Abstract
Background
Type 2C protein phosphatase (PP2C) is a negative regulator of ABA signaling pathway, which plays important roles in stress signal transduction in plants. However, little research on the PP2C genes family of cucumber (Cucumis sativus L.), as an important economic vegetable, has been conducted.
Results
This study conducted a genome-wide investigation of the CsPP2C gene family. Through bioinformatics analysis, 56 CsPP2C genes were identified in cucumber. Based on phylogenetic analysis, the PP2C genes of cucumber and Arabidopsis were divided into 13 groups. Gene structure and conserved motif analysis showed that CsPP2C genes in the same group had similar gene structure and conserved domains. Collinearity analysis showed that segmental duplication events played a key role in the expansion of the cucumber PP2C genes family. In addition, the expression of CsPP2Cs under different abiotic treatments was analyzed by qRT-PCR. The results reveal that CsPP2C family genes showed different expression patterns under ABA, drought, salt, and cold treatment, and that CsPP2C3, 11–17, 23, 45, 54 and 55 responded significantly to the four stresses. By predicting the cis-elements in the promoter, we found that all CsPP2C members contained ABA response elements and drought response elements. Additionally, the expression patterns of CsPP2C genes were specific in different tissues.
Conclusions
The results of this study provide a reference for the genome-wide identification of the PP2C gene family in other species and provide a basis for future studies on the function of PP2C genes in cucumber.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08734-y.
Keywords: Cucumber, Protein phosphatase 2C, Stress response
Background
Plants have developed multiple response mechanisms in response to salt, drought and cold stress, among which the regulation of related gene expression and protein modification are two important pathways. Protein reversible phosphorylation, a protein modification process that regulates multiple physiological responses in plants, is catalyzed by protein kinases (PKs) and phosphatases (PPs) [1, 2]. PKs primarily phosphorylate serine (Ser), threonine (Thr) and tyrosine (Tyr), while PPs can reverse this function by eliminating phosphate groups [3]. Based on substrate specificity, protein phosphatases (PPs) can be divided into three groups: Ser/Thr phosphatases (STPs), protein Tyr phosphatases (PTPs), and dual-specificity phosphatases (DSPTPs). Moreover, PTPs are further classified into phosphoprotein metal phosphatases (PPM) and phosphoprotein phosphatases (PPP) based on crystal structure, amino acid sequence and specific response to inhibitors [4]. The PPP family contains a variety of protein phosphatases, including PP1, PP4, PP5, PP6, PP7, PP2A and PP2B [5, 6]. At present, very little is known about the PPP family. However, we know that PP2A changes cell morphology in the PPP family during elongation, affecting root growth and root hair growth [7]. PP1 is a highly conserved and ubiquitous phosphatase in eukaryotes [8]. Nine members of the PP1 gene family have been identified in Arabidopsis [9, 10]. In addition, a PP1 gene up-regulated by biological stress was found in Phaseolus vulgaris [11].
Type 2C protein phosphatases (PP2C) are Mg2+/Mn2+ dependent serine/threonine phosphatases that are closely related to the PPP family, but have no sequence homology, thus forming a unique group belonging to the PPM family. The PP2C gene family is evolutionarily conserved, significantly regulates stress signaling pathways, and is primarily present in bacteria, fungi, plants and animals [12]. Plant PP2C family proteins have unique structures that contain a conserved catalytic region at the N- or C-terminus and an unconserved extension region of varying length at the other end [13]. The diversity of their structures indicates that PP2C family proteins have different functions in the signal transduction mechanism [14, 15]. A total of 80 PP2C genes were identified in Arabidopsis, which were divided into 13 subsets (A-J) according to their evolutionary relationships. Six of the nine members of subgroup A (ABI1, ABI2, AHG1, AHG3/ ATPP2CA, HAB1 and HAB2) negatively regulate ABA signaling, and the remaining three members (HAI1, HAI2/AIP1 and HAI3) respond differently to stress than the other six members [16–18]. PP2C of subgroup A inactivated PP2C through dephosphorylation, inhibited ABA receptor (PYR/PYL/RCRA) activity, and negatively regulated ABA signaling [19]. The identification of two PP2C mutants, abi1 and abi2, provides favorable support for their negative regulatory function of ABA signaling [20]. Group B participated in the mitogen-activated protein kinase (MAPK) signaling pathway and catalyzed the dephosphorylation of MAPKs, leading to their inactivation [21, 22]. Group C is involved in the regulation of flower development [23]. Group D members can respond to salt and alkali stress [24]. Group E PP2C is involved in the regulation of stomatal signaling, for example, as a response to dehydration, PP2C interacts with MPK3 and MPK6 to induce stomatal closure and inhibit water loss due to transpiration [25, 26]. Group F PP2C is involved in the induction of bacterial stress response [27]. Currently, the PP2C gene family has been identified and studied in a variety of plants other than Arabidopsis, such as rice [28], maize [29], and Brachypodium distachyon [12]. Rice OsPP108 belonging to subgroup A could be highly expressed under ABA, drought, and salt stress; when overexpressed in Arabidopsis, transgenic Arabidopsis showed stronger stress resistance [30]. Overexpression of the maize ZmPP2C gene in Arabidopsis reduced the sensitivity to ABA and the tolerance to drought and salt treatment [31]. FaABI1 in strawberry had negative regulatory effects on fruit ripening [32]. The expression level of the PP2C gene regulated by endogenous ABA in Arabidopsis was significantly up-regulated under exogenous ABA and stress [19]. In conclusion, the PP2C gene family plays a critical role in plant development and environmental stress.
Although the PP2C gene family has been extensively studied in other species, there are few reports on the PP2C gene family in cucumber. Therefore, identifying the PP2C gene family in cucumber and analyze its expression under stress is of great significance. In this study, 56 PP2C genes were identified at the genome-wide level in cucumber by bioinformatics methods. The physicochemical properties, protein secondary structure, chromosome location, phylogenetic analysis, gene structure, conserved motifs and cis-acting elements, and relative expression of 56 PP2C genes were systematically analyzed, providing reference for the function of abundant PP2C gene families. This study will lay the foundation for understanding the potential molecular mechanism of PP2C in stress signal transduction.
Results
Identification and basic information of the CsPP2C gene family
In this study, we determined that 56 putative CsPP2C were present in the cucumber genome through BLASTp using 80 AtPP2C protein sequences as references. From the analysis of their physical and chemical properties (Table 1), the 56 CsPP2C genes identified encode proteins varying from 233 to 813 amino acids in length, isoelectric point (PI) value ranging from 4.5 to 9.61, and the molecular weight ranging from 30 kDa to 90 kDa. The total average hydrophobic index of the 56 CsPP2C gene family members were all less than zero and therefore encoded hydrophilic proteins. The subcellular localization prediction indicated that most of the CsPP2C proteins might be located in the nucleus, chloroplast, or cytoplasm, while only CsPP2C48 might be located in the endoplasmic reticulum and CsPP2C50 in the cytoskeleton.
Table 1.
