Abstract
SARS-CoV-2 is the causative agent behind the ongoing COVID-19 pandemic. This virus is a cumulative outcome of mutations, leading to frequent emergence of new variants and their subvariants. Some of them are a matter of high concern, while others are variants of interest for studying the mutational effect. The major five variants of concern (VOCs) are Alpha (B.1.1.7), Beta (B.1.315), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529.*/BA.*). Omicron itself has >100 subvariants at present, among which BA.1 (21K), BA.2 (21L), BA.4 (22A), BA.5 (22B), and BA.2.12.1 (22C) are the dominant ones. Undoubtedly, these variants and sometimes their progeny subvariants have significant differences in their spike region that impart them the unique properties they harbor. But alongside, the mutations in their non-spike regions could also be responsible elements behind their characteristics, such as replication time, virulence, survival, host immune evasion, and such. There exists a probability that these mutations of non-spike proteins may also impart epistatic effects that are yet to be brought to light. The focus of this review encompasses the non-spike mutations of Omicron, especially in its widely circulating subvariants (BA.1, BA.2, BA.4, BA.5, and BA.2.12.1). The mutations such as in NSP3, NSP6, NSP13, M protein, ORF7b, and ORF9b are mentioned few of all, which might have led to the varying properties, including growth advantages, higher transmission rate, lower infectivity, and most importantly better host immune evasion through natural killer cell inactivation, autophagosome-lysosome fusion prevention, host protein synthesis disruption, and so on. This aspect of Omicron subvariants has not yet been explored. Further study of alteration of expression or interaction profile of these non-spike mutations bearing proteins, if present, can add a great deal of knowledge to the current understanding of the viral properties and thus effective prevention strategies.
Keywords: Variants of concern, Omicron, Subvariants, Non-spike mutations, Non-structural protein, Immune-evasion, Epistasis, Accessory proteins
1. Introduction
COVID-19's first case appeared on December 31, 2019 in China, and the world health organization (WHO) declared this a global pandemic on March 11, 2020. The causative agent behind this pandemic is severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1]. This is a single-stranded positive-sense RNA virus of ∼30 kb genome size and consists of ORF1a, ORF1b, Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N), ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10. ORF1a and OFR1b encode non-structural proteins NSP1 – NSP11 and NSP12 – NSP16, respectively [2,3]. These proteins have individual and combined roles in viral replication, pathogenicity, and transmission [4].
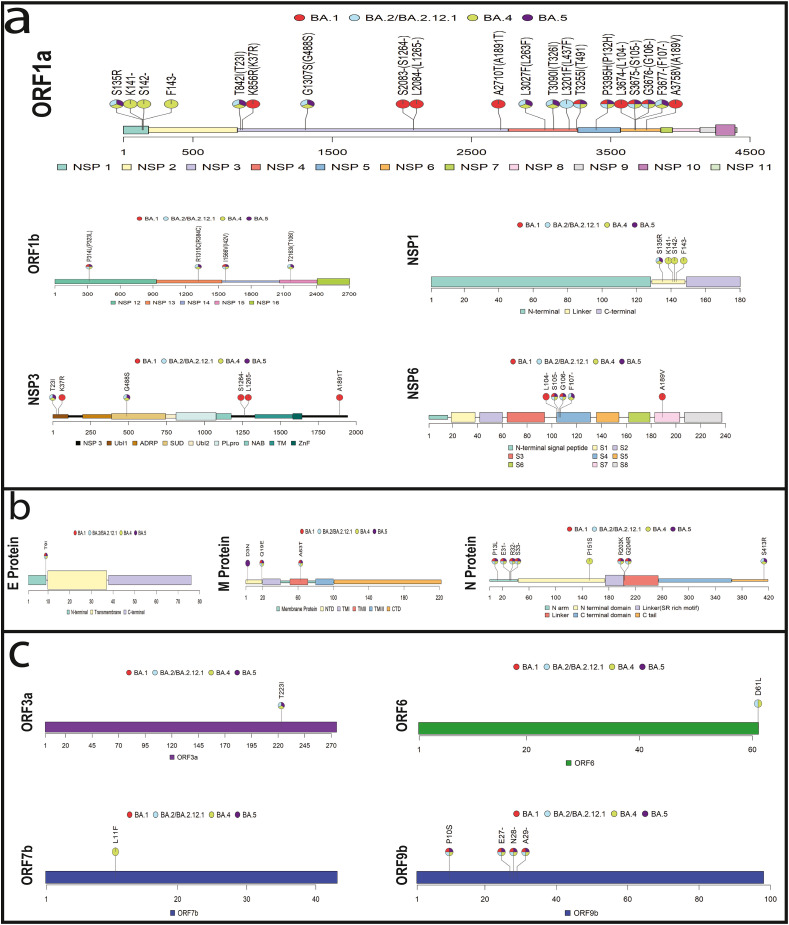
In the Omicron variant, we observe featured mutations in those non-spike proteins (i.e., considering all proteins of the virus apart from spike protein), which are not studied in other variants (Table 1 ). These mutations thus could impart significant effects pertinent to its pathogenicity, immune-evasion, and thus fitness [5]. To realize whether the mutations might or might not have remarkable impacts, the study and analysis must be composed of the followings: the mutations' residing domain, an individual function of that domain, the function of any interactive motifs within that domain, and the protein-protein or protein-RNA interaction of that domain or motif, targeting the mutation (Fig. 1 ). The findings of the study can be further verified through in-depth in silico analysis.
Table 1.
The table comprises of all the non-spike mutations in all VOCs so that addressing the unique mutations in Omicron becomes convenient. The positions of mutations are expressed according to their individual protein positions. The mutations were singled out from Nextstrain site and literature review. The non-spike proteins and ORF which do not contain any mutation in any variant (NSP7, NSP8, NSP9, NSP10, NSP11, NSP16, ORF10) are not mentioned in the table for simplicity.
