Abstract
Listeria monocytogenes (L. monocytogenes) is frequently detected in ruminants, especially dairy cattle, and associated with the sporadic and epidemic outbreak of listeriosis in farms. In this epidemiological study, the prevalence, virulence, antibiotic resistance profiles, and genetic diversity of L. monocytogenes in three Egyptian dairy cattle farms were investigated. The risk factors associated with the fecal shedding of L. monocytogenes were analyzed. The L. monocytogenes strains from the three farms were categorized into distinct genotypes based on sampling site and sample type through enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR). A total of 1896 samples were collected from animals, environments, and milking equipment in the three farms. Results revealed that 137 (7.23%) of these samples were L. monocytogenes positive. The prevalence of L. monocytogenes in the animal samples was high (32.1%), and the main environmental source of prevalent genotypes in the three farms was silage. For all sample types, L. monocytogenes was more prevalent in farm I than in farms II and III. Risk factor analysis showed seasonal variation in production hygiene. For all sample types, L. monocytogenes was significantly more prevalent in winter than in spring and summer. The level of L. monocytogenes fecal shedding was high likely because of increasing age, number of parities, and milk yield in dairy cattle. Two virulence genes, namely, hlyA & prfA, were also detected in 93 strains, whereas only one of these genes was found in 44 residual strains. Conversely, iap was completely absent in all strains. The strains exhibited phenotypic resistance to most of the tested antibiotics, but none of them was resistant to netilmicin or vancomycin. According to sample type, the strains from the animal samples were extremely resistant to amoxicillin (95.2%, 80/84) and cloxacillin (92.9%, 78/84). By comparison, the strains from the environmental samples were highly resistant to cefotaxime (86.95%, 20/23). Furthermore, 25 multi-antibiotic resistance (MAR) patterns were observed in L. monocytogenes strains. All strains had a MAR index of 0.22–0.78 and harbored antibiotic resistance genes, including extended-spectrum β-lactamase (blaCTX-M [92.7%] and blaDHA-1 [66.4%]), quinolones (qnrS [91.2%], qnrA [58.4%], parC [58.4%], and qnrB [51%]), macrolides (erm[B] [76.6%], erm(C) [1.5%], and msr(A) [27%]), trimethoprim (dfrD [65.7%]), and tetracyclines (tet(M) [41.6%], tet(S) [8%], and int-Tn [26.3%]). ERIC-PCR confirmed that the strains were genetically diverse and heterogeneous. A total of 137 isolated L. monocytogenes strains were classified into 22 distinct ERIC-PCR groups (A–V). Among them, ERIC E (10.2%) was the most prevalent group. These results indicated that environment and milking equipment served as reservoirs and potential transmission ways of virulent and multidrug-resistant L. monocytogenes to dairy animals, consequently posing threats to public health. Silage is the main environmental source of prevalent genotypes on all three farms. Therefore, hygienic measures at the farm level should be developed and implemented to reduce L. monocytogenes transmission inside dairy cattle farms.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11356-022-19495-2.
Keywords: Antibiotic resistance, Dairy cattle farm, ERIC-PCR, Listeria monocytogenes, Molecular epidemiology, Risk factors
Introduction
Listeria monocytogenes is a facultative intracellular Gram-positive bacterium, which has been widely explored because of its association with numerous outbreaks of listeriosis worldwide (Vazquez-Boland et al. 2001). It is included in the World Health Organization list of foodborne pathogens and mostly related to raw milk, unpasteurized milk, and other dairy products (Shamloo et al. 2019). L. monocytogenes infections in humans and animals include eye infections, uveitis, keratitis (Nightingale et al. 2004), septicemia, encephalitis, uterine infections (causing abortion and still birth) (Papić et al. 2019), and subclinical mastitis (Constable et al. 2016).
Listeria monocytogenes has been extensively characterized in many animal species although farm livestock have the greatest pretentious (Chow et al. 2021). It is shed by most diseased ruminants, which are asymptomatic carriers, into their environment via their feces (Nightingale et al. 2004). It is a global environmental bacterium present in diverse farm environments, including water, silage, and feces, which are the main resources and potential reservoirs of L. monocytogenes in dairy farms. Consequently, the dairy cattle farms have different genotypes of L. monocytogenes associated with human listeriosis outbreaks (Castro et al. 2018).
The ecology of L. monocytogenes in the farm environment is complicated and inadequately appreciated. Furthermore, factors influencing the persistence of genotypes in dairy farms are unknown. Therefore, our knowledge about the persistence patterns and contamination methods of L. monocytogenes in dairy cattle farms should be improved to elucidate the contaminant source and provide crucial data for reducing transmission and developing intervention strategies against L. monocytogenes at the level of farms and from animals to humans (Walland et al. 2015; Castro et al. 2018; Chow et al. 2021).
Fecal shedding of L. monocytogenes has many risk factors, including inadequate hygiene, sanitation, housing conditions, silage, antibiotic therapy (approved by veterinarians), and atmospheric season (Bandelj et al. 2018). In addition, bulk tank milk (BTM), milk filter, milking machine, milk handler, fecal contamination, poor on-farm hygiene during milking, storage, and transportation are considered contamination sources (Pantoja et al. 2012).
The virulence potential of L. monocytogenes is determined by several molecular determinants (Camejo et al. 2011), which are classified as follows: listeriolysin O (encoded by hlyA), internalins (coded by inlA, inlC, and inlJ), virulence regulator (represented by prfA), actin assembly (coded by actA), phosphatidylinositol-phospholipase C (encoded by plcA), and invasion-associated protein (represented by iap) (Liu et al. 2007). Listeriolysin O (LLO) is a pore-forming toxin produced by hlyA that contributes to the lysis of bacterium-containing phagocytic vacuoles; as a result, bacterial cells are released into the host cytoplasm.
The persistent increase in antibiotic resistance among Listeria spp., notably L. monocytogenes, has been linked to ongoing selective pressure from widespread antibiotic usage in agriculture, animals, and humans during the last few decades (Baquero et al. 2020). L. monocytogenes is resistant to β-lactam, fosfomycin, third-generation cephalosporins, quinolones, erythromycin, tetracycline-minocycline, and trimethoprim (Iwu and Okoh 2020). Multi-antibiotic-resistant (MAR) L. monocytogenes strains are determined in food, environmental, and clinical samples (Baquero et al. 2020; Iwu and Okoh 2020; Swetha et al. 2021). Antibiotic resistance develops in these pathogens through gene mutations or acquisition of mobile genetic elements. Antibiotic resistance genes are rapidly being acquired by L. monocytogenes, which may have originated from commensal organisms prevalent in foods and food-processing areas. Some antibiotic resistance genes identified in L. monocytogenes are β-lactamase genes (blaSHV, blaCTX-M, blaOXA, blaIMP, blaCMY, and blaTEM), quinolone-resistant genes (qnrA, qnrB, qnrS, gyrA, and parC), macrolide-resistant genes (erm (A), erm (B), erm(C), erm (TR), mef (A), and msr (A)), and tetracycline-resistant genes (tetA, tetK, tetL, tetM, tetS, and int-Tn) (Baquero et al. 2020). However, additional information about the mechanisms of antibiotic resistance among L. monocytogenes strains should be obtained to design strategies that can prevent the emergence and spread of resistance and to develop innovative therapeutic approaches against multidrug-resistant organisms.
One of the critical points to avoid the distribution of healthcare-associated infections and develop infection control is distinguishing the genetic relatedness among different pathogenic strains. Repetitive element sequence-based polymerase chain reaction (PCR) is a molecular approach used to study repetitive nucleotide sequences within a bacterial genome and to cluster bacterial strains. Enterobacterial repetitive intergenic consensus (ERIC) is one of the repetitive basics characterized by the diverse patterns and numbers of bacterial genomes. L. monocytogenes strains possess short ERIC sequences (Jersek et al. 1999). Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) is a rapid, dependable, and cost-effective technique for molecular typing that distinguishes the genetic diversity among strains (Parsaie Mehr et al. 2017).
Few epidemiological surveys have been performed on L. monocytogenes in dairy cattle farms in Egypt and its resistance to various antibiotics. Hence, this research was designed to (a) systematically survey the prevalence and distribution of L. monocytogenes in three dairy cattle farms, (b) categorize the risk factors associated with L. monocytogenes fecal shedding in the three dairy farms, (c) determine the virulence and phenotypic and genotypic antibiotic resistance profiles of distinct types of L. monocytogenes strains and (d) establish the genetic diversity of L. monocytogenes strains through Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR).
Materials and methods
Study areas
A cross-sectional study was performed to investigate the prevalence of L. monocytogenes and the associated risk factors in the environment of dairy cattle farms in Dakahlia Governorate, Egypt. Three dairy cattle farms were chosen on the basis of their owners’ willingness to permit recurrent sample collection. The map of Dakahlia Governorate was constructed to highlight the location of the three selected dairy cattle farms in relation to the rest of Dakahlia (Supplementary Fig. 1).
Questionnaire preparation and data collection
For basic purposes, a structured questionnaire was prepared. The schedule was pretested with necessary adjustment to confirm the importance of the questions and the nature of the sample producers. The dairy farms were selected on the basis of owners’ willingness to allow frequent sample collection. The following data were obtained using the questionnaire: general farm information, animal movement, purchase of animals, visitors and staff, health status, and milking process, as shown in Supplementary Table 1. The farmers were asked questions, and their responses were recorded in the questionnaire. Other details, such as the sanitary condition of the farms, were collected through direct observation.