List of 56 CsPP2C genes and their basic characterizations
| Gene identifier | Gene name | Size (aa) | Mass (kDa) | pI | Instability index | Aliphatic index | Grand average of hydropathicity | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CsaV3_1G004080.1 | CsPP2C1 | 348 | 37.934 | 7.67 | 28.63 | 89.97 | −0.299 | Nuclear |
| CsaV3_1G034580.1 | CsPP2C2 | 426 | 46.884 | 5.06 | 57.49 | 73.66 | −0.45 | Nuclear |
| CsaV3_1G035680.1 | CsPP2C3 | 358 | 39.549 | 5.26 | 69.22 | 72.4 | −0.499 | Chloroplast |
| CsaV3_1G036330.1 | CsPP2C4 | 392 | 42.073 | 5.97 | 52.12 | 83.93 | −0.194 | Chloroplast |
| CsaV3_1G038430.1 | CsPP2C5 | 370 | 41.493 | 8.01 | 38.05 | 84.84 | −0.254 | Nuclear |
| CsaV3_1G038460.1 | CsPP2C6 | 428 | 46.271 | 7.96 | 32.07 | 87.01 | −0.173 | Chloroplast |
| CsaV3_1G039750.1 | CsPP2C7 | 380 | 41.393 | 5.24 | 54.5 | 78.47 | −0.323 | Chloroplast |
| CsaV3_2G003570.1 | CsPP2C8 | 357 | 39.617 | 5.27 | 48.44 | 76.78 | −0.341 | Nuclear |
| CsaV3_2G006810.1 | CsPP2C9 | 367 | 40.752 | 6.08 | 40.34 | 78.88 | −0.283 | Nuclear |
| CsaV3_2G010540.1 | CsPP2C10 | 484 | 53.551 | 5.74 | 42.41 | 74.32 | −0.508 | Chloroplast |
| CsaV3_2G012660.1 | CsPP2C11 | 275 | 31.235 | 9.51 | 43.29 | 76.22 | −0.471 | Chloroplast |
| CsaV3_2G016210.1 | CsPP2C12 | 397 | 44.019 | 8.96 | 46.3 | 87.41 | −0.313 | Chloroplast |
| CsaV3_2G024970.1 | CsPP2C13 | 424 | 46.836 | 8.25 | 53.61 | 85.94 | −0.302 | Cytoplasmic |
| CsaV3_2G033210.1 | CsPP2C14 | 309 | 34.579 | 6.29 | 46.91 | 82.33 | −0.384 | Cytoplasmic |
| CsaV3_3G000550.1 | CsPP2C15 | 390 | 42.936 | 7.7 | 46.19 | 90.44 | −0.219 | Nuclear |
| CsaV3_3G001890.1 | CsPP2C16 | 813 | 89.039 | 5.29 | 46.54 | 76.17 | −0.473 | Nuclear |
| CsaV3_3G003600.1 | CsPP2C17 | 523 | 57.737 | 5.39 | 50.39 | 75.3 | −0.409 | Nuclear |
| CsaV3_3G013890.1 | CsPP2C18 | 414 | 45.372 | 5.27 | 33.49 | 84.69 | −0.282 | Chloroplast |
| CsaV3_3G014600.1 | CsPP2C19 | 521 | 56.883 | 5.31 | 42.17 | 89.08 | −0.227 | Nuclear |
| CsaV3_3G016530.1 | CsPP2C20 | 421 | 45.192 | 5.61 | 68.05 | 74.8 | −0.302 | Nuclear |
| CsaV3_3G019720.1 | CsPP2C21 | 387 | 42.372 | 5.39 | 42 | 90.03 | −0.105 | Chloroplast |
| CsaV3_3G022030.1 | CsPP2C22 | 349 | 38.668 | 5.54 | 36.64 | 78.51 | −0.406 | Chloroplast |
| CsaV3_3G027970.1 | CsPP2C23 | 233 | 26.258 | 6.76 | 36.69 | 81.2 | −0.488 | Chloroplast |
| CsaV3_3G035300.1 | CsPP2C24 | 370 | 41.113 | 8.84 | 39.84 | 91.68 | −0.326 | Nuclear |
| CsaV3_3G038810.1 | CsPP2C25 | 390 | 43.875 | 7.18 | 7.18 | 88.46 | −0.299 | Chloroplast |
| CsaV3_3G043720.1 | CsPP2C26 | 424 | 46.135 | 5.61 | 41.4 | 91.46 | −0.119 | Chloroplast |
| CsaV3_3G047510.1 | CsPP2C27 | 281 | 31.084 | 6.51 | 36.8 | 88.86 | −0.351 | Chloroplast |
| CsaV3_4G009460.1 | CsPP2C28 | 236 | 26.175 | 6.07 | 40.16 | 87.63 | −0.453 | Cytoplasmic |
| CsaV3_4G025000.1 | CsPP2C29 | 712 | 79.584 | 5.76 | 40.03 | 72.46 | −0.574 | Nuclear |
| CsaV3_4G026720.1 | CsPP2C30 | 446 | 49.889 | 8.55 | 39.04 | 71.41 | −0.591 | Nuclear |
| CsaV3_4G033570.1 | CsPP2C31 | 283 | 31.385 | 5.88 | 37.1 | 81.98 | −0.398 | Cytoplasmic |
| CsaV3_4G034220.1 | CsPP2C32 | 428 | 46.643 | 5.49 | 48.53 | 79.25 | −0.303 | Chloroplast |
| CsaV3_4G035500.1 | CsPP2C33 | 325 | 35.755 | 5.32 | 44.62 | 86.4 | −0.26 | Nuclear |
| CsaV3_4G036320.1 | CsPP2C34 | 293 | 31.576 | 5.07 | 43.56 | 78.26 | −0.313 | Cytoplasmic |
| CsaV3_4G036470.1 | CsPP2C35 | 364 | 39.767 | 5.22 | 33.4 | 73.46 | −0.374 | Chloroplast |
| CsaV3_4G037450.1 | CsPP2C36 | 386 | 41.981 | 5.31 | 54.25 | 82.9 | −0.199 | Nuclear |
| CsaV3_5G006460.1 | CsPP2C37 | 389 | 43.513 | 7.24 | 42.56 | 93.7 | −0.23 | Chloroplast |
| CsaV3_5G010270.1 | CsPP2C38 | 363 | 39.279 | 6.68 | 58.25 | 80.83 | −0.269 | Chloroplast |
| CsaV3_5G034510.1 | CsPP2C39 | 433 | 46.590 | 8.64 | 39.69 | 85.36 | −0.194 | Chloroplast |
| CsaV3_6G000080.1 | CsPP2C40 | 372 | 40.917 | 5.27 | 54.21 | 91.48 | −0.177 | Cytoplasmic |
| CsaV3_6G001340.1 | CsPP2C41 | 402 | 44.641 | 5.91 | 61.87 | 73.71 | −0.533 | Chloroplast |
| CsaV3_6G003780.1 | CsPP2C42 | 400 | 44.337 | 8.83 | 32.12 | 78 | −0.416 | Cytoplasmic |
| CsaV3_6G005520.1 | CsPP2C43 | 403 | 44.260 | 4.5 | 31.29 | 83.4 | −0.268 | Cytoplasmic |
| CsaV3_6G016880.1 | CsPP2C44 | 715 | 79.492 | 5.73 | 37.16 | 82.64 | −0.473 | Nuclear |
| CsaV3_6G022710.1 | CsPP2C45 | 349 | 38.860 | 8.59 | 42.43 | 77.59 | −0.406 | Nuclear |
| CsaV3_6G028560.1 | CsPP2C46 | 377 | 42.560 | 6.34 | 41.69 | 89.2 | −0.294 | Cytoplasmic |
| CsaV3_6G031110.1 | CsPP2C47 | 275 | 30.192 | 5.24 | 36.02 | 81.93 | −0.268 | Chloroplast |
| CsaV3_6G032130.1 | CsPP2C48 | 553 | 59.388 | 4.7 | 48.54 | 85.17 | −0.158 | Endoplasmic reticulum |
| CsaV3_6G047490.1 | CsPP2C49 | 398 | 44.604 | 7.27 | 53.52 | 88.87 | −0.24 | Chloroplast |
| CsaV3_6G052400.1 | CsPP2C50 | 275 | 30.748 | 4.9 | 51.66 | 84.73 | −0.446 | Cytoskeleton |
