Abstract
Centromeres are epigenetically specified by the histone H3 variant CENP-A and typically associated with highly repetitive satellite DNA. We previously discovered natural satellite-free neocentromeres in Equus caballus and Equus asinus. Here, through ChIP-seq with an anti-CENP-A antibody, we found an extraordinarily high number of centromeres lacking satellite DNA in the zebras Equus burchelli (15 of 22) and Equus grevyi (13 of 23), demonstrating that the absence of satellite DNA at the majority of centromeres is compatible with genome stability and species survival and challenging the role of satellite DNA in centromere function. Nine satellite-free centromeres are shared between the two species in agreement with their recent separation. We assembled all centromeric regions and improved the reference genome of E. burchelli. Sequence analysis of the CENP-A binding domains revealed that they are LINE-1 and AT-rich with four of them showing DNA amplification. In the two zebras, satellite-free centromeres emerged from centromere repositioning or following Robertsonian fusion. In five chromosomes, the centromeric function arose near the fusion points, which are located within regions marked by traces of ancestral pericentromeric sequences. Therefore, besides centromere repositioning, Robertsonian fusions are an important source of satellite-free centromeres during evolution. Finally, in one case, a satellite-free centromere was seeded on an inversion breakpoint. At 11 chromosomes, whose primary constrictions seemed to be associated with satellite repeats by cytogenetic analysis, satellite-free neocentromeres were instead located near the ancestral inactivated satellite-based centromeres; therefore, the centromeric function has shifted away from a satellite repeat containing locus to a satellite-free new position.
Keywords: neocentromere, Robertsonian fusion, centromere repositioning, genome evolution, ChIP-seq, CENP-A
Introduction
Centromeres are essential nucleoprotein structures of eukaryotic chromosomes responsible for the correct segregation of sister chromatids during cell division. Centromeres are epigenetically specified by the histone H3 variant CENP-A, the hallmark of a functional centromere (Allshire and Karpen 2008). Mammalian centromeres are typically associated with extended arrays of tandemly iterated sequences (satellite DNA), which are divergent and represent the most rapidly evolving components of genomes (Plohl et al. 2014). The presence of such sequences has hampered comprehensive molecular analysis of these intriguing loci. Despite their typical presence, satellite sequences are not sufficient nor required for centromere function. Direct evidence originally came from the examination of human chromosomal abnormalities (Choo 2000; Amor and Choo 2002; Cleveland et al. 2003; Kalitsis and Choo 2012). Pseudodicentric chromosomes contain two identical, well-separated regions of centromeric DNA, but only one retains the centromeric function, suggesting that the centromeric sequence is not sufficient for centromere establishment (Earnshaw and Migeon 1985; Choo 2000; Marshall et al. 2008). The identification of sporadic human chromosomes carrying centromere function in hitherto noncentromeric chromosomal regions devoid of canonical centromeric repeated DNA (Voullaire et al. 1993; Choo 2000; Marshall et al. 2008) demonstrated that satellite DNA is not necessary for centromere establishment. The discovery of natural satellite-free centromeres fixed in some animal and plant species clearly proves that satellite DNA is not strictly required for the centromeric function. The first satellite-free centromere, fixed in a vertebrate species, was discovered by our group in one horse chromosome (Wade et al. 2009). Our discovery that equids are characterized by the existence of centromeres completely devoid of satellite DNA made these species an exceptional model system for dissecting the molecular architecture of mammalian centromeres as well as understanding the mechanisms driving centromere birth and maturation during evolution (Wade et al. 2009; Piras et al. 2010, 2022; Purgato et al. 2015; Giulotto et al. 2017; Nergadze et al. 2018; Roberti et al. 2019).
The family of Equidae, with its only extant genus, the Equus (horses, asses, and zebras), belongs to the order Perissodactyla together with Tapiridae (tapirs) and Rhinocerotidae (rhinoceroses). Although the karyotypes of Tapiridae and Rhinocerotidae remained quite stable during evolution and resemble the putative perissodactyl ancestral karyotype, characterized by high chromosomal number and a prevalence of acrocentric chromosomes (Trifonov et al. 2008), Equus karyotypes underwent a rapid evolution after their divergence from the common ancestor, dated around 4 million years ago (Ma) (Orlando et al. 2013). The most recent radiation events, which differentiated asses and zebra species, date back to <1 Ma, and many species and subspecies emerged in this short evolutionary time (Trifonov et al. 2008, 2012; Jónsson et al. 2014). The extensive karyotype reshuffling that occurred during equids speciation is due to chromosome rearrangement and centromere repositioning, which is the shift of the centromeric function without sequence rearrangement (Carbone et al. 2006; Piras et al. 2010; Trifonov et al. 2012). These phenomena led to the evolution from the ancestral karyotype, with the majority of chromosomes being acrocentric, to karyotypes with a reduced chromosomal number, mainly comprised of meta- and submetacentrics. Our previous cytogenetic analyses suggested that several of these chromosomes harbor satellite-free centromeres (Carbone et al. 2006; Piras et al. 2010) while displaying blocks of satellite DNA sequences at noncentromeric chromosome ends, as relics of the inactivated centromeres from the ancestral acrocentrics. We then characterized, at the molecular level, satellite-free centromeres deriving from centromere repositioning events in horses and donkeys (Wade et al. 2009; Nergadze et al. 2018). The position of these peculiar centromeres, identified as CENP-A binding domains, can slide within a relatively wide region, probably limited by epigenetic boundaries. These domains were defined as epialleles and the phenomenon was called centromere sliding (Purgato et al. 2015; Nergadze et al. 2018).
ChIP-seq experiments performed with an anti-CENP-A antibody on individuals from families composed of donkeys, horses, and their hybrid offspring (mule/hinny) revealed that such epialleles are inherited as Mendelian traits, but their position can slide in one generation (Nergadze et al. 2018). Conversely, the position of the centromere is stable during mitotic propagation of cultured cells grown for several population doublings, suggesting that the sliding can presumably take place during meiosis (Nergadze et al. 2018).
Sequence analysis of the donkey satellite-free centromeric domains demonstrated that five satellite-free centromeres were characterized by novel tandem repetitions. These amplified genomic sequences are chromosome-specific, with amplified units ranging in size from a few to tens of kilobases (Nergadze et al. 2018). The repeat copy number was variable in different individuals, suggesting the existence of polymorphism in the population. We suggested that these amplified DNA may represent an intermediate evolutionary stage toward satellite DNA formation during the process of centromere maturation (Nergadze et al. 2018). According to this model, after centromere inactivation, associated satellite sequences are maintained at the original site while the newly born centromere is completely devoid of satellite sequences. Subsequently, satellite DNA is gradually lost at the locus of the ancestral centromere, whereas, at the functional satellite-free centromere, sequence amplification may occur before complete maturation via satellite DNA acquisition.
Besides centromere repositioning, it is well described that Robertsonian translocations, which involve centric fusion of two acrocentric chromosomes, marked the rapid karyotype reshaping of zebras (Piras et al. 2010; Musilova et al. 2013), whose centromeres still lack a molecular characterization. According to current taxonomy, there are three zebra species, namely, plains zebra (Equus quagga, also known as Equus burchelli), Grevy’s zebra (Equus grevyi), and mountain zebra (Equus zebra) (Ransom and Kaczensky 2016).
In previous work, we described the localization of satellite DNA families by FISH in Burchell’s zebra and Grevy’s zebra, showing that several centromeres are devoid of satellite DNA at the cytogenetic resolution (Piras et al. 2010). The aim of the present work was to identify and characterize at the molecular level satellite-free centromeres in the two zebras and to investigate the mechanisms leading to the emergence of satellite-free centromeres during evolution.
Results
Burchell’s Zebra Reference Genome
In 2020, a de novo chromosome-length assembly for the Burchell’s zebra (Equus_quagga_HiC) was released by the DNA Zoo team (https://www.dnazoo.org). Since this assembly contains scaffolds corresponding to entire zebra chromosomes but lacking chromosome assignment, we performed a whole-genome alignment of the Equus_quagga_HiC assembly to the horse genome and assigned the scaffolds to specific horse chromosomes using EquCab3.0 as reference. Since the horse and the Burchell’s zebra genomes share high-sequence identity (Jónsson et al. 2014) and chromosome orthologies are well described (Musilova et al. 2013), taking advantage of the subchromosomal comparative maps between horse and Burchell’s zebra (Musilova et al. 2013), we assigned chromosome numbers to the 22 Burchell’s zebra chromosomal scaffolds. In supplementary figure S1, Supplementary Material online, the pairwise whole-genome alignment plot between EquCab3.0 and the Equus_quagga_HiC assembly is shown. The results showed that 14 scaffolds were correctly oriented compared with the direction previously determined by comparative cytogenetics (Musilova et al. 2013), whereas 9 scaffolds, corresponding to chromosomes 2, 6, 9, 12, 13, 14, 16, 21, and X, had a reverse orientation.
Identification and Sequence Assembly of Satellite-free Centromeres in Burchell’s Zebra
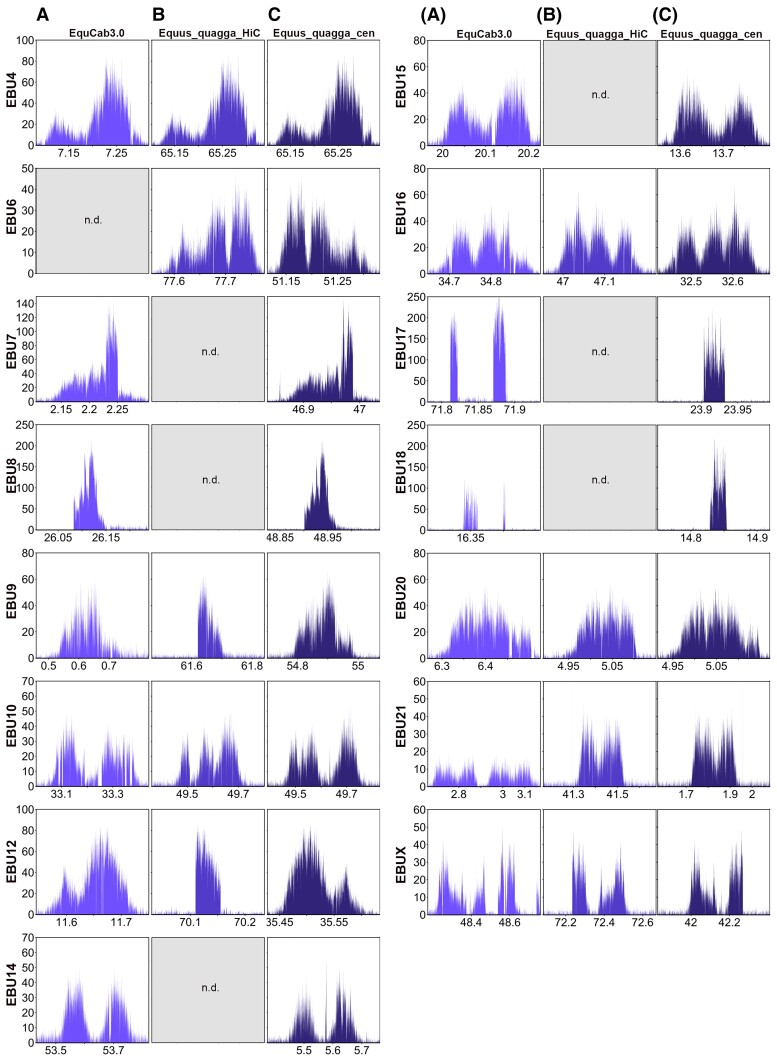
In previous work, we described several centromeres from E. burchelli (Burchell’s zebra) lacking detectable satellite repeats at the cytogenetic level (Piras et al. 2010). To identify satellite-free CENP-A binding domains of Burchell’s zebra at the molecular level, a ChIP-seq experiment with an antibody against CENP-A was carried out on primary skin fibroblasts. The ChIP-seq reads were mapped both on the Burchell’s zebra (Equus_quagga_HiC) assembly and on the horse (EquCab3.0) reference genome, which remains the best assembled genome sequence among equids (Wade et al. 2009; Kalbfleisch et al. 2018). Using the pipeline that we developed for satellite-free centromeres in the donkey (Nergadze et al. 2018), genomic regions, enriched for CENP-A binding, were identified. In figure 1AandB, a graphical representation of the CENP-A enrichment peaks found using as reference EquCab3.0 or Equus_quagga_HiC, respectively, is shown. Map positions of the enrichment peaks are reported in supplementary table S1, Supplementary Material online. These peaks, which correspond to satellite-free centromeres, were located on 15 out of the 22 chromosomes. Thirteen of these chromosomes (4, 6, 7, 8, 9, 10, 12, 14, 15, 16, 17, 18, and X) are meta- or submetacentric, whereas two of them (20 and 21) are acrocentric. As expected, CENP-A binding peaks were not identified at satellite-based centromeres since, given the repetitive nature of these regions, they are not assembled. Therefore, the reads deriving from these loci map on unplaced scaffolds.