| Protein | BA.1 (21K) | BA.2 (21L) | BA.4 (22A) | BA.5 (22B) | BA.2.12.1 (22C) | Delta 21A | Gamma 20J | Beta 20H | Alpha 20I |
|---|---|---|---|---|---|---|---|---|---|
| NSP1 | S135R*# | S135R,K141-**#,S142-**#,F143-**# | S135R*# | S135R*# | |||||
| NSP2 | T85I | ||||||||
| NSP3 | K38R** #, S1265- **#,L1266I**#, A1892T**# |
T24I *#, G489S*# | T24I*#,G489S*# | T24I*#,G489S*# | T24I* #, G489S*# | S370L, K977Q | K837 N | T183I, A890D,I1412T | |
| NSP4 | T492I* | T492I *, L264F* #, T327I*#, L438F* | L264F*#,T327I*#,T492I* | L264F*#,T327I*#,T492I* | T492I*, L264F *#, T327I *#, L438F* | ||||
| NSP5 | P132H*# | P132H*# | P132H*# | P132H*# | P132H*# | K90R | |||
| NSP6 | L105- ** #,S106-,G107-,I189V**# | S106 -,G107-,F108- | S106 -,G107-,F108- | S106 -,G107-,F108- | S106 -,G107-,F108- | S106 -,G107-,F108- | S106-,G107-,F108- | S106-,G107-,F108- | |
| NSP12 | P323L | P323L | P323L | P323L | P323L | P323L,G671S | P323L | P323L | P323L |
| NSP13 | R392C*# | R392C*# | R392C*# | R392C*# | P77L | E341D | |||
| NSP14 | I42V*# | I42V*# | I42V*# | I42V*# | I42V*# | ||||
| NSP15 | T112I* | T112I* | T112I* | T112I* | |||||
| E | T9I*# | T9I*# | T9I*# | T9I*# | T9I*# | P71L | |||
| M | Q19E *#,A63T*# D3G**# |
Q19E* #,A63T*# | Q19E*#,A63T*# | D3N**#,Q19E*#,A63T*# | Q19E*#,A63T*# | I82T | |||
| N | P13L*#, E31-*#, R32- *#,S33-*#, R203K, G204R | P13L *#, E31- *#, R32-*#,S33-*#, R203K, G204R,S413R* | P13L*#,E31-*#,R32-*#,S33-*#,P151S**#,R203K,G204R,S413R* | P13L*#, E31-* #, R32-*#,S33-*#, R203K, G204R,S413R* | P13L*#, E31-* #, R32-*#,S33- *#, R203K, G204R,S413R* | D63G, R203 M, D377Y | P80R, R203K, G204R | T205I | D3L, R203K, G204R, N235F |
| ORF3a | T223I* | T223I* | T223I* | T223I* | S26L | S253P | Q57H | ||
| ORF6 | D61L*# | D61L*# | D61L*# | ||||||
| ORF7a | V82A, T120I | ||||||||
| ORF7b | L11F**# | ||||||||
| ORF8 | D119-, F120- | E92K | Q27-, R52I, Y73C | ||||||
| ORF9b | P10S* #, E27-*#,N28- *#, A29-*# | P10S* #, E27-*#,N28- *#, A29-*# | P10S*#, E27-* #,N28- *#, A29-*# | P10S*#, E27-*#,N28- *#, A29-*# | P10S*#, E27-*#,N28- *#, A29-*# | T60A |
‘*’ represents unique mutations in Omicron that are absent in Alpha, Beta, Gamma, and Delta variants.
‘**’ represents unique mutations in each sub variant of Omicron (BA.1, BA.2, BA.4, BA.5, and BA.2.12.1).
‘#’ denotes unique mutation of omicron in functionally important domains.
Fig. 1.
Unique amino acid substitution in the non-spike protein for SARS-CoV-2 B.1.1.529 (Omicron variant), the aa substitution shown here resides in the interacting domains, which might have an impact on the functionality for the Omicron variant; a) The ORF1ab polyprotein has 19 substitutions for the Omicron variant, the mutations for NSP1, NSP3, and NSP6 are shown; b) The non-spike structural protein, Membrane, Nucleocapsid, and Envelope protein have 3, 5, and 1 aa substitution; and c) The accessory proteins ORF3a, ORF6, and ORF9b have 1, 1 and 4 aa substitution, respectively. The red in the labelling circle represents BA.1 subvariant, the sky blue, green and violet is for BA.2/BA.2.12.1, BA.4 and BA.5 subvariants, respectively.
In general, the most explored reasons that differentiate Omicron from other variants are however the novel spike mutations [[6], [7], [8], [9]]. The major Omicron subvariants (BA.1, BA.2, BA.4, BA.5, and BA.2.12.1) that circulated before or are circulating worldwide now have a distinct mutation package for the spike as well than its other minor sporadically spread sister subvariants. BA.4 and BA.5 are one exception, however; they have broadly identical spike mutations (https://www.ncbi.nlm.nih.gov/activ). The spike protein has thus always been the target of studying the variants, as well as now subvariants, to delineate their uniqueness in fitness. No difference is seen for Omicron as, among 50 mutations from wild type, more than 30 residues are in the spike region, being one of the obvious reasons behind its higher transmissibility, better immune escape, and lower lethality [[10], [11], [12], [13]].
The main concern about the Omicron spike has been the antibody escape, especially against clinically used monoclonal antibodies, effective vaccines including boosters, and previous natural infections, even with slightly older subvariants BA.1 and BA.2 [[13], [14], [15]]. However, the role of other immune systems, such as T-cell, killer cell, the intracellular immune response, and the cytokine response, should also be considered. Besides, spike protein alone is not the only immune eliciting entity [6]. It interacts with multiple regions of M protein, and the C terminal of M protein has close interaction with the N protein. The transmembrane and the cytoplasmic domain of M protein also interact with E [[16], [17], [18]]. Therefore, mutations in M, N, and E proteins must be parallelly considered to understand the holistic effect in terms of the immune responses against Omicron.
Considering other immune evasion aspects, NSP6 interacts with ATP6AP1 to evade autophagosome fusion with the lysosome, and this mechanism helps the virus's survival [19,20]. It is known that the accessory proteins from ORF9b are potent interferon antagonists as they target TOM70 and multiple components of signaling pathways [[21], [22], [23], [24], [25], [26]]. These are independent of spike mutations; thus, their mutations can impart independent effects yet not explored. Again, Omicron's more efficient evasion of the host immune response can not only be credited to less recognition by neutralizing antibodies but also should be attributed to the reduction in natural killer (NK) cell activation. This mechanism is mainly driven by antibodies against ORF3a, M protein, and N protein rather than spike protein [[27], [28], [29]].
In terms of successful viral replication and further pathogenesis, NSP1 has its own way of stopping host protein translation by binding to ribosomal subunits [30,31]. Efficient viral assembly and release depend on the interaction amidst NSP3, NSP4, and NSP6, which modifies endoplasmic reticulum (ER) into double-membrane vesicles and promotes budding [[32], [33], [34], [35], [36], [37]]. Post-transcriptional modifications relying upon the NSP10-14-16 sub-complex can be altered due to mutation of NSP14, which might change the dynamics of interaction and complex formation [[38], [39], [40]].
Spike mutations of Omicron certainly enhance receptor binding and ease antibody escape, thus adding to a higher transmission and replication fitness alongside better immune escape. But to support this fitness property of Omicron, non-spike proteins must have an individual or cumulative role, resulting in advantageous replication capacity, better immune evasion, and such. To know the epistatic effect or to relate the cumulative outcome of the mutations, it is essential to bring all the non-spike unique mutations into one frame, which is the main aim of this review. Another purpose of this review is to derive the causes behind the higher transmissibility, growth advantage, differential immune response, and ability to cause plausibly distinct host response by discussing on non-spike proteins' mutations of Omicron.