| CsaV3_7G001180.1 | CsPP2C51 | 287 | 31.871 | 9.61 | 46.22 | 97.42 | −0.137 | Chloroplast |
| CsaV3_7G004290.1 | CsPP2C52 | 471 | 51.927 | 5.12 | 50.33 | 69.94 | −0.417 | Chloroplast |
| CsaV3_7G005840.1 | CsPP2C53 | 291 | 31.923 | 8.76 | 30.42 | 90.1 | −0.331 | Cytoplasmic |
| CsaV3_7G007970.1 | CsPP2C54 | 382 | 42.507 | 8.51 | 47.81 | 89.06 | −0.277 | Chloroplast |
| CsaV3_7G030300.1 | CsPP2C55 | 393 | 43.077 | 4.64 | 52.99 | 89.52 | −0.161 | Cytoplasmic |
| CsaV3_7G031840.1 | CsPP2C56 | 367 | 40.317 | 5.05 | 38.99 | 68.88 | −0.496 | Chloroplast |
Chromosome distribution and collinearity analysis of the PP2C gene family in cucumber
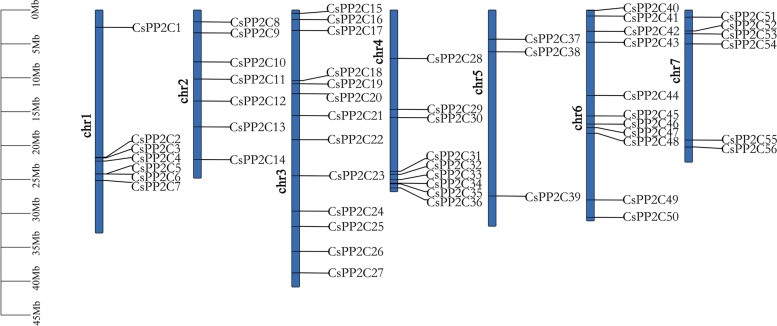
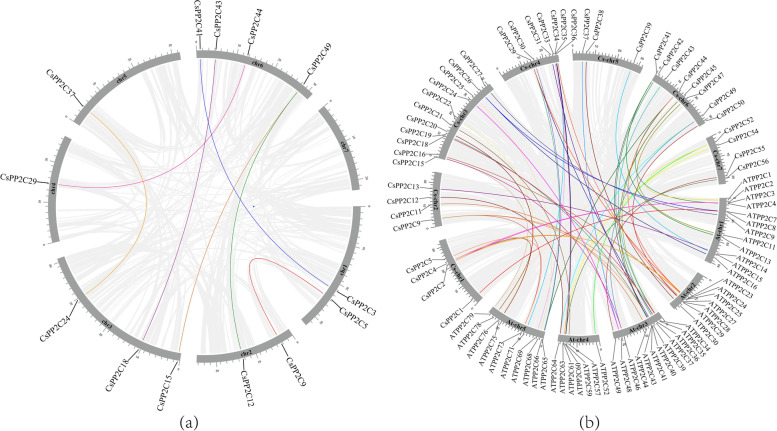
To obtain information on the position of CsPP2C genes on the chromosome, we used TBtools to map the chromosomal location (Fig. 1). A total of 56 PP2C genes were anchored to corresponding chromosomes and designated as CsPP2C1–CsPP2C56 according to their order on the chromosomes, among which chromosomes 3 and 6 were more distributed, and chromosome 5 was the least distributed, with only 3 PP2C genes. Closely related genes located within a distance of less than 200 kb on the same chromosome were defined as tandem duplications, otherwise they were defined as segmental duplications [33]. To further understand the expansion mechanism of the CsPP2Cs, we examined segmental and tandem duplications within the cucumber genome. Our results showed that the PP2C gene family had no tandem duplication gene pairs, but that there were seven fragment repeat gene pairs (Fig. 2 a). In the seven pairs of collinear relationships, CsPP2C49 was paired with CsPP2C15 and CsPP2C12, respectively, while the others were one-to-one paired.
Fig. 1.
Chromosomal distribution and localization of CsPP2Cs. The chromosome names are shown at the left of each chromosome. The chromosome scale is in millions of bases (Mb) on the left
Fig. 2.
Collinearity analysis of the PP2C gene family in cucumber. a Chromosomes 1–7 are represented by gray rectangles. The gray lines indicate synteny blocks in the cucumber genome, while the lines of different colors between chromosomes delineate segmental duplicated gene pairs. b Synteny analysis of PP2Cs between cucumber and Arabidopsis. Gray lines denote the collinear blocks between cucumber and Arabidopsis genomes and the lines of different colors denote the syntenic gene pairs of PP2Cs. Gray rectangles represent respectively the cucumber chromosomes (1–7) and Arabidopsis chromosomes (1–5)
In addition, we also detected homologous PP2C gene pairs between cucumber and Arabidopsis. There were 59 collinear gene pairs between 41 CsPP2Cs and 48 AtPP2Cs (Fig. 2 b). The maximum number of homologous genes in cucumber was 11 pairs on chromosome 3, while the minimum number was three pairs on chromosome 5. According to this result, we speculated that cucumber and Arabidopsis may have high homology.
Analysis of dN/ds values between collinear gene pairs
To further investigate the divergence and selection in duplication of PP2C genes, the non-synonymous substitution rate (dN), synonymous substitution rate (ds), and dN/ds values were evaluated for the homologous gene pairs among cucumber and Arabidopsis (Table S1). Where dN/dS > 1 is the positive selection, dN/dS = 1 is the neutral selection, and 0 < dN/dS < 1 is the purifying selection [34]. The dN/ds value of all cucumber gene pairs was less than 1. Similarly, the dN/ds value of all collinear gene pairs in cucumber and Arabidopsis was less than 1. These data suggest that these genes were primarily under purifying selection during evolution and could help to maintain the basic function of this gene.
Phylogenetic analysis of CsPP2C genes
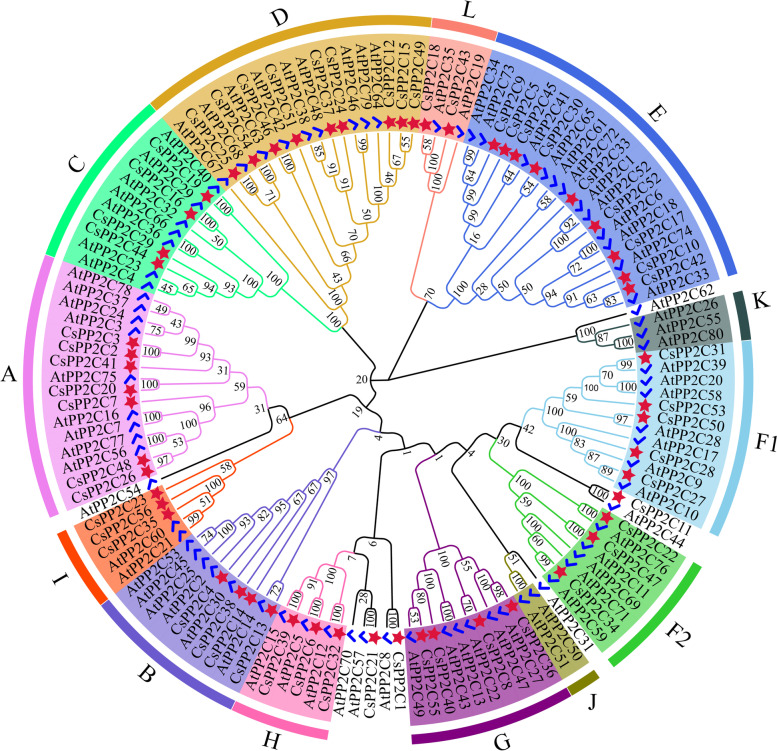
To investigate the phylogenetic relationships of PP2C genes between cucumber and Arabidopsis, we used the maximum likelihood method to construct a phylogenetic tree based on 80 PP2C genes in Arabidopsis and 56 in cucumber (Fig. 3). The phylogenetic analyses indicated that each subfamily includes PP2C proteins from cucumber and Arabidopsis, and that the genes of cucumber and Arabidopsis tended to form independent branches in each subgroup, that is, cucumber genes clustered together, and Arabidopsis genes clustered together. The 56 CsPP2C proteins were divided into 13 subgroups (A-L), while CsPP2C1, 21, and 11 were not clustered with any other group. This was similar to the grouping of PP2C in Arabidopsis and rice. Each group included seven, four, four, nine, nine, five, three, four, three, three, zero, zero, and two CsPP2C genes. Aside from subgroup J and K (only the AtPP2C gene), the distribution of PP2Cs in cucumber and Arabidopsis subgroups was similar. This suggests that the PP2C gene family may have evolved from a common ancestor.
Fig. 3.
