Abstract
MUTYH carriers have an increased colorectal cancer (CRC) risk in case-control studies, with loss of heterozygosity (LOH) as the presumed mechanism. We evaluated cancer risk among carriers in a prospective, population-based cohort of older adults. Additionally, we assessed if cancers from carriers demonstrated mutational signatures (G:C>T:A transversions) associated with early LOH. We calculated incident risk of cancer and CRC among 13,131 sequenced study participants of the ASPirin in Reducing Events in the Elderly (ASPREE) cohort, stratified by sex and adjusting for age, smoking, alcohol use, BMI, polyp history, history of cancer, and aspirin use. MUTYH carriers were identified among 13,033 participants in The Cancer Genome Atlas and International Cancer Genome Consortium, and somatic signatures of cancers were analyzed. Male MUTYH carriers demonstrated an increased risk for overall cancer incidence (multivariable HR 1.66, 95% CI [1.03, 2.68]; P = 0.038) driven by increased CRC incidence (multivariable HR 3.55, 95% CI [1.42, 8.78]; P = 0.007), as opposed to extracolonic cancer incidence (multivariable HR 1.40, 95% CI [0.81, 2.44]; P = 0.229). Female carriers did not demonstrate increased risk of cancer, CRC, or extracolonic cancers. Analysis of mutation signatures from cancers of MUTYH carriers revealed no significant contribution toward early mutagenesis from widespread G:C>T:A transversions among gastrointestinal epithelial cancers. Among cancers from carriers, somatic transversions associated with base-excision repair deficiency are uncommon, suggestive of diverse mechanisms of carcinogenesis in carriers compared to those who inherit biallelic MUTYH mutations.
Keywords: MUTYH, somatic signature, colorectal cancer, 8-oxoG
Introduction.
The creation of 8-Oxoguanine (8-oxoG) after oxidative damage to guanine is one of the most common forms of DNA damage from reactive oxygen species. 8-oxoG binds with adenine and can result in G:C>T:A transversion mutations if not corrected (1). The MYH DNA glycosylase enzyme, encoded by MUTYH, is a critical component of base excision repair (BER) of 8-oxoG related mutagenesis (2). Inheritance of two inactive alleles of MUTYH results in a colonic polyposis phenotype (MUTYH-associated polyposis, MAP) associated with a high lifetime risk of colorectal cancer (CRC), as well as hypermutant cancers occurring in other tissues such as the stomach and duodenum (3-6).
Given the 1-2% prevalence of MUTYH carriers among the European population, multiple investigations have been performed to estimate the cancer risk among monoallelic carriers (7,8). These studies have identified a modest increase in CRC incident risk among older MUTYH carriers, or those with family histories of CRC (9,10). However, previous prospective and population-based studies represent case-control analyses derived from cancer registries and/or kin-cohorts (11,12). Often, such genetic studies overestimate risk estimates through ascertainment bias (13). Risk estimates presently remain lacking from population-based, prospective cohorts of unrelated individuals yet to develop cancer. As a result, present National Comprehensive Cancer Network (NCCN) guidelines only recommend more frequent screening for monoallelic carriers with personal or first-degree family histories of CRC (14). In addition, many practitioners default to current United States Preventative Task Force (USPTF) guidelines for older adults, which suggest individualized recommendations according to general health and prior screening results (15). Thus, robust risk estimates to inform screening strategies in older carriers are lacking, but needed given the increased CRC risk and diminished responses to DNA-damage in older persons (16,17). This uncertainty also extends to extracolonic cancer risk among older carriers, with several studies demonstrating a non-statistically significant trend for increased risk for several other tumor types (18,19).
An unresolved aspect remains; the mechanism by which monoallelic MUTYH mutations predispose to cancer. Tumors of individuals with biallelic mutations arise from accumulation of genome-wide somatic G:C>T:A mutations in oncogenes and tumor suppressors. Among carriers, it remains unclear whether loss of heterozygosity (LOH) mediates this risk, and if these mechanisms are uniform among different tumor types. With the increasing availability of cancer genomic sequencing data, analyses of single base substitutions (SBS) have identified mutational signatures associated with MUTYH deficiency (1,20). Colorectal cancers lacking MUTYH function demonstrate an abundance of COSMIC SBS signature 36, a measure of G:C>T:A mutagenesis found in colorectal and extracolonic cancers from those with biallelic MUTYH loss. COSMIC SBS signature 18, which is closely related to COSMIC Signature 36, has been noted in extracolonic malignancies possessing an enrichment of G:C>T:A transversions in the setting of NpCpA or NpCpT sequences due to oxidative damage (21). Somatic LOH has been reported in case series from tumor samples of MUTYH carriers affected by colorectal cancer, PNET, adrenocortical carcinoma, pancreatic ductal adenocarcinoma, and breast cancer, resulting in detection of COSMIC Signatures 18 and 36 (18,20,22).
To address some of these outstanding questions regarding monoallelic MUTYH risk and mechanisms of tumor evolution, we performed targeted high coverage germline DNA sequencing of a large population-based prospective cohort and evaluated the risk of monoallelic pathogenic mutations in MUTYH on cancer incidence. We analyzed Australian participants enrolled in the ASPirin in Reducing Events in the Elderly (ASPREE) clinical trial who represent healthy, community-dwelling individuals age ≥ 70 without dementia, disability, or pre-existing disease expected to cause mortality within 5 years of enrollment (23). To elucidate possible mechanisms by which cancers from MUTYH monoallelic carriers evolved, we calculated SBS signatures from an initial pool of 13,033 cancers (The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC)) and compared the relative contributions of SBS18 and SBS36 among monoallelic MUTYH carriers and those without such germline alterations.
Materials and Methods.
Study Population.
Details of the Aspirin in Reducing Events in the Elderly (ASPREE) cohort and clinical trial have been described previously (23,24). Briefly, ASPREE was a double-blind, randomized, placebo-controlled trial aimed at determining the effect of low-dose aspirin on all-cause mortality, disability-free survival, and cardiovascular events in healthy older (≥70 years of age) individuals (23-26). A total of 19,114 individuals in the ASPREE study were followed prospectively after enrollment and monitored for a number of primary and secondary outcomes, including cancer incidence and cancer-related mortality. Demographic, lifestyle, clinical and other health data from ASPREE participants were collected at baseline and at annual in-person study visits. At study enrollment and updated annually, participants provided self-reported personal medical history related to colon polyps (consisting of yes/no/unsure and number of polyps), cancer history (consisting of yes/no/unsure and cancer type), and cancer screening tests (by type).
