Abstract
There is a massive demand to identify alternative methods to detect new cases of COVID-19 as well as to investigate the epidemiology of the disease. In many countries, importation of commercial kits poses a significant impact on their testing capacity and increases the costs for the public health system. We have developed an ELISA to detect IgG antibodies against SARS-CoV-2 using a recombinant viral nucleocapsid (rN) protein expressed in E. coli. Using a total of 894 clinical samples we showed that the rN-ELISA was able to detect IgG antibodies against SARS-CoV-2 with high sensitivity (97.5%) and specificity (96.3%) when compared to a commercial antibody test. After three external validation studies, we showed that the test accuracy was higher than 90%. The rN-ELISA IgG kit constitutes a convenient and specific method for the large-scale determination of SARS-CoV-2 antibodies in human sera with high reliability.
Keywords: SARS-CoV-2, COVID-19, Diagnosis, Serological assay, ELISA, Prototyping
1. Background
SARS-CoV-2, a member of the Coronaviridae family, is the causative agent of COVID-19, a disease marked by the occurrence of severe acute respiratory syndrome in many of the infected patients. Emergence of virus variants has brought more complexity to this scenario, leading to an increase in the number of cases challenging the effectiveness of vaccines that are in use against COVID-19 [1], [2], [3].
As of July 2022, the disease reached the astonishing mark of 555 million people affected worldwide and has caused more than 6.3 million deaths [4]. Taking into account that the detection of antibodies is particularly relevant during later stages of infection and the rate of asymptomatic infections and the extent of the disease transmission, serologic assays are needed for epidemiologic studies and surveillance [5].
SARS-CoV-2 has four structural proteins: Spike (S,) Envelope (E), Membrane (M), and Nucleocapsid (N). The S protein, which is cleaved into S1 (containing the receptor binding domain, RBD) and S2 (further cleaved into S2’ to form the viral fusion peptide) subunits is critical for viral entry and has been described as a neutralizing target [6,7]. Besides the S protein, the SARS-CoV-2 N protein has been described as an immunodominant antigen in studies with COVID-19 patients. This antigen is highly expressed during infection [8], [9], [10], making it a suitable antigen to be included in serology-based diagnostics [11], [12], [13].
The different vaccines being applied in Brazil have varying degrees of immunogenicity and provisional protective efficacy, and their durability over time is not yet known. Therefore, serological tests may be relevant as an assessment tool after vaccination and, in the cases of vaccines targeting spike glycoprotein from SARS-CoV-2 (mRNA and adenovirus vectors encoding the S protein of SARS-CoV-2), an anti-N ELISA can differentiate immune response between vaccinated and convalescent individuals [14].
Due to the urgency and high demand, many serological tests to diagnose COVID-19 have been rapidly developed-and made available on the market, with validation often limited to testing relatively few clinical samples [15], and some commercial ELISA kits have been validated worldwide [16] and approved by health regulatory agencies. However, none of the currently available serological kits are fully nationalized in several countries, which makes the products more expensive, less accessible, and subject to availability fluctuation due to international demands and importation difficulties. This is particularly critical in Brazil, considering that the country is the third in the world in number of cases, and the second in terms of death toll [4].
We aimed to developed a new ELISA kit to help COVID-19 diagnosis (henceforth named EIE COVID-19 IgG kit) through the detection of IgG antibodies against the SARS-CoV-2 nucleocapsid (N) protein.
2. Study design
2.1. Antigen production
The full length coding region of the nucleocapsid (N) gene of SARS-CoV-2 (Genebank accession number: MT126808.1) was codon-optimized, subcloned into pET-24a-(+) expression vector and used to transform E.coli BL21(DE3) strain. Plasmid-positive clones were cultivated in LB medium, cultures were induced with IPTG (0,5 mM, 4 h) and the recombinant protein was purified by affinity chromatography using nickel columns in an AKTAprime plus system following manufacturer's instructions (GE Healthcare, USA).