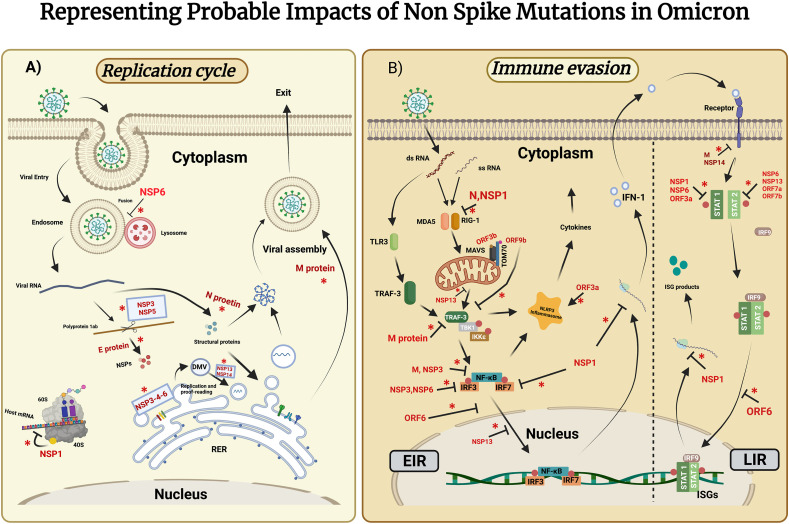
The non-spike proteins are classified into three categories for ease of discussion: 1) Non-structural proteins (NSP), 2) Structural proteins (E, M, N), and 3) Accessory proteins (Fig. 1). A schematic diagram also represents the plausible impacts of these mutated proteins on pathogenesis and immune evasion (Fig. 2 ). Finally, we explain these plausible impacts of featured mutations by representing five major Omicron subvariants BA.1 (21K), BA.2 (21L), BA.4 (22A), BA.5 (22B), and BA.2.12.1 (22C) to ease discussion.
Fig. 2.
Probable impacts of non-spike mutations in Omicron in the replication cycle (A) and immune evasion (B). Here, NSP, N protein, E protein, M protein, ORF denotes Non-Structural protein, nucleocapsid protein, Envelope protein, and Membrane protein. (A)Schematic representation of the role of NSPs, E protein, M protein, and N protein harboring significant mutations in the replicative pathway. Viral NSP1 inhibits host mRNA from translating, which can be interfered with by S135R mutation in Omicron (K141-,S142-, F143- mutations are present only in BA.4). NSP3 possesses some unique mutations (T24I, G489S, K38R) in the region involved in binding ss RNA and nucleocapsid. Viral polyproteins cleavage is conducted by NSP3 and NSP5(P132H). Also, NSP3(S1265-, L1266I, A1892T) along with NSP4(L264F, T327I) and NSP6 forming a subcomplex mediates Double Membrane Vesicle (DMV) formation, which is required for viral RNA replication. NSP6 (L105-, I189V) inhibits autophagosome formation. NSP13(R392C) is the RNA helicase has role in double stranded RNA unwinding. The proofreading mechanism is facilitated by NSP14 (I42V). Structural proteins such as E protein (T9I), N protein (P13L, E31-, R32-, S33-), M protein (D3G, D3N, Q19E, A63T) have a role in post-translational modification, nucleosome formation, and viral assembly respectively. (B)Schematic representation of possible effects of proteins of interest in distinctive immune evasion pathways. Cytokine storm is mediated by ORF3a (T223I), whereas ORF6(D61L), ORF7b (L11F), and ORF9b (P10S, E27-, N28-, A29-) are potent Interferon (IFN) antagonists. NSP1, NSP3, NSP6, N protein, and M protein also show some role in immune evasion through distinctive pathways. ‘*’ denotes mutations in these proteins may alter their corresponding functions. Abbreviation- RER: Rough Endoplasmic Reticulum, TLR3: Toll-Like Receptor 3, TRAF-3: Tumor Necrosis Factor Receptor-Associated Factor-3, TBK1: TANK Binding Kinase 1, IKKε: IkappaB kinase ε, MDA5: Melanoma Differentiation -Associated protein 5, RIG-1: Retinoic acid-inducible Gene-1, MAVs: Mitochondrial Antiviral signaling proteins, IRF: Interferon Regulatory factor, ISG: Interferon-Stimulated Gene, TOM70: Translocase of Outer Membrane 70, EIR: Early IFN Response, LIR: Late IFN Response.
2. Non-structural proteins
Non-structural proteins (NSPs) are proteins that are not components of virion but are transcribed and translated upon infection of the host cell. These include enzymes, such as replicase, protease, methyltransferase, and transcription factors. The 16 NSPs of SARS-CoV-2 from ORF1ab are not different and mediate viral replication, translation, post-translational modification, assembly, host immune evasion, and such essential functions. Here, we discuss the non-structural proteins that possess presumably significant mutations in Omicron, as shown in Table 1 and Fig. 1.
NSP1, the leader protein, abrogates host protein synthesis by binding to the 40S ribosomal subunit and inhibiting the binding of host mRNA. It also causes endo-nucleolytic RNA cleavage in 5′-UTR of host mRNA. In the presence of inhibitory action of NSP1, less abundant but strong and efficient transcripts belonging to the virus are favored to be translated over host mRNAs [34,41,42]. NSP1 consists of the N terminal domain (1–128 amino acid residue), the linker region (128–148), and the C terminal domain (148–180). The C terminal domain carries out the main translation inhibition activity. The N terminal domain regulates cellular mRNA stability or suppresses host innate immune functions. Besides, the N terminal interacts with the 40S subunit, and the linker region helps this interaction. They also interact with host initiation factors, especially eIF3 [19,43,44]. The linker region connects them and provides flexibility [30,45,46]. All Omicron subvariants but BA.2 have a unique mutation at the 135 position of NSP1 (S135R: Serine→Arginine) in the linker region, which might thus affect the overall interaction and function of N and C terminal domains (Fig. 2).
NSP3, a papain-like protease cleaving the polyproteins to its component proteins, is the largest protein of the virus. NSP3 has eight domains: the ubiquitin-like domain 1 (Ubl1), the Glu-rich acidic domain (also called "hypervariable region"), a macrodomain (also named "X domain"), the ubiquitin-like domain 2 (Ubl2), the papain-like protease 2 (PL2pro), the NSP3 ectodomain (3Ecto, also called "zinc-finger domain"), as well as the domains Y1 and CoV–Y of unknown functions along with the two transmembrane regions, TM1 and TM2 [2]. Being a large protein, the NSP3 of Omicron expectedly showed several unique mutations. T24I(Threonine→Isoleucine) in the Ubl1 domain and G489S(Glycine→Serine) in the macrodomain (Mac1/ADRP) were observed in four subvariants of Omicron except in BA.1. In the Omicron BA.1 subvariant, K38R(Lysine→Arginine) in Ubl1, S1265-(Serine→deletion) and L1266I(Leucine→Isoleucine) in the βSM region, and A1892T(Alanine→Threonine) in the C terminal were observed (Table 1 and Fig. 1). The Ubl1 domain binds to ssRNA and nucleocapsid. Macrodomain (Mac1) has a role in de-MARylation, de-PARylation, and ADPR binding. The domain of βSM is disordered; no definite functional role has yet been observed. Co-expression of NSP3 with NSP4 and NSP6 can induce double-membrane vesicle (DMV) formation in host cells. However, lacking the C-terminal third (residues 1319–1922) of the NSP3 will produce different results while co-expressing with NSP4 and NSP6. Correspondingly, co-expression of only the C-terminal third of NSP3 (residues 1256–1922) together with NSP4 may enhance DMV formation by inducing the occurrence of the zippered endoplasmic reticulum (ER) and membrane curvature [32,35,36,[47], [48], [49], [50], [51], [52], [53], [54], [55]]. Since the featured mutations of Omicron subvariants resided in these important domains, they might impart eventful outcomes (Fig. 2).
