Abstract
The deoR gene located just upstream the dra-nupC-pdp operon of Bacillus subtilis encodes the DeoR repressor protein that negatively regulates the expression of the operon at the level of transcription. The control region upstream of the operon was mapped by the use of transcriptional lacZ fusions. It was shown that all of the cis-acting elements, which were necessary for full DeoR regulation of the operon, were included in a 141-bp sequence just upstream of dra. The increased copy number of this control region resulted in titration of the DeoR molecules of the cell. By using mutagenic PCR and site-directed mutagenesis techniques, a palindromic sequence located from position −60 to position −43 relative to the transcription start point was identified as a part of the operator site for the binding of DeoR. Furthermore, it was shown that a direct repeat of five nucleotides, which was identical to the 3′ half of the palindrome and was located between the −10 and −35 regions of the dra promoter, might function as a half binding site involved in cooperative binding of DeoR to the regulatory region. Binding of DeoR protein to the operator DNA was confirmed by a gel electrophoresis mobility shift assay. Moreover, deoxyribose-5-phosphate was shown to be a likely candidate for the true inducer of the dra-nupC-pdp expression.
Bacillus subtilis is able to use deoxyribonucleosides and deoxyribose (dRib) as the sole carbon and energy source. After transport into the B. subtilis cell, deoxyribonucleosides are cleaved by nucleoside phosphorylases to dRib-1-phosphate (dRib-1-P) and the respective pyrimidine or purine base. dRib-1-P can be converted to dRib-5-P in a reaction catalyzed by phosphodeoxyribomutase. Finally, dRib-5-P is cleaved to acetaldehyde and glyceraldehyde-3-phosphate by deoxyriboaldolase. dRib is converted to dRib-5-P by the action of deoxyribokinase after transport into the cell. The syntheses of nucleoside phosphorylase, deoxyribomutase, and deoxyriboaldolase, as well as deoxyribokinase, are increased when B. subtilis cells grow in the presence of dRib or deoxyribonucleosides (15).
In B. subtilis, three genes, dra, nupC, and pdp, which encode deoxyriboaldolase, nucleoside uptake protein, and pyrimidine nucleoside phosphorylase, respectively, are organized as an operon. The operon is transcribed from a promoter located upstream the first gene dra. The gene deoR, encoding a repressor protein, is found immediately upstream of the operon. DeoR is a negative regulator for the expression of the operon. A DeoR-deficient mutant has a high level of deoxyriboaldolase and pyrimidine nucleoside phosphorylase, even in the absence of dRib or deoxyribonucleosides (15). The operon is also subject to catabolite repression by glucose, and a consensus-like catabolite responsive element (CRE) sequence (6, 15) is located in the start of dra.
In Escherichia coli, the genes encoding deoxyriboaldolase (deoC), phosphodeoxyribomutase (deoB), and thymidine phosphorylase (deoA) are organized in the deo operon (11). In addition to these genes, the deo operon also contains deoD, which encodes purine nucleoside phosphorylase. The regulation of the expression of the deo operon in E. coli has been studied in great detail. The expression is regulated negatively by the DeoR and CytR repressors and positively by the cyclic AMP (cAMP)-cAMP receptor protein activator complex (1, 3, 18). The presence of deoxyribonucleosides in the growth medium induces deo operon expression through the intracellular formation of dRib-5-P, which is the effector molecule for the DeoR repressor (11, 15). DeoR repression has been illustrated by the chelate model, in which the cooperative binding of the protein to several operator sites generates a high degree of repression, even though the intrinsic affinity of the repressor for each site is relatively low (3).
Here we report the identification of the operator site for the binding of DeoR upstream of the dra-nupC-pdp operon. We also report the interaction of DeoR repressor protein with the operator DNA and indicate that dRib-5-P may be the true inducer of the dra-nupC-pdp expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this work are listed in Table 1. B. subtilis was grown in Spizizen minimal salt medium (15), supplemented with 50 μg of l-tryptophan per ml and with either 0.4% glucose or 0.4% succinate as carbon source. L broth (Difco Laboratories, Detroit, Mich.) was used as a rich medium for both E. coli and B. subtilis. Culturing of cells was performed at 37°C. For selection of antibiotic resistance, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 6 μg/ml for B. subtilis and 20 μg/ml for E. coli; neomycin, 5 μg/ml; erythromycin, 1 μg/ml; lincomycin, 25 μg/ml; and tetracycline, 8 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 168 | trpC | C. Anagnostopoulos |
| HH232 | trpC2 deoR:::erm | 15 |
| HH246 | metB 10 lys-3 trpC2 amyE::drm-lacZ drm::cat::erm | H. H. Saxild |
| XM11–-16 | trpC2 amyE::dra-lacZ (pJOY1 through -6)b | Tfc of 168 by pJOY1–-6, Neor |
| XM25 and -26 | trpC2 amyE::dra-lacZ (pJOY5 and -6) deoR::erm | Tf of XM15 and -16 by HH232, Err |
| XM33 | trpC2 amyE::dra-lacZ (pJOY5) + pJOY13 | Tf of XM15 by pJOY13, Cmr |
| XM34 | trpC2 amyE::dra-lacZ (pJOY6) + pJOY12 | Tf of XM16 by pJOY12, Cmr |
| XM35 | trpC2 amyE::dra-lacZ (pJOY5) + pJOY12 | Tf of XM15 by pJOY12, Cmr |
| XM36 | trpC2 amyE::dra-lacZ (pJOY6) + pJOY13 | Tf of XM16 by pJOY13, Cmr |
| XM37 | trpC2 amyE::dra-lacZ (pJOY5) + pAMB22 | Tf of XM15 by pAMB22, Cmr |
| XM38 | trpC2 amyE::dra-lacZ (pJOY6) + pAMB22 | Tf of XM15 by pAMB22, Cmr |
| XM40–-45 | trpC2 amyE::dra-lacZ (pJOY5) + pJOY40–-42 | Tf of XM15 by pJOY40–-42, Cmr |
| XM47 and -48 | trpC2 amyE::dra-lacZ (pJOY5) + pJOY47–-48 | Tf of XM15 by pJOY47 and -48, Cmr |
| XM116–-118 | trpC2 amyE::dra-lacZ (pJOY16–-18) | Tf of 168 by pJOY16–-18, Neor |
| XM123 | trpC2 amyE::dra-lacZ (pJOY23) | Tf of 168 by pJOY23, Neor |
| XM129–-131 | trpC2 amyE::dra-lacZ (pJOY29–-31) | Tf of 168 by pJOY29–-31, Neor |
