Abstract
Objective:
Autism spectrum disorder (ASD) is heritable and younger siblings of ASD probands are at higher likelihood of developing ASD themselves. Prospective magnetic resonance imaging (MRI) studies of siblings report atypical brain development precedes ASD diagnosis, though the link between brain maturation and genetic factors is unclear. Given that familial recurrence of ASD is predicted by higher levels of ASD traits in the proband, we investigated associations between proband ASD traits and brain development among younger siblings.
Methods:
In a sample of n = 384 proband-sibling pairs (89 pairs concordant for ASD), we examined associations between proband ASD traits and sibling brain development at 6, 12, and 24 months in key MRI phenotypes: total cerebral volume and cortical surface area, extra-axial cerebrospinal fluid, occipital cortical surface area, and splenium white matter microstructure. Results from primary analyses led us to implement a data-driven approach using functional connectivity MRI at 6 months.
Results:
Greater levels of proband ASD traits was associated with larger total cerebral volume and surface area, and larger surface area and reduced white matter integrity in components of the visual system in siblings who developed ASD. This aligned with weaker functional connectivity between several networks and the visual system among all siblings during infancy.
Conclusions:
Findings provide evidence that specific early brain MRI phenotypes of ASD reflect quantitative variation familial ASD traits. Multimodal anatomical and functional convergence on cortical regions, fiber pathways, and functional networks involved in visual processing suggest inherited liability has a role in shaping the prodromal development of visual circuitry in ASD.
Introduction
Autism spectrum disorder (ASD) is a highly heritable (1) neurodevelopmental disorder diagnosed in 1 in 54 children in the United States (2). Younger siblings of children with ASD have a higher likelihood of developing ASD, where 1 in 5 siblings followed from infancy will receive a diagnosis of ASD by three years of age (3). Prospective magnetic resonance imaging (MRI) and behavioral studies of infant siblings have revealed that brain changes in ASD precede the onset of core diagnostic features and are temporally associated with behavioral changes that emerge in the latter part of the first and second years of life (4). Aberrant white matter integrity (5), altered morphology of the corpus callosum (6) and increased extra-axial cerebrospinal fluid (CSF) volumes (7, 8) are detectable as early as 6 months of age in infants who go on to develop ASD. Cortical surface area hyper-expansion from 6 to 12 months, particularly in regions in the occipital cortex, precedes and is correlated with brain volume overgrowth from 12 to 24 months (9), coinciding with the emergence and consolidation of ASD symptoms in the second year. Brain features, and in particular regional surface area, measured from MRIs taken in the first year of life predicted the later diagnosis of ASD in toddlerhood (9). Less is known about functional brain development in early autism, and no functional connectivity MRI (fcMRI) studies to date have demonstrated group differences in connectivity patterns between infants who develop ASD and infants who do not. However, a subset of behavior-related 6-month region-to-region connections has been shown to accurately predict diagnostic outcome at 24 months of age in infant siblings (10). To date, brain imaging markers from the first year of life remain some of the strongest predictors of later diagnosis among infants (11).
While it is evident that brain development is atypical in infants who are later diagnosed with ASD, the link between brain maturation and inherited genetic factors is unclear. Common polygenic variation accounts for the majority of genetic liability for autism (12), especially in multiplex families where more than one child has an ASD diagnosis (13), though the current predictive utility of molecular genetic markers of polygenic liability for ASD is limited (14). In the context of the infant sibling study design, family traits may serve as useful, cost-effective early markers of inherited genetic liability for autism. Autistic traits aggregate in families and are heritable(15–19), with numerous studies reporting higher likelihood for recurrence (3, 20) and greater levels of ASD traits (21, 22) in multiplex families relative to single-incidence (simplex) families. In line with this work, we previously reported that elevated levels of autistic traits in older ASD siblings (probands) increase the likelihood of an ASD diagnosis in younger siblings (23). Linking family traits to individual variation in early brain imaging markers of autism in infants has important implications for both etiology and prediction. If family traits account for significant variation in infant brain development, this would not only identify which brain phenotypes track with familial liability for ASD and warrant further molecular genetic dissection and mechanistic study, but it could also yield insight into individualized areas of concern relevant to early intervention. This type of family-design approach has already exhibited great promise for predicting clinical severity in rare genetic disorders affecting brain development (24, 25).
To address the gap in our understanding of how inherited liability for ASD impacts infant brain development, we tested whether autistic traits in ASD probands explain variation in ASD-associated brain phenotypes in their younger siblings during the period of time preceding and coinciding with the onset of symptoms. First, we examined whether proband autistic traits were associated with brain phenotypes in their younger siblings which have been shown to differ in infants later diagnosed with ASD with independent lab/cohort replication: cerebral volume (7, 9), cortical surface area (9, 26), and extra-axial CSF (7, 8). We hypothesized that higher levels of ASD traits in probands – indicative of increased genetic liability for ASD (23) – would be associated with larger brain volume, cortical surface area, and extra-axial CSF volumes in siblings. Next, we took a targeted approach to study occipital cortical surface area and splenium white matter microstructure based on evidence that (i) occipital cortical regions have been shown to exhibit hyper-expansion during infancy in infants who develop ASD (9) and (ii) splenium microstructure at 6 months of age predicts autism diagnosis at 24 months (27), and has been implicated in the development of visual orienting, a behavior that is aberrant as early as 6 months of age in infants who later develop ASD (28). Given that these global and regional brain phenotypes have been shown to differ in ASD and controls, we were also interested in whether proband traits may have differential associations with brain development in siblings later diagnosed with ASD versus those without a diagnosis. Finally, results from analyses of occipital surface area and splenium microstructure led us to investigate associations between proband ASD traits and functional connectivity during infancy. Findings reported herein provide evidence that specific early brain phenotypes of ASD reflect quantitative variation in familial ASD traits, revealing insights into the developmental nature of gene-brain associations in autism during the presymptomatic period leading up to diagnosis.
Materials and Methods
Participants
Younger siblings of ASD probands were recruited and enrolled as part of the Infant Brain Imaging Study (IBIS). Proband ASD diagnosis was verified by medical records and the Autism Diagnostic Interview Revised (ADI-R) (29). All siblings met study inclusion criteria (see online supplement). A total of 384 proband-sibling pairs were included (Table S1), 89 (23%) of whom were concordant for a diagnosis of ASD, meaning the younger sibling received a diagnosis of ASD at 24 months of age based on DSM-IV-TR criteria; this rate of diagnosis among younger siblings is similar to previously published reports(3). ASD probands were phenotyped using a behavioral battery, and siblings were phenotyped with a behavioral battery and neuroimaging at multiple timepoints during infancy and toddlerhood. Parents provided written informed consent prior to participating in this study, and procedures were approved by the Institutional Review Boards at each data collection site: University of North Carolina at Chapel Hill, University of Washington, Children’s Hospital of Philadelphia, and Washington University. The LORIS data management platform (30) served as the behavioral, clinical, and imaging hub for this study for data collection, curation, preparation for analysis, and archiving.
Proband Phenotyping
Probands were phenotyped with a battery (23) including the Social Communication Questionnaire (31) (SCQ; n = 345, available on 90% of the sample) and ADI-R (n = 371, 97%). A subset of probands (n = 329, 86%) were phenotyped with the Vineland Adaptive Behavior Scales (VABS) (32). Higher scores on the SCQ and ADI-R are indicative of the endorsement of more ASD symptoms and behaviors; lower scores on the VABS are indicative of lower adaptive functioning. The primary measure of proband ASD traits used in this study is the total score from the SCQ, which was selected for two main reasons. First, in our prior work, we demonstrated that SCQ total scores in probands were predictive of recurrence in younger siblings(23) suggesting it captures some aspect of shared genetic liability among sibling pairs. This interpretation of the SCQ as a marker of genetic liability for autism is supported by data from twins and molecular genetic investigations. Frazier and colleagues(17) report that the heritability of SCQ scores above 21 or higher is 99%, with heritability gradually increasing for SCQ scores above 12; in our sample, 97% of probands scored ≥ 12 and 55% scored ≥ 21. A genome-wide QTL study linked the SCQ total score and domain scores to loci on five chromosomes(18), and there is evidence of SNP-based heritability for the SCQ(19). Second, the SCQ is a short screening questionnaire that can be completed by a caregiver in a matter of minutes and scored easily by a clinician, as opposed to the ADI-R which is an clinician-administered interview with a caregiver that typically takes more than an hour to complete; thinking forward to application, the SCQ would be easier to administer while screening families at higher likelihood for autism recurrence. As described below, we also test for associations between proband ADI-R scores and sibling brain phenotypes to ensure our results are not restricted to the SCQ, but generalize to another measure of ASD traits.
Sibling Image Acquisition and Processing
Image acquisition and analysis procedures have been described previously (8, 9, 33) and are detailed in the online supplement. Siblings underwent MRI scans at 6, 12, and 24 months of age using 3T Siemens Tim Trio scanners with 12-channel head coils. The scanning protocol included a high-resolution T1- and T2-weighted scan (1mm3 resolution) and a diffusion weighted imaging sequence with 25 gradient directions. BOLD functional sequences were collected on a subset of the sample, as this modality was added later in the course of the study. All scans were collected during natural sleep with the exception of three ASD children who were scanned under sedation at the request of their parents at 24 months; no functional connectivity data in the present study was collected under sedation. MRI sample sizes by age are shown Table S1.
Brain volumes were obtained using a pediatric-specific, atlas-based multi-modal pipeline for probabilistic tissue classification including co-registration of multi-modal (T1/T2) MRI, bias correction, brain masking, noise reduction, and multivariate classification (9). Total cerebral volume (reported in mm3) is defined as the summation of gray and white matter volumes of the cerebrum, including a portion of the midbrain/brainstem.
Cortical surface area measurements for 12 and 24-month images were obtained via a CIVET workflow adapted for pediatric images and using an age-corrected automated anatomical labeling (AAL) atlas(34, 35). For 6-month datasets, measurements were extracted from surfaces propagated via deformable multi-modal, within-subject co-registration(35) of MRI datasets from the 12-month scan. We utilized total surface area (reported in mm2) and surface area measurements in targeted regions of interest. These regions included occipital regions shown in Hazlett et al.(9) to be hyper-expanding in infants who develop ASD (bilateral cuneus, right lingual gyrus, and bilateral middle occipital gyrus). To speak to specificity, we included the right middle frontal gyrus and left inferior temporal gyrus, which were also found to hyper-expand in ASD(9), as well as two additional bilateral control regions (precentral gyrus, supramarginal gyrus) selected to not overlap with any regions shown to have differential development in infants who develop ASD versus those who do not, or any regions shown to contribute to a prediction of ASD (see Figures 2 and 3 from Hazlett et al.,(9)).
Figure 2. Greater levels of proband autism traits are associated with larger surface area in the occipital and frontal cortices in infant siblings who develop autism spectrum disorder.
