Abstract
Purpose:
To evaluate genetic testing platforms used to aid in the diagnosis of inherited retinal degenerations (IRDs).
Design:
Evaluation of diagnostic test or technology
Subjects:
Targeted genetic panel testing for IRDs
Methods, Intervention, or Testing:
Data collected regarding targeted genetic panel testing for IRDs offered by different labs were investigated for inclusion of coding and non-coding variants in disease genes. Both large IRD panels and smaller, more focused disease specific panels were included in the analysis.
Main Outcome Measures:
Number of disease genes tested as well as the commonality and uniqueness across testing platforms in both coding and non-coding variants of disease.
Results:
Across the three IRD panel tests investigated, 409 unique genes are represented, of which 269 genes are tested by all three panels. The top 20 genes known to cause over 70% of all IRDs are represented in the 269 common genes tested by all three panels. In addition, 138 noncoding variants are assayed across the three platforms in 50 unique genes. Focused disease specific panels exhibited significant variability across 5 testing platforms that were studied.
Conclusions:
Ordering genetic testing for IRDs is not straightforward, as evidenced by the multitude of panels available to providers. It is important that there is coverage of both coding and non-coding regions in IRD genes to offer a diagnosis in these patients. This paper details the diversity of testing platforms currently available to clinicians and provides a thorough explanation of genes tested in the different IRD panels. In a time of increased importance for clinical genetic testing of IRD patients, knowledge of the proper test to order is paramount.
Approximately 1 in 1000 people are affected with genetic eye disease worldwide1. Inherited retinal degenerations (IRDs) are a heterogeneous group of predominantly monogenic disorders that feature loss or dysfunction of photoreceptor cells as a primary or secondary event and have a prevalence of ~1 in 2,000 to ~1 in 3,000 individuals2,3. In the pediatric population, IRDs are a major cause of visual impairment4 and can be one of the first presenting features of a syndromic condition5. Early genetic diagnosis of such conditions minimizes potential extraocular morbidity6. Nearly 300 genes are implicated in IRDs7, however approximately 70% of those with IRDs can be attributed to 20 genes8,9. Compared to adults, there is a higher proportion of those with X-linked inheritance in the pediatric population, and there are distinct genes involved in adult onset versus pediatric onset IRDs8.
Genetic testing using next generation sequencing (NGS) can deliver a definitive diagnosis, which provides patients and families with accurate recurrence risks. Furthermore, genetic testing provides a molecular diagnosis to guide treatment decisions10 and potential access to FDA approved treatment or clinical trials or approved treatments in the future. The American Academy of Ophthalmology advocates for genetic testing in patients with presumed genetically caused retinal degeneration, with potential benefit to at-risk family members as well11. Genetic testing is carried out with patient DNA, typically isolated from saliva or a peripheral blood sample (Figure 1). NGS methods range from targeted gene panel sequencing (TS), to exome sequencing (ES), to genome sequencing (GS). NGS approaches sequence multiple short DNA fragments12, which are then bioinformatically aligned to the human reference genome13. Each sequence variant compared to the reference genome is identified by bioinformatic processing and analyzed to establish its possible association with the disease phenotype and classified accordingly14.
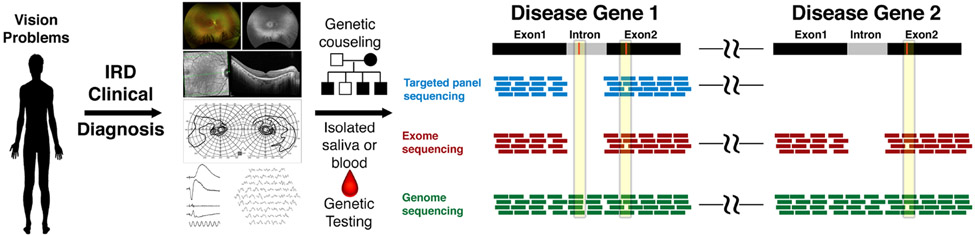
Figure 1. Genetic testing paradigm for patients with IRDs.
This schematic highlights the process of genetic testing patients with suspected IRDs. The process starts with careful clinical diagnosis by the clinician. Once a diagnosis of IRD is suspected, the patient should be referred for genetic counseling for detailed family history and genogram construction as well as for discussion of the implications of genetic testing. Isolated blood or saliva can then be taken for genetic testing. The type of genetic testing will determine the coverage and identification of pathogenic genetic variants. In targeted gene panel sequencing, or panel testing, reads map to exons of targeted genes. In this example, the pathogenic variant (red) in exon 2 of disease gene 1 would be identified, but not the intronic variant in disease gene 1. Since the targets are pre-set, exonic variants in a distant (denoted by spacers) disease gene 2 would not be captured with this test. In exome sequencing, reads map to exons of all genes so both exonic variants would be detected, but not the intronic variant. Genome sequencing, which has reads encompassing the entire region, would be expected to identify both intronic and exonic variants in both disease genes.
