Abstract
Despite substantial observational and experimental evidence that aspirin use can provide protection against the development of colorectal neoplasia, our understanding of the molecular mechanisms involved is inadequate and limits our ability to use this drug effectively and safely for chemoprevention. We employed an untargeted plasma metabolomics approach using liquid chromatography with high-resolution mass spectroscopy to explore novel metabolites that may contribute to the chemopreventive effects of aspirin. Associations between levels of metabolic features in plasma and aspirin treatment were investigated among 523 participants in a randomized placebo-controlled clinical trial of two doses of aspirin (81 or 325 mg/day) and were linked to risk of colorectal adenoma occurrence over 3 years of follow-up. Metabolic pathways that were altered with aspirin treatment included linoleate and glycerophospholipid metabolism for the 81 mg dose and carnitine shuttle for both doses. Metabolites whose levels increased with 81 mg/day aspirin treatment and were also associated with decreased risk of adenomas during follow-up included certain forms of lysophosphotidylcholine (LysoPC) and lysophosphotidylethanolamine (LysoPE) as well as trihydroxyoctadecenoic acid (TriHOME), which is a derivative of linoleic acid and is upstream of cyclooxygenase inhibition by aspirin in the linoleate and arachidonic acid metabolism pathways. In conclusion, our findings regarding lysophospholipids and metabolites in the linoleate metabolism pathway may provide novel insights into the chemopreventive effects of aspirin in the colorectum, although they should be considered hypothesis-generating at this time.
Keywords: Colorectal Cancer, Chemoprevention, Untargeted Metabolomics, Aspirin, Colorectal Adenoma
INTRODUCTION
Colorectal cancer is one of the most common and lethal cancers in the US (1) and worldwide (2). Aspirin, which is used clinically for the secondary prevention of cardiovascular disease, is also a potential cancer chemopreventive agent (3). In particular, findings from observational studies and clinical trials provide support for aspirin use in the prevention of colorectal neoplasia (4, 5). Based on a systematic evidence review in 2016, the US Preventive Services Task Force (USPSTF) recommended the use of low dose aspirin to prevent colorectal cancer in addition to cardiovascular disease in some individuals who are at increased risk of cardiovascular disease (6). In 2021 however, an updated USPSTF draft statement downgraded its prior recommendations based on new data suggesting that the potential risks from bleeding may outweigh the potential benefits (7).
Despite the large amount of research conducted to date, a fundamental question still to be resolved is aspirin’s mechanism of action in the chemoprevention of colorectal neoplasia. A clearer understanding of the key biological effects may aid in the identification of biomarkers that could be used to optimize the dose and duration of treatment as well as to appropriately target specific populations or individuals who are most likely to benefit and least likely to be harmed from using aspirin for colorectal neoplasia prevention (8). Although inhibition of cyclooxygenase (COX) enzymes and reductions in downstream prostanoids, especially prostanglandin E2, have been implicated, aspirin has a multitude of effects and the role of COX-1 versus COX-2 and of numerous non-COX mechanisms are not clear.
In the present work, we use a global untargeted high-resolution metabolomics approach to investigate aspirin’s biological effects on the development of colorectal adenomas, the most common precursor lesion to colorectal cancer. Metabolomics represents a powerful, but still evolving tool for evaluating effects of environmental exposures, including drug therapies, on complex biological outcomes such as colorectal cancer (9). Our untargeted approach is ideal for exploring mechanisms in a hypothesis-generating manner to help address the existing uncertainty. Here, we analyze archived blood plasma samples that were collected at baseline and after three years of treatment in the Aspirin/Folate Polyp Prevention Study, a randomized placebo-controlled trial conducted between 1994–2001 in which low dose aspirin reduced adenoma recurrence (10). To our knowledge, our previous analysis in colon tissue (11) and our present analysis in blood plasma are the first to use metabolomics to study aspirin’s mechanism of action in colorectal chemoprevention.
METHODS
Clinical Trial Study Population and Design
The Aspirin/Folate Polyp Prevention Study was a multicenter, double-blind, placebo-controlled, randomized trial of aspirin and/or folic acid for the chemoprevention of colorectal adenomas, as described previously (NCT00272324)(10, 12). The study was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki). All participants provided written informed consent and institutional review boards approved the study at all participating institutions. Participants between 21 to 80 years old were recruited from nine clinical centers in North America between 1994–1998. For eligibility, a recent diagnosis of colorectal adenoma was required and a colonoscopy within 3 months before study entry. Major exclusion criteria included a history of colorectal cancer or familial colorectal cancer syndromes. At enrollment, participants provided information on demographics and health history and were told to avoid using aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) for the duration of study treatment. Dietary information was collected using a food frequency questionnaire that assessed the average consumption of 109 food items during the past year (13). Following a short run-in period, eligible and willing participants who took 80% or more of their study pills were independently randomized to aspirin (placebo, 81 mg/day, or 325 mg/day) and folic acid (placebo or 1 mg/day). However, due to a delay in funding for the folate component of the study, the first 100 participants were only randomized to aspirin. Thereafter at 4 months intervals, participants provided updated information about their study pill taking habits, use of over-the-counter and prescription drugs, and recent medical diagnoses or procedures. Treatment ended at the time of a surveillance colonoscopy about three years after the pre-enrollment colonoscopy. Adenoma occurrence was assessed via central pathological review and diagnosis of all large bowel lesions. By definition, advanced adenomas included those with cancer, high-grade dysplasia, villous histology (more than 25%), or large size (at least 1 cm). By definition, high-risk findings included the occurrence of at least one advanced adenoma or 3 or more adenomas of any type.
