Abstract
Monoclonal antibodies are being used to treat a remarkable breadth of human disorders. Nevertheless, there are several key challenges at the earliest stages of antibody drug development that need to be addressed using simple and widely accessible methods, especially related to generating antibodies against membrane proteins and identifying antibody candidates with drug-like biophysical properties (high solubility and low viscosity). Here we highlight key bionanotechnologies for preparing functional and stable membrane proteins in diverse types of lipoparticles that are being used to improve antibody discovery and engineering efforts. We also highlight key bionanotechnologies for high-throughput and ultra-dilute screening of antibody biophysical properties during antibody discovery and optimization that are being used for identifying antibodies with superior combinations of in vitro (formulation) and in vivo (half-life) properties.
Keywords: mAb, membrane protein, virus-like particle, self-interaction, non-specific interaction, viscosity, opalescence
Introduction
The success of antibodies as drugs is directly attributable to their unique combination of properties, including high affinity, potent effector functions, low toxicity, long half-life, and favorable biophysical properties (e.g., high specificity, stability and solubility). The FDA approval of >100 antibody drugs and related products and ongoing clinical trials for hundreds more has motivated a tremendous amount of effort to improve the discovery and development of these molecules [1]. Despite great advances in this field, there are several key challenges that continue to limit the pace and success of antibody drug development [2–4]. In this review, we highlight two such challenges and illustrate how bionanotechnology holds great promise to address these challenges in ways that are increasingly simple and widely accessible.
Bionanotechnology for improved antibody drug discovery
One outstanding challenge for therapeutic antibody development is the discovery of antibodies against membrane proteins. As key mediators of myriad pathological conditions, membrane proteins represent a large fraction of promising therapeutic targets. As such, there is significant interest in establishing reliable methodologies to identify antibodies against these biomolecules. However, there are few approved antibody drugs specific for membrane proteins (<10) relative to the great number of unique membrane protein drug targets (thousands) [5–7]. The fundamental challenge for discovering antibodies against these targets is the ability to prepare membrane proteins in functional and soluble formats that can be used for either initial selection or secondary screening or both [8]. Unfortunately, membrane proteins are extremely difficult to produce as soluble antigens in stable, biologically relevant conformations. Transmembrane proteins, with highly hydrophobic midsections, are heavily reliant on an orderly lipid bilayer for stability and functionality, an environment that is difficult to recapitulate in a manner conducive to experimental selections of therapeutic antibodies. Several approaches have been reported for addressing this challenge, including DNA and RNA immunization [9, 10], surfactant-solubilized membrane lysates [11] and whole cells [11–13] for generating antibodies against membrane proteins. However, these methods are limited by several common challenges, which are often linked to the low concentrations, purities and/or stabilities of membrane proteins either in lysates or displayed on whole cells. Moreover, DNA and RNA immunizations do not obviate the need for membrane protein reagents during secondary screening. Finally, in vitro antibody screening against whole cells is challenging due to the presence of many irrelevant protein antigens on the cell surface and the complexity of interfacing relatively large mammalian cells with yeast cells or even phage particles in a highly specific manner.
Encouragingly, several powerful bionanotechnology methods have emerged as promising general strategies for the controlled presentation of membrane proteins in a biologically-relevant manner. These methods have facilitated the production of soluble, stabilized, and functional membrane protein antigens for use in immunizations and in vitro biopanning. Such methodologies have enabled both direct biological production of functional antigens (e.g., virus-like lipoparticles) as well as synthetic creation of functional antigens (e.g., nanodiscs and peptidiscs).
Virus-like lipoparticles are spheroidal assemblies (~150 nm in diameter) surrounded by intact plasma membrane [14, 15]. These biological particles have been produced using cellular systems overexpressing a desired cell surface antigen and Gag proteins. The Gag proteins localize to membranes and cause curvatures, which bud off as lipoparticles which can be purified via ultracentrifugation, PEGylation precipitation, or chromatographic separation [16, 17]. Recently, these lipoparticles were employed for isolating antibodies against insulin-responding glucose transporter (GLUT4) [18]. This transporter, containing 12 transmembrane spanning segments, is implicated in disease states including diabetes and obesity. Notably, virus-like lipoparticles were generated with human GLUT4 levels that were 10–100 times higher than that of conventional membrane preparations. For antibody discovery, human GLUT4 lipoparticles were first used for immunization, and then the resulting antibody variable genes were cloned and reformatted as single-chain variable fragments (scFvs) for phage display screening (Figure 2A). Next, the scFv library was panned for binding to GLUT4 lipoparticles, and isolated scFvs were further evaluated by ELISA for binding to GLUT4 lipoparticles and lack of binding to control lipoparticles lacking GLUT4. Several antibody clones from unique sequence families were characterized and observed to bind to cells overexpressing GLUT4. The antibodies did not bind GLUT4 in western blot analysis, suggesting engagement of conformational GLUT4 epitopes. Notably, several of the identified mAbs showed unique selectivity for GLUT4 conformational states (Figure 2B and 2C) and mAbs specific for the inward-open conformation also demonstrated inhibitory bioactivity, locking the transporters in an inactive state. More broadly, similar types of lipoparticles have been employed in several discovery campaigns at various stages, including as biotinylated antigens for biosensor-based affinity measurements [9, 19–21]. Overall, virus-like lipoparticles may offer several advantages over alternative membrane protein formulations in that they are highly immunogenic, contain intact membrane lipid bilayers, and are relatively straightforward to produce.
Figure 2. Discovery of conformation-specific antibodies against the membrane protein GLUT4 using lipoparticles.