The most commonly used approach to genetically diagnose IRD patients is panel-based targeted sequencing. These panels are custom designed to target exons and flanking intronic regions of genes implicated in IRDs. The benefit of this approach is that it is an economical method of focusing sequencing capacity in smaller genomic regions, therefore maximizing the coverage of clinically relevant genes. Panel-based NGS techniques have a detection rate of 60-70%15, but these rates can vary depending on the type of IRD16. Exome sequencing approaches can capture all coding regions from gene panels and can aid in diagnosis of complex monogenic phenotypes17. However, both NGS and exome sequencing approaches have the ability to identify secondary or incidental medically actionable genetic risk, such as increased risk for hereditary breast cancer or cardiac arrhythmia which can also impact other family members such as parents or siblings without retinal disease. Providing accurate and complete pre-test genetic counseling for all genetic testing approaches, including the option to decline unrelated secondary findings is essential to obtaining written informed patient consent for NGS, exome or genome sequencing in both the clinical and research setting18. Comparatively, genome sequencing approaches are superior in detecting a wider spectrum of genetic changes including large structural variations, non-coding variants excluded from targeted NGS analysis regions, and variants in genes excluded from targeted capture19. However, the cost is approximately five times higher for GS on a per-sample basis than exome-based approaches 20. A tiered testing strategy, starting with panel-based testing, can reduce cost and false genotype rate21. In some cases, however, exome and genome sequencing is now recommended as a first line (or second line, according to clinical judgment) genetic test for patients with intellectual disability or multiple congenital anomalies22 or complex phenotypes for which the differential diagnosis is broad23.
Unfortunately, genetic testing is not a straightforward process for eye care providers such as retina specialists who care for IRD patients. There are multiple targeted NGS panels for IRDs as well as numerous smaller, more focused panels for specific IRDs (i.e. retinitis pigmentosa). Blueprint Genetics, Invitae, and Prevention Genetics each offer large retinal dystrophy-specific panels. Blueprint Genetics has partnered with the Foundation Fighting Blindness (FFB) to offer a 351-gene retinal dystrophy panel as part of the My Retina Tracker open access, no-cost testing program. Establishment of the My Retina Tracker Genetic Testing Study24 sponsored by the FFB has increased access to genetic testing for patients25. Invitae has a similar no-cost testing program with ID your IRD through Spark Therapeutics that encompasses a 329-gene retinal disorders panel. Prevention Genetics offers a 321-gene comprehensive inherited retinal dystrophy panel. Each of these three companies, along with others such as GeneDx and Molecular Vision Lab (MVL), also offer focused panels for specific retinal diseases. These numerous testing options can create difficulty for clinicians and genetic counselors to decide on the best course of action for their patients. This paper details each of the comprehensive IRD panels and disease-specific panels available to highlight the commonalities and differences that separate these tests. We also discuss the importance of partnering with genetic counseling as genetic testing should only be provided in the context of pre- and post-test genetic counseling. This information should be used as a resource for the vision science community in a time of increased importance for clinical genetic testing of IRD patients.
Methods
The study adhered to the tenents of the Decleration of Helsinki. This study did not require IRB approval.
Analysis of IRD panels
The three retinal dystrophy-specific panels offered by Blueprint Genetics, Invitae, and Prevention Genetics were investigated. The list of genes assayed by each IRD panel as well as inclusion of any non-coding variants was documented. The genomic location and name of each disease gene was documented across each panel. If non-coding variants were tested it was documented for the gene in question. This curated list was then compared to examine overlapping and unique genes across testing platforms.
Analysis of focused disease specific panels
Analysis of focused disease panels included those offered by Blueprint Genetics, GeneDx, Invitae, Molecular Vision Labs, and Prevention Genetics. These panels were chosen as they utilize next generation sequencing and are most commonly selected by our clinics due to their broad coverage. However, these panels do not represent a complete list of labs that offer excellent disease specific panels. Similar to the overarching IRD panel analysis, the genomic location and name of each disease gene was documented across each panel. This curated list was then compared to examine overlapping and unique genes across testing platforms.
Results
Genetic variant identification in targeted IRD panels.
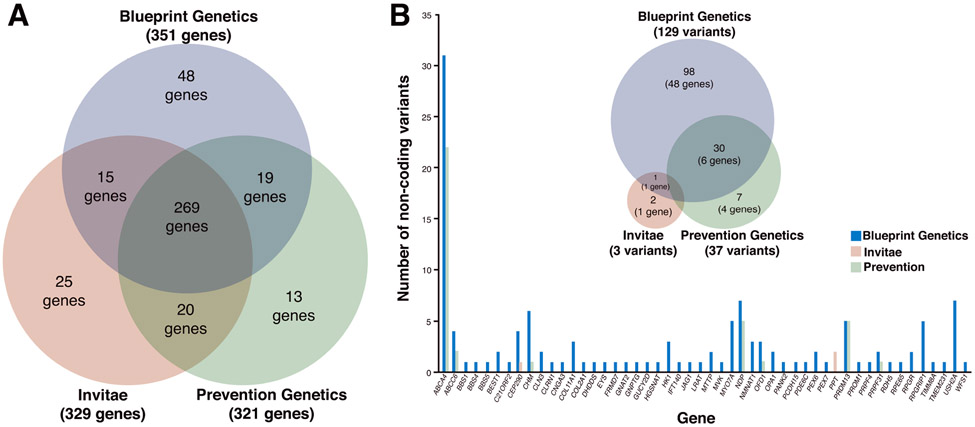
The three retinal dystrophy panels offered by Blueprint Genetics, Invitae, and Prevention Genetics together test 409 unique genes, of which 269 genes are tested by all three panels (Figure 2A). Although the top 20 genes known to cause over 70% of all IRDs are represented in the 269 common genes tested by all three panels, it should be noted that the coverage of RPGR in the Invitae panel is not appropriate for diagnosis of X-linked retinitis pigmentosa as only specific transcripts are assayed. It is important to understand genes that are unique to each panel. Blueprint Genetics has 48 unique genes, 37 of them are mitochondrial genes, which have been implicated in inherited eye disease26, and specifically in retinal degenerations27. The Invitae panel contains 25 unique genes with a subset of genes implicated in rare retinal degenerative phenotypes as well as albinism and related ocular disorders28. Prevention Genetics panel has 13 unique genes with a few responsible for rare forms of retinitis pigmentosa due to structural variants29. Overall, all three panels provide excellent coverage of retinal disease genes listed on RetNet7. There are 43 loci listed on RetNet that are not included in any of the panels, but these loci either have limited evidence for harboring retinal disease variants or are not known to cause an IRD (such as RB1, which causes retinoblastoma30).