Assessment of production hygiene
The production hygiene of all farms was assessed depending on the premises hygiene, which was evaluated at each visit during the study period. Aspects assessed included milk room, milking station, waiting area, manure passage, resting area, cow cleanliness, feed troughs, and water troughs. Full scores of 1 to 3 were assigned for each of the evaluated sites in the three farms, where “1” referred to a major deficit in production hygiene, “2” denoted a minor deficit in production hygiene, and “3” indicated no notable deficit in production hygiene according to Castro et al. (2018). During farm visits, the production hygiene of each farm was evaluated on the basis of the cleanliness of the premises, particularly overall cleanliness, hygienic design and condition of materials, drainage, air quality, lighting, and insect control.
Collection and processing of samples
A total of 1896 samples were collected from the three examined dairy cattle farms. Among them, 858, 432, and 606 samples were obtained from farms I, II, and III, respectively. Each farm was visited three times (in February, April, and June 2021) so that seasonal variation could be examined. At each farm level, the samples were collected from animals (milk and feces), environment (water, silage, manure, and soil), and milking equipment (teat cup swabs, milk filters, BTM samples, and floor swabs in storage areas). In farm I, 300 animal samples were obtained for each type; 15 environmental samples were acquired for each type; and 150, 30, 12, and 6 samples were obtained from teat cup swabs, milk filters, BTM samples, and floor swabs in storage areas, respectively. In farm II, 150 animal samples were collected for each type; 9 environmental samples were obtained for each type; and 75, 12, 6, and 3 samples were gathered from teat cup swabs, milk filters, BTM samples, and floor swabs in storage areas, respectively. In farm III, 210 animal samples were obtained for each type; 12 environmental samples were collected for each type; and 105, 18, 9, and 6 samples were acquired from teat cup swabs, milk filters, BTM samples, and floor swabs in storage areas, respectively.
Animal samples
A. Milk samples
Milk samples were collected before milking early in the morning in accordance with previously described methods (Scha et al. 1971). Briefly, before the samples were collected, udders, particularly teats, were cleansed and dried. Each teat end was cleaned with a pledget of cotton wet with 70% ethyl alcohol. A separate pledget of cotton was used for each teat. The first few streams of milk were discarded. About 10 ml of milk was collected into 15-ml sterile glass vials and labeled as RF (right front), and RR (right rear), LF (left front), LR (left rear).
B. Fecal samples
Fecal samples were obtained directly from the rectum of each cow by using a separate clean plastic sleeve for each sample. The plastic sleeves were inverted, and the content was aseptically transferred into sterile plastic vials (conical 50-ml propylene screw top; VWR International, Inc., West Chester, PA).
Environmental samples
A. Water
Water samples were collected from common water troughs in a 50-cm sterile glass bottle (American Public Health Association 1971).
B. Soil
Soil samples were obtained from various sites of yards, especially from the wetted region with high moisture and organic matter load at a depth of 5 cm in a sterile glass bottle fitted with a sterile glass stopper (Clegg et al. 1983).
C. Silage
Silage samples were gathered from silage bunkers of the three dairy farms and placed in a sterile Whirl–Pak bag (Vongkamjan et al. 2012).
D. Manure
Manure composite samples were aseptically obtained from various sites in each pen by using a sterile plastic sleeve. The plastic sleeve content was homogenized, and an aliquot was aseptically transferred into 50-ml plastic vials.
Milking equipment
Bulk tank milk samples (100 ml) were aseptically collected. Milk filter samples were obtained and aseptically transported into a sterile sealable plastic bag. For the milking equipment, a sponge was utilized to wipe the inner side of a certain area. Aseptic cotton swabs were used to collect teat cups and floor swabs in the storage area when the routine washing cycle was complete. The sponges were deposited in sterilized bags containing neutralizing buffer, and the cotton swabs were placed in aseptic tubes with 3 ml of neutralizing buffer. Each sample was loaded in coolers with ice packs and transferred to the laboratory for bacterial examination.
Sample analysis for L. monocytogenes
The soil, silage, fecal, and manure composite samples were weighed, diluted with 1% buffered peptone water (BPW), and pummeled (BagMixer; Weymouth, MA, Interscience Laboratories, Inc.) for bacterial analysis as formerly pronounced by Latorre et al. (2010). All samples were pre-enriched by adding 5 ml of BPW or 5 ml of milk to 5 ml of half Fraser broth (Oxoid, Basingstoke, UK) and incubated for 24 h at 37 °C. For the enrichment, 1 ml of the pre-enriched half Fraser broth was added to 9 ml of Fraser broth (Oxoid, Basingstoke, UK) for each sample and incubated at 37 °C for 24 h. Each enriched Fraser broth culture was cultured onto Palcam agar (Oxoid, Basingstoke, UK) and incubated for 48 h at 37 °C. Presumptive L. monocytogenes colonies (gray with black center) were biochemically identified using the following tests: catalase test, oxidase test, evaluation of hemolysis type, motility at 25 °C and 37 °C, in addition to sugar fermentation test (Van Kessel et al. 2004). The strains expressing these standard features were further tested using the API Listeria test (BioMerieux) for confirmation.
Molecular characterization of L. monocytogenes
The genomic DNA was extracted from overnight culture of brain heart infusion broth using the boiled lysate method (Agersborg et al. 1997). Amplification of 16S RNA gene, virulence genes (listeriolysin O (hlyA), positive regulatory factor (prfA)), and adherence (iap) genes for the L. monocytogenes species identification was done by the standard PCR assays in Applied Biosystem, 2720 Thermal Cycler (USA), in a total volume of 25 μL consisted of 12.5 μL of 2 × PCR master mix (Promega, Madison, USA), 1 μL of individual primer (Metabion, Germany), 4.5 μL PCR-grade water, and 6 μL DNA template. The primers and PCR conditions were used as earlier described (Wang et al. 1992; Bohnert et al. 1992; Germini et al. 2009; Soni et al. 2014). The amplified PCR products were organized on a 1.5% agarose gel that was tainted by 1% ethidium bromide and photo-documented under UV illumination. L. monocytogenes ATCC 35,152 strain was utilized as a positive control.
Antibiotic resistance of L. monocytogenes
Antibiotic-resistant L. monocytogenes strains were obtained via the agar disk diffusion approach on Mueller–Hinton agar (Difco), as endorsed by the Clinical and Laboratory Standards Institute (CLSI 2020). Commonly used antibiotics for humans and animals were selected. The following antimicrobial discs (Oxoid, Ltd.) were used: penicillin G (P/10 IU), amoxicillin (AML/10 μg), cloxacillin (OB/5 μg) belonging to β-lactams, cefotaxime (CTX/30 μg), cefoxitin (FOX/30 μg) belonging to cephalosporines, tetracycline (TE/30 μg) belonging to tetracyclines, streptomycin (S/10 μg), gentamicin (CN/10 μg), amikacin (AK/30 μg), neomycin (N), netiL. monocytogenesicin (NET/30 μg) belonging to aminoglycosides, chloramphenicol (C/30 μg) belonging to phenicols, ciprofloxacin (CIP/5 μg), norfloxacin (NOR/10 μg), nalidixic acid (NA/30 μg) belonging to fluoroquinolones, sulfamethoxazole/trimethoprim (SXT/25 μg) belonging to sulfonamides, vancomycin (VA/30 μg) belonging to glycopeptides, and erythromycin (E/15 μg) belonging to macrolides. L. monocytogenes strains were assessed as susceptible, intermediate, or resistant in accordance with the CLSI (2020) guidelines for Staphylococcus aureus ATCC 25,923 and Escherichia coli ATCC 25,922. The strains displaying resistance to at least three classes of the antimicrobial agents tested were considered MAR strains. The MAR index of each resistant pattern was calculated using the formula provided by Singh et al. (2010). MAR index = number of resistance (strains classified as intermediate based on inhibition zone were considered as sensitive for MAR index) antibiotics/total number of antibiotics tested.
Molecular detection of antibiotic-resistant genes
The resistant L. monocytogenes strains were investigated in terms of the following resistance genes through simplex polymerase chain reaction: blaCTX-M, blaDHA-1, and blaSFO-1 for extended-spectrum β-lactamases (ESBL); qnrA, qnrB, qnrS, gyrA, and parC for quinolones; erm (A), erm (B), erm (C), erm (TR), mef (A), and msr (A) for macrolides; dfrD for trimethoprim; and tet (K), tet (L), tet (M), tet (S), and int-Tn, which encodes the Tn916-Tn1545 integrase of the transposon family, for tetracyclines (Morvan et al. 2010). The primer sequence, cycling conditions, and expected amplicon size are illustrated in Supplementary Table 2. As previously stated, PCR and electrophoresis were performed.
Genetic diversity analysis using ERIC-PCR
The L. monocytogenes strains were genotyped via the enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) fingerprinting assay as described in previous study (Bilung et al. 2018). Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Germany). The oligonucleotide forward primer sequence was ERIC1 primer 5′ATGTAAGCTCCTGGGGATTCAC-3′, and the reverse primer sequence was ERIC1 primer 5′ AAGTAAGTGACTGGGGTGAGCG-3′ (Versalovic et al. 1991). Each 25 μL of the PCR mixture was composed of 12.5 μL of 2 × PCR master mix (Promega, Madison, USA), 1 μL of individual primer (Metabion, Germany), 4.5 μL of PCR-grade water, and 6 μL of DNA template. PCR amplification was performed with the following thermal cycles (Biometra): primary denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 1 min, and extension at 72 °C for 1 min; and a final cycle of 72 °C for 12 min. The amplified PCR products were resolved by electrophoresis on 1.5% agarose gels and photo-documented under UV illumination (Alpha Innotech). ERIC fingerprinting data were converted into a binary code based on the presence or deficiency of each band. Dendrogram was created by the unweighted pair group approach with arithmetic average (UPGMA) and Ward’s hierarchical clustering routine. Cluster analysis and dendrogram construction were presented with SPSS, version 22 (IBM 2013) (Hunter 1990). Similarity index (Jaccard/Tanimoto coefficient and number of intersecting elements) among all samples was analyzed by the online tool (https://planetcalc.com/1664/).