2.2. Sera bank and ethical considerations
The use of sera samples from patients and healthy volunteers was approved by the UFMG's Ethics Committee and by the National Research Ethics’ Committee (CAAE: 1686320.0.0000.5149). Negative sera obtained before 2020 are from healthy donors, and sera obtained after 2020 are from individuals who tested negative for the viral RNA screening test qRT-PCR (nasal swab). Positive samples were selected based on a reported history of a positive SARS-CoV-2 PCR nasal swab, or on results from a rapid test-dual path platform (DPP) COVID-19 IgM/IgG, according to the supplier's instructions (Bio-Manguinhos, Fiocruz, Brazil). The TR DPP® COVID-19 IgM/IgG - Bio-Manguinhos uses a combination of SARS-CoV-2 antigens bound to a membrane (solid phase), specific antibodies and protein A conjugate with colloidal gold particles.
Considering samples from patients confirmed by positive qPCR result (+qPCR) and used during the internal validation (n=54), 20.4% (n=11) were from hospitalized individuals and 79,6% (n=43) were from mildly symptomatic, non-hospitalized patients. No clinical information about the hospitalized patients were provided. Information on positive mildly symptomatic patients is available (Supplementary data Table S1). The median time between the PCR results and the first blood collection (out of serial collections) was 7 (± 1) days.
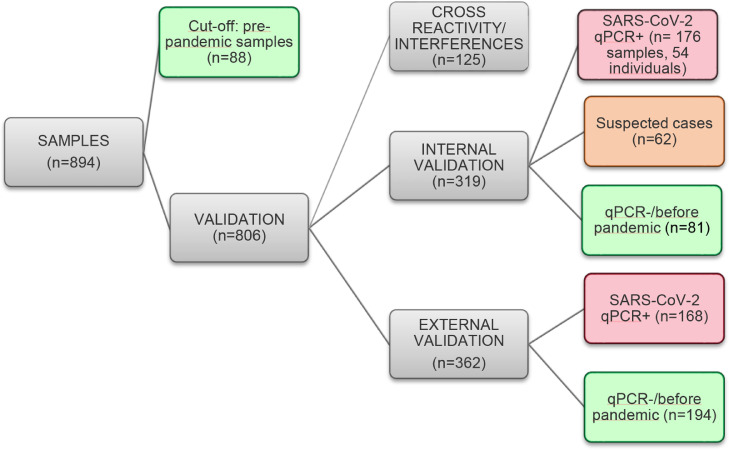
In all, 894 samples were tested, 362 from SARS-CoV-2-positive patients (240 individuals), 407 from SARS-CoV-2-negative donors and 125 samples from patients bearing other different conditions (patients that were seropositive for other viruses, homolyzed sera, icteric sera and sera with increased levels of rheumatoid factors). Fig. 1 summarizes how samples were used for the rN-ELISA kit validation.
Fig. 1.
Sera samples used during rN-ELISA validation. Experiments are categorized and showed in gray, positive samples (qPCR or DPP) in red, negative samples (negative PCR or pre-pandemic) in green and suspected cases (negative or no PCR data, but that had contact with confirmed COVID-19 individuals) in orange. The rN-ELISA sensitivity and specificity during internal validation included samples from healthy donors (n = 81) and qPCR positive samples from hospitalized (n = 11) and non-hospitalized individual (n = 43). Agreement between rN-ELISA and DPP included: all qPCR+ samples (n = 176), suspected cases (n = 62) and 23 healthy donors tested by DPP. The positive agreement to qPCR was calculated using samples from 43 individuals (n = 157 samples, with information about the time after the qPCR confirmation).
2.3. ELISA
The ELISA was performed by coating plates of polystyrene (Costar, USA) with the recombinant antigen diluted in carbonate buffer to a final concentration of 4 μg/mL (overnight at 4°C). Wells were blocked (PBS with 1% bovine serum albumin) for 2 h at room temperature (25 ± 2°C). For each assay, samples diluted (1:101) in PBS-T (0.05% Tween-20) were added and incubated for 30 min at 37°C. After five washes (PBS-T, 1% Tween-20), the conjugate (1:5.000 in diluent Moss, horseradish peroxidase HRP-conjugated anti-human IgG goat immunoglobulin, Fapon, China) was added and the plates were further incubated (30 min at 37°C). After further washing, reactions were revealed using TMB (3,3′,5,5;-tetramethylbenzidine, Moss, USA) for 15 min, and H2SO4 (0,5 M) was added to stop reactions. Plates were analyzed in a Microplate Reader at optical density (OD) of 450 nm.