| XM133 and -134 | trpC2 amyE::dra-lacZ (pJOY33 and -34) | Tf of 168 by pJOY33 and -34, Neor |
| XM246 | trpC2 amyE::dra-lacZ drm::cat::erm | Tf of XM15 by HH246, Neor, Emr |
| XM311–-319 | trpC2 amyE::dra (pJOY311–-319)-lacZd | Tf of 168 by a plasmid library containing random mutations in the dra control region, Neor |
| XM502 and -503 | trpC2 amyE::dra-lacZ (pJOY502 and -503) | Tf of 168 by pJOY502 and -503, Neor |
| XM512 and -513 | trpC2 amyE::dra-lacZ (pJOY512 and -513) | Tf of 168 by pJOY512 and -513, Neor |
| E. coli | ||
| JOY99 | MC1061 + pTCR99A + pRI952 | This work |
| JOY100 | MC1061 + pJOY66 + pRI952 | This work |
| MC1061 | F− araD139 Δ(ara-leu)7696 galE 15 galK 16Δ(lac)X74 rpsl (Strr) hsdR2(r− m−) mcrA mcrB | Laboratory stock |
| Plasmids | ||
| pAMB22 | Tcr (E. coli) Cmr (B. subtilis). Multiple-copy shuttle vector containing the pBR322 replication origin for replication in E. coli and pC194 replication origin for replication in B. subtilis | 19 |
| pDG268neo | Apr (E. coli) Neor (B. subtilis). Integration vector for construction of transcriptional lacZ fusion integrated into the amyE gene of B. subtilis | 2 |
| pHH1002 | 15 | |
| pHH1011 | 15 | |
| pJOY1–-6 | 676-, 582-, 416-, 322-, 235-, and 141-bp EcoRI-BamHI PCR fragments generated by using primers 2+1, 3+1, 2+4, 3+4, 2+5, and 3+5, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY12–-13 | 235- and 141-bp EcoRI-KpnI PCR fragments generated by primers 5+27 and 5+28, ligated to pAMB22 digested with EcoRI and KpnI | This work |
| pJOY16–-18 | 221-bp EcoRI-BamHI PCR fragments generated by one of the primers 8–10 + primer 2, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY23 | 221-bp EcoRI-BamHI PCR fragments generated by primers 2 + 11, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY29–-31 | 221-bp EcoRI-BamHI PCR fragments generated by one of the primers 12–14 + primer 2, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY33 and -34 | 221-bp EcoRI-BamHI PCR fragments generated by primers 15 or 16 + primer 2, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY40–-42 | 36-, 22-, and 18-bp EcoRI-KpnI fragments generated by annealing primer 17+18, 19+20, 21+22, ligated to pAMB22 digested with EcoRI and KpnI | This work |
| pJOY43–-45 | 51-, 43-, and 41-bp EcoRI-KpnI PCR fragments generated by primers 26+23, 26+24, and 26+25, ligated to pAMB22 digested with EcoRI and KpnI | This work |
| pJOY47 and -48 | 221-bp EcoRI-KpnI PCR fragments generated by primers 2+9 and 2+10, ligated to pAMB22 digested with EcoRI and KpnI | This work |
| pJOY66 | 942-bp NcoI-BamHI PCR fragment containing deoR generated by primers 29 and 30, ligated to pTCR99A digested with NcoI and BamHI | This work |
| pJOY502 and -503 | 203- and 109-bp EcoRI-BamHI PCR fragments generated by primers 2+6 and 3+6, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pJOY512 and -513 | 195- and 101-bp EcoRI-BamHI PCR fragments generated by primers 2+7 and 3+7, ligated to pDG268 digested with EcoRI and BamHI | This work |
| pRI952 | Cmr | 14, 17 |
| pTCR99A | Apr | H. H. Saxild |
Ap, ampicillin; Cm, chloramphenicol; Neo, neomycin; Er, erythromycin; Tc, tetracycline.
A plasmid in parentheses indicates the plasmid (or plasmids) which is integrated into the amyE locus on the chromosome.
Tf, transformation of the first strain either with the noted plasmid or with DNA from the second strain.
Contains random mutations in the dra control region (see Materials and Methods).
DNA manipulations and genetic techniques.
Chromosomal DNA from B. subtilis was isolated as described previously (15). Plasmid DNA was isolated by the alkaline-sodium dodecyl sulfate (SDS) method (15). Transformations of E. coli and B. subtilis were performed as described previously (15). Treatment of DNA with restriction enzymes and T4 DNA ligase was performed as recommended by the supplier. DNA sequences were obtained by using the Amersham (Cleveland, Ohio) Thermo Sequenase radiolabeled termination cycle sequencing kit. All sequencing analysis was done with double-stranded plasmid or PCR product DNA templates and was performed as described by the supplier. A standard PCR was performed as follows: 5 μl of 10× reaction buffer (Pharmacia), 2 μl of 5 mM deoxynucleoside triphosphate (dNTP) mix, 10 μl of template DNA (1 to 10 ng), 10 μl of each primer (40 μg/ml), 12.5 μl of H2O and 0.5 μl (2 U) of Taq polymerase (Pharmacia) were mixed. The PCR mixture was incubated for 30 cycles of 1 min at 94°C, 45 s at 55°C, and 1 min at 72°C in a Biometra Trio 48 Thermocycler (Biometra, Hamburg, Germany).
Construction of the dra-lacZ fusions for deletion analysis.
Different deletion derivatives of the dra control region were amplified by PCR by using plasmid pHH1011 containing the 3′ end of deoR and the 5′ end of dra as the template DNA (15). The forward and reverse oligonucleotide primers were synthesized with EcoRI and BamHI 5′-linked restriction sites, respectively (Table 2). The amplifications were performed with the following primers: primer 1 plus primer 2 (676-bp product), primer 1 plus primer 3 (582-bp product), primer 4 plus primer 2 (416-bp product), primer 4 plus primer 3 (322-bp product), primer 5 plus primer 2 (235-bp product), primer 5 plus primer 3 (141-bp product), primer 6 plus primer 2 (203-bp product), primer 6 plus primer 3 (109-bp product), primer 7 plus primer 2 (195-bp product), and primer 7 plus primer 3 (101-bp product). The PCR products were digested with EcoRI and BamHI, ligated to EcoRI- and BamHI-digested plasmid pDG268, and transformed into E. coli MC1061, selecting for Apr. Plasmids extracted from E. coli (pJOY1 through -6, -502, and -503, and -512 and -513 [Table 1]) were linearized by KpnI and recombined into the amyE locus of B. subtilis 168 by transformation following subsequent selection for Neor. The correct integration into the amyE locus was confirmed by the amylase-negative phenotype of the transformants. The resulting B. subtilis strains were XM11 through -16, -502, -503, -512, and -513 (Table 1).
