Zhang Z.X., et al. New bioactive compounds from aquatic endophyte Chaetomium globosum. Nat Prod Res. 2018;32(9):1050–1055. doi: 10.1080/14786419.2017.1378210. [DOI] [PubMed] [Google Scholar]
- 39.Jiang C., Song J.Z., Zhang J.Z., Yang Q. Identification and characterization of the major antifungal substance against Fusarium sporotrichioides from Chaetomium globosum. World J Microbiol Biotechnol. 2017;33:108. doi: 10.1007/s11274-017-2274-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D., Ge H.L., Xie D., Chen R.D., Zou J.H., Tao X.Y., et al. Periconiasins A-C, New cytotoxic cytochalasans with an unprecedented 9/6/5 tricyclic ring system from endophytic fungus Periconia sp. Org Lett. 2013;15:1674–1677. doi: 10.1021/ol400458n. [DOI] [PubMed] [Google Scholar]
- 41.Yin W.B., Amaike S., Wohlbach D.J., Gasch A.P., Chiang Y.M., et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol Microbiol. 2012;83(5):1024–1034. doi: 10.1111/j.1365-2958.2012.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou S., Zhang P., Zhou H., Liu X., Li S.M., Guo L., et al. A new regulator RsdA mediating fungal secondary metabolism has a detrimental impact on asexual development in Pestalotiopsis fici. Environ Microbiol. 2019;21(1):416–426. doi: 10.1111/1462-2920.14473. [DOI] [PubMed] [Google Scholar]
- 43.Hui, L., Sang S., Hui W., Ren X., Tan Y.. Comparative proteomic analysis reveals the regulatory network of the veA gene during asexual and sexual spore development of Aspergillus cristatus. doi:10.1042/BSR-20180067_RET. [DOI] [PMC free article] [PubMed] [Retracted]
- 44.Pandit S.S., Lohmar J.M., Ahmed S., Etxebeste O., Espeso E.A., Calvo A.M. UrdA controls secondary metabolite production and the balance between asexual and sexual development in Aspergillus nidulans. Genes. 2018;9(12):570. doi: 10.3390/genes9120570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cary J.W., Han Z., Yin Y., Lohmar J.M., Shantappa S., et al. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot Cell. 2015;14(10):983–997. doi: 10.1128/EC.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhingra S., Lind A.L., Lin H.C., Tang Y., Rokas A., et al. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.