Phylogenetic analysis of PP2C proteins among cucumber and Arabidopsis, red asterisks represent CsPP2C, black circles represent Arabidopsis. The phylogenetic tree was constructed by MEGA 7 using the Maximum Likelihood Method (1000 bootstrap)
Gene structural and conserved domain analyses of CsPP2Cs
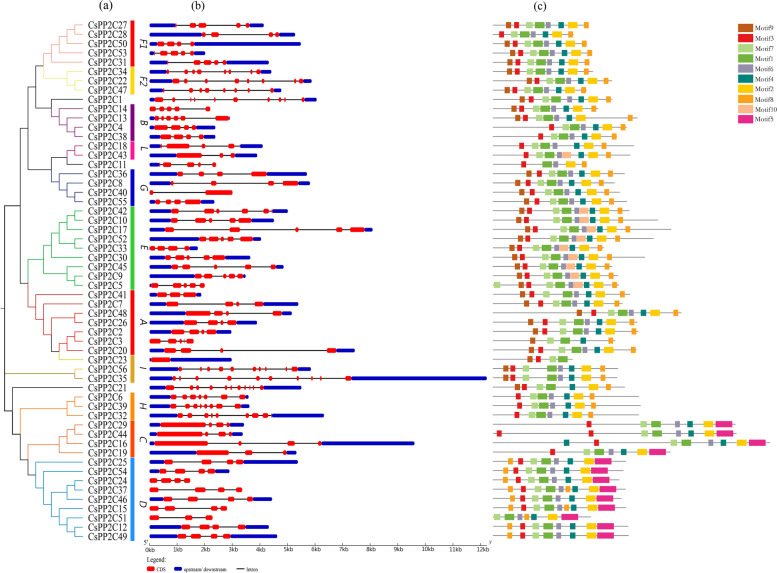
Since the pattern diversity of exon/intron structure and protein domain plays an important role in the evolution of gene families, we studied the exon/intron structure patterns of CsPP2C genes and conserved domain based on their phylogenetic relationships (Fig. 4 a). Studies on exon/intron structure showed that most members of the same subfamily have similar exon/intron numbers but differ in length (Fig. 4 b). The structure of the CsPP2C gene in each group was mostly similar, but there were differences in the exon/intron arrangement of some genes. For example, in group F, the 3′-end of CsPP2C50 has the longest non-coding region and the number of exons in group F2 (8 exons) was almost twice that in group F1 (4–5 exons). In addition, CsPP2C35 in group I has 10 exons, and its longest gene fragment is more than 12 kb long. CsPP2C40 in group G had no non-coding regions, and only two exons, the lowest number of exons in all PP2C genes. CsPP2C14, 40, 3, 24, 37, 15, and 51 have no upstream and downstream gene sequences, and most were located in group D. This indicated that the CsPP2C gene was relatively conservative in its evolution, ensuring the integrity of its gene structure so that there is little change in its function.
Fig. 4.
Phylogenetic tree, gene structure and motif analysis of CsPP2Cs. a The phylogenetic tree of CsPP2C is divided into ten groups. b Exon–intron structures of CsPP2C genes. c Distribution of all motifs identified by MEME
To identify common motifs among different groups of CsPP2C proteins, we used the MEME motif search tool to identify 10 conserved motifs (Table 2). As shown in Fig. 4. c, proteins in the same group exhibited similar motif distribution patterns. Motifs 1, 2 (except CsPP2C3, 23), 3 (except CsPP2C51), 4, 6, and 7 were found in all CsPP2C genes. In addition to the common motif, there were specific motifs in each group. For example, motif 8 was not present in group C, but was present in all other groups. Motif 5 was present in group C and D, but not in the other groups. Motif 9 was not found in group C, D, and H, while it was found in all other groups. According to these results, the CsPP2C genes in the same subgroup had similar conserved motif composition and distribution, suggesting that the CsPP2C members in the same cluster likely share similar functions.
Table 2.
Conserved motifs in the amino acid sequences of CsPP2C proteins
| Motif | Width Multilevel | consensus sequence |
|---|---|---|
| 1 | 29 | SGSTALVALIQGDTLYVANVGDSRAVLAR |
| 2 | 29 | LTPEDEFLILASDGLWDVLSNZEAVDIVR |
| 3 | 15 | AFFGVFDGHGGPGAA |
| 4 | 17 | GGLAVSRAIGDFYLKQY |
| 5 | 50 | PRNGSAKRLVKAALQEAAKKREMRYSDLKKIDRGVRRHFHDDITVIVVFL |
| 6 | 15 | AIQLSVDHKPSREDE |
| 7 | 20 | WEKAJKKAFLKTDEEFLSLV |
| 8 | 15 | RGSKDBISVIVVQFK |
| 9 | 16 | QGKRGEMEDAHIVWED |
| 10 | 27 | AERIKQCKGRVFALQDEPEVYRVWLPN |
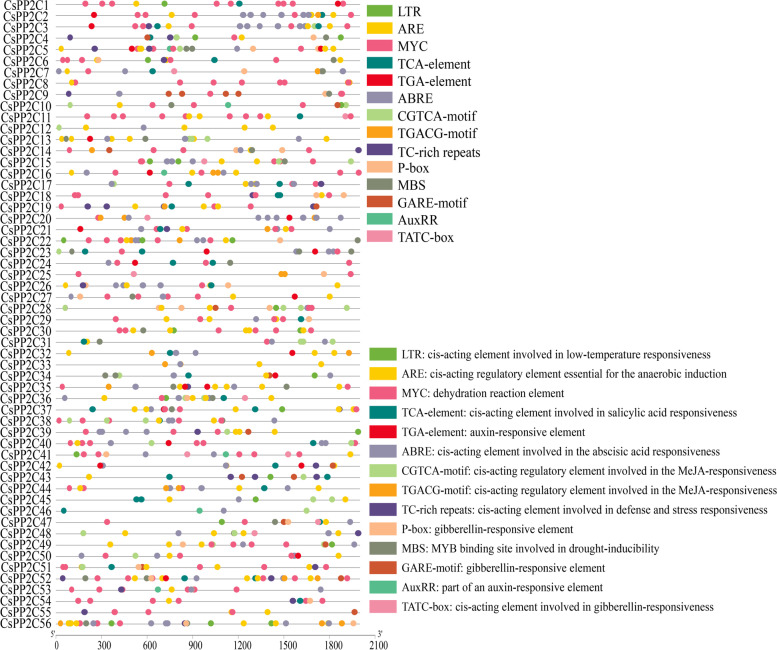
Cis-element analysis of the CsPP2Cs promoter in cucumber
Abundant responsive regulatory elements were found in the promoter regions of CsPP2Cs through PlantCARE analysis (Fig. 5). The cis-elements screened were divided into two categories. The first type of element was hormone response, such as TCA-element (salicylic acid response element), the ABRE (ABA response element), the TGA-element (auxin response element), the CGTCA-motif and TGACG-motif (MeJA-responsiveness response element), and the P-box (gibberellin response element), among others. The second type of element was stress response, such as LTR, MYC, TC-rich repeats and MBS. The abscisic acid ABA-responsive (ABRE) elements were identified abundantly in the promoter regions of CsPP2Cs, among which CsPP2C2 and CsPP2C3 contained 12 ABREs, which was the largest number, followed by CsPP2C7, CsPP2C20, CsPP2C26, CsPP2C40, and CsPP2C56; it was the most abundant element in the promoter region. The second was the MYC element, which was not present in only CsPP2C12–13, CsPP2C32–34, CsPP2C42–43, CsPP2C45, 46, and 48, and was most abundant in CsPP2C1. This suggests that most CsPP2C genes may respond to various abiotic stresses.
Fig. 5.