Study participants were enrolled from March 2010 through December 2014, with follow-up data collected until June 2017 (mean follow-up 4.7 years). Of the total cohort 13,806 Australian study participants consented for genetic study, of those 13,131 germline DNA samples were available for sequencing (via stored biospecimens collected through the ASPREE Healthy Ageing Biobank). Differences between the sequenced cohort and the larger ASPREE cohort are presented in Supplementary Table S1. This study was approved by the human research ethics committee at the Alfred Hospital in Melbourne, Australia in accordance with Declaration of Helsinki. Informed written consent was obtained for all enrolled study participants.
Genomic Sequencing and Variant Calling of ASPREE Participants.
Sample DNA was diluted to 5ng/μL and libraries were prepared with the included Ion AmpliSeq Library Kit Plus reagents and the Ion Xpress Barcode Adapters 1-16 Kit (Thermo Fisher Scientific # 4471250) to barcode each pair of samples, following standard manufacturer’s instructions. Briefly, 10 ng of DNA was added to two master amplicon mixes, one for each primer pool of the DNA panel and the two primer pool reactions 54 were amplified in different wells of the same plate following the specified cycling conditions in the user guide. Following PCR amplification, the DNA amplification reactions from the two DNA primer pool reactions were56 combined in wells and processed as follows. The primers were partially digested with the proprietary FuPa enzyme and each sample was barcoded with a unique IonExpress barcode. Finally, a 1.5X bead purification was performed with Agencourt AMPure XP Reagent (Beckman Coulter, Brea, CA, #A63880) following the instructions in the protocol to clean up the sample and remove adapter dimers. At the end of the cleanup protocol, the samples were eluted and amplified with a master mix of library amplification mix and Equalizer primers provided in the Ion Library Equalizer Kit (Thermo Fisher Scientific, #4482298). The amplified libraries were then incubated and washed using Equalizer kit reagents following the protocol, resulting in all the libraries normalized to ~100pMol.
After priming the Ion Torrent chip per manufacturer’s protocol, samples were sequenced on the Ion Torrent S5TM XL System with library read length set at 400 bp and 850 flows per chip, with all other instrument settings set to the manufacturer’s default for the Ion 510/520/530 Kit. Analyses of sequencing raw data were performed with Ion Torrent Suite (version 5.6.0) using 81 the “coverageAnalysis,” “sampleID,” “variantCaller” (with somatic/low stringency settings for the “variantCaller”), and custom BED file plugins, with all other settings for the run report set to the manufacturer’s default.
Torrent Suite 5.2 on Ion Torrent S5XL produced raw reads (average 200x coverage) and exported them as fastQ format. These raw reads on a per sample basis, were aligned to the human genome reference version GRCh37. Aligned bam files were passed onto the Torrent Variant Calling Suite for low-stringency variant calls. Variant calling was performed on each sample separately, which produced a single sample VCF file. Post variant calling, all per-sample VCF files were merged using bcftools merge option to produce a single multi-sample vcf file. Variants were flagged for average quality scores <30 across the dataset, low reads (<50x), minor allele frequency (MAF) and/or homozygous genotype frequency >10x higher than gnomAD (Non-Finish Europeans), visible evidence of misalignment and/or errors around the variant site, evidence of allele-specific amplification bias, imbalanced stand coverage or variant allele frequency (<25%), direct adjacency to homopolymers, and evidence of multi-allelic base calls. Variants with three or more flags were excluded as technical sequencing artifacts. Variants called as borderline were flagged for validation by Sanger sequencing. Study participants harboring rare, loss-of-function variants (nonsense, splice-site, or frameshift mutations presumed to cause nonsense mediated decay) or missense variants labeled as pathogenic in ClinVar were considered as MUTYH carriers.
Identification of Pathogenic Germline Mutations Among TCGA and ICGC Samples.
BAM slices encompassing the MUTYH gene were individually downloaded, and analyzed with the Genome Analysis Toolkit Best Practices.(27) TCGA samples were analyzed with the reference genome GRCh38, while ICGC were analyzed with GRCh37. Annotation was performed using SNPeff.(28) Loss-of-function variants were defined as nonsense or frameshift mutations occuring 50 bp prior to the last exon-exon junction. Pathogenic missense variants were identified by those annotated in ClinVar.(29) All qualifying variants were inspected through IGV to ensure no sequencing artifacts or homopolymer errors were evident.(30)
Single Base Substitution Signature Analyses.
Somatic mutational signatures were calculated from whole-exome and whole-genome sequencing data processed by the MuTect somatic variant caller (31). Mutect2 processed VCF files of The Cancer Genome Atlas (TCGA) samples were downloaded from the National Cancer Institute Genomic Data Commons, and International Cancer Genome Consortium (ICGC) samples were downloaded from the Ontario Institute for Cancer Research Collaboratory Repository and analyzed through a non-negative matrix factorization algorithm to decompose mutational spectra. The DeconstructSigs package in R version 3.4 was used to compare components in the context of the reference signatures identified from the Wellcome Trust Sanger Institute (WTSI) Mutational Signature Framework version 3 (32,33). For TCGA and ICGC samples the germline variants in MUTYH were called from BAM file slices of the gene downloaded from their respective repository. The tumor types analyzed, and their respective repositories, are presented in Supplementary Table S2. OncoKB was used to identify catalogued driver somatic mutations that have been previously described as oncogenic (34).
Statistical Analysis.
Comparison of continuous and categorical variables between MUTYH gene variant carriers and non-carriers were tested with the Student’s t-test and Chi-squared test, respectively. Odds Ratios (OR) and 95% CIs were calculated for the association between pathogenic MUTYH mutations and history of all cancers, colonic polyps, and colorectal cancer. The crude and the adjusted OR were estimated using univariate and multivariable logistic regression analysis. Hierarchical logistic regression methods were used to explore the associations between history of prior cancers and MUTYH carrier status. Model 1 included pathogenic MUTYH variant status as a risk factor. Model 2 included pathogenic MUTYH variant status, as well as age, sex, smoking status, and alcohol use.