NSP4 consists of four transmembrane domains and luminal coils. Large luminal loop (35–280) and transmembrane domains (13–34, 281–302,315-337, 365–387) play an essential role in membrane conformational change when combined with NSP3 and NSP6. A large luminal loop plays a major role in binding with NSP3C. Those proteins collectively initiate membrane scaffold formation with a role in the assembly and viral release [33,37,47,56,57]. T492I(Threonine→Isoleucine) mutation is present in all of the five subvariants of Omicron, whereas L438F(Leucine→Phenylalanine) is only specific for BA.2 and its descendant lineage BA.2.12.1. T492I and L438F reside in the cytoplasmic coil region between the 4th transmembrane domain and the H domain of NSP4. Unique mutations L264F(Leucine→Phenylalanine) (large luminal loop) and T327I(Threonine→Isoleucine) (3rd transmembrane domain) have been found in all except BA.1 subvariant (Table 1). These two mutations (L264F and T328I) might have functional significance (Fig. 2).
NSP5, as the main protease, has domain I (residues 10–99) and domain II (residues 100–182), which harbor the binding site between them within a six-stranded antiparallel beta-barrel. In Omicron's all subvariants, the mutation P132H (Proline→Histidine) thus resides in domain II. The NSP5's substrate-binding site is located between domains I and II as a catalytic pocket with four subsites (S1, S2, S3, and S4), which exhibit a bridging dyad between the nucleophilic sulfur atom of Cys145 and the imidazole ring of Histidine residue 41 [58,59]. Thus, NSP5 P132H mutation might affect enzyme activity.
NSP6 consists of eight pore-lining transmembrane domains S1(19–38), S2(42–60), S3(64–94), S4(104–131), S5(136–154), S6(162–179), S7(183–203) and S8(207–237). Possibly, NSP6 interacts with ATP6AP1 (an ATPase proton pump), interfering with digestive autophagosome formation [19,20,46,52,60,61]. The sequential three amino acid deletion S106-(Serine→deletion), G107-(Glycine→deletion), and F108-(Phenylalanine→deletion) was observed in all the VOCs except the Delta variant, and this deletion resides in the S4 domain. But in the BA.1 subvariant of Omicron, 108 (F108-)(Phenylalanine→deletion) deletion was absent, but 105 deletion (L105-)(Leucine→deletion) was observed along with I189V(Arginine→Valine) substitution. The L105, S106, and G107 triad forms a very short alpha helix connecting the transmembrane loop with an extra membranous alpha helix. The triad mutation in Omicron was considered significant using in silico analysis as it facilitates autophagosome evasion by favoring interaction with membrane [62]. The global structure of NSP6 is not altered by this triad deletion, particularly concerning the transmembrane core. However, the structure and dynamics of the peripheral helices of the deleted region (L105-F108) perturbs. These peripheral helices in the Omicron variant are more deeply buried in the polar head region than in the wild type (WT), hence suggesting stronger interaction and an increased capacity to recruit specific lipids. This differential modulation of the lipid interaction and thus autophagy may partly explain the immune system resistance of the Omicron variant and its different pathological evolution [62]. The significance of these sequential deletions (LSG105-107 or SGF106-108) and the effect of the variation in Omicron can be a significant target for future study.
NSP13 (1–601 amino acid residues) is the NTP-dependent helicase enzyme that unwinds double-stranded RNA in a 5′–3′ direction [63]. This protein also plays a role in immune evasion by interfering with the MAVS-associated signaling pathway for IFN production or the JAK/STAT signaling pathway for ISGs expression [64] (). NSP13 is a triangular pyramid-shaped protein with five domains: an N-terminal Zinc binding domain (ZBD), a helical "stalk" domain, a beta-barrel 1B domain, and two "RecA like" helicase subdomains 1A and 2A that contain the residues responsible for nucleotide binding and hydrolysis [63]. The unique mutation R392C(Arginine→Cysteine) found in Omicron (except BA.1 subvariant) is located in the Rec1A domain. This domain is responsible for NTPase (RNA Nucleoside Triphosphatase) activity. Six key residues in the cleft of that domain are involved in NTP hydrolysis. The R392C mutation is located close to three of those six residues (Asp374, Glu375, Gln404) [65]. Thus, the mutation might impact the NTPase activity of NSP13. Considering NSP13 as the potent antiviral drug target, this mutation might be crucial in interfering with one of the fragments to nearly resided Arg382.
NSP14 has proofreading ability with the N-terminal exoribonuclease domain. It also has N7-guanine methyltransferase activity at its C terminal domain [38,40,[66], [67], [68], [69], [70], [71], [72]]. Three conserved motifs have been structurally defined at the active site of NSP14: DXE, W(X)4 EL, and DAIMTR. Omicron's I42V(Isoleucine→valine) mutation was resides in the exon domain (DXE). NSP14 also interacts with NSP10 to boost its exoribonuclease activity using V40 and N41, and one of the two small hydrophobic side chains, due to this mutation, might makecontact with them and may affect binding with NSP10 and enzyme activity [73].
3. Structural proteins
Structural proteins are virion components and play a role in assembly, nucleic acid binding, virion formation, etc. The major structural proteins of SARS-CoV-2 include Envelope protein (E), Membrane Protein (M), Nucleocapsid protein (N), and Spike protein (S). Our point of discussion includes the E, M, and N proteins.
The smallest structural protein, E protein, consists of three domains where there is hydrophilic amino terminus (7–12 amino acids), a hydrophobic transmembrane domain (TMD), and a hydrophilic carboxyl terminus that comprises the majority region [74]. The most significant domain is the C terminal, but the N terminus contains additional Golgi-targeting information [75]. The unique mutation in all subvariants of Omicron (T9I: Threonine → Isoleucine) was observed in that region. The SARS-CoV doesn't undergo myristoylation, but this might occur in SARS-CoV-2 as a separate study for Omicron wasn't conducted. Besides, NSP3 interacts with E protein for ubiquitination and glycosylation, a substantial post-translational modification in this region [76]. Thus, the mutation residing in this region can be studied further if any post-translational modification alteration occurs due to the amino acid isoleucine (Fig. 2).