Fig. 1.
Satellite-free centromeres in Burchell’s zebra. ChIP-seq reads from primary fibroblasts of Burchell’s zebra were mapped on EquCab3.0 (A), Equus_quagga_HiC (B), and Equus_quagga_cen (C). CENP-A enriched domains are visualized as peaks. The y-axis reports the normalized read count, whereas the x-axis reports the coordinates on the reference genome. For each chromosome, genomic windows of the same length are plotted.
Satellite-free CENP-A binding domains on chromosomes 4, 9, 10, 12, 16, 20, 21, and X were identified on orthologous positions in both reference genomes (fig. 1AandB and supplementary table S1, Supplementary Material online). For chromosome 6, a CENP-A binding domain was detected only in the zebra assembly, suggesting that the orthologous horse locus is highly rearranged. A detailed analysis of this centromere, which is reported in a following paragraph, confirms this hypothesis. On the contrary, the centromeric domains of chromosomes 7, 8, 14, 15, 17, and 18 were detected only in the horse reference genome, suggesting that these loci are not properly assembled in the DNA Zoo Equus_quagga_HiC scaffolds.
Although some peaks showed a fairly regular Gaussian-like shape (such as those on chromosomes 4 and 20), some of them were irregular and contained several gaps on both reference genomes (such as those on chromosomes 10 and 18). In addition, for several centromeres (such as 9 and 12), the shape and size of the peak were different between the two assemblies, reflecting sequence variation between the two species and/or sequence gaps.
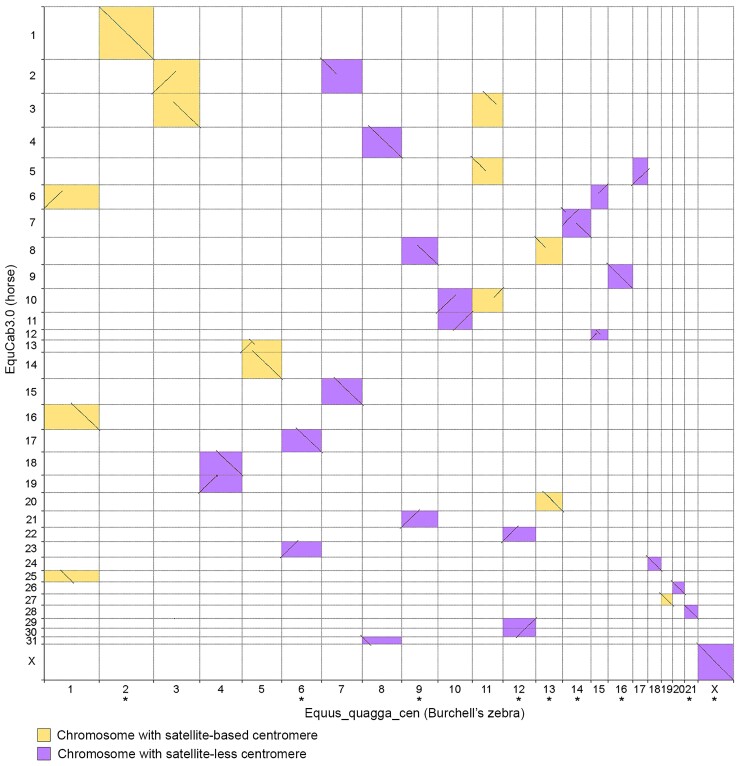
To determine more precisely the organization of the CENP-A binding domains at satellite-free centromeres, the actual DNA sequence corresponding to the 15 Burchell’s zebra centromeres was assembled from our Illumina reads (see Materials and Methods). For each centromeric region, genomic segments ranging in size between 67 and 445 kb, containing the CENP-A binding domain, were obtained (accession numbers: OM643400-OM643414). We then corrected the Equus_quagga_HiC sequence by replacing the incomplete or misassembled centromeric loci with the newly assembled genomic segments of Burchell’s zebra. In addition, we adjusted the direction of the chromosomes that were incorrectly oriented in the DNA Zoo assembly (2, 6, 9, 12, 13, 14, 16, 21, and X) (fig. 2 and supplementary table S2, Supplementary Material online). Two of the newly oriented chromosomes (2 and 13) contain satellite-based centromeres. The resulting genome sequence is, from now on, called Equus_quagga_cen. In figure 2, the pairwise genome comparison between EquCab3.0 and Equus_quagga_cen chromosomes is shown. The ChIP-seq reads from Burchell’s zebra were then mapped on this new reference genome and enrichment peaks, corresponding to CENP-A binding domains, were obtained (fig. 1C and supplementary table S3, Supplementary Material online). The peak profiles visualized on the new reference genome showed that several gaps and irregular shapes were no longer detected and their profiles (compare fig. 1A–C) as well as mapping statistics were improved (supplementary tables S2 and S4 and fig. S2, Supplementary Material online). It is worth noticing that, when Equus_quagga_cen was used as reference, some CENP-A binding peaks (6, 12, 16, and X) showed a specular shape compared with the peaks obtained with the horse and Burchell’s zebra reference genomes. This effect is a consequence of the opposite orientation of these chromosomes in the Equus_quagga_cen assembly compared with the other reference genomes. In a few cases, such as EBU10, EBU14, and EBU15, two well-separated or partially overlapping CENP-A binding domains are visible. They probably correspond to different epialleles on the two homologous chromosomes in the individual analyzed here and suggest positional variation of satellite-free centromeres in the population, as previously demonstrated in the horse and in the donkey (Purgato et al. 2015; Nergadze et al. 2018).
Fig. 2.
Pairwise genome comparison between EquCab3.0 and Equus_quagga_cen. Chromosome numbers were assigned to Equus_quagga_HiC scaffolds corresponding to entire zebra chromosomes. Aligned segments between horse (y-axis) and Burchell’s zebra (x-axis) chromosomes are represented as lines. The orientation of the chromosomes marked with an asterisk was corrected according to the direction previously determined by cytogenetic analysis (Musilova et al. 2013).
Identification and Sequence Assembly of Satellite-free Centromeres in Grevy’s Zebra
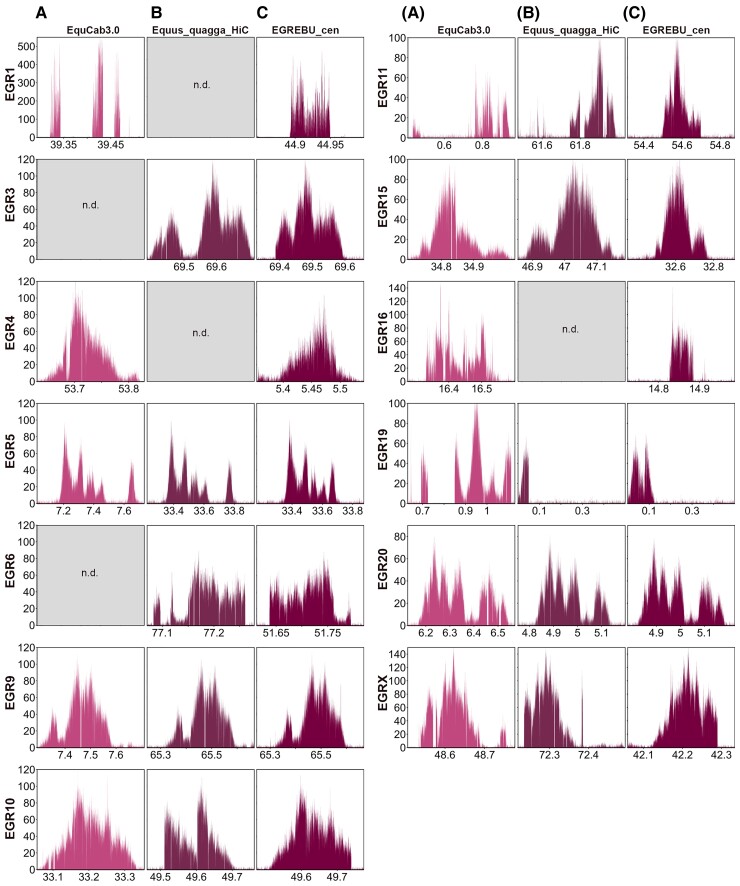
To describe at the molecular level Grevy’s zebra satellite-free centromeres, a ChIP-seq experiment was carried out using the anti-CENP-A antibody with Grevy’s zebra fibroblasts. Since a reference genome for this species is not available, given the high karyotype (Musilova et al. 2013) and sequence (Jónsson et al. 2014) identity with the Burchell’s zebra, we mapped the reads on the horse genome (EquCab3.0) and on the Equus_quagga_HiC assembly. As for Burchell’s zebra, we obtained CENP-A enrichment peaks that correspond to satellite-free centromeres (fig. 3 and supplementary table S1, Supplementary Material online). These centromeres were identified on 13 out of the 23 chromosomes. Eleven of these chromosomes (1, 3, 4, 5, 6, 9, 10, 11, 15, 16, and X) are meta- or submetacentric, whereas two of them (19 and 20) are acrocentric. Eight satellite-free centromeres (5, 9, 10, 11, 15, 19, 20, and X) were identified on orthologous positions in both reference genomes (fig. 3AandB). The centromeric domains of chromosomes 1, 4, and 16 were detected only in the horse reference genome, suggesting that these loci are not properly assembled in the Equus_quagga_HiC scaffolds, whereas the CENP-A binding domains of chromosomes 3 and 6 were detected only in the zebra assembly, suggesting that the orthologous horse loci are highly rearranged.
Fig. 3.
Satellite-free centromeres in Grevy’s zebra. ChIP-seq reads from primary fibroblasts of Grevy’s zebra were mapped on EquCab3.0 (A), Equus_quagga_HiC (B), and EGR_EBU_cen (C). CENP-A enriched domains are visualized as peaks. The y-axis reports the normalized read count, whereas the x-axis reports the coordinates on the reference genome. For each chromosome, genomic windows of the same length are plotted.
Several enrichment peaks display irregular shapes due to sequence variation between the Grevy’s zebra and the reference genomes. To obtain a more precise reference for the centromeric loci of the Grevy’s zebra, the sequence corresponding to the 13 CENP-A binding peaks was assembled from our Illumina reads (see Materials and Methods), and contigs spanning between 60 and 370 kb were obtained (accession numbers: OM643415–OM643427). Using the approach previously applied by us to the donkey genome (Nergadze et al. 2018), we constructed a chimeric reference genome by replacing the Equus_quagga_cen contigs with the orthologous newly assembled Grevy’s zebra sequences (supplementary table S5, Supplementary Material online). The result was a virtual hybrid reference genome, called EBU_EGR_cen, which was then used for mapping ChIP-seq reads to obtain refined CENP-A enrichment peaks (fig. 3C and supplementary table S6, Supplementary Material online). As shown in figure 3C, several peak profiles were improved compared with those obtained using EquCab3.0 or Equus_quagga_HiC as reference genomes. In addition, mapping statistics at the regions of interest were generally improved (supplementary tables S4 and S5 and fig. S2, Supplementary Material online).