A cut-off value was determined using 88 samples from healthy donors (obtained before 2020) according to equation 1:
| (1) |
Average of negative samples (OD)
standard deviation of negative samples (OD)
Positive and negative controls were prepared using heat-inactivated samples (56°C, 30 min) [17].
The cut-off of the following tests was based on a positive control (C+). This standard consisted of pooled plasma from 10 seropositive individuals confirmed by DPP and was used as a calibrant according to equation 2:
| (2) |
The internal evaluation of accuracy included the testing of 135 serum samples, from patients admitted to a hospital in Belo Horizonte (Minas Gerais, Brazil) and presenting positive PCR for COVID-19 (n = 11); sera from non-hospitalized individuals (symptomatic or oligosymptomatic) presenting positive PCR for COVID-19 (n = 43, 13 ± 2,2 days post PCR confirmation); sera from healthy donors (taken after 2020) who tested negative for SARS-CoV-2 nasal swab PCR (n = 38); and sera from healthy donors who sampled blood before the COVID19 emergence (before 2020) (n = 43). P values were determined through unpaired, two-sided Mann–Whitney U-test. To all validation assays, an index (I) for each sample was calculated, according to equation 3:
| (3) |
The results were classified as: non-reactive (I < 0.8), borderline (0.8 ≤ I < 1.1) or reactive (I ≥ 1.1).
2.4. Statistical analysis
The analyses were done using the GraphPad Prism program (version 8.0 for Windows). ROC curves were constructed with indexes values of the positive versus negative samples. The diagnostic performance was evaluated by estimation of sensitivity, specificity, area under the curve (AUC). Confidence intervals (CIs) were defined with 95% confidence level (95% CI). P values were determined through unpaired, two-sided Mann–Whitney U-test, and P < 0.05 values were considered significant.
3. Results
3.1. rN-ELISA accuracy
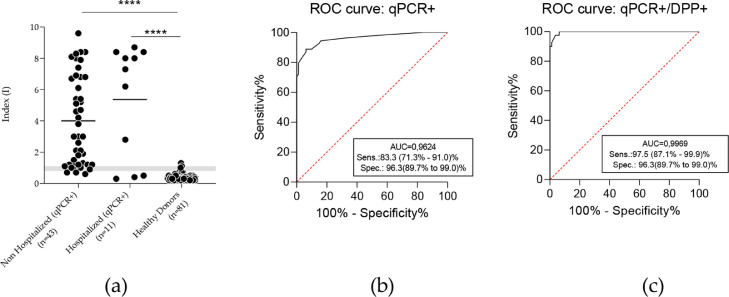
The full-length recombinant N protein (MW 48 KDa), was expressed at high yields in E. coli BL21(DE3) (60 mg of purified protein/mL of bacterial culture), and obtained at high purity grade (data not shown). The cut-off value for the optical density obtained with the rN-ELISA test was defined as three standard deviations greater than the average OD450 of 88 negative samples (obtained before 2020, from healthy individuals) (cut-off=0.426, Supplementary materials, Table S2). Besides these samples, 81 negative sera were tested (before 2020, or after 2020 with PCR negative results with two-weeks intervals) (Supplementary materials, Table S3), 76 of them presented a non-reactive result (Index<0.8, with Index, or I=OD450nm/cut-off), two presented a reactive result (I>1.1) and three were classified as borderline samples (0.8≤ I <1.1), corresponding to a 96.3% (95% CI: 89.7-99.0%) specificity for the test. Of all samples that tested positive by qRT-PCR (n = 54, 13 ± 2 days post molecular confirmation), 44 presented positive results (I ≥ 1.1), four were classified as borderline samples, and six presented negative results (Supplementary materials, Table S4). These resulted on a sensitivity of 83,3% (CI: 71.3-91.0%) for the assay. Sensitivity increased to 97.5% (87.1-99.9) % when we considered rapid test DPP-confirmed positive samples (38/40, two borderline samples). Results were further supported by ROC curves (Fig. 2 ).
Fig. 2.
Evaluation of the accuracy of anti-SARS-CoV-2 rN-ELISA IgG kit. (a) The sensitivity and specificity of the rN antigen were calculated according to the index in ELISA and confirmed by ROC curve. A comparison between qPCR positive patients and healthy donors showed significant differences between groups (p < 0,0001, by Mann Whitney test). Grey zone (Index ranging from 0.8 to 1,09) indicates borderline results. (b) ROC curve considering all PCR positive results, including antibody non detected by DPP. (c) Analysis considering PCR positive patients with positive antibody confirmation by DPP (serological reference test).