Figure 2. Examination of genes included in retinal dystrophy panels offered by Blueprint Genetics, Invitae and Prevention Genetics.
(A) Of the 409 unique coding regions of genes assayed across the three different retinal dystrophy panels, there are 269 genes in common including the top 20 genes known to cause over 70% of IRDs. In the Blueprint Genetics Retinal Dystrophy panel, there are 48 unique genes of which 37 are mitochondrial genes. Prevention genetics has 13 unique genes with some indicated in rare forms of RP associated with structural variants. Invitae’s panel has 25 unique genes with a subset of those implicated in oculocutaneous albinism. (B) Of the 138 non-coding variants assayed across all platforms, 98 (across 49 genes) are unique to Blueprint, 2 are unique to Invitae in the gene PPT, whereas 7 (across 4 genes) are unique to Prevention Genetics, reflected in the proportionate Venn diagram. There is 1 variant in common between Blueprint Genetics and Invitae in the gene CEP290 and Blueprint and Prevention Genetics have 30 non-coding variants in common across 6 genes. The plot shows the number of non-coding variants in the targeted genes, with ABCA4 having the most variants.
Non-coding variant testing in IRDs.
An emerging consideration in the genetic diagnosis of IRDs is investigation of non-coding variants31. Non-coding variants can be considered pathogenic if it results in a loss or gain of function resulting in a disease phenotype. Unlike coding regions, in which the effects of a genetic variation can generally be predicted based on amino acid structure, such hallmarks of pathogenicity are limited for non-coding elements32. Prioritization and characterization of non-coding variants is particularly important in patients with an autosomal recessive IRD and whose genetic studies have revealed only one disease-causing coding variant. In such populations, non-coding intronic regions of the disease gene can harbor the other allelic variant and account for the missing heritability33. The three dystrophy panels described earlier are all based on an exome backbone and target the coding exons and adjacent 10-20 base pairs from the exon-intron boundary for each gene assayed. This limits their ability to capture deep intronic variants. To overcome this limitation, panels can include specifically targeted known pathogenic deep intronic variants in a subset of genes in their panels. For example, the Blueprint Genetics IRD panel captures 129 non-coding variants across 49 IRD genes, the Invitae IRD panel captures 3 non-coding variants across 2 genes, and the Prevention Genetics IRD panel captures 37 variants across 7 genes (Figure 2B). The gene with the most non-coding variants assayed was in adenosine triphosphate-binding cassette subfamily A, member 4 (ABCA4)34, which causes Stargardt disease35-37. This disease is the most common macular dystrophy that is inherited in an autosomal recessive fashion. Unsolved cases carrying no ABCA4 pathogenic alleles or one such allele can often be explained by the presence of deep intronic variants38,39. However, testing for these non-coding variants differs greatly among the panels, with Blueprint Genetics targeting 31 non-coding variants and Prevention Genetics targeting 22 non-coding variants (18 common to both panels), whereas Invitae does not provide coverage for any ABCA4 non-coding variants. For the USH2A gene, which accounts for greater than 75% of all cases of autosomal recessive Usher disease40, approximately 20% of patients with well-defined clinical features consistent with the disease have only one identifiable mutation in USH2A. Blueprint Genetics targets 7 non-coding variants whereas neither Invitae nor Prevention Genetics target any non-coding variants in USH2A.
Disease specific panels in IRDs.