TABLE 2.
Oligonucleotides used for PCR amplifications
| Primer no. | 5′- or 3′-linked restric-tion site sequence | Nucleotide sequencea | Coordinate(s)b |
|---|---|---|---|
| Deletion analysis | |||
| 1 | 5′ EcoRI site | 5′-GCCGGAATTC-CGATGCCGCGCTATCTC-3′ | −522 |
| 2 | 5′ BamHI site | 5′-CGCGGATCC-CCATGTCGGATTGACAC-3′ | +154 |
| 3 | 5′ BamHI site | 5′-CGCGGATCC-GCTGTATGATCAATTATG-3′ | +60 |
| 4 | 5′ EcoRI site | 5′-GCCGGAATTC-GCAGCGCCATCAATGAC-3′ | −261 |
| 5 | 5′ EcoRI site | 5′-GCCGGAATTC-GTGACACGTTCAAACCTT-3′ | −80 |
| 6 | 5′ EcoRI site | 5′-GCCGGAATTC-TTCAATTACCAATTTACATATG-3′ | −48 |
| 7 | 5′ EcoRI site | 5′-GCCGGAATTC-CCAATTTACATATGTTCAAAAG-3′ | −40 |
| Site-directed mutagenesis | |||
| 8 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATCGAACAAAATTTC-3′ | −66 |
| 9 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTCAACAAAATTTC-3′ | −66 |
| 10 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGCACAAAATTTC-3′ | −66 |
| 11 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGAACAAAATTCCAATTACC-3′ | −66 |
| 12 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGAACCAAATTTCAATTACC-3′ | −66 |
| 13 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGAACACAATTTCAATTACC-3′ | −66 |
| 14 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGAACAACATTTCAATTACC-3′ | −66 |
| 15 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATTGAACAAAACTTCAATTACC-3′ | −66 |
| 16 | 5′ EcoRI site | 5′-GCCGGAATTC-CCTTTCATCGAACAAAATTTTAATTACC-3′ | −66 |
| Operator fragments | |||
| 17 | None | 5′-AAACCTTTCATTGAACAAAATTTCAATTACCAATTT-3′ | −69 |
| 18 | 5′ EcoRI overhang, 3′ KpnI overhang | 5′-AATT-AAATTGGTAATTGAAATTTTGTTCAATGAAAGGTTT-GTAC-3′ | −34 |
| 19 | None | 5′-TCATTGAACAAAATTTCAATTA-3′ | −62 |
| 20 | 5′ EcoRI overhang, 3′ KpnI overhang | 5′-AATT-TAATTGAAATTTTGTTCAATGA-GTAC-3′ | −41 |
| 21 | None | 5′-ATTGAACAAAATTTCAAT-3′ | −60 |
| 22 | 5′ EcoRI overhang, 3′ KpnI overhang | 5′-AATT-ATTGAAATTTTGTTCAAT-GTAC-3′ | −45 |
| 23 | 5′ EcoRI | 5′-GCCGGAATTC-AAACCTTTCATTGAACAAAATTTCAATTACC-3′ | −69 |
| 24 | 5′ EcoRI | 5′-GCCGGAATTC-TCATTGAACAAAATTTCAATTACC-3′ | −62 |
| 25 | 5′ EcoRI | 5′-GCCGGAATTC-ATTGAACAAAATTTCAATTACC-3′ | −60 |
| 26 | 5′ KpnI | 5′-CGGCGGTACC-CTTTTGAACATATGTAAATTGGTAATTG-3′ | −19 |
| 27 | 5′ KpnI | 5′-CGGCGGTACC-CCATGTCGGATTGACAC-3′ | +154 |
| 28 | 5′ KpnI | 5′-CGGCGGTACC-GCTGTATGATCAATTATG-3′ | +60 |
| Amplification of deoR | |||
| 29 | 5′ NcoI | 5′-GCCGCCATGG-ATCGGGAAAAACAGC-3′ | 4052107–4052089 |
| 30 | 5′ BamHI | 5′-GCGGGATCC-TCACAAATCATTAACAAGCG-3′ | 4051166–4051185 |
| Mobility shift assay | |||
| 31 | 5′ KpnI | 5′-GCCGGGTACC-ATCCTTCGCACACTTCC-3′ | +30 |
| 32 | 5′ KpnI | 5′-CGGCGGTACC-CATATGTAAATTGGTAATTG-3′ | −27 |
| 33 | 5′ EcoRI | 5′-GCCGGAATTC-CCTTTCATTGAACAAAATTTCAATTACC-3′ | −66 |
Letters in italics indicate linker sequences. Underlined letters indicate the position of the restriction site sequence or restriction site overhang in the linker region. Boldface letters indicate nucleotides that differ from the wild-type sequence.
Mutagenic PCR.
To obtain random mutations in the dra control region, a mutagenic PCR protocol was derived from the standard PCR. The following changes were used. The MgCl2 concentration was increased to 7 mM, 0.5 mM MnCl2 was added to diminish the template specificity of the polymerase, the concentration of dCTP and dTTP was increased to 1 mM to promote misincorporation, and, finally, the amount of Taq polymerase was increased to 5 U. Primers 5 and 2 were used as the primers, and plasmid pHH1011 was used as the template DNA. The mutated 234-bp PCR product was digested with EcoRI and BamHI, ligated to EcoRI- and BamHI-digested pDG268, and transformed into E. coli MC1061, selecting for Apr. Approximately 10,000 transformants were pooled, and plasmids were extracted from the pooled culture. The plasmids were transformed into B. subtilis 168 as described above, and operator-negative candidate clones were selected by screening for blue Neor transformants on X-Gal (5-bromo-4-chloro-3-indolyl-β-galactoside)-containing minimal succinate plates without inducer. Nine mutant candidates (XM311 through -319 [Table 1]) with phenotypes ranging from dark blue to light blue were isolated.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by using oligonucleotides with a single nucleotide mismatch as PCR primers. The primer nucleotide sequences are shown in Table 2. A standard PCR was performed with one of these primers together with primer 2. The resulting 221-bp PCR products were cloned into pDG268 and transformed into B. subtilis as described above. The resulting strains are listed in Table 1.