Conservation of Centromeric Domains in the two Species
Following a comparative analysis of the position of CENP-A binding domains in the two species, we observed that the centromeric regions of EGR4, EGR10, EGR15, EGR16, EGR20, and EGRX were overlapping with those of the orthologous chromosomes EBU14, EBU10, EBU16, EBU18, EBU20, and EBUX, respectively (supplementary fig. S3, Supplementary Material online). We then compared the Grevy’s zebra sequences, assembled from our ChIP-seq reads, with the orthologous loci of the Equus_quagga_cen genome assembly (supplementary fig. S3, Supplementary Material online). All orthologous loci shared a high-sequence identity (supplementary table S7, Supplementary Material online). In EGR10, EGR15, EGR16, and EGR20, a few species-specific deletions and insertions were detected with respect to the orthologous Burchell’s zebra loci. In EGR4 and EGRX, we detected a few species-specific deletions/insertions and small translocations.
We did not include the centromeres from EGR6 and EGR11 since the CENP-A binding domains of their satellite-free centromeres are located very close, but not overlapping, to those of EBU6 and EBU9, respectively (supplementary tables S3 and S5, Supplementary Material online).
Satellite-free Centromeres with Tandem Repetitions
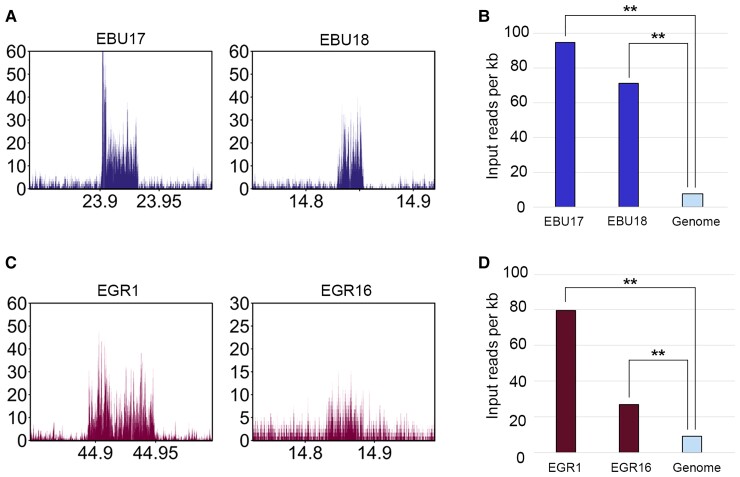
Similar to what was previously described for some donkey satellite-free centromeres (Nergadze et al. 2018), two Burchell’s zebra (EBU17 and EBU18) and two Grevy’s zebra (EGR1 and EGR16) centromeres display enrichment peaks with a spike-like shape (figs. 1C and 3C). We previously proved that, in the donkey, such peaks correspond to centromeres characterized by tandem repetitions of a sequence that was single copy in the horse reference genome (Nergadze et al. 2018). In particular, the EBU17 centromere has the same shape and resides in the same position of the centromere of donkey chromosome 16, which was previously shown to contain tandem repetitions (Nergadze et al. 2018). Thus, we hypothesized that, also in the two zebras, the shape of such peaks indicates the presence of centromeric loci with DNA amplification. These loci could not be entirely assembled because of their repetitive nature.
To confirm the presence of tandem repetitions at these centromeres, we analyzed the Burchell’s zebra input reads mapped on the Equus_quagga_cen reference genome and the Grevy’s zebra input reads mapped on the EBU_EGR_cen reference genome. The peaks shown in figure 4AandC demonstrate that, at the genomic loci corresponding to the satellite-free centromeres of EBU17, EBU18, EGR1, and EGR16, multiple copies of the underlying genomic sequences are present. To approximately quantify the copy number of these repeats, we compared the number of input reads mapping at the two Burchell’s zebra loci with the average genome coverage. As shown in figure 4B, the number of reads at EBU17 and EBU18 centromeres is about 12.5 and 9.5 times, respectively, compared with genome average. The same analysis was carried out for the Grevy’s zebra loci, and the number of reads at the EGR1 and at the EGR16 centromeres was about 8.3 and 2.8 times, respectively, compared with genome average (fig. 4D).
Fig. 4.
DNA sequence amplification at Burchell’s zebra and Grevy’s zebra satellite-free centromeres. (A) Profiles of input (not immunoprecipitated) reads from Burchell’s zebra at EBU17 and EBU18 centromeric regions. The y-axis reports the RPKM count, whereas the x-axis reports the coordinates on the reference genome. (B) Input read counts per kilobase (y-axis) are shown for EBU17 and EBU18 satellite-free centromeres and for the whole genome (Equus_quagga_cen). The double asterisks indicate statistically significant differences, with a P-value of <0.01. (C) Profile of input reads from Grevy’s zebra at EGR1 and EGR16 centromeres. The y-axis reports the RPKM count, whereas the x-axis reports the coordinates on the reference genome. (D) Input read counts per kilobase (y-axis) are shown for EGR1 and EGR16 satellite-free centromeres and the whole genome (EGR_EBU_cen). The double asterisks indicate statistically significant differences, with a P-value of <0.01.
The presence of sequence amplifications was also observed in the contigs that we assembled. However, given the repetitive nature of these regions, we could not obtain a complete assembly of these centromeres. In the EBU18, EGR1, and EGR16, the CENP-A binding domains (accession numbers: OM643411, OM643415, and OM643424) contain several subrepeats (supplementary fig. S4, Supplementary Material online). Some of these subrepeats are shared between EBU18 and EGR16, in agreement with their evolutionarily common origin.
In EBU17, we did not identify subrepeats. A possible interpretation is that, at this centromere, amplification may involve the entire 30 kb region.
Sequence Analysis of the Satellite-free Centromeres
DNA sequence features of the satellite-free centromeres of the two zebra species were compared with the average genome-wide values obtained from the Burchell’s zebra genome. The four centromeres containing tandem repetitions were excluded from this analysis because we cannot precisely define their complete underlying sequences. As shown in supplementary figure S5, Supplementary Material online, in both species, the satellite-free centromeres are significantly enriched in LINE-1 whereas they are depleted in LINE-2 elements, SINEs, and DNA transposons. On the other hand, the abundance of LTRs does not differ from the genome average.
Finally, Burchell’s and Grevy’s zebra centromeres showed 37% and 36.6% GC content, respectively, which are lower than the genome-wide average of 41.43%. These differences are statistically significant, proving that these centromeres are AT-rich (supplementary fig. S5, Supplementary Material online).
To identify, at the genome-wide level, Burchell’s zebra loci containing arrays of sequences homologous to known equine satellite DNA, we carried out a RepeatMasker analysis on the chromosome scaffolds of Equus_quagga_cen. We also analyzed, with the same tool, the centromeric contigs of Grevy zebra. As shown in supplementary table S8, Supplementary Material online, this analysis revealed a great number of loci containing satellite arrays of variable length. In particular, we found arrays corresponding to the three satellite DNA sequences previously described in the genus Equus, namely, 37cen (221 bp units), 2PI (23 bp units), and EC137 (137 bp units) (Piras et al. 2010; Nergadze et al. 2014). The great majority of these sequences are located at fusion regions or at putative pericentromeric regions of satellite-based centromeres. Since the centromeric satellites are not well assembled, the majority of satellite DNA repeats are comprised in the unplaced scaffolds and are not included in this table. Relatively short sequences homologous to satellite repeats are also located at several interstitial noncentromeric sites. None of these loci homologous to satellite DNA is located within satellite-free CENP-A binding domains. The only exceptions are the centromeres of EBUX and EGRX chromosomes, where short 2PI arrays were detected within the CENP-A binding domain (supplementary table S8, Supplementary Material online). These sequences are well assembled and do not flank gaps, suggesting that they are not a part of larger unassembled satellite array. The length of these stretches is negligible with respect to the length of the overall centromeric domains (302 and 176 kb, respectively), and therefore, also the centromeres of EBUX and EGRX can be considered satellite-free.
To verify whether novel satellite repeats may be present at the Burchell’s zebra and the Grevy’s zebra satellite-free centromeres, we performed a de novo search of tandem repeats using ULTRA (Olson and Wheeler 2018). This analysis showed that no satellite repeats were present at these centromeres. For EBUX and EGRX centromeres, we identified repeated units corresponding to 2PI satellite stretches, which were identified in the RepeatMasker analysis as well. These units have low copy number and are inserted into contiguous well-assembled sequences (supplementary table S9, Supplementary Material online). Thus, they cannot be considered arrays of novel satellite repeats. These findings confirm that the zebra satellite-free centromeres do not contain satellite DNA, that is extended tandem arrays, typical of mammalian centromeres.
Mechanisms of Satellite-free Centromeres Formation in Burchell’s Zebra
With the goal of determining the mechanisms of neocentromere formation during evolution, we carried out whole chromosome alignments between zebra chromosomes, harboring satellite-free centromeres, and the orthologous elements from the horse, here used as an outgroup (supplementary fig. S6, Supplementary Material online). Indeed, the horse is considered the closest extant species to the equid ancestor, and all the centromeres, with the only exception of the one on chromosome 11, are satellite-based. The position of the satellite-free centromeres and of satellite DNA arrays, together with the previous cytogenetic observations, is now summarized in supplementary table S10, Supplementary Material online.
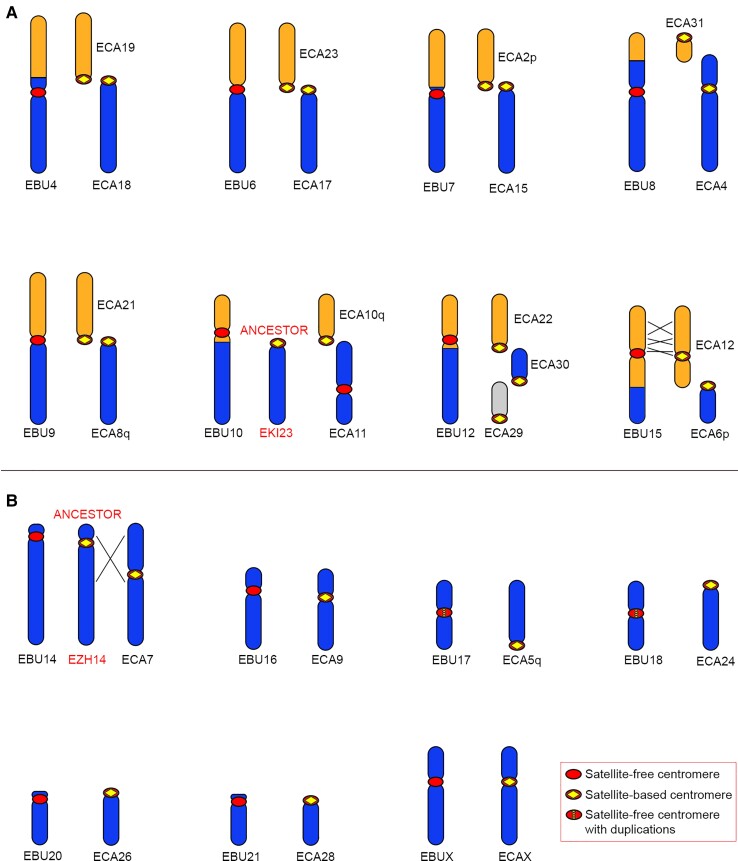
Eight of the chromosomes (4, 6, 7, 8, 9, 10, 12, and 15) containing a satellite-free centromere originated by a fusion of ancestral chromosomes (fig. 5A). In all these chromosomes, the ancestral centromeres were inactivated. To clarify the relationship between fusion and satellite-free centromere formation, we measured the distance between each satellite-free centromere and the fusion point. To this purpose, since in the horse, as in the equid ancestor, the elements involved in the fusion are separated, we aligned the chromosomes from the Equus_quagga_cen assembly with the orthologous chromosomes from the EquCab3.0 horse assembly. Following this procedure, we were able to determine the coordinates of the fusion regions (supplementary table S10, Supplementary Material online). These regions correspond to sequences placed on specific chromosomes of the Equus_quagga_cen assembly, range in size between about 5 kb and 1.5 Mb, and are not present as such in the horse genome assembly. A detailed description of all fusion events, as depicted in figure 5A, and of the fusion regions is reported below.