The viral M protein of SARS-CoV-2 promotes the assembly of new viral particles by the integration into Endoplasmic Reticulum (ER). The predicted structure of M protein (222-amino acid) has been shown to contain three closely packed N-terminal transmembrane alpha-helix domains essential for the assembly of viral particles with a long endo-domain at the C terminal end [17,34,[77], [78], [79]]. Q19E (Glutamine→Glutamic acid)and A63T(Arginine→Threonine) mutations were observed for all the major Omicron subvariants. The Q19Emutation resides in the N terminal domain, and A63T resides in the TMII(transmembrane) domain (Fig. 2). The A63T might impact the stabilization of the M protein dimer since it is close to one of the crucial residues (Valine: V66) [80]. The N-terminal D3G(Aspartic acid→Glycine) and D3N(Aspartic acid→Asparagine) mutations were present only in BA.1 and BA.5, respectively, possibly resulting in the N-myristoylation site at the 3–8 position [81]. Therefore, whether membrane integrity and post-translational modification are altered due to mutation can be a study target [82,83].
The primary function of N protein is RNA encapsidation, but they are also potent immunogens and interferon antagonists. The N protein with 419 amino acids has two conserved domains: N terminal domain (NTD) and C terminal domain (CTD), and three intrinsically disordered regions named N-arm, central Ser/Arg-rich flexible linker region (LKR), and C-tail [84]. The Omicron variant was observed to have P13L(Proline→Leucine), E31-(Glutamic acid→deletion), R32-(Arginine→deletion), and S33-(serine→deletion) unique mutations, each of which resides in the N arm (1–44 residues) preceding the NTD. The function of the N arm is the modulation of RNA binding. Hence, the mutation can significantly affect regulating RNA binding (Fig. 2). The mutation S413R(serine→arginine) of all but BA.1 subvariants resides in the C-terminal end with no apparent effect on dimerization [85]. The R203K(Arginine→Lysine)+G204R(Glycine→Arginine) substitutions occur most frequently during N protein evolution [86]. The mutations together show more significant inhibition of RNA-induced interferon expression, and they can increase SARS-CoV-2 replication, pathogenicity, and fitness by modulating host-virus interactions [87,88].
4. Accessory proteins
Accessory protein plays a role in viral pathogenesis, acting as the virulence factor of the virus. It is known that the major role played by accessory proteins is in immune evasion mechanisms enhancing better viral survival in host system. In SARS-CoV-2, eleven accessory proteins (ORF3a, ORF3b, ORF3c, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, ORF10) have been translated whose detailed functions are not still clear. Some have a role in viral pathogenesis by impairing host immune responses. For instance, ORF3b, ORF6, ORF7a, ORF7b, and ORF8 are potent interferon antagonists. ORF9b and ORF9c interacting with cellular organelles reduce the antiviral responses. The mutations within the protein, along with their likely impacts on the virus/host, are stated as follows:
Upon viral infection in the presence of viral double-stranded RNA, three major pathways for interferon production have been activated: the RIG-I-like receptors (RLRs) pathway, the 2′,5′-oligoadenylate synthetase (OAS)/RNAseL pathway, and the PKR pathway. RLRs binding RNA through the C terminal domain of helicase lead to eventual oligomerization and activation of mitochondrial antiviral signaling (MAVS) protein onto the mitochondrial membrane. Then TANK Binding Kinase 1(TBK1), or inhibitor of κ-B kinase ε (IKKε), promotes autophosphorylation of kinase and downstream activation of transcription factors, IRF3 and NF-κB ultimately promote the expression of IFN type I and III and virus stress-inducible genes. OAS proteins (OAS1, OAS2, OAS3) and PKR are IFN-stimulated genes (ISGs). The late antiviral response is the induction of IFN Stimulated Genes (ISGs) induced through the JAK/STAT signaling pathway (Beyer, 2022). Viral accessory proteins and some non-spike proteins mainly interfere in these pathways.
ORF3a is the largest accessory protein (275-amino acid) with pro-apoptotic ability. The domains include the N terminus domain with signal peptide involved in subcellular localization of ORF3a protein itself, a (TRAF-3) binding motif to activate the NF-kB, and NLRP3 regulating viral uptake and trafficking of protein to the plasma membrane or intracellular membranes using a cysteine-rich caveolin binding motif, a tyrosine-based sorting motif YXXφ responsible for Golgi to plasma membrane transport, and an SGD motif [60,61,89,90]. Products of ORF3a also inhibit STAT1 phosphorylation and thus interfering in the late antiviral response pathway [64]. Omicron's T223I(Threonine→Isoleucine) mutation is present all but in BA.1 subvariant doesn't reside in any of the mentioned domains and is towards the C terminus end. The mutation thus mightn't have any significant impact (Fig. 2).
ORF6 is a 61 amino acid long protein localized on the ER and Golgi membranes. ORF6 protein has IFN-antagonistic activity and interacts with the NUP98-RAE1 complex at the nuclear pore [[91], [92], [93], [94], [95]]. This protein inhibits cytoplasm-membrane trafficking by forming a complex and also blocks nuclear transportation of TF STAT and NF-kB complex. The methionine at 58 number residue is necessary for ORF6 functionality. In Omicrons BA.2, BA.2.12.2, and BA.4 subvariant, unique mutation D61L(Aspartic acid→Leucine) was observed (Table 1), close to key amino acid Met58 [92]. Hence, the mutation region might also be part of the functionality (Fig. 2). The mutation was even absent in BA.1 and BA.5 subvariants (Table 1).
ORF7b is a 43-amino acid long single transmembrane protein localized into the membrane of the Golgi apparatus. It has a role in immune modulation and can induce apoptosis [96]. Though the function of this protein is unknown, some literature states that this protein inhibits STAT signaling phosphorylation and ultimately interferes with downstream ISGs expression (Yang R, 2021). A unique mutation, L11F(Leucine→Phenylalanine), has been found in the transmembrane region of the Omicron protein (BA.4) that might impact signal modulation (Table 1 and Fig. 2).
ORF9b localizes on the membrane of mitochondria and suppresses type I interferon (IFN–I) responses through association with TOM70. It suppresses innate immunity by targeting MAVS signalosomes. It antagonizes type I and III interferons by targeting multiple components of the RIG‐I/MDA‐5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways [[23], [24], [25],49,[97], [98], [99]]. The region involved in interaction with TOM70 is 43–78 residues, but the N terminus has a role in stabilizing the TOM70-ORF9b structure. Omicron possesses a P10S(Proline→Serine) unique mutation and a sequential deletion E27-(Glutamic acid→deletion), N28-(Asparagine→deletion), A29-(Alanine→deletion) at the N-terminal site (Table 1), which may impact the protein's stability or interaction with other proteins in other signaling pathways (Fig. 2).