Statistical analysis
Data was recorded using Microsoft Excel spreadsheet (version 15.0), and the investigation was conducted using SPSS (Statistical Set for Social Science) software version 22. Accordingly, descriptive statistics such as percentages and frequency distribution were utilized to determine the prevalence. The antibiotic sensitivity patterns were presented in percentages. Probability level (P) was calculated by using non-parametric test (Chi-square test).
Results
Prevalence and ecology of L. monocytogenes in the dairy cattle farms
Of 1896 samples that were collected, 7.23% (farm I, 3.4%; farm II, 15.2%; and farm III, 6.9%) were positive for L. monocytogenes (Table 1). L. monocytogenes (7.23%) was more predominant in animal samples (32.1%) than in milking equipment (21.9%) and environmental samples (16.8%). L. monocytogenes was determined in all the sampled areas of the three investigated farms except water and floor swabs from the storage area of farm I which were L. monocytogenes negative. Overall, L. monocytogenes was more prevalent in farm I (for all types of samples) than in farms II and III. The prevalence of L. monocytogenes in fecal samples was slightly higher than that in milk samples and varied among the three farms (I, 3% and 2.7%; II, 14.7% and 12%; III, 6.7% and 6.2%, respectively). In environmental samples, L. monocytogenes was found most commonly in silage (farm I, 20%; farm II, 44.4%; and farm III, 25%) followed by manure (farm I, 13.3%; farm II, 33.3%; farm III, 16.6%), water (farm II, 22.2%, and farm III, 8.3%) and soil (farm I, 6.7%; farm II, 11.1%; and farm III, 8.3%). In milking equipment, the prevalence of L. monocytogenes was the highest in teat cup swabs (farm I, 8.3%; farm II, 50%; farm III, 22.2%) and floor swabs in the storage area (farm I, 0%; farm II, 66.7%; and farm III, 16.7%) followed by milk filters (farm I, 3.3%; farm II, 16.7%; and farm III, 5.6%) and BTM samples (farm I, 2.7%; farm II, 12%; and farm III, 3.8%).
Table 1.
Prevalence and distribution of Listeria monocytogenes in the three dairy cattle farms
| Samples | Farm I | Farm II | Farm III | Total no. of positive (%) | |||
|---|---|---|---|---|---|---|---|
| Total no. of samples | Isolate no. (%) | Total no. of samples | Isolate no. (%) | Total no. of samples | Isolate no. (%) | ||
| Animal samples | |||||||
| Fecal samples | 300 | 9 (3) | 150 | 22 (14.7) | 210 | 14 (6.7) | 45 (6.8) |
| Milk samples | 300 | 8 (2.7) | 150 | 18 (12) | 210 | 13 (6.2) | 39 (5.9) |
| Environmental samples | |||||||
| Water | 15 | 0 | 9 | 2 (22.2) | 12 | 1 (8.3) | 3 (8.3) |
| Silage | 15 | 3 (20) | 9 | 4 (44.4) | 12 | 3 (25) | 10 (27.8) |
| Manure | 15 | 2 (13.3) | 9 | 3 (33.3) | 12 | 2 (16.6) | 7 (19.4) |
| Soil | 15 | 1(6.7) | 9 | 1 (11.1) | 12 | 1 (8.3) | 3 (8.3) |
| Milking equipment | |||||||
| Bulk tank milk samples | 150 | 4 (2.7) | 75 | 9 (12) | 105 | 4 (3.8) | 17 (5.2) |
| Milk filters | 30 | 1 (3.3) | 12 | 2 (16.7) | 18 | 1 (5.6) | 4 (6.7) |
| Teat cups swabs | 12 | 1 (8.3) | 6 | 3 (50) | 9 | 2 (22.2) | 6 (22.2) |
| Floor swabs in the storage area | 6 | 0 | 3 | 2 (66.7) | 6 | 1 (16.7) | 3 (20) |
| Total | 858 | 29 (3.4) | 432 | 66 (15.2) | 606 | 42 (6.9) | 137 (7.3) |
Associated risk factors of the fecal shedding of L. monocytogenes
The prevalence of L. monocytogenes showed seasonal variation in different samples collected from the animals, environment, and milking equipment of the three dairy cattle farms (Table 2). Analyses revealed that L. monocytogenes was significantly (P = 0.000) more prevalent in all samples obtained in winter (farm I, 6.6%; farm II, 25.7%; and farm III, 13.7%) than in spring (farm I, 3%; farm II, 13.9%; and farm III, 6.8%) and summer (farm I, 0.35%; farm II, 6.3%; farm III, 0%). The seasonal prevalence of L. monocytogenes in all sample categories for each farm was significantly higher (P = 0.043–0.000) in winter than in spring and summer. In winter, the prevalence of L. monocytogenes was the highest in the environmental samples followed by the milking equipment. Conversely, the lowest prevalence was observed in the animal samples, on the level of three farms.
Table 2.
Seasonal variation in the prevalence of Listeria monocytogenes in the three dairy cattle farms
| Samples | Season | P value | ||||||
|---|---|---|---|---|---|---|---|---|
| Winter | Spring | Summer | ||||||
| Total no. of samples | No. of positive samples (%) | Total no. of samples | No. of Positive samples (%) | Total no. of samples | No. of Positive samples (%) | |||
| Farm I | Animal samples | 20 | 9 (4.5) | 200 | 7 (3.5) | 200 | 1 (0.5) | 0.043 |
| Environmental samples | 20 | 5 (25) | 20 | 1 (5) | 20 | 0 | 0.018 | |
| Milking equipment | 6 | 5 (7.6) | 66 | 1 (1.5) | 66 | 0 | 0.027 | |
| Total | 286 | 19 (6.6) | 286 | 9 (3) | 286 | 1 (0.35) | 0.000** | |
| Farm II | Animal samples | 100 | 22 (22) | 100 | 12 (12) | 100 | 6 (6) | 0.004 |
| Environmental samples | 12 | 6 (50) | 12 | 2 (16.7) | 12 | 2 (16.7) | 0.1 | |
| Milking equipment | 32 | 9 (28.1) | 32 | 6 (18.8) | 32 | 1 (3.1) | 0.025 | |
| Total | 144 | 37 (25.7) | 144 | 20 (13.9) | 144 | 9 (6.3) | 0.000** | |
| Farm III | Animal samples | 140 | 17 (12.1) | 140 | 10 (7.1) | 140 | 0 | 0.000 |
| Environmental samples | 16 | 6 (37.5) | 16 | 1 (6.3) | 16 | 0 | 0.006 | |
| Milking equipment | 49 | 5 (10.2) | 49 | 3 (6.1) | 49 | 0 | 0.016 | |
| Total | 205 | 28 (13.7) | 205 | 14 (6.8) | 205 | 0 | 0.000** | |
**Superscripts indicate highly significant difference for seasonal prevalence of L. monocytogenes across all sample categories within each farm at p ≤ 0.05
The three farms had different hygiene levels (Table 3). The overall hygiene scores (2.8 to 2.5) of farms I and III were higher than that of farm II (1.4). The rank of each farm according to hygiene score had a strong inverse connection with the prevalence of L. monocytogenes. Differences in production hygiene stated in the questionnaire might be due to the considerably greater incidence of L. monocytogenes in farm II (15.2%) than in farms I (6.9%) and III (3.4%). The fecal shedding of L. monocytogenes was positively associated (P < 0.05) with age, parity, and milk yield (Table 4). At the farm level, the higher levels of L. monocytogenes shedding in the feces of dairy cattle were associated with increasing age of > 8 years (farm I, 12%; farm II, 27.8%; and farm III, 17.4%), number of parities of > 4 (farm I, 6.9%; farm II, 29.3%; and farm III, 33.3%), and milk yield of > 18 kg/day (farm I, 4%; farm II, 40%; and farm III, 14.3%).
Table 3.
Evaluation of hygiene on the investigated dairy cattle farms
| Area | Hygiene score by farm | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Farm I | Farm II | Farm III | |||||||
| Winter | Spring | Summer | Winter | Spring | Summer | Winter | Spring | Summer | |
| Milk room | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 3 | 3 |
| Milking station a | 3 | 3 | 3 | 1 | 1 | 1 | 2 | 2 | 2 |
| Waiting area | 3 | 3 | 3 | 2 | 2 | 2 | 3 | 3 | 3 |
| Manure passage | 2 | 2 | 3 | 1 | 1 | 1 | 2 | 2 | 3 |
| Resting area | 2 | 3 | 3 | 2 | 1 | 2 | 2 | 2 | 3 |
| Cow cleanliness | 2 | 3 | 3 | 1 | 1 | 1 | 2 | 2 | 2 |
| Feed troughs | 2 | 3 | 3 | 1 | 1 | 2 | 2 | 2 | 3 |
| Water troughs | 3 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | 3 |
| Mean score of evaluation overall | 2.5 | 2.9 | 3 | 1.4 | 1.3 | 1.6 | 2.3 | 2.4 | 2.8 |
| Mean hygiene score | 2.8 | 1.4 | 2.5 | ||||||
a: Refers to the milking parlor in farms I and III or the milking unit of an automatic milking system in farm II
Each area received a score from 1 to 3, where 1 is major deficits in production hygiene, 2 is minor deficits in production hygiene, and 3 is no notable deficits in production hygiene
Table 4.