Cloning of operator DNA in pAMB22.
Three short operator-containing double-stranded DNA fragments were produced by annealing three pairs of oligonucleotides with complementary sequence. Equimolar amounts of primers 17 and 18, 19 and 20, and 21 and 22 (Table 2), respectively, were mixed in a volume of 20 μl. After incubation for 10 min at 80°C, the mixture was cooled down slowly to room temperature. Since one of the oligonucleotides of a pair was synthesized with 5′ and 3′ sequences complementary to the single-stranded overhangs produced by EcoRI and KpnI, respectively, the double-stranded annealed products could be directly ligated to pAMB22 digested with EcoRI and KpnI. The ligation mixture was transformed into E. coli MC1061, selecting for Tcr. Plasmids with the three annealed operators cloned on them were then extracted and transformed into B. subtilis XM15, selecting for Cmr. The resulting strains were XM40 through -42 (Table 1). To produce slightly longer operator fragments, primer 23, 24, or 25 was used together with primer 26 in a standard PCR, except that no template DNA was added. Because primers 23, 24, and 25 have eight nucleotides in their 3′ end which are complementary to the 3′ end of primer 26, a double-stranded DNA fragment could be produced in the absence of template DNA. The PCR products were cloned in pAMB22 and transformed into B. subtilis XM15 as just described. The resulting strains were XM43 through -45 (Table 1). Two longer control DNA fragments similar to the one fused in front of lacZ as in strains XM15 and XM16 were produced in the standard PCR mixtures containing primers 5 and 27 (235-bp product) and 5 and 28 (141-bp product) (Table 2). The fragment was cloned in pAMB22 as described above (Table 1). Finally, two DNA fragments of 221 bp containing single-base-pair mutations in the operator sequence were produced by PCR by using primer 9 or 10 with primer 28. These fragments were cloned in pAMB22 and transformed into XM15, resulting in strains XM47 and XM48 (Table 1).
β-Galactosidase assay.
β-Galactosidase activity was measured according to the method of Miller (10). Specific enzyme activities were expressed in units per milligram of protein. One unit is defined as 1 nmol of substrate converted per min. The presented values are means of at least two different experiments. The variation was less than 10%. Total protein was determined by the method of Lowry et al. (9).
Overproduction of DeoR repressor protein.
The deoR gene was amplified in a standard PCR with plasmid pHH1002 (15) containing the 5′ end of the dra and 1.5 kb of upstream DNA as the template. As primers, primer 29 synthesized with a 5′ NcoI site and primer 30 synthesized with a BamHI site were used (Table 2). The deoR gene was inserted in the NcoI and BamHI sites of plasmid pTRC99A, resulting in pJOY66. pTRC99A has a trc promoter upstream of the NcoI site and an rrnB transcription termination signal downstream of the BamHI site. Cloning in the NcoI site next to the trc promoter provided direct expression of inserts possessing the start codon ATG. pJOY66 was transformed (Apr) into E. coli MC1061 containing pRI952 (Cmr), resulting in strain JOY100. pRI952 was used to optimize the production of DeoR protein in E. coli. pRI952 contains tRNA genes for the rare arginine codons AGG and AGA and for the isoleucine codon AUA. Several of these rare E. coli codons are encoded by the B. subtilis deoR gene. E. coli cells were cultured in L broth medium at 37°C to an optical density at 450 nm of 0.1. Production of DeoR protein was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM, and incubation was continued for 2 h. Judging from the SDS-polyacrylamide gel electrophoresis of the cleared lysate of the induced E. coli culture, less than 1% of the total protein appeared to be DeoR.
Mobility shift assay.
After cultivation and induction of the E. coli culture, cells were harvested, resuspended in a mixture of 10 mM Tris-HCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT [pH 7.5]), and sonically disrupted. Cell debris was removed by centrifugation at 15,000 × g for 10 min. The standard PCR mixtures containing 25 μmol of [α-33P]dATP (25 μCi) were used to produce the radiolabeled operator-containing DNA fragments. The following labeled fragments were generated: primers 5 plus 31 (111-bp product), 25 plus 26 (42-bp product), 25 plus 32 (34-bp product), 33 plus 26 (49 bp product), and 9 plus 26 (48-bp product). Each binding reaction mixture contained 0.4 to 15 μg of protein, approximately 1 ng of the labeled DNA fragment, and 1 μg of nonspecific DNA. Binding was carried out in a mixture of 10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1 mM DTT (pH 7.5), and 5% glycerol in a final volume of 10 μl. Before addition of the labeled DNA, the reaction mixture was preincubated for 10 min at room temperature, and incubation was continued for 20 min at room temperature after the addition of the labeled DNA. All of the 10-μl sample was then loaded onto a preelectrophoresed 4% polyacrylamide gel and electrophoresed at 7 V/cm for 1 h. The gel was dried, and bands were visualized by autoradiography.
RESULTS
Deletion analysis of the dra control region.
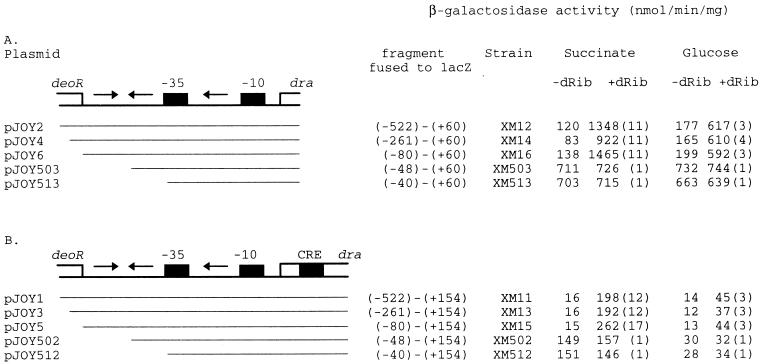
It has been shown that all of the cis-acting elements, which are necessary for full DeoR regulation of the dra-nupC-pdp operon, are included in a 650-bp sequence just upstream of the dra gene (15). To determine the minimal sequence for full regulation of the operon, 10 fusions of the dra control region to lacZ were inserted into the amyE gene on the chromosome as described in Materials and Methods (Fig. 1). β-Galactosidase activity was measured for these strains grown in minimal medium with glucose or succinate as a carbon source (Fig. 2). For strains XM12, -14, and -16, the levels of induction by dRib were approximately 10-fold in succinate-grown cells and 3-fold in glucose-grown cells. Further deletion of the upstream sequence (XM503 and XM513) caused the loss of dRib induction (Fig. 2A), which indicated that the region between −80 and −40 bp upstream of the transcription start point was necessary for the DeoR regulation. When the CRE sequence was present in the fusions, the same expression pattern was seen, but at a lower level of expression, probably because of the different downstream fusion positions (Fig. 2B). Catabolite repression was clearly observed for the CRE-including strains, although we noticed that there was no catabolite repression of the strains in which the upstream fusion point extended −48 bp, when growing without inducer.