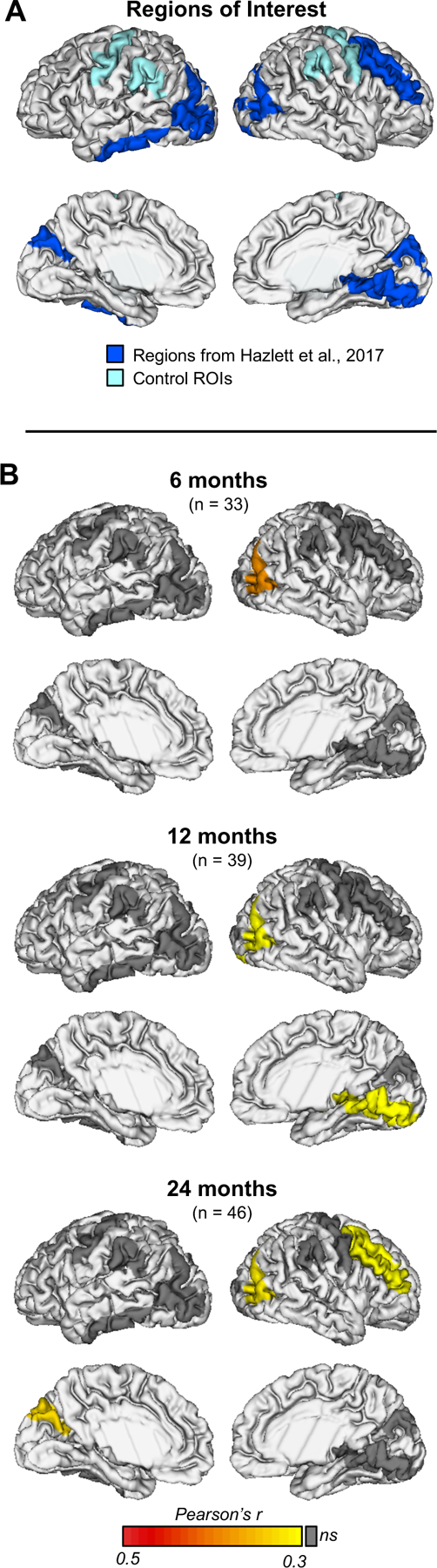
(A) Primary regions of interest included occipital cortical regions shown to exhibit hyper-expansion in autism spectrum disorder (ASD). To speak to specificity, we included the right middle frontal gyrus and left inferior temporal gyrus, which were also found to hyper-expand in ASD. Additionally, two control regions were selected to not overlap with any regions shown to have differential development in ASD during infancy or to contribute to prediction of ASD in Hazlett et al., 2017. Hyper-expanding regions are shown in royal blue, control regions are shown in light blue. (B) Proband SCQ score was positively correlated with surface area in the right middle occipital gyrus at 6 months (r = 0.44, P = 0.010), 12 months (r = 0.38, P = 0.017), and 24 months (r = 0.39, P = 0.007) in the ASD group. This aligned with longitudinal model results indicating that higher proband SCQ score was significantly associated with greater surface area in the right middle occipital cortex in the ASD group during this developmental window (β= 26.82, 95% CI 11.15 to 42.5, df = 267, P < 0.001). Additional correlations were found between proband Social Communication Questionnaire (SCQ) scores and sibling surface area in the right lingual gyrus at 12 months (r = 0.38, P = 0.017), and the left cuneus (r = 0.40, P = 0.006) and right middle frontal gyrus (r = 0.38, P = 0.010) at 24 months. No significant correlations were found between proband SCQ scores and control regions at any age.
Figure 3. Proband autism traits have an age-specific association with splenium microstructure in ASD siblings.
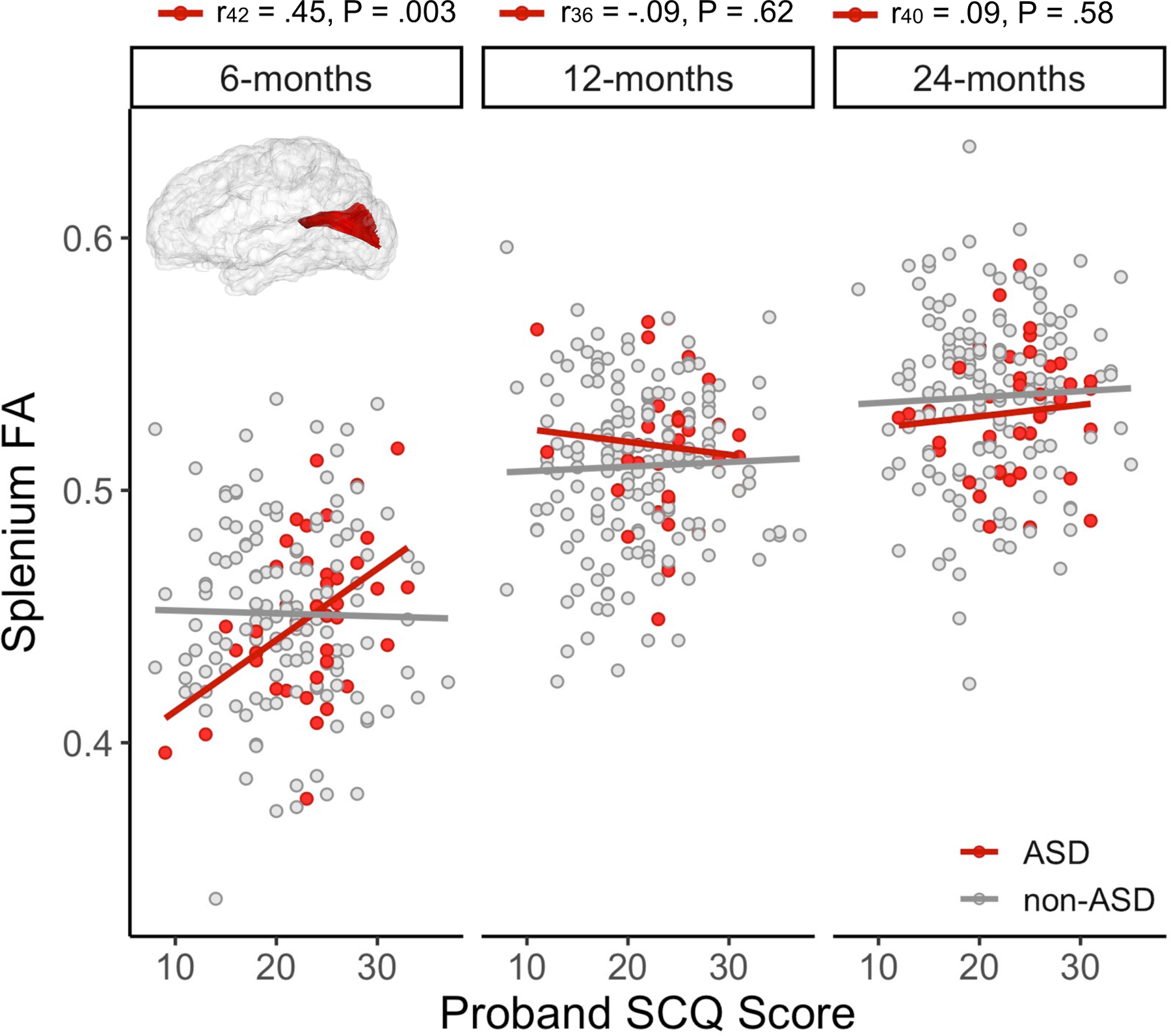
The reconstructed splenium tract in atlas space is inset into scatterplots depicting bi-variate associations between proband autism traits (Social Communication Questionnaire (SCQ) scores) and sibling splenium fractional anisotropy (FA) at 6, 12, and 24 months. Pearson’s correlation coefficients and associated sample size and p-value are reported above the label for each time point for the ASD group (red). Higher autism trait levels (indicated by higher SCQ scores) in probands was associated with increased splenium fractional anisotropy (FA) at 6 months, but not after. Proband SCQ scores were not correlated splenium FA in the non-ASD group (gray); see Table S6.
Extra-axial CSF volumes (reported in mm3) were calculated via an automated multi-modal processing stream involving distortion correction, mutual registration, transformation to stereotactic space, and CSF/brain tissue segmentation (8). Diffusion weighted images (DWIs) were pre-processed for appropriate quality using DTI prep(36). Visual quality control was performed by expert raters to remove additional images with residual artifacts. Tractography was performed using the UNC-Utah NA-MIC framework(37) using a study-specific template. White matter fractional anisotropy (FA) values were computed via quantitative tractography and averaged across each fiber tract (27, 33); tract-average values from selected fibers (splenium, and control tracts: genu, and body of the corpus callosum) were analyzed.
The functional MRI data were processed following methods described in previous publications(38, 39) using the 4dfp suite of tools (http://4dfp.readthedocs.io). Images were compensated for slice-dependent time shifts; head movement was quantified for spatial realignment both within and across runs; whole brain image intensity was normalized to a mode value of 1000(40); and images were registered into standardized 3-mm isotropic atlas space through an affine transformation. Global signal regression, nuisance signal regression, spatial and temporal bandpass filtering, and motion scrubbing were applied (41). Motion scrubbing was implemented at a frame-to-frame displacement (FD) of 0.2 mm(42); there are no differences in mean FD between the ASD and non-ASD sibling sample in this study (FD = 0.089 mm and 0.096 mm, respectively; P = 0.160). All infants included in this analysis provided a minimum of 7 minutes of scrubbed fcMRI (mean = 10.45min, SD = 2.5m). Functional connectivity values were calculated as Pearson correlations between pairs of regions’ time-series and Fisher r-to-Z transformed for analyses; regions of interest included 230 previously published regions (43–45) and four recently added cerebellar regions(46). The full set of 234 regions of interest (described in Extended Data Table 1) were sorted into functional networks by applying the Infomap community detection algorithm(47) at edge densities ranging from 0.02 to 0.10 (steps of 0.01). An automated procedure was used to generate a single consensus model of network structure, and structure-specific thresholding was used to integrate subcortical and cerebellar regions into whole-brain networks(48). Unassigned regions (n = 4) were excluded from network analyses. The network solution is shown in Figure S1. Infant network nomenclature largely reflects naming of canonical networks found in adults; some network structures, however, appear to be specific to infants (e.g. pDMN was named as such because it contains the posterior cingulate cortex, the adult DMN does not fracture into anterior and posterior components).
Additional information for each image processing pipeline can be found in the online supplement. All imaging data underwent visual quality control and only data which passed inspection were used in analyses.
Statistical Analysis
Pearson correlations were computed between proband SCQ score and each sibling brain phenotype at each time point (6, 12, and 24 months) to generate interpretable effect sizes, correcting for multiple comparisons using FDR (49). Correlations were computed separately for infants who received a diagnosis of ASD (ASD group) and those who did not (non-ASD group), given that our prior behavioral study(23) suggested that proband ASD traits may have differential predictive utility in ASD and non-ASD siblings and that the selected brain phenotypes have been shown to differentiate infants who later develop ASD from other control groups. P-values were adjusted using false discovery rate (FDR)(49) within each sibling group (ASD vs non-ASD) separately for global (cerebral volume, total surface area, extra-axial CSF volumes) and regional phenotypes (regional surface area, white matter FA in fiber tracts of interest). Brain phenotypes shown to exhibit significant correlations with proband SCQ scores (for global phenotypes q < 0.05, for regional phenotypes P < 0.05) were carried forward for further investigation using longitudinal mixed effects models for repeated measures, incorporating a subject-specific intercept and slope and modeling proband SCQ score, sibling diagnosis, age, sex, and study site as fixed effects. Interaction terms between sibling diagnosis and proband SCQ score, and sibling diagnosis and age were also modeled. In secondary analyses, potential time-varying associations (e.g., splenium FA) were modeled with a three-way interaction term (proband SCQ score x sibling diagnosis x time). Model parameters and diagnostics, and analyses including other covariates are described in the online supplement. To assess the generalizability of our results beyond a single parent report measure of ASD traits (SCQ), we also computed correlations using proband behavior derived from the Autism Diagnostic Interview-Revised (ADI-R), administered to a parent by a clinical expert(29). We tested two domain scores from the ADI-R with varying degrees of construct overlap and correlation with the proband SCQ: reciprocal social interaction (RSI; r331 = 0.56, P < 0.0001) and restricted and repetitive behaviors (r331 = 0.19, P < 0.001). While proband and sibling autism severity were not found to correlate in our prior report(23), we also included a supplemental analysis adding sibling autism symptoms measured by the Autism Diagnostic Observation Schedule(50) calibrated severity score at 24 months of age to the models to test whether our findings with proband ASD traits were driven by any potential associations between proband and sibling phenotype.