Animal’s risk factors related to shedding of Listeria monocytogenes in feces on the level of three dairy cattle farms
| Risk factor | Farm I | Farm II | Farm III | |||
|---|---|---|---|---|---|---|
| Total no. of samples | Positive no. (%) | Total no. of samples | Positive no. (%) | Total no. of samples | Positive no. (%) | |
| Age (years) | ||||||
| 3–5 | 136 | 0 | 62 | 2 (3.2) | 110 | 4 (3.6) |
| 6–8 | 89 | 0 | 34 | 5 (14.7) | 77 | 6 (7.8) |
| > 8 | 75 | 9 (12) | 54 | 15 (27.8) | 23 | 4 (17.4) |
| P value | 0.000 | 0.001 | 0.05 | |||
| Parity (no.) | ||||||
| Up to 2 | 18 | 0 | 49 | 2 (4) | 69 | 2 (2.9) |
| 2–4 | 180 | 2 (1.1) | 60 | 8 (13.3) | 120 | 5 (4.2) |
| > 4 | 102 | 7 (6.9) | 41 | 12 (29.3) | 21 | 7 (33.3) |
| P value | 0.01 | 0.003 | 0.000 | |||
| Milk yield (kg/day) | ||||||
| Low (< 12) | 0 | 0 | 50 | 2 (4) | 8 | 0 |
| Medium (12–18) | 80 | 0 | 70 | 8 (11.4) | 114 | 0 |
| High (over 18) | 220 | 9 (4) | 30 | 12 (40) | 98 | 14 (14.3) |
| P value | 0.000 | 0.000 | 0.000 | |||
Factors statistically significant at p ≤ 0.05
Virulence genes in L. monocytogenes strains
All strains (n = 137) from various farm samples were examined for the existence of virulence genes, as demonstrated in Table 5. A total of 93 L. monocytogenes strains were positive for both hlyA and prfA genes, i.e., 56 animal strains (60.2%), 22 milking equipment strains (23.7%), and 15 environmental strains (16.1%). The 44 remaining strains carried only one virulence gene: 30 strains with hlyA and 14 strains with prfA. Conversely, none of the strains had iap.
Table 5.
Ecological distribution of antibiotic resistance of Listeria monocytogenes strains in the three examined dairy cattle farms (n = 137)
| Samples | Total no. of isolates | Antibiotics | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | N | FOX | NA | AML | OB | CTX | AK | E | NOR | TE | CN | S | SXT | CIP | C | NET | VA | |||
| Farm I | Animal samples | 17 | 17 | 17 | 17 | 17 | 13 | 11 | 9 | 5 | 5 | 0 | 0 | 9 | 4 | 0 | 0 | 0 | 0 | 0 |
| Environmental samples | 6 | 6 | 6 | 6 | 4 | 4 | 3 | 5 | 3 | 3 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Milking equipment | 6 | 6 | 6 | 6 | 4 | 4 | 3 | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Total | 29 | 29 | 29 | 29 | 21 | 21 | 17 | 16 | 10 | 10 | 0 | 0 | 11 | 8 | 0 | 0 | 0 | 0 | 0 | |
| Farm II | Animal samples | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 36 | 29 | 31 | 30 | 25 | 17 | 0 | 0 | 0 |
| Environmental samples | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 7 | 6 | 6 | 8 | 5 | 8 | 0 | 0 | 0 | |
| Milking equipment | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 12 | 11 | 10 | 10 | 7 | 7 | 0 | 0 | 0 | |
| Total | 66 | 66 | 66 | 66 | 66 | 66 | 66 | 66 | 66 | 66 | 55 | 46 | 47 | 48 | 37 | 32 | 0 | 0 | 0 | |
| Farm III | Animal samples | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 21 | 23 | 23 | 21 | 17 | 9 | 10 | 19 | 16 | 5 | 0 | 0 |
| Environmental samples | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 5 | 6 | 6 | 5 | 7 | 1 | 2 | 4 | 4 | 1 | 0 | 0 | |
| Milking equipment | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 6 | 8 | 8 | 6 | 3 | 5 | 3 | 7 | 5 | 3 | 0 | 0 | |
| Total | 42 | 42 | 42 | 42 | 42 | 42 | 42 | 32 | 37 | 37 | 32 | 27 | 15 | 15 | 30 | 25 | 9 | 0 | 0 | |
| Total | 137 | 137 | 137 | 137 | 137 | 129 | 125 | 114 | 113 | 113 | 87 | 73 | 73 | 71 | 67 | 57 | 9 | 0 | 0 | |
| Resistance (%) | 100 | 100 | 100 | 100 | 100 | 94.2 | 91.2 | 83.2 | 82.5 | 82.5 | 62.8 | 53.3 | 53.3 | 50.4 | 48.9 | 32 | 6.6 | 0 | 0 | |
P, penicillin; N, neomycin; FOX, cefoxitin; NA, nalidixic acid; AML, amoxicillin; OB, cloxacillin; CTX, cefotaxime; AK, amikacin; E, erythromycin; NOR, norfloxacin; TE, tetracycline; CN, gentamicin; S, streptomycin; SXT, sulphamethazole/trimethoprim; CIP, ciprofloxacin; C, chloramphenicol; NET, netilmicin; VA, vancomycin
Antibiotic resistance of Listeria monocytogenes strains
The antibiotic susceptibility test results of 137 L. monocytogenes strains screened for 18 relevant antimicrobial agents which belong to eight various classes of antibiotics including beta-lactams, tetracyclines, quinolones, phenicols, sulfonamides, macrolides, aminoglycoside, and glycopeptides are illustrated in Table 6. The highest resistance of the L. monocytogenes strains was observed in penicillin, neomycin, cefoxitin, and nalidixic acid (100%), followed by amoxicillin (94.2%), cloxacillin (91.2%), cefotaxime (83.2%), amikacin (82.5%), erythromycin (82.5%), and norfloxacin (62.8%). By contrast, the resistance of the L. monocytogenes strains to tetracycline, gentamicin, streptomycin, and sulfamethoxazole-trimethoprim was low (53.3%, 53.3%, 50.4%, and 48.9%, respectively). According to sample type, the strains from the animal samples were highly resistant to amoxicillin (95.2%, 80/84) and cloxacillin (92.9%, 78/84). The strains from the environmental samples were also highly resistant to cefotaxime (86.95%, 20/23).
Table 6.
ERIC-PCR, antibiotic resistance and virulence of Listeria monocytogenes isolated from dairy cattle farms (n = 137)
| Farm | Source | Type of sample | Isolates no. or ID | ERIC-PCR Type | *Antibiotics pattern | Virulence genes | Antibiotics resistance genes |
|---|---|---|---|---|---|---|---|
| Farm I | Animals | Milk | 14 | F | 4 | prfA | CTX-M |
| 21 | F | 8 | prfA | CTX-M, dfrD | |||
| 25 | K | 9 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA | |||
| 15 | L | 4 | hlyA | DHA, qnrS, dfrD | |||
| 24 | M | 9 | hlyA,prfA | CTX-M, qnrA, qnrS, parC, ermB, msrA, dfrD | |||
| 1 | Q | 1 | prfA | DHA, qnrS, dfrD | |||
| 19 | Q | 5 | hlyA | CTX-M, dfrD | |||
| 22 | T | 14 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA | |||
| Feces | 11 | E | 3 | hlyA | DHA, qnrS, dfrD | ||
| 17 | I | 4 | hlyA | DHA, qnrS | |||
| 5,6 | F | 2 | hlyA | CTX-M, DHA, qnrS | |||
| 9 | F | 3 | hlyA, | DHA, qnrS | |||
| 2 | G | 1 | hlyA, prfA | DHA, qnrS, dfrD | |||
| 18 | G | 5 | prfA | CTX-M, dfrD | |||
| 23 | G | 14 | hlyA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| 13 | L | 4 | hlyA | DHA, qnrS, dfrD | |||
| Environment | Silage | 20 | G | 8 | hlyA | CTX-M, qnrS, ermB, dfrD | |
| 12 | F | 3 | hlyA | CTX-M | |||
| 29 | J | 9 | prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB | |||
| Manure | 8 | J | 2 | prfA | CTX-M, qnrS, dfrD | ||
| 26 | M | 9 | hlyA, prfA | CTX-M, qnrA, qnrS, parC, ermB, dfrD | |||
| Soil | 7 | N | 2 | hlyA | CTX-M, qnrS, dfrD | ||
| Milking equipment | BTM | 16 | A | 4 | hlyA | CTX-M, qnrS | |
| 10 | E | 3 | hlyA | CTX-M, qnrS, dfrD | |||
| 4 | F | 1 | prfA | qnrS | |||
| 27 | M | 9 | hlyA, prfA | CTX-M, qnrA, qnrS,parC, ermB, dfrD | |||
| Teat cups swab | 28 | S | 9 | hlyA | CTX-M, qnrA, qnrB,qnrS, ermB, msrA, parC | ||
| Milk filter | 3 | Q | 1 | prfA | DHA, qnrS, dfrD | ||
| Farm II | Animals | Milk | 70 | A | 11 | hlyA, prfA | CTX-M, qnrA, qnrB,qnrS, parC, ermB, msrA, dfrD |
| 76 | A | 12 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD | |||
| 40,56 | G | 24 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 88 | K | 7 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD | |||
| 80 | L | 12 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, msrA | |||
| 35 | M | 20 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| 59 | M | 24 | prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 51 | N | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM | |||
| 39 | Q | 20 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, dfrD, tetM, int-Tn | |||
| 41 | Q | 18 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, dfrD, tetM | |||
| 45 | Q | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, dfrD, tetM, int-Tn | |||
| 65 | Q | 19 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM | |||
| 66 | S | 19 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, tetM | |||
| 34 | T | 20 | prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, dfrD, tetM | |||
| 47 | T | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, dfrD, tetM | |||
| 38 | U | 20 | hlyA, prfA | CTX-M, DHA, dfrD, tetM | |||
| 44 | U | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM | |||
| Feces | 91, 93 | A | 15 | hlyA, prfA | CTX-M, DHA, qnrS, ermB | ||
| 94, 95 | A | 24 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 32 | A | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM, int-Tn | |||
| 89 | C | 7 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| 92 | C | 15 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD | |||
| 64 | C | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 30 | F | 9 | hlyA, prfA | CTX-M, qnrS, ermB | |||
| 31,67,68 | F | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 52 | F | 23 | hlyA | CTX-M, tetM, int-Tn | |||
| 79 | I | 16 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| 81 | I | 12 | hlyA, prfA | CTX-M, qnrS | |||
| 90 | I | 15 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD | |||