Incidence of any cancer (excluding non-melanoma skin cancers) and colorectal cancer were calculated by dividing the number of individuals who had developed the incident case by years at risk during the study follow-up. Factors associated with incident case were analyzed with Cox regression model, which included only those factors that were significant in the univariate analysis. We used Schoenfeld residual tests to evaluate the proportional hazard assumption and checked interactions between variables. This model generates regression coefficients for the independent variables, the exponents of which reflect hazard ratios. Model 1 consisted of a univariate analysis of pathogenic MUTYH variant status stratified by sex. Model 2 also incorporated age, smoking status, and alcohol use; Model 3 additionally incorporated baseline BMI and randomization to aspirin use; and Model 4 further incorporated history of cancer (for all cancer and extracolonic cancers), history of CRC (for incident CRC), and history of colonic polyp (for incident CRC). Aspirin use was incorporated into the latter models given that the results of ASPREE demonstrated an increase in cancer-related mortality associated with randomization to aspirin (23). All epidemiological data were analyzed with Stata software, version 15.1 and 2-sided P values less than .05 were considered statistically significant.
Outliers for SBS18 and SBS36 were prospectively designated as cancers with either greater than 50% of mutational fractions attributable to SBS18/SBS36, or with contributions of these signatures within the top quartile of tumor type and absolute contribution of at least 10%.
Data Availability.
The Cancer Genome Atlas dataset is available through dbgap accession phs000178.v11.p8 . International Cancer Genome Consortium datasets can be accessed through https://daco.icgc.org/ The human sequence data generated in this study are not publicly available due to patient privacy requirements but are available upon reasonable request from the corresponding authors. Other data generated in this study are available within the article and its supplementary data files.
Results.
Characteristics of Study Participants
Two-hundred and sixteen individuals among the 13,131 sequenced study participants (1.64%) harbored monoallelic pathogenic variants in MUTYH as detailed in Supplementary Table S3. No individuals with biallelic pathogenic mutations were discovered. No monoallelic carriers harbored concurrent mutations in Lynch Syndrome (35). Table 1 gives the characteristics of sequenced participants stratified by monoallelic MUTYH carrier status, which were balanced among carriers and non-carriers. Similar percentages were observed for carriers and non-carriers with prior history of colonic polyps (16.7% vs. 20.2%), prior history of colon cancer (1.8% vs. 2.6%), and prior history of any cancer (19.4% vs. 19.8%). Given the varied colorectal and extracolonic cancer risk estimates up to the age of 70 previously reported for MUTYH carriers, we performed a multivariable logistic regression analysis adjusting for age, sex, smoking status, alcohol use, and baseline BMI as presented in Supplementary Table S4. Despite this adjustment, no significant differences were observed between carriers and non-carriers for prior histories of colonic polyps, past history of colorectal cancer, or any cancer type.
Table 1:
Characteristics of ASPREE Participants by MUTYH Carrier Status (n=13,131).
| Clinical Characteristic |
MUTYH non- carriers (n=12,915) |
MUTYHcarriers (n=216) |
P value |
|---|---|---|---|
| Age at randomization (years), mean (SD) | 75.0 (4.2) | 74.7 (3.9) | 0.202 |
| Baseline BMI | 28.0 (4.5) | 28.5 (4.7) | 0.104 |
| Gender, n (%) | |||
| Female | 6938 (53.7) | 119 (55.1) | 0.688 |
| Male | 5977 (46.3) | 97 (44.9) | |
| Baseline smoking status, n (%) | |||
| Never | 7190 (55.7) | 124 (57.4) | 0.860 |
| Current | 403 (3.1) | 7 (3.2) | |
| Former | 5322 (41.2) | 85 (39.3) | |
| Baseline alcohol use, n (%) | |||
| Never | 2014 (15.6) | 37 (17.1) | 0.526 |
| Current | 10291 (79.7) | 166 (76.9) | |
| Former | 610 (4.7) | 13 (6.0) | |
| Patient’s history of all cancer type, n (%) | |||
| No | 10312 (80.2) | 174 (80.6) | 0.896 |
| Yes | 2546 (19.8) | 42 (19.4) | |
| Patient’s history of colorectal cancer, n (%) | |||
| No | 12522 (97.4) | 212 (98.2) | 0.489 |
| Yes | 335 (2.6) | 4 (1.8) | |
| Patient’s history of colon polyp, n (%) | |||
| No | 10167 (79.8) | 179 (83.3) | 0.210 |
| Yes | 2574 (20.2) | 36 (16.7) | |
| Aspirin use, n (%) | 6421 (49.7) | 117 (54.2) | 0.195 |
| Total person-years (all cancers) | 55635 | 897 | NA |
| Total person-years for Colorectal Cancer | 57206 | 937 | NA |
Carriers were defined as those individuals harboring a loss-of-function mutation (nonsense, splice-site, or frameshift) or missense mutation labeled as pathogenic in ClinVar. Of note, 0.4% (58/13,131) participants had missing data in baseline BMI and history of colorectal cancer, 0.4% (57/13, 131) in the patients' history of cancer, and 1.3% (175/13,131) in history of colon polyp.
Incident Cancer Risk Among MUTYH carriers.
During the study follow-up period, 1,328 incident cancers developed among 10% of sequenced study participants, of which 177 (13.3%) were colorectal cancers. The distribution of extracolonic cancers by sex and genotype are presented in Table 2. Among male MUTYH carriers, we observed a significantly increased risk for incidence of any cancer type (HR 1.66, 95% CI, [1.04, 2.66]; P = 0.035; Table 3) and colorectal cancer incidence (HR 3.50, 95% CI, [1.43, 8.79]; P = 0.006) in univariate analysis. Adjustment for age, smoking status, alcohol use, baseline BMI, cancer history, colorectal cancer history, polyp history, and randomization to aspirin did not alter the increased risk seen with any cancer type (multivariable HR 1.66, 95% CI [1.03, 2.68]; P = 0.038) or colorectal cancer (multivariable HR 3.55, 95% CI [1.42, 8.78]; P = 0.007). To determine if the total incident cancer risk was primarily driven by incident CRCs, we calculated the incident risk of extracolonic cancers and observed no significant increase among monoallelic carriers (multivariable HR 1.40, 95% CI [0.81, 2.44]; P = 0.229). In contrast, female MUTYH carriers did not demonstrate any increased risk in overall cancer incidence (multivariable HR 1.45, 95% CI [0.84, 2.51]; P = 0.187), colorectal cancer incidence (multivariable HR 0.81, 95% CI [0.11, 5.82]; P = 0.113), or extracolonic cancer incidence (multivariable HR 1.55, 95% CI [0.88, 2.75]; P = 0.130).
