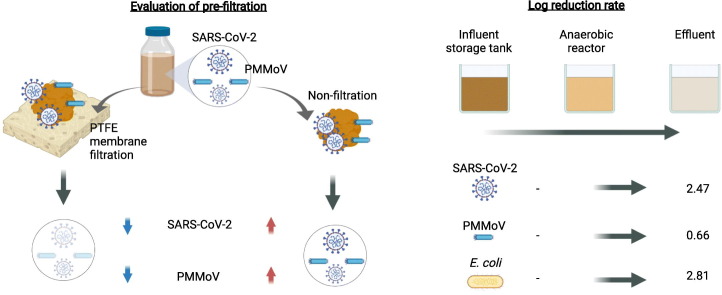
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is known to be present in sewage, and wastewater-based epidemiology has attracted much attention. However, the physical partitioning of SARS-CoV-2 in wastewater and the removal efficiency of treatment systems require further investigation. This study aimed to investigate the detectability and physical partitioning of SARS-CoV-2 in wastewater and assess its removal in a large-scale septic tank employing anaerobic, anoxic, and oxic processes in a sequential batch reactor, which was installed in a coronavirus disease 2019 (COVID-19) quarantine facility. The amount of SARS-CoV-2 RNA in wastewater was determined with polyethylene glycol (PEG) precipitation followed by quantitative polymerase chain reaction (qPCR), and the association of SARS-CoV-2 with wastewater solids was evaluated by the effect of filtration prior to PEG precipitation (pre-filtration). The amount of SARS-CoV-2 RNA detected from pre-filtered samples was substantially lower than that of samples without pre-filtration. These results suggest that most SARS-CoV-2 particles in wastewater are associated with the suspended solids excluded by pre-filtration. The removal efficiency of SARS-CoV-2 in the septic tank was evaluated based on the SARS-CoV-2 RNA concentrations in untreated and treated wastewater, which was determined by the detection method optimized in this study. Escherichia coli and pepper mild mottle virus (PMMoV) were also quantified to validate the wastewater treatment system's performance. The mean log10 reduction values of SARS-CoV-2, E. coli, and PMMoV were 2.47 (range, 2.25–2.68), 2.81 (range, 2.45–3.18), and 0.66 (range, 0.61–0.70), respectively, demonstrating that SARS-CoV-2 removal by the wastewater treatment system was comparable to or better than the removal of fecal indicators. These results suggest that SARS-CoV-2 can be readily removed by the septic tank. This is the first study to determine the removal efficiency of SARS-CoV-2 in a facility-level sequencing batch activated sludge system.
Keywords: SARS-CoV-2, Wastewater-based epidemiology, Wastewater treatment, Activated sludge, Quantification method
Abbreviations: CDC, Centers for Disease Control and Prevention; HRT, Hydraulic retention time; MHV, Murine hepatitis virus; MNV, Murine norovirus; MPC, Molecular process control; PCR, polymerase chain reaction; PMMoV, Pepper mild mottle virus; PTFE, Polytetrafluoroethylene; RT, Reverse transcription; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; SBR, Sequencing batch reactor; WBE, Wastewater-based epidemiology; WPC, Whole process control; WWTP, Wastewater treatment plant
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of the ongoing global pandemic of coronavirus disease 2019 (COVID-19). A substantial proportion of COVID-19 patients are asymptomatic or mildly symptomatic (Lavezzo et al., 2020), which leads to rapid and unrecognized spreading of SARS-CoV-2 and makes it difficult to control the transmission. As almost half of the infected individuals, including asymptomatic carriers, shed the virus through their feces (Cheung et al., 2020), wastewater-based epidemiology (WBE) has attracted much attention as an unbiased and cost-effective approach for community-level COVID-19 surveillance. The detection of SARS-CoV-2 RNA in wastewater has been reported in a number of studies (Shah et al., 2022), but it is not clear whether the viral RNA in wastewater is derived from infectious virus particles.
An epidemiological study in China identified a potential transmission of SARS-CoV-2 associated with aerosols derived from wastewater caused by sealing failures in a building's wastewater pipes (Kang et al., 2020). This epidemiological evidence implies that the wastewater discharged from a facility where patients are present may contain infectious viruses, and transmission caused by exposure to inadequately treated wastewater could occur. Reducing SARS-CoV-2 in wastewater treatment systems is an important factor in protecting public health, especially in decentralized systems (e.g., facility-level septic tanks) where the wastewater is relatively “fresh” compared to centralized ones, namely, wastewater treatment plants (WWTPs). Some studies have reported that SARS-CoV-2 reduction efficiencies in upflow anaerobic sludge blanket (UASB) systems were >1 log10 (Espinosa et al., 2022; Kumar et al., 2021). Another study (Hong et al., 2021) reported that the SARS-CoV-2 reduction from untreated wastewater to an open air biological activated sludge tank was 0.3 to 0.5 log10. However, the SARS-CoV-2 reduction efficiency in a whole system of the activated sludge process remains unknown.