| 33 | J | 24 | hlyA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 43,49,50 | J | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 58 | G | 24 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 57 | Q | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM | |||
| Environment | Water | 84 | I | 12 | hlyA, prfA | CTX-M, qnrS | |
| 53 | J | 23 | hlyA, prfA | qnrS, ermB, dfrD, tetM, int-Tn | |||
| Silage | 54 | A | 23 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, tetM, int-Tn | ||
| 61 | C | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM, int-Tn | |||
| 82 | E | 17 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM | |||
| 71 | F | 11 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB | |||
| Manure | 75 | M | 11 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | ||
| 86 | M | 7 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| 55 | Q | 23 | prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetS | |||
| Soil | 62 | M | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC | ||
| Milking equipment | BTM | 77,78 | L | 16 | hlyA, prfA | CTX-M, DHA, dfrD, tetM, int-Tn | |
| 36 | M | 20 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| 60 | M | 21 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, tetM | |||
| 73,74 | M | 11 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | |||
| 42 | Q | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 63 | I | 24 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 37 | J | 20 | hlyA, prfA | CTX-M, dfrD, tetM | |||
| Teat cups swab | 48 | E | 25 | hlyA, prfA | CTX-M, dfrD, tetM | ||
| 46 | J | 25 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| 83 | K | 17 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| Milk filter | 72 | S | 11 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | ||
| 69 | T | 24 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, msrA, dfrD, tetM, int-Tn | |||
| Floor swabs | 85 | H | 12 | hlyA, prfA | CTX-M, qnrS, ermB, dfrD | ||
| 87 | K | 7 | hlyA, prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| Farm III | Animals | Milk | 115,116 | C | 21 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, tetS |
| 96,97,98 | L | 24 | hlyA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | |||
| 135,136 | O | 13 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, msrA, tetM, int-Tn | |||
| 101 | P | 24 | hlyA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetM | |||
| 102,103 | R | 22 | hlyA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| 137 | T | 6 | hlyA.prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| 128 | V | 10 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD, tetM | |||
| 127 | E | 6 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | |||
| Feces | 131 | B | 6 | PrfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | ||
| 124,125 | C | 10 | hlyA, PrfA | CTX-M, DHA, qnrS, ermB, dfrD | |||
| 106, 107, 109 | D | 22 | hlyA | CTX-M, DHA, qnrS, ermB, dfrD, tetS | |||
| 111, 112, 113, 114 | E | 21 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, tetM | |||
| 120, 122, 123 | E | 6 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD | |||
| 129 | V | 10 | prfA | CTX-M, DHA, dfrD | |||
| Environment | Water | 104 | D | 22 | hlyA | CTX-M, DHA, qnrS, ermB, dfrD, tetS | |
| Silage | 134 | B | 13 | hlyA, prfA | CTX-M, qnrS, ermC, tetS | ||
| 118 | C | 21 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermC, tetS | |||
| 119 | E | 21 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, tetM | |||
| Manure | 130 | V | 6 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | ||
| 126 | V | 10 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD, tetM | |||
| Soil | 133 | S | 6 | prfA | CTX-M, qnrA, qnrB, qnrS, parC, ermB, dfrD | ||
| Milking equipment | BTM | 99 | L | 24 | hlyA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB | |
| 108 | P | 22 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD, tetS | |||
| 105 | R | 22 | hlyA | CTX-M, DHA, qnrS, ermB, msrA, dfrD, tetM, int-Tn | |||
| 117 | T | 21 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD, tetS | |||
| Teat cup swab | 100 | I | 24 | hlyA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD | ||
| 110 | P | 22 | hlyA, prfA | CTX-M, DHA, qnrS, ermB, dfrD, tetS | |||
| Milk filter | 121 | E | 6 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB, dfrD | ||
| Floor swab | 132 | V | 6 | hlyA, prfA | CTX-M, DHA, qnrA, qnrB, qnrS, parC, ermB |
*Antibiotic patterns: 1;P, N, FOX, NA; 2;P, N, FOX, NA, CTX; 3;P, N, FOX, NA, AML; 4;P, N, FOX, NA, AML, OB, CN; 5;P, N, FOX, NA, AML, OB, CTX, CN; 6;P, N, FOX, NA, OB, AK, SXT, CIP; 7;P, N, FOX, NA, OB, CTX, AK, CIP; 8;P, N, FOX, NA, AML, OB, CTX, AK, CN; 9; P, N, FOX, NA, AML, OB, CTX, AK, S; 10; P, N, FOX, NA, AML, OB, CTX, NOR, SXT; 11; P, N, FOX, NA, OB, CTX, AK, S, CIP; 12;P, N, FOX, NA, OB, CTX, NOR, AK, CN; 13; P, N, FOX, NA, OB, CTX, NOR, AK, TE; 14; P, N, FOX, NA, AML, OB, CTX, AK, CN, S; 15; P, N, FOX, NA, AML, OB, CTX, NOR, AK, S; 16; P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN; 17; P, N, FOX, NA, OB, CTX, NOR, AK, TE, S; 18;P, N, FOX, NA, AML, OB, CTX, NOR, AK, TE, S; 19;P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, S; 20; P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, SXT; 21; P, N, FOX, NA, AML, OB, CTX, NOR, AK, TE, S, CIP; 22;P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, SXT, C; 23;P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, S, CIP; 24; P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, SXT, S, CIP; 25;P, N, FOX, NA, OB, CTX, NOR, AK, TE, CN, SXT, S
Listeria monocytogenes strains showed 25 multi-resistance patterns to antibiotics ranging from 4 to 14 (Supplementary Table 3). The predominant multi-resistance pattern was P, N, FOX, NA, OB, CTX, NOR, AK, E, TE, CN, SXT, S, CIP (16.8%, 23/137). This pattern was followed by P, N, FOX, NA, OB, AK, E, SXT, CIP; P, N, FOX, NA, AML, OB, CTX, NOR, AK, E, TE, S, CIP and P, N, FOX, NA, OB, CTX, NOR, AK, E, TE, CN, SXT, S (7.3%, 10/137) for each. These four resistance patterns were found in 38.7% (53/137) of all strains. As for the strains from the farms, the dominant multi-resistance patterns from farm I strains were P, N, FOX, NA, AML, OB, CN (20.7%, 6/29) and P, N, FOX, NA, AML, OB, CTX, AK, E, S (17.2%, 5/29). The dominant patterns of the strains from farm II were P, N, FOX, NA, OB, CTX, NOR, AK, E, TE, CN, SXT, S, CIP (25.8%, 17/66) and P, N, FOX, NA, OB, CTX, NOR, AK, E, TE, CN, SXT, S (15.2%, 10/66). The most common pattern of the strains from farm III was P, N, FOX, NA, OB, AK, E, SXT, CIP (23.8%, 10/42).
All L. monocytogenes strains (100%) had MAR to at least 4 of the 18 antibiotics examined. Their MAR index ranged from 0.22 to 0.78. The highest MAR index of 0.78 was recorded in 24.1% (33/137) isolates from which 27 strains were found in farm II. Furthermore, a MAR index of 0.72 was detected in 16.8% (23/137) of isolates.
Antibiotic resistance genes
The PCR screening of the antibiotic resistance genes in MAR L. monocytogenes strains showed that all examined strains (n = 137) contained at least one antibiotic resistance gene (Table 5). In particular, 127 (92.7%) L. monocytogenes strains had blaCTX-M gene, and only 91 (66.4%) strains were positive for blaDHA-1 resistance gene. Conversely, no strain had blaSFO-1 gene. Genotyping analysis revealed the existence of quinolone resistance genes: qnrS in 125 (91.2%) strains, qnrA and parC in 80 (58.4%) strains, and qnrB in 70 (51%) strains. By contrast, no strain had gyrA gene. Macrolide resistance genes were also detected: erm (B) (76.6%, 105/137), erm (C) (1.5%, 2/137), and msr (A) genes (27%, 37/137). However, erm (A), erm (TR), and mef (A) genes were not present in the examined strains. trimethoprim dfrD gene was greatly abundant in 65.7% (n = 90) of strains. tet(M) gene as a determinant of resistance to tetracyclines through ribosome protection was detected in 41.6% (57/137) of the examined strains, and the tet(S) gene was found in 8% (11/137). The existence of int-Tn gene for the integrase of Tn916-Tn1545 was observed in 26.3% (36/137) of strains harboring tet(M). Other tetracycline resistance genes (tetK and tetL) were not found in the L. monocytogenes strains.
Most (82.8%, 24/29) strains isolated from farm I harbored qnrS gene, whereas most strains from farms II (98.5%, 65/66) and III (100%, 42/42) had blaCTX-M gene.
Genetic diversity of L. monocytogenes strains
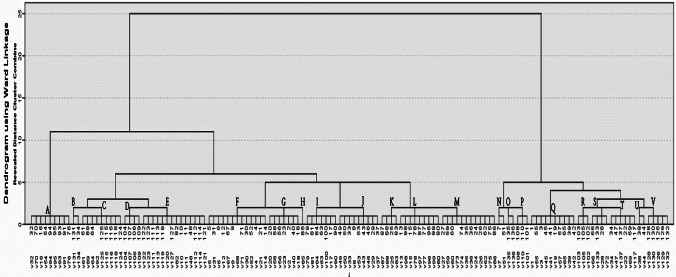
The electrophoretic profile of DNA fragments obtained from 137 L. monocytogenes strains following ERIC-PCR amplification produced 1–8 bands, whose size ranged from 163 to 3074 bp. The visual comparison of the banding patterns showed 22 distinct ERIC profiles (A–V). The most common ERIC type (Table 5) was ERIC E (10.2%, 14), followed by ERIC F (9.5%, 13), ERIC M (8.8%, 12), ERIC Q (7.3%, 10), ERIC A,C,J,L (6.6%, 9), and ERIC G,I (5.1%, 7), ERIC T (4.4%, 6), ERIC V (3.6%, 5), ERIC D,K,S (2.9%, 4), ERIC P,R (2.2%, 3), ERIC B,N,O,U (1.4%, 2), and ERIC H (0.7%, 1).