Table 2:
Cancers among ASPREE Participants Stratified by MUTYH Genotype and Sex
| Primary Cancer Site | Number of MUTYH Carriers |
Percent of All MUTYHCarriers |
Number of Male MUTYH Carriers |
% Male of Cancers |
Number of Non-carriers | Percentof All Non-carriers |
Number of Male Non- Carriers |
% Male of Cancers |
|---|---|---|---|---|---|---|---|---|
| Bladder | 0 | 0.0% | 0 | - | 47 | 0.4% | 40 | 85.1% |
| Blood | 3 | 1.4% | 1 | 33% | 128 | 1.0% | 67 | 52.3% |
| Brain | 0 | 0.0% | 0 | - | 16 | 0.1% | 7 | 43.8% |
| Breast | 6 | 2.8% | 0 | 0% | 164 | 1.3% | 0 | 0.0% |
| Cervical | 0 | 0.0% | 0 | - | 1 | 0.0% | 0 | 0.0% |
| Colorectal | 6 | 2.8% | 5 | 83.3% | 173 | 1.3% | 92 | 53.2% |
| Gallbladder or bile duct | 3 | 1.4% | 0 | 0% | 13 | 0.1% | 7 | 53.8% |
| Kidney | 0 | 0.0% | 0 | - | 19 | 0.1% | 10 | 52.6% |
| Liver | 0 | 0.0% | 0 | - | 3 | 0.0% | 2 | 66.7% |
| Lung | 2 | 0.9% | 2 | 100% | 103 | 0.8% | 62 | 60.2% |
| Melanoma | 2 | 0.9% | 1 | 50% | 139 | 1.1% | 87 | 62.6% |
| Ovary or Endometrium | 1 | 0.5% | 0 | 0% | 42 | 0.3% | 0 | 0.0% |
| Pancreas | 0 | 0.0% | 0 | - | 38 | 0.3% | 23 | 60.5% |
| Prostate | 9 | 4.2% | 9 | 100% | 253 | 2.0% | 253 | 100.0% |
| Stomach | 1 | 0.5% | 1 | 100% | 21 | 0.2% | 13 | 61.9% |
| Thyroid | 0 | 0.0% | 0 | - | 6 | 0.0% | 2 | 33.3% |
| Unknown primary | 0 | 0.0% | 0 | - | 19 | 0.1% | 10 | 52.6% |
| Other cancer - Anal | 0 | 0.0% | 0 | - | 7 | 0.1% | 1 | 14.3% |
| Other cancer - Neck | 0 | 0.0% | 0 | - | 25 | 0.2% | 20 | 80.0% |
| Other cancer - mesothelioma | 0 | 0.0% | 0 | - | 9 | 0.1% | 9 | 100.0% |
| Other cancer - Neuroendocrine | 0 | 0.0% | 0 | - | 7 | 0.1% | 5 | 71.4% |
| Esophageal and Gastroesophageal junction | 0 | 0.0% | 0 | - | 23 | 0.2% | 17 | 73.9% |
| Other cancer - miscellaneous | 0 | 0.0% | 0 | - | 15 | 0.1% | 6 | 40.0% |
| Other cancer - gastrointestinal | 0 | 0.0% | 0 | - | 6 | 0.0% | 5 | 83.3% |
| Other cancer -genitourinary | 0 | 0.0% | 0 | - | 3 | 0.0% | 1 | 33.3% |
| Other cancer - skin | 0 | 0.0% | 0 | - | 5 | 0.0% | 5 | 100.0% |
| Other cancer - sarcoma | 0 | 0.0% | 0 | - | 10 | 0.1% | 8 | 80.0% |
Table 3:
Association between MUTYH carrier status and Incident Cancer and Colorectal Risks by Sex
| All Cancer | Colorectal Cancer | Extracolonic Cancer* |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Non- carrier |
MUTYH Carrier |
Non- carrier |
MUTYH
carrier |
Non- carrier |
MUTYH
carrier |
||||
| HR (95% CI) |
HR (95% CI) |
P | HR (95% CI) |
HR (95% CI) |
P | HR (95% CI) |
HR (95% CI) |
P | |
| Female | |||||||||
| Model 1a | 1 | 1.53 (0.90, 2.58) | 0.114 | 1 | 0.71 (0.10, 5.11) | 0.736 | 1 | 1.67 (0.97, 2.88) | 0.065 |
| Model 2b | 1 | 1.57 (0.93, 2.66) | 0.092 | 1 | 0.79 (0.11, 5.66) | 0.844 | 1.71 (0.99, 2.95) | 0.054 | |
| Model 3c | 1 | 1.47 (0.85, 2.54) | 0.170 | 1 | 0.81 (0.11, 5.79) | 0.831 | 1 | 1.58 (0.89, 2.80) | 0.115 |
| Model 4d | 1 | 1.45 (0.84, 2.51) | 0.187 | 1 | 0.81 (0.11, 5.82) | 0.113 | 1 | 1.55 (0.88, 2.75) | 0.130 |
| Male | |||||||||
| Model 1a | 1 | 1.66 (1.04, 2.66) | 0.035 | 1 | 3.50 (1.41, 8.66) | 0.007 | 1 | 1.41 (0.82, 2.43) | 0.216 |
| Model 2b | 1 | 1.68 (1.05, 2.68) | 0.032 | 1 | 3.54 (1.43, 8.79 | 0.006 | 1 | 1.41 (0.82, 2.45) | 0.209 |
| Model 3c | 1 | 1.67 (1.04, 2.68) | 0.035 | 1 | 3.53 (1.42, 8.78) | 0.007 | 1 | 1.41 (0.81, 2.44) | 0.220 |
| Model 4d | 1 | 1.66 (1.03, 2.68) | 0.038 | 1 | 3.55 (1.43, 8.79) | 0.006 | 1 | 1.40 (0.81, 2.44) | 0.229 |
All cancers excluding colorectal cancer and non-melanoma skin cancers.
Univariate Cox regression analysis.
Multivariable Cox regression analysis adjusting for age, smoking status, and alcohol use
Multivariable Cox regression analysis adjusting for age, smoking status, alcohol use, baseline BMI, and randomization to aspirin.