To evaluate the efficiency of wastewater treatments for SARS-CoV-2, appropriate methods for virus detection in wastewater should be employed. Although several studies have detected SARS-CoV-2 RNA in solid fractions from wastewater, suggesting that SARS-CoV-2 tends to be present in wastewater solids (Kitamura et al., 2020; Peccia et al., 2020), a quantitative analysis of the liquid-solid partitioning of indigenous SARS-CoV-2 particles in wastewater has not yet been conducted. When evaluating the efficiency of SARS-CoV-2 detection methods, wastewater samples artificially contaminated with surrogate viruses, such as heat-inactivated SARS-CoV-2, murine hepatitis virus (MHV), Φ6, and MS2, have been widely used (Ahmed et al., 2020; Torii et al., 2021a; Torii et al., 2021b). However, the physical properties, such as liquid-solid partitioning, of exogenously spiked virus particles could be substantially different from indigenous ones, and this difference could affect the interpretation of experimental results and the evaluation of detection methods. Therefore, using wastewater that contains a high concentration of indigenous SARS-CoV-2 is highly desirable for accurately evaluating detection methods.
In Japan, SARS-CoV-2 RNA was detected in municipal wastewater collected from WWTPs (Haramoto et al., 2020; Kitamura et al., 2020; Torii et al., 2021a), but the SARS-CoV-2 RNA concentrations were not high enough to assess the ability of wastewater treatment to reduce SARS-CoV-2. The relatively low concentrations of SARS-CoV-2 in wastewaters in Japan are probably due to the lower prevalence of COVID-19 there than in other countries. As of March 1, 2022, a total of 213 cumulative COVID-19 deaths per million people had been reported in Japan, which is the lowest among the G7 countries (World Health Organization, 2022). To mitigate the burden of medical facilities, the Ministry of Health, Labour and Welfare, Japan issued a guideline that mildly symptomatic or asymptomatic COVID-19 patients must stay at home or in facilities such as quarantine hotels. Based on this background, we presumed that wastewater from a quarantine facility accommodating COVID-19 patients might contain high concentrations of indigenous SARS-CoV-2 and therefore serve as ideal samples for evaluating detection methods and the removal efficiency of SARS-CoV-2 in the wastewater treatment process.
In the present study, we aimed to investigate the detectability and physical partitioning of SARS-CoV-2 in wastewater and assess its removal in a large-scale septic tank employing anaerobic, anoxic, and oxic processes, which was installed in a COVID-19 quarantine facility in Japan. We also measured the E. coli and pepper mild mottle virus (PMMoV), a potential viral indicator of human fecal contamination and one of the most abundant viruses in wastewater (Kitajima et al., 2014; Kitajima et al., 2018), to validate the wastewater treatment system's performance.
2. Materials and methods
2.1. Characteristics of the septic tank and wastewater sampling
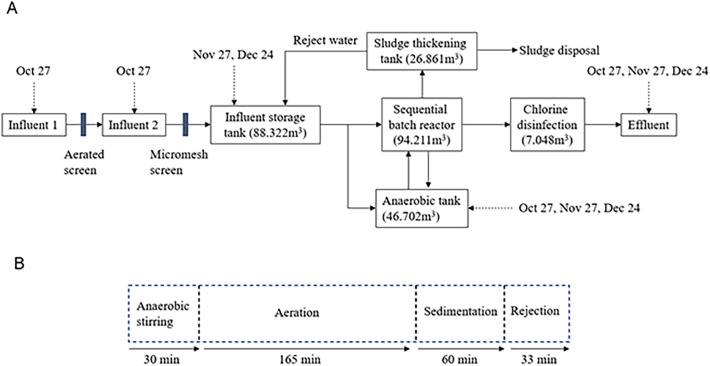
We collected wastewater samples from a large-scale septic tank in a COVID-19 quarantine facility with 461 rooms in the Tokyo metropolitan area. Because this quarantine facility is located in a cavity of a public wastewater network, its own wastewater treatment system (i.e., a large-scale septic tank, NPKB-II, Nikko Company, Hakusan, Ishikawa, Japan) was installed in the facility. This septic tank was designed to treat wastewater from 607 people and remove phosphorous, organic matter, nitrogen, and pathogens, with levels of biochemical oxygen demand (BOD), total nitrogen, and total phosphorous in effluent water set at 10, 10, and 1 mg/L, respectively (Fig. 1 ). Phosphorous removal was employed in the treatment system to prevent eutrophication because the treated wastewater was directly discharged to an open water body. The septic tank's treatment system was basically an activated sludge process with nutrient removal, consisting of an anaerobic tank and sequential batch reactor (SBR) with anaerobic, anoxic, and oxic processes followed by chlorination. To enhance phosphorous removal, the suspended solids were coagulated with polyaluminum chloride (PAC) and settled with gravity in the SBR and thickened in a sludge thickening tank. The hydraulic retention time of the influent storage tank (HRT inf) was calculated as follows:
where V inf and Q inf represent the volume influent storage tank and flow rate into the influent storage tank, respectively. As the actual flow rate was not measured, the volume of daily water usage in this facility was used as a proxy of the flow rate (Q inf). Based on the average water usage between October 6 and December 5, 2020, the HRT inf was calculated to be 6.86 days (derived from V inf of 88.32 m3 and Q inf of 12.89 m3/day), which was much longer than the originally designed HRT inf of 0.63 days.
Fig. 1.
Characteristics of the large-scale septic tank with a sequential batch reactor installed in the quarantine facility. (A) The treatment flow, tank volumes, and sampling points and dates. The solid arrows indicate the flow of wastewater or sludge, and the dashed arrows indicate the collected samples and sampling dates. (B) The operation flow of the sequential batch reactor indicated with processing time for each treatment step.