FIG. 1.
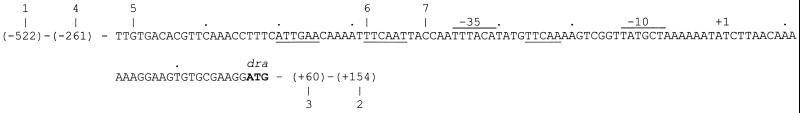
Nucleotide sequence of the regulatory region upstream of the dra-nupC-pdp operon. Underlining indicates a palindromic sequence and a direct repeat of one of the palindromic halves. Horizontal lines above the sequence indicate the positions of the −10 and −35 promoter elements. Vertical lines and numbers above the sequence indicate the positions and numbers of forward primers, respectively. Vertical lines and numbers beneath the sequence indicate the positions and numbers of reverse primers, respectively. Only the positions of primers used in the deletion analysis illustrated in Fig. 2 are shown. Letters in boldface indicate the translational start codon for the deoxyriboaldolase (dra) reading frame. Numbers in parentheses indicate the positions of primers mapping outside the nucleotide sequence shown in the figure. The transcription start point is identical to genome nucleotide no. 4051089 (7).
FIG. 2.
Deletion analysis of the regulatory region upstream of the dra-nupC-pdp operon. Only a part of the deoR and dra genes is shown. The arrows indicate a palindromic sequence or a direct repeat sequence to the 3′ half of the palindrome. The black boxes indicate the −35 to −10 region of the promoter and a CRE sequence located in the start of dra. The lines beneath the map show the fragments cloned in front of the lacZ reporter gene of plasmid pDG268 (the lines are not drawn to scale with respect to their relative length in nucleotides). (A) Fusion series without CRE. (B) Fusion series with CRE. The numbers in the parentheses indicate folds of regulation. The fold regulation is determined by dividing the enzyme activity of cells induced by dRib by the enzyme activity of cells without inducer. Cells were grown in minimal medium with glucose or succinate as a carbon source. Inducer was added to a final concentration of 1 mg/ml.
deoR-dependent induction.
The lacZ fusions in XM15 and XM16 were introduced into the deoR mutant strain HH232 (15). As expected, β-galactosidase activity in the resulting deoR strains XM25 and XM26 was increased and could no longer be induced by dRib (Table 3).
TABLE 3.
β-Galactosidase level in B. subtilis wild-type and deoR strains carrying different dra-lacZ fusionsa
| Strain | Region fused to lacZ | deoR status | CRE status | Inducer added | Enzyme activity (nmol/min/mg)b |
|---|---|---|---|---|---|
| XM15 | −80–+154 | + | + | None | 15 |
| dRib | 262 | ||||
| XM25 | −80–+154 | − | + | None | 107 |
| dRib | 110 | ||||
| XM16 | −80–+60 | + | − | None | 138 |
| dRib | 1,465 | ||||
| XM26 | −80–+60 | − | − | None | 1,133 |
| dRib | 1,029 |
Cells were grown in succinate minimal medium. Inducer was added to a final concentration of 1 mg/ml.
The enzyme levels of XM15 and XM16 were taken from Fig. 2.
Titration of the DeoR molecules by multiple copies of the control region.
To investigate whether the DeoR molecules in the cell could be titrated by increasing the copy number of the dra control region, two fragments of 235 bp (from −80 to +154) and 141 bp (from −80 to +60), both containing the dra promoter region, were cloned into plasmid pAMB22 as described in Materials and Methods. The resulting plasmids were transformed into both XM15 and XM16. As a control, plasmid pAMB22 was also transformed into XM15 and XM16. As shown in Table 4, lacZ was constitutively expressed in strains containing multiple copies of the dra control region.
TABLE 4.
β-Galactosidase level in B. subtilis dra-lacZ fusion strains carrying pAMB22 derivatives containing different parts of the dra control regiona
| Strain | Region fused to lacZ | Control region on plasmid | Inducer added | Enzyme activity (nmol/min/mg) | Fold induction |
|---|---|---|---|---|---|
| XM35 | −80–+154 | −80–+154 | None | 195 | |
| dRib | 226 | 1.1 | |||
| XM33 | −80–+154 | −80–+60 | None | 206 | |
| dRib | 204 | 1.0 | |||
| XM36 | −80–+60 | −80–+154 | None | 1,215 | |
| dRib | 1,606 | 1.3 | |||
| XM34 | −80–+60 | −80–+60 | None | 1,489 | |
| dRib | 1,490 | 1.0 | |||
| XM37 | −80–+154 | None | None | 17 | |
| dRib | 228 | 13 | |||
| XM38 | −80–+154 | None | None | 91 | |
| dRib | 1,289 | 14 |
Cells were grown in succinate minimal medium. Inducer was added to a final concentration of 1 mg/ml.
Site-directed and random mutagenesis of the dra control region.
A palindromic sequence, 5′-ATTGAA-6 nt (nucleotides) TTCAAT-3′, was found just upstream of the −35 region of the dra promoter (Fig. 1). The role of this sequence in the control of dra expression was analyzed by determining the β-galactosidase levels in different strains containing dra-lacZ fusions with a single-base-pair mutation in the palindromic sequence. Nine mutant strains were constructed as described in Materials and Methods, and β-galactosidase activity was measured for cultures grown in succinate minimal medium with or without inducer (Fig. 2). All strains showed various degrees of derepressed β-galactosidase levels when growing in the absence of inducer. It was observed that changes in the nonpalindromic core sequence could also result in a derepressed β-galactosidase level. The variation in the degree of derepression among the mutants could indicate that some nucleotide positions in the palindromic region are more important for the control mechanism than others. Furthermore, we observed that the nature of the substitution also plays a significant role (data not shown).