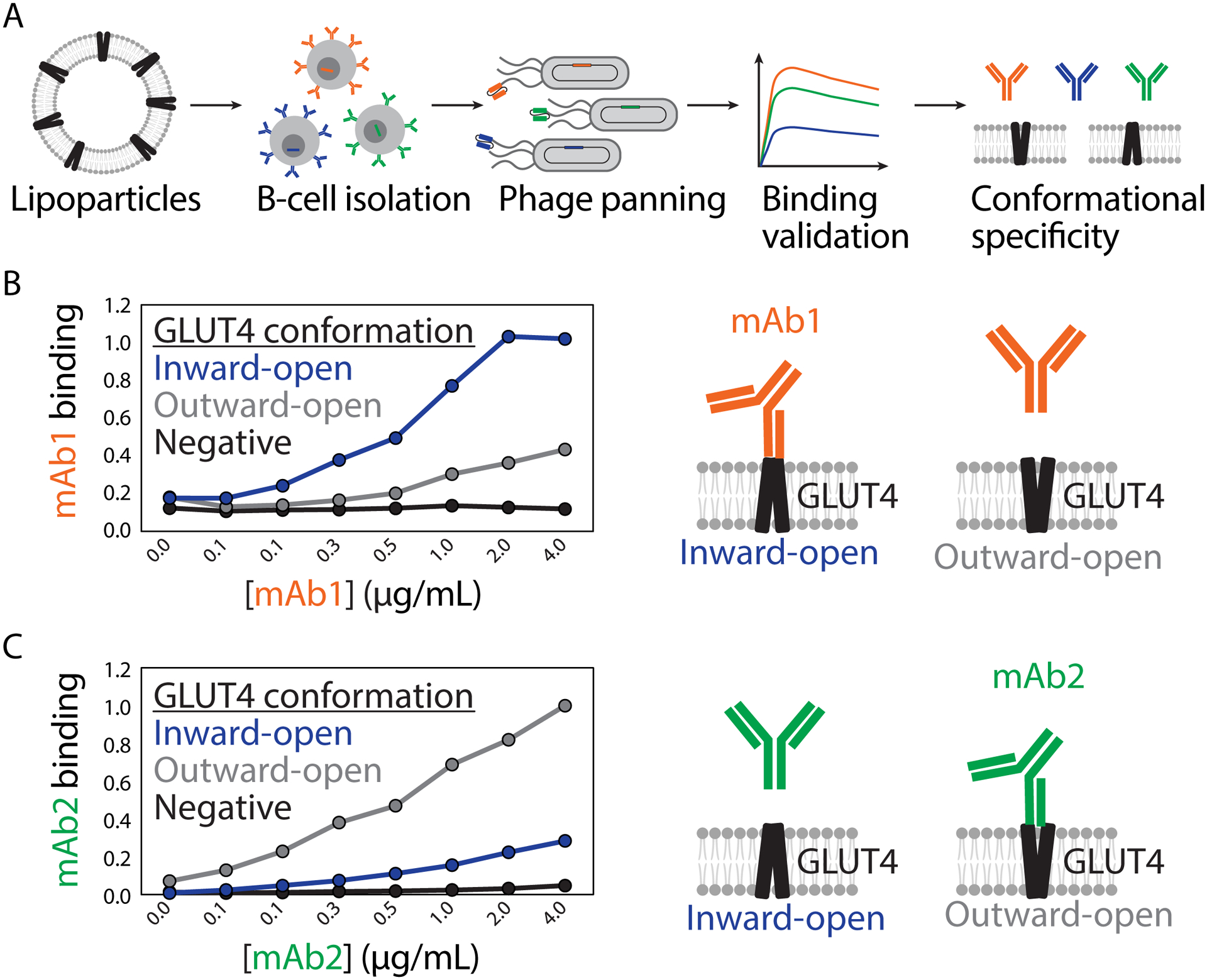
(A) Presentation of the native conformations of GLUT4 (inward-open and outward-open) for immunization and B-cell isolation was achieved using lipoparticle (also known as virus-like particle) technology. Phage panning with GLUT4 lipoparticles led to the isolation of several high affinity antibodies. (B-C) Two selected GLUT4 antibodies (mAb 1 and mAb 2) display conformational specificity for either the (B) inward-open (mAb 1) or (C) outward-open (mAb 2) conformations. The figure is adapted from a previous publication [18].
Despite the many strengths of biological lipoparticles, there has also been much progress in generating different types of synthetic nanoparticles, which also contain lipids, for presenting membrane proteins for antibody discovery applications [22–27]. Initial efforts sought to solubilize intact membrane proteins via harsh detergents that often disturb antigen structure and stability. To obviate the need for such detergents, inspiration from high-density human lipoproteins led to the development of nanodiscs, colloidal assemblies of lipid bilayer with modified membrane scaffold proteins wrapped around the hydrophobic midsection [28]. Nanodiscs have been modified to present a wide range of complex antigens and were recently used to identify high affinity antibodies against a model transmembrane protein, human Nav1.7 ion channel [29•]. A chimeric surrogate (VSD4-NavAb) containing a portion of the bacterial homolog NavAb was selected for the campaign, which has previously been found to enhance expression and stability. Structurally, VSD4-NavAb is a complex transmembrane protein, assembling into a homotetramer with a single ion pore flanked by four voltage-sensing domains. After expression of VSD4-NavAb in insect cells, the protein was solubilized with detergent and purified via chromatography. Addition of stoichiometric amounts of lipid and membrane scaffold protein resulted in displacement of detergent and the formation of nanodiscs, which were confirmed via SDS-PAGE after size-exclusion chromatography purification. Notably, biotinylated nanodiscs were employed for flow cytometry and fluorescence activated cell-sorting (FACS) applications, which showed improved signal relative to sorting with proteoliposome formulations of the antigen. Encouragingly, the nanodiscs were successfully used to isolate high affinity B-cells via single-cell sorting of antigen-specific hybridoma cells after immunization of mice with VSD4-NavAb liposomes (Figure 3).
Figure 3. Isolation of antibodies against the membrane-pass membrane protein, VSD4-NavAb, using nanodiscs.