3.2. Cross-reactivity (analytical specificity) and interferences
A cross-reactivity study was performed with a panel of clinical samples from patients infected with other, non-SARS-CoV2, relevant human viruses. Low cross-reactivity (1/19) was observed with influenza antibody-positive samples. Cross-reactions to other human pathogenic viruses were not observed. Negative samples with high levels of rheumatoid factors, hemoglobin and bilirubin were also tested and did not cause any detectable interference in the rN-ELISA IgG kit detection capability (Table 1 ).
Table 1.
Sera bank tested with rN-ELISA IgG kit in cross-reactivity and interference studies.
| rN-ELISA IgG kit |
|||
|---|---|---|---|
| PANEL | N | Negatives | % Negatives |
| Fresh vaccination against Influenzaa | 8 | 8 | 100% |
| Influenza antibody positiveb | 19 | 18 | 95% |
| Measles antibody positiveb | 15 | 15 | 100% |
| Measles PCR positivec | 10 | 10 | 100% |
| Human Parvovirus antibody positivec | 6 | 6 | 100% |
| Chikungunya antibody positivea | 8 | 8 | 100% |
| Dengue antibody positivea | 8 | 8 | 100% |
| Zika antibody positivea | 8 | 8 | 100% |
| Yellow Fever antibody positivec | 13 | 13 | 100% |
| Rheumatoid factorsa | 8 | 8 | 100% |
| Hemolytica | 11 | 11 | 100% |
| Icterica | 11 | 11 | 100% |
Tested at CT-Vacinas (UFMG).
Tested at Laboratory of Respiratory Viruses and Measles (Fiocruz-RJ).
Tested at Laboratory of Virology (USP).
3.4. Clinical performance evaluation of the rN-ELISA IgG kit
Forty-three individuals who tested positive for SARS-CoV-2 RNA through qRT-PCR were followed for up to 80 days and assayed by the DPP rapid test and our rN-ELISA IgG kit. Of these, 19 presented a positive result for one or both serological test at the first blood collection and 24 patients were initially seronegative and then seroconverted during the follow up period. The median day of seroconversion for rN-ELISA IgG kit and DPP (IgG) was 14 days post PCR confirmation. All patients presented seroconversion as evaluated by the developed the rN-ELISA IgG kit within a maximum of 21 days after PCR confirmation (Supplementary materials, Table S5).
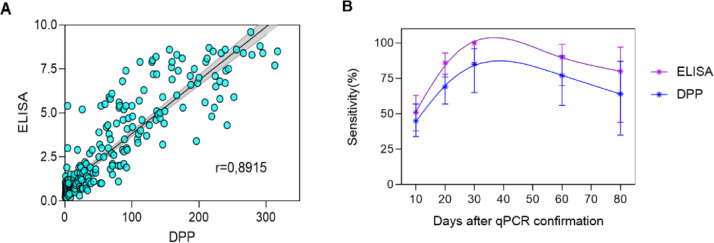
The positive agreement to PCR was evaluated using 157 serum samples, collected at different time-points. Considering borderline results as negative, the rN-ELISA IgG kit displayed a 48% (32/66) positive agreement to PCR before 10 days post molecular confirmation, 82% (54/66) from 11 to 20 days, and 100.0% (25/25) after 21 days. The DPP rapid test (serological reference method) displayed a 47% (31/66) positive agreement to PCR before 10 days post confirmation, from 11 to 20 days post-PCR confirmation, the positive agreement was 73% (48/66) and after 21 days, the positive agreement to PCR was 84% (21/25) (Table 2 ).
Table 2.