Putative cis-elements existed in the 2 kb upstream region of cucumber PP2C genes. The elements which respond to hormones are displayed in differently coloured boxes. The homeopathic elements represented by different color boxes and their names and functions
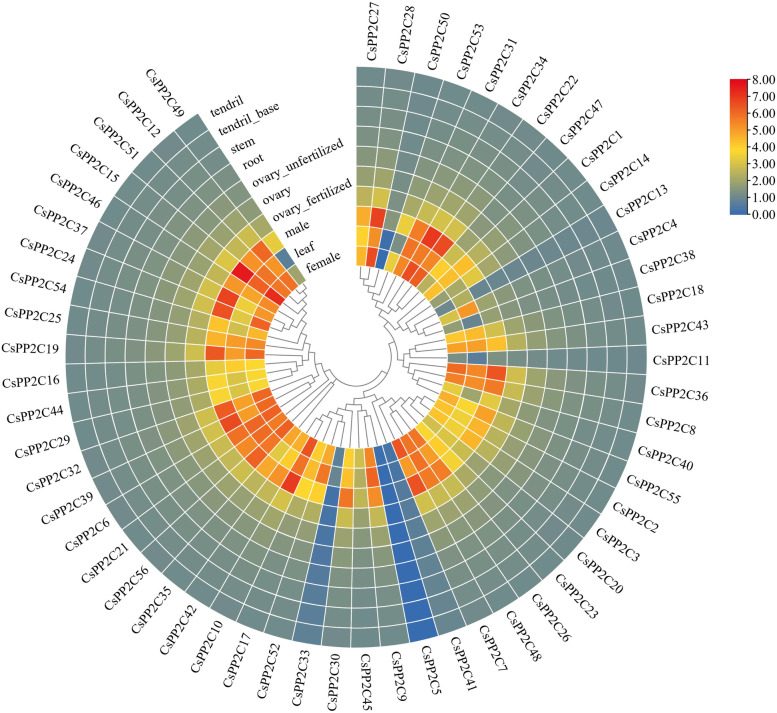
Tissue-specific expression profiles of CsPP2C genes
To better understand the role of CsPP2C genes in cucumber growth and development, the temporal and spatial expression patterns of CsPP2C genes was analyzed using RNA-Seq data of different tissues of the cucumber cultivar Chinese long ‘9930’ (Fig. 6). Only CsPP2C11, 41, 5, 33, and 50 had low expression in all 10 tissues. On the contrary, the expression levels of other CsPP2C genes were high in the fertilized ovaries, male, female, and leaf but low in other organs, such as CsPP2C12, 51, 15, 46, 37, 31, 22, and 47. Moreover, the expression level of CsPP2C49 was medium in males but low in other tissues. Similarly, the expression level of CsPP2C53 was medium in females and males but low in other tissues. In general, most cucumber CsPP2C genes showed similar expression patterns in different tissues.
Fig. 6.
Expression patterns of CsPP2C genes in various wheat tissues. Heatmap of CsPP2C RNA-seq data in five tissues at three different developmental stages was created by TBTools. The expression values mapped to a color gradient from low (blue) to high expression (orange) are shown at the right of the figure
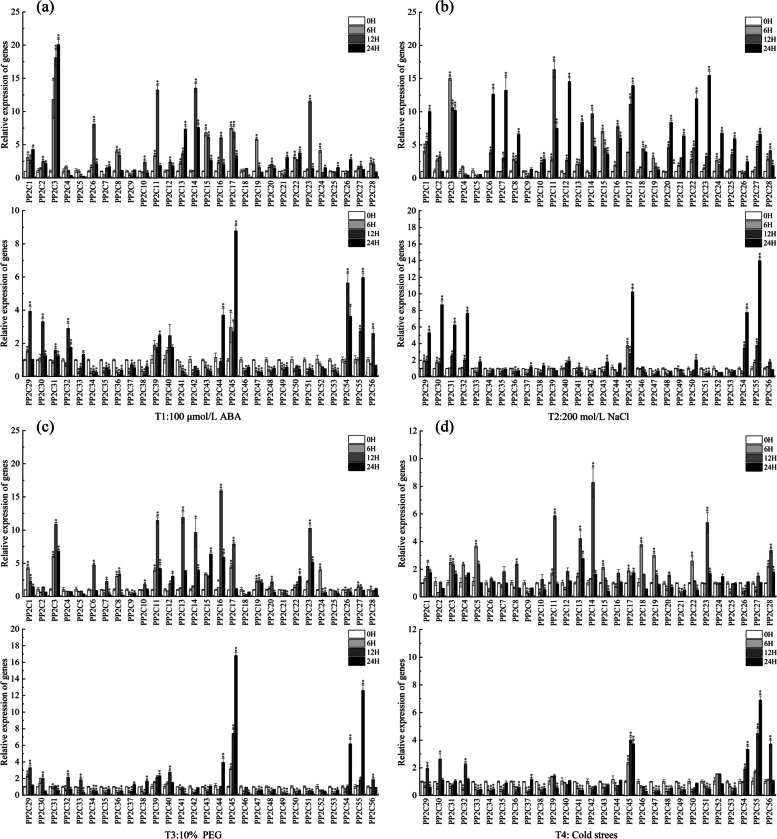
Changes in relative expression of the CsPP2C gene under osmotic stress and ABA treatment
Several members of group A PP2Cs have been shown to function as negative regulators of the ABA signaling pathway in Arabidopsis. The expression of seven PP2C genes in Arabidopsis was suppressed by ABA treatment, two of which are members of subfamily D. The PP2C genes in different subfamilies might play different functional roles in distinct signaling pathways. Therefore, the expression of 56 CsPP2C genes under ABA, salt, drought, and cold treatment was analyzed by qRT-PCR. Under ABA treatment (Fig. 7 a), we observed that the expression of CsPP2C1–3, 13, 22, 29, 39, 45, 54, and 55 continually increased with the extension of ABA treatment time. Among them, the expression level of CsPP2C3 was the highest at 24 h, 20 times that at 0 h. On the contrary, the expression levels of CsPP2C4, 5, 9, 33–38, 41–43, and 46–53 had decreasing trends. The expression levels of CsPP2C6, 8, 11, 14–17, 19, 20, 23, 27–29, and 40 were increased at 6 and 12 h but decreased at 24 h; among them, CsPP2C14–17 was up-regulated most significantly, and although their expression decreased at 24 h, was still higher than that at 0 h. Compared with 0 h, the expression of CsPP2C19, 24 was up-regulated at 6 h, the expression of CsPP2C10, 12, 24, 30, and 32 was up-regulated at 12 h, while the expression of CsPP2C25 and 31 was almost unchanged. As shown in Fig. 7 b, under 10% PEG treatment, CsPP2C1, 3, 11–17, 23, 45, and 55 were significantly up-regulated, among which CsPP2C3, CsPP2C11–17 and CsPP2C45 were up-regulated most noticeably and were more than eight times higher than that at 0 h after 6 h treatment, especially CsPP2C45, which was more than 16 times higher than that at 0 h after 24 h treatment. The relative expression levels of other genes such as CsPP2C9, 18, 21, 34–36, and 46–53 were down-regulated. It is worth noting that after 200 mmol/L NaCl treatment (Fig. 7 c), the expression of many genes increased significantly after 12 and 24 h treatment and were higher than other treatments, especially CsPP2C1, CsPP2C6–8, CsPP2C11, CsPP2C12, CsPP2C17, and CsPP2C20–32. The highest was more than 15 times higher than that at 0 h after 24 h treatment (CsPP2C23). The relative expression of other genes was similar to that of drought treatment. Interestingly, the expression of almost all CsPP2C genes under cold stress (Fig. 7 d) was lower than that of other treatments, except for the genes whose expression was down-regulated, such as CsPP2C1, 3, 11–17, 23, 45, and 54–55. Compared with that at 0 h, the up-regulation of these genes was not high, and CsPP2C14 had the highest up-regulation range at 12 h, more than eight times that of 0 h. Under ABA, drought, and salt treatments, the expression level of CsPP2C3, 11–17, 23, 45, 54, and 55 was significantly up-regulated. This may play an important role in abiotic stress. These results provide a basis for the functional study of the PP2C gene in the future.
Fig. 7.