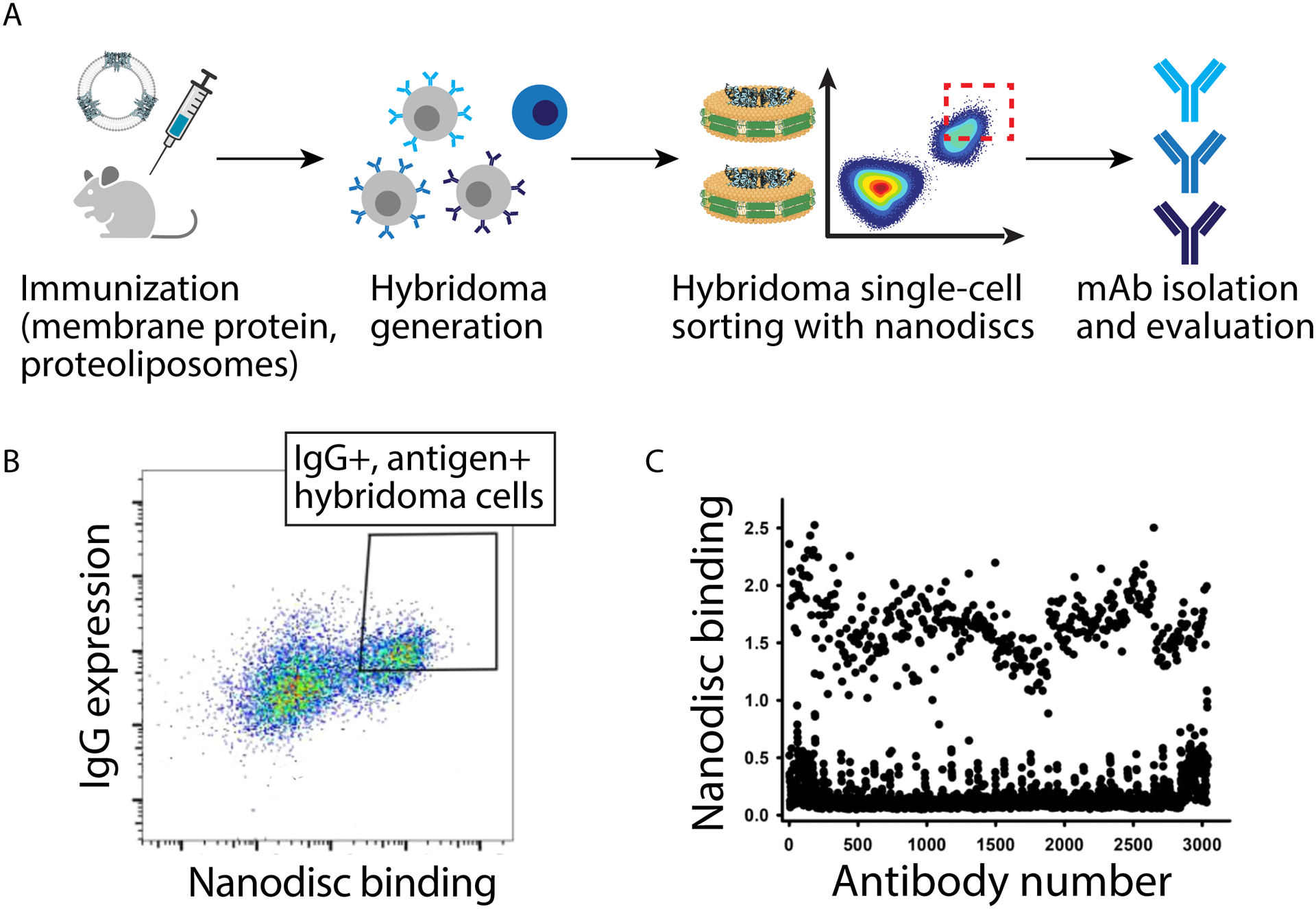
(A) Mice were immunized using the membrane protein (VSD4-NavAb) prepared in proteoliposomes, hybridomas were generated, and then hybridomas were single-cell FACS sorted using biotinylated nanodiscs presenting VSD4-NavAb. (B) Single-cell sorting of hybridoma cells using nanodiscs resulted in the selection of large numbers (>400) of antibodies with high binding activity, as judged by an ELISA assay with immobilized nanodiscs. (C) Selected VSD4-NavAb antibodies generally display similar or higher levels of binding to nanodiscs than proteoliposomes. The figure is adapted from a previous publication [29].
Despite successful applications of nanodiscs in antibody discovery campaigns, they are relatively challenging to produce, and often require optimization of molecular formulation components (e.g., lipid length and rigidity) on an antigen-specific basis [30]. To address this challenge, two alternative peptide-based formulations have been developed, namely Peptidisc and Salipro particles [31, 32]. Peptidiscs are assemblies of transmembrane proteins stabilized by surrounding amphipathic bi-helical scaffold peptides [32], and, unlike nanodiscs, do not require additional lipids for stabilization. This scaffold peptide has proven amenable to the stabilization of multiple different antigens while requiring minimal optimization. Similarly, saposin-lipoprotein (Salipro) particles, which are based on a class of lipid-binding proteins containing amphipathic helices (saposins), facilitate the stabilization of transmembrane proteins in lipid nanoparticles [31]. In Salipro particles, saposin-lipid complexes form a stabilizing belt around the hydrophobic region of transmembrane proteins. While the method of stabilization is similar to that for nanodiscs, the stabilizing components of Salipro particles are smaller and more flexible than nanodisc components, enabling natural adaptation of the number of stabilizing saposins to the size of complexed transmembrane proteins and lipids. This simplifies the optimization of Salipro particle production relative to nanodisc production. The Salipro technology has been used to stabilize multiple membrane proteins, including a bacterial peptide transporter and the HIV-1 spike protein, for structural and functional studies.
The generation of related synthetic lipoparticles has also been reported using polymers. In particular, styrene-maleic acid lipoparticles (SMALPs) have been validated for the synthesis of membrane protein formulations that are amenable to high-throughput antibody selections using cell sorting methods and exhibit improved thermostability over detergent-solubilized antigens [33]. These discoidal lipoparticles are generated from pores formed by the scaffold co-polymer (styrene-maleic acid) that surrounds target membrane protein within a native lipid bilayer. SMALPs have advantages over Salipro and Peptidisc particles because they do not require initial detergent extraction of membrane proteins and lack scaffold polypeptides, thereby reducing the risk of antibody isolation against non-target polypeptides [17]. Peptidisc, Salipro, and SMALP particles have not yet been reported as antigen formulations for the isolation of therapeutic antibodies against membrane proteins, but these unique particles hold great promise to improve such antibody generation and warrant future investigation.