Positive agreement to PCR of anti-SARS-CoV-2 ELISA according to time.
| r-N ELISA IgG | |||||
|---|---|---|---|---|---|
| Days post PCR confirmation | n | Neg | Pos | Bord* | Positive (%) agreement to PCR (95% CI) |
| ≤10 | 66 | 30 | 32 | 4 | 48 % (35.99-61.12%) |
| (11-20) | 66 | 5 | 54 | 7 | 82 % (70.39-0.24%) |
| ≥21 | 25 | 0 | 25 | 0 | 100 % (82.28-100.0%) |
| DPP IgG | |||||
| Days post PCR confirmation | n | Neg | Pos | Positive (%) agreement to PCR (95% CI) | |
| ≤10 | 66 | 35 | 31 | 47% (34.56-59.66%) | |
| (11-20) | 66 | 18 | 48 | 73% (60.36-82.97%) | |
| ≥21 | 25 | 4 | 21 | 84% (63.92-95.46%) | |
Borderline counted as negative.
A total of 261 samples were used to evaluate the agreement between the rN-ELISA IgG and DPP (serological reference), including samples at different times post-infection from non-hospitalized individuals (n = 157, from 43 patients after positive nasal swab PCR); hospitalized patients (n = 19, from 11 individuals after positive nasal swab PCR); samples from healthy donors with a negative nasal swab PCR (n = 23) and suspected cases (negative or no qPCR data, but that had contact with confirmed COVID-19 individuals, n = 62) (Supplementary materials, Tables S5 and S6). A strong correlation (r = 0,8915) was observed when we compared ELISA index and DPP signal, with the ELISA presenting a higher sensitivity in all timepoints (Fig. 3 ).
Fig. 3.
Comparison between ELISA and DPP (serologial reference). A: Linear regression of ELISA index and DPP signal with 261 samples, including COVID-19 patients and healthy donors. B: Follow up of 43 patients after confirmation of COVID-19 by nasal swab PCR using ELISA and DPP.
Considering samples with a positive result from both PCR and the serological reference test (DPP), the rN-ELISA IgG presented a sensitivity of 98.4% (CI:94.3-99.8%) (123/125, three borderline results, 0,8≤Index<1,1) and specificity of 100% (CI:88.8-00%) (31/32, one borderline result) based on PCR negative nasal swab or non-reagent DPP. Moreover, among the qPCR+/DPP-, the rN-ELISA IgG reacted against 20 samples. No false-positive tests (considering healthy donor with negative PCR) were observed. These results returned a K = 0.775 (95% CI: 0.697-0.854), indicating a substantial agreement between both tests [18]. rN-ELISA IgG borderline results (n = 19, three reactive by DPP and 16 non-reactive by DPP) were not included (Table 3 ).
Table 3.
Agreement between the rN-ELISA and DPP (serological reference) to detect IgG against SARS-CoV-2.
| rN-ELISA IgG | ||||
|---|---|---|---|---|
| Neg | Pos | Total | ||
| DPP COVID-19 (IgG) |
Neg | 92 | 25 | 117 |
| Pos | 2 | 123 | 125 | |
| Total | 94 | 148 | 242 | |
| Kappa: 0.775 (95% CI: 0.697-0.854), observed agreements: 215 (88,8% of the observations). | ||||
3.6. Independent clinical agreement validation study
The rN-ELISA IgG kit was independently tested at three external laboratories: Laboratory of Virology, at the University of São Paulo (USP), Laboratory of Respiratory Viruses and Measles and Laboratory of Diagnostic Technology, both at the Oswaldo Cruz Foundation (Fiocruz-RJ). The test was validated against a panel of previously characterized samples consisting of SARS-CoV-2 PCR and/or antibody-positive samples (n = 168) and PCR negative (or prepandemic samples) (n = 194). The rN-N ELISA sensitivity ranged from 84.6% to 95.7% and specificity from 97.7% to 100%. The test accuracy was higher than 90% in all three studies (Table 4 ).
Table 4.
Independent clinical agreement validation study using rN-ELISA IgG.
| Positivesamples | Negativesamples | Sensitivity %(CI 95%) | Specificity %(CI 95%) | Accuracy %(CI 95%) | |
|---|---|---|---|---|---|
| Studya | qPCR + (n = 52) | Before pandemic (n = 36) | 84.6 (72.5-92.0) % | 100(90.4-100.0)% | 93.2(85.8-97.5)% |
| Studyb | qPCR + (n = 68) | PCR- (n = 30) | 88.2 (78.5-93.9) % | 100(88.7-100.0)% | 91.3(82.8-96.4)% |
| Studyc | qPCR+/DPP+ (n = 48) | Before pandemic (n = 128) | 95.7 (85.7-99.5) % | 97.7(93.3-99.5)% | 96.0(92.0-98.4)% |
Laboratory of Virology (USP).