By using the PCR procedure described in Materials and Methods, random mutagenesis was performed on a 235-bp fragment composed of the −80 to +154 nucleotides of the dra control region. Nine mutants (XM311 through -319) that constitutively expressed the lacZ reporter gene were isolated, and the DNA sequence of the control regions was determined. All of the mutants had a mutation or mutations in the palindromic sequence or in the core sequence (Fig. 3). The level of β-galactosidase in these mutants was determined in succinate-grown cells in both the presence and absence of dRib. Again we observed that mutations in the palindrome or the core sequence lead to constitutive lacZ expression. From these results and the results from site-directed mutagenesis, we concluded that the palindrome immediately upstream of the −35 region of the dra promoter is at least a part of the operator for DeoR.
FIG. 3.
Nucleotide sequences of the operator region of the wild type and mutants isolated by site-directed mutagenesis and mutagenic PCR. Underlining indicates the palindrome. For mutants, only the mutant nucleotides are shown. Fragments containing the control region with either wild-type sequence or with the indicated mutations were fused to lacZ in pDG268 and integrated into the amyE locus. β-Galactosidase activity is shown for succinate-grown cells. Enzyme levels of XM15 were taken from Fig. 1. The inducer dRib was added to a final concentration of 1 mg/ml.
Titration of DeoR by multiple copies of synthetic operators.
Three synthetic operators of 36, 22, and 18 bp, respectively, each including the palindromic sequence, were cloned in pAMB22 as described in Materials and Methods. The levels of β-galactosidase in strains XM40 through -42 were determined and compared to those in XM35, which had the 235-bp control region cloned in pAMB22, and XM37, which contains pAMB22 (Fig. 4 and Table 4). Surprisingly, none of the DNA fragments cloned in XM40 through -42 could titrate DeoR. Obviously, the palindrome alone was not enough for the binding of DeoR. After a further analysis of the nucleotide sequences of the control region, a sequence of five nucleotides, 5′-TTCAA-3′, identical to the 3′ half of the palindrome was found between the −35 and −10 regions of the promoter (Fig. 1). To test whether this direct repeat sequence is involved in the regulatory mechanism, for instance, by acting as a half binding site for DeoR, three DNA fragments with the same 5′ end as those cloned in strains XM40 through -42, but including the repeated sequence in the 3′ end, were generated and cloned in pAMB22 (strains XM43 through -45 [Table 1]). As shown in Fig. 4, these fragments were able to titrate DeoR. The β-galactosidase levels for these strains were not different from that of the control strain, XM35 (Table 4). This result supported the idea that there might be more than one operator site or half site in the dra-nupC-pdp operon which could be recognized by the DeoR repressor protein and that binding of DeoR to the operator DNA might be cooperative.
FIG. 4.
Nucleotide sequences of the synthetic operators. Underlining indicates the palindrome or the direct repeat to the 3′ half of the palindrome. β-Galactosidase activity was measured for strains XM40 through -45, which contain pAMB22 with the indicated operator sequences cloned in it. Cells were grown in succinate minimal medium. The inducer dRib was added to a final concentration of 1 mg/ml.
Interaction of DeoR with operator DNA.
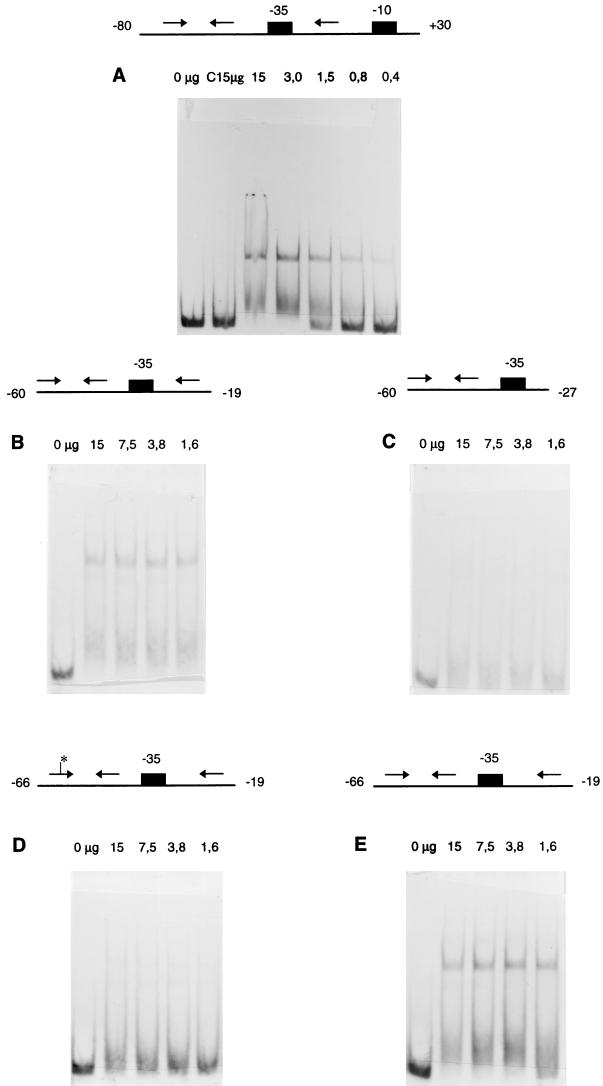
Binding of DeoR repressor protein to the operator DNA of the dra-nupC-pdp operon was analyzed in a gel mobility shift assay by using a 33P-labeled 111-bp DNA fragment containing the region between −80 and +30. As sources of protein, cell extracts of strain JOY100 (DeoR+) or JOY99 (DeoR−) were employed. As shown in Fig. 5A, the mobility of the labeled DNA was retarded by cell extracts of strain JOY100 (15 to 0.4 μg of protein), but not by cell extracts of the control strain, JOY99 (lane C [15 μg]). Variations in the repressor concentration resulted in the different mobilities of the retarded DNA probe. At the lowest protein concentration, almost all of the labeled DNA was free, and a very weak retarded band was observed. As the concentration of protein was increased, one or two retarded bands were formed. At the highest protein concentrations, two clear bands were observed. The different retarded bands probably corresponded to different protein-DNA complexes of one, two, or more binding sites saturated with DeoR molecules.