Bionanotechnology for improved antibody developability analysis
A second outstanding challenge in the field of antibody drug development is the early-stage and ultra-dilute screening of antibody biophysical properties, especially antibody self-association because of its strong impact on concentrated antibody formulation properties such as viscosity and solubility [4, 34–38]. There is intense interest in preparing antibody drugs as concentrated liquid formulations for subcutaneous administration, which increases patient compliance and quality of life and reduces costs associated with administration. However, antibody therapeutic candidates possess variable and difficult-to-predict properties in concentrated liquid formulations, including high viscosity, opalescence and aggregation, and these problems are typically discovered too late to address via selection of alternative candidates or protein engineering [39–41]. Therefore, there is a critical need for technologies that enable screening of antibody colloidal interactions at early stages of discovery, which are compatible with the large numbers of candidates (thousands) that are extremely dilute (<0.1 mg/mL) and only partially purified (e.g., one-step Protein A purification).
Encouragingly, bionanotechnology is enabling key advances in early-stage screening of antibody self-interactions. For example, a nanoparticle-based method has been reported for evaluating antibody self-association in physiological solution conditions, namely Affinity-Capture Self-Interaction Nanoparticle Spectroscopy (AC-SINS) [42]. This approach involves immobilization of anti-human IgG capture antibodies on gold nanoparticles (10–20 nm), and then capture of human mAbs at dilute concentrations (0.001–0.05 mg/mL). The colloidal interactions between the immunogold conjugates are evaluated in terms of the plasmon wavelength redshift via measurement of the absorbance spectra, which can be performed using standard absorbance plate readers and 96- or 384-well plates. This approach has been used for early-stage screening for identifying antibodies with superior solubilities [43] and viscosities [41]. In particular, this approach was used to evaluate the self-association of 87 mAbs generated against a common antigen using unpurified cell culture supernatants at ultra-dilute concentrations (0.001 mg/mL) [43]. Strikingly, the mAbs displayed a remarkably wide range of self-interactions, ranging from highly repulsive to highly attractive. Moreover, these self-interaction measurements were strongly predictive of the solubility of the human antibodies, as those mAbs with repulsive self-association displayed solubilities up to 200 mg/mL while those with strongly attractive self-association displayed solubilities below 10 mg/mL. This and other studies [44, 45] demonstrate the significant potential of early-stage screening of antibody self-association to reduce the risk for suboptimal solution properties in concentrated antibody formulations.
Nevertheless, one key limitation of AC-SINS is that it is generally incompatible with common formulation conditions at acidic pH values (pH ~5–6.5) and extremely low ionic strengths (e.g., 10 mM histidine or acetate). Therefore, there has been significant interest in adapting this assay to be compatible with common formulation conditions. A recent study demonstrated both the origin of the problem and a surprisingly simple solution [39•]. After gold nanoparticles are coated with anti-human IgG capture antibodies, the investigators found that conjugates aggregated between pH ~5 and 6.5 regardless of the buffer (acetate or histidine), which was due to the zeta potential crossing from positive charge due to the highly positively charged antibodies to negative charge due to the negatively charged gold nanoparticles (Figure 4A). This led to the simple solution of co-adsorbing polylysine and capture antibodies to prepare gold-antibody conjugates that retained their positive charge in the key pH range of interest (pH ~5–6.5; Figure 4B). This modified assay, Charge-Stabilized Self-Interaction Nanoparticle Spectroscopy (CS-SINS), enabled ultra-dilute measurements of human mAb self-association (0.01 mg/mL), which was predictive of antibody solution properties at four orders of magnitude higher antibody concentrations (150 mg/mL). Antibodies with CS-SINS scores >0.35 displayed high probability of displaying abnormally high viscosity (>30 cP) or opalescence (>12 NTU; Figure 4C and 4D). This approach and other complementary approaches using functionalized nanoparticles with different charged and hydrophobic properties to identify aggregation-resistant antibodies [46, 47] hold great potential to identify antibody candidates with drug-like properties early in the discovery and optimization process.
Figure 4. Charge-stabilized affinity-capture nanoparticle spectroscopy (CS-SINS) enables ultra-dilute screening of antibody self-association for identifying candidates with low viscosity and opalescence in concentrated antibody formulations.