Quantitative real-time PCR analysis of CsPP2C genes in response to ABA (a), NaCl (b), PEG (c), cold treatments (d). he gene relative expression was calculated using the 2-∆∆Ct method with CsActin as an internal control, and value represents mean ± SE of three biological replicates. Statistical analyses were carried out by student’s t-test to determine the differences in gene abundances between 0 h and other treatment times. Asterisks indicated values that are significantly different from CK (0 h) (* p < 0.05, ** p < 0.01, one-way ANOVA)
Discussion
Plant PP2Cs play a role in plant development as well as in drought, salt, alkali, and fungal pathogen stress resistance [24, 35] and are important PKs in the ABA signaling pathway [36]. In recent years, they have been identified in several plants, such as Arabidopsis [16], rice [16], maize [29], and wheat [37]. In this study, we comprehensively analyzed CsPP2C genes in cucumber, including genome-wide identification, chromosome location, collinear relationship, gene structure, conserved motifs, and expression patterns. Fifty-six cucumber PP2C genes were identified by homology comparison. Compared with Arabidopsis (80) [16], wheat (95) [37], maize (97) [16, 38], rice (78) [16] and B. distachyon (86) [12], the amount of PP2C in cucumber was much less. This indicated that the increase and expansion of PP2C genes are diverse in different species and may also be related to their adaptation to complex environments or to the few chromosomes and small genome of cucumber (2n = 2x = 14) [39].
Chromosome mapping (Fig. 1) found that the PP2C genes located on Chr 3, 4, and 6 were the most distributed, with the number of 13, 9 and 11, respectively. The CsPP2C gene does not cluster on the chromosome, and we did not find tandem duplicate gene pairs. Collinearity results (Fig. 2 a) showed that seven homologous gene pairs existed among CsPP2Cs, and there were 59 pairs of collinear genes in Arabidopsis and cucumber (Fig. 2 b), indicating that CsPP2C genes expand primarily through segmental duplication of the chromosome. Compared with tandem repeats, segmental duplication is more conducive to the maintenance of gene function in the process of gene replication [40]. In addition, the amplification of AtPP2C genes in Arabidopsis is also through fragment replication [41], which was consistent with our results. Furthermore, the dN/dS values of these gene replication pairs were all less than 1, indicating that they underwent purification selection, and their high conservatism in the evolution process was predicted (Table S1).
According to the phylogenetic tree, the PP2C genes of cucumber and Arabidopsis were divided into 13 groups (Fig. 3). This grouping is the same as that in Arabidopsis, rice, and wheat [16, 37]. Phylogenetic tree analysis showed that most subfamilies contained both cucumber and Arabidopsis genes. Cucumber and Arabidopsis had a similar number of members in the same subfamily, and the members of the two species tended to cluster separately. In addition, we analyzed the gene structure and conserved motifs of CsPP2C genes. The exon/intron structure of genes is an important marker of the evolutionary relationship among gene family members [42]. Our results indicated that cucumber PP2C genes in the same group had similar exon/intron structures (Fig. 4 b). While the distribution pattern of exon/intron structures was similar to most genes in the same group, there were some exceptions, which may be due to a variety of reasons [43]. Unlike other genes, CsPP2C14, 40, 3, 24, 37, 15 and 51 have no non-coding region. In addition, all CsPP2C genes contain different numbers of exons and introns, of which CsPP2C35 has the longest coding sequence and CsPP2C21 has the largest number of exons. A total of 10 conserved motifs were identified in the amino acid sequence of CsPP2C genes. The results (Fig. 4 c) showed that CsPP2C genes in the same group showed similar motif distribution, and motifs 1, 2 (except CsPP2C3, 23), 3 (except CsPP2C51), 4, 6 and 7 existed in all cucumber PP2C genes. This motif pattern was closely related to the catalytic core domain of the PP2C protein [44].
Understanding the subcellular location of proteins (Table 1) may provide us with necessary information to infer the biological function of proteins. CsPP2Cs were primarily located in the nucleus, cytoplasm and chloroplast, thus it is speculated that they are related to photosynthesis, respiration, and cell growth and development. At the same time, the CsPP2C gene also showed specific expression in different tissues (Fig. 6). The expression of all CsPP2C genes in female, leaf, male, and fertilized ovaries was higher (except CsPP2C5, 33, and 41), but lower in other tissues. In cotton, the majority of the GhPP2CAs was predominantly expressed in flowers [45]. In Arabidopsis and rice, most AtPP2C genes (84%) and OsPP2C genes (72%) are expressed in more than two tissues [16]. In maize, most ZmPP2Cs had a very broad expression spectrum; only ZmPP2C51, ZmPP2C87, ZmPP2C86, ZmPP2C80, ZmPP2C58 and ZmPP2C41 were low or not expressed in different tissues [29]. This shows that most PP2C gene family members in different plants play a role in multiple processes of plant growth and development, and that only a small number of members act in tissue-specific biological processes.
Cis-acting elements are important regulators of hormone response in plant development and resistance to various stresses [46]. In this study, ABRE elements were most abundant in all CsPP2C gene promoter regions, followed by MYC elements (Fig. 5). qRT-PCR results (Fig. 7) showed that CsPP2C1, 3, 6, 8, 11–17, 22, 23, 45, 54, and 55 responded significantly to ABA and drought treatment; CsPP2C1, 3, 6, 8, 11, 14–17, 54 and 55 contain many MYC elements; and CsPP2C3, 13, 15–17, 22, and 23 contain many ABA response elements. These results indicated that the number of homeopathic elements in the promoter region of CsPP2C genes may be related to its response to stress.
Studies on Arabidopsis and rice suggest that family A PP2C plays an important role in plant responses to abiotic stress, especially in ABA signaling [47]. Overexpression of group E PP2C gene, AtPP2CF1, increased plant biomass in Arabidopsis [48]. AtPP2CG1 in group G is an ABA-dependent positive regulator of salt tolerance [49]. In addition, the expression patterns of PP2C genes in maize [29], rice [16] and B. distachyon [12] under various stresses have been examined. In B.distachyon, BdPP2C70 from subgroup D, BdPP2C13 from subgroup F and BdPP2C32 from subgroup G exhibited strongly increased expression levels in response to ABA and abiotic treatments [12]. In rice, the expression levels of all OsPP2C genes in subgroup A increased after ABA and salt treatment [16]. Our study showed that (Fig. 7) the relative expressions of CsPP2C3 from subgroup A, CsPP2C14, CsPP2C13, from subgroup B, CsPP2C12, CsPP2C15, CsPP2C54 from subgroup D, CsPP2C17, CsPP2C45 from subgroup E, CsPP2C55 from subgroup G, and CsPP2C23 from subgroup I were up-regulated under the four treatments. In addition, the expression of many CsPP2C genes showed a similar trend under four different treatments, which was consistent with research results for Medicago truncatula [50]. In cucumber, subgroup A CsPP2Cs includes seven members (CsPP2C2, CsPP2C3, CsPP2C7, CsPP2C20, CsPP2C26, CsPP2C41, and CsPP2C48). The qRT-PCR results suggested that only CsPP2C3 was highly induced by exogenous ABA treatment, and that CsPP2C41 and CsPP2C48 showed a downward trend, while other genes were up-regulated but not noticeable relative to 0 h. In eight members of B.distachyon subgroup A, BdPP2C27 and BdPP2C34 were also insensitive to ABA treatment. These results showed that not all CsPP2C genes homologous to group A members of Arabidopsis responded to ABA treatment as well as CsPP2C genes in other groups, and that there may be different expression patterns of PP2C genes in different species. Under cold stress, the relative expression level of most CsPP2C genes was lower (Fig. 7d), which was not significant, compared with that of other treatments. Many genes are silenced or reduced in expression. This indicated that the PP2C gene of cucumber had a weak response to cold stress. In conclusion, CsPP2C genes in different groups have different responses to different stresses, and there were CsPP2CAs genes that did not respond to ABA treatment. The expression of CsPP2C3, 11–17, 23, 45, 54, and 55 was significantly up-regulated under the four treatments, suggesting that these genes from different subgroups may play an important role in cucumber resistance to abiotic stress. These results provide a reference for the study of PP2C under different stresses; their functions need to be further explored.
Conclusion
In this study, the whole genome of the cucumber CsPP2C gene family was identified, and its expression level was analyzed. Fifty-six CsPP2C genes were highly similar in gene structure and had conserved motifs. Collinearity and selection pressure indicated that CsPP2C genes were amplified by fragment replication and underwent purifying selection during evolution, ensuring the stability of their functions. In addition, qRT-PCR results showed that in subgroup A, only CsPP2C3 responded significantly to ABA and other treatments, other genes were also up-regulated but not significant, while the expressions of CsPP2C41 and CsPP2C48 were down-regulated. In addition, the members of other subgroups also have genes that respond significantly to abiotic stress, such as CsPP2C11–17, CsPP2C23, 45, 54, and 55. These genes may play an important role in cucumber growth and development. This study provided relevant information for follow-up study of PP2C gene function in cucumber.