Fig. 5.
Comparison between Burchell’s zebra chromosomes with satellite-free centromeres and the orthologous horse chromosomes. For each Burchell’s zebra chromosome carrying a satellite-free centromere, the corresponding orthologous horse chromosomes are shown. For EBU10 and EBU14, the ancestral chromosomes, represented by kiang chromosome 23 (EKI23) and Hartmann’s zebra chromosome 14 (EZH14), respectively, are shown. Inverted segments are indicated with crossed lines. (A) EBU chromosomes deriving from fusions. (B) EBU chromosomes corresponding to entire horse chromosomes or chromosome arms.
EBU4 is a metacentric chromosome deriving from the Robertsonian fusion of two ancestral acrocentric elements that correspond to horse chromosomes 18 and 19 (fig. 5A). The satellite-free centromeric region of EBU4 is orthologous to a noncentromeric locus on ECA18 and is located at about 6.5 Mb from the fusion region (supplementary table S10, Supplementary Material online), suggesting that a centromere repositioning event moved the zebra centromere far from the fusion point. From these data, it is not possible to determine whether centromere repositioning occurred before, during, or after the fusion event. Interestingly, on the fusion region, we detected the presence of an array of 2PI, a satellite that we previously localized at most horse pericentromeres (Piras et al. 2010; Cerutti et al. 2016), of transposable elements and duplicons shared by different horse pericentromeres. All these sequences likely represent relics of the sequences associated with the ancestral centromeres that were involved in the Robertsonian fusion and are now inactive.
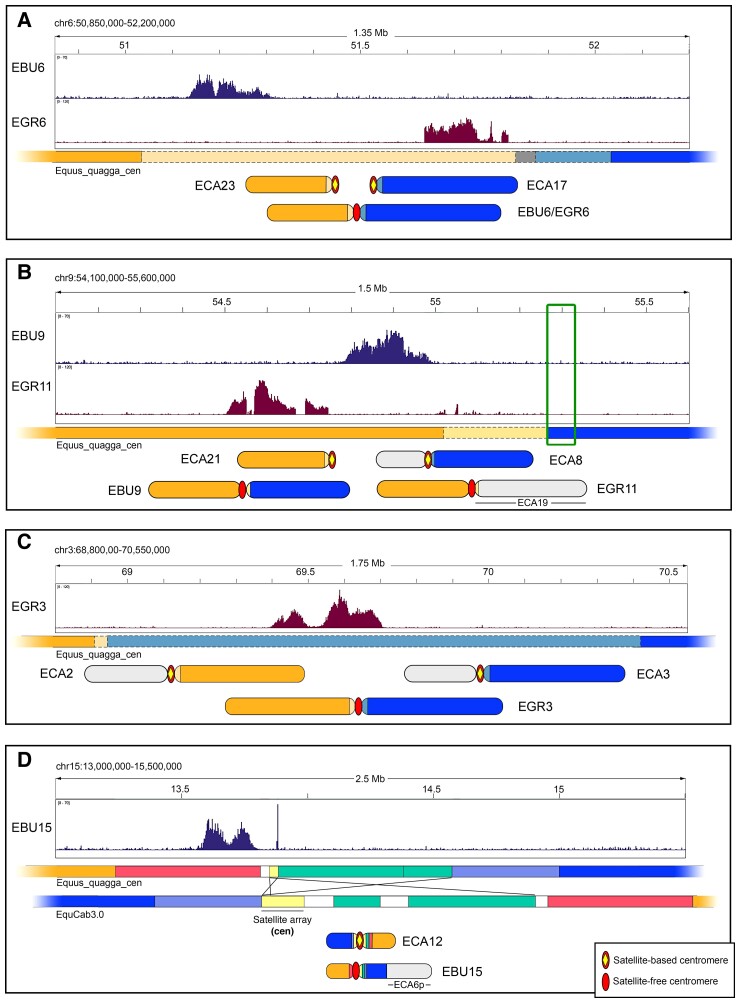
EBU6 derived from a fusion between ancestral acrocentrics orthologous to horse chromosomes 23 and 17 (fig. 5A). The CENP-A binding domain of EBU6 lays in a region that is unique in the Burchell’s zebra assembly but shares up to 80% identity with different horse pericentromeric sequences. This is the reason why this CENP-A binding domain was detected only when the ChIP-seq reads were mapped on the Burchell’s zebra genome, whereas it was not detected when EquCab3.0 was used as reference (fig. 1). The organization of this fusion region is depicted in figure 6A and shows that the CENP-A binding peak lays on a sequence sharing high identity with several horse pericentromeres, including the one from ECA 23 (light orange line in fig. 6A). The gray region, which contains the fusion point, is a highly rearranged 52 kb sequence with no good alignment with ECA23 nor ECA17 pericentromeres. On the other side of the gray region, a sequence sharing high identity with the pericentromere of ECA17 was detected (light blue line in fig. 6A). In conclusion, in EBU6, the CENP-A binding domain resides in the fusion region, within sequences deriving from an ancestral pericentromere. In figure 6A, the position of the CENP-A binding peak on Grevy’s zebra chromosome 6 is also reported.
Fig. 6.
Sequence organization of satellite-free centromeric regions deriving from chromosomal rearrangements. (A) The CENP-A binding peaks of EBU6 and EGR6 on the Equus_quagga_cen reference are shown on top. The colored bar schematically represents this Equus_quagga_cen genomic region, with colors referring to orthologous sequences in the EquCab3.0 horse genome: orange, orthology with ECA23; blue, orthology with ECA17; light orange and light blue, orthology with horse pericentromeres, including ECA23 and ECA17, respectively; gray, orthology with several horse pericentromeric sequences not including ECA23 and ECA17. The zebra submetacentric chromosomes deriving from the fusion between ancestral elements corresponding to ECA23 (orange) and ECA17 (blue) are sketched on the bottom. (B) The CENP-A binding peaks of EBU9 and EGR11 on the Equus_quagga_cen reference are shown on top. The colored bar schematically represents this Equus_quagga_cen genomic region with colors referring to orthologous sequences in the EquCab3.0 horse genome: orange, orthology with ECA21; blue, orthology with ECA8q; light orange, orthology with horse pericentromeres, including ECA21; the green box indicates satellite DNA. The EBU9 submetacentric chromosome, deriving from the fusion between ancestral elements corresponding to ECA21 (orange) and ECA8q (blue), together with the EGR11 submetacentric chromosome, deriving from the fusion between ancestral elements corresponding to ECA21 (orange) and ECA19 (light gray), are sketched on the bottom. The ECA8 p element, which did not participate in this fusion, is also in light gray. (C) The CENP-A binding peak of EGR3 on the Equus_quagga_cen reference is shown on top. The colored bar schematically represents this genomic region, with colors referring to orthologous sequences in the EquCab3.0 horse genome: orange, orthology with ECA2q; blue, orthology with ECA3q; light orange and light blue, orthology with horse pericentromeres, including ECA2q and ECA3q, respectively. The zebra submetacentric chromosome deriving from the fusion between ancestral elements corresponding to ECA2q (orange) and ECA3q (blue) are sketched on the bottom. ECA2p and ECA3p, which did not participate in this fusion, are in light gray. (D) The CENP-A binding peak of EBU15 on the Equus_quagga_cen reference is shown on top. The colored bars represent the inverted regions in Equus_quagga_cen and in EquCab3.0. The colors mark locally collinear blocks, that is, orthologous segments between the Burchell’s zebra and the horse genome. The white blocks correspond to species-specific sequences. The crossed lines indicate inverted segments. Satellite arrays from the ECA12 pericentromere, which are partially assembled in EquCab3.0 reference, are in light yellow.
EBU7 derived from the fusion of the ancestral acrocentrics corresponding to ECA2p and ECA15 (fig. 5A). Its CENP-A binding domain lays in a region orthologous to a noncentromeric locus of ECA15 located 2 Mb away from the fusion region, where the relics of ancestral pericentromeric sequences, including the blocks of 2PI and EC137 (Piras et al. 2010; Nergadze et al. 2014; Cerutti et al. 2016), could be observed (supplementary table S10, Supplementary Material online). Given the distance between the CENP-A binding domain and the fusion point, we hypothesize that the satellite-free centromere of EBU7, similar to EBU4, could be the result of a centromere repositioning event that occurred before, during, or after the fusion.
EBU8 derived from a telomere–telomere fusion between elements orthologous to the acrocentric ECA31 and the submetacentric ECA4 (fig. 5A). At the cytogenetic level, the centromere of EBU8 appears at the same position of the ECA4 centromere; however, at the molecular level, the CENP-A binding domain of EBU8 corresponds, in the horse, to a noncentromeric locus that is located at a distance of about 500 kb from 2PI pericentromeric satellites on ECA4. These observations suggest that the EBU8 centromere is repositioned relative to the ancestral ECA4-derived element. At the fusion site, located at 24 Mb from the CENP-A binding domain, no pericentromeric-type sequences were detected (supplementary table S10, Supplementary Material online) and, although EBU8 derived from a telomere–telomere fusion, no telomeric repeats were detected either.
EBU9 resulted from the centric fusion between the ancestral segments corresponding to ECA21 and ECA8q (fig. 5A). The organization of this fusion region is depicted in figure 6B. The EBU9 CENP-A binding domain is orthologous to a satellite-free noncentromeric locus of ECA21 (orange line in fig. 6B) and immediately adjacent to a region sharing high identity with ECA21 pericentromeric sequences (light orange line in fig. 6B). The fusion point is located at about 260 kb from the CENP-A binding domain, at the junction between the pericentromeric sequences derived from the ECA21 (light orange) and the ECA8q (blue in fig. 6B) ancestral elements. Moreover, in the junction region, 2PI and EC137 satellites (boxed in fig. 6B) can still be detected as traces of the ancient pericentromeres (supplementary table S10, Supplementary Material online). Similar to EBU6, this satellite-free centromere resides very close to the fusion point. In figure 6B, the position of the CENP-A binding peak on Grevy’s zebra chromosome 11 is also reported.
EBU10 derived from the fusion between ancestral elements corresponding to horse chromosomes 10q and 11 (fig. 5A). We previously demonstrated that, contrary to all other horse centromeres, the centromere of horse chromosome 11 is devoid of satellite DNA and evolutionarily recent (Wade et al. 2009; Piras et al. 2010). Therefore, as shown in figure 5A, ECA11 does not represent the ancestral configuration of this chromosomal element. As previously suggested through cytogenetic analysis, the acrocentric chromosome 23 from E. kiang retained the ancestral configuration (Musilova et al. 2013). Therefore, EBU10 derived from a centric fusion between two acrocentric ancestral elements (orthologous to ECA10q and EKI23). Following fusion, a repositioning event moved the centromere away from the fusion point. At the fusion site, which is located 4 Mb away from the CENP-A binding domain, arrays of 2PI and EC137 satellites were found as relics of the ancestral centromeres involved in the fusion (supplementary table S10, Supplementary Material online).