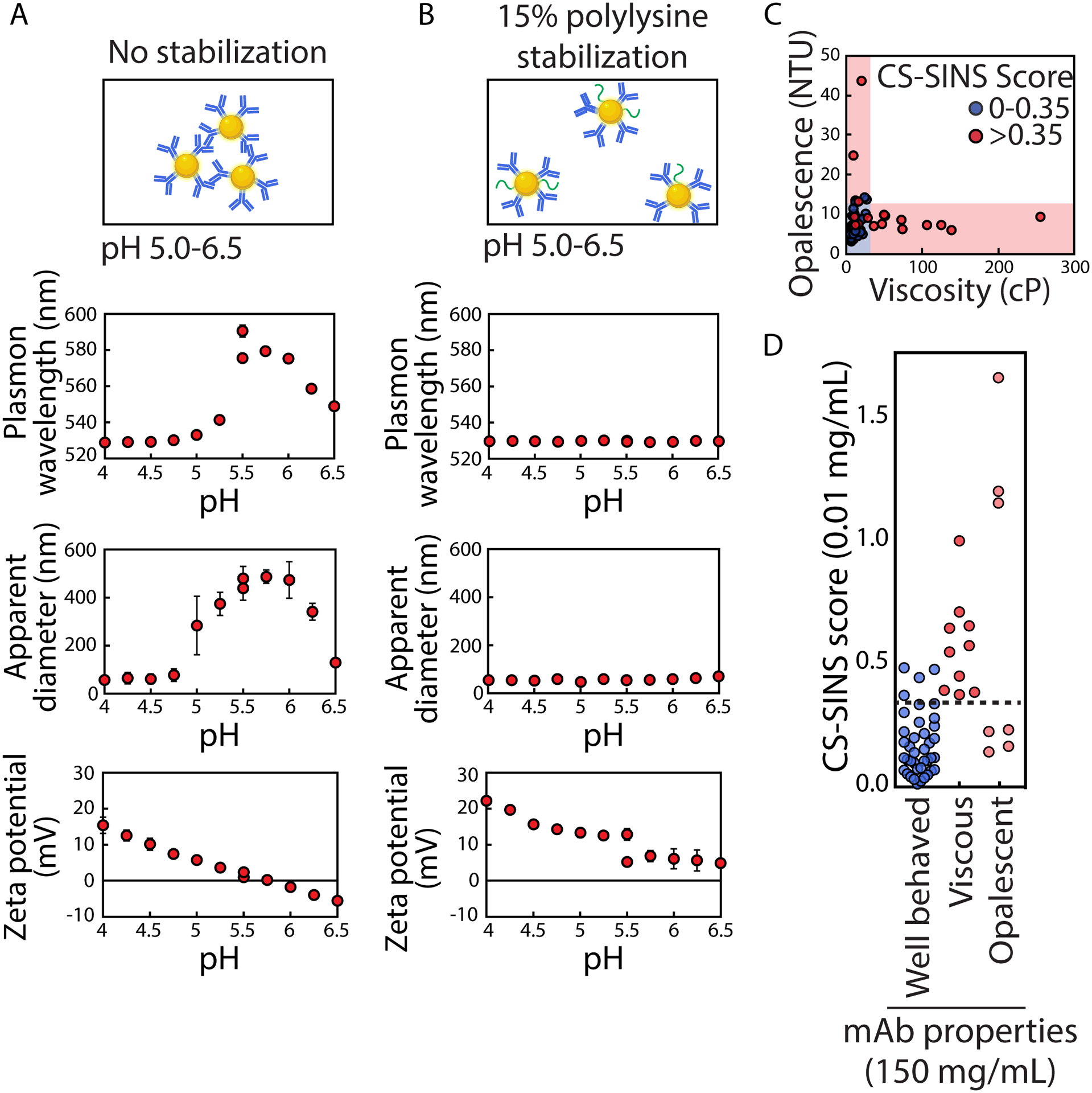
(A) Gold nanoparticles coated with anti-human Fc capture antibodies aggregate at weakly acid pHs (e.g., pH ~5–6.5) and low ionic strengths (e.g., 10 mM histidine or acetate) because the zeta potential of the conjugates is low and crosses zero net charge around pH ~5.5. (B) Gold nanoparticles co-adsorbed with anti-human Fc capture antibodies and positively-charged polymers (polylysine) fail to aggregate at weakly acidic pH values and low ionic strengths because of the increased charge of the conjugates. (C and D) CS-SINS measurements of >0.35, which are measured at a mAb concentration of 0.01 mg/mL, display low risk for high viscosity (>30 cP) or high opalescence (>12 NTU) when formulated at 150 mg/mL. In (C) and (D), the solution conditions were pH 6 and 10 mM histidine. In (D), well-behaved mAbs refer to those that display both viscosity values <30 cP and opalescence values <12 NTU. The figure is adapted from a previous publication [39].
In addition to the need to evaluate weakly attractive antibody self-association during early stages of drug development, it is also critical to evaluate weak non-specific colloidal interactions between antibodies and non-antigen molecules [48]. Antibodies with high levels of non-specific interactions have been linked to fast antibody clearance in vivo [49, 50], which is due to several clearance mechanisms such as increased cellular internalization and degradation. There have been a wide range of proposed assays for evaluating antibody non-specific binding, including ELISAs [44, 50, 51], surface plasmon resonance [52], cross-interaction chromatography [53–55], and flow cytometry assays using various types of cells [56, 57]. Despite the usefulness of these assays, they generally suffer from one or more common limitations, including low sensitivity for detecting non-affinity (non-specific) antibody interactions, low throughput and/or lack of compatibility with existing soluble, full-length IgGs.
Recently, a magnetic particle-based assay, namely the PolySpecificity Particle (PSP) assay, has been developed for sensitively and rapidly evaluating antibody non-specific interactions [40•]. The PSP assay involves capturing IgGs and other Fc-fusion proteins on micron-sized beads functionalized with Protein A, and then evaluating the binding of biotinylated reagents such as complex mixtures of proteins (e.g., soluble membrane proteins from mammalian cells) and proteins with diverse physicochemical properties (e.g., ovalbumin) to immobilized IgGs via flow cytometry. The use of flow cytometry results in high signal-to-noise ratios and a strong ability to differentiate between antibodies with different levels of non-specific binding. Encouragingly, the PSP measurements are strongly correlated with previously reported levels of non-specific binding measured using a proprietary technology for displaying full-length IgGs on yeast [44], which is significant given that PSP uses soluble IgGs and is amenable to diverse types of antibodies and Fc-fusion proteins. Moreover, PSP is highly sensitive at ultra-dilute antibody concentrations (0.01–0.015 mg/mL) and simple to perform in a high-throughput manner using a standard flow cytometer with a microplate sampler [40]. This approach holds great potential for improving early assessment of antibody non-specific binding and generation of antibodies with reduced risk for abnormal pharmacokinetics.