Methods
Plant materials and growth conditions
The germinated seeds (Cucumis sativus L, “L306” cultivar) were treated by 5% NaClO for 10 min and washed with deionized water 3–5 times. Afterwards, the cucumber seeds were put into an artificial climate incubator to promote germination. The cultivation conditions were as follows: relative humidity 80%, temperature 25 °C/18 °C (day/night), with a light intensity during the day of 250 μmol m− 2 s− 1. After 4 days of culture, cucumber seedlings were soaked in Yamazaki nutrient solution for hydroponics. When cucumber seedlings grew to three true leaves, four treatments were set, T1: 100 μmol/L ABA; T2: 10% PEG; T3: 200 mmol/L NaCl; T4: cold stress. Cucumber leaves after 0, 6, 12 and 24 h of different treatments were collected, and the samples were put into liquid nitrogen and immediately stored in a refrigerator at − 80 °C.
Identification of PP2C genes in cucumber
The protein sequences of 80 PP2C genes in Arabidopsis were downloaded from the Arabidopsis information resource website (https://www.arabidopsis.org/). Then, cucumber genome data (http://cucurbitgenomics.org/) was used for BLASTp. Next, screening of candidate genes was performed, and the retrieval threshold was set as E-value < E− 10. Redundant results were removed manually. At the same time, the protein sequences of the Arabidopsis PP2C gene were compared in TBtools to further screen candidate genes. Then, the Pfam (http://pfam.xfam.org/search#tabview=tab1) and NCBI-CDD databases (https://www.ncbi.nlm.nih.gov/cdd/) were used for domain identification of candidate gene sequence structures. Candidate genes that did not contain the specific domain of the PP2C gene (registration number: PF00481) were manually removed [51].
Sequence analysis and basic information of the cucumber PP2C gene family
The subcellular localization of the PP2C protein in cucumber was predicted by Wolf PSORT (https://wolfpsort.hgc.jp/) [52]. The number of amino acids and isoelectric genes were identified using ExPASy (websitehttps://web.expasy.org/protparam/) [53]. Additionally, the secondary structure of the cucumber PP2C protein were predicted through the online web site (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl? page = npsa_sopma. html).
Analysis of chromosome location and collinearity analysis
The Chinese long ‘9930’ v3 gff3 file was downloaded from the cucumber genome database, and the chromosome location and length information of 56 PP2C genes were screened for and classified as CsPP2Cs according to their distribution on chromosomes. MG2C (http://mg2c.iask.in/mg2c_v2.0/) was used to map the chromosomal location. Using MCScanX search for homology, the protein-coding genes from the cucumber genome were compared against themselves and those from Arabidopsis genomes using BLASTp, and the retrieval threshold was set as E-value < E− 5. Others were not modified by default parameters. Whole-genome BLASTp results were used to compute collinear blocks for all possible pairs of chromosomes and scaffolds [54]. Subsequently, TBtools was used to highlight the identified PYL collinear pairs and their collinear pairs with Arabidopsis [55].
Selective pressure analysis of the cucumber PP2C genes family
PAL2NAL (http://www.bork.embl.de/pal2nal/index.cgi?) was used to calculate the non-synonymous/synonymous (dN/ds) value of duplicate gene pairs [56].
Construction of phylogenetic tree
The phylogenetic tree of the PP2C gene family of cucumber and Arabidopsis thaliana was constructed using MEGA 7 with muscle-sequence alignment methods selected and the Bootstrap value set as 1000 [57]. Finally, a stable minimum neighbor tree was selected to represent their evolutionary relationship and the constructed tree was beautified using iTOL (https://itol.embl.de/).
Analysis of gene exon-intron structures and protein conserved motifs
The structure of the PP2C gene in cucumber was analyzed by GSDS (http://gsds.gao-lab.org/) [58]. The conserved motif of the cucumber PP2C gene was analyzed by MEME (http://meme-suite.org/tools/meme) [59]. The maximum motif number was set as 10, and the remaining parameters were set as default values.
Analysis of cis-acting elements in CsPP2C gene promoters
The first 2000 bp upstream sequence of 56 identified PP2C gene initiation codons (ATG) was extracted using TBtools. The 2000 bp sequence was then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/ webtools/plantcare/html/) for cis role element forecast analysis [60].
Tissue expression analysis of PP2C genes
To study the gene-specific expression of CsPP2C genes in different tissues of cucumber, the accession number PRJNA80169 was used from cucumber genome data (http://cucurbitgenomics.org/) to obtain cucumber RNA samples from different tissue and organ (tendril-base, tendril, root, leaf, stems, ovary-unfertilized, ovary-fertilized, ovary) RNA-Seq data [61]. Finally, the log2 method was used for data conversion and TBtools was used to draw the expression heat map of the CsPP2C genes.
RNA extraction and real-time PCR (qRT-PCR)
Total RNA was isolated from collected samples using a Plant RNA Extraction kit (Tiangen, China). The complementary DNA was synthesized using the fastking cDNA dispersing RT supermaxs kit (Tiangen, China) with 2 μL RNA as the template. The coding sequences of CsPP2C genes were input into the homepage of Shanghai biology company (Shanghai, China) for online primer design (Table S2). The SYBR Green kit (Tiangen, China) was used for fluorescence quantitative analysis. The volume of the reaction system was 20 μL, containing 2 μL cDNA solution, 10 μL 2*SuperReal PreMix Plus, 0.6 μL of 10 μM forward and reverse primers, 0.4 μL 50*ROX Reference Dye, and 6.4 μL of distilled deionized water. Next, qRT-PCR was performed using the LightCycler® 480 II real-time fluorescence quantitative PCR instrument. The amplification program conditions were as follows: 95 °C for 15 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Each sample was replicated three times. The relative expressions levels of the PP2C genes were calculated using the 2-∆∆CT method [62]. SPSS 20.0 was used to analyze the relative expressions and Origin 9.0 was used to complete the histogram of relative expressions.
Supplementary Information
Additional file 1: Table S1. Selective pressure analysis of PP2C genes family.
Additional file 2: Table S2. Conserved motifs in the amino acid sequences of CsPP2C proteins.
Additional file 3: Mean value, SE and T-test of relative expression of PP2C genes.
Acknowledgments
This experiment thanks the support and help of the Key Laboratory of Arid Habitat Crop Science, China.
Authors’ contributions
GZ and SL conceived of the study, performed the experiments. ZW and XL performed the bioinformatic analysis. ZZ and JL analyzed the data, wrote the manuscript. ZL prepared figures and/or Tables. JY contributed reagents/materials/analysis tools. The author(s) read approved the final manuscript.
Funding
This research was supported by Director Foundation of Key Laboratory of Arid Habitat Crop Science, China (GHSJ-2020-Z3).
Availability of data and materials
The qRT-PCR data supporting the gene relative expression results of this study can be found in supplementary material 1. The login numbers of all CsPP2C genes identified in the experiment can be obtained in the cucumber (Chinese Long) v3 genome database (http://cucurbitgenomics.org/organism/20).
Declarations
All methods were carried out in accordance with relevant guidelines and regulations.