Multivariable Cox regression analysis adjusting for age, smoking status, alcohol use, baseline BMI, randomization to aspirin, history of cancer (all cancer and extracolonic risk only), history of CRC (CRC risk only), and history of colon polyps (CRC risk only).
Mutation Signature Analysis of Cancers from MUTYH carriers.
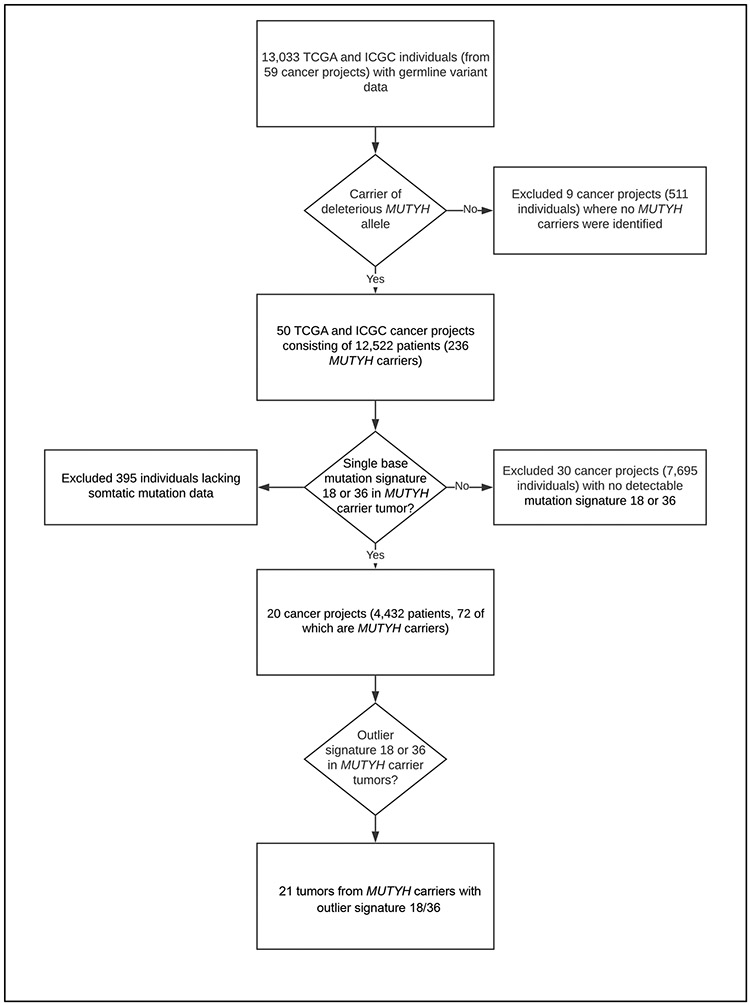
Given the sex-dependent difference observed in risk of incident overall cancer and colorectal cancer, we next hypothesized that MUTYH loss of heterozygosity resulting in the widespread G:C>T:A transversions, as seen in CRCs associated with MAP, was unlikely the predominant mechanism of tumorigenesis seen in cancers from MUTYH carriers. To test this hypothesis, we analyzed single base substitution signatures in MUTYH carriers from TCGA and ICGC cancer samples (Figure 1). Among 10,483 individuals from 33 different TCGA cancer projects, and 2,550 participants from 26 ICGC cancer projects (Supplementary Table S2), MUTYH monoallelic carriers were found among 185 TCGA participants (1.76%) and 51 ICGC participants (2.0%). In total, 41 different TCGA and ICGC cancer projects with MUTYH carriers lacked any measurable contribution from SBS18 or SBS36, and did not depend upon sex. Of the 72 tumors from MUTYH carriers with appreciable SBS18 or SBS36 components, only 21 cancers were found to either have greater than half of the mutations attributable to SBS18/36 or have mutation fractions of SBS18/36 of at least 10% that were in the top quartile in their respective tumor type. The age distributions of TCGA and ICGC study participants in the context of SBS18/36 contributions are presented in Supplementary Figure 1.
Figure 1: Analytical Pipeline Used to Identify MUTYH carriers with Outlier Single Base Substitution Signature 18 or 36 fractions.
Workflow of Exomes and Genomes from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC).
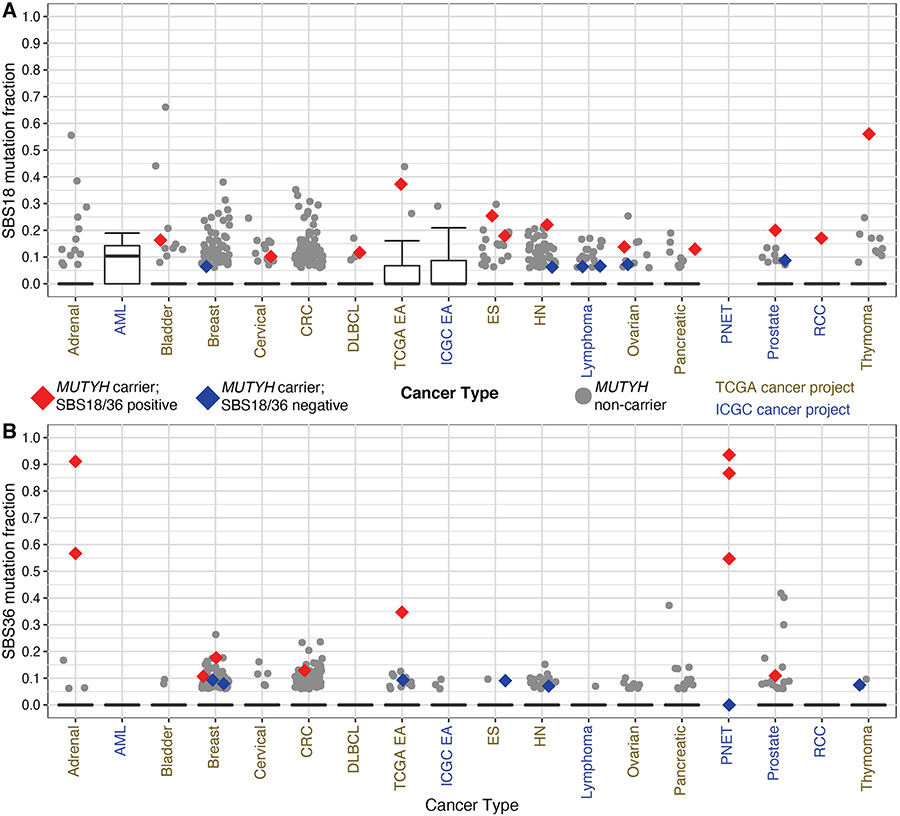
No colorectal or gastric cancers from MUTYH carriers displayed widespread G:C>T:A alterations similar to those seen in MAP or possessed key early oncogenic events driven by such mutations. Furthermore, no differences in SBS18 or SBS36 were noted between males and females in cancer types, including colorectal or gastric cancers (Supplementary Figure 2 and Supplementary Figure 3). Among the cancer types with detectable SBS18 or SBS36, considerable inter-tumor heterogeneity existed among the mutation fractions attributable to these signatures (Figure 2A and Figure 2B). While some breast, colon, prostate, pancreatic adenocarcinoma, and ovarian cancers from MUTYH carriers demonstrated outlier contributions of SBS18 or SBS36 in their respective tumor type, none of these cancer types demonstrated genome-wide G:C>T:A transversions to the magnitude seen in CRCs arising from individuals who inherit biallelic mutations. However, a few tumor types displayed consistent and widespread G:C>T:A transversions to such levels. Three out of four MUTYH carriers with pancreatic neuroendocrine tumors, and 2/2 MUTYH carriers with adrenocortical carcinomas demonstrated high fractions. Cases with high fractions consisted of males and females in each tumor type. In addition, all recognizable oncogenic events for these tumors, as identified by OncoKB, occurred via G:C>T:A mutations (Supplementary Table S5). Among gastrointestinal epithelial tumors, only one esophageal adenocarcinoma demonstrated extensive G:C>T:A alterations, with large contributions from both SBS18 and SBS36. Examination of the oncogenic mutations allowed us to infer that LOH likely occurred after transformation into adenocarcinoma (Supplementary Figure 4). The oncogenic events associated with the development of Barrett’s esophagus with high-grade dysplasia and early adenocarcinoma (KRAS and TP53) were not induced by G:C>T:A alterations, whereas a pathogenic mutation in SMAD4 that typically arises after the development of invasive adenocarcinoma appeared to be induced by such a transversion (36).
Figure 2: Single Base Substitution Signature 18/36 fractions Across TCGA and ICGC.