Conclusions
The significant improvements in antibody discovery and developability analysis afforded by bionanotechnology raise several intriguing possibilities for future research and implementation. The advances in preparing soluble and functional versions of membrane proteins in various types of lipoparticles make it much simpler to perform sophisticated antibody selections using in vitro display technologies and generate a much greater array of antibody hits than has been possible previously. For example, routine generation of lipoparticles displaying functional membrane proteins, and the use of positive and negative antibody selections using phage or yeast surface display, would greatly simplify the selection of antibodies that recognize different membrane protein conformations, diverse epitopes and cross-species reactive epitopes. The advances in discovery-stage biophysical characterization of antibody self-association and non-specific binding now makes it possible to screen dramatically larger numbers of antibody candidates and antibody mutants during antibody discovery and optimization, which greatly simplifies the identification of antibodies with globally superior properties. This is particularly important because antibody selections against specific types of targets or specific epitopes, such as those that are hydrophobic and/or negatively-charged, commonly results in panels of antibodies with antigen-binding sites that are over enriched in hydrophobic and/or positively-charged residues. These types of molecular features are key risk factors for poor developability properties and increases the need for extensive screening to identify rare antibody variants with drug-like biophysical properties. Finally, the importance of these advances in antibody developability analysis is expected to be even more important for the rapidly expanding class of multispecific antibodies and non-conventional biologics (e.g., antibody fragments, Fc fusion proteins, nanobodies and cytokines), which demand even greater emphasis on screening for drug-like biophysical properties given their unusual and non-natural formats that are much more prone to poor biophysical properties than conventional IgGs.
Figure 1. Overview of emerging bionanotechnologies that are improving antibody drug discovery and early-stage developability analysis.
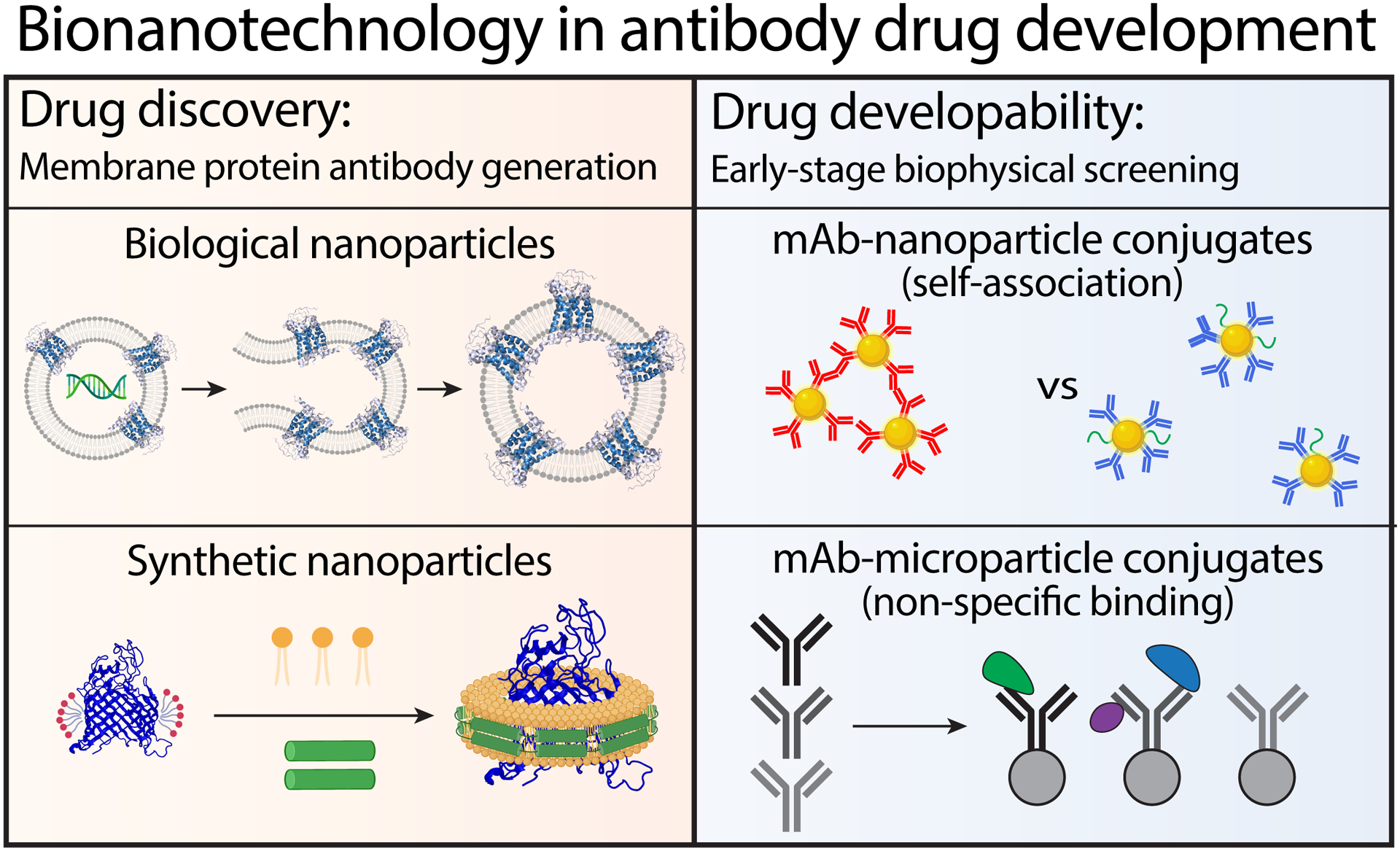
One of the key challenges in antibody drug development is the generation of antibodies specific for membrane proteins given the difficulty in preparing soluble and functional versions of membrane proteins that can be used for immunization and in vitro antibody selections. Advances in generating diverse types of biological and synthetic lipoparticles displaying functional membrane proteins is simplifying the discovery of antibodies against a myriad of membrane proteins as well as diverse panels of epitopes and cross-specifies reactive epitopes. A second key challenge in antibody drug development is the assessment of antibody biophysical properties such as self-association and non-specific binding at the earliest stages of antibody drug discovery. This is particularly important because the complementarity-determining regions of antibodies, which govern antibody affinity and specificity, also mediate antibody self-association and non-specific binding. Advances in biophysical screening methods using bionanotechnologies is enabling the unusually large-scale screening of antibody self-association and non-specific binding in addition to affinity and specificity to identify antibodies with global superior properties.
Acknowledgements
We thank members of the Tessier lab for their helpful suggestions. This work was supported by the National Institutes of Health (R01GM104130 and R01AG050598 to P.M.T., 1T32GM140223-01 to E.K.M., and F32GM137513 to J.S.S.), National Science Foundation (CBET 1813963, 1605266 and 1804313 to P.M.T.), and the Albert M. Mattocks Chair (to P.M.T).