FIG. 5.
Binding of the DeoR repressor protein to the operator DNA of the dra-nupC-pdp operon. An autoradiograph of a 4% polyacrylamide gel is shown for each assay (A to E). As sources of protein, cell extracts of E. coli strains JOY99 (DeoR−) and JOY100 (DeoR+) were employed. The arrows indicate the palindromic sequence or the direct repeat sequence located 3′ to the palindrome. The black boxes indicate the −35 or −10 region of the dra promoter. The nucleotide numbering relative to the transcription start point is given at the ends of the DNA fragment. An asterisk indicates a mutation. The amount of protein (micrograms) used in each binding assay is given in the figure. C 15μg, 15 μg of control protein from cell extract of strain JOY99.
To characterize further the interaction of DeoR with the operator DNA, the gel mobility shift assays were performed with four other DNA fragments. Figure 5B and C show the interaction of DeoR with two DNA fragments of 42 and 34 bp, corresponding to whether the earlier-mentioned direct repeat sequence 5′-TTCAA-3′ was included or not. Two retarded bands were observed for the 42-bp fragment, but no clear retarded band was observed for the 34-bp fragment, although the lanes in which protein was added were different from those containing the free DNA fragment. The breadth of the shifted bands when the 34-bp fragment was used might suggest relatively weak binding of DeoR. Figure 5D and E show the interaction of DeoR with two DNA fragments of 48 bp of either mutant or wild-type DNA. The mutant DNA fragment could not be retarded, while two retarded bands were observed for the wild-type DNA. These results indicate that binding of the DeoR repressor protein in vitro is dependent on both the palindromic sequence and the direct repeat sequence.
Effect of dRib-5-P and dRib-1-P on DeoR-operator interaction.
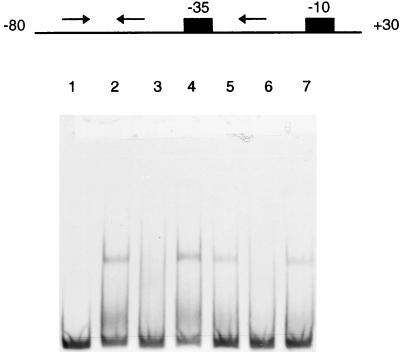
Both dRib and deoxyribonucleosides induce the expression of the dra-nupC-pdp operon. The metabolite dRib-1-P formed by the degradation of deoxyribonucleosides has been suggested as the true inducer molecule (14). However, from the in vitro data obtained by the mobility shift assay as shown Fig. 6, we conclude that dRib-5-P is also a likely candidate for the internal inducer. When dRib-5-P was present in the assay mixture, the retarded bands disappeared (lanes 3 and 6), while one or two retarded bands were observed in assays in which dRib-5-P was not added (lanes 2 and 5) or dRib was added (lanes 4 and 7). No retardation is what should be expected when the potential inducer dRib-5-P binds to the repressor protein and leads to the release of protein from the protein-DNA complex. This would induce the expression of the dra-nupC-pdp operon in vivo.
FIG. 6.
Effect of dRib-5-P on DeoR-operator interaction. An autoradiograph of a 4% polyacrylamide gel is shown. As sources of protein, cell extracts of E. coli JOY100 containing pTCR99A with deoR cloned in it were employed. The arrows indicate the palindromic sequence or the 3′-located direct repeat sequence. The boxes indicate the −35 or the −10 region of the promoter. The nucleotide numbering of the genome sequence is given at the ends of the DNA fragment. Lanes: 1, free DNA fragment; 2 to 4, 1.5 μg of protein; 5 to 7, 0.8 μg of protein. dRib-5-P and dRib (10 mM) were added for lanes 3 and 6 and lanes 4 and 7, respectively.
Unfortunately, dRib-1-P was not available to us, so we could not analyze its effect on the binding of DeoR to the operator DNA in vitro. However, in vivo data from the β-galactosidase activity of the dra-lacZ fusion in a drm background (XM246) suggested that dRib-1-P functioned as an alternative inducer, although it did not induce the expression of the operon as efficiently as dRib-5-P did (the drm mutant cannot convert dRib-1-P to dRib-5-P). When growing in succinate minimal medium, the level of induction by dRib was 10-fold in XM246, the same as in wild-type XM15 cells, but only 5-fold by thymidine in XM246 compared to 10-fold in XM15 (data not shown).
DISCUSSION
The expression of the B. subtilis dra-nupC-pdp operon is controlled in two ways: by deoxyribonucleoside induction and by glucose repression. The region involved in glucose repression was mapped to a region between positions −80 and +154 relative to the transcription start point. This region has already been reported to contain a CRE sequence (15), and in this report, we show that the CRE sequence is indeed involved in glucose repression. A more detailed study of the catabolite repression control of the dra-nupC-pdp operon is not the scope of this report, but will soon be communicated in another work.
The necessary region for deoxyribonucleoside induction was contained in a 141-bp region between positions −80 and +60 covering the dra promoter. When this region was fused to lacZ and introduced into a deoR genetic background, constitutive β-galactosidase production was observed. This confirms the previous results which indicated that DeoR is a negative regulator of the dra promoter activity (15). It was also demonstrated that the 141-bp sequence was able to titrate the DeoR molecules in the cells, when present in multiple copies. We therefore conclude that B. subtilis contains only a limited number of functional DeoR molecules. The previous observation that deoR is expressed at a very low level supports this conclusion (15).
The deoR gene product from B. subtilis (DeoR) has been shown to be the regulator protein responsible for deoxyribonucleoside induction of the dra-nupC-pdp operon (15). B. subtilis DeoR contains the classical α-helix–turn–α-helix DNA binding domain as found in many other DNA binding proteins (12). B. subtilis DeoR shows no similarity to the DeoR repressor protein from E. coli, which belongs to the LacI-GalR family of repressors. In the database, only two repressor proteins have significant similarity to B. subtilis DeoR. These two proteins are SorC and DalR from Klebsiella pneumoniae, which are the regulators for sorbitol and arabinitol catabolism, respectively (5).