Influent 1 and 2 (before and after an aerated screen, respectively) samples were collected on one occasion (October 27, 2020), and untreated wastewater (influent storage tank) samples were collected on two occasions (November 27 and December 24, 2020), and anaerobic tank and treated wastewater (effluent) samples were collected on three occasions (October 27, November 27, and December 24, 2020) (Fig. 1). All the samples (1 L each) were collected via grab sampling at the fixed time of day (i.e., 3 PM to 4 PM) with proper personal protective equipment. These samples were frozen immediately and transported to the laboratory on dry ice, except for small aliquots for E. coli enumeration, which were transported on ice. Removal efficiencies were defined as the difference between the concentrations in the influent storage tank and effluent collected on the same occasion, which were calculated as follows:
where C 0 and C e represent the viral RNA concentration in the influent storage tank and effluent (copies/L), respectively.
2.2. Enumeration of Escherichia coli
E. coli in wastewater samples were enumerated by a culture-based method using a CHROMagar ECC (Kanto Chemical, Tokyo, Japan) following the instructions of the manufacturer. After incubation at 37 °C for 24 h, the number of blue colonies was counted, and the E. coli concentration was calculated using colony-forming units (CFU)/mL.
2.3. Virus concentration, RNA extraction, and reverse transcription (RT)
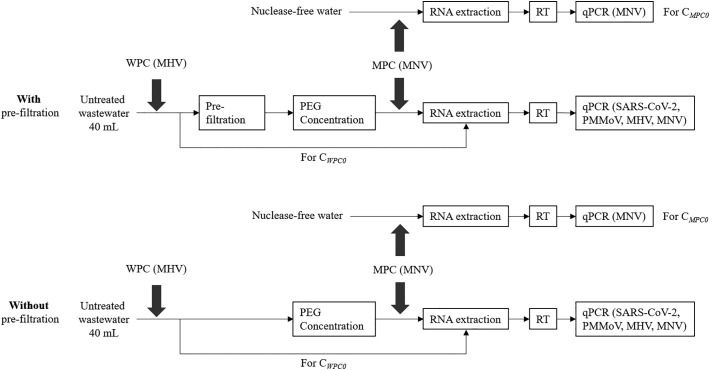
Fig. 2 shows the experimental design and flow. A set of 40-mL aliquots of wastewater was subjected to virus concentration with polyethylene glycol (PEG) precipitation. Another set of 40-mL aliquots of wastewater was filtered with a disposable hydrophilic PTFE membrane filter unit (pore size, 0.20 μm; diameter, 25 mm; Advantec, Tokyo, Japan) prior to PEG precipitation (pre-filtration). The 40-mL samples with and without pre-filtration were supplemented with 4.0 g of PEG 6000 (Wako, Osaka, Japan; cat. no.169-09125) and 0.8 g of NaCl (Wako). The samples were agitated at 4 °C overnight and then centrifuged at 12,000 ×g for 1 h (Jones and Johns, 2009). The supernatant was discarded, and the resultant pellet was resuspended in 1.0 mL of TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Viral RNA was extracted from 140 μL of the virus concentrate with a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) to obtain a 60-μL RNA extract according to the manufacturer's protocol. A High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) was used to synthesize cDNA from viral RNA via reverse transcription (RT), according to the manufacturer's protocol.
Fig. 2.
Design of the murine hepatitis virus (MHV) and murine norovirus (MNV) spike experiments as a whole process control (WPC) and a molecular process control (MPC) respectively, to evaluate the effect of pre-filtration on SARS-CoV-2 RNA detection. CWPC0 represents the observed MHV RNA concentration in the seeded wastewater sample and CMPC0 represents the observed MNV RNA concentration in the seeded nuclease-free water sample. PEG, polyethylene glycol, RT, reverse transcription, PMMoV, pepper mild mottle virus.
2.4. Sample process controls
MHV (A59 strain), propagated in DBT cells, and murine norovirus (MNV, S7-PP3 strain), kindly provided by Dr. Y. Tohya (Nihon University, Kanagawa, Japan) and propagated in RAW 264.7 (ATCC TIB-71) cells (American Type Culture Collection, Manassas, VA, USA), were used as a whole process control (WPC) and a molecular process control (MPC), respectively (Haramoto et al., 2018). MHV was spiked into 40-mL aliquots of the original wastewater samples, and 140 μL of the MHV-spiked wastewater sample was subjected to RNA extraction followed by RT-quantitative polymerase chain reaction (qPCR) described above to determine the initial concentration of MHV in the wastewater samples. The % WPC recovery efficiency (E WPC) was calculated as follows:
where C WPC represents the observed MHV RNA concentration in a concentrate after PEG precipitation (copies/mL), and C WPC0 represents the observed MHV RNA concentration in the seeded wastewater sample (copies/mL). The WPC was used to evaluate the viral RNA detection efficiency of the whole detection process including pre-filtration and PEG precipitation.
MNV was also spiked into 140 μL of nuclease-free water, which was used as a non-inhibition control to determine extraction-RT-qPCR efficiency. The % MPC recovery efficiency (E MPC; i.e., extraction-RT-qPCR efficiency) was calculated as follows:
where C MPC represents the observed MNV RNA concentration in a wastewater sample (copies/140 μL), and C MPC0 represents copy numbers in the non-inhibition control (copies/140 μL in nuclease-free water). The MPC was used to identify the viral RNA loss during extraction and the occurrence of RT-qPCR inhibition.