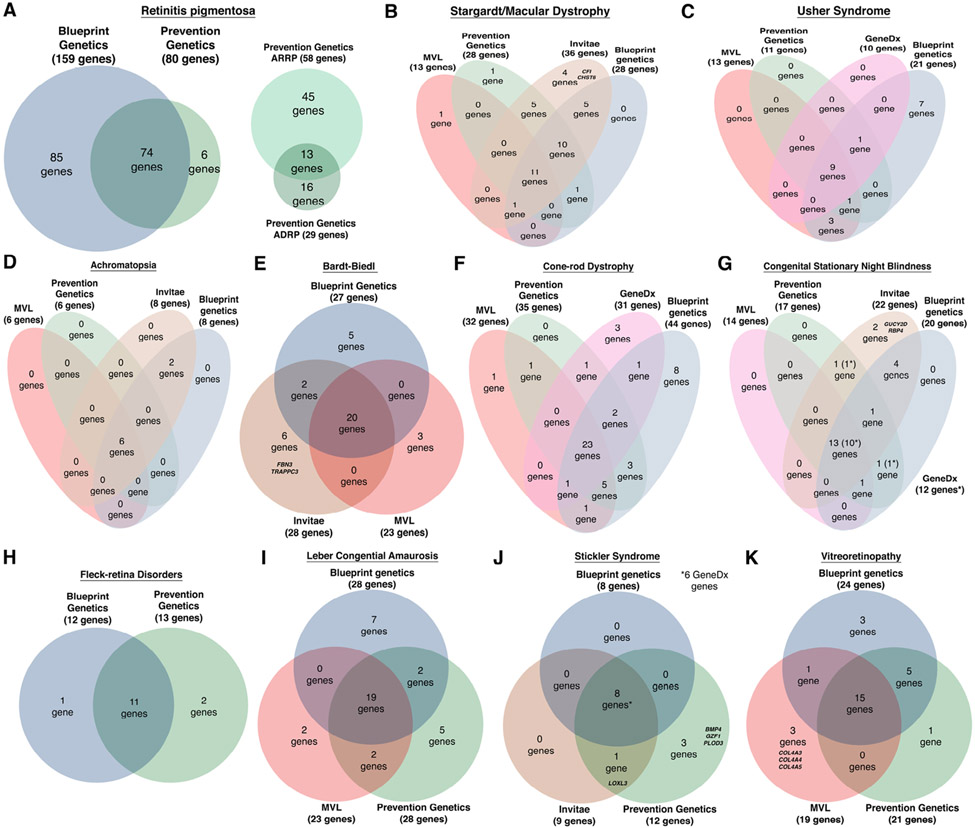
In patients with clear clinical phenotypes or known family history of a specific IRD, a subset of possible causative genes can be high on the clinician’s differential. In such cases, disease-specific panels are an option for the ordering provider. Whereas many companies provide disease specific panels, this work investigated those commonly ordered by our clinics: Blueprint Genetics has 13 IRD disease-specific panels, whereas Invitae has 3, Prevention Genetics has 13, GeneDx has 6, and MVL has 8. The panels, however, are composed of a varying number of genes unique to each panel (Figure 3). For retinitis pigmentosa41 (Figure 3A), the most common IRD42, Blueprint Genetics offers a more comprehensive targeted panel of 159 genes compared to that of 80 genes in Prevention Genetics. There is some variability in coverage of disease-specific panels as well. For example, Blueprint Genetics offers a panel to test for Usher disease (Figure 3C) that contains 21 genes including non-coding variants, 7 of which are not tested by any other panel, whereas the other 3 panels target fewer genes and do not contain any targets unique to their panel. It is important to note that some genes are included in a panel to aid in the diagnosis of phenotypically similar diseases, and not necessarily for the disease itself. An example is the inclusion of genes implicated in the peroxisome biogenesis disorder Heimler syndrome (PEX1, PEX6)43,44 in the Blueprint Genetics Usher syndrome panel. Interestingly, there are 4 genes in the Invitae targeted panels (Bardt-Biedl and Stargardt/Macular Dystrophy), 3 genes in a Prevention Genetics panel (Stickler syndrome), and 1 shared between the two in the Stickler syndrome panel that are not included in their more comprehensive IRD specific panels, possibly because these genes have been implicated as rare causes of disease. Of the platforms from GeneDx and MVL, only MVL targeted 3 genes in the Vitreoretinopathy panel that was not in the larger IRD panels offered by Blueprint Genetics, Invitae, or Prevention Genetics. These three collagen genes (COL4A3, COL4A4, and COL4A5) in the MVL panel were likely included due to their involvement in Alport syndrome to help distinguish this disorder.
Figure 3. Not all retinal disease panels are created equal.
A survey of different retinal disease panels highlight the differences that exist between the different platforms surveyed (Blueprint Genetics, GeneDx, Invitae, MVL, and Prevention Genetics). In the most prevalent IRD, retinitis pigmentosa (A), the larger Blueprint Genetics panel, as indicated by the proportionate Venn diagram, encompasses the majority of retinitis pigmentosa genes. There are panels specific to different inheritance patterns that exist with Prevention Genetics, but the overarching disease specific panels include these genes. Survey of focused panels for more common IRDs of (B) Stargardt and (C) Usher syndrome illustrate some specific platforms may be better suited for each disease. In Stargardt disease Invitae is the most comprehensive whereas for Usher Syndrome the Blueprint Genetics platform is most inclusive of disease genes. Other panels available for (D) Achromatopsia, (E) Bardt-Biedl, (F) Cone-rod dystrophy, (G) Congenital stationary night blindness (the 12 GeneDx genes are marked by asteriks in parenthesis in the diagram) , (H) Fleck-retina disorders, (I) Leber's congenital amaurosis, (J) Stickler syndrome, and (K) Vitreoretinopathy are shown. Interestingly, Invitae and Prevention Genetics disease specific panels included genes that were not in their larger retinal dystrophy panels. These 8 genes are listed by symbol in their respective disease specific diagram ((E) Bardt-Biedl, (B) Stargardt/Macular Dystrophy, (J) Stickler syndrome). The only other genes unique to these panels were the 3 collagen genes listed from MVL in the (K) Vitreoretinopathy panel.