Collection and Selection of Blood Plasma Samples for Metabolomics Analyses
In the clinical trial, baseline blood samples were collected at enrollment and year three blood samples were collected a median of 16 days (interquartile range 10–33 days) prior to participant’s end-of-treatment colonoscopy. Non-fasting blood was collected in 7-ml EDTA Vacutainer brand tubes, put on ice and then centrifuged at 1,100xg for 10 min at 4oC. The plasma fraction was withdrawn and kept frozen at ≤−20oC for 12 months or less at the local clinical center before shipment with dry ice to the study biorepository at Dartmouth for long term storage at ≤−70oC.
Participants with paired baseline and year three blood plasma samples were selected for inclusion in this metabolomics analysis (Figure 1). Among all randomized participants, a total of 1007 had both baseline and year three plasma samples available. Of those, 92 were excluded due to lack of consent or IRB approval, or missing data on adenoma outcomes. We further excluded 392 participants who reported aspirin or NSAID use more than 24 days/year at baseline or more than 2 days/month during study participation, or who reported less than 85% adherence to study pills. This left 523 participants with paired baseline and year three plasma samples.
Figure 1.
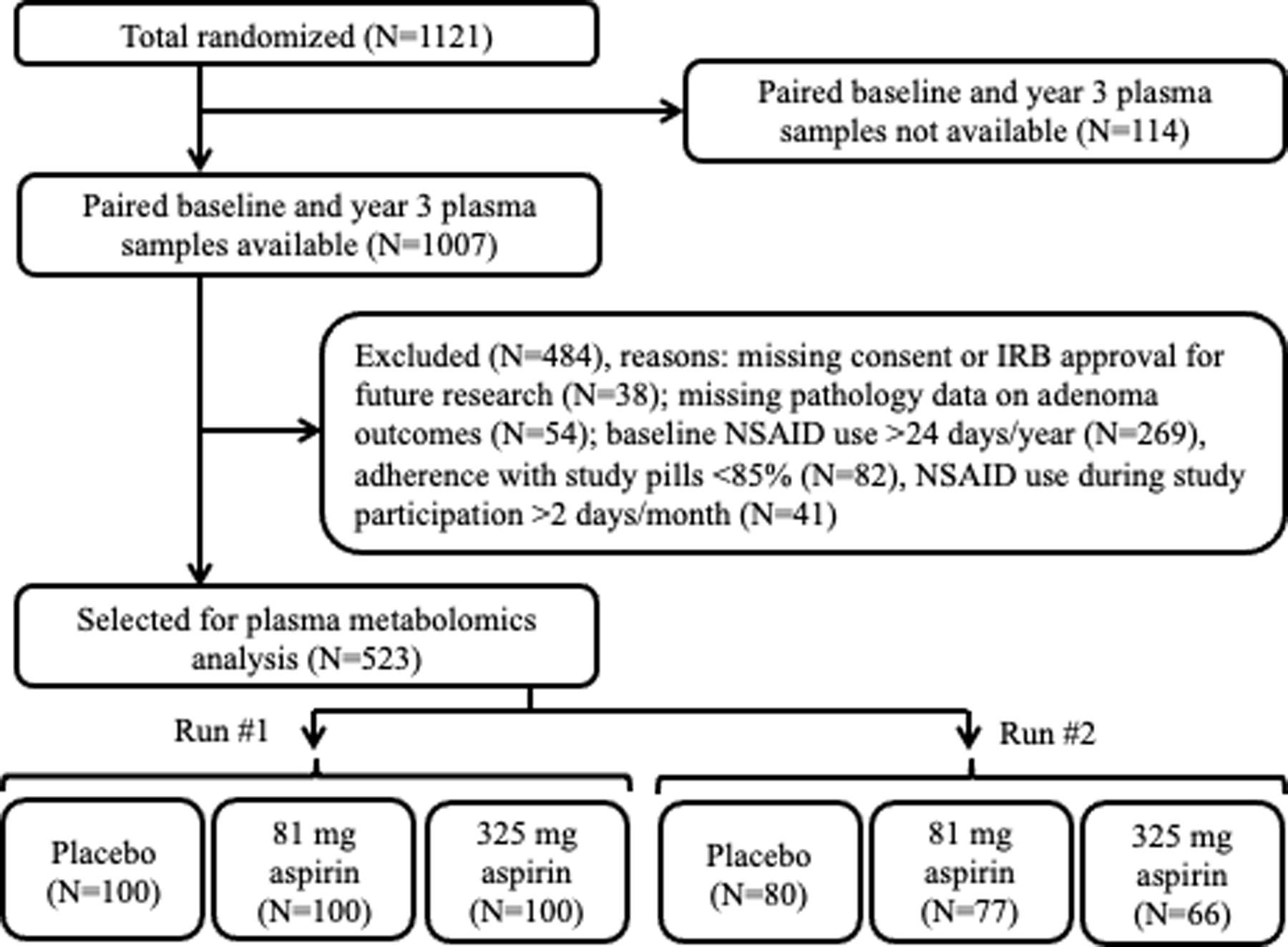
Participant Flow Chart. Selection of participants for inclusion in plasma metabolomics analysis. In Run 1, 600 paired baseline and year 3 plasma samples from 300 participants were analyzed. In Run 2 (replication run), 446 paired baseline and year 3 plasma samples from 223 participants were analyzed.