Results from a priori analyses above led us to utilize our fcMRI data on a subset of our sample with available fcMRI data (n = 58 infants, n = 13 ASD) to test for associations between proband ASD traits and sibling functional connectivity at 6 months of age. Based on our findings from the structural and diffusion analyses described above, we hypothesized that connections within and between visual networks would be associated with proband ASD trait level. If our hypotheses were correct, this would suggest multimodal anatomical and functional convergence on cortical regions, fiber pathways, and functional networks involved in visual processing. Rather than test visual circuitry directly, which could leave us unable to speak to the specificity of findings, we conducted fcMRI enrichment analyses. Enrichment is a data-driven whole-brain approach that identifies clusters of strong brain-behavior relationships within and between functional networks(38, 46). Analyses proceeded by identifying the top 5% of sibling brain-proband behavior associations, followed by the computation of enrichment p-values for every network-network pair in relation to proband behavior, and finally, a correction for family-wise error rate (P-values < .001 approximate a 5% experiment-wide false-positive rate; p-values < .01 are trend-level), improving upon our prior fcMRI publications(46). Additional details on the enrichment approach are described in the online supplement.
Enrichment analyses were performed separately with the proband SCQ, ADI-R RSI, and VABS Socialization (VABS-Soc) scores. The VABS-Soc provides a complementary measure of social behavior available for a subset of probands. We used all three proband scores as inputs to our enrichment analysis to increase the search space. Owing to the limited number of subjects with fcMRI data available at 6 months of age, enrichment analyses were conducted across the combined infant sibling sample (i.e., ASD and non-ASD groups), as in our prior published fcMRI work (38, 46). This approach aligns with findings that functional connectivity did not differentiate infants based on diagnosis in cohort of a similar size (51).
RESULTS
Proband ASD traits correlate with ASD sibling cerebral volume, cortical surface area but not extra-axial CSF
Bivariate Pearson correlations revealed significant positive associations between proband SCQ score and cerebral volume (6-, 12-, 24-months) and surface area (12 and 24-months) in the ASD group, but not in non-ASD group. Proband scores explained 12.3% (r48 = 0.35) and 16.8% (r46 = 0.41) of the variance in cerebral volume and surface area, respectively, at 24 months of age among ASD siblings (Figure 1, Table S3). As shown in Table 1, linear mixed model analyses adjusting for covariates revealed a significant proband SCQ score x sibling diagnostic group interaction for both cerebral volume (β = 6110.88, 95% CI 2282.61 to 9939.15, df = 293, P = 0.002) and cortical surface area (β = 384.43, 95% CI 119.11 to 649.74, df = 267, P = 0.005), consistent with correlation results. Plots visualizing longitudinal trajectories of total cerebral volume and total surface area as a function of proband SCQ are shown in Figure S2.
Figure 1. Proband autism traits explain significant variation in total cerebral volume and cortical surface area in infant siblings who later develop autism.
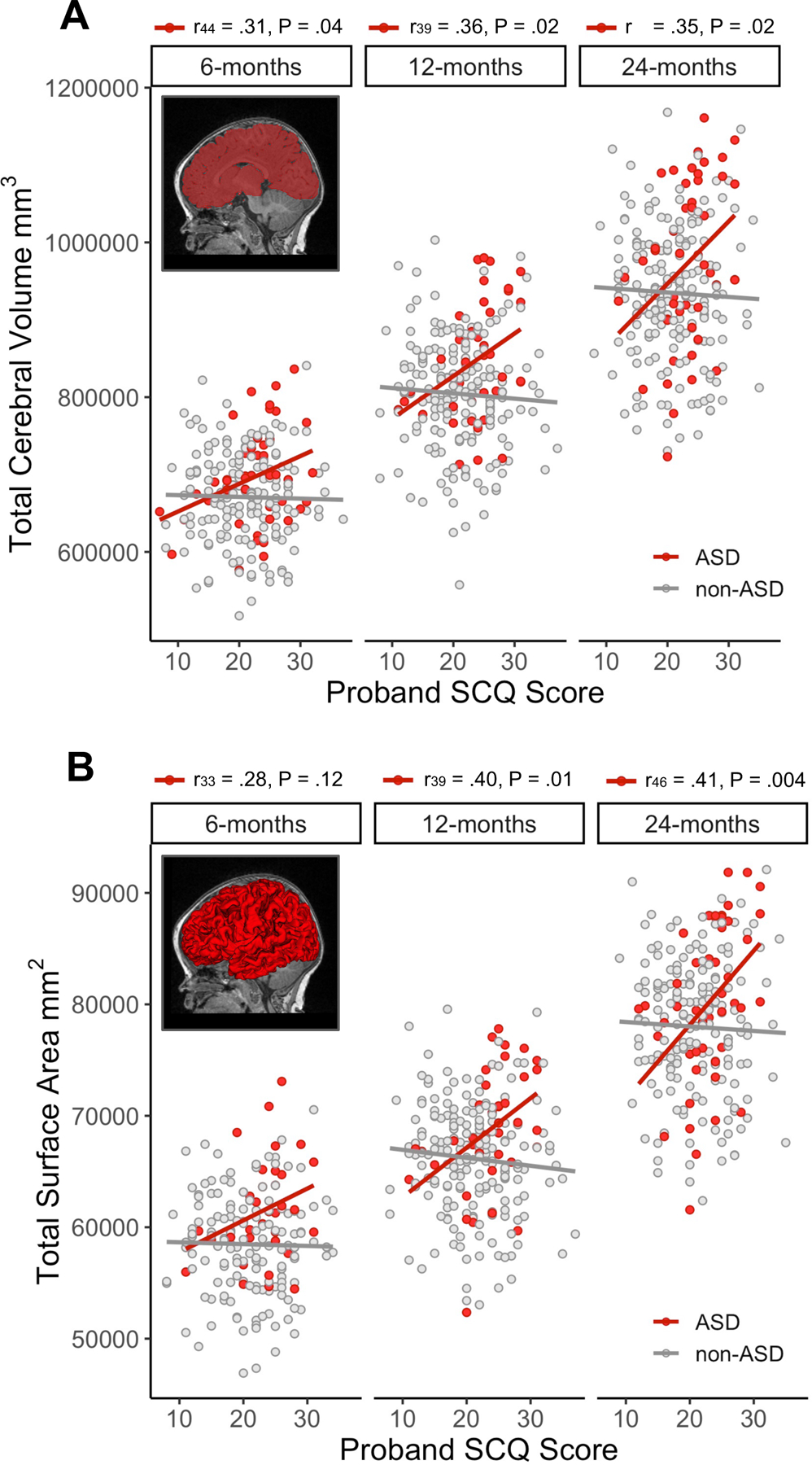
Example cerebral volumetric segmentations (A) and surface reconstructions (B) for 6-month structural MRIs are inset into scatterplots depicting bi-variate associations between proband autism traits (Social Communication Questionnaire (SCQ) scores) and each sibling brain feature at 6, 12, and 24 months. Pearson’s correlation coefficients and associated sample size and p-value are reported above the label for each time point for the autism spectrum disorder group (ASD group; red); higher levels of autism traits (indicated by higher SCQ scores) in probands was associated with increased cerebral volume and surface area. Proband SCQ scores were not correlated with measures of cerebral volume or surface in the non-ASD group (gray); see Table S3.
Table 1.
Longitudinal models predicting total cerebral volume and surface areaa
| Estimateb | 95% CI | df | P value | |
|---|---|---|---|---|
| Cerebral Volume Model (mm 3 ) | ||||
| Intercept | 764514 | (744590, 784439) | 374 | <0.001 |
| Proband SCQ | −1008 | (−2546, 528) | 293 | 0.198 |
| Sibling Sex - Male | 73631 | (57103, 90160) | 293 | <0.001 |
| Sibling Group - High-risk ASD | 4661 | (−18343, 27667) | 293 | 0.690 |
| Group x Proband SCQ | 6110 | (2282, 9939) | 293 | 0.002 |
| Group x Age MRI | 263 | (−905, 1433) | 374 | 0.657 |
| Site 2 | −15933 | (−39438, 7570) | 293 | 0.183 |
| Site 3 | −12093 | (−33470, 9283) | 293 | 0.266 |
| Site 4 | 2109 | (−20900, 25119) | 293 | 0.857 |
| Sibling Age at MRI | 6503 | (5987, 7018) | 374 | <0.001 |
| Surface Area Model (mm 2 ) | ||||
| Intercept | 56059 | (54801, 57318) | 369 | <0.001 |
| Proband SCQ | −71.82 | (−166, 22.46) | 267 | 0.135 |
| Sibling Sex - Male | 4501 | (3465, 5538) | 267 | <0.001 |
| Sibling Group - High-risk ASD | 312 | (−1068, 1692) | 267 | 0.656 |
| Group x Proband SCQ | 384 | (119, 649) | 267 | 0.005 |
| Group x Age MRI | 8.14 | (−46.18, 62.46) | 369 | 0.768 |
| Site 2 | −71.07 | (−1559, 1416) | 267 | 0.925 |
| Site 3 | −581 | (−1947, 785) | 267 | 0.403 |
| Site 4 | 15.98 | (−1442, 1474) | 267 | 0.983 |
| Sibling Age at MRI | 1056 | (1032, 1080) | 369 | <0.001 |
Linear mixed models were used to predict sibling total cerebral volume and surface area from proband Social Communication Questionnaire (SCQ) total scores, adjusting for sibling age and sex, study site, and including a SCQ x sibling diagnostic group interaction and group x sibling age interaction.
Estimates generated using linear mixed effects models. Reference groups for sex and sibling diagnostic group are female and non-ASD, respectively. Estimates for main effects reflect the change in cerebral volume in mm3 (top section) or surface area in mm2 (bottom section) per one-unit increase in the predictor variable (each row in the table) for the non-ASD group, holding other predictors constant. Interaction effects reflect the difference in the slope of the association between cerebral volume (top) or surface area (bottom) and the predictor variable (e.g., proband SCQ, Age at MRI) between the non-ASD and ASD groups, holding other predictors constant.
The effect of proband autism traits appeared specific to infant brain size and structure, as Pearson correlations revealed no significant association between proband SCQ score and sibling extra-axial CSF volumes at 6 months (ASD: r42 = −0.02, P = 0.905; non-ASD: r149 = −0.02, P = 0.821), 12 months (ASD: r39 = 0.16, P = 0.319; non-ASD: r182 = −0.07, P = 0.370), or 24 months (ASD: r42 = 0.20, P = 0.202; non-ASD: r159 = −0.10, P = 0.222).
Similarly to findings from the SCQ, proband ADI-R RSI score was significantly positively correlated with cerebral volume and cortical surface area at 12 months (volume: r42 = 0.40, P = 0.008; surface area: r42 = 0.47, P = 0.002) and 24 months (volume: r52 = 0.33, P = 0.016; surface area: r50 = 0.33, P = 0.021) in the ASD group. We found no significant correlations between sibling brain volume or surface area and proband restricted and repetitive behaviors as measured by the ADI-R in either group (Table S4). As with the SCQ, no associations were found between proband ADI-R RSI scores and sibling cerebral volume or surface area in the non-ASD group. Mixed model analyses with proband ADI-R scores aligned with correlation results and are presented in the online supplement (Table S5). Including the sibling’s autism severity measured at 24 months in models of total cerebral volume and total surface area growth (Tables S6) did not improve model fit or change the interpretation of our results; the same was observed with phenotypes tested later in the analysis (regional cortical surface area, splenium FA; Tables S7, S8). This suggests that proband severity explains significant variation in brain phenotypes in ASD siblings above and beyond what can be explained by their own ASD symptoms measured later in development at two years of age.