Discussion
With the advent of gene therapy for biallelic RPE6545 variants implicated in Leber congenital amaurosis, knowledge of the specific disease-causing genetic variants in patients plays an increasingly important role in the diagnosis and management of disease. As clinical trials for other IRDs progress to treatment46,47, genetic testing will be essential to properly identify patients who may benefit from intervention. However, ordering genetic testing for IRDs is not straightforward, as evidenced by the multitude of panels available to providers. In addition to the IRD panels available from Blueprint Genetics, Invitae, and Prevention Genetics, there are larger panels for general ocular disorders offered by GeneDx and MVL. The MVL Vision Panel v6 includes 935 genes and the GeneDx Retinal Dystrophy Xpanded Panel consists of 779 genes. In addition to inherited retinal disease genes, these panels encompass other hereditary ocular conditions such as developmental eye diseases and anterior segment anomalies which are outside the scope of testing required for patients suspected to have an IRD. These larger panels may not add substantially to IRD diagnostics. For example, of the 935 genes in the MVL Vision panel v6, there are 284 retinal dystrophy genes, and only 5 are unique compared to those in Blueprint Genetics, Invitae, and Prevention Genetics panels. However, these 5 genes (CKAP4, CLN13, CLN14, DGKB, and GJB2) have not been implicated as retinal disease causing genes in humans. Thus, ordering larger gene panels does not necessarily equate to improved clinical diagnostic success and may increase the false discovery rate21. The main consideration should be accurate representation of genes known to cause IRD in humans. Panels offered by Blueprint Genetics, Invitae, and Prevention Genetics offer coverage of the 20 genes known to affect greater than 70% of those with IRDs.
Another consideration when ordering genetic testing is coverage of both coding and noncoding regions in IRD genes. The majority of human genetic variation falls within noncoding regions of the genome48 and genome-wide epigenomic maps have highlighted the emerging importance of cis-regulatory elements (CREs)49 in retinal development and diseases50-53. Moving forward there will be greater importance in screening pathogenic non-coding variants in IRDs to identify the variants in IRD patients whose genetic diagnoses are still a mystery. The IRD panels investigated in this study did include specific non-coding variants, albeit to varying degrees. The Blueprint Genetics panel had the most robust non-coding variant targeting with 129 unique variants across 49 genes. While machine learning approaches have been developed to interpret non-coding variants and possible pathogenic effects54 in the context of human disease55, these must be confirmed experimentally as the majority of non-coding variants have not been validated for having any functional consequence56. As more pathogenic variants are identified in noncoding regions, it will be important for these sequences to be validated before being incorporated into gene panels. Proper curation and incorporation of known pathogenic non-coding variants will be crucial in helping to definitively diagnose patients with autosomal recessive IRDs whose testing has only revealed monoallelic coding variants.
Another important consideration for ordering providers is to decide between ordering the larger IRD panels or focused disease-specific panels. The focused panels have significant variability and choosing a more robust one that includes coverage of disease-specific genes as well those of phenotypically similar diseases can help result in a clear diagnosis. Otherwise, the clinician risks an inconclusive test and the potential need to order a more comprehensive panel down the line. All large panels and genomic testing have the potential to identify one or more variants of uncertain significance (VUS). These are non-diagnostic, and should not be interpreted as causative of an IRD. However, the clinician must also be cognizant of the criteria for reclassification of VUSs, and that over time, most VUSs will be reclassified as benign/likely benign, but a few will be reclassified as likely pathogenic/pathogenic57. Clinicians should also keep in mind that the presentation of IRDs can be heterogeneous in families. There have been reports of multiple IRDs in the same family with similar phenotypic presentation. This has been seen in families with RP with causative mutations in different IRD genes58,59. Even more complex are cases where a single family has causative mutations in multiple different genes with distinct inheritance patterns60. Thus, it is possible that the same disease-causing genetic variant within one family can manifest in a wide variety of clinical phenotypes or even more rarely, multiple genetic diseases may exist within the same family. Therefore, if the genetic diagnosis is ambiguous, it may be preferable to order the more encompassing IRD panels rather than focused diseased-specific panels. Genetic testing for patients with presumed genetically caused retinal degeneration performed by a physician or genetic counselor who is knowledgeable about IRDs aligns with the stance on genetic testing by the AAO11,61.