In E. coli, the regulatory region of the deo operon contains three DeoR operator sites where deoO1 and deoO2 overlap the −10 region of deoP1 and deoP2, respectively, while the third site, deoOE1, is located several hundred base pairs upstream of deoP1 (11). The deoOE1 and deoO1 operators are separated by a distance of 599 bp, and deoO1 and deoO2 are separated by a distance of 278 bp (1, 3). The native DeoR repressor exists as an octamer and may interact with two or all three deo operator sites at the same time, thereby forming a loop in the intervening DNA. Once bound to one of the operator sites, the DeoR repressor protein, which is present in very low intracellular concentrations, locally increases its effective concentration for secondary sites, thus pushing the equilibrium toward loop formation and efficient repression. This cooperative binding presumably generates a high degree of repression, even though the intrinsic affinity of the repressor for each site is very low (1, 3).
Deletion analysis as well as random and site-directed mutagenesis experiments showed that the palindromic sequence 5′-ATTGAACAAAATTTCAAT-3′ functions as a part of a potential binding site for B. subtilis DeoR. There is no similarity between this sequence and the deo operator of E. coli (3). The B. subtilis operator contains a 6-bp adenine-rich sequence between the palindromic halves. Adenine-rich sequences usually bend or increase the flexibility of the DNA (8, 13, 16). We observed that changes in this part also affected the binding of DeoR. Both the titration experiment and mobility shift assay showed that the direct repeat sequence 5′-TTCAA-3′ located between the −35 and −10 regions is essential for the binding of DeoR. The distance between the palindrome and the direct repeat is 22 bp, which corresponds to 2.1 helical turns, or an angular spacing of approximately 41°.
It is worth mentioning that another 5′-TTCAA-3′ sequence was found 12 bp upstream of the palindrome. However, neither our data obtained in vivo (deletion analysis and titration experiment) nor in vitro (gel shift assay) indicate that this sequence is necessary for the binding of DeoR repressor. No other nucleotide sequence identical to the palindromic halves has been found in the region around the operon.
The operator DNA for the DeoR repressor of B. subtilis consists of a palindrome and a direct repeat sequence located 3′ to the palindrome. Since at least two subunits are needed for binding to symmetrical operator sites (4, 12), we expect the functional DeoR molecule in the cell to be a multimer.
One important characteristic of the DeoR repression in E. coli is that long-range cooperative regulation occurs in the deo operon (3). Although we cannot rule out the possibility of long-distance regulation of B. subtilis DeoR, no more than 141 bp of DNA was found to be enough for full DeoR regulation of the dra-nupC-pdp operon expression.
The metabolite dRib-1-P has been suggested as the low-molecular weight effector for the expression of the dra-nupC-pdp operon (15); however, our data obtained both in vitro and in vivo showed that dRib-5-P is also a likely internal inducer. In fact we observed that in a drm mutant which is unable to convert dRib-1-P to dRib-5-P, dRib induced the expression of the operon two times better than thymidine, whereas dRib and thymidine induced operon expression equally well in the wild type. This indicates that both dRib-1-P and dRib-5-P induce the expression of the dra-nupC-pdp operon and that dRib-5-P induces expression twice as effectively as dRib-1-P does. The in vitro gel shift assay strengthens our suggestion that dRib-5-P also acts as a internal inducer of dra-nupC-pdp operon expression. When dRib-5-P was present in the binding mixtures, only the free DNA band was observed. We should mention that the cell extracts of the E. coli strain we used in the assay were Drm+; in theory, conversion of dRib-5-P to dRib-1-P in the assay mixture was possible.
dRib-5-P has been shown to be the low-molecular-weight effector for the B. subtilis drm-pupG operon expression, but DeoR is not responsible for the regulation of the drm-pupG operon. The drm-lacZ fusion in the drm background was found not to be induced by deoxyribonucleosides but by dRib (11a). This suggested that the conversion of dRib-1-P to dRib-5-P is required for the induction of the drm-pupG operon, which is not the case for the dra-nupC-pdp operon, although dRib-5-P seems to be the preferred inducer.
ACKNOWLEDGMENTS
We thank Mogens Kilstrup for careful reading of the manuscript.
This research received financial support from the Novo Nordic Foundation and from the Saxild Family Foundation.
REFERENCES
- 1.Amouyal M, Mortensen L, Buc H, Hammer K. Single and double loop formation when DeoR repressor binds to its natural operator sites. Cell. 1989;58:545–551. doi: 10.1016/0092-8674(89)90435-2. [DOI] [PubMed] [Google Scholar]
- 2.Christiansen L C, Schou S, Nygaard P, Saxild H H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dandanell G, Valentin-Hansen P, Larsen J E L, Hammer K. Long-range cooperativity between gene regulatory sequences in a prokaryote. Nature (London) 1987;325:823–826. doi: 10.1038/325823a0. [DOI] [PubMed] [Google Scholar]
- 4.Harrison S C, Aggarwal A. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 5.Heuel H, Shakeri-Garakani A, Turgut S, Lengeler W. Genes for D-arabinitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology. 1998;144:1631–1639. doi: 10.1099/00221287-144-6-1631. [DOI] [PubMed] [Google Scholar]
- 6.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 7.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 8.Lefstin J A, Yamamoto K R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 9.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 10.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 11.Mortensen L, Dandanell G, Hammer K. Purification and characterization of the DeoR repressor of Escherichia coli. EMBO J. 1989;8:325–331. doi: 10.1002/j.1460-2075.1989.tb03380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Nygard, P. Personal communication.
- 12.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg A H, Goldman E, Dunn J J, Studier F W, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxild H H, Andersen L N, Hammer K. dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J Bacteriol. 1996;178:424–434. doi: 10.1128/jb.178.2.424-434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleif R. DNA looping. Science. 1988;240:127–240. doi: 10.1126/science.3353710. [DOI] [PubMed] [Google Scholar]
- 17.Tito B J D, Jr, Ward J M, Hodgson J, Gershater C J L, Edwards H, Wysocki L A, Watson F A, Sathe G, Kane J F. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentin-Hansen P, Søgaard-Andersen L, Pedersen H. A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol Microbiol. 1996;20:461–466. doi: 10.1046/j.1365-2958.1996.5341056.x. [DOI] [PubMed] [Google Scholar]
- 19.Zukowski M M, Miller L. Hyperproduction of an intracellular heterologous protein in a SacU mutant of Bacillus subtilis. Gene. 1986;46:247–255. doi: 10.1016/0378-1119(86)90409-9. [DOI] [PubMed] [Google Scholar]