Proband ASD traits explain variation in ASD sibling occipital cortical surface area
Pearson correlations revealed that proband SCQ score explained significant variation in surface area measurements for ASD siblings in the occipital and frontal cortices, but not bilateral control regions in the premotor and parietal cortices (Figure 2, Table S9).
Proband SCQ score was correlated with ASD sibling surface area in the right middle occipital gyrus at 6 months (r33 = 0.44, P = 0.010), 12 months (r39 = 0.38, P = 0.017), and 24 months (r46 = 0.39, P = 0.007), explaining 14.4% to 19.4% of the variance in regional surface area across this developmental window in ASD siblings. No significant associations were found between proband SCQ and sibling regional cortical surface area in non-ASD siblings (Figure 2, Table S9). This aligns with longitudinal mixed model results reporting a significant proband SCQ score x sibling group interaction (β= 26.82, 95% CI 11.15 to 42.5, df = 267, P < 0.001; Table S10). Plots visualizing longitudinal trajectories of right middle occipital cortical SA as a function of proband SCQ are shown in Figure S2. Bi-variate scatterplots showing associations between proband SCQ and sibling regional surface area are shown in Figure S3.
Correlations were also found between proband SCQ score and sibling surface area in the right lingual gyrus at 12 months (r39 = 0.38, P = 0.017), the left cuneus (r46 = 0.40, P = 0.006), and right middle frontal gyrus (r46 = 0.38, P = 0.010) at 24 months (Table S9, Figure 2); however, mixed models revealed no significant association between proband SCQ score and the developmental trajectories of these three cortical regions after correction for multiple comparisons (Table S10).
Proband ASD traits have an age-specific association with ASD sibling splenium FA
Pearson correlations revealed that proband SCQ score explained 20.3% of the variance in FA in the splenium in ASD infants at 6 months (r42 = 0.45, P = 0.003; Figure 3), but not at 12 or 24 months. The time varying nature of these results was explored using a three-way interaction term in the linear mixed model (β= −0.14×10−3, 95% CI −0.29×10−3 to 0.10×10−4, df = 295, P = 0.062; Table S11). FA values in the genu and body of the corpus callosum were not significantly correlated with proband ASD trait level in either group (Table S9).
Proband ASD traits are associated with sibling functional connectivity at 6 months
Out of a total of 196 possible network pairs per experiment, enrichment analyses identified four 6-month network pairs that were associated with proband trait level on the SCQ, ADI-R RSI, and/or VABS-Soc (Figure 4; Table S12) at a level approaching experiment-wide significance (P-values < 0.01). Consistent with our structural and diffusion findings, the visual (Vis) and medial visual (mVis) networks were well represented among enriched network pairs (3 of 4). The posterior default mode network (pDMN) was similarly well-represented (3 of 4).
Figure 4. Proband autism traits are associated with functional connectivity in networks including visual cortical regions among siblings at 6 months of age.
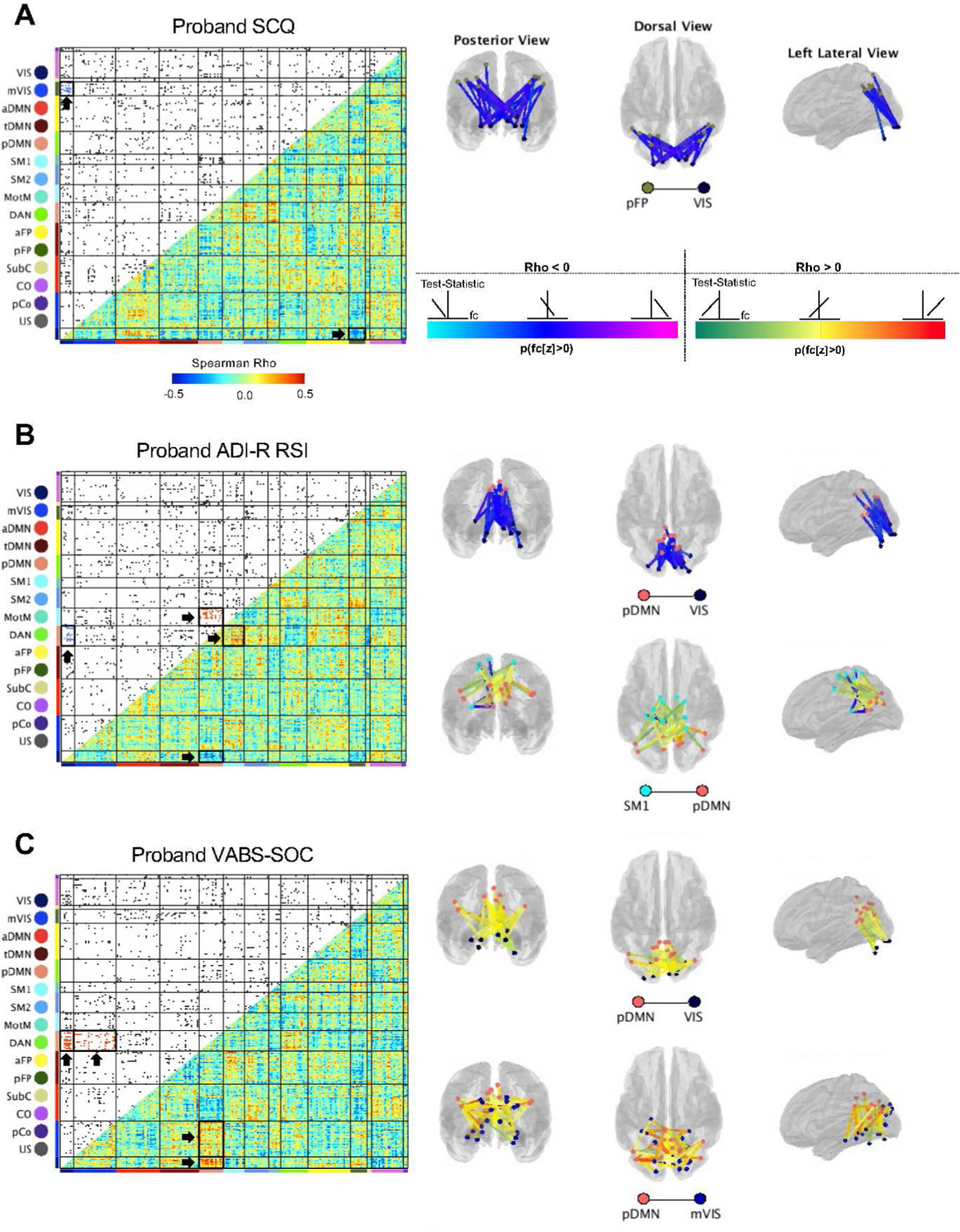
Functional connectivity enrichment analyses identified four 6-month network pairs that were associated with proband trait level as measured by the Social Communication Questionnaire (SCQ) score (a), Autism Diagnostic Interview-Revised Reciprocal Social Interaction (ADI-R RSI) score (b), and/or the Vineland Adaptive Behavior Scales Socialization (VABS-Soc) score (c). Lower triangles of each matrix depict heatmaps of Spearman Rho (ρ) correlations between functional connectivity (fc) values and proband behavioral scores; upper triangles are thresholded to display the strongest 5% of sibling brain-proband behavior correlations, with region-to-region connections visualized as dots and colored within enriched network pairs by the direction of effect (blue = negative, red = positive). Network pairs approaching experiment-wide significance (p-values < .001 maintain 5% experiment-wide false-positive rate; p-values < 0.01 are considered trending) are surrounded by bold black boxes and indicated by arrows in the correlation matrices and are depicted in brain space. The color of the lines connecting pairs of regions of interest reflects the proportion of individual functional connectivity values that are above zero (a). Light blue to dark blue to magenta colors represent negative brain-behavior correlations and green to yellow to red colors denote positive brain-behavior correlations. Light blue and green denote that the region-to-region pair contains predominantly negative functional connectivity values; magenta and red reflect region-to-region pairs with predominantly positive functional connectivity. Blue and yellow reflect functional connectivity values that are distributed across zero. High proband scores on the SCQ and ADI-R RSI are indicative of greater levels of autism traits; lower scores on the VABS-Soc are indicative of poorer social functioning. Stronger correlations between pDMN-Vis, pDMN-mVis and pFP-Vis were associated with lower proband trait levels, while stronger correlations between pDMN-SM1 were associated with increased autism traits in probands.
In relation to the SCQ, a cluster of sibling brain and proband behavior associations was observed for functional connections between Vis and pFP (posterior frontoparietal network; P = 0.0077). In relation to the ADI-R RSI, clusters of brain-behavior associations were observed for functional connections between pDMN and Vis (P = 0.0097) and pDMN and SM1 (somatomotor network 1; P = 0.0031). Finally, in relation to the VABS-Soc, clusters of brain-behavior associations were observed for functional connections between pDMN and Vis (P = 0.0012) and pDMN and mVis (P = 0.0094). Increased levels of ASD traits (higher SCQ, higher ADI-R RSI, lower VABS-Soc) in probands were associated with weaker functional connectivity correlation values between pDMN-Vis (ADI-R RSI, Figure 4B, Spearman’s Rho < 0; VABS-Soc, Figure 4C, Spearman’s Rho > 0), pDMN-mVis (VABS-Soc, Figure 4C, Spearman’s Rho > 0), and Vis-pFP (SCQ, Figure 4A, Spearman’s Rho < 0). Conversely, increased levels of ASD traits in probands were associated with stronger functional connectivity between pDMN-SM1 (ADI-R RSI, Figure 4C, Spearman’s Rho > 0). Notably, enrichment of pDMN-Vis was observed across two measures of proband ASD traits, suggesting a robustness of brain-behavior associations between these networks. Within each pair of enriched networks, region-to-region functional connectivity values spanned zero (e.g., including both positive and negative connections), as depicted in brain visualizations in Figure 4.
DISCUSSION
In this report we utilized a family-study design to demonstrate that autistic traits in ASD probands, as indices of familial genetic liability for ASD, correlate with neurodevelopment in their infant siblings who were later diagnosed with ASD. Proband autism traits – and, in particular, social behavior captured by multiple instruments – explained variation in sibling cerebral volume, cortical surface area, and splenium white matter microstructure during the presymptomatic period leading up to diagnosis. Our structural and diffusion MRI findings included cortical regions and fiber pathways involved in processing visual information at 6 months of age, and were consistent with our fcMRI enrichment results, demonstrating convergence across multiple imaging modalities. Together, these findings suggest a role for heritable ASD liability in shaping the development of visual circuitry during infancy when aberrant visual behaviors in autism are evident. Results also indicate that ASD traits in older siblings may foreshadow the emergence of ASD in their younger siblings, and may be useful as markers of family-level liability.
Brain volume overgrowth is well documented in ASD and becomes apparent in the second year of life, following the hyper-expansion of cortical surface area (9). Common ASD genetic variants are predicted to regulate corticogenesis (14), and gene expression profiles in postmortem cortico-cortical projection neurons have been found to correlate with symptom severity in ASD (52). Our findings build upon this work to link familial indices of genetic liability with variations in the early postnatal development of cerebral volume and cortical surface area, suggesting that autistic traits in families may serve as markers for ASD-associated brain overgrowth in infants.
Regional analyses revealed that greater levels of autism traits in probands was associated with larger surface area in a subset of cortical areas that exhibit hyper-expansion and contribute to individual-level diagnostic prediction in infants who develop ASD (9). Distinct sets of genes are involved in the development of specific cortical regions in humans, with strong genetic correlations for surface area among occipital cortical regions surrounding major early-forming sulci (53). Canonical Wnt signaling, which has been implicated in the pathophysiology of ASD (54), modulates regional surface area with functional links to genes involved in Wnt signaling enriched in occipital cortical areas (53). This aligns with our finding that surface area in the occipital cortex is influenced by proband traits that index genetic liability for ASD, implicating mechanisms governing occipital cortical areal expansion in the pathophysiology of ASD.