2.5. Quantification of viral genomes by qPCR
TaqMan-based qPCR assays (Table S1 in the Supplementary Material) for SARS-CoV-2, PMMoV, MHV, and MNV were performed with an ABI PRISM 7500 or ABI PRISM 7500 Fast sequence detection system (Thermo Fisher Scientific). The qPCR reaction mixtures (25 μL) consisted of 12.5 μL of QuantiTect Probe PCR Master Mix (Qiagen), forward and reverse primers, TaqMan probe, and 2.5 μL of cDNA template. The sequences of primers and probes are shown in Table S1. The reaction mixtures were subjected to thermal cycling, and fluorescence readings were collected and analyzed with Sequence Detector Software version 2.0 (Thermo Fisher Scientific). The threshold value of relative fluorescence intensity (ΔRn) was adjusted to be 0.01 according to the instructions from manufacturer of the PCR master mix (Qiagen), and the cycle threshold (C T) value was determined as the PCR cycle number where relative fluorescence intensity exceeded the threshold value. The genome copy numbers of each virus were determined based on the standard curve prepared with 10-fold serial dilutions of positive control plasmid DNA or gBlocks (IDT, Iowa, USA) at a concentration of 105 to 100 copies per reaction, which showed the linear relationship between the log10 concentration of the positive control and the C T value (R 2 = 0.942–0.999). Efficiency (E) of the PCR was determined to be 96.5–117.5 from the slope (S) of the linear regression curve (−2.96 to −3.41), according to the formula E = 10–1/S − 1.
Rigorous attention for quality assurance and quality control (QA/QC) was maintained throughout the qPCR experiment process. All qPCR reactions were performed in duplicate; two PCR tubes were used for all samples and standards and the average copy numbers obtained from the two tubes were used for subsequent calculations. To avoid laboratory contamination from different steps, RNA extraction and RT-qPCR sample preparation were performed in Class II biological hazard hood and PCR clean bench, respectively, in separate rooms, which were also separated from the room where PCR amplification and gel electrophoresis of PCR products were performed. The biological hazard hood and clean bench were cleaned with 70 % ethanol and DNA Away (Thermo Scientific), respectively, and exposed to UV light for at least 20 min prior to and after usage. Positive controls were always included in the qPCR reaction plates to ensure that false-negative qPCR results were avoided. Negative controls were also included to avoid false-positive results from cross-contamination, and no false-positive qPCR signals were observed.
2.6. Next generation sequencing
A PCR assay partially targeting the first protein-coding open reading frame (ORF1a) region of SARS-CoV-2 (Shirato et al., 2020) was used to confirm the presence of SARS-CoV-2 in wastewater. The PCR reaction was performed in 50 μL of reaction volume containing 2.5 μL of cDNA, 25 μL of KOD one PCR Master Mix (TOYOBO, Osaka, Japan) and 15 pmol each of forward and reverse primers. The PCR was performed under the following conditions: 35 cycles of denaturation at 98 °C for 10 s, primer annealing at 60 °C for 10 s, and extension reaction at 68 °C for 5 s. The PCR products were separated by electrophoresis on a 2 % agarose gel and visualized under ultraviolet light after ethidium bromide staining. The PCR products of the expected size were excised from the gel and purified using the FastGene Gel/PCR Extraction Kit (Nippon Genetics).
The purified PCR products (10 ng) were subjected to library preparation using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA). The quality and quantity of the libraries were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and a Qubit 4 Fluorometer (Thermo Fisher Scientific), respectively. Finally, the libraries were pooled and sequenced on the Illumina MiSeq platform using a MiSeq Reagent Kit v3, 600 Cycles (Illumina, San Diego, CA), according to the manufacturer's instructions.
The FASTQ files were generated using Local Run Manager software (Illumina). For detecting single nucleotide variants (SNVs) and short insertions/deletions (Indels), we used our in-house pipeline, HGC_CovidPipeLine (ver.2.1.0 https://www.hgc.jp/~ppl_ht/CovidPipeLine/2.1.0/docs/#), constructed at the Human Genome Center, The University of Tokyo. In the HGC_CovidPipeLine, we removed truncated reads (≤300 bp) using Trimmomatic (ver. 0.39) and aligned them to the SARS-CoV-2 reference genome (NC_045512.2) using Burrows-Wheeler Aligner (ver. 0.7.17) (Li and Durbin, 2009). SNVs and Indels were identified with the following procedure: (i) remove low quality reads (Q < 20); (ii) excluding variants with a low read depth (minimum depth ≥ 50); (iii) excluding variants with a low variant allele frequency (minimum VAF ≥ 0.1). The SnpEff (ver. 5.0) (Cingolani et al., 2012) was then used to annotate the variants.
3. Results
3.1. Effect of pre-filtration on the detectability of SARS-CoV-2 RNA in wastewater
To investigate the partitioning of SARS-CoV-2 particles between water and solid phases in wastewater, we assessed the effect of pre-filtration with a 0.20-μm pore size PTFE membrane on the detectability of SARS-CoV-2 RNA using the untreated wastewater samples (influent 1 and 2) collected on October 27, 2020. The wastewater samples were processed with or without pre-filtration prior to PEG precipitation and then examined for SARS-CoV-2 and PMMoV using RT-qPCR. Table 1 shows the result of SARS-CoV-2 (both CDC-N1 and CDC-N2) and PMMoV detection. The observed SARS-CoV-2 RNA concentrations in the wastewater samples without pre-filtration were significantly higher than those in pre-filtered wastewater for both influent 1 and 2 (paired t-test, p < 0.01) (Table 1). The levels of PMMoV RNA detected in samples without pre-filtration were comparable to or higher than pre-filtrated wastewater samples in influent 1 and 2, respectively. For subsequent analyses, wastewater samples were processed without pre-filtration because of its higher detectability for both SARS-CoV-2 and PMMoV than that of samples without pre-filtration. Regarding the SARS-CoV-2 qPCR assay comparison, the CDC-N1 assay exhibited a higher detection ratio and observed concentrations than the CDC-N2 assay (Table 1); and therefore, the CDC-N1 assay was used for qPCR for SARS-CoV-2 RNA detection.