The ERIC-PCR dendrogram (Fig. 1) showed that L. monocytogenes obtained from the three farms were genetically diverse and heterogeneous, as indicated by their categorization into specific genotype by sampling site and sample type. The ecological distribution of the 22 ERIC types of L. monocytogenes in the examined farms is shown in Table 5. Furthermore,13, 16, and 12 ERIC types were observed in farm I (A, E–G, I–N, M, Q, S, and T), farm II (A, C, E–N, Q, and S–U), and farm III (B–E, I, L, O, P, R–T, and V). Only ERIC E, I, L, S, and T were common in the three farms. ERIC A, E, F, G, I–N, Q, and S–T were present in farms I and II. ERIC C, E, I, L, S, and T were found in farms II and III. In farm I, the predominant type was ERIC F (24%, 7/29), followed by ERIC G (13.8%, 4/29) at which ERIC F was common in milk, feces, silage, and BTM isolates, whereas ERIC G was common in the isolates of feces and silage only. The predominant type in farm II was ERIC M (13.6%, 9/66), followed by ERIC A (12.1%, 8/66), ERIC J (10.6%, 7/66), ERIC Q (10.6%, 7/66), ERIC F (9%, 6/66), ERIC I (7.6%, 5/66), and ERIC C (6%, 4/66), which accounted for 69.7% (46/66) of the isolates in farm II (Table 5). ERIC A was common in isolates of milk, feces, and silage; ERIC F and C were common in feces and silage isolates; and ERIC M was common in milk, manure, and BTM isolates. The five predominant types in farm III were ERIC E (23.8%, 10/42), ERIC C (11.9%, 5/42), ERIC V (11.9%, 5/42), ERIC D (9.5%, 4/42), and ERIC L (9.5%, 5/42). ERIC E and C were evident in the isolates of faces and silage. ERIC V was found in the isolates of milk, manure, and floor swabs of the milking room. It was also common in fecal and water isolates.
Fig. 1.
Dendrogram representing genetic relationships between L. monocytogenes isolates based on ERIC-PCR fingerprints. Twenty-two ERIC profile represented by A–V and the isolates ID represented by 1–136
Discussion
The epidemiology of L. monocytogenes in clinical human, animal, and food specimens has been widely explored (Castro et al. 2018; Swetha et al. 2021). To our knowledge, this study was the first to elucidate the ecology of L. monocytogenes in dairy cattle farms and identify the sources of the pathogen and risk factors in the environment of dairy cattle farms. Various samples were collected from clinically normal cattle, the environment, and the milking system. The results showed that the prevalence of L. monocytogenes was 7.23% (farm I, 3.4%; farm II, 15.2%; farm III, 6.9%) in all the three studied farms. Mohammed and Abdel Aziz (2017) found a greater prevalence (28.1%) in Egypt possibly because of differences in farm size, management practices, and hygiene ranking. Conversely, the prevalence of L. monocytogenes was low in the USA (4.48%) (Van Kessel et al. 2011), Iran (2.02%) (Sohrabi et al. 2013), and Italy (1.6%) (Bianchi et al. 2013).
The prevalence of L. monocytogenes was higher in the environment than in dairy cattle. Moreover, silage (27.8%) and manure (19.4%) followed by water and soil (8.3%, each) were considered the principal source of L. monocytogenes in the dairy farm environment. These values confirmed the importance of environmental sources of L. monocytogenes in dairy farms, especially silage (Mohammed and Abdel Aziz 2017). The prevalence of L. monocytogenes in milk filters was higher (6.7%) than that in the BTM samples (5.2%) partly because of the concentrations of the bacteria in the filter and dilution in the tanks (Vilar et al. 2007; Bandelj et al. 2018).
Data analysis showed that five risk factors were significantly (P < 0.05) associated with the increased shedding of L. monocytogenes in dairy cattle farms. These risk factors included two environmental factors (season and hygienic condition in the farms) and three animal factors (age of cows, parity, and milk yield). The prevalence of L. monocytogenes had significant seasonal variation in all types of samples (animal, environmental, and milking equipment samples) from the three examined farms. In particular, the prevalence was higher in winter than in spring and summer. Although the analysis of seasonal variation was limited by the short study period, this finding was consistent with those of other investigations (Dalzini et al. 2016; Bandelj et al. 2018) which identified significant seasonal differences and revealed the higher prevalence of L. monocytogenes in cold seasons (winter and early spring) than in other seasons. This high prevalence in cold months could be due to several factors, including the crowding of cattle in indoor facilities, difficulty in maintaining excellent hygiene practices under such circumstances, and the ingestion of spoiled silage (Ryser and Marth, 2007). Other studies have identified insignificant seasonal differences (Hassan et al. 2001; Mohammed et al. 2010). This disparity in seasonal prevalence among different studies might be attributed to variations in study design, climate, and management practices in various geographical regions.
The environmental prevalence of L. monocytogenes in farm II was higher than that in farms I and III. As such, it could be due to poor milking hygiene and contamination pressure from the environment of farms to dairy cattle in our study (Castro et al. 2018).
In the three farms studied, the fecal shedding of L. monocytogenes had a positive significant relationship with the age, parity, and milk yield of dairy cattle. Dairy cattle are affected by several stressors, such as pregnancy, parturition, and lactation, which may suppress host immunity, increase L. monocytogenes fecal shedding in healthy ruminants, and increase their susceptibility to pathogen infection (Roberts and Wiedmann 2003).
The presence of key virulence factors, hlyA and prfA, confers pathogenicity to L. monocytogenes strains (Poimenidou et al. 2018). The hlyA, which encodes listeriolysin O (LLO), is a basic L. monocytogenes pathogenicity gene that helps release bacterial cells through the host cell vacuole (Roberts et al. 2005). PrfA is a protein required for the transcription of the prfA-regulated virulence gene cluster and prfA itself. The iap is a surface protein that acts as a murein hydrolase. In our study, the majority of L. monocytogenes strains (93) were positive for hlyA and prfA although 44 tested strains were positive for only one virulence gene (hlyA or prfA). Conversely, all the tested bacterial strains were negative for iap. These virulence genes are linked to L. monocytogenes strains from clinical and food samples in Ireland (Poimenidou et al. 2018), animals in Egypt (Elbar et al. 2020), and the environment in South Africa (Iwu and Okoh 2020). Consistently, the rates of prfA and hly in L. monocytogenes strains obtained from environmental water in South Africa are high (Kayode et al. 2021). Therefore, the presence of the virulence genes strongly suggested that L. monocytogenes strains from the studied dairy cattle farms could cause listeriosis in humans.
MAR L. monocytogenes from different sources, such as human, food, and environmental samples, has been widely described (Castro et al. 2018; Swetha et al. 202). In this work, all L. monocytogenes strains from dairy farms were resistant to penicillin, neomycin, cefoxitin, and nalidixic acid. They were also resistant to amoxicillin, cloxacillin, cefotaxime, amikacin, erythromycin, norfloxacin, tetracyclines, and gentamicin, which are frequently useful in the treatment of human listeriosis. However, they were highly susceptible to chloramphenicol and ciprofloxacin. These findings confirmed previous observations, which demonstrated that L. monocytogenes strains have variable resistance to commonly used antibiotics in the medication of clinical and veterinary infections (Su et al. 2016; Swetha et al. 2021). Tahoun et al. (2017) reported high tetracycline, clindamycin, and rifampicin resistance in L. monocytogenes isolated from an Egyptian dairy farm. However, a previous investigation showed that L. monocytogenes isolated from environmental water has high resistance rates against sulfamethoxazole, oxytetracycline, and amoxicillin but not against ampicillin (Kayode et al. 2021). These differences in the susceptibility patterns of L. monocytogenes strains could be dependent on geographical variations and antibiotic use for humans and animals.
All L. monocytogenes strains exhibited multiple resistances to four classes of antibiotics, namely, β-lactam (particularly second-generation cephalosporins), aminoglycosides, quinolones, and macrolides, which pose risks to public health because of challenges in the treatment of listeriosis. Furthermore, 25 antimicrobial resistance patterns were observed among the MAR L. monocytogenes strains isolated from dairy farms, viewing resistance to antibiotics ranging from 4 to 14. This observation was consistent with the findings of Iwu and Okoh (2020) on higher multiple resistance than single resistance. The observed resistance might be attributed to medication use or feed additives in the livestock industry (Zeitoun et al. 2015). It is significant in the context of the incidence of temporal and spatial changes in antibiotic resistance (Yan et al. 2010). Thus, the emergence of antibiotic resistance should be continuously monitored, and other treatment methods should be developed. In the current work, all L. monocytogenes had a MAR index of > 0.20, indicating that the strains isolated from the three dairy farms originated from high-risk sources in which they were constantly exposed to antibiotics and had a high-risk potential (Bilung et al. 2018).
The mechanisms conferring resistance to different classes of antibiotics were explored to determine whether each MAR L. monocytogenes strain in this investigation had at least one antimicrobial resistance gene. The results showed more prevalence of blaCTX-M gene responsible to produce CTX-M β-lactamases among cefotaxime-resistant strains in comparison to the gene encoding DHA-type β-lactamases (Iwu and Okoh 2020). The first L. monocytogenes strains isolated from dairy farms harboring the plasmid-mediated AmpC β-lactamase DHA-1 gene were designed a few years after the statement of gram-negative bacteria harboring the blaDHA-1 gene. The widespread use of β-lactam antibiotics has resulted in a surge in the occurrence rate of ESBLs because of their low toxicity and effectiveness in the treatment of various infectious diseases, thereby posing a severe threat to global health (Livermore 1996).