Unlike brain volume and surface area, we found no associations between proband traits and sibling extra-axial CSF volumes. Prior reports indicate that extra-axial CSF volumes are increased in toddlers with ASD regardless of familial liability (55), and thus, extra-axial CSF may represent a non-specific marker of vulnerability to atypical neurodevelopment. Our family study paradigm may help to identify brain phenotypes most strongly linked to inherited polygenic variants (cortical volume, surface area) versus those which may arise through a separate pathophysiology (extra-axial CSF).
Proband ASD traits explained 20% of the variance in splenium microstructure at 6 months of age, but not at 12 or 24 months, such that greater levels of proband ASD traits was associated with an age-specific increase in FA in ASD siblings. This is consistent with evidence that infants who develop ASD have higher values of FA at 6 months of age in white matter tracts spanning the brain, followed by a period of slowed growth and ultimately lower FA at 24 months of age (5). Our results suggest that initially higher FA in ASD siblings – reflective of an over-abundance of axons, increased myelination, or both – are driven by genetic liability for autism, but that the slower maturation of white matter FA thereafter may be modified to a greater degree by experience-dependent mechanisms.
Greater levels of autism traits in probands were associated with weaker connectivity between the pDMN-Vis, pDMN-mVis, and pFP-Vis networks at 6 months of age in siblings, suggesting that greater familial genetic liability for ASD confers weaker functional connectivity between visual and DMN regions, and visual and task-control regions. Weaker connectivity between DMN and visual networks has been linked to initiating fewer bids for joint attention in our sample (38) and observed in ASD toddlers with deficits in visual-social engagement (56). Differences in the interhemispheric connectivity of the posterior cingulate cortex (a hub of the DMN) and extrastriate cortex have also recently been reported among infants at 9 months of age compared to control infants without a family history of ASD (51). These results align with our findings linking proband traits to other aspects of the visual system, including the middle occipital gyrus, which lies along the dorsal stream(57) and is involved in the processing of visual information including object recognition(58, 59), and the splenium, which is critical for interhemispheric communication between visual areas(60) and has been shown to reflect visual orienting latencies in infants(28).
Together, these results suggest that genetic liability for autism plays a role in shaping the development of neural circuitry relevant for visual processing at 6 months of age, prior to the emergence of the defining behavioral features of ASD. Eye-looking and gaze behavior (61), viewing of social scenes (62), and visual orienting (28) are aberrant during infancy and toddlerhood in ASD. Patterns of visual preference to social stimuli (e.g., eyes, mouth) during infancy are under strong genetic control (62), suggesting that genetic background may play a pivotal role in shaping an infant’s experience of the environment around them, generating a dynamic gene-environment developmental system for social learning (63). Thus, we posit that the early, atypical structure and function of visual circuitry related to a genetic predisposition for autism may initiate a developmental cascade whereby altered visual circuitry subserves atypical visually-guided behaviors, which in turn shapes visual experience and experience-dependent circuit refinement and contributes to the emergence of the defining symptoms of ASD (4).
Findings linking proband ASD traits to sibling structural and diffusion brain imaging phenotypes were specific to the ASD group and not observed in the non-ASD group. The association between proband traits and sibling brain phenotypes is indicative of a shared genetic liability among sibling pairs who develop ASD, while the lack of associations in non-ASD siblings could be explained by non-shared genetics, phenotypic heterogeneity(64), or both. These hypotheses will need to be tested through genetic investigation. It also warrants mention that the SCQ was designed to capture trait variation at the diagnostic end of the continuum, and therefore may not index characteristics that are qualitatively similar but milder than those seen in the diagnostic category of ASD which are known to aggregate in first degree relatives without a diagnosis (e.g., non-ASD siblings. Finally, using the ADI-R, we found that proband symptoms in the social domain appear to be more strongly associated with ASD sibling brain development than restricted and repetitive behaviors. This aligns with evidence that social and non-social domains of autism symptomology are both heritable(19) and genetically dissociable(65), suggesting that their underlying genetic architecture may have different impacts on the developing brain.
There are limitations of the current study. Familial autistic traits are not direct measures of the genetic architecture of ASD and likely capture some degree of environmental influences; future studies should seek to expand these findings to molecular genetic investigations (i.e., polygenic risk scores) that may have more relevance for early brain development. Limited sample sizes prevented us from testing for diagnostic group interactions with proband traits in the fcMRI enrichment analyses; we were unable to determine whether the associations between proband trait level and fcMRI were driven by the ASD group (as was the case with the structural and diffusion findings) or were similar in nature across the entire sample. Finally, the published work identifying regions in the occipital lobe(9) and the splenium(27, 28) as phenotypes of interest drew from the same cohort of infants studied in this report, and thus the specificity of findings to these brain areas could be influenced by cohort-specific effects. The ongoing collection of another infant cohort will help to address sample size concerns in future investigations and allow for studies seeking to replicate findings from this study and much of our prior work identifying brain biomarkers in emerging ASD. It will be critical to determine whether these findings generalize to other infant cohorts.
These limitations notwithstanding, the results from this study provide a proof-of-principle for utilizing heritable, familial autistic traits to identify neural signatures of ASD that may be impacted by genetic liability prior to the onset of symptoms. This sets the stage for parsing phenotypic heterogeneity and polygenicity of idiopathic autism by mapping genes to neural signatures, or endophenotypes, of ASD that may more closely reflect the underlying biology. Large-scale imaging genetics studies have demonstrated that common genetic variation is more strongly associated with brain structure than categorical neuropsychiatric diagnosis, and that brain imaging phenotypes show reduced polygenicity and increased discoverability relative to diagnostic categories (66). It follows that by focusing efforts on gene discovery using phenotypes such as visual cortical surface area during infancy, for example, we reduce the search space for possible underlying pathogenic processes, ultimately accelerating the discovery of causal mechanisms and therapeutic targets. Coupling such a methodological approach with detailed longitudinal investigation in human infants has great potential to inform our understanding of the links between polygenic variants, brain development, and behavior during a period when autistic symptoms are first unfolding. Targeting studies to the early postnatal period is likely to be critical, as growing evidence suggests that clinical symptomology after onset may be more driven by environmental and stochastic effects, blurring the line between initial genetic pathogenic mechanisms and cumulative changes in the environment, or one’s experience of their environment, that may subsequently shape clinical presentation.
Extended Data
Extended Data Table.
| Key | Talairach_X | Talairach_Y | Talairach_Z | MNI_x | MNI_y | MNI_z | 6M_Network_Name |