EBU12 resulted from the fusion of three acrocentric chromosomes corresponding to horse chromosomes 22, 30, and 29 (fig. 5A). Its CENP-A binding domain corresponds to a noncentromeric locus on ECA22 and is located 10 Mb away from the ECA22/ECA30 fusion site, suggesting that this satellite-free centromere emerged from repositioning. Also in this case, the fusion region contains arrays of the 2PI pericentromeric satellite (supplementary table S10, Supplementary Material online).
EBU15 derives from the fusion between ancestral elements corresponding to horse chromosomes 12 and 6p (fig. 5A). The organization of this fusion region is depicted in figure 6D. The CENP-A binding domain of EBU15 is orthologous to a noncentromeric horse locus that is located at a distance of about 1.1 Mb from the pericentromeric satellite of ECA12 (fig. 6D). The p-arm of EBU15 and the q-arm of ECA12 are not collinear, but several rearrangements, including two previously undescribed inversions, occurred (fig. 5A). In addition, in the region immediately surrounding the centromere, two relatively small inversions differentiate the Burchell’s zebra and the horse genomes (fig. 6D). Interestingly, one of the breakpoints of this complex rearrangement falls exactly at the border of the satellite-free CENP-A binding domain where a 10 kb stretch of 2PI and 37cen satellites is present (yellow in fig. 6D and supplementary table S8, Supplementary Material online). It is tempting to speculate that this break was involved in the inactivation of the ancestral centromere and in the formation of the new one. It is likely that these arrays are relics of the ancestral pericentromeric satellites corresponding to the satellite-based centromere of horse chromosome 12.
In figure 5B, the Burchell’s zebra chromosomes that carry satellite-free centromeres and did not derive from fusions (chromosomes 14, 16, 17, 18, 20, 21, and X) are depicted. They correspond to entire horse chromosomes or chromosome arms.
In the case of EBU14, previous cytogenetic comparative maps showed that the ancestral configuration is not represented by the horse chromosome 7 but rather by the Hartmann’s zebra chromosome 14 (Musilova et al. 2013). Indeed, EBU14 is collinear with the ancestral chromosome but carries a repositioned satellite-free centromere, whereas ECA7 derived from a pericentromeric inversion.
EBU16 and EBUX are submetacentric chromosomes entirely collinear with ECA9 and ECAX, respectively. In a previous cytogenetic study, the two Burchell’s zebra centromeres were considered at the same position of the horse ones (Musilova et al. 2013). Here, sequence analysis allowed us to demonstrate that EBU16 and EBUX carry satellite-free centromeres that are orthologous to horse noncentromeric loci (supplementary table S1, Supplementary Material online).
EBU17 and EBU18 are entirely collinear with ECA5q and with the acrocentric chromosome ECA24, respectively (Piras et al. 2009; Musilova et al. 2013). Their centromeres moved from the ancestral terminal position to a new position void of satellite DNA, leading to the formation of metacentric chromosomes.
Finally, from previous cytogenetic analysis (Musilova et al. 2013), EBU20 and EBU21 were considered identical to their horse acrocentric orthologs, ECA26 and ECA28, respectively. Here, we found that a satellite-free centromere is present on these two Burchell’s zebra chromosomes and that their CENP-A binding domains are located 4.9 and 1.7 Mb away from their p-arm terminus, respectively (supplementary table S3, Supplementary Material online). The horse loci orthologous to these two satellite-free centromeres do not contain functional centromeres. Satellite repeats, remnants of the ancestral satellite-based centromeres, are now located on the short arm (Piras et al. 2010). These sequences are orthologous to noncentromeric sequences located on the proximal portion of the q arms of horse chromosomes 26 and 28 (supplementary fig. S6, Supplementary material online). Therefore, the short arms of EBU20 and EBU21 are a few megabases longer than the horse ones.
Mechanisms of Satellite-free Centromeres Formation in Grevy’s Zebra
We investigated the mechanisms of neocentromere formation in Grevy’s zebra. As mentioned above, a reference genome was not available for this species, but, given the high karyotype (Musilova et al. 2013) and sequence (Jónsson et al. 2014) identity with Burchell’s zebra, we used our Burchell’s zebra genome assembly (Equus_quagga_cen) and the hybrid genome (EBU_EGR_cen) as reference.
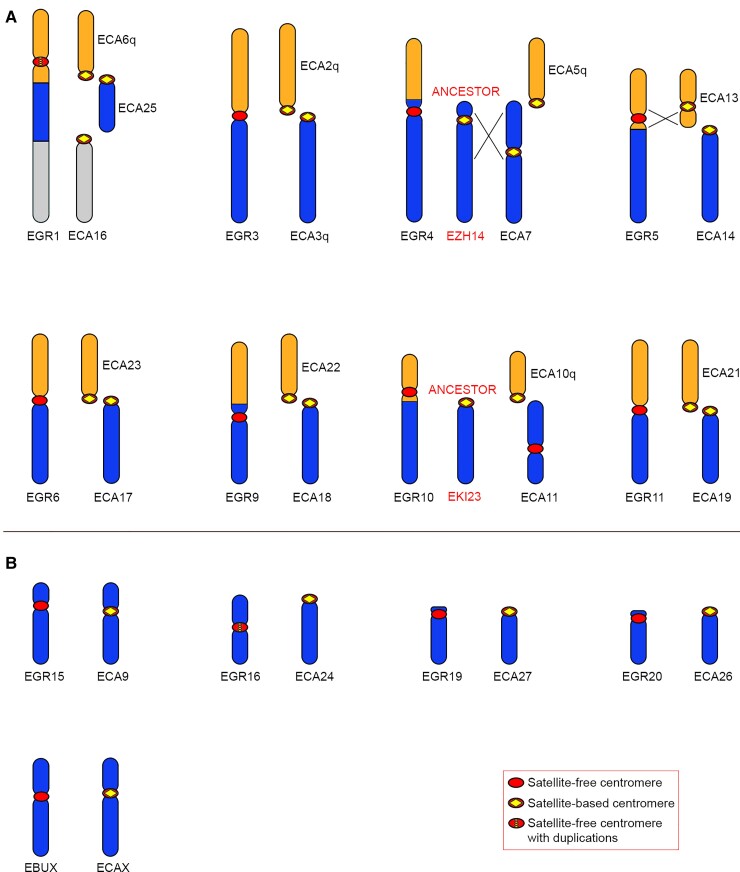
In Grevy’s zebra, chromosomes 1, 3, 4, 5, 6, 9, 10, and 11 originated by fusions and carried a satellite-free centromere (fig. 7A). In all these chromosomes, the ancestral centromeres were inactivated. At the cytogenetic level, chromosomes 1, 3, 5, 6, and 10 are collinear with Burchell’s zebra chromosomes 1, 3, 5, 6, and 10, respectively, suggesting that these fusion events occurred in the common ancestor of the two species (Musilova et al. 2013). Differently, Grevy’s zebra chromosomes 4, 9, and 11 derived from lineage-specific fusion events. A description of all fusion events, as depicted in fig. 7A, is reported below.
Fig. 7.
Comparison between Grevy’s zebra chromosomes with satellite-free centromeres and the orthologous horse chromosomes. For each Grevy’s zebra chromosome carrying a satellite-free centromere, the corresponding orthologous horse chromosomes are shown. For EGR4 and EGR10, the ancestral chromosomes, represented by Hartmann’s zebra chromosome 14 (EZH14) and kiang chromosome 23 (EKI23), respectively, are reported. Inverted segments are indicated with crossed lines. (A) EGR chromosomes deriving from fusions. (B) EGR chromosomes corresponding to entire horse chromosomes or chromosome arms.
EBU1 and EGR1 are collinear and derived from the fusion of ancestral elements corresponding to horse chromosomes 6q, 25, and 16 (Musilova et al. 2013) (fig. 7A). At the cytogenetic level, the EBU1 and EGR1 centromeres appear at the same position; however, EGR1 is characterized by a satellite-free centromere, whereas the centromere of EBU1 is satellite-based. To evaluate the distance between the CENP-A binding domain of EGR1 and the fusion region, we aligned the EBU1 chromosome from the Equus_quagga_cen assembly with the orthologous chromosomes from the horse reference genome. This analysis shows that the satellite-free centromere of EGR1 is located at about 10 Mb from the 6q/25 fusion region (supplementary table S10 and fig. S6, Supplementary Material online), suggesting that a repositioning event moved this centromere far from the fusion point. The sequence of the EGR1 centromere, which is characterized by the presence of amplifications, corresponds to a gap in the Burchell’s zebra genome assembly (fig. 3).
EGR3 derived from a centric fusion involving two ancestral acrocentric chromosomes orthologous to horse chromosomes 2q and 3q (Myka et al. 2003) and is collinear with EBU3 (supplementary table S10 and fig. S6, Supplementary Material online). The organization of this fusion region is depicted in figure 6C. Although EBU3 contains a satellite-based centromere, the CENP-A binding domain of EGR3 resides on a sequence that is unique in Burchell’s zebra but shares high identity with several horse pericentromeres, including the one from horse chromosome 3q (fig. 6C). This is the reason why this satellite-free centromere was detected in the Burchell’s zebra assembly but not in the EquCab3.0 reference genome (fig. 3). The fusion point lays about 500 kb upstream of the centromeric domain where a good alignment with ECA2q pericentromeric sequences was observed (light orange in fig. 6C). The presence of the CENP-A binding domain within the ancestral pericentromeric sequences suggests that the centric fusion contributed to satellite-free centromere formation.
EGR4 derived from the fusion of two ancestral elements corresponding to horse chromosome 5q and Hartmann’s zebra chromosome 14, which is orthologous to Burchell’s zebra chromosome 14 (figs. 5A and 7A). The satellite-free centromere of EGR4 lies in the same position of the one on EBU14 (supplementary table S1, Supplementary Material online), suggesting that a centromere repositioning event occurred in the common ancestor of the two zebra species before the fusion with the ECA5q element in the Grevy’s zebra lineage.
EGR5 is orthologous to EBU5 (Musilova et al. 2013); however, although the centromere of EGR5 is satellite-free, the one from EBU5 is satellite-based (Piras et al. 2010) (supplementary table S10, Supplementary Material online). EGR5 originated by the fusion between ancestral elements corresponding to horse chromosomes 13 and 14. From previous cytogenetic analysis, an inversion was identified in the EGR5 segment orthologous to ECA13 (Musilova et al. 2013). Our sequence analysis confirmed that the EGR5 neocentromere is contained in the inverted region (fig. 7A). The EGR5 CENP-A binding domain is located 4.8 Mb away from the fusion site, which contains 2PI satellite arrays (supplementary table S10, Supplementary Material online).
EGR6, deriving from the centric fusion between elements corresponding to ECA17 and ECA23 (fig. 7A), is orthologous to EBU6, which was described above (fig. 5A and supplementary table S10, Supplementary Material online). Notably, the CENP-A binding domain of EGR6 is only 13 kb away from the 52 kb region containing the fusion point. As shown in figure 6A, the CENP-A binding domains of the satellite-free centromeres on EGR6 and EBU6 are located only 278 kb apart on the reference genome (fig. 6A and supplementary table S1, Supplementary Material online).
EGR9 is partially orthologous to EBU4 (Musilova et al. 2013) (figs. 5A and 7A and supplementary table S10, Supplementary Material online). These two chromosomes derive from different fusion events, both involving an ancestral element corresponding to horse chromosome 18. In EGR 9, this element was fused with an element orthologous to horse chromosome 22 (fig. 7A), whereas in EBU4, it was fused with an element orthologous to horse chromosome 19 (fig. 5A). In EGR9 and in EBU4, the satellite-free centromere is localized in the same genomic region on the ECA18-derived element (supplementary table S1, Supplementary Material online) at about 6.5 Mb from the fusion region.