Footnotes
Conflict of interest
None
References
Papers of special interest (•)
Papers of outstanding interest (••)
- 1.Mullard A, FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov, 2021. 20(7): p. 491–495. [DOI] [PubMed] [Google Scholar]
- 2.Weiner GJ, Building better monoclonal antibody-based therapeutics. Nat Rev Cancer, 2015. 15(6): p. 361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mould DR and Meibohm B, Drug Development of Therapeutic Monoclonal Antibodies. BioDrugs, 2016. 30(4): p. 275–93. [DOI] [PubMed] [Google Scholar]
- 4.Makowski EK, et al. , Discovery-stage identification of drug-like antibodies using emerging experimental and computational methods. mAbs, 2021. 13(1): p. 1895540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arinaminpathy Y, et al. , Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today, 2009. 14(23–24): p. 1130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolgin E, First GPCR-directed antibody passes approval milestone. Nat Rev Drug Discov, 2018. 17(7): p. 457–459. [DOI] [PubMed] [Google Scholar]
- 7.Mullard A, FDA approves second GPCR-targeted antibody. Nat Rev Drug Discov, 2018. 17(9): p. 613. [DOI] [PubMed] [Google Scholar]
- 8.Dodd RB, Wilkinson T, and Schofield DJ, Therapeutic Monoclonal Antibodies to Complex Membrane Protein Targets: Antigen Generation and Antibody Discovery Strategies. BioDrugs, 2018. 32(4): p. 339–355. [DOI] [PubMed] [Google Scholar]
- 9.van der Woning B, et al. , DNA immunization combined with scFv phage display identifies antagonistic GCGR specific antibodies and reveals new epitopes on the small extracellular loops. mAbs, 2016. 8(6): p. 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S, Wang S, and Lu S, DNA immunization as a technology platform for monoclonal antibody induction. Emerg Microbes Infect, 2016. 5(4): p. e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tillotson BJ, Cho YK, and Shusta EV, Cells and cell lysates: a direct approach for engineering antibodies against membrane proteins using yeast surface display. Methods, 2013. 60(1): p. 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfaleh MA, et al. , Strategies for Selecting Membrane Protein-Specific Antibodies using Phage Display with Cell-Based Panning. Antibodies, 2017. 6(3): p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissel K, et al. , Immunohistochemical localization of the murine transferrin receptor (TfR) on blood-tissue barriers using a novel anti-TfR monoclonal antibody. Histochem Cell Biol, 1998. 110(1): p. 63–72. [DOI] [PubMed] [Google Scholar]
- 14.Haynes JR, Influenza virus-like particle vaccines. Expert Review of Vaccines, 2009. 8(4): p. 435–445. [DOI] [PubMed] [Google Scholar]
- 15.Thompson CM, et al. , Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnology, 2015. 15(1): p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balliet JW and Bates P, Efficient infection mediated by viral receptors incorporated into retroviral particles. J Virol, 1998. 72(1): p. 671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodd R, et al. , Generating therapeutic monoclonal antibodies to complex multi-spanning membrane targets: Overcoming the antigen challenge and enabling discovery strategies. Methods, 2020. 180: p. 111–126. [DOI] [PubMed] [Google Scholar]
- 18.Tucker DF, et al. , Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proc Natl Acad Sci U S A, 2018. 115(22): p. E4990–e4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths K, et al. , i-bodies, Human Single Domain Antibodies That Antagonize Chemokine Receptor CXCR4. Journal of Biological Chemistry, 2016. 291(24): p. 12641–12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mettler Izquierdo S, et al. , High-efficiency antibody discovery achieved with multiplexed microscopy. Microscopy, 2016. 65(4): p. 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saville JW, Troman L, and Duong Van Hoa F, PeptiQuick, a One-Step Incorporation of Membrane Proteins into Biotinylated Peptidiscs for Streamlined Protein Binding Assays. J Vis Exp, 2019(153). [DOI] [PubMed] [Google Scholar]
- 22.Ren H, et al. , Function-based high-throughput screening for antibody antagonists and agonists against G protein-coupled receptors. Commun Biol, 2020. 3(1): p. 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, et al. , Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Science Advances, 2020. 6(3): p. eaax7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda H, et al. , Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Scientific Reports, 2015. 5(1): p. 11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominik PK, et al. , Conformational Chaperones for Structural Studies of Membrane Proteins Using Antibody Phage Display with Nanodiscs. Structure, 2016. 24(2): p. 300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suharni, et al. , Proteoliposome-based selection of a recombinant antibody fragment against the human M2 muscarinic acetylcholine receptor. Monoclon Antib Immunodiagn Immunother, 2014. 33(6): p. 378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto Y, et al. , Engineered membrane protein antigens successfully induce antibodies against extracellular regions of claudin-5. Scientific Reports, 2018. 8(1): p. 8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayburt TH, Grinkova YV, and Sligar SG, Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters, 2002. 2(8): p. 853–856. [Google Scholar]
- 29.Gardill B, et al. , Nanodisc technology facilitates identification of monoclonal antibodies targeting multi-pass membrane proteins. Scientific Reports, 2020. 10(1): p. 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the use of a complex target antigen, namely an ion channel membrane protein (Nav1.7), stabilized within synthetic nanodiscs to isolate antibodies. Interestingly, antigen presentation using nanodiscs was observed to be superior to presentation via proteoliposomes for single-cell sorting of hydridomas (derived from proteoliposome immunized mice). Further, this investigation highlights the potential broad utility of early-stage single-cell screening against stabilized membrane-protein antigens for improved antibody discovery.