Distribution of Single Base Substitution Signature 36 (SBS36) (A) and SBS 18 (B) by genomic project and cancer type. Tumor types displayed possessed at least one MUTYH carrier with a detectable mutation signature 18 or 36. Red markings designate samples from MUTYH carriers that had detectable components greater than 0.10 and in the top quartile in their respective tumor type (SBS18/36 positive). Blue markings designate those samples from carriers that failed to achieve a minimum component of 0.10 (SBS18/36 negative), but with a detectable fraction. TCGA-ESCA project was subdivided into esophageal adenocarcinoma (EA) and esophageal squamous cell carcinoma (ES). The TCGA-COAD and TCGA-READ projects were combined into colorectal cancer (CRC), and the ICGC PRAD-CA and PRAD-UK were combined into prostate cancer. Four pancreatic neuroendocrine tumors (PNETs) from MUTYH carriers were plotted irrespective of fractions given known association with MUTYH-associated signatures. HN = Head and Neck squamous cell carcinoma, AML = acute myeloid leukemia, RCC = renal cell carcinoma.
Discussion.
In this study, we demonstrate through a large population-based, prospective cohort of community-dwelling adults that older, male MUTYH carriers are at an increased risk for the development of cancer driven primarily by an increased risk of colorectal cancer. To further investigate the mechanisms of this sex-dependent risk, we examined the mutational signatures of cancers from MUTYH carriers deposited in the TCGA and ICGC repositories. We discovered that the widespread G:C>T:A transversions seen in CRCs arising from biallelic inheritance of pathogenic MUTYH mutations are largely absent in the cancers of carriers, particularly of those of gastrointestinal epithelial origin. These novel epidemiologic and genomic findings provide further insight into the risks, as well as the heterogeneous mechanisms of carcinogenesis, associated with inherited monoallelic disruption of MUTYH.
Previously reported CRC risk estimates in carriers have been based on case-control analyses derived from individuals and family members enrolled in colon cancer registries with matched controls or kin-cohort studies. While risk estimates derived from these studies have prompted the NCCN to issue enhanced CRC screening recommendations for only those carriers with family histories of colorectal cancer, there is little guidance for monoallelic carriers at age 75 or over. In this study, we present the first reported risk estimates from a population-based, prospective cohort selected prior to the development of cancers, and our data suggest that healthy, older male MUTYH carriers represent a high-risk group that may benefit from continued colorectal cancer screening beyond age 75.
While we report a strong sex-dependent cancer risk among older MUTYH carriers, a trend of much smaller magnitude has been observed in prior case-control studies (10). Moreover, the elevated risk for gastrointestinal cancers among males has been noted in other cancer-predisposing syndromes such as Lynch Syndrome, and observed in esophageal adenocarcinoma that typically afflict older adults (37-39). In our study, we incorporated several extrinsic exposures in our analyses that are known to differ by sex, including history of polyps, BMI, smoking, and alcohol use. Adjusting for these covariates, we nevertheless observed a sex-dependent risk.
Given these results, we next hypothesized that the mechanisms for carcinogenesis among MUTYH carriers were likely pleiotropic, and not uniform to base-excision repair. To begin to answer this question, we identified and analyzed cancers from all MUTYH carriers present in two large cancer genome repositories (TCGA and ICGC), thereby performing the largest analysis known to date. Cancers from those who inherit biallelic mutations display G:C>T:A transversions early in development. In the cases of colorectal cancers from MAP patients, APC and KRAS are uniformly mutated by these transversions (3,40). LOH has been the presumed mechanism in monoallelic carriers, given the frequent loss of chromosome 1p observed in colorectal cancers; however these prior studies were contaminated by analyzing large numbers of non-pathogenic, common variants in MUTYH prior to large-scale genetic epidemiology studies (41,42).