|---|---|---|---|---|---|---|---|
| 1 | −23.00 | −96.00 | −15.00 | −24.66 | −97.84 | −12.33 | VIS |
| 2 | 26.00 | −96.00 | −15.00 | 26.68 | −97.30 | −13.49 | VIS |
| 3 | 23.00 | 27.00 | −12.00 | 23.96 | 31.94 | −17.78 | aFP |
| 4 | −53.00 | −45.00 | −24.00 | −56.16 | −44.76 | −24.23 | DAN |
| 5 | 8.00 | 36.00 | −18.00 | 8.13 | 41.12 | −24.31 | aFP |
| 6 | −20.00 | −24.00 | −18.00 | −21.38 | −22.22 | −19.97 | mVIS |
| 7 | −35.00 | −30.00 | −24.00 | −37.26 | −28.80 | −25.58 | DAN |
| 8 | 62.00 | −27.00 | −15.00 | 64.60 | −24.41 | −18.57 | tDMN |
| 9 | 50.00 | −36.00 | −24.00 | 51.79 | −34.17 | −27.23 | DAN |
| 10 | 53.00 | −33.00 | −14.00 | 55.18 | −30.80 | −16.93 | tDMN |
| 11 | 32.00 | 33.00 | −6.00 | 33.55 | 38.46 | −12.03 | aFP |
| 12 | −8.00 | −54.00 | 57.00 | −7.12 | −52.22 | 60.71 | SM1 |
| 13 | 8.00 | −6.00 | 45.00 | 9.50 | −1.84 | 44.73 | SM1 |
| 14 | −8.00 | −24.00 | 63.00 | −6.90 | −20.59 | 65.21 | SM1 |
| 15 | −8.00 | −36.00 | 69.00 | −6.79 | −33.09 | 72.27 | SM1 |
| 16 | −52.00 | −25.00 | 41.00 | −53.52 | −22.54 | 43.10 | SM2 |
| 17 | 8.00 | −48.00 | 69.00 | 9.94 | −45.52 | 72.63 | SM1 |
| 18 | −39.00 | −22.00 | 52.00 | −39.63 | −19.04 | 54.21 | SM2 |
| 19 | 26.00 | −42.00 | 57.00 | 28.54 | −39.24 | 59.17 | SM2 |
| 20 | 47.00 | −24.00 | 42.00 | 50.24 | −20.37 | 41.74 | SM2 |
| 21 | 18.00 | −32.00 | 58.00 | 20.21 | −28.80 | 59.80 | SM1 |
| 22 | −29.00 | −45.00 | 57.00 | −29.10 | −43.00 | 60.66 | SM2 |
| 23 | 20.00 | −45.00 | 66.00 | 22.45 | −42.29 | 68.99 | SM1 |
| 24 | −44.00 | −34.00 | 44.00 | −45.10 | −31.85 | 46.63 | SM2 |
| 25 | −21.00 | −34.00 | 58.00 | −20.66 | −31.33 | 60.85 | SM1 |
| 26 | 39.00 | −24.00 | 54.00 | 42.14 | −20.24 | 54.59 | SM2 |
| 27 | 35.00 | −21.00 | 45.00 | 37.74 | −17.30 | 45.01 | SM2 |
| 28 | −48.00 | −14.00 | 34.00 | −49.47 | −11.06 | 34.95 | MotM |
| 29 | 34.00 | −13.00 | 16.00 | 36.04 | −9.44 | 13.95 | MotM |
| 30 | 48.00 | −10.00 | 34.00 | 51.14 | −5.80 | 32.42 | MotM |
| 31 | −51.00 | −13.00 | 24.00 | −52.84 | −10.23 | 24.41 | MotM |
| 32 | 62.00 | −12.00 | 27.00 | 65.64 | −7.88 | 24.83 | MotM |
| 33 | −4.00 | −2.00 | 53.00 | −2.88 | 2.38 | 53.21 | CO |
| 34 | 51.00 | −31.00 | 34.00 | 54.22 | −27.83 | 33.64 | SM2 |
| 35 | 17.00 | −12.00 | 63.00 | 19.33 | −7.71 | 63.88 | SM1 |
| 36 | −11.00 | −6.00 | 42.00 | −10.48 | −2.10 | 42.02 | SM1 |
| 37 | 35.00 | −3.00 | 0.00 | 36.73 | 0.78 | −3.57 | CO |
| 38 | 5.00 | 3.00 | 51.00 | 6.52 | 7.69 | 50.58 | CO |
| 39 | −43.00 | −3.00 | 10.00 | −44.76 | 0.10 | 8.83 | CO |
| 40 | 47.00 | 4.00 | 3.00 | 49.40 | 8.32 | −1.12 | CO |
| 41 | −33.00 | 0.00 | 6.00 | −34.37 | 3.29 | 4.19 | CO |
| 42 | −6.00 | 13.00 | 36.00 | −5.33 | 17.80 | 34.41 | CO |
| 43 | 34.00 | 6.00 | 5.00 | 35.83 | 10.32 | 1.18 | CO |
| 44 | 62.00 | −36.00 | 21.00 | 65.43 | −33.20 | 19.97 | SM2 |
| 45 | 55.00 | −19.00 | 10.00 | 57.88 | −15.62 | 7.49 | pCO |
| 46 | −37.00 | −35.00 | 16.00 | −38.43 | −33.34 | 16.98 | pCO |
| 47 | −58.00 | −27.00 | 13.00 | −60.48 | −25.22 | 13.82 | pCO |
| 48 | −47.00 | −28.00 | 5.00 | −49.14 | −26.30 | 5.18 | tDMN |
| 49 | 41.00 | −26.00 | 21.00 | 43.45 | −22.93 | 19.85 | SM2 |
| 50 | −48.00 | −36.00 | 24.00 | −49.77 | −34.36 | 25.74 | SM2 |
| 51 | −51.00 | −24.00 | 22.00 | −52.92 | −21.83 | 22.97 | SM2 |
| 52 | −53.00 | −12.00 | 12.00 | −55.22 | −9.42 | 11.73 | MotM |
| 53 | 53.00 | −9.00 | 16.00 | 55.96 | −5.03 | 13.25 | MotM |
| 54 | 56.00 | −21.00 | 30.00 | 59.40 | −17.34 | 28.69 | SM2 |
| 55 | −29.00 | −29.00 | 12.00 | −30.12 | −27.02 | 12.20 | SM1 |
| 56 | −39.00 | −75.00 | 22.00 | −40.50 | −75.27 | 25.80 | pDMN |
| 57 | 5.00 | 60.00 | 3.00 | 5.55 | 66.69 | −3.55 | aDMN |
| 58 | 8.00 | 42.00 | −9.00 | 8.36 | 47.59 | −15.18 | aDMN |
| 59 | −17.00 | 57.00 | −3.00 | −17.65 | 63.19 | −9.17 | aFP |
| 60 | −44.00 | −61.00 | 18.00 | −45.79 | −60.69 | 20.85 | tDMN |
| 61 | 41.00 | −73.00 | 26.00 | 43.43 | −72.21 | 28.00 | pDMN |
| 62 | −41.00 | 9.00 | −30.00 | −43.58 | 11.99 | −34.15 | tDMN |
| 63 | 44.00 | 12.00 | −24.00 | 45.64 | 16.20 | −30.02 | tDMN |
| 64 | −55.00 | −27.00 | −14.00 | −57.97 | −25.69 | −14.73 | tDMN |
| 65 | 26.00 | 12.00 | −12.00 | 27.06 | 16.22 | −16.93 | aDMN |
| 66 | −43.00 | −65.00 | 31.00 | −44.45 | −64.64 | 34.78 | pDMN |
| 67 | −7.00 | −56.00 | 25.00 | −6.84 | −54.90 | 27.05 | pDMN |
| 68 | 5.00 | −60.00 | 33.00 | 5.91 | −58.82 | 35.45 | pDMN |
| 69 | −11.00 | −57.00 | 14.00 | −11.29 | −56.20 | 15.60 | pDMN |
| 70 | −3.00 | −50.00 | 12.00 | −2.94 | −48.79 | 12.87 | pDMN |
| 71 | 7.00 | −50.00 | 29.00 | 7.94 | −48.37 | 30.57 | pDMN |
| 72 | 14.00 | −64.00 | 24.00 | 15.12 | −63.09 | 25.98 | pDMN |
| 73 | −3.00 | −39.00 | 42.00 | −2.20 | −36.68 | 43.85 | pDMN |
| 74 | 10.00 | −55.00 | 16.00 | 10.77 | −53.83 | 17.09 | pDMN |
| 75 | 49.00 | −61.00 | 34.00 | 52.04 | −59.37 | 35.52 | tDMN |
| 76 | 21.00 | 27.00 | 50.00 | 23.33 | 33.07 | 47.68 | aDMN |
| 77 | −17.00 | 23.00 | 54.00 | −16.40 | 28.52 | 53.05 | aDMN |
| 78 | 20.00 | 33.00 | 42.00 | 22.11 | 39.21 | 38.90 | aDMN |
| 79 | −20.00 | 39.00 | 42.00 | −19.78 | 45.07 | 39.48 | aDMN |
| 80 | 5.00 | 48.00 | 21.00 | 5.94 | 54.42 | 16.18 | aDMN |
| 81 | −7.00 | 45.00 | 4.00 | −7.04 | 50.82 | −1.29 | aDMN |
| 82 | 8.00 | 48.00 | 9.00 | 8.80 | 54.23 | 3.45 | aDMN |
| 83 | −3.00 | 39.00 | −4.00 | −3.06 | 44.41 | −9.46 | aDMN |
| 84 | 7.00 | 37.00 | 0.00 | 7.51 | 42.49 | −5.35 | aDMN |
| 85 | −11.00 | 39.00 | 12.00 | −11.06 | 44.62 | 7.61 | aDMN |
| 86 | −3.00 | 32.00 | 39.00 | −2.06 | 37.85 | 36.34 | aDMN |
| 87 | −3.00 | 36.00 | 20.00 | −2.50 | 41.70 | 16.05 | aDMN |
| 88 | −8.00 | 42.00 | 27.00 | −7.55 | 48.08 | 23.18 | aDMN |
| 89 | 62.00 | −15.00 | −15.00 | 64.64 | −11.80 | −19.30 | tDMN |
| 90 | −53.00 | −15.00 | −9.00 | −55.72 | −12.96 | −10.24 | tDMN |
| 91 | −55.00 | −31.00 | −4.00 | −57.75 | −29.70 | −3.94 | tDMN |
| 92 | 62.00 | −33.00 | −6.00 | 64.80 | −30.55 | −8.70 | tDMN |
| 93 | 11.00 | 30.00 | 24.00 | 12.25 | 35.63 | 20.30 | aDMN |
| 94 | 50.00 | −6.00 | −12.00 | 52.16 | −2.43 | −16.40 | tDMN |
| 95 | −25.00 | −41.00 | −8.00 | −26.44 | −39.95 | −8.26 | mVIS |
| 96 | 26.00 | −39.00 | −11.00 | 26.94 | −37.34 | −12.76 | mVIS |
| 97 | −32.00 | −39.00 | −15.00 | −33.93 | −38.06 | −15.60 | mVIS |
| 98 | 28.00 | −76.00 | −31.00 | 28.46 | −76.56 | −31.64 | VIS |
| 99 | 50.00 | 3.00 | −24.00 | 51.90 | 6.81 | −29.61 | tDMN |
| 100 | −50.00 | 0.00 | −24.00 | −52.89 | 2.55 | −27.06 | tDMN |
| 101 | 44.00 | −52.00 | 28.00 | 46.68 | −50.08 | 28.76 | pDMN |
| 102 | −47.00 | −43.00 | 0.00 | −49.30 | −42.15 | 0.83 | tDMN |
| 103 | −29.00 | 15.00 | −15.00 | −30.63 | 18.71 | −18.98 | aDMN |
| 104 | −3.00 | −37.00 | 30.00 | −2.47 | −34.80 | 31.07 | pDMN |
| 105 | −7.00 | −72.00 | 38.00 | −6.58 | −71.47 | 41.74 | pDMN |
| 106 | 10.00 | −67.00 | 39.00 | 11.27 | −66.01 | 42.09 | pDMN |
| 107 | 3.00 | −50.00 | 48.00 | 4.20 | −48.06 | 50.71 | pDMN |
| 108 | −44.00 | 27.00 | −9.00 | −46.17 | 31.26 | −13.03 | aFP |
| 109 | 47.00 | 30.00 | −6.00 | 49.26 | 35.47 | −12.20 | aFP |
| 110 | 8.00 | −90.00 | −9.00 | 7.98 | −91.08 | −7.10 | VIS |
| 111 | 17.00 | −90.00 | −15.00 | 17.27 | −91.09 | −13.64 | VIS |
| 112 | −11.00 | −93.00 | −15.00 | −12.08 | −94.56 | −12.80 | VIS |
| 113 | 17.00 | −48.00 | −9.00 | 17.53 | −46.86 | −9.88 | mVIS |
| 114 | 38.00 | −73.00 | 13.00 | 39.98 | −72.49 | 14.36 | mVIS |
| 115 | 8.00 | −72.00 | 9.00 | 8.45 | −71.84 | 10.79 | mVIS |
| 116 | −8.00 | −80.00 | 5.00 | −8.43 | −80.50 | 7.44 | mVIS |
| 117 | −27.00 | −79.00 | 16.00 | −28.07 | −79.45 | 19.43 | mVIS |
| 118 | 19.00 | −66.00 | 1.00 | 19.81 | −65.56 | 1.72 | mVIS |
| 119 | −23.00 | −90.00 | 15.00 | −23.94 | −90.98 | 18.96 | mVIS |
| 120 | 26.00 | −60.00 | −9.00 | 26.93 | −59.37 | −9.36 | mVIS |
| 121 | −14.00 | −72.00 | −9.00 | −15.02 | −72.42 | −7.68 | mVIS |
| 122 | −17.00 | −68.00 | 3.00 | −17.87 | −68.03 | 4.81 | mVIS |
| 123 | 41.00 | −78.00 | −12.00 | 42.52 | −78.17 | −11.78 | DAN |
| 124 | −44.00 | −75.00 | −12.00 | −46.54 | −75.95 | −9.95 | DAN |
| 125 | −14.00 | −90.00 | 27.00 | −14.22 | −90.66 | 31.40 | mVIS |
| 126 | 14.00 | −87.00 | 33.00 | 15.27 | −87.09 | 36.89 | mVIS |
| 127 | 27.00 | −77.00 | 23.00 | 28.68 | −76.62 | 25.42 | mVIS |
| 128 | 19.00 | −85.00 | −4.00 | 19.64 | −85.62 | −2.39 | VIS |
| 129 | 14.00 | −77.00 | 28.00 | 15.18 | −76.68 | 31.00 | mVIS |
| 130 | −15.00 | −53.00 | −2.00 | −15.85 | −52.34 | −1.43 | mVIS |