- 30.Hagn F, Nasr ML, and Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nature Protocols, 2018. 13(1): p. 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frauenfeld J, et al. , A saposin-lipoprotein nanoparticle system for membrane proteins. Nature Methods, 2016. 13(4): p. 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson ML, et al. , The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. eLife, 2018. 7: p. e34085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luna VM, et al. , Generation of membrane proteins in polymer-based lipoparticles as flow cytometry antigens. European Polymer Journal, 2018. 109: p. 483–488. [Google Scholar]
- 34.Sawant MS, et al. , Toward Drug-Like Multispecific Antibodies by Design. International journal of molecular sciences, 2020. 21(20): p. 7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann M and Gieseler H, Predictive Screening Tools Used in High-Concentration Protein Formulation Development. J Pharm Sci, 2018. 107(3): p. 772–777. [DOI] [PubMed] [Google Scholar]
- 36.Shan L, et al. , Developability Assessment of Engineered Monoclonal Antibody Variants with a Complex Self-Association Behavior Using Complementary Analytical and in Silico Tools. Mol Pharm, 2018. 15(12): p. 5697–5710. [DOI] [PubMed] [Google Scholar]
- 37.Mieczkowski C, et al. , Characterization and Modeling of Reversible Antibody Self-Association Provide Insights into Behavior, Prediction, and Correction. Antibodies, 2021. 10(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kingsbury JS, et al. , A single molecular descriptor to predict solution behavior of therapeutic antibodies. Science Advances, 2020. 6(32): p. eabb0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starr CG, et al. , Ultradilute Measurements of Self-Association for the Identification of Antibodies with Favorable High-Concentration Solution Properties. Mol Pharm, 2021. 18(7): p. 2744–2753. [DOI] [PubMed] [Google Scholar]; This study pioneers the methodology of Charge-Stabilized Interaction Nanoparticle Spectroscopy (CS-SINS), which enables early-stage antibody developability analysis of self-interactions at important antibody formulation conditions (pH ~6 and low ionic strength) and with ultra-dilute antibody concentrations (0.01 mg/mL). To address a key limitation of the parent technology (known as AC-SINS), the investigators co-adsorbed positively-charged polymers and captured IgGs on the surface of gold nanoparticles to enhance electrostatic repulsion and thereby prevent aggregation that typically occurs at mildly acidic pH conditions (pH ~5–6.5). Overall, CS-SINS is a valuable demonstration of an applied nanotechnology to simplify early-stage, high-throughput antibody development analysis.
- 40.Makowski EK, et al. , Highly sensitive detection of antibody nonspecific interactions using flow cytometry. MAbs, 2021. 13(1): p. 1951426. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports a novel flow cytometry approach for sensitive, robust and ultra-dilute early-stage antibody developability analysis of antibody polyspecificity, which addresses limitations of existing technologies including the use of proprietary antibody cell-based display technologies and complex polyspecificity reagents. This simple approach uses micron-sized magnetic beads coated with Protein A to capture extremely dilute levels of test antibodies (0.001–0.015 mg/mL), and enables flow cytometry screening for binding to polyspecificity reagents. The investigators demonstrate the utility of this approach by validating a panel of clinical-stage antibodies with known polyspecificity levels using conventional (e.g., soluble membrane proteins) and novel (e.g., ovalbumin) polyspecificity reagents.
- 41.Geng SB, et al. , Improving monoclonal antibody selection and engineering using measurements of colloidal protein interactions. J Pharm Sci, 2014. 103(11): p. 3356–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. , High-throughput screening for developability during early-stage antibody discovery using self-interaction nanoparticle spectroscopy. MAbs, 2014. 6(2): p. 483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, et al. , Discovery of highly soluble antibodies prior to purification using affinity-capture self-interaction nanoparticle spectroscopy. Protein Engineering, Design and Selection, 2015. 28(10): p. 403–414. [DOI] [PubMed] [Google Scholar]
- 44.Jain T, et al. , Biophysical properties of the clinical-stage antibody landscape. Proceedings of the National Academy of Sciences, 2017. 114(5): p. 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estep P, et al. , An alternative assay to hydrophobic interaction chromatography for high-throughput characterization of monoclonal antibodies. MAbs, 2015. 7(3): p. 553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopp MRG, Capasso Palmiero U, and Arosio P, A Nanoparticle-Based Assay To Evaluate Surface-Induced Antibody Instability. Molecular Pharmaceutics, 2020. 17(3): p. 909–918. [DOI] [PubMed] [Google Scholar]
- 47.Kopp MRG, et al. , An accelerated surface-mediated stress assay of antibody instability for developability studies. mAbs, 2020. 12(1): p. 1815995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starr CG and Tessier PM, Selecting and engineering monoclonal antibodies with drug-like specificity. Curr Opin Biotechnol, 2019. 60: p. 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dostalek M, Prueksaritanont T, and Kelley RF, Pharmacokinetic de-risking tools for selection of monoclonal antibody lead candidates. MAbs, 2017. 9(5): p. 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hötzel I, et al. , A strategy for risk mitigation of antibodies with fast clearance. MAbs, 2012. 4(6): p. 753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouquet H, et al. , Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature, 2010. 467(7315): p. 591–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avery LB, et al. , Establishing in vitro in vivo correlations to screen monoclonal antibodies for physicochemical properties related to favorable human pharmacokinetics. MAbs, 2018. 10(2): p. 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobs SA, et al. , Cross-interaction chromatography: a rapid method to identify highly soluble monoclonal antibody candidates. Pharm Res, 2010. 27(1): p. 65–71. [DOI] [PubMed] [Google Scholar]
- 54.Hedberg SHM, et al. , Cross-interaction chromatography as a rapid screening technique to identify the stability of new antibody therapeutics. Eur J Pharm Biopharm, 2018. 133: p. 131–137. [DOI] [PubMed] [Google Scholar]
- 55.Tessier PM, Sandler SI, and Lenhoff AM, Direct measurement of protein osmotic second virial cross coefficients by cross-interaction chromatography. Protein science : a publication of the Protein Society, 2004. 13(5): p. 1379–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, et al. , Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel, 2013. 26(10): p. 663–70. [DOI] [PubMed] [Google Scholar]
- 57.Dyson MR, et al. , Beyond affinity: selection of antibody variants with optimal biophysical properties and reduced immunogenicity from mammalian display libraries. mAbs, 2020. 12(1): p. 1829335. [DOI] [PMC free article] [PubMed] [Google Scholar]