In our analysis, we observed that in carriers, no cancers arising from a gastrointestinal epithelial origin demonstrated widespread G:C>T:A transversions that were initiated early in tumorigenesis due to loss of heterozygosity. However, in select cancer types, such as adrenocortical carcinomas, pancreatic neuroendocrine tumors, and thymomas, widespread G:C>T:A transversions occurred early in carcinogenesis, and contributed to a similar fraction of the overall mutagenesis seen in MAP-associated CRCs. These biallelic alterations were seen in men and women, and at variable ages. The large differences in G:C>T:A transversions among tumor types suggests that cell intrinsic factors, along with functions other than base-excision repair are likely relevant in the ability of MUTYH to suppress tumor formation. MUTYH has been linked to DNA-damage signaling, PARP1 activation, and SIRT6 activity; many of these processes have notable sex-dependent effects.(43,44) Additional studies are required to examine the mechanisms by which aging, sex, and MUTYH monoallelic disruption interact to promote carcinogenesis.
Our study has several strengths. The incident risk estimates associated with MUTYH carrier status are derived from a large, healthy, older, population-based, prospective cohort with detailed covariate data. In addition, the genomic analyses of cancers tissues from MUTYH gene carriers are the largest yet performed given the size of TCGA and ICGC cohorts. The genomic analyses identified which oncogenic driver changes were specifically induced by the G:C>T:A transversions on a genome-wide scale; prior studies have used LOH of MUTYH to infer pathogenicity, which fails to distinguish its impact as an initiating, driver event or later-stage passenger alteration. Several limitations of this study should also be noted. Medical history prior to enrollment was self-reported. Second, ASPREE participants represent a healthy subset of the older population, and thus any lack of association with history of prior cancers before enrollment may reflect selection bias of healthy volunteers. Also, we lack granular data on colon cancer screening prior to enrollment. As a result, it may be possible that the lack of risk seen in elderly, female MUTYH may be attributable to higher overall higher rates of participation in colorectal cancer screening in women. Finally, while the number of the incident number of colorectal cancers among MUTYH carriers may appear small, the deep phenotypic and longitudinal characterization of participants and cohort selection prior to the development of cancers ensure that risk estimates are less likely affected by ascertainment and selection biases.
The decision to pursue and cease colorectal cancer screening in older individuals is a complex, decision-making process that balances patient preferences, medical history, functional status, life expectancy, and the risk of colorectal cancer that increases with age (45,46). In this study, we identified a promising genetic biomarker that is strongly associated with increased colorectal cancer risk among otherwise healthy, older men. Given its prevalence in the European population, monoallelic, pathogenic MUTYH mutations are among the most frequent genetic alterations identified during genetic testing for cancer predisposition. Additional studies will be required to establish if enhanced or continued colorectal cancer screening for older, male MUTYH carriers results in decreased cancer incidence and mortality.
Supplementary Material
Prevention Relevance:
Despite absence of loss of heterozygosity in colorectal cancers, elderly male MUTYH carriers appeared to be at increased of colorectal cancer.
Acknowledgements.
We thank the trial staff in Australia and the United States, the participants who volunteered for this trial, and the general practitioners and staff of the medical clinics who cared for the participants.
Financial Support:
This study was funded by the National Institutes of Health (U01AG029824 to J.J. McNeil and A.M. Murray, K23DK103119 to M. Gala, and U19AG062682 to R.L. Woods, A.M. Murray, and J.J. McNeil), American College of Gastroenterology (Resident Clinical Research Award to J.M. Downie), the National Health and Medical Research Council of Australia (grant numbers 334047 to J.J. McNeil and 1127060 to R.L. Woods and J.J. McNeil), Monash University (Australia) to R.L. Woods, J.J. McNeil, and P. Lacaze and the Victorian Cancer Agency (Australia) to R.L. Woods, J.J. McNeil, and P. Lacaze. The ASPREE Healthy Ageing Biobank was supported by a Flagship cluster grant (including the Commonwealth Scientific and Industrial Research Organization, Monash University, Menzies Research Institute, Australian National University, and University of Melbourne) to J.J. McNeil and P. Lacaze.
Footnotes
Disclosures: Dr. Manish Gala has equity in New Amsterdam Genomics, Inc. Dr. Robert Sebra also serves as vice-president of technology development at Sema4. Dr. Eric Schadt also functions as chief executive officer at Sema4. No other conflicts were reported.
References.
- 1.Viel A, Bruselles A, Meccia E, Fornasarig M, Quaia M, Canzonieri V, et al. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. EBioMedicine 2017;20:39–49 doi 10.1016/j.ebiom.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Molecular and cellular biology 1995;15(2):989–96 doi 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nature genetics 2002;30(2):227–32 doi 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 4.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. The New England journal of medicine 2003;348(9):791–9 doi 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 5.Baglioni S, Melean G, Gensini F, Santucci M, Scatizzi M, Papi L, et al. A kindred with MYH-associated polyposis and pilomatricomas. American journal of medical genetics Part A 2005;134A(2):212–4 doi 10.1002/ajmg.a.30585. [DOI] [PubMed] [Google Scholar]
- 6.Vogt S, Jones N, Christian D, Engel C, Nielsen M, Kaufmann A, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 2009;137(6):1976–85 e1-10 doi 10.1053/j.gastro.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 7.Cleary SP, Cotterchio M, Jenkins MA, Kim H, Bristow R, Green R, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology 2009;136(4):1251–60 doi 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Win AK, Jenkins MA, Dowty JG, Antoniou AC, Lee A, Giles GG, et al. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2017;26(3):404–12 doi 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrington SM, Tenesa A, Barnetson R, Wiltshire A, Prendergast J, Porteous M, et al. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. American journal of human genetics 2005;77(1):112–9 doi 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Win AK, Dowty JG, Cleary SP, Kim H, Buchanan DD, Young JP, et al. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology 2014;146(5):1208–11 e1-5 doi 10.1053/j.gastro.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balaguer F, Castellvi-Bel S, Castells A, Andreu M, Munoz J, Gisbert JP, et al. Identification of MYH mutation carriers in colorectal cancer: a multicenter, case-control, population-based study. Clin Gastroenterol Hepatol 2007;5(3):379–87 doi 10.1016/j.cgh.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins MA, Croitoru ME, Monga N, Cleary SP, Cotterchio M, Hopper JL, et al. Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population-based case-family study. Cancer Epidemiol Biomarkers Prev 2006;15(2):312–4 doi 10.1158/1055-9965.EPI-05-0793. [DOI] [PubMed] [Google Scholar]