This paper details the diversity of testing platforms currently available to clinicians. Patients with clinical features suggestive of an IRD should be offered clinical genetic testing. In addition, IRD patients with a negative or uncertain genetic test result in whom a genetic etiology is supported by their clinical findings, age of onset in childhood or young adulthood, or with a positive family history should be considered for updated genetic testing. Our practice is to recommend genetic re-evaluation in 3 to 5 years after negative results as there is evidence in this time span the diagnostic yield doubles with re-examination62 in line with the rapid pace of improvement in knowledge of genetic causes of disease and advances in sequencing technology. However, in addition to weighing genetic coverage between panels, clinicians should also consider out-of-pocket costs to patients when deciding on appropriate testing. Many insurance companies will not cover genetic testing of IRDs, and while patients and families have the desire to pursue testing, the cost may be prohibitive. For example, Blueprint Genetics has a list price of $1,650 for their large panels, such as the Retinal Dystrophy panel, and Prevention Genetics quotes a list price of $1,740 for their comprehensive IRD panel. Of note, many genetic testing labs have their own form of financial assistance programs or policies that have been established with the understanding that genetic testing is expensive. These programs can greatly reduce the cost of many of these panels to under $400. One commercial lab in particular, Invitae, offers a reduced self-pay price of $250 for all of their diagnostic panels, regardless of financial assistance status or need. There are opportunities for sponsored panels that offer no-cost genetic testing, but there are eligibility criteria that must be met for these panels. These criteria include no previous comprehensive panel, exome, or genome test in the recent few years or a diagnosis of a non-genetic ocular or retinal condition in the patient to explain their phenotypuc presentation such as optic neuropathy, age-related macular degeneration or diabetic eye disease. It also is important to note that sponsored genetic testing programs are limited to the cost of genetic testing, and do not cover the cost of the associated medical or genetic counseling visit, which is billed separately. However, many of these sponsored programs have the option of genetic counseling services through contracted companies. It is important to consider these resources when discussing the best testing options for IRD patients. If insurance authorization is being pursued, thorough and accurate documentation of the patient’s phenotype (visual acuity, refraction, dilated fundus exam and imaging findings, electrophysiological findings, etc) is needed in order to claim medical necessity for the testing. However, it is important to note that insurance authorization for an IRD panel, or any type of genetic testing, does not necessarily mean the testing is fully covered for the patient.
Regardless of the choice of panel, an essential resource during the process of genetic testing is partnering with genetic counselors. Genetic testing should be provided only in the context of pre- and post-testing genetic counseling, and should be done by clinical providers with expertise in clinical genetics and genetic counseling63. Some ophthalmologists have undergone fellowship training to gain this expertise, others refer to centers of excellence which have access to Genetics clinics for consultation with certified master’s level genetic counselors and physicians who are board-certified in Clinical Genetics and Genomics. Because of the time and expertise required for counseling, and because genetic testing requires understanding and applying genetic concepts not required in all ophthalmology training programs including inheritance, penetrance, variable expression, segregation, somatic mosaicism, and variant interpretation and reinterpretation, patients benefit from interdisciplinary care with genetic professionals. In addition, patients with complex, multi-organ syndromic retinal dystrophies such as Bardet-Biedl syndrome, Joubert syndrome, or Usher syndrome, are often followed in Pediatric Genetics clinics as their medical home. Thus, it is important to ensure there is proper genetic counseling available to the patient to not only discuss the post-test results and implications, but to also meet pre-test to prepare the patient and family for the possible results and answer any questions before proceeding with the test. The significant burden on IRD patients and their families can be improved with not only greater access to testing, but also to genetic counseling64. Patients and families value the role of genetic counseling during diagnosis65. As the field progresses, additional causative genes and specific regions of interest, such as in non-coding regions, will be identified. The analyzed target genes and non-coding variants assayed in IRD panels will have to be updated. This work provides a snapshot in time of the state of IRD testing platforms. Current online resources, such as the NCBI Genetic Testing Registry lists available genetic test by IRD or a specific gene, but does not provide information about commonality or uniqueness between different panels. As testing evolves and panels expand, we will continuously update this online resource to provide the community with an up-to-date snapshot of the state of genetic testing for IRDs (Online Resource).
The next step in the field of genetic testing for IRDs is the more widespread use of GS. As GS becomes more accessible in a clinical setting, we will be able to overcome the major limitation in exome-based sequencing approaches in resolving structural or intronic variants as well as regulatory regions to explain causative changes in rare diseases66 to potentially provide a superior solve rate of genetic disorders22,67. This effort will require continued prioritization and functional annotation of non-coding variants in IRD genes. The key will be proper annotation of variants as potentially pathogenic as it can then provide diagnoses to previously unsolved cases. Refined variant classification has been shown to provide molecular diagnoses in previously undiagnosed pediatric patients68. The rise of artificial intelligence is a potential solution to the big data problem in genomics and can enhance detection and diagnosis of disease69. Integrating DNA sequencing data with electronic health records can elucidate loss-of-function variants and uncover undiagnosed phenotype-genotype associations of rare disease variants. In this way, very rare causes of IRDs can be studied in aggregate and not in a univariate fashion70 in the future.
Financial Support:
An unrestricted grant from Research to Prevent Blindness, NIH grant (5R01EY026030) to JRC
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: No conflicting relationship exists for any author.
Clinical genetic testing of inherited retinal disease (IRD) patients is essential for proper diagnosis and management. This work details the comprehensive and disease-specific IRD panels available to highlight the commonalities and differences among these tests.