Ethics approval and consent to participate
The experimental research and field studies on plants, including the collection of plant material, were carried out in accordance with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang L, Liang S, Lu Y-T. Characterization, physical location and expression of the genes encoding calcium/calmodulin-dependent protein kinases in maize (Zea mays L.) Planta. 2001;213(4):556–564. doi: 10.1007/s004250100540. [DOI] [PubMed] [Google Scholar]
- 2.Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013;14(1):1–15. doi: 10.1186/1471-2164-14-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58(1):453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 5.Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54(1):63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 6.MacKintosh C, Coggins J, Cohen P. Plant protein phosphatases. Subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem J. 1991;273(3):733–738. doi: 10.1042/bj2730733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H-W, Nussbaumer C, Chao Y, DeLong A. Disparate roles for the regulatory a subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell. 2004;16(3):709–722. doi: 10.1105/tpc.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith RD, Walker JC. Expression of multiple type 1 phosphoprotein phosphatases in Arabidopsis thaliana. Plant Mol Biol. 1993;21(2):307–316. doi: 10.1007/BF00019946. [DOI] [PubMed] [Google Scholar]
- 9.Takemiya A, Kinoshita T, Asanuma M, Shimazaki K-I. Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc Natl Acad Sci. 2006;103(36):13549–13554. doi: 10.1073/pnas.0602503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Q, Li J, Smith RD, Walker JC. Molecular cloning and chromosomal mapping of type one serine/threonine protein phosphatases in Arabidopsis thaliana. Plant Mol Biol. 1998;37(3):471–481. doi: 10.1023/A:1005912413555. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerlin A, Jupe SC, Bolwell GP. Molecular cloning of the cDNA encoding a stress-inducible protein phosphatase 1 (PP1) catalytic subunit from French bean (Phaseolus vulgaris L.) Plant Mol Biol. 1995;28(3):363–368. doi: 10.1007/BF00020386. [DOI] [PubMed] [Google Scholar]
- 12.Cao J, Jiang M, Li P, Chu Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genomics. 2016;17(1):1–17. doi: 10.1186/s12864-016-2526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song S-K, Hofhuis H, Lee MM, Clark SE. Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell. 2008;15(1):98–109. doi: 10.1016/j.devcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9(5):236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem. 2003;278(21):18945–18952. doi: 10.1074/jbc.M300878200. [DOI] [PubMed] [Google Scholar]
- 16.Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9(1):1–21. doi: 10.1186/1471-2164-9-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell. 2007;19(7):2213–2224. doi: 10.1105/tpc.106.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagne JM, Clark SE. The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell. 2010;22(3):729–743. doi: 10.1105/tpc.109.068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics. 2012;100(2):110–115. doi: 10.1016/j.ygeno.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25(3):295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 21.Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279(40):41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 22.Smékalová V, Doskočilová A, Komis G, Šamaj J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol Adv. 2014;32(1):2–11. doi: 10.1016/j.biotechadv.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Wu P, Wang W, Li Y, Hou X. Divergent evolutionary patterns of the MAPK cascade genes in Brassica rapa and plant phylogenetics. Hortic Res. 2017;4:17079. doi: 10.1038/hortres.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Yu Y, Ding X, Liu B, Duanmu H, Zhu D, Sun X, Cao L, Li Q, Zhu Y. Genome-wide analysis and expression profiling of PP2C clade D under saline and alkali stresses in wild soybean and Arabidopsis. Protoplasma. 2018;255(2):643–654. doi: 10.1007/s00709-017-1172-2. [DOI] [PubMed] [Google Scholar]
- 25.Haider MS, Kurjogi MM, Khalil-Ur-Rehman M, Fiaz M, Pervaiz T, Jiu S, Haifeng J, Chen W, Fang J. Grapevine immune signaling network in response to drought stress as revealed by transcriptomic analysis. Plant Physiol Biochem. 2017;121:187–195. doi: 10.1016/j.plaphy.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Galbiati M, Simoni L, Pavesi G, Cominelli E, Francia P, Vavasseur A, Nelson T, Bevan M, Tonelli C. Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. Plant J. 2008;53(5):750–762. doi: 10.1111/j.1365-313X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee MW, Jelenska J, Greenberg JT. Arabidopsis proteins important for modulating defense responses to pseudomonas syringae that secrete HopW1-1. Plant J. 2008;54(3):452–465. doi: 10.1111/j.1365-313X.2008.03439.x. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11(1):1–18. doi: 10.1186/1471-2164-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan K, Yuan S, Chen J, Chen Y, Li Z, Lin W, Zhang Y, Liu J, Lin W. Molecular evolution and lineage-specific expansion of the PP2C family in Zea mays. Planta. 2019;250(5):1521–1538. doi: 10.1007/s00425-019-03243-x. [DOI] [PubMed] [Google Scholar]
- 30.Singh A, Jha SK, Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One. 2015;10(4):e0125168. doi: 10.1371/journal.pone.0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Hu X, Song J, Zong X, Li D, Li D. Over-expression of a Zea mays L. protein phosphatase 2C gene (ZmPP2C) in Arabidopsis thaliana decreases tolerance to salt and drought. J Plant Physiol. 2009;166(5):531–542. doi: 10.1016/j.jplph.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Jia H-f, Lu D, Sun J-H, Li C-I, Xing Y, Qin L, Shen Y-Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot. 2013;64(6):1677–1687. doi: 10.1093/jxb/ert028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui L-C, Scherer SW. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003;4(4):1–10. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 2015;22(1):79–90. doi: 10.1093/dnares/dsu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akimoto-Tomiyama C, Tanabe S, Kajiwara H, Minami E, Ochiai H. Loss of chloroplast-localized protein phosphatase 2Cs in Arabidopsis thaliana leads to enhancement of plant immunity and resistance to Xanthomonas campestris pv. Campestris infection. Mol Plant Pathol. 2018;19(5):1184–1195. doi: 10.1111/mpp.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Tsz-fung FC. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X, Han J, Wang E, Xiao J, Hu R, Yang G, He G. Genome-wide identification and homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum L.) Front Genet. 2019;10:561. doi: 10.3389/fgene.2019.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei K, Pan S. Maize protein phosphatase gene family: identification and molecular characterization. BMC Genomics. 2014;15(1):1–20. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Koo DH, Li Y, Zhang X, Luan F, Havey MJ, Jiang J, Weng Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012;71(6):895–906. doi: 10.1111/j.1365-313X.2012.05017.x. [DOI] [PubMed] [Google Scholar]
- 40.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 41.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):1–21. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4(11):865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 43.Rogozin IB, Sverdlov AV, Babenko VN, Koonin EV. Analysis of evolution of exon-intron structure of eukaryotic genes. Brief Bioinform. 2005;6(2):118–134. doi: 10.1093/bib/6.2.118. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Lu T, Zhang G, Wang Y, He S, Sun L, Hao F. Genome-wide characterization and expression analysis of PP2CA family members in response to ABA and osmotic stress in Gossypium. PeerJ. 2019;7:e7105. doi: 10.7717/peerj.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61(2):290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade a protein phosphatase 2Cs. Plant Physiol. 2012;160(1):379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto H, Kondo S, Tanaka T, Imamura C, Muramoto N, Hattori E, Ki O, Mitsukawa N, Ohto C. Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J Exp Bot. 2014;65(18):5385–5400. doi: 10.1093/jxb/eru297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Zhu Y, Zhai H, Cai H, Ji W, Luo X, Li J, Bai X. AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem Biophys Res Commun. 2012;422(4):710–715. doi: 10.1016/j.bbrc.2012.05.064. [DOI] [PubMed] [Google Scholar]
- 50.Yang Q, Liu K, Niu X, Wang Q, Wan Y, Yang F, Li G, Wang Y, Wang R. Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci Rep. 2018;8(1):1–14. doi: 10.1038/s41598-018-29627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(suppl_2):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong E, Zheng C, Wu X, Wang W. Protein subcellular location: the gap between prediction and experimentation. Plant Mol Biol Report. 2016;34(1):52–61. doi: 10.1007/s11105-015-0898-2. [DOI] [Google Scholar]
- 53.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49–e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11(5):725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 57.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(suppl_2):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, Zhang Z, Yan P, Huang S, Fei Z, Lin K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genomics. 2011;12(1):1–11. doi: 10.1186/1471-2164-12-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Selective pressure analysis of PP2C genes family.
Additional file 2: Table S2. Conserved motifs in the amino acid sequences of CsPP2C proteins.
Additional file 3: Mean value, SE and T-test of relative expression of PP2C genes.
Data Availability Statement
The qRT-PCR data supporting the gene relative expression results of this study can be found in supplementary material 1. The login numbers of all CsPP2C genes identified in the experiment can be obtained in the cucumber (Chinese Long) v3 genome database (http://cucurbitgenomics.org/organism/20).