- 13.Ranola JMO, Tsai GJ, Shirts BH. Exploring the effect of ascertainment bias on genetic studies that use clinical pedigrees. Eur J Hum Genet 2019;27(12):1800–7 doi 10.1038/s41431-019-0467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S, Provenzale D, Llor X, Halverson AL, Grady W, Chung DC, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 2.2019. Journal of the National Comprehensive Cancer Network : JNCCN 2019;17(9):1032–41 doi 10.6004/jnccn.2019.0044. [DOI] [PubMed] [Google Scholar]
- 15.Force USPST, Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr., et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. Jama 2016;315(23):2564–75 doi 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe K, Ikuno Y, Kakeya Y, Ikeno S, Taniura H, Kurono M, et al. Age-related dysfunction of the DNA damage response in intestinal stem cells. Inflammation and regeneration 2019;39:8 doi 10.1186/s41232-019-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriwaki S, Ray S, Tarone RE, Kraemer KH, Grossman L. The effect of donor age on the processing of UV-damaged DNA by cultured human cells: reduced DNA repair capacity and increased DNA mutability. Mutation research 1996;364(2):117–23 doi 10.1016/0921-8777(96)00029-8. [DOI] [PubMed] [Google Scholar]
- 18.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017;543(7643):65–71 doi 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 19.Win AK, Reece JC, Dowty JG, Buchanan DD, Clendenning M, Rosty C, et al. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. International journal of cancer 2016;139(7):1557–63 doi 10.1002/ijc.30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibodeau ML, Zhao EY, Reisle C, Ch'ng C, Wong HL, Shen Y, et al. Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harbor molecular case studies 2019;5(2) doi 10.1101/mcs.a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nature genetics 2015;47(12):1402–7 doi 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilati C, Shinde J, Alexandrov LB, Assie G, Andre T, Helias-Rodzewicz Z, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. The Journal of pathology 2017;242(1):10–5 doi 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
- 23.McNeil JJ, Nelson MR, Woods RL, Lockery JE, Wolfe R, Reid CM, et al. Effect of Aspirin on All-Cause Mortality in the Healthy Elderly. N Engl J Med 2018;379(16):1519–28 doi 10.1056/NEJMoa1803955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil JJ, Woods RL, Nelson MR, Murray AM, Reid CM, Kirpach B, et al. Baseline Characteristics of Participants in the ASPREE (ASPirin in Reducing Events in the Elderly) Study. The journals of gerontology Series A, Biological sciences and medical sciences 2017;72(11):1586–93 doi 10.1093/gerona/glw342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeil JJ, Woods RL, Nelson MR, Reid CM, Kirpach B, Wolfe R, et al. Effect of Aspirin on Disability-free Survival in the Healthy Elderly. The New England journal of medicine 2018;379(16):1499–508 doi 10.1056/NEJMoa1800722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeil JJ, Wolfe R, Woods RL, Tonkin AM, Donnan GA, Nelson MR, et al. Effect of Aspirin on Cardiovascular Events and Bleeding in the Healthy Elderly. The New England journal of medicine 2018;379(16):1509–18 doi 10.1056/NEJMoa1805819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303 doi 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92 doi 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic acids research 2018;46(D1):D1062–D7 doi 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics 2013;14(2):178–92 doi 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome biology 2016;17:31 doi 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500(7463):415–21 doi 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO precision oncology 2017;2017 doi 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacaze P, Sebra R, Riaz M, Tiller J, Revote J, Phung J, et al. Medically actionable pathogenic variants in a population of 13,131 healthy elderly individuals. Genet Med 2020;22(11):1883–6 doi 10.1038/s41436-020-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver JMJ, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nature genetics 2014;46(8):837–43 doi 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider R, Schneider C, Jakobeit C, Furst A, Moslein G. Gender-Specific Aspects of Lynch Syndrome and Familial Adenomatous Polyposis. Viszeralmedizin 2014;30(2):82–8 doi 10.1159/000360839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcaino AP, Moreno V, Lambert R, Parkin DM. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries, 1973-1995. International journal of cancer 2002;99(6):860–8 doi 10.1002/ijc.10427. [DOI] [PubMed] [Google Scholar]
- 39.Rutegard M, Shore R, Lu Y, Lagergren P, Lindblad M. Sex differences in the incidence of gastrointestinal adenocarcinoma in Sweden 1970-2006. European journal of cancer 2010;46(6):1093–100 doi 10.1016/j.ejca.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 40.Jones S, Lambert S, Williams GT, Best JM, Sampson JR, Cheadle JP. Increased frequency of the k-ras G12C mutation in MYH polyposis colorectal adenomas. British journal of cancer 2004;90(8):1591–3 doi 10.1038/sj.bjc.6601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kambara T, Whitehall VL, Spring KJ, Barker MA, Arnold S, Wynter CV, et al. Role of inherited defects of MYH in the development of sporadic colorectal cancer. Genes Chromosomes Cancer 2004;40(1):1–9 doi 10.1002/gcc.20011. [DOI] [PubMed] [Google Scholar]
- 42.Croitoru ME, Cleary SP, Di Nicola N, Manno M, Selander T, Aronson M, et al. Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst 2004;96(21):1631–4 doi 10.1093/jnci/djh288. [DOI] [PubMed] [Google Scholar]
- 43.Raetz AG, David SS. When you're strange: Unusual features of the MUTYH glycosylase and implications in cancer. DNA repair 2019;80:16–25 doi 10.1016/j.dnarep.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer KE, Riddle NC. Sex Differences in Aging: Genomic Instability. The journals of gerontology Series A, Biological sciences and medical sciences 2018;73(2):166–74 doi 10.1093/gerona/glx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Hees F, Saini SD, Lansdorp-Vogelaar I, Vijan S, Meester RG, de Koning HJ, et al. Personalizing colonoscopy screening for elderly individuals based on screening history, cancer risk, and comorbidity status could increase cost effectiveness. Gastroenterology 2015;149(6):1425–37 doi 10.1053/j.gastro.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cenin DR, Tinmouth J, Naber SK, Dube C, McCurdy BR, Paszat L, et al. Calculation of Stop Ages for Colorectal Cancer Screening Based on Comorbidities and Screening History. Clin Gastroenterol Hepatol 2021;19(3):547–55 doi 10.1016/j.cgh.2020.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Cancer Genome Atlas dataset is available through dbgap accession phs000178.v11.p8 . International Cancer Genome Consortium datasets can be accessed through https://daco.icgc.org/ The human sequence data generated in this study are not publicly available due to patient privacy requirements but are available upon reasonable request from the corresponding authors. Other data generated in this study are available within the article and its supplementary data files.