Table 1.
| Sample type | Pre-filtrationd | PMMoV RNA concentratione (log10 copies/L) |
SARS-CoV-2 RNA concentratione (log10 copies/L) |
|
|---|---|---|---|---|
| CDC-N1 | CDC-N2 | |||
| Influent 1 | + | 6.96 ± 0.60 | 3.09 | N.D.f |
| − | 7.35 ± 0.10 | 5.20 ± 0.19 | 5.05 ± 0.17 | |
| Influent 2 | + | 6.14 ± 0.28 | 3.73 ± 0.39 | 3.86 ± 0.10 |
| − | 7.15 ± 0.22 | 5.44 ± 0.38 | 5.24 ± 0.27 | |
PMMoV, pepper mild mottle virus.
All wastewater samples were collected on October 27, 2020.
Triplicate wastewater samples (n = 3) were analyzed for each condition.
+, sample processed with pre-filtration; −, sample processed without pre-filtration.
When SARS-CoV-2 RNA was detected from all samples (n = 3), standard deviations (SD) were calculated. The values are indicated as mean ± SD.
N.D., not detected.
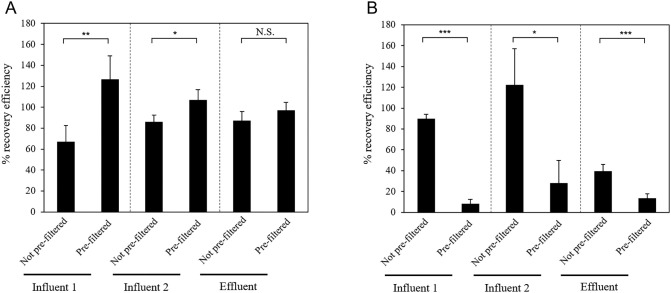
3.2. Recovery efficiencies of process controls
To assess the loss of SARS-CoV-2 RNA during the whole detection process (i.e, concentration, RNA extraction, and RT-qPCR), we determined the recovery efficiencies of MHV (WPC) and MNV (MPC) spiked in the original wastewater samples and virus concentrates, respectively (Fig. 2). As shown in Fig. 3 , the mean MPC (MNV) recovery efficiencies were >67 % for all samples (Fig. 3A), suggesting that no significant loss occurred during the extraction-RT-qPCR steps. However, the recovery efficiencies of WPC (MHV) from the pre-filtered samples were significantly lower than those from the samples without pre-filtration (paired t-test, p < 0.05) (Fig. 3B). These results suggest that MHV is likely to be physically trapped in the PTFE membrane, consistent with the result of SARS-CoV-2 partitioning.
Fig. 3.
Results of recovery of (A) murine norovirus (MNV) as a molecular process control (MPC) and (B) murine hepatitis virus (MHV) as a whole process control with or without polytetrafluoroethylene (PTFE) pre-filtration +, sample processed with pre-filtration; −, sample processed without pre-filtration. Recovery efficiencies (%) respectively were measured in influent 1, influent 2 and effluent samples (n = 3). Error bars indicate standard deviations. *p < 0.05, **p < 0.01, ***p < 0.005, N.S., no significant difference.
3.3. Removal of SARS-CoV-2 and microbial indicators by the wastewater treatment
To evaluate the reduction of SARS-CoV-2 and microbial indicators by the wastewater treatment system, wastewater samples were collected from an influent storage tank, anaerobic reactor, and effluent tank on two occasions (November 27 and December 24, 2020). Because no sample was collected from the influent storage tank on October 27, 2020 (Fig. 1), removal efficiencies could be calculated only for the two occasions. The samples were analyzed for SARS-CoV-2 RNA concentrations, together with E. coli and PMMoV RNA, which were determined by a culture-based method and RT-qPCR, respectively. In untreated wastewater, SARS-CoV-2 RNA was detected with a mean concentration of 6.50 log10 copies/L (Table 2 ). In effluent wastewater samples, SARS-CoV-2 RNA was also successfully quantified, which allowed for determining the removal efficiencies of the wastewater treatment system installed in the facility. The mean log10 reduction value (LRV) of SARS-CoV-2 for the whole treatment process was determined to be 2.47 log10 (Table 2). The mean LRVs of E. coli and PMMoV for the whole treatment process were calculated to be 2.81 log10 and 0.65 log10, respectively. The LRV of SARS-CoV-2 was substantially higher than that of PMMoV, indicating that the removal characteristics of SARS-CoV-2 are different from those of PMMoV, which is a non-enveloped virus known to be persistent in the environment (Kitajima et al., 2018).
Table 2.