EGR10 is orthologous to EBU10 (Musilova et al. 2013) (figs. 5A and 7A and supplementary table S10, Supplementary Material online). These two chromosomes carry their satellite-free centromeres in the same genomic locus at about 4 Mb from the fusion site (supplementary tables S1 and S10, Supplementary Material online), suggesting that chromosome fusion and satellite-free centromere formation occurred in their common ancestor.
EGR11 is partially orthologous to EBU9 (Musilova et al. 2013) (figs. 5A and 7A and supplementary table S10, Supplementary Material online). EGR11 and EBU9 derive from different fusion events involving an ancestral element corresponding to ECA21. The organization of this fusion region is depicted in figure 6B. In EGR11, this element was fused with an element corresponding to ECA19 (fig. 7A), whereas in EBU9, it was fused with an element corresponding to ECA8q (fig. 5A). The EGR11 CENP-A binding domain is orthologous to a satellite-free noncentromeric locus on ECA21 and lies in the same chromosomal locus of the satellite-free centromere of EBU9 (fig. 6B). These satellite-free centromeres are adjacent to the fusion region (fig. 6B).
The remaining Grevy’s zebra chromosomes carrying a satellite-free centromere (EGR15, 16, 19, 20, and X) correspond to entire horse chromosome or chromosome arms and are entirely collinear with them (fig. 7B). All these centromeres reside in genomic regions orthologous to noncentromeric regions in the horse. The CENP-A binding domains of EGR15, 16, 20, and X are located in the same genomic position of the ones of their Burchell’s zebra orthologs (EBU16, 18, 20 and X) (supplementary table S10, Supplementary Material online), suggesting that they originated before the separation of the two lineages.
Discussion
Identification of Satellite-free Centromeres in Burchell’s and Grevy’s Zebra
The DNA Zoo team recently released the first genome assembly for the Burchell’s zebra (Equus_quagga_HiC). This genome sequence contains scaffolds that correspond to entire zebra chromosomes but lack chromosome assignment. In this work, thanks to the availability of subchromosomal comparative maps between the Burchell’s zebra and the horse (Musilova et al. 2013), we improved the DNA Zoo assembly by assigning chromosome numbers to scaffolds and adjusting the direction of some chromosomes. To further improve the Burchell’s zebra reference genome, we replaced the regions surrounding the 15 satellite-free centromeres with sequences that we assembled from our ChIP-seq reads.
The new assembly was also used as reference to identify the 13 satellite-free centromeres of Grevy’s zebra, a species closely related to Burchell’s zebra but still lacking a reference genome.
This work allowed us to demonstrate, at the DNA sequence level, that Burchell’s zebra and Grevy’s zebra are characterized by an extraordinary high number of centromeres completely devoid of satellite DNA. Indeed, as for the donkey (Nergadze et al. 2018), more than half of their chromosomes have satellite-free centromeres, further proving that the absence of satellite DNA at centromeric domains is compatible with genome stability and species survival.
Using this approach, we could not obtain ChIP-seq peaks from the satellite-based centromeres because their corresponding reads could not be mapped on specific chromosomes since each satellite-based centromere contains megabases of unassembled satellite arrays. These centromeres are probably organized similarly to the typical mammalian centromeres, as already shown for satellite-based horse centromeres (Nergadze et al. 2014; Cerutti et al. 2016).
Previous work from our laboratory demonstrated that the position of satellite-free centromeric domains can vary in the horse and donkey populations (Purgato et al. 2015; Nergadze et al. 2018; Cappelletti et al. 2019). Here, although we analyzed only one individual from each species, we were able to observe, in a few cases, two well-separated CENP-A binding domains, corresponding to different epialleles on the two homologs. This observation confirms that the phenomenon of centromere sliding that we previously observed in the horse and donkey is also occurring in the two zebras.
Seven Burchell’s zebra centromeres (from EBU6, EBU7, EBU8, EBU9, EBU16, EBU17, and EBU18 chromosomes) and nine Grevy’s zebra centromeres (from EGR1, EGR3, EGR4, EGR5, EGR6, EGR10, EGR11, EGR15, and EGR16 chromosomes) were shown to lack satellite DNA at the FISH resolution level (Piras et al. 2010). Here we confirmed, at the DNA sequence level, that they are completely satellite-free. However, not all the satellite-free centromeres identified in the present work coincide with those described in our previous cytogenetic analysis (Piras et al. 2010). The first discrepancy concerns several chromosomes whose primary constrictions seemed to be associated to satellite repeats (EBU4, EBU10, EBU12, EBU14, EBU15, EBU20, EBU21, EBUX, EGR9, EGR19, and EGR20) (Piras et al. 2010). In the present work, we found that the CENP-A binding domains of these chromosomes lay on single-copy regions. We propose that these satellite-free neocentromeres formed relatively close to highly repetitive tandem repeats cytogenetically coinciding with primary constrictions. The satellite repeats previously observed by FISH may correspond to ancestral centromeric domains that are now inactive, whereas the CENP-A binding domains lay on single-copy regions. These observations indicate that the centromeric function has shifted away from a satellite repeat containing locus to a satellite-free new position and confirm that satellite DNA sequences are not sufficient nor necessary for specifying centromere function.
In addition, at seven Grevy’s zebra chromosomes (EGR2, EGR8, EGR13 EGR14, EGR17, EGR18, and EGR22), although no satellite FISH signals were detected in our previous cytogenetic analysis, we did not identify, in the present study, satellite-free centromeres by ChIP-seq. It is possible that, at these “elusive” centromeres, short or chromosome-specific tandem arrays, undetectable at the FISH resolution level, are present. Alternatively, since a Grevy’s zebra reference genome is not available, it is possible that their CENP-A binding domains lay on single copy regions that may be absent both in the Burchell’s zebra and in the horse assemblies, suggesting that the number of Grevy’s zebra satellite-free centromeres may be underestimated. In the future, the availability of good assemblies for these species will allow a more comprehensive comparative analysis of centromeres and chromosome rearrangements.
DNA Sequence Composition of Satellite-free Centromeres
The extraordinary high number of sequenced satellite-free centromeres allowed us to investigate whether any conserved sequence feature is present at these genomic regions. As previously shown for the donkey satellite-free centromeres (Nergadze et al. 2018), the Burchell’s and Grevy’s zebra centromeres are LINE-1 rich. Although not universal, the presence of retroelements is common at the centromeres of many mammals, insects, plants, and fungi (Plohl et al. 2008; Longo et al. 2009; Klein and O’Neill 2018; Yadav et al. 2018; Chang et al. 2019), whereas it is still under debate whether these elements might promote genome instability of these regions.
It is well described that LINE-1 elements are preferentially inserted into AT-rich sequences that display low nucleosome occupancy (Sultana et al. 2019). AT richness is a typical feature of centromeres in a number of organisms (Clarke and Carbon 1985; Marshall et al. 2008; Talbert and Henikoff 2020) and might favor the adoption of a non-B DNA configuration that is usually found at centromeres (Kasinathan and Henikoff 2018; Talbert and Henikoff 2020). It was recently shown that AT-rich exogenous DNA is also capable of functioning as a centromere in the model organism Schizosaccharomyces pombe (Barbosa et al. 2022). As previously shown for the 16 satellite-free donkey centromeres (Nergadze et al. 2018), the neocentromeres of the two zebras are AT-rich as well, suggesting that such a sequence feature may favor CENP-A binding and other epigenetic modifications.
Sequence analysis revealed that, in two Burchell’s zebra (from EBU17 and EBU18) and in two Grevy’s zebra (from EGR1 and EGR16) satellite-free centromeres, amplification of DNA sequences occurred. We previously proposed that evolutionary new centromeres, initially devoid of satellite DNA, can undergo a process of “maturation” during their evolution through the acquisition of satellite DNA (Piras et al. 2010). In this view, the presence of duplications, that we previously observed also at a subset of donkey satellite-free neocentromeres, has been interpreted as the first step of centromere “satellitization” (Nergadze et al. 2018). At EBU18 and EGR16, which are orthologous and completely collinear, the CENP-A binding domains are partially overlapping on the reference genomes, suggesting that centromere formation and sequence amplification occurred before the separation of the two lineages. Although in the commonly accepted phylogenetic tree of equids (Jónsson et al. 2014), the lineages of donkey and Burchell’s zebra are separated, EBU17 is orthologous and completely collinear with donkey chromosome 16 and their centromeres are both characterized by the presence of amplifications (Nergadze et al. 2018). A possible explanation of this finding is that centromere repositioning occurred in a common ancestor of asses and zebras and that lineage sorting or chromosome rearrangement was then responsible for maintaining this centromere only in these two lineages. Alternatively, we can hypothesize that independent repositioning events occurred in the two lineages at a hotspot for neocentromere formation similar to what was already observed for human clinical neocentromeres (Marshall et al. 2008). Another observation supporting this hypothesis is the formation of clinical neocentromeres at positions corresponding to ancestral nonhuman primate centromeres, where presumably a latent centromere forming potentiality persisted (Ventura et al. 2004; Capozzi et al. 2009; Rocchi et al. 2012).
Conservation of Centromeric Domains
The Burchell’s and the Grevy’s zebras share 13 orthologous chromosomes that are identical at the cytogenetic level (Trifonov et al. 2012; Musilova et al. 2013). In the present work, we demonstrated that in only one of the couples of orthologs (EBU2/EGR2), both centromeres are satellite-based, whereas six out of these couples (EBU6/EGR6, EBU10/EGR10, EBU16/EGR15, EBU18/EGR16, EBU20/EGR20, and EBUX/EGRX) harbor a satellite-free centromere in the same locus, suggesting that neocentromere formation occurred in the common ancestor of the two species. These evolutionary new centromeres did not have the time to acquire the typical complexity of mammalian centromeres. This situation is consistent with the evolutive youth of these species, which emerged about 1.4 Ma (Jónsson et al. 2014). On the other hand, six orthologous pairs (EBU1/EGR1, EBU3/EGR3, EBU7/EGR7, EBU8/EGR8, EBU15/EGR14, and EBU19/EGR19) are cytogenetically collinear, but we could detect a satellite-free centromere in one of the two zebras only. The EBU19 and EGR19 orthologs are small chromosomes that were described as acrocentric (Musilova et al. 2013). In EGR19, the centromere might have been repositioned very close to the terminal position, whereas the ancestral satellite-based centromere was maintained in EBU19. All the other pairs of orthologs with divergent centromeres derived from fusion events. Two alternative explanations can be proposed to explain these observations: (1) independent fusion events involving the same ancestral elements and inactivation of the old centromeres occurred in the two species. In this scenario, only one of the two fusion events was successively accompanied by the formation of a satellite-free centromere; (2) chromosome fusion and satellite-free centromere formation, occurred in the common ancestor of the two zebras, but then centromere maturation took different routes in the two species generating new satellite repeats in one species only. In this scenario, for each pair of orthologous chromosomes, the centromere of a species acquired new satellite DNA sequences, reaching the typical complexity of mammalian centromeres, whereas in the other lineage, the “immature” satellite-free centromere was maintained. The case of EBU1 and EGR1 may support the second possibility: in EBU1, a satellite-based centromere is present, whereas in EGR1, a satellite-free centromere with amplified sequences may represent an intermediate maturation step.
There are other pairs of chromosomes that are only partially collinear, in which both species carry a satellite-free neocentromere in the same genomic locus (EBU4/EGR9, EBU9/EGR11, and EBU14/EGR4). A possible explanation of this finding is that these satellite-free centromeres appeared in a common ancestor and that lineage-specific fusion events occurred after neocentromere formation. Alternatively, two different Robertsonian fusions occurred in the two species and then satellite-free centromeres arose independently in the same “centromerization” hot spot.