References
- 1.Stone EM. Genetic testing for inherited eye disease. Arch Ophthalmol. 2007;125:205–212. [DOI] [PubMed] [Google Scholar]
- 2.Bessant DA, Ali RR, Bhattacharya SS. Molecular genetics and prospects for therapy of the inherited retinal dystrophies. Curr Opin Genet Dev. 2001;11:307–316. [DOI] [PubMed] [Google Scholar]
- 3.Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–315. [DOI] [PubMed] [Google Scholar]
- 4.Solebo AL, Teoh L, Rahi J. Epidemiology of blindness in children. Arch Dis Child. 2017;102:853–851. [DOI] [PubMed] [Google Scholar]
- 5.Hamblion EL, Moore AT, Rahi JS, British Childhood Onset Hereditary Retinal Disorders Network. Incidence and patterns of detection and management of childhood-onset hereditary retinal disorders in the UK. Br J Ophthalmol. 2012;96:360–365. [DOI] [PubMed] [Google Scholar]
- 6.Khan AO. PHENOTYPE-GUIDED GENETIC TESTING OF PEDIATRIC INHERITED RETINAL DISEASE IN THE UNITED ARAB EMIRATES. Retina. 2020; 40: 1829–1837. [DOI] [PubMed] [Google Scholar]
- 7.Daiger S, Rossiter BJF, Greenberg J, et al. Data services and software for identifying genes and mutations causing retinal degeneration. Invest Ophthalmol Vis Sci. 1998;39. [Google Scholar]
- 8.Pontikos N, Arno G, Jurkute N, et al. Genetic Basis of Inherited Retinal Disease in a Molecularly Characterized Cohort of More Than 3000 Families from the United Kingdom. Ophthalmology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz KE, Reeves MJ, Gagadam S, et al. Genetic testing for inherited eye conditions in over 6,000 individuals through the eyeGENE network. Am J Med Genet C Semin Med Genet. 2020;184:828–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splinter K, Adams DR, Bacino CA, et al. Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N Engl J Med. 2018;379:2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan JL, Bernstein PS, Birch DG, et al. Recommendations on Clinical Assessment of Patients with Inherited Retinal Degenerations-2016. 2016. [Google Scholar]
- 12.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider VA, Graves-Lindsay T, Howe K, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisschuh N, Mayer AK, Strom TM, et al. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS One. 2016;11:e0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisschuh N, Obermaier CD, Battke F, et al. Genetic architecture of inherited retinal degeneration in Germany: A large cohort study from a single diagnostic center over a 9-year period. Hum Mutat. 2020;41:1514–1527. [DOI] [PubMed] [Google Scholar]
- 17.Dillon OJ, Lunke S, Stark Z, et al. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet. 2018;26:644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller DT, Lee K, Gordon AS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23:1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellingford JM, Barton S, Bhaskar S, et al. Whole Genome Sequencing Increases Molecular Diagnostic Yield Compared with Current Diagnostic Testing for Inherited Retinal Disease. Ophthalmology. 2016;123:1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20:1122–1130. [DOI] [PubMed] [Google Scholar]
- 21.Stone EM, Andorf JL, Whitmore SS, et al. Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmology. 2017;124:1314–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manickam K, McClain MR, Demmer LA, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021. [DOI] [PubMed] [Google Scholar]
- 23.Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med. 2018;379:1353–1362. [DOI] [PubMed] [Google Scholar]
- 24.Shaberman B, Durham T. The Foundation Fighting Blindness Plays an Essential and Expansive Role in Driving Genetic Research for Inherited Retinal Diseases. Genes. 2019;10:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao PY, Branham K, Schlegel D, et al. Association of No-Cost Genetic Testing Program Implementation and Patient Characteristics With Access to Genetic Testing for Inherited Retinal Degenerations. JAMA Ophthalmol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu-Wai-Man P, Newman NJ. Inherited eye-related disorders due to mitochondrial dysfunction. Hum Mol Genet. 2017;26:R12–R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birtel J, von Landenberg C, Gliem M, et al. Mitochondrial Retinopathy. Ophthalmol Retina. 2021. [DOI] [PubMed] [Google Scholar]
- 28.Grønskov K, Ek J, Brondum-Nielsen K. Oculocutaneous albinism. Orphanet J Rare Dis. 2007;2:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bruijn SE, Fiorentino A, Ottaviani D, et al. Structural Variants Create New Topological-Associated Domains and Ectopic Retinal Enhancer-Gene Contact in Dominant Retinitis Pigmentosa. Am J Hum Genet. 2020;107:802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, Lupski JR. Non-coding genetic variants in human disease. Hum Mol Genet. 2015;24:R102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, He L, Goggin SM, et al. High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. Nat Commun. 2018;9:5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Scipio M, Tavares E, Deshmukh S, et al. Phenotype Driven Analysis of Whole Genome Sequencing Identifies Deep Intronic Variants that Cause Retinal Dystrophies by Aberrant Exonization. Invest Ophthalmol Vis Sci. 2020;61:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allikmets R, Singh N, Sun H, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Starqardt macular dystrophy. Nature Genetics. 1997;15:236–246. [DOI] [PubMed] [Google Scholar]
- 35.Strauss RW, Ho A, Muñoz B, et al. The Natural History of the Progression of Atrophy Secondary to Stargardt Disease (ProgStar) Studies: Design and Baseline Characteristics: ProgStar Report No. 1. Ophthalmology. 2016;123:817–828. [DOI] [PubMed] [Google Scholar]
- 36.Fishman GA, Stone EM, Grover S, et al. Variation of Clinical Expression in Patients With Stargardt Dystrophy and Sequence Variations in the ABCRGene. Arch Ophthalmol. 1999;117:504–510. [DOI] [PubMed] [Google Scholar]