| 131 | 40.00 | −66.00 | −8.00 | 41.60 | −65.50 | −8.27 | DAN |
| 132 | 23.00 | −87.00 | 21.00 | 24.41 | −87.21 | 24.01 | mVIS |
| 133 | 5.00 | −72.00 | 21.00 | 5.59 | −71.65 | 23.52 | mVIS |
| 134 | −40.00 | −73.00 | −2.00 | −42.10 | −73.62 | 0.38 | DAN |
| 135 | 25.00 | −79.00 | −16.00 | 25.66 | −79.47 | −15.56 | mVIS |
| 136 | −16.00 | −77.00 | 30.00 | −16.21 | −76.97 | 33.82 | mVIS |
| 137 | −3.00 | −81.00 | 18.00 | −2.88 | −81.25 | 21.10 | mVIS |
| 138 | −38.00 | −87.00 | −9.00 | −40.21 | −88.44 | −6.19 | DAN |
| 139 | 35.00 | −84.00 | 11.00 | 36.76 | −84.11 | 12.99 | mVIS |
| 140 | 6.00 | −81.00 | 4.00 | 6.21 | −81.41 | 6.11 | mVIS |
| 141 | −25.00 | −89.00 | 0.00 | −26.39 | −90.23 | 3.12 | VIS |
| 142 | −31.00 | −78.00 | −15.00 | −33.00 | −79.02 | −13.24 | DAN |
| 143 | 35.00 | −81.00 | 0.00 | 36.51 | −81.16 | 1.20 | DAN |
| 144 | −43.00 | −2.00 | 45.00 | −43.93 | 1.80 | 45.70 | CO |
| 145 | 45.00 | 19.00 | 30.00 | 47.98 | 24.56 | 26.50 | aFP |
| 146 | −45.00 | 7.00 | 24.00 | −46.50 | 10.85 | 23.04 | aFP |
| 147 | −51.00 | −50.00 | 39.00 | −52.60 | −48.83 | 42.50 | pFP |
| 148 | 56.00 | −54.00 | −12.00 | 58.31 | −52.79 | −13.61 | DAN |
| 149 | 23.00 | 39.00 | −9.00 | 24.07 | 44.61 | −15.35 | aFP |
| 150 | 32.00 | 48.00 | −6.00 | 33.60 | 54.22 | −12.95 | aFP |
| 151 | 17.00 | −79.00 | −34.00 | 16.85 | −79.89 | −34.36 | aDMN |
| 152 | 34.00 | −67.00 | −33.00 | 34.72 | −67.08 | −34.45 | aDMN |
| 153 | 44.00 | 5.00 | 35.00 | 47.01 | 9.93 | 32.66 | aFP |
| 154 | −40.00 | 2.00 | 33.00 | −41.06 | 5.81 | 32.72 | aFP |
| 155 | −41.00 | 33.00 | 24.00 | −42.23 | 38.21 | 21.35 | aFP |
| 156 | 36.00 | 37.00 | 20.00 | 38.37 | 43.18 | 15.06 | aFP |
| 157 | 46.00 | −45.00 | 44.00 | 49.18 | −42.41 | 45.16 | pFP |
| 158 | −28.00 | −59.00 | 44.00 | −28.40 | −57.93 | 47.78 | pFP |
| 159 | 41.00 | −55.00 | 45.00 | 43.93 | −52.95 | 46.95 | pFP |
| 160 | 35.00 | −66.00 | 38.00 | 37.45 | −64.70 | 40.38 | pFP |
| 161 | −41.00 | −56.00 | 41.00 | −42.09 | −54.98 | 44.74 | pFP |
| 162 | 37.00 | 13.00 | 42.00 | 39.87 | 18.39 | 39.72 | aFP |
| 163 | −33.00 | 49.00 | 9.00 | −34.16 | 54.83 | 4.36 | aFP |
| 164 | −40.00 | 40.00 | 2.00 | −41.68 | 45.16 | −2.31 | aFP |
| 165 | 31.00 | −55.00 | 42.00 | 33.38 | −53.12 | 44.02 | pFP |
| 166 | 41.00 | 43.00 | 4.00 | 43.25 | 49.25 | −2.31 | aFP |
| 167 | −41.00 | 20.00 | 31.00 | −42.10 | 24.68 | 29.53 | aFP |
| 168 | −4.00 | 21.00 | 46.00 | −2.98 | 26.41 | 44.42 | aDMN |
| 169 | 9.00 | −41.00 | 48.00 | 10.51 | −38.54 | 50.02 | SM1 |
| 170 | 52.00 | −47.00 | 36.00 | 55.27 | −44.59 | 36.70 | pFP |
| 171 | 39.00 | −5.00 | 48.00 | 42.05 | −0.39 | 47.10 | CO |
| 172 | 29.00 | 27.00 | 30.00 | 31.24 | 32.79 | 26.39 | aFP |
| 173 | 45.00 | 17.00 | 14.00 | 47.60 | 22.16 | 9.74 | aFP |
| 174 | −34.00 | 16.00 | 3.00 | −35.44 | 20.03 | 0.07 | CO |
| 175 | 34.00 | 17.00 | 7.00 | 35.91 | 21.91 | 2.62 | CO |
| 176 | 35.00 | 27.00 | 3.00 | 36.89 | 32.35 | −2.24 | CO |
| 177 | 32.00 | 12.00 | −3.00 | 33.56 | 16.45 | −7.58 | aDMN |
| 178 | −2.00 | 10.00 | 45.00 | −0.94 | 14.86 | 43.99 | aDMN |
| 179 | −27.00 | 46.00 | 25.00 | −27.50 | 52.04 | 21.28 | aFP |
| 180 | 4.00 | 18.00 | 39.00 | 5.23 | 23.22 | 37.03 | aDMN |
| 181 | 9.00 | 17.00 | 30.00 | 10.26 | 22.06 | 27.48 | aDMN |
| 182 | 29.00 | 49.00 | 20.00 | 31.07 | 55.71 | 14.49 | aFP |
| 183 | 24.00 | 43.00 | 31.00 | 26.07 | 49.56 | 26.58 | aFP |
| 184 | −10.00 | −21.00 | 8.00 | −10.28 | −18.48 | 7.04 | SubC |
| 185 | 11.00 | −20.00 | 9.00 | 11.75 | −17.18 | 7.54 | SubC |
| 186 | −21.00 | 4.00 | −2.00 | −21.97 | 7.48 | −4.78 | CO |
| 187 | 29.00 | −17.00 | 4.00 | 30.50 | −13.92 | 1.65 | CO |
| 188 | 22.00 | 6.00 | 5.00 | 23.26 | 10.19 | 1.46 | CO |
| 189 | 27.00 | −3.00 | 7.00 | 28.52 | 0.82 | 4.01 | CO |
| 190 | −30.00 | −14.00 | 1.00 | −31.38 | −11.48 | −0.30 | MotM |
| 191 | 51.00 | −45.00 | 22.00 | 53.90 | −42.76 | 21.83 | tDMN |
| 192 | −54.00 | −51.00 | 8.00 | −56.47 | −50.48 | 9.92 | tDMN |
| 193 | −53.00 | −41.00 | 12.00 | −55.30 | −39.89 | 13.51 | tDMN |
| 194 | 49.00 | −35.00 | 9.00 | 51.52 | −32.52 | 7.55 | tDMN |
| 195 | 49.00 | −31.00 | −2.00 | 51.28 | −28.52 | −4.30 | tDMN |
| 196 | 53.00 | −48.00 | 12.00 | 55.75 | −46.07 | 11.42 | tDMN |
| 197 | 50.00 | 27.00 | 6.00 | 52.68 | 32.58 | 0.57 | aFP |
| 198 | −47.00 | 21.00 | 2.00 | −49.07 | 25.13 | −0.98 | aFP |
| 199 | 22.00 | −58.00 | −22.00 | 22.43 | −57.55 | −23.11 | mVIS |
| 200 | 1.00 | −62.00 | −18.00 | 0.51 | −61.91 | −18.14 | SubC |
| 201 | 32.00 | −15.00 | −30.00 | 32.85 | −12.41 | −34.41 | UNA |
| 202 | −29.00 | −12.00 | −33.00 | −31.13 | −9.99 | −36.32 | UNA |
| 203 | 47.00 | −6.00 | −33.00 | 48.52 | −2.85 | −38.49 | tDMN |
| 204 | −47.00 | −9.00 | −36.00 | −50.06 | −7.09 | −39.24 | tDMN |
| 205 | 8.00 | −63.00 | 57.00 | 9.61 | −61.50 | 60.88 | UNA |
| 206 | −50.00 | −63.00 | 3.00 | −52.44 | −63.14 | 5.29 | tDMN |
| 207 | −44.00 | −51.00 | −21.00 | −46.68 | −50.91 | −20.91 | DAN |
| 208 | 44.00 | −48.00 | −15.00 | 45.68 | −46.67 | −16.85 | DAN |
| 209 | 44.00 | −33.00 | 48.00 | 47.21 | −29.75 | 48.70 | SM2 |
| 210 | 20.00 | −66.00 | 45.00 | 21.90 | −64.74 | 48.12 | pFP |
| 211 | 44.00 | −60.00 | 4.00 | 46.09 | −58.93 | 3.93 | DAN |
| 212 | 23.00 | −60.00 | 57.00 | 25.34 | −58.18 | 60.34 | SM2 |
| 213 | −32.00 | −48.00 | 44.00 | −32.56 | −46.42 | 47.20 | DAN |
| 214 | −26.00 | −71.00 | 33.00 | −26.60 | −70.72 | 36.86 | pFP |
| 215 | −32.00 | −5.00 | 53.00 | −32.23 | −1.08 | 54.06 | CO |
| 216 | −40.00 | −60.00 | −10.00 | −42.26 | −60.12 | −8.85 | DAN |
| 217 | −17.00 | −60.00 | 60.00 | −16.50 | −58.57 | 64.46 | pFP |
| 218 | 26.00 | −9.00 | 54.00 | 28.56 | −4.62 | 53.99 | CO |
| 219 | 48.00 | 10.00 | 22.00 | 50.91 | 14.99 | 18.54 | aFP |
| 220 | 26.00 | 4.00 | −4.00 | 27.23 | 7.96 | −8.00 | CO |
| 221 | −8.00 | −12.00 | 58.00 | −6.98 | −8.08 | 59.20 | SM1 |
| 222 | −9.00 | 10.00 | 10.00 | −9.10 | 14.14 | 7.23 | aDMN |
| 223 | −48.00 | −66.00 | −8.00 | −50.61 | −66.47 | −6.18 | DAN |
| 224 | −28.00 | 42.00 | −8.00 | −29.34 | 47.21 | −13.27 | aFP |
| 225 | −20.00 | 2.00 | 52.00 | −19.65 | 6.39 | 52.29 | aDMN |
| 226 | 20.00 | −70.00 | −9.00 | 20.61 | −69.94 | −8.61 | mVIS |
| 227 | 12.00 | −78.00 | 38.00 | 13.31 | −77.56 | 41.66 | UNA |
| 228 | 56.00 | −8.00 | −2.00 | 58.68 | −4.28 | −5.87 | tDMN |
| 229 | 39.00 | −39.00 | −20.00 | 40.35 | −37.36 | −22.56 | DAN |
| 230 | −20.00 | −22.00 | 64.00 | −19.44 | −18.61 | 66.42 | SM1 |
| 231 | 34.00 | −84.00 | −39.00 | 34.53 | −85.05 | −39.74 | aDMN |
| 232 | 31.00 | −75.00 | −54.00 | 31.06 | −75.90 | −56.04 | CO |
| 233 | −14.00 | −78.00 | −24.00 | −15.39 | −79.00 | −23.14 | VIS |
| 234 | −35.00 | −51.00 | −45.00 | −37.82 | −51.25 | −46.45 | aDMN |
Supplementary Material
Acknowledgements:
This study was supported by grants from the National Institutes of Health (K01-MH122779, R01-HD055741, T32-HD040127, P30-HD003110, R01-MH118362, MH118362–02S1, P30-NS098577) and the Simons Foundation (#140209). We are sincerely grateful to all the families and children who participated in the IBIS study.
IBIS Network: The Infant Brain Imaging Study (IBIS) Network is an NIH funded Autism Centers of Excellence project and consists of a consortium of 9 universities in the U.S. and Canada. Members and components of the IBIS Network include: J. Piven (IBIS Network PI), Clinical Sites: University of North Carolina: H.C. Hazlett, C. Chappell, M.D. Shen, J.B. Girault, R.L. Grzadzinski; University of Washington: S. Dager, A. Estes, D. Shaw, T. St. John; Washington University: K. Botteron, J. Constantino; Children’s Hospital of Philadelphia: R. Schultz, J. Pandey; Behavior Core: University of Washington: A. Estes; University of Alberta: L. Zwaigenbaum; University of Minnesota: J. Elison, J. Wolff; Imaging Core: University of North Carolina: M. Styner; New York University: G. Gerig; Washington University in St. Louis: R. McKinstry, J. Pruett; Data Coordinating Center: Montreal Neurological Institute: A.C. Evans, D.L. Collins, V. Fonov, L. MacIntyre; S. Das; Statistical Analysis Core: University of North Carolina: K. Truong; Environmental risk core: John Hopkins University: H. Volk; Genetics Core: John Hopkins University: D. Fallin; UNC: M.D. Shen.
Footnotes
Previous Presentations: This work was previously presented virtually at the International Society for Autism Research and the Seaver Autism Center Conference in May of 2021.