5. Remarks and future prospects on immune evasion, pathogenesis, and epistasis
The non-spike proteins have multiple significant roles in immune regulation, transcriptional regulation, and viral pathogenesis. But, there reside gaps in knowledge regarding their specific mode of action and roles, especially in terms of unique mutations of Omicron. However, there have been some reports on the effect of non-spike protein mutations on viral fitness and disease severity in both experimental and clinical settings [87,[100], [101], [102]]. The studied mutations however belong to other variants, such as the beta variant (B.1.351), except for the R203K + G204R mutation of N protein that is also found in the Omicron variant. Mutations in non-spike proteins may also lead to less clinical disease by impacting immune regulation and pathogenesis that can eventually increase transmission by extending time toward knowing an infection exists in a person. Here, we focus on unique mutations present in or close to the significant domain in the non-spike proteins of Omicron based on accessible pieces of literature and theoretical perspectives. One good ground of this approach is that detailed functions of different domains or regions of most non-spike proteins are still not well-studied. Thus, we cannot confidently comment on the exact impact of those mutations but rather mention the possible effects.
Epistasis occurs when more than one mutation in the same or different protein works together to create a new virus feature. Sometimes these mutations arise sequentially, and other times they co-emerge within a population. This event is evident for the Omicron variant regarding spike protein mutations [103]. With non-spike proteins, very few reports demonstrated the epistasis in experiments, albeit not with mutations of the Omicron variant. For example, with N and ORF8 proteins, ORF3a elicits the strongest antibody responses measured in sera from COVID-19 patients [100]. The Delta R203 M mutation in N protein has a statistically significant synergistic effect with L452R Spike mutation, and the growth rate of the 203K(N)/484K(S)/501Y(S) is consistently higher than the 203R(N)/484K(S)/501Y(S) variants. Supported by clinical data, the Spike D614G + NSP12 L323P may have increased the replication rate [104]. More importantly, this association of non-spike mutations found in other variants thus opens windows for more similar studies for the Omicron variant. The presence of unique mutations such as in NSP6, in M protein, and in ORFs, which sets the two subvariants apart, may also contribute to these impacts through a chain of epistatic effects [ [105]s]. Notably, there are however robust literatures on the plausible epistasis of non-spike mutations based on computational studies [88,[106], [107], [108]].
6. Conclusion
The factors determining the virus-host interaction should not only be confined to spike protein mutations that we observe in the SARS-CoV-2 variants and their subvariants. We should also consider mutations of non-spike proteins. The evasion of host immunity after natural infection or vaccination needs to escape from T-cell and other effector cells related mainly to the non-spike mutations. To understand the viruses' differential infectiousness and immune evasion strategies, we thus need to focus on the role of non-spike protein mutations. This review has summarized the non-spike mutations of the Omicron variant and its major dominant subvariants and then discussed the plausible impacts of those mutations on viral pathogenicity and immune evasion. Our review will initially help other researchers get all the necessary information regarding Omicron non-spike mutations with their probable functions. This review will also shed light on future research by exploiting the underlying mechanisms of non-spike mutations about their interaction with the host. Recently, infection cases due to particular subvariants are also gradually increasing compared to other subvariants, especially for BA.4 and BA.5 subvariants. Since they have no difference in spike proteins, the effects of their distinct non-spike mutations can not be neglected. Further research and review work on this aspect of fitness variation among not only variants but also subvariants should answer our questions more specifically. These studies and reviews on non-spike mutations will be critical for developing and deploying more effective therapeutic and preventive tools such as novel antivirals and vaccines, thus impacting public health.
CRediT authorship contribution statement
Anamica Hossain: Writing – original draft, Investigation, Formal analysis. Shammi Akter: Investigation, Visualization, Writing – original draft. Alfi Anjum Rashid: Investigation, Formal analysis. Sabik Khair: Software, Methodology, Visualization. A.S.M. Rubayet Ul Alam: Validation, Supervision, Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., Li B., Zhang W., Wang L.F., Shi Z.L., Daszak P. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19 . The COVID-19 resource centre is hosted on Elsevier Connect , the company' s public news and information. BBA - Mol. Basis Dis. 2020:1–17. [Google Scholar]
- 3.Lu R., Wang Y., Wang W., Nie K., Zhao Y., Su J., Deng Y., Zhou W., Li Y., Wang H., Wang W., Ke C., Ma X., Wu G., Tan W. Complete genome sequence of Middle East respiratory syndrome coronavirus (MERS-CoV) from the first imported MERS-CoV case in China. Genome Announc. 2015;3 doi: 10.1128/genomeA.00818-15. 2014–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., Prashar V., Gupta G.D., Panicker L., Kumar M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung C., Kmiec D., Koepke L., Zech F., Jacob T., Sparrer K.M.J., Kirchhoff F. Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J. Virol. 2022;96 doi: 10.1128/jvi.02077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martins V., Amorim D.F., De Souza R.F., De Souza A.S. 2022. Molecular Dynamics Simulations of the Spike Trimeric Ectodomain of the SARS-CoV-2 Omicron Variant : Structural Relationships with Infectivity , Evasion to Immune System and Transmissibility. 0–16. [DOI] [PubMed] [Google Scholar]
- 7.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., Menachery V.D., Weaver S.C., Dormitzer P.R., Shi P.Y. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Wang T., Zhang J., Shao B., Gong H., Wang Y., He X., Liu S., Liu T.Y. Exploring the regulatory function of the N-terminal domain of SARS-CoV-2 spike protein through molecular dynamics simulation. Adv. Theory Simulations. 2021;4:1–13. doi: 10.1002/adts.202100152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., Ren Y., Quan L., Zhao W., Seto D., Chodosh J., Luo Z., Wu J., Zhang Q. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Targeted Ther. 2022;7 doi: 10.1038/s41392-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan L., Zheng Q., Zhang H., Niu Y., Lou Y. The SARS-CoV-2 spike glycoprotein biosynthesis , structure , function , and antigenicity : implications for the design of spike-based vaccine. Immunogens. 2020;11:1–12. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaway E. Omicron variant puts. Nature. 2021;600:21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 13.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Q., Meng M., Kumar R., Wu Y., Huang J., Lian N., Deng Y., Lin S., Chow N., Fleming-Dutra K., Gierke R., Hall A., Hughes M., Pilishvili T., Ritchey M., Roguski K., Skoff T., Ussery E., Faure E., Kipnis E., Bortolotti P., Salik J., Cai G., Bossé Y., Xiao F., Kheradmand F., Mph E.M.A., Jong G.W., Yang C.L. C or r e sp ondence Crystallopathies. J. Med. Virol. 2020;69:2016–2017. http://www.ncbi.nlm.nih.gov/pubmed/32293753 [Google Scholar]
- 15.Wang Q., Guo Y., Iketani S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chan J.Y., Shah J.G., Nguyen N., Chen Z., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D. BioRxiv; 2022. Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. 2022.05.26.493517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P., Kumar A., Garg N., Giri R. An insight into SARS-CoV-2 membrane protein interaction with spike, envelope, and nucleocapsid proteins. J. Biomol. Struct. Dyn. 2021 doi: 10.1080/07391102.2021.2016490. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh Y.C., Li H.C., Chen S.C., Lo S.Y. Interactions between M protein and other structural proteins of severe, acute respiratory syndrome-associated coronavirus. J. Biomed. Sci. 2008;15:707–717. doi: 10.1007/s11373-008-9278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X., Liu Y., Huang Z., Xu W., Hu W., Yi L., Liu Z., Chan H., Zeng J., Liu X., Chen H., Yu J., Chan F.K.L., Ng S.C., Wong S.H., Wang M.H., Gin T., Joynt G.M., Hui D.S.C., Zou X., Shu Y., Cheng C.H.K., Fang S., Luo H., Lu J., Chan M.T.V., Zhang L., Wu W.K.K. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022 doi: 10.1038/s41418-021-00916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. vol. 1. 2011. pp. 1335–1347. (Coronavirus Nsp6 Proteins Generate Autophagosomes from the Endoplasmic Reticulum via an Omegasome Intermediate). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.wei Jiang H., nan Zhang H., feng Meng Q., Xie J., Li Y., Chen H., xiao Zheng Y., ning Wang X., Qi H., Zhang J., Wang P.H., Han Z.G., ce Tao S. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70, Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stukalov A., Girault V., Grass V., Bergant V., Karayel O., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., Hamad S.M., Piras A., Tanzer M., Hansen F.M., Engleitner T., Reinecke M., Lavacca T.M., Ehmann R., Wölfel R., Jores J., Küster B., Protzer U., Rad R., Ziebuhr J., Thiel V., Scaturro P., Mann M., Pichlmair A. BioRxiv; 2020. Multi-level Proteomics Reveals Host-Perturbation Strategies of SARS-CoV-2 and SARS-CoV. 2020.06.17.156455. [DOI] [PubMed] [Google Scholar]
- 23.Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., Zhong T., Tang H., Du W., Wang L., Wo J., Mu J., Qiu Y., Yang K., Zhang L.K., Ye B.C., Qi N. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L., Zhuang M.W., Deng J., Zheng Y., Zhang J., Nan M.L., Zhang X.J., Gao C., Wang P.H. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. J. Med. Virol. 2021;93:5376–5389. doi: 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Hadjadj, N. Yatim, L. Barnabei, A. Corneau, J. Boussier, E (2 , 6 , 7)., 724 (2020) 718–724.