Mechanisms for Satellite-free Centromeres Formation
The rapid equid evolution was characterized by extensive karyotype reshaping. The formation of satellite-free centromeres and inactivation of old satellite-based centromere played a key role in this process. In the horse and donkey, the mechanism for satellite-free centromere formation was the shift of the centromeric function from an ancestral terminal position to a new interstitial position not involving chromosome rearrangement (Carbone et al. 2006; Wade et al. 2009; Piras et al. 2010; Nergadze et al. 2018). In Burchell’s and Grevy’s zebras, we observed several satellite-free centromeres that were generated through this mechanism; however, chromosome rearrangements, particularly chromosome fusions, were also involved. In supplementary figure S7, Supplementary Material online, models for the role of repositioning (panel A) and fusion (panel B) in satellite-free centromere formation are proposed. These models involve successive steps of centromere maturation during evolution. In the karyotype of the two zebra species, chromosomes corresponding to each evolutionary step were identified and reported in the figure.
Interestingly, the CENP-A binding domain often resides in the fusion regions, on pericentromeric sequences derived from the ancestral inactivated centromeres, strongly suggesting that the Robertsonian fusion directly triggered the formation of a satellite-free centromere within the fusion region.
Our sequence analysis revealed that, in zebras, the centromeres of chromosomes derived from Robertsonian fusions are different from the ones previously described in mouse (Garagna et al. 2001, 2014). Cytogenetic analysis showed that, in the mouse, the fusion region maintains long stretches of satellite repeats with an antiparallel symmetry and an equal contribution of the two ancestral telocentric chromosomes in the formation of the new satellite-based centromere (Garagna et al. 2001). On the contrary, the molecular analysis presented here showed that, in most zebra Robertsonian fusions, the new centromere is satellite-free, whereas the ancestral satellite repeats are lost.
A peculiar satellite-free centromere is the one on EBU15, where an inversion was probably involved in its formation. Indeed, this satellite-free centromere was seeded on an inversion breakpoint. The repair of the DNA double-strand break within the ancestral centromere may have caused loss and/or rearrangement of sequences which, in turn, may have inactivated the centromeric function. In this scenario, the formation of the novel satellite-free centromere on the acentric chromosome would have been essential to rescue proper segregation potential of the chromosome.
It has been suggested that centromere strength is directly correlated with the length of satellite arrays. According to this view, the fixation of fused chromosomes can result from meiotic drive and occur when the centromere of a bi-armed chromosome is “stronger” than the original centromeres, ending in its preferential segregation into the egg during female meiosis (Fishman and Saunders 2008; Chmátal et al. 2014; Iwata-Otsubo et al. 2017; Wei et al. 2017; Kursel and Malik 2018; Talbert and Henikoff 2020). According to this model, extended satellite arrays would allow CENP-A nucleosome expansion, resulting in the formation of a stronger kinetochore compared with short arrays. On the contrary, the results presented here show that, in zebras, neocentromeres completely devoid of satellite DNA are present in fusion chromosomes, indicating that the contribution of repeated DNA in strengthening the centromere is not universal and that other genetic or epigenetic features may guarantee the stability of centromeres. It is also possible that factors such as small population size and bottle-neck during evolution may have contributed to the fixation of satellite-free centromeres in Robertsonian chromosomes (Trifonov et al. 2008; Ransom and Kaczensky 2016).
Materials and Methods
Cell Lines
Primary fibroblast cell lines from Burchell’s zebra and Grevy’s zebra were previously described (Piras et al. 2010). The cells were cultured in high-glucose DMEM medium supplemented with 20% fetal bovine serum, 2 mM l-glutamine, 1% penicillin/streptomycin, and 2% non-essential amino acids. The cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
ChIP-seq
Chromatin from primary fibroblasts was cross-linked with 1% formaldehyde, extracted, and sonicated to obtain DNA fragments ranging from 200 to 800 bp. Immunoprecipitation was performed as previously described (Nergadze et al. 2018) by using an anti-CENP-A serum (Cappelletti et al. 2019). Paired-end sequencing was performed with an Illumina HiSeq2500 platform by IGA Technology Services (Udine, Italy). In supplementary figure S8, Supplementary Material online, the MultiQC plots resulting from FastQC (Ewels et al. 2016) analysis, describing the quality of raw reads, are reported. As shown in these figures, the quality of these reads is very high: per base mean sequence quality always higher than 30 (base accuracy >99.9%).
Comparative Genomic Analysis
Pairwise alignments between whole genomes were performed with Chromeister (version 1.5a) (Pérez-Wohlfeil et al. 2019) available at the European Galaxy web platform (https://usegalaxy.eu/) (Afgan et al. 2016) using default parameters. The aligned genomes were the EquCab3.0 horse genome, the Equus_quagga_HiC Burchell’s zebra genome, and the modified Burchell’s zebra genome that we produced, Equus_quagga_cen. The Equus_quagga_HiC draft assembly, available at https://www.dnazoo.org/assemblies/Equus_quagga, was generated by the DNA Zoo consortium from short insert-size PCR-free DNA-Seq data using w2rap-contigger (Clavijo et al. 2017) (see Dudchenko et al. 2017, 2018 for details). The blood sample for in situ Hi-C preparation was donated by a female individual named Zena and obtained from Nancy Nunke (Hearts & Hands Animal Rescue), Greg Barsh (Stanford, Hudson Alpha), and Ren Larison (UCLA). Global statistics for EquCab3.0, Equus_quagga_HiC, and Equus_quagga_cen genome assemblies are shown in supplementary table S11, Supplementary Material online. Metrics were obtained using Quast Genome assembly Quality (Galaxy Version 5.0.2 + galaxy4) (Gurevich et al. 2013).
Pairwise alignments between single chromosomes were run locally using the same Chromeister version (https://github.com/estebanpw/chromeister) with default parameters. The resulting plots were used to evaluate chromosome orthologies and Burchell’s zebra chromosome orientations. Fine-scaled orthologies were resolved by BLAT-searching a sequence of interest from the Burchell’s zebra genome against the horse EquCab3.0 reference using UCSC Genome Browser (https://genome.ucsc.edu/index.html).
Bioinformatic Analysis of ChIP-seq Data
Reads were aligned with paired-end mode to the reference genomes with Bowtie2 aligner using default parameters (version 2.4.2) (Langmead et al. 2009; Langmead and Salzberg 2012). Normalization of read coverage of the ChIP data sets against the input data sets was performed using bamCompare available in the deepTools suite (3.5.0 version) (Ramírez et al. 2016) using RPKM (Reads Per Kilobase per Million mapped reads) normalization in subtractive mode. Normalization of read coverage of the input data sets for amplification analysis was performed using bamCoverage available in the deepTools suite (Ramírez et al. 2016) using RPKM (reads per kilobase per million mapped reads) normalization. The resulting coverage files were visualized using Integrative Genomics Viewer (IGV) software (http://software.broadinstitute.org/software/igv/home). Peaks were obtained with the R software package Sushi (Phanstiel et al. 2014). Peak calling was performed with MACS2 (version 2.2.7.1) (Zhang et al. 2008; Feng et al. 2012) using –broad option to identify large enriched genomic regions. We did not include in our analysis enriched regions containing satellite arrays. Enriched regions from Equus_quagga_HiC were BLAT-searched against EquCab3.0 using UCSC Genome Browser (https://genome.ucsc.edu/index.html) to confirm the correspondence with the orthologous enriched loci identified in the horse genome.
Since the Bowtie2 default parameters are not optimized for aligning repetitive sequences, we also aligned the reads using two alternative settings (alternative 1: -k10 option; alternative 2: -end-to-end -very-sensitive -no-unal -no-mixed -no-discordant -overlap –dovetail -I10 -X700 as used by Kasinathan and Henikoff (2018)) and compared the results. The plots obtained with R from the ChIP samples in the regions of interest are reported in supplementary figures S9 and S10, Supplementary Material online. The shape of the CENP-A enrichment peaks, obtained with these parameters, is largely identical to those obtained with the default parameters. We subsampled 1% of the ChIP reads aligned with Bowtie2 default parameters in the centromeric regions. These reads were searched in the BAM files obtained with the parameters optimized for repetitive sequences. Mapping conservation of these reads was evaluated using CompareBams (Lindenbaum 2015) (supplementary table 12, Supplementary Material online).
Assembly of Centromeric Regions, Improvement of the Burchell’s Zebra Reference Genome and Construction of a Chimeric Genome
The assembly of centromeric regions from Burchell’s and Grevy’s zebras was performed using an iterative chromosome walking approach based on the paired-end ChIP-seq reads. We previously used the same method to assemble the centromeric regions of the donkey satellite-free centromeres (Nergadze et al. 2018). Peaks corresponding to CENP-A binding domains were visualized using Integrative Genome Viewer (IGV, Version 2.9.2) on EquCab3.0 and/or Equus_quagga_HiC reference genomes. The reads mapping in these regions have high mapping quality and coverage (supplementary figs. S2, S9, and S10, Supplementary Material online). We retrieved the consensus of the mapped reads using the “Copy consensus sequence” function of IGV. Consensus bases other than ACGTN were corrected by a visual inspection of reads aligned to the reference genome. We then proceeded to refine these draft sequences, resolving misassembled regions by de novo assembly of raw paired-end reads using a walking approach. To this end, we joined read pairs of ChIP and Input data sets using FASTQ joiner (Galaxy Version 2.0.1.1 + galaxy0) (Blankenberg et al. 2010). Queries of 60–95 bp, flanking gaps or misassembled regions of the draft sequences, were searched in the joined paired-end reads using the Grep command. Retrieved reads were aligned using MultAlin (Corpet 1988) and a new query was designed on the newly obtained sequence. This procedure was reiterated to resolve gaps and misassembled regions that were present in the draft consensus.
To obtain a refined reference genome for Burchell’s zebra, we first run BLAT (v. 36) (Kent 2002) using the assembled contigs as query to identify their incomplete and/or misassembled counterparts in the Equus_quagga_HiC scaffolds. The incomplete and/or misassembled sequences were then removed and substituted with the newly assembled centromeric contigs using SAMtools (version 1.11) (Danecek et al. 2021). Finally, when the orientation of the Equus_quagga_HiC scaffolds identified through the Chromeister tool was reverse, we adjusted it using the Reverse.seq tools (Galaxy Version 1.39.5.0) (Schloss et al. 2009) available at the Galaxy web platform (https://usegalaxy.org/) (Afgan et al. 2016).
To obtain a more accurate reference for Grevy’s zebra, the regions orthologous to the newly assembled contigs were identified by BLAT (v. 36) in the Equus_quagga_cen genome. These regions were removed and substituted with the Grevy’s zebra centromeric contigs to obtain a chimeric reference genome, as previously described (Nergadze et al. 2018).
The resulting reference genomes were used to re-map ChIP-seq reads and obtain enrichment peaks, as described above. Mapping statistics were obtained using QualiMap BamQC Galaxy Version 2.2.2c + galaxy1 (García-Alcalde et al. 2012; Okonechnikov et al. 2016) (supplementary table S4, Supplementary Material online). The number of N’s per 100 kb in the original sequences and in the newly assembled contigs was obtained using Quast Genome assembly Quality (Galaxy Version 5.0.2 + galaxy4 or online version available at http://cab.cc.spbu.ru/quast/).
Sequence Analysis of Centromeric Domains
The presence of DNA amplification at centromeres was evaluated by comparing the read counts per kilobase in the centromeric domains with the genome-wide average using input reads aligned to the corrected Burchell’s zebra reference genome or to the chimeric reference genome. Counts were obtained using SAMtools suite (version 1.11).