Plasmid-mediated quinolone resistance genes (qnrS, qnrA, and qnrB) and parC were observed in quinolone-resistant strains. Though quinolones are not recommended treatment options for Listeria infections, they can indirectly disseminate the emergence of resistant L. monocytogenes strains because of their massive usage for the medication of multiple infections (Godreuil et al. 2003). The resistance of Gram-positive bacteria to quinolones is due to adjustments in the quinolone resistance–determining regions of the intracellular targets of quinolones, DNA gyrase encoded by gyrA and gyrB, and topoisomerase IV encoded by parC and parE (Hooper and Jacoby 2016). The erm (erythromycin ribosome methylase) encodes a 23S rRNA methyltransferase responsible for the modification of the macrolide–lincosamide–streptogramin B (MLSB) antibiotic binding site (Leclercq 2002).
Three MLSB resistance genes, namely, ermB, ermC, and msrA, were determined in the erythromycin-resistant strains. Among them, ermB is the most predominant (Morvan et al. 2010). However, erm (A), erm (TR), and mef (A) encoding the recorded efflux pumps in Gram-positive bacteria were not observed in the tested strains (Leclercq 2002; Granier et al. 2011). The high occurrence of dfrD, encoding a resistant dihydrofolate reductase, has been reported in L. monocytogenes strains from the environment and humans (Morvan et al. 2010). In our study, the prevalence of tet (M) was higher than that of tet (S) that was also noticed by Escolar et al. (2017). The MAR strains exhibited resistance to tetracycline, indicating ribosome protection because of tet(M) or tet(S) (Charpentier and Courvalin 1999). As a determinant of resistance to tetracyclines, tet (M) is highly prevalent in Gram-positive bacteria resistant to tetracyclines and commonly related to conjugative elements of the Tn916 family (Leclercq et al. 2005). In our study, the existence of int-Tn gene in the strains harboring tet (M) confirmed that L. monocytogenes was partly resistant to tetracycline because of the acquisition of conjugative transposons (Poyart-Salmeron et al., 1989).
ERIC-PCR analysis revealed that the L. monocytogenes strains from the three dairy farms were genetically diverse and heterogeneous. Heterogeneity was indicated by the various origins of the strains (animal, environment, and milking equipment) and sampling locations. The potential contamination routes of L. monocytogenes in the dairy cattle farm environment were detected by investigating the prevalence of genotypes among the three farms and sampling areas inside each farm through ERIC-PCR. In the three farms, the sources of predominant genotypes were milk, feces, silage, manure, floor swabs of the milking room, and water, suggesting that various locations in the farm environment could be considered ecological niches for the persistence of L. monocytogenes inside dairy cattle farms (Morvan et al. 2010). This study suggested that the consumption of contaminated silage was the potential source of L. monocytogenes in the investigated farms (Nightingale et al. 2005; Ho et al. 2007). L. monocytogenes contamination in BTM was likely attributed to the milking system (teat cups and milking filters), as observed in other studies (Terentjeva et al. 2021). Our study also demonstrated low similarity in the L. monocytogenes genotypes among the three farms possibly because of the geographical location of the three farms (Dakahlia Governorate). Moreover, strains with the same genotype had diverse antibiotic susceptibility. For example, the strains (n = 7) with a similar ERIC A type had different antibiotic resistance patterns, antibiotic resistance genes, and virulence genes.
Conclusion
For the first time, this work investigated the prevalence of virulent and multi-antibiotic-resistant L. monocytogenes strains in dairy cattle farms in Egypt. Genotyping analysis through ERIC-PCR revealed that all L. monocytogenes strains were diverse and heterogenous, as they were categorized into specific genotypes in terms of sampling sites and sample type. Our study confirms the importance of hygienic practices with respect to silage production and milking hygiene should be developed and implemented to prevent not only the introduction and spread of L. monocytogenes into herds but also its entry in milk. These findings elucidated the epidemiology of L. monocytogenes in dairy cattle farms and served as a basis for implementing control strategies to reduce the risks of L. monocytogenes dissemination in dairy cattle and the environment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This paper is based upon work supported by Science, Technology & Innovation Funding Authority (STDF) under research support grant (ID:30196). The authors acknowledge the Ministry of Higher Education and Mansoura University. Also, we would like to thank all the members of the animal farms especially the farm workers (livestock contact) for helping us in sample collection.
Author contribution
Mona Elsayed conceived the study and was involved in the design and coordination of the study. Mona Elsayed and Rasha Elkenany were involved in practical part, data analysis, manuscript drafting, and editing. Basma Badawy and Amira Zakaria were involved in the practical part. All authors read and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work was financially supported by Science, Technology & Innovation Funding Authority (STDF). Grant type is research support: project ID (30196).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
This protocol was performed by following the animal ethics guidelines and approved by Medical Research Ethics Committee of Mansoura University with code number (R/75).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agersborg A, Dahl R, Martinez I. Sample preparation and DNA extraction procedures for polymerase chain reaction identification of Listeria monocytogenes in sea foods. Int J Food Microbiol. 1997;35:275–280. doi: 10.1016/s0168-1605(97)01245-2. [DOI] [PubMed] [Google Scholar]
- American Public Health Association . Standard methods for examination of waters and wastewater. 14. Washington: D. C; 1971. [Google Scholar]
- Bandelj P, Ciglenecki UJ, Ocepek M, Blagus R, Vengust M (2018) Risk factors associated with fecal shedding of Listeria monocytogenes by dairy cows and calves. Vet Intern Med 1–7. 10.1111/jvim.15234 [DOI] [PMC free article] [PubMed]
- Baquero F, Lanza F, V, Duval M, Coque TM, Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol Microbiol. 2020;113(3):570–579. doi: 10.1111/mmi.14454. [DOI] [PubMed] [Google Scholar]
- Bianchi DM, Barbaro A, Gallina S, Vitale N, Chiavacci L, Caramelli M, Decastelli L. Monitoring of foodborne pathogenic bacteria in vending machine raw milk in Piedmont, Italy. Food Control. 2013;32:435–439. doi: 10.1016/j.foodcont.2013.01.004. [DOI] [Google Scholar]
- Bilung LM, Chai LS, Tahar AS, Ted CK, Apun K (2018) Prevalence, genetic heterogeneity, and antibiotic resistance profile of Listeria spp. and Listeria monocytogenes at farm level: a highlight of ERIC- and BOX-PCR to reveal genetic diversity. Biomed Res Int 12. 10.1155/2018/3067494 [DOI] [PMC free article] [PubMed]
- Bohnert M, Dilasser F, Dalet C, Mengaud J, Cossart P. Use of specific oligonucleotides for direct enumeration of Listeria monocytogenes in food samples by colony hybridization and rapid detection by PCR. Res Microbiol. 1992;143:271–280. doi: 10.1016/0923-2508(92)90019-k. [DOI] [PubMed] [Google Scholar]
- Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–394. doi: 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- Castro H, Jaakkonen A, Hakkinen M, Korkeala H, Lindström M. Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three Finnish dairy cattle farms: a longitudinal study. Appl Environ Microbiol. 2018;84:e02000–e2017. doi: 10.1128/aem.02000-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother. 1999;43(9):2103–2108. doi: 10.1128/AAC.43.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JTH, Gall AR, Johnson AK, Huynh TN. Characterization of Listeria monocytogenes isolates from lactating dairy cows in a Wisconsin farm: antibiotic resistance, mammalian cell infection, and effects on the fecal microbiota. J Dairy Sci. 2021;104(4):4561–4574. doi: 10.3168/jds.2020-18885. [DOI] [PubMed] [Google Scholar]
- Clegg FG, Chiejina SN, Duncan AL, Kay RN, Wray C (1983) Outbreak of Salmonella Newport infection in dairy herds and their relationship to management and contamination of the environment. Vet Rec 112. 10.1136/vr.112.25.580 [DOI] [PubMed]
- Clinical and laboratory standards institute, CLSI (2020) Performance standards for antimicrobial susceptibility testing; Thirty Informational Supplement, 950 West Valley Road, Suite 1400 Wayne, PA 19087 USA.
- Constable PD, Hinchcliff KW, Done SH, Gruenberg W (2016) Mastitis. Chapter 20. in: Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats. Elsevier Health Sciences, Amsterdam, the Netherlands, pp. 2113–2208.