Removal efficiencies of SARS-CoV-2a, PMMoVa and E. colib in large-scale septic tank in the COVID-19 quarantine facility.
| Sampling date | SARS-CoV-2 |
PMMoV |
E. coli |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA concentrationc (log10 copies/L) |
Log10 reductiond | RNA concentrationb (log10 copies/L) |
Log10 reductiond | Concentration (CFU/mL) |
Log10 reductiond | |||||||
| Influent storage tank | Anaerobic reactor | Effluent | Influent storage tank | Anaerobic reactor | Effluent | Influent storage tank | Anaerobic reactor | Effluent | ||||
| Nov. 27, 2020 | 6.39 ± 0.14 | 4.45 ± 0.20 | 4.14 | 2.25 | 7.66 ± 0.05 | 6.70 ± 0.01 | 6.96 ± 0.01 | 0.70 | 18,000 | N.T.e | 64 | 2.45 |
| Dec. 24, 2020 | 6.62 ± 0.07 | 4.42 ± 0.05 | 3.94 ± 0.10 | 2.68 | 7.71 ± 0.06 | 6.78 ± 0.03 | 7.10 ± 0.01 | 0.61 | 18,000 | N.T.e | 12 | 3.18 |
Triplicate wastewater samples (n = 3) were analyzed for SARS-CoV-2 and pepper mild mottle virus (PMMoV).
E. coli was assayed in duplicate (n = 2) for each sample.
When SARS-CoV-2 RNA was detected in all the samples (n = 3), standard deviations were calculated. The values are indicated as mean ± SD.
The log10 reduction was calculated for the whole treatment process based on the concentrations in the influent storage tank and the effluent.
N.T., not tested.
3.4. Confirmation of the detected SARS-CoV-2 sequences
To validate the qPCR results by confirming the presence of SARS-CoV-2 sequences in the wastewater samples, one region (501–913) within ORF1a was amplified by PCR and the PCR products were sequenced. The ORF1a sequence obtained from the influent storage tank collected on December 24, 2020 completely matched with the SARS-CoV-2 genomes available in the GenBank database (NC_045512.2).
4. Discussion
As of March 18, 2022, Japan had experienced roughly sixty thousand cumulative confirmed cases of COVID-19 per million people, a relatively small figure compared to countries in Europe and North America (World Health Organization, 2022). A low per-capita COVID-19 cases leads to low concentrations of SARS-CoV-2 in wastewater, meaning that using municipal wastewater samples in Japan to evaluate the virus detection methods has been challenging. In this study, the observed SARS-CoV-2 RNA concentration in untreated wastewater from a COVID-19 quarantine facility was 3.1 × 106 copies/L (Table 2; average of SARS-CoV-2 RNA concentration in an influent storage tank), whereas Kitamura et al. (Kitamura et al., 2020) reported SARS-CoV-2 RNA concentrations of 2.5 × 102–1.3 × 104 copies/L (from June 9, 2020, to August 10, 2020) in untreated municipal wastewater in Japan. Wastewater samples containing high concentrations of indigenous SARS-CoV-2 were needed to evaluate the detection methods and removal during wastewater treatment. We demonstrated here that wastewater from a COVID-19 quarantine facility could serve as ideal samples for those purposes.
Our results indicate that the majority of indigenous SARS-CoV-2 was associated with wastewater solids, which corroborates with previous reports suggesting that enveloped viruses, such as SARS-CoV-2, MHV and Pseudomonas phage Φ6, tend to be associated with solid materials (Kitamura et al., 2020; Ye et al., 2016). This finding means that solids containing SARS-CoV-2 are likely to be trapped upon membrane filtration, suggesting that previous studies using membrane pre-filtration might have underestimated the number of virus particles (Anderson-Coughlin et al., 2022). These results highlighted the importance of extracting viral RNA from solid fractions, and further research to maximize the recovery efficiency of SARS-CoV-2 RNA from solid fractions is highly awaited.
Due to the stringent biosafety requirements for handling SARS-CoV-2, surrogate viruses such as MHV and Φ6 have been used to evaluate detection methods (Ahmed et al., 2020; Torii et al., 2021a). However, exogenously seeded surrogate viruses might have different physical partitioning from indigenous SARS-CoV-2 in wastewater. SARS-CoV-2 is excreted in feces and is more likely to be embedded inside solid material coagulated with organic material; however, based on our previous investigation (data not shown), exogenously added viruses are more likely than indigenous viruses to be detected in liquid fractions. This difference might affect viral RNA recovery efficiency from a wastewater sample and, consequently, the viral RNA amount detected by RT-qPCR. The present study highlights the importance of assessing solid-liquid partitioning of indigenous SARS-CoV-2 virus rather than making assumptions based on the results of exogenously seeded surrogate viruses.
The present study revealed the LRV of SARS-CoV-2 in a sequencing batch activated sludge system. It has been extremely challenging to determine the reduction of SARS-CoV-2 by full-scale wastewater treatment systems in Japan, because the concentration of SARS-CoV-2 RNA in municipal wastewater has been a few orders of magnitude lower than in other countries. The wastewater in the quarantine facility contained high concentrations of SARS-CoV-2 RNA, which enabled us to determine the efficiency of SARS-CoV-2 reduction by the individual wastewater treatment processes installed in the facility. The LRV of SARS-CoV-2 (2.47) was greater than that of PMMoV (0.65), suggesting that SARS-CoV-2 was efficiently removed in the biological treatment system. Although Hong et al. (Hong et al., 2021) evaluated the reduction of SARS-CoV-2 (LRV of 0.3 to 0.5) for a part of the wastewater treatment process (from untreated wastewater to open air activated sludge tank), the reduction rate during the whole process remained unknown. In the present study, wastewater containing a high concentration of SARS-CoV-2 and an improved detection method enabled us to determine the LRV during the whole process in the sequencing batch activated sludge system. We observed the similarities in concentrations at the different stages of treatment and log10 reductions of SARS-CoV-2, PMMoV, and E. coli on the two sampling occasions, implying stable SBR operation and consequent constant microbial removal. It should be noted that the wastewater treatment process of the COVID-19 quarantine facility, surveyed in this study, has different characteristics than that of municipal wastewater treatment plants, where virus concentrations in influent fluctuate over time depending on infection rates in the service area. The microbial concentrations in the influent storage tank of the COVID-19 quarantine facility are expected to be uniform due to the long HRT inf, and in fact, the difference in microbial concentrations between the two occasions was not substantial. Given that quarantine facility wastewater tends to be fresh and contain a higher amount of SARS-CoV-2 than municipal wastewater, the public health risks associated with exposure to the quarantine facility wastewater should be carefully assessed. Further studies are required to assess public health risks and to evaluate the infectivity of SARS-CoV-2 in wastewater using fresh wastewater samples that contains a high concentration of SARS-CoV-2.