- 26.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderven H.A., Ana-Sosa-Batiz F., Jegaskanda S., Rockman S., Laurie K., Barr I., Chen W., Wines B., Hogarth P.M., Lambe T., Gilbert S.C., Parsons M.S., Kent S.J. What lies beneath: antibody dependent natural killer cell activation by antibodies to internal influenza virus proteins. EBioMedicine. 2016;8:277–290. doi: 10.1016/j.ebiom.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribero M.S., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:1–22. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ca F., Sabberwal P., Jc W., Ejd G., Twm C., Zelek W., Ir H., Merrick B., Doores K., Sj W., Pj L., Ecy W., Rj S. 2021. ADNKA Overcomes SARS-CoV2-Mediated NK Cell Inhibition through Non-spike Antibodies. [Google Scholar]
- 30.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 31.Weisser M., Schäfer T., Leibundgut M., Böhringer D., Aylett C.H.S., Ban N. Structural and functional insights into human Re-initiation complexes. Mol. Cell. 2017;67:447–456. doi: 10.1016/j.molcel.2017.06.032. e7. [DOI] [PubMed] [Google Scholar]
- 32.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai Y., Kawachi K., Terada Y., Omori H., Matsuura Y., Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorkhali R., Koirala P., Rijal S., Mainali A., Baral A., Bhattarai H.K. Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinf. Biol. Insights. 2021;15 doi: 10.1177/11779322211025876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/jvi.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/jvi.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadlage M.J., Sparks J.S., Beachboard D.C., Cox R.G., Doyle J.D., Stobart C.C., Denison M.R. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 2010;84:280–290. doi: 10.1128/jvi.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saramago M., Bárria C., Costa V.G., Souza C.S., Viegas S.C., Domingues S., Lousa D., Soares C.M., Arraiano C.M., Matos R.G. New targets for drug design: importance of nsp14/nsp10 complex formation for the 3’-5' exoribonucleolytic activity on SARS-CoV-2. FEBS J. 2021;288:5130–5147. doi: 10.1111/febs.15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/jvi.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/jvi.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graziadei A., Schildhauer F., Spahn C., Kraushar M. 2022. SARS-CoV-2 Nsp1 N-Terminal and Linker Regions as a Platform for Host Translational Shutoff; pp. 1–28. [Google Scholar]
- 44.Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., Hobman T.C., Ptak C., Gao B., Minna J.D., Chen Z., García-Sastre A., Ren Y., Wozniak R.W., Fontoura B.M.A. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7:1–13. doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown A., Baird M.R., Yip M.C.J., Murray J., Shao S. Structures of translationally inactive mammalian ribosomes. Elife. 2018;7:1–18. doi: 10.7554/eLife.40486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J.M., Reggiori F., De Haan C.A.M. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/jvi.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/jvi.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan J., Kusov Y., Mutschall D., Tech S., Nagarajan K., Hilgenfeld R., Schmidt C.L. The "SARS-unique domain" (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 2007;364:877–882. doi: 10.1016/j.bbrc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eriksson K.K., Cervantes-Barragán L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/jvi.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lécine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/jvi.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J.M., de Haan C.A.M. Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 2011;85:4572–4577. doi: 10.1128/jvi.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bensussen A., Álvarez-Buylla E.R., Díaz J. SARS-CoV-2 Nsp5 protein causes acute lung inflammation, A dynamical mathematical model. Front. Syst. Biol. 2021;1:1–16. doi: 10.3389/fsysb.2021.764155. [DOI] [Google Scholar]
- 59.Nakagawa K., Lokugamage K.G., Makino S. first ed. Elsevier Inc.; 2016. Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bignon E., Marazzi M., Grandemange S., Monari A. Autophagy and evasion of the immune system by SARS-CoV-2. Structural features of the non-structural protein 6 from wild type and Omicron viral strains interacting with a model lipid bilayer. Chem. Sci. 2022;13:6098–6105. doi: 10.1039/d2sc00108j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newman J.A., Douangamath A., Yadzani S., Yosaatmadja Y., Aimon A., Brandão-Neto J., Dunnett L., Gorrie-stone T., Skyner R., Fearon D., Schapira M., von Delft F., Gileadi O. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beyer D.K., Forero A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J.C., Debarnot C., Borg J.P., Bouvet M., Canard B., Morelli X., Lécine P. Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 2010;285:33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin X., Chen Y., Sun Y., Zeng C., Wang Y., Tao J., Wu A., Yu X., Zhang Z., Tian J., Guo D. Characterization of the guanine-N7 methyltransferase activity of coronavirus nsp14 on nucleotide GTP. Virus Res. 2013;176:45–52. doi: 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saramago M., Peregrina A., Robledo M., Matos R.G., Hilker R., Serrania J., Becker A., Arraiano C.M., Jiménez-Zurdo J.I. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res. 2017;45:1371–1391. doi: 10.1093/nar/gkw1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mers-cov R., Ogando N.S., Zevenhoven-dobbe J.C., Van Der Meer Y., Bredenbeek P.J. Vol. 94. 2020. crossm The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical; pp. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solis-moreira B.J. 2022. Study Identi Fi Es Current Mechanism of Action behind Reduced Severity of Omicron Variant COVID - 19 Infection; pp. 1–4. [Google Scholar]