Laboratory of Respiratory Viruses and Measles (Fiocruz-RJ).
Laboratory of Diagnostic Technology (Fiocruz-RJ).
3.7. Long term stability study
To assess the rN-ELISA IgG kit stability, plates coated with the N recombinant protein and all the reagents were stored at -5°C for twelve months and tested every three months. At the end of the storage period, the overall drop rate of the kit was 5% (95% CI: 2-8%) and the samples showed no change to their previous classification (Figure S7).
4. Discussion
Serological tests, such as Enzyme-linked immunosorbent assay (ELISA) and rapid tests available in Brazil are dependent on imported technology. This is also true for many developing countries where the pandemic has hit hard. The main imported raw material is the antigen, which makes the diagnosis more expensive, less accessible and subject to availability fluctuation due to international demands. Here, we described the development of a serological assay to detect IgG antibodies against SARS-CoV-2 infection. For the production of the recombinant antigen, a crucial step in terms of performance and costs, we used a prokaryotic system because, in terms of scale production, bacteria-made proteins present cost-effective advantages when compared to eukaryotic systems, including higher productivity and reduced costs [19] Indeed, commercial kits range from 25 to 60 times more expensive than our kit. Most commercial COVID-19 serological assays available to date use the Spike protein as the antigen [8,[20], [21], [22]]. However, to maintain its antigenic properties, this antigen must be produced in eukaryotic expression systems. The N protein has been described an immunodominant antigen generated during SASR-CoV-2 replication, and a superior antigenicity of N over S has been suggested [12,23,24]. Indeed, our rN-ELISA IgG kit based on the rN protein displayed similar performances when compared to tests from well-established manufacturers using the S1 antigen [25,26] . Furthermore, the developed rN-ELISA IgG kit presents improved sensitivity when compared to a commercially available test (DPP COVID-19 IgM/IgG, from Bio-Manguinhos, Fiocruz, Brazil).
Levels of antibodies in COVID-19 sera samples vary throughout the course of infection, and sensitivity data for serodiagnosis should be interpretated accordingly. In general, patients infected with SARS-CoV-2 display antibody responses between day 10 and 21 after the initial infection. Detection of antibodies in mild cases can take even longer (four weeks or more) and in a small number of cases, antibodies (i.e., IgM, IgG) were not detected at all (at least during the studies’ time scale). Based on the currently available data, the IgM and IgG antibodies to SARS-CoV-2 develop between 6 and 15 days post disease onset [6], [7], [8], [9]. In our study, the median day of seroconversion for both EIE COVID-19 IgG kit and DPP (IgG) was 14 days post PCR confirmation.
Technology for the production of the rN-ELISA was transferred to the Laboratory of Diagnostic Technology, at Bio-Manguinhos Institute, where it was further developed into industrial batches and registered at the Brazilian Sanitary and Healthcare Authority (ANVISA, Register number 1169789210).
In addition, the real-time stability assay demonstrated that the rN-ELISA kit was stable for at least 12 months, in storage at 2-8°C. As demonstrated (Fig. S7), the absorbance values of the tested sera remained stable over time, with the percentage drop being lowert than that recommended by regulatory agencies (± 20%; FDA, 2018) [27], which allows the detection of reliable results throughout the product shelf life.
Although widelly validated and consistent, our kit was not tested with samples from other human coronaviruses such as 229E, NL63, OC43, and HKU1, and that is an important limitation of the work. Other studies showed that cross-reactivity against certain SARS-CoV-2 antigens may occour, with N being detected at 11-16% overall prevalence [28,29].
The rN-ELISA was adapted to evaluate the presence of antibodies in the urine of patients post COVID-19 infection. Some adjustments have been done, such as incubation time and conjugated dilution and the results were published [30].
Considering the easy production of the antigen, the robustness of the test, the technical familiarity and the wide use of ELISA in clinical laboratory settings, this product represents an important addition to the currently available serodiagnosis toolbox for diagnosis of COVID-19.