- 37.Fujinami K, Lois N, Mukherjee R, et al. A longitudinal study of Stargardt disease: quantitative assessment of fundus autofluorescence, progression, and genotype correlations. Invest Ophthalmol Vis Sci. 2013;54:8181–8190. [DOI] [PubMed] [Google Scholar]
- 38.Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Human Molecular Genetics. 2013;22:5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sangermano R, Garanto A, Khan M, et al. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet Med. 2019;21:1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baux D, Larrieu L, Blanchet C, et al. Molecular and in silico analyses of the full-length isoform of usherin identify new pathogenic alleles in Usher type II patients. Hum Mutat. 2007;28:781–789. [DOI] [PubMed] [Google Scholar]
- 41.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. [DOI] [PubMed] [Google Scholar]
- 42.Chen T-C, Huang D-S, Lin C-W, et al. Genetic characteristics and epidemiology of inherited retinal degeneration in Taiwan. NPJ Genom Med. 2021;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratbi I, Falkenberg KD, Sommen M, et al. Heimler Syndrome Is Caused by Hypomorphic Mutations in the Peroxisome-Biogenesis Genes PEX1 and PEX6. Am J Hum Genet. 2015;97:535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaki MS, Heller R, Thoenes M, et al. PEX6 is Expressed in Photoreceptor Cilia and Mutated in Deafblindness with Enamel Dysplasia and Microcephaly. Hum Mutat. 2016;37:170–174. [DOI] [PubMed] [Google Scholar]
- 45.Maguire AM, Russell S, Wellman JA, et al. Efficacy, Safety, and Durability of Voretigene Neparvovec-rzyl in RPE65 Mutation-Associated Inherited Retinal Dystrophy: Results of Phase 1 and 3 Trials. Ophthalmology. 2019;126:1273–1285. [DOI] [PubMed] [Google Scholar]
- 46.Lam BL, Davis JL, Gregori NZ, et al. Choroideremia Gene Therapy Phase 2 Clinical Trial: 24-Month Results. Am J Ophthalmol. 2019;197:65–13. [DOI] [PubMed] [Google Scholar]
- 47.MacLaren RE, Groppe M, Barnard AR, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. The Lancet. 2014;383:1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nord AS, Blow MJ, Attanasio C, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andzelm MM, Cherry TJ, Harmin DA, et al. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghiasvand NM, Rudolph DD, Mashayekhi M, et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:518–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nathans J, Davenport CM, Maumenee IH, et al. Molecular genetics of human blue cone monochromacy. Science. 1989;245:831–838. [DOI] [PubMed] [Google Scholar]
- 53.Cherry TJ, Yang MG, Harmin DA, et al. Mapping the cis-regulatory architecture of the human retina reveals noncoding genetic variation in disease. Proc Natl Acad Sci U S A. 2020;117:9001–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells A, Heckerman D, Torkamani A, et al. Ranking of non-coding pathogenic variants and putative essential regions of the human genome. Nat Commun. 2019;10:5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moyon L, Berthelot C, Louis A, et al. Classification of non-coding variants with high pathogenic impact. bioRxiv. 2021:2021.05.03.442347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biggs H, Parthasarathy P, Gavryushkina A, Gardner PP. ncVarDB: a manually curated database for pathogenic non-coding variants and benign controls. Database . 2020;2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fung JLF, Yu MHC, Huang S, et al. A three-year follow-up study evaluating clinical utility of exome sequencing and diagnostic potential of reanalysis. NPJ Genom Med. 2020;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dockery A, Carrigan M, Wynne N, et al. A Novel FLVCR1 Variant Implicated in Retinitis Pigmentosa. Adv Exp Med Biol. 2019;1185:203–207. [DOI] [PubMed] [Google Scholar]
- 59.Jones KD, Wheaton DK, Bowne SJ, et al. Next-generation sequencing to solve complex inherited retinal dystrophy: A case series of multiple genes contributing to disease in extended families. Mol Vis. 2017;23:470–481. [PMC free article] [PubMed] [Google Scholar]
- 60.Birtel J, Gliem M, Hess K, et al. Comprehensive Geno- and Phenotyping in a Complex Pedigree Including Four Different Inherited Retinal Dystrophies. Genes . 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stone EM, Aldave AJ, Drack AV, et al. Recommendations for genetic testing of inherited eye diseases: report of the American Academy of Ophthalmology task force on genetic testing. Ophthalmology. 2012;119:2408–2410. [DOI] [PubMed] [Google Scholar]
- 62.Liu P, Meng L, Normand EA, et al. Reanalysis of Clinical Exome Sequencing Data. N Engl J Med. 2019;380:2478–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li AS, MacKay D, Chen H, et al. Challenges to Routine Genetic Testing for Inherited Retinal Dystrophies. Ophthalmology. 2019;126:1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galvin O, Chi G, Brady L, et al. The Impact of Inherited Retinal Diseases in the Republic of Ireland (ROI) and the United Kingdom (UK) from a Cost-of-Illness Perspective. Clin Ophthalmol. 2020;14:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strait S, Loman R, Erickson L, DeBenedictis M. Inherited retinal degeneration current genetics practices - a needs assessment. Ophthalmic Genetics. 2020;41:533–538. [DOI] [PubMed] [Google Scholar]
- 66.Turro E, Astle WJ, Megy K, et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.James KN, Clark MM, Camp B, et al. Partially automated whole-genome sequencing reanalysis of previously undiagnosed pediatric patients can efficiently yield new diagnoses. NPJ Genom Med. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park J, Lucas AM, Zhang X, et al. Exome-wide evaluation of rare coding variants using electronic health records identifies new gene-phenotype associations. Nat Med. 2021;27:66–12. [DOI] [PMC free article] [PubMed] [Google Scholar]