- 74.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85:5794–5803. doi: 10.1128/jvi.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Álvarez E., DeDiego M.L., Nieto-Torres J.L., Jiménez-Guardeño J.M., Marcos-Villar L., Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mahtarin R., Islam S., Islam M.J., Ullah M.O., Ali M.A., Halim M.A. Structure and dynamics of membrane protein in SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1861983. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas S. The structure of the membrane protein of sars-cov-2 resembles the sugar transporter semisweet. Pathog. Immun. 2020;5:342–363. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y.Z., Wang S.Y., Zheng Z.Q., Huang Yi, Li W.W., Xu Z.S., Wang Y.Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 2021;18:613–620. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marques-Pereira C., Pires M.N., Gouveia R.P., Pereira N.N., Caniceiro A.B., Rosário-Ferreira N., Moreira I.S. SARS-CoV-2 membrane protein: from genomic data to structural new insights. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23062986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jakhmola S., Indari O., Kashyap D., Varshney N., Das A., Manivannan E., Jha H.C. Mutational analysis of structural proteins of SARS-CoV-2. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ingraham N.E., Ingbar D.H. The omicron variant of SARS‐CoV‐2: understanding the known and living with unknowns. Clin. Transl. Med. 2021;11:1–6. doi: 10.1002/ctm2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kannan S.R., Spratt A.N., Sharma K., Chand H.S., Byrareddy S.N., Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022;126 doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bai Z., Cao Y., Liu W., Li J. Structure , biological functions , and a potential target for drug. Viruses. 2021;13:1–13. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang M., He S., Chen X., Huang Z., Zhou Z., Zhou Z., Chen Q., Chen S., Kang S. Structural insight into the SARS-CoV-2 nucleocapsid protein C-terminal domain reveals a novel recognition mechanism for viral transcriptional regulatory sequences. Front. Chem. 2021;8:1–12. doi: 10.3389/fchem.2020.624765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rahman M.S., Islam M.R., Alam A.S.M.R.U., Islam I., Hoque M.N., Akter S., Rahaman M.M., Sultana M., Hossain M.A. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021;93:2177–2195. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- 87.Mourier T., Shuaib M., Hala S., Mfarrej S., Alofi F., Naeem R., Alsomali A., Jorgensen D., Subudhi A.K., Ben Rached F., Guan Q., Salunke R.P., Ooi A., Esau L., Douvropoulou O., Nugmanova R., Perumal S., Zhang H., Rajan I., Al-Omari A., Salih S., Shamsan A., Al Mutair A., Taha J., Alahmadi A., Khotani N., Alhamss A., Mahmoud A., Alquthami K., Dageeg A., Khogeer A., Hashem A.M., Moraga P., Volz E., Almontashiri N., Pain A. SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-28287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson B.A., Zhou Y., Lokugamage K.G., Vu M.N., Bopp N., Crocquet-Valdes P.A., Kalveram B., Schindewolf C., Liu Y., Scharton D., Plante J.A., Xie X., Aguilar P., Weaver S.C., Shi P.-Y., Walker D.H., Routh A.L., Plante K.S., Menachery V.D. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minakshi R., Padhan K. The YXXΦ motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport. Virol. J. 2014;11:1–10. doi: 10.1186/1743-422X-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K.W., Chan H.Y.E. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajani K.R., Pettit Kneller E.L., McKenzie M.O., Horita D.A., Chou J.W., Lyles D.S. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Addetia A., Lieberman N.A.P., Phung Q., Hsiang T.Y., Xie H., Roychoudhury P., Shrestha L., Loprieno M.A., Huang M.L., Gale M., Jerome K.R., Greninger A.L. Sars-cov-2 orf6 disrupts bidirectional nucleocytoplasmic transport through interactions with rae1 and nup98. mBio. 2021;12:1–16. doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pillon M.C., Frazier M.N., Dillard L.B., Williams J.G., Kocaman S., Krahn J.M., Perera L., Hayne C.K., Gordon J., Stewart Z.D., Sobhany M., Deterding L.J., Hsu A.L., Dandey V.P., Borgnia M.J., Stanley R.E. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-020-20608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., García-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang R., Zhao Q., Rao J., Zeng F., Yuan S., Ji M., Sun X., Li J., Yang J., Cui J., Jin Z., Liu L., Liu Z. SARS-CoV-2 accessory protein ORF7b mediates tumor Necrosis factor-α-induced apoptosis in cells. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.654709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao X., Zhu K., Qin B., Olieric V., Wang M., Cui S. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-23118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li J., Liu Y., Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/jvi.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGrath M., Xue Y., Oldfield L., Assad-Garcia N., Zaveri J., Singh N., Baracco L., Taylor L., Vashee S., Frieman M. SARS-CoV-2 Variant Spike and accessory gene mutations alter pathogenesis. BioRxiv Prepr. Serv. Biol. 2022 doi: 10.1073/pnas.2204717119. https://biorxiv.org/cgi/content/short/2022.05.31.494211v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kern D.M., Sorum B., Mali S.S., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Brohawn S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zinzula L. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:116–124. doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moulana A., Dupic T., Phillips A.M., Chang J., Nieves S., Roffler A.A., Greaney A.J., Starr T.N., Bloom J.D., Desai M.M. vol. 1. BioRxiv; 2022. https://www.biorxiv.org/content/10.1101/2022.06.17.496635v1%0Ahttps://www.biorxiv.org/content/10.1101/2022.06.17.496635v1.abstract (Compensatory Epistasis Maintains ACE2 Affinity in SARS-CoV-2 Omicron BA). 2022.06.17.496635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garvin M.R., Prates E.T., Pavicic M., Jones P., Amos B.K., Geiger A., Shah M.B., Streich J., Felipe Machado Gazolla J.G., Kainer D., Cliff A., Romero J., Keith N., Brown J.B., Jacobson D. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020;21:1–26. doi: 10.1186/s13059-020-02191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Statement on Omicron Sublineage BA.2. WHO; 2022. https://www.who.int/news/item/22-02-2022-statement-on-omicron-sublineage-ba.2 [Google Scholar]
- 106.Rodriguez-Rivas J., Croce G., Muscat M., Weigt M. Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2113118119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Obermeyer F., Jankowiak M., Barkas N., Schaffner S.F., Pyle J.D., Yurkovetskiy L., Bosso M., Park D.J., Babadi M. vol. 1332. 2022. pp. 1327–1332. (Analysis of 6.4 Million SARS-CoV-2 Genomes Identifies Mutations Associated with Fitness). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Velazquez-Salinas L., Zarate S., Eberl S., Gladue D.P., Novella I., Borca M.V. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.550674. [DOI] [PMC free article] [PubMed] [Google Scholar]