Disclosures: Dr. John Constantino receives royalties from Western Psychological Services for the commercial distribution of the Social Responsiveness Scale. Dr. Robert McKinstry serves on the advisory board of Nous Imaging, Inc. and receives funding for meals and travel from Siemens Healthineers and Philips Healthcare. All other authors report no financial relationships with commercial interests.
References
- 1.Bai D, Yip BHK, Windham GC, et al. : Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA psychiatry 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maenner MJ, Shaw KA, Baio J, et al. : Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. Mmwr Surveill Summ 2020; 69:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozonoff S, Young GS, Carter A, et al. : Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics 2011; 128:e488–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piven J, Elison JT, Zylka MJ: Toward a conceptual framework for early brain and behavior development in autism. Molecular psychiatry 2017; 22:1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolff JJ, Gu H, Gerig G, et al. : Differences in White Matter Fiber Tract Development Present From 6 to 24 Months in Infants With Autism. American Journal of Psychiatry 2012; 169:589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff JJ, Gerig G, Lewis JD, et al. : Altered corpus callosum morphology associated with autism over the first 2 years of life. Brain 2015; 138:2046–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen MD, Nordahl CW, Young GS, et al. : Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain 2013; 136:2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen MD, Kim SH, McKinstry RC, et al. : Increased Extra-axial Cerebrospinal Fluid in High-Risk Infants Who Later Develop Autism. Biological psychiatry 2017; 82:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazlett HC, Gu H, Munsell BC, et al. : Early brain development in infants at high risk for autism spectrum disorder. Nature 2017; 542:348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerson RW, Adams C, Nishino T, et al. : Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Science translational medicine 2017; 9:eaag2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen MD, Piven J: Brain and behavior development in autism from birth through infancy. Dialogues in clinical neuroscience 2017; 19:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaugler T, Klei L, Sanders SJ, et al. : Most genetic risk for autism resides with common variation. Nature genetics 2014; 46:881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klei L, Sanders SJ, Murtha MT, et al. : Common genetic variants, acting additively, are a major source of risk for autism. Molecular autism 2012; 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grove J, Ripke S, Als TD, et al. : Identification of common genetic risk variants for autism spectrum disorder. Nature genetics 2019; 51:431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Constantino JN, Zhang Y, Frazier T, et al. : Sibling recurrence and the genetic epidemiology of autism. The American journal of psychiatry 2010; 167:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Constantino JN, Todd RD: Autistic traits in the general population: a twin study. Archives of general psychiatry 2003; 60:524–530 [DOI] [PubMed] [Google Scholar]
- 17.Frazier TW, Thompson L, Youngstrom EA, et al. : A Twin Study of Heritable and Shared Environmental Contributions to Autism. J Autism Dev Disord 2014; 44:2013–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijmeijer JS, Arias-Vásquez A, Rommelse NNJ, et al. : Identifying Loci for the Overlap Between Attention-Deficit/Hyperactivity Disorder and Autism Spectrum Disorder Using a Genome-wide QTL Linkage Approach. J Am Acad Child Adolesc Psychiatry 2010; 49:675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas TR, Koomar T, Casten L, et al. : Clinical autism subscales have common genetic liability that is heritable, pleiotropic, and generalizable to the general population. Medrxiv 2021; 2021.08.30.21262845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald NM, Senturk D, Scheffler A, et al. : Developmental Trajectories of Infants With Multiplex Family Risk for Autism: A Baby Siblings Research Consortium Study. JAMA Neurology 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virkud YV, Todd RD, Abbacchi AM, et al. : Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2009; 150B:328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Losh M, Childress D, Lam K, et al. : Defining key features of the broad autism phenotype: a comparison across parents of multiple- and single-incidence autism families. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 2008; 147B:424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girault JB, Swanson MR, Meera SS, et al. : Quantitative trait variation in ASD probands and toddler sibling outcomes at 24 months. Journal of neurodevelopmental disorders 2020; 12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno-De-Luca A, Evans DW, Boomer KB, et al. : The Role of Parental Cognitive, Behavioral, and Motor Profiles in Clinical Variability in Individuals With Chromosome 16p11.2 Deletions. Jama Psychiat 2015; 72:119. [DOI] [PubMed] [Google Scholar]
- 25.Klaassen P, Duijff S, Veye HS de, et al. : Explaining the variable penetrance of CNVs: Parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 2016; 171:790–796 [DOI] [PubMed] [Google Scholar]
- 26.Ohta H, Nordahl CW, Iosif A-M, et al. : Increased Surface Area, but not Cortical Thickness, in a Subset of Young Boys With Autism Spectrum Disorder. Autism Research 2016; 9:232–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff JJ, Swanson MR, Elison JT, et al. : Neural circuitry at age 6 months associated with later repetitive behavior and sensory responsiveness in autism. Molecular autism 2017; 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elison JT, Paterson SJ, Wolff JJ, et al. : White matter microstructure and atypical visual orienting in 7-month-olds at risk for autism. The American journal of psychiatry 2013; 170:899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutter M, Couteur AL, Lord C: Autism diagnostic interview-revised Western Psychological Services, 2003 [Google Scholar]
- 30.Das S, Glatard T, MacIntyre LC, et al. : The MNI data-sharing and processing ecosystem. Neuroimage 2016; 124:1188–1195 [DOI] [PubMed] [Google Scholar]
- 31.Rutter M, Bailey A, Berument S, et al. : Social Communication Questionnaire (SCQ) Western Psychological Services, 2003 [Google Scholar]
- 32.Sparrow SS, Cicchetti DV, Balla DA: Vineland adaptive behavior scales: Second Edition (Vineland II), The expanded interview form Pearson Assessments, 2008 [Google Scholar]
- 33.Swanson MR, Wolff JJ, Elison JT, et al. : Splenium development and early spoken language in human infants. Developmental Science 2015; n/a-n/a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. : Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 2002; 15:273–289 [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Fonov VS, Dietrich C, et al. : Adaptive prior probability and spatial temporal intensity change estimation for segmentation of the one-year-old human brain. J Neurosci Meth 2012; 212:43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oguz I, Farzinfar M, Matsui J, et al. : DTIPrep: quality control of diffusion-weighted images. Frontiers in neuroinformatics 2014; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verde AR, Budin F, Berger J-B, et al. : UNC-Utah NA-MIC framework for DTI fiber tract analysis. Frontiers in neuroinformatics 2014; 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eggebrecht AT, Elison JT, Feczko E, et al. : Joint Attention and Brain Functional Connectivity in Infants and Toddlers. Cerebral cortex (New York, NY : 1991) 2017; 27:1709–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emerson RW, Gao W, Lin W: Longitudinal Study of the Emerging Functional Connectivity Asymmetry of Primary Language Regions during Infancy. The Journal of neuroscience : the official journal of the Society for Neuroscience 2016; 36:10883–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojemann JG, Akbudak E, Snyder AZ, et al. : Anatomic Localization and Quantitative Analysis of Gradient Refocused Echo-Planar fMRI Susceptibility Artifacts. NeuroImage 1997; 6:156–167 [DOI] [PubMed] [Google Scholar]
- 41.Power JD, Mitra A, Laumann TO, et al. : Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage 2014; 84:320–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen AN, Greene DJ, Gratton C, et al. : Evaluating the Prediction of Brain Maturity From Functional Connectivity After Motion Artifact Denoising. Cereb Cortex 2018; 29:2455–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dosenbach NUF, Fair DA, Miezin FM, et al. : Distinct brain networks for adaptive and stable task control in humans. Proc National Acad Sci 2007; 104:11073–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marek S, Dosenbach NUF: The frontoparietal network: function, electrophysiology, and importance of individual precision mapping. Dialogues Clin Neurosci 2018; 20:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair A, Jolliffe M, Lograsso YSS, et al. : A Review of Default Mode Network Connectivity and Its Association With Social Cognition in Adolescents With Autism Spectrum Disorder and Early-Onset Psychosis. Frontiers Psychiatry 2020; 11:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawks ZW, Todorov A, Marrus N, et al. : A prospective evaluation of infant cerebellar-cerebral functional connectivity in relation to behavioral development in autism. Biological Psychiatry Global Open Sci 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosvall M, Bergstrom CT: Maps of random walks on complex networks reveal community structure. Proc National Acad Sci 2008; 105:1118–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seitzman BA, Gratton C, Marek S, et al. : A set of functionally-defined brain regions with improved representation of the subcortex and cerebellum. Neuroimage 2020; 206:116290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing [Internet]. Journal of the Royal Statistical Society 1995; 57:289–300Available from: http://www.jstor.org/stable/2346101 [Google Scholar]
- 50.Gotham K, Risi S, Pickles A, et al. : The Autism Diagnostic Observation Schedule: revised algorithms for improved diagnostic validity. Journal of autism and developmental disorders 2007; 37:613–627 [DOI] [PubMed] [Google Scholar]
- 51.Rolison M, Lacadie C, Chawarska K, et al. : Atypical Intrinsic Hemispheric Interaction Associated with Autism Spectrum Disorder Is Present within the First Year of Life. Cereb Cortex New York N Y 1991 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velmeshev D, Schirmer L, Jung D, et al. : Single-cell genomics identifies cell type-specific molecular changes in autism. Science 2019; 364:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grasby KL, Jahanshad N, Painter JN, et al. : The genetic architecture of the human cerebral cortex. Science 2020; 367:eaay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchetto MC, Belinson H, Tian Y, et al. : Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Molecular psychiatry 2017; 22:820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen MD, Nordahl CW, Li DD, et al. : Extra-axial cerebrospinal fluid in high-risk and normal-risk children with autism aged 2–4 years: a case-control study. The Lancet Psychiatry 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombardo MV, Eyler L, Moore A, et al. : Default mode-visual network hypoconnectivity in an autism subtype with pronounced social visual engagement difficulties. Elife 2019; 8:e47427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wandell BA, Dumoulin SO, Brewer AA: Visual Field Maps in Human Cortex. Neuron 2007; 56:366–383 [DOI] [PubMed] [Google Scholar]
- 58.Grill-Spector K, Kourtzi Z, Kanwisher N: The lateral occipital complex and its role in object recognition. Vision Res 2001; 41:1409–1422 [DOI] [PubMed] [Google Scholar]
- 59.Malach R, Reppas JB, Benson RR, et al. : Object-related activity revealed by functional magnetic resonance imaging in human occipital cortex. Proc National Acad Sci 1995; 92:8135–8139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Putnam MC, Steven MS, Doron KW, et al. : Cortical Projection Topography of the Human Splenium: Hemispheric Asymmetry and Individual Differences. J Cognitive Neurosci 2010; 22:1662–1669 [DOI] [PubMed] [Google Scholar]
- 61.Jones W, Klin A: Attention to eyes is present but in decline in 2–6-month-old infants later diagnosed with autism. Nature 2013; 504:427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Constantino JN, Kennon-McGill S, Weichselbaum C, et al. : Infant viewing of social scenes is under genetic control and is atypical in autism. Nature 2017; 547:340–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scarr S, McCartney K: How People Make Their Own Environments: A Theory of Genotype Environment Effects. Child Dev 1983; 54:424–435 [DOI] [PubMed] [Google Scholar]
- 64.Ozonoff S, Young GS, Belding A, et al. : The broader autism phenotype in infancy: when does it emerge? Journal of the American Academy of Child and Adolescent Psychiatry 2014; 53:398–407.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warrier V, Toro R, Won H, et al. : Social and non-social autism symptoms and trait domains are genetically dissociable. Commun Biology 2019; 2:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matoba N, Love MI, Stein JL: Evaluating brain structure traits as endophenotypes using polygenicity and discoverability. Hum Brain Mapp 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