Author contributions
Conceptualization, F.F.B., R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F.; methodology, F.F.B., R.T.G., S.M.R.T, L.A.F.A., N.C.S., A.P.S.M.F. and F.G.F.; validation, F.F.B., S.A.R.S., M.M.F., L.C.G., N.C.S., E.D.S., C.P.S., A.A, F. J.A., A.G.P.F, and E.L.D.; formal analysis, F.F.B., M.M.F., L.C.G., N.C.S., R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F.; writing—original draft preparation, F.F.B., R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F.; writing—review and editing, F.F.B., A.G.P.F, E.L.D., R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F.; project administration, R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F.; funding acquisition, R.T.G., S.M.R.T, A.P.S.M.F. and F.G.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study received funds from CAPES, CNPq, FAPEMIG and from the Brazilian Ministry of Science, Technology and Innovation (MCTI) through the “Rede Virus” initiative (and its many individual projects). Further funding came from the Instituto Nacional de Ciência e Tecnologia de Vacinas (INCTV) and from the Minas Gerais Secretary of Health (SES-MG).
Declaration of Competing Interest
The authors declare no conflict of interest. Funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
We thank all personnel at UFMG's Vaccine Technology Center (CT-Vacinas) Belo Horizonte, Brazil, for their support and teamwork. We also thank Dr. Marilda A. M. T. de Siqueira, from Laboratory of Respiratory Viruses and Measles (Fiocruz-RJ, Brazil); teams from Laboratory of Virology (USP, Brazil); and Laboratory of Diagnostic Technology (Fiocruz-RJ, Brazil) for their support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2022.100101.
Appendix. Supplementary materials
References
- 1.CDC COVID-19 Response Team SARS-CoV-2 B.1.1.529 (Omicron) Variant - United States, December 1-8, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70(50):1731–1734. doi: 10.15585/mmwr.mm7050e1. PMID: 34914670; PMCID: PMC8675659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V, da Silva Peixoto P., Kraemer M.U.G., Oikawa M.K., Salomon T., Cucunuba Z.M., Castro M.C., de Souza Santos A.A., Nascimento V.H., Pereira H.S., Ferguson N.M., Pybus O.G., Kucharski A., Busch M.P., Dye C., Faria N.R. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/s0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naveca FG, Nascimento V, de Souza VC, Corado A, Nascimento F, Silva G, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021 doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 4.Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, Coronavirus COVID-19 global cases, (2021). https://coronavirus.jhu.edu/map.html (Accessed July 07, 2022).
- 5.Traugott M., Aberle S.W., Aberle J.H., Griebler H., Karolyi M., Pawelka E., Puchhammer-Stöckl E., Zoufaly A., Weseslindtner L. Performance of severe acute respiratory syndrome coronavirus 2 antibody assays in different stages of infection: comparison of commercial enzyme-linked immunosorbent assays and rapid tests. J. Infect. Dis. 2020;222:362–366. doi: 10.1093/infdis/jiaa305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carattoli A., Di Bonito P., Grasso F., Giorgi C., Blasi F., Niedrig M., Cassone A. Recombinant protein-based ELISA and immuno-cytochemical assay for the diagnosis of SARS. J. Med. Virol. 2005;76:137–142. doi: 10.1002/jmv.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saijo M., Ogino T., Taguchi F., Fukushi S., Mizutani T., Notomi T., Kanda H., Minekawa H., Matsuyama S., Long H.T., Hanh N.T.H., Kurane I., Tashiro M., Morikawa S. Recombinant nucleocapsid protein-based IgG enzyme-linked immunosorbent assay for the serological diagnosis of SARS. J. Virol. Methods. 2005;125:181–186. doi: 10.1016/j.jviromet.2005.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo P.C.Y., et al. Relative rates of non-pneumonic sars coronavirus infection and SARS coronavirus pneumonia. Infect. Dis. Clin. Pract. 2004;12:275. doi: 10.1097/01.idc.0000130890.12611.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Hachim, N. Kavian, C.A. Cohen, A.W. Chin, D.K. Chu, C.K.P. Mok, O.T. Tsang, Y.C. Yeung, R.A. Perera, L.L. Poon, M.J. Peiris, S.A. Valkenburg, Beyond the Spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients, MedRxiv. (2020) 2020.04.30.20085670. https://doi.org/10.1101/2020.04.30.20085670.