We calculated standardized Z-score for the coverage values of EBU17, EBU18, EGR1, and EGR6 with respect to genome-wide coverage. To test whether the differences between coverages are statistically significant, we calculated the P-values using Z-score calculator (available at https://www.socscistatistics.com/pvalues/normaldistribution.aspx).
An analysis of the organization of duplicated sequences was performed manually using MultAlin (Corpet 1988) to identify and align repeated sequences.
An analysis of the content in repetitive elements in centromeric domains was carried out with RepeatMasker (Galaxy Version 4.0.9) (http://www.repeatmasker.org) available at the Galaxy web platform. To analyze satellite DNA content, RepeatMasker was run using a combined database made by Dfam-Dfam_3.0 and RepBase library (release October 26, 2018), which contains the consensus sequences of the major equid satellite DNA families and a custom library made by the consensus sequence of the EC137 pericentromeric satellite DNA family (Nergadze et al. 2014) which is not present in the RepBase library. The same procedure was used to analyze interspersed repeats. Masked bp and number of repeats are reported in supplementary table S13, Supplementary Material online. The same analysis was carried out on the entire Burchell’s zebra genome in order to obtain genome-wide values (supplementary table S14, Supplementary Material online). Statistical significance was assessed by t-test using the VassarStats website by comparing the values of centromeric genomic regions with the values of genome scaffolds (http://vassarstats.net/). Centromeres containing DNA amplifications were excluded from this analysis.
To test whether the sequences underlying the CENP-A binding domains are truly satellite-less and do not contain novel satellite arrays, we ran ULTRA (Olson and Wheeler 2018) using -p 1000 option to also detect large period repeats. Identified repeats were selected for array length (>1000 bp) and P-value (<0.05).
Supplementary Material
Acknowledgments
We would like to thank Marco Corbo, Simone Faravelli, Riccardo Gamba, and Francesco Gozzo for their contribution to some bioinformatic analyses preceding the present work and Kevin Sullivan for helpful suggestions during the revision of the manuscript. This work was supported by the Italian Ministry of Education, University and Research (MIUR) [Dipartimenti di Eccellenza Program (2018-2022)—Department of Biology and Biotechnology “L. Spallanzani”, University of Pavia], PRIN grant no. 2015RA7XZS_002 and from the Consiglio Nazionale delle Ricerche (CNR-Progetto Bandiera Epigenomica). The Galaxy server that was used for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research [BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR (de.NBI)].
Contributor Information
Eleonora Cappelletti, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Francesca M Piras, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Lorenzo Sola, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Marco Santagostino, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Wasma A Abdelgadir, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Elena Raimondi, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Francesco Lescai, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Solomon G Nergadze, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Elena Giulotto, Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia, 27100 Pavia, Italy.
Supplementary material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
E.G. conceived and supervised the study. E.C., F.M.P., S.G.N., and E.G. contributed to experimental design. E.C., F.M.P., and S.G.N. carried out molecular and cell biology experiments and bioinformatic analyses. E.C., F.M.P., S.G.N., and E.G. wrote the manuscript. F.L., L.S., M.S., W.A.A., and E.R. contributed to analyses. All authors read and approved the final manuscript.
Data Availability
Raw sequencing data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA804871. The assembled centromeric regions of Burchell’s zebra and Grevy’s zebra from this study have been submitted to the NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) under accession numbers OM643400-OM643427. A corrected version of the Equus_quagga_HiC assembly is available at 10.6084/m9.figshare.19145453.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. 2016. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 9:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Choo KH. 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet. 71:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Xu Z, Karari K, Williams W, Hauf S, Brown WRA. 2022. Mutation and selection explain why many eukaryotic centromeric DNA sequences are often A + T rich . Nucleic Acids Res. 50:579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy Team . 2010. Manipulation of FASTQ data with galaxy. Bioinformatics 26:1783–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Purgato S, D’Addabbo P, Archidiacono N, Battaglia P, Baroncini A, Capucci A, Stanyon R, Della Valle G, Rocchi M. 2009. Evolutionary descent of a human chromosome 6 neocentromere: a jump back to 17 million years ago. Genome Res. 19:778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti E, Piras FM, Badiale C, Bambi M, Santagostino M, Vara C, Masterson TA, Sullivan KF, Nergadze SG, Ruiz-Herrera A, et al. 2019. CENP-A binding domains and recombination patterns in horse spermatocytes. Sci Rep. 9:15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, et al. 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87:777–782. [DOI] [PubMed] [Google Scholar]
- Cerutti F, Gamba R, Mazzagatti A, Piras FM, Cappelletti E, Belloni E, Nergadze SG, Raimondi E, Giulotto E. 2016. The major horse satellite DNA family is associated with centromere competence. Mol Cytogenet. 9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen CC, Erceg J, Beliveau BJ, Wu CT, et al. 2019. Islands of retroelements are major components of drosophila centromeres. PLoS Biol. 17:e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 24:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH. 2000. Centromerization. Trends Cell Biol. 10:182–188. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. 1985. The structure and function of yeast centromeres. Annu Rev Genet. 19:29–55. [DOI] [PubMed] [Google Scholar]
- Clavijo B, Garcia Accinelli G, Wright J, Heavens D, Barr K, Yanes L, Di-Palma F. 2017. W2RAP: a pipeline for high quality, robust assemblies of large complex genomes from short read data. bioRxiv 110999.
- Cleveland DW, Mao Y, Sullivan KF. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421. [DOI] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes Aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Shamim S, Batra S, Durand N, Musial N, Mostofa R, Pham M, Hilaire B, Yao W, Stamenova E, et al. 2018. The juicebox assembly tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. biorxiv:254797.
- Earnshaw WC, Migeon BR. 1985. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92:290–296. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. 2016. Multiqc: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322:1559–1562. [DOI] [PubMed] [Google Scholar]
- Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA. 2001. Pericentromeric organization at the fusion point of mouse robertsonian translocation chromosomes. Proc Natl Acad Sci U S A. 98:171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna S, Page J, Fernandez-Donoso R, Zuccotti M, Searle JB. 2014. The robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma 123:529–544. [DOI] [PubMed] [Google Scholar]
- García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- Giulotto E, Raimondi E, Sullivan KF. 2017. The unique DNA sequences underlying equine centromeres. Prog Mol Subcell Biol. 56:337–354. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE. 2017. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27:2365–2373.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, Schubert M, Seguin-Orlando A, Ginolhac A, Petersen L, Fumagalli M, Albrechtsen A, Petersen B, Korneliussen TS, Vilstrup JT, et al. 2014. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci U S A. 111:18655–18660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch TS, Rice ES, DePriest MS, Walenz BP, Hestand MS, Vermeesch JR, Connell BLO, Fiddes IT, Vershinina AO, Saremi NF, et al. 2018. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun Biol. 1:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Choo KH. 2012. The evolutionary life cycle of the resilient centromere. Chromosoma 121:327–340. [DOI] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S. 2018. Non-B-form DNA is enriched at centromeres. Mol Biol Evol. 35:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SJ, O’Neill RJ. 2018. Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 26:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS. 2018. The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol. 52:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbaum P. 2015. Jvarkit: java-based utilities for bioinformatics. Figshare. [Google Scholar]
- Longo MS, Carone DM, Green ED, O’Neill MJ, O’Neill RJ, Program NCS. 2009. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics 10:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 82:261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova P, Kubickova S, Vahala J, Rubes J. 2013. Subchromosomal karyotype evolution in equidae. Chromosome Res. 21:175–187. [DOI] [PubMed] [Google Scholar]
- Myka JL, Lear TL, Houck ML, Ryder OA, Bailey E. 2003. Homologous fission event(s) implicated for chromosomal polymorphisms among five species in the genus equus. Cytogenet Genome Res. 102:217–221. [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Belloni E, Piras FM, Khoriauli L, Mazzagatti A, Vella F, Bensi M, Vitelli V, Giulotto E, Raimondi E. 2014. Discovery and comparative analysis of a novel satellite, EC137, in horses and other equids. Cytogenet Genome Res. 144:114–123. [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Piras FM, Gamba R, Corbo M, Cerutti F, McCarter JGW, Cappelletti E, Gozzo F, Harman RM, Antczak DF, et al. 2018. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 28:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F. 2016. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson D, Wheeler T. 2018. ULTRA: a model based tool to detect tandem repeats. ACM BCB 2018:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, et al. 2013. Recalibrating equus evolution using the genome sequence of an early middle pleistocene horse. Nature 499:74–78. [DOI] [PubMed] [Google Scholar]
- Pérez-Wohlfeil E, Diaz-Del-Pino S, Trelles O. 2019. Ultra-fast genome comparison for large-scale genomic experiments. Sci Rep. 9:10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Araya CL, Snyder MP. 2014. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30:2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Cappelletti E, Santagostino M, Nergadze SG, Giulotto E, Raimondi E. 2022. Molecular dynamics and evolution of centromeres in the genus equus. Int J Mol Sci. 23:4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E. 2010. Uncoupling of satellite DNA and centromeric function in the genus equus. PLoS Genet. 6:e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Poletto V, Cerutti F, Ryder OA, Leeb T, Raimondi E, Giulotto E. 2009. Phylogeny of horse chromosome 5q in the genus equus and centromere repositioning. Cytogenet Genome Res. 126:165–172. [DOI] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrović N, Mantovani B. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (Hetero)chromatin. Gene 409:72–82. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B. 2014. Centromere identity from the DNA point of view. Chromosoma 123:313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purgato S, Belloni E, Piras FM, Zoli M, Badiale C, Cerutti F, Mazzagatti A, Perini G, Della Valle G, Nergadze SG, et al. 2015. Centromere sliding on a mammalian chromosome. Chromosoma 124:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom J, Kaczensky P. 2016. Wild equids: ecology, management, and conservation. Baltimore: (MD: ): JHU Press. [Google Scholar]
- Roberti A, Bensi M, Mazzagatti A, Piras FM, Nergadze SG, Giulotto E, Raimondi E. 2019. Satellite DNA at the centromere is dispensable for segregation fidelity. Genes (Basel) 10:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R. 2012. Centromere repositioning in mammals. Heredity (Edinb) 108:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 75:7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Zine El Aabidine A, Pioger L, Nigumann P, Saccani S, Andrau JC, et al. 2019. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell. 74:555–570.e7. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2020. What makes a centromere? Exp Cell Res. 389:111895. [DOI] [PubMed] [Google Scholar]
- Trifonov VA, Musilova P, Kulemsina AI. 2012. Chromosome evolution in perissodactyla. Cytogenet Genome Res. 137:208–217. [DOI] [PubMed] [Google Scholar]
- Trifonov VA, Stanyon R, Nesterenko AI, Fu B, Perelman PL, O’Brien PC, Stone G, Rubtsova NV, Houck ML, Robinson TJ, et al. 2008. Multidirectional cross-species painting illuminates the history of karyotypic evolution in perissodactyla. Chromosome Res. 16:89–107. [DOI] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D’Addabbo P, Wandall A, Björck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res. 14:1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH. 1993. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH, Reddy HM, Rathnam C, Lee J, Lin D, Ji S, Mason JM, Clark AG, Barbash DA. 2017. A pooled sequencing approach identifies a candidate meiotic driver in Drosophila. Genetics 206:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Sun S, Billmyre RB, Thimmappa BC, Shea T, Lintner R, Bakkeren G, Cuomo CA, Heitman J, Sanyal K. 2018. RNAi is a critical determinant of centromere evolution in closely related fungi. Proc Natl Acad Sci U S A. 115:3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA804871. The assembled centromeric regions of Burchell’s zebra and Grevy’s zebra from this study have been submitted to the NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) under accession numbers OM643400-OM643427. A corrected version of the Equus_quagga_HiC assembly is available at 10.6084/m9.figshare.19145453.