The SARS-CoV-2 concentrations in the influent storage tank in the present study ranged from 2.5 × 106 to 4.2 × 106 copies/L, which is comparable to those in untreated municipal wastewater observed in other countries, such as the United States and Nepal (Weidhaas et al., 2021; Wu et al., 2020; Tandukar et al., 2022). Although most of the residents in the quarantine facility were COVID-19 patients, the SARS-CoV-2 RNA concentration in the samples was lower than expected. There are four possible reasons that explain this observation. First, SARS-CoV-2 RNA might have been degraded in the influent storage tank. The HRT inf of the influent storage tank on the sampling day was calculated to be 6.86 days, which is much longer than the designed HRTinf of 0.63 days. This is because the number of guests during the sampling period ranged from 5 to 34, resulting in a significantly low wastewater inflow into a treatment system with a capacity for 607 people. Indeed, previous studies observed substantial degradation of SARS-CoV-2 RNA in untreated wastewater under room temperature (approximately 2 log10 reduction within 7 days) ((Ahmed et al., 2020): (Bivins et al., 2020)), which may support our observation. Moreover, suspended solids possibly containing a large amount of SARS-CoV-2 may have already been sedimented in such a long HRT, leading to lower levels of observed SARS-CoV-2 RNA concentrations in the untreated wastewater samples. Second, asymptomatic or mildly symptomatic patients, who were residents of the quarantine facility, might shed a significantly lower amount of SARS-CoV-2 into wastewater than symptomatic patients. Third, country-specific factors may exist, such as per-capita water usage, racial differences in the amount of viral shedding, and changes in the time course between symptom onset and viral concentrations in feces (Miura et al., 2021). Fourth, some residents may have been non-infected individuals who were falsely diagnosed as positive by quantitative antigen screening that is less specific than PCR.
Our study had a few limitations. First, we obtained the sample from only one COVID-19 quarantine facility. Since the number of COVID-19 quarantine facilities equipped with a facility-level wastewater treatment system is limited, we collected samples at multiple times to increase the study's validity. Second, the sensitivity of the SARS-CoV-2 RNA detection method that we used needs further improvement. Third, our sampling strategy did not consider the HRTs of each treatment process, which were not available at the time of sampling. This means that the influent storage tank and effluent samples were not collected from the same body of wastewater on each sampling occasion. Further studies taking into account HRTs with increased sampling occasions are desired. Forth, our results on viral RNA detection obtained in the present study themselves alone do not imply the infectivity and transmissibility of the virus. In the future, we plan to use additional samples to evaluate the infectivity of SARS-CoV-2 in wastewater via cell culture (Ando and Kitajima, 2021).
5. Conclusion
We performed a quantitative analysis of SARS-CoV-2 RNA in wastewater samples from a COVID-19 quarantine facility in Japan, where asymptomatic and mildly symptomatic patients were isolated. The results have revealed that most of the indigenous SARS-CoV-2 in wastewater existed in solid fractions, from which viral RNA should be extracted. Moreover, indigenous SARS-CoV-2 is effectively removed during the SBR process. To the best of our knowledge, this is the first study to successfully analyze the SARS-CoV-2 reduction rate during a whole system of sequential batch activated sludge employing anaerobic, anoxic, and oxic processes. The use of quarantine facility wastewater is suitable to improve detection methodology and understand the behavior of indigenous SARS-CoV-2 in wastewater treatment systems.
The following is the supplementary data related to this article.
Primers and probes used in the present study.
CRediT authorship contribution statement
Wastewater sampling was conducted under permission from the facility. All study data from this study were anonymized and did not require any ethical approval. All the authors have approved the final version of the manuscript.
Ryo Iwamoto: Conceptualization, Formal analysis, Investigation, Methodology, Writing – Original draft, Kiyoshi Yamaguchi: Investigation, Methodology, Chisato Arakawa: Investigation, Validation, Hiroki Ando: Investigation, Validation, Eiji Haramoto: Investigation, Writing – Review and editing, Ken-ichi Setsukinai: Writing – Review and editing, Takuya Yamagishi: Supervision, Sumire Sorano: Conceptualization, Michio Murakami: Conceptualization, Writing – Review and editing, Shigeru Kyuwa: Resource, Kotoe Katayama: Methodology, Hiroyuki Kobayashi: Supervision, Funding acquisition, Satoshi Okabe: Supervision, Writing – review and editing, Seiya Imoto: Supervision, Conceptualization, Methodology, Writing – review and editing, Masaaki Kitajima: Supervision, Conceptualization, Investigation, Methodology, Funding acquisition, Writing – review and editing.