- Dalzini E, Bernini V, Bertasi B, Daminelli P, Losio MN, Varisco G. Survey of prevalence and seasonal variability of Listeria monocytogenes in raw cow milk from northern Italy. Food Control. 2016;60:466–470. doi: 10.1016/j.foodcont.2015.08.019. [DOI] [Google Scholar]
- Elbar S, Elkenany R, Elhadidy M, Younis G (2020) Prevalence, virulence and antibiotic susceptibility of Listeria monocytogenes isolated from sheep. Mansoura Veterinary Med J 21(2): 48–52. 10.35943/mvmj.2020.21.2.207
- Escolar C, Gómez D, del Carmen Rota García M, Conchello P, Herrera A. Antimicrobial resistance profiles of Listeria monocytogenes and Listeria innocua isolated from ready-to-eat products of animal origin in Spain. Foodborne pathogens and disease. 2017;14(6):357–363. doi: 10.1089/fpd.2016.2248. [DOI] [PubMed] [Google Scholar]
- Germini A, Masola A, Carnevali P, Marchelli R. Simultaneous detection of Escherichia coli O175:H7, Salmonella spp., and Listeria monocytogenes by multiplex PCR. Food Control. 2009;20:733–738. doi: 10.1016/j.foodcont.2008.09.010. [DOI] [Google Scholar]
- Godreuil S, Galimand M, Gerbaud G, Jacquet C, Courvalin P. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob Agents Chemother. 2003;47:704–708. doi: 10.1128/AAC.47.2.704-708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao TT, Courvalin P, Brisabois A. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl Environ Microbiol. 2011;77(8):2788–2790. doi: 10.1128/AEM.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Wang P, Ma Y, Yang Y, Ye X, Wang M. Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J Antimicrob Chemother. 2012;67(10):2361–2366. doi: 10.1093/jac/dks244. [DOI] [PubMed] [Google Scholar]
- Hassan L, Mohammed HO, McDonough PL. Farm-management and milking practices associated with the presence of Listeria monocytogenes in New York state dairy herds. Prev Vet Med. 2001;51:63–73. doi: 10.1016/S0167-5877(01)00207-0. [DOI] [PubMed] [Google Scholar]
- Ho AJ, Ivanek R, Gröhn YT, Nightingale KK, Wiedmann M. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev Vet Med. 2007;80:287–305. doi: 10.1016/j.prevetmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci. 2016;1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwu CD, Okoh AI. Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples. PLoS ONE. 2020;15(2):e0228956. doi: 10.1371/journal.pone.0228956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jersek B, Gilot P, Gubina M, Klun N, Mehle J, Tcherneva E, Rijpens N, Herman L. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J Clin Microbiol. 1999;37(1):103–109. doi: 10.1128/JCM.37.1.103-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayode A, Semerjian L, Osaili T, Olapade O, Okoh A. Occurrence of multidrug-resistant Listeria monocytogenes in environmental waters: a menace of environmental and public health concern. Front Environ Sci. 2021;12:373. doi: 10.3389/fenvs.2021.737435. [DOI] [Google Scholar]
- Latorre AA, et al. Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with Listeria monocytogenes. J Dairy Sci. 2010;93:2792–2802. doi: 10.3168/jds.2009-2717. [DOI] [PubMed] [Google Scholar]
- Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- Leclercq R, Huet C, Picherot M, Trieu-Cuot P, Poyart C. Genetic basis of antibiotic resistance in clinical isolates of Streptococcus gallolyticus (Streptococcus bovis) Antimicrob Agents Chemother. 2005;49(4):1646–1648. doi: 10.1128/aac.49.4.1646-1648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lawrence ML, Austin FW, Ainsworth AJ. A multiplex PCR for species-and virulence-specific determination of Listeria monocytogenes. J Microbiol Methods. 2007;71:133–140. doi: 10.1016/j.mimet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Livermore DM. Are all beta-lactams created equal? Scandinavian journal of infectious diseases. Supplementum. 1996;101:33–43. doi: 10.1016/j.mimet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Lópe M, Tenorio C, Del Campo R, Zarazaga M, Torres C (2011) Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enterococci of different origins. J Chemother 23(2):87–91. 10.1179/joc.2011.23.2.87 [DOI] [PubMed]
- Mohammed AN, Abdel Aziz SA. Ecological study on Listeria monocytogenes and the extent of its resistance to different disinfectants in dairy farm for improving animal health. Asian Journal of Animal and Veterinary Advances. 2017;12:302–310. doi: 10.3923/ajava.2017.302.310. [DOI] [Google Scholar]
- Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, Courvalin P, Le Monnier A. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother. 2010;54:2728–2731. doi: 10.1128/AAC.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani T, Kobayashi T, Matsumoto T. Emergence and prevalence of β-lactamase-producing Klebsiella pneumoniae resistant to cephems in Japan. Int J Antimicrob Agents. 2006;27(6):491–499. doi: 10.1016/j.ijantimicag.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Nightingale KK, Fortes ED, Ho AJ, Schukken YH, Gröhn YT, Wiedmann M. Evaluation of farm management practices as risk factors for clinical listeriosis and fecal shedding of Listeria monocytogenes in ruminants. J Am Vet Med Assoc. 2005;227:1808–1814. doi: 10.2460/javma.2005.227.1808. [DOI] [PubMed] [Google Scholar]
- Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl Environ Microbiol. 2004;70(8):4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojdana D, Sacha P, Wieczorek P, Czaban S, Michalska A, Jaworowska J (2014) The occurrence of blaCTX-M, blaSHV, and blaTEM genes in extended-spectrum β-lactamase-positive strains of Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis in Poland. Intern J Antibi 7. 10.1155/2014/935842
- Pantoja JCF, Rodrigues ACO, Hulland C, Reinemann DJ, Ruegg PL. Investigating contamination of bulk tank milk with Listeria monocytogenes on a dairy farm. Food Prot Trends. 2012;32(5):512–521. [Google Scholar]
- Papić B, Pate M, Félix B, Kušar D. Genetic diversity of Listeria monocytogenes strains in ruminant abortion and rhombencephalitis cases in comparison with the natural environment. BMC Microbiol. 2019;19:299. doi: 10.1186/s12866-019-1676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsaie Mehr V, Shokoohizadeh L, Mirzaee M, Savari M (2017) Molecular typing of Klebsiella pneumoniae isolates by enterobacterial repetitive intergenic consensus (ERIC)–PCR. Infect Epidemiol Microbiol 3(4): 112–116. 10.18869/modares.iem.3.4.112
- Poimenidou SV, Dalmasso M, Papadimitriou K, Fox EM, Skandamis PN, Jordan K. Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front Microbiol. 2018;9:1103. doi: 10.3389/fmicb.2018.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Chan Y, Wiedmann M. Definition of genetically distinct attenuation mechanisms in naturally virulence attenuated Listeria monocytogenes by comparative cell culture and molecular characterization. Appl Environ Microbiol. 2005;71:3900–3910. doi: 10.1128/aem.71.7.3900-3910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Wiedmann M. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell Mol Life Sci. 2003;60:1–15. doi: 10.1007/s00018-003-2225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser ET, Marth EM. Listeria, listeriosis, and food safety. 3. Boca Raton: Taylor & Francis Group; 2007. [Google Scholar]
- Scha OW, Carrol E, Jain NC. Bovine mastitis. 1. Philadelphia: Lea and Febiger; 1971. [Google Scholar]
- Shamloo E, Hosseini H, Abdi Moghadam Z, Halberg Larsen M, Haslberger A, Alebouyeh M. Importance of Listeria monocytogenes in food safety: a review of its prevalence, detection, and antibiotic resistance. Iran J Vet Res. 2019;20(4):241–254. [PMC free article] [PubMed] [Google Scholar]
- Singh S, Yadav A, Singh S, Bharti P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res Int. 2010;43(8):2027–2030. doi: 10.1016/j.foodres.2010.06.001. [DOI] [Google Scholar]
- Sohrabi R, Rashedi M, Dorcheh MP, Shahbazi AM. Prevalence of Listeria species in raw milk in Esfahan Province. Iran Afr J Microbiol Res. 2013;7:2057–2060. doi: 10.5897/AJMR12.1463. [DOI] [Google Scholar]
- Soni DK, Singh M, Singh DV, Dubey SK. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014;14:241. doi: 10.1186/s12866-014-0241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Zhang S, Shi W, Yang X, Li Y, Pan H, Kuang D, Xu X, Shi X, Meng J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control. 2016;70:96–102. doi: 10.1016/j.foodcont.2016.04.020. [DOI] [Google Scholar]
- Swetha CS, Porteen K, Elango A, Ronald BSM, Kumar TS, Milton AP, Sureshkannan S (2021) Genetic diversity, virulence and distribution of antimicrobial resistance among Listeria monocytogenes isolated from milk, beef, and bovine farm environment. Iran J Vet Res 22(1):1–8. 10.22099/ijvr.2020.37618.5472 [DOI] [PMC free article] [PubMed]
- Tahoun AB, Abou Elez RM, Abdelfatah EN, Elsohaby I, El-Gedawy AA, Elmoslemany AM. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. Journal of Global Antimicrobial Resistance. 2017;10:264–270. doi: 10.1016/j.jgar.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Terentjeva M, Šteingolde Ž, Meister I, Elferts D, Avsejenko J, Streikiša M, Gradovska S, Alksne L, Kibilds J, Bērziņš A. Prevalence, genetic diversity and factors associated with distribution of Listeria monocytogenes and other Listeria spp in cattle farms in Latvia. Pathogens. 2021;10(7):851. doi: 10.3390/pathogens10070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel JS, Karns JS, Gorski L, McCluskey BJ, Perdue ML. Prevalence of Salmonellae, Listeria monocytogenes, and fecal coliforms in bulk tank milk on US dairies. J Dairy Sci. 2004;87:2822–2830. doi: 10.3168/jds.s0022-0302(04)73410-4. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongkamjan K, Andrea MS, Henk C, Bakker D, Esther DF, Martin W. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl Environ Microbiol. 2012;78(24):8666–8675. doi: 10.1128/AEM.01859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Neogi SB, Mo Z, Guan W, Shen Z, Zhang S, Li L, Yamasaki S, Shi L, Zhong N. Prevalence and characterization of antimicrobial resistance of foodborne Listeria monocytogenes isolates in Hebei province of Northern China, 2005–2007. Int J Food Microbiol. 2010;144:310–316. doi: 10.1016/j.ijfoodmicro.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Wang RF, Cao WW, Johnson MG. 16S rRNA-based probes and polymerase chain reaction method to detect Listeria monocytogenes cells added to foods. Appl Environ Microbiol. 1992;58:2827–2831. doi: 10.1128/aem.58.9.2827-2831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitoun H, Kassem M, Raafat D, AbouShlieb H, Fanaki N. Microbiological testing of pharmaceuticals and cosmetics in Egypt. BMC Microbiol. 2015;15(1):1–13. doi: 10.1186/s12866-015-0609-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.