- 12.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Wajnberg, M. Mansour, E. Leven, N.M. Bouvier, G. Patel, A. Firpo, R. Mendu, J. Jhang, S. Arinsburg, M. Gitman, J. Houldsworth, I. Baine, V. Simon, J. Aberg, F. Krammer, D. Reich, C. Cordon-Cardo, Humoral immune response and prolonged PCR positivity in a cohort of 1343 SARS-CoV 2 patients in the New York City region, MedRxiv. (2020) 2020.04.30.20085613. https://doi.org/10.1101/2020.04.30.20085613.
- 14.Castillo-Olivares J., et al. Analysis of serological biomarkers of SARS-CoV-2 infection in convalescent samples from severe, moderate and mild COVID-19 cases. Front. Immunol. 2021;12:4711. doi: 10.3389/fimmu.2021.748291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R. Lassaunière, A. Frische, Z. Harboe, A.C. Nielsen, A. Fomsgaard, K. Krogfelt, C. Jørgensen, Evaluation of nine commercial SARS-CoV-2 immunoassays, (2020) 1–15. https://doi.org/10.1101/2020.04.09.20056325.
- 16.FDA (U.S. Food and Drug Administration), EUA Authorized Serology Test Performance, (2020).
- 17.Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Heat inactivation of different types of SARS-CoV-2 samples: what protocols for biosafety, molecular detection and serological diagnostics? Viruses. 2020;12 doi: 10.3390/v12070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bland J.M., Altman D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 19.Rosano G.L., Ceccarelli E.A. Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 2014;5:1–17. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Hingrat Q.Le, Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. SARS-CoV-2 specific antibody responses in COVID-19 patients. MedRxiv. 2020:1–18. doi: 10.1101/2020.03.18.20038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perera R.A.P.M., Mok C.K.P., Tsang O.T.Y., Lv H., Ko R.L.W., Wu N.C., Yuan M., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Leung K., Chan K.H., Chan K.C.K., Li K.C., Wu J.T., Wilson I.A., Monto A.S., Poon L.L.M., Peiris M. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv H., Wu N.C., Tsang O.T.Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H., Wang Y., Choi C.Y.C., Lin Y., Ng W.W., Zhao J., Poon L.L.M., Peiris J.S.M., Wilson I.A., Mok C.K.P. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 24.Touma M. COVID-19: molecular diagnostics overview. J. Mol. Med. 2020;98:947–954. doi: 10.1007/s00109-020-01931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro R., Luz P.M., Wakimoto M.D., Veloso V.G., Grinsztejn B., Perazzo H. COVID-19: a meta-analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz. J. Infect. Dis. 2020;24:180–187. doi: 10.1016/j.bjid.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P., Gao Q., Wang T., Ke Y., Mo F., Jia R., Liu W., Liu L., Zheng S., Liu Y., Li L., Wang Y., Xu L., Hao K., Yang R., Li S., Lin C., Zhao Y. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19) MedRxiv. 2020:1–28. doi: 10.1101/2020.03.17.20036954. [DOI] [Google Scholar]
- 27.FDA F., A D. U.S. Department of Health and Human Services, Food and Drug Administration; 2018. Bioanalytical Method Validation Guidance for Industry; pp. 1–41. n. May. [Google Scholar]
- 28.Galipeau Y, Siragam V, Laroche G, Marion E, Greig M, McGuinty M, Booth RA, Durocher Y, Cuperlovic-Culf M, Bennett SAL, Crawley AM, Giguère PM, Cooper C, Langlois MA. Relative ratios of human seasonal coronavirus antibodies predict the efficiency of cross-neutralization of SARS-CoV-2 spike binding to ACE2. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103700. Epub 2021 Nov 30. PMID: 34861490; PMCID: PMC8629681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson EM, Goodwin EC, Verma A, Arevalo CP, Bolton MJ, Weirick ME, et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184(7):1858–1864. doi: 10.1016/j.cell.2021.02.010. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludolf F., Ramos F.F., Bagno F.F., Oliveira-da-Silva J.A., Reis T.A.R., Christodoulides M., Vassallo P.F., Ravetti C.G., Nobre V., da Fonseca F.G., Coelho E.A.F. Detecting anti-SARS-CoV-2 antibodies in urine samples: a noninvasive and sensitive way to assay COVID-19 immune conversion. Sci Adv. 2022;8(19):eabn7424. doi: 10.1126/sciadv.abn7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.