Funding
This study was partly funded by the Ministry of Health, Labor and Welfare of Japan (grant numbers JPMH20HA2007 and JPMH20HA2009) and Shionogi & Co., Ltd. The employees of Shionogi & Co., Ltd. involved in the study design, data collection, analysis and interpretation, and the writing of the report made the decision to serve as authors.
Declaration of competing interest
Ryo Iwamoto, Ken-ichi Setsukinai, and Hiroyuki Kobayashi are employees of Shionogi & Co., Ltd. Masaaki Kitajima received research funding and patent royalties from Shionogi & Co., Ltd. Satoshi Okabe received research funding from Shionogi & Co., Ltd. Kiyoshi Yamaguchi, Chisato Arakawa, Hiroki Ando, Eiji Haramoto, Kotoe Katayama, Takuya Yamagishi, Sumire Sorano, Michio Murakami, Shigeru Kyuwa, and Seiya Imoto have no competing interests to declare.
Acknowledgements
The authors would like to thank the anonymous quarantine facility for providing wastewater samples and relevant data. We thank Yumiko Isobe at the University of Tokyo for her support in the sequencing experiment and Yoshinori Ando and Kazuya Okada from Shionogi &Co., Ltd. for their technical assistance in wastewater sampling. The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo. Editorial support, in the form of medical writing, assembling tables, and creating high-resolution images based on authors' detailed directions; collating author comments; copyediting; fact-checking; and referencing, was provided by Editage.
Editor: Kevin V. Thomas
Data availability
Data will be made available on request.
References
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-Coughlin B.L., Shearer A.E.H., Omar A.N., Litt P.K., Bernberg E., Murphy M., Anderson A., Sauble L., Ames B., Damminger O., Jr., Ladman B.S., Dowling T.F., Wommack K.E., Kniel K.E. Coordination of SARS-CoV-2 wastewater and clinical testing of university students demonstrates the importance of sampling duration and collection time. Sci. Total Environ. 2022;830 doi: 10.1016/j.scitotenv.2022.154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando Y., Kitajima M. Establishment of an infectivity assay for SARS-CoV-2 to manage risks derived from wastewater during COVID-19 pandemic. J. Jpn. Soc. Civ. Eng. Ser. G Environ. Res. 2021 doi: 10.2208/jscejer.77.7_iii_191. 77-7, III_191 – III_197 (in Japanese) [DOI] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.-H., Fung A.Y.-F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.-H., Yuen K.-Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa M.F., Verbyla M.E., Vassalle L., Leal C., Leroy-Freitas D., Machado E., Fernandes L., Rosa-Machado A.T., Calábria J., Chernicharo C., Mota Filho C.R. Reduction and liquid-solid partitioning of SARS-CoV-2 and adenovirus throughout the different stages of a pilot-scale wastewater treatment plant. Water Res. 2022;212(118069) doi: 10.1016/j.watres.2022.118069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P.-Y., Rachmadi A.T., Mantilla-Calderon D., Alkahtani M., Bashawri Y.M., Al Qarni H., O’Reilly K.M., Zhou J. Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: to what extent of the outbreak can surveillance of wastewater tell us? Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.H., Johns M.W. Improved detection of F-specific RNA coliphages in fecal material by extraction and polyethylene glycol precipitation. Appl. Environ. Microbiol. 2009;75(19):6142–6146. doi: 10.1128/AEM.00436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., Qu Y., Qian H., Zhuang Y., Chen X., Peng X., Shi T., Wang J., Wu J., Song T., He J., Li Y., Zhong N. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann. Intern. Med. 2020;173(12):974–980. doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes–identification of potential viral indicators. Sci. Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. Npj Clean Water. 2018;1(1):19. doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2020;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., Joshi C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with upflow anaerobic sludge blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., Rossi L., Manganelli R., Loregian A., Navarin N., Abate D., Sciro M., Merigliano S., De Canale E., Vanuzzo M.C., Besutti V., Saluzzo F., Onelia F., Pacenti M., Parisi S.G., Carretta G., Donato D., Flor L., Cocchio S., Masi G., Sperduti A., Cattarino L., Salvador R., Nicoletti M., Caldart F., Castelli G., Nieddu E., Labella B., Fava L., Drigo M., Gaythorpe K.A.M., Imperial College COVID-19 Response Team, Brazzale A.R., Toppo S., Trevisan M., Baldo V., Donnelly C.A., Ferguson N.M., Dorigatti I., Crisanti A. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584(7821):425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kitajima M., Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021;769 doi: 10.1016/j.scitotenv.2020.144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J., Pang J. Wastewater surveillance to infer COVID-19 transmission: a systematic review. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for detection for novel Coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Tandukar S., Sthapit N., Thakali O., Malla B., Sherchan S.P., Shakya B.M., Shrestha L.P., Sherchand J.B., Joshi D.R., Lama B., Haramoto E. Detection of SARS-CoV-2 RNA in wastewater, river water, and hospital wastewater of Nepal. Sci. Total Environ. 2022;824 doi: 10.1016/j.scitotenv.2022.153816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Oishi W., Zhu Y., Thakali O., Malla B., Yu Z., Zhao B., Arakawa C., Kitajima M., Hata A., Ihara M., Kyuwa S., Sano D., Haramoto E., Katayama H. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Sci. Total Environ. 2021;807 doi: 10.1016/j.scitotenv.2021.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO Coronavirus (COVID-19) dashboard. 2022. https://covid19.who.int/
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b0087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and probes used in the present study.
Data Availability Statement
Data will be made available on request.