Due to limitations on the number of batches that could be analyzed without instrument re-calibration, samples were designated for analysis in two separate runs about 6 months apart. Three hundred samples were randomly selected for analysis in run 1, 100 from each of the three aspirin treatment groups, after ensuring equal numbers with and without folate treatment in each group. The remaining 223 participants were analyzed in run 2 for replication. Of the 1046 samples to be analyzed in total, 31.6% (331) had never been thawed previously, 67.0% (701) had been thawed once, 1.2% (13) had been thawed twice and one sample had an unknown thaw history. Samples were shipped on dry ice to the Emory Department of Medicine Clinical Biomarkers Laboratory and stored at ≤−70oC prior to metabolomics analysis.
High Resolution Metabolomics Analyses and Data Processing
The global metabolic effects of aspirin were assessed in blood plasma using liquid chromatography coupled with high-resolution mass spectrometry. Metabolomics methods were similar to those described previously (14–16). Briefly, batches of 20 experimental samples were thawed daily on ice and 50 μl aliquots were removed for analysis. Acetonitrile (100 ul) containing 2.5 μl of internal standards was added to each aliquot followed by 30 minutes on ice, centrifugation at 14,000 rpm at 4°C for 10 minutes, and removal of 100 ul of the resulting supernatant. For each sample three technical replicates of 10 μl each were chromatographed using a 100 X 2.1 mm C18 column (Thermo Accucore) followed by mass spectrometry using a Thermo QExactive High Field mass spectrometer (Thermo Fisher Scientific) operated in positive electrospray ionization mode at a mass resolution of 120,000. Participants’ paired baseline and year 3 samples were analyzed in random order in the same analytical batch and identified using only eight-digit barcoded labels without information on treatment classification. Reference samples analyzed in each batch for quality control purposes included pooled human plasma samples from the National Institute of Standards and Technology (NIST, Gaithersburg, MD; SRM #1950) and from Equitech Bio (Kerrville, TX). Average pairwise Pearson correlation coefficients for all features across all samples and across all pooled plasma reference samples were, respectively, 0.95 and 0.91 in run 1 and 0.98 and 0.97 in run 2.
Raw datasets were processed using apLCMS v6.3.3 (17) with enhancements by xMSanalyzer v2.0.7 (18). Unique metabolic features (ions) were identified by accurate mass (±5 ppm), mass-to-charge ratio (m/z) and retention time. Batch correction was implemented with Combat (19) and ion intensity values were averaged across triplicates. Following initial data extraction, there were 8,458 and 7,425 metabolic features in the two runs, respectively (Supplementary Figure S1, step 1). Two samples from run 1 with mean overall pairwise Pearson correlation <70% between replicates were omitted from the analyses. Metabolic features were omitted due to high variability between replicates (median coefficient of variation >50%) or low abundance (undetectable in >50% of samples in each of the three treatment groups), leaving 4,667 and 6,117 features to be analyzed in the two runs, respectively (Supplementary Figure S1, step 2).
Detection of Aspirin Catabolites in Blood Plasma Samples
Levels of two common aspirin catabolites (salicylic acid and salicyluric acid) detected in baseline and year three plasma samples were used to validate participants’ adherence to randomized aspirin treatments. Identities of these aspirin catabolites were confirmed at level 1 per the Metabolomics Standards Initiative (MSI)(20) by co-elution using tandem mass spectrometry (MS/MS) relative to authentic standards: m/z = 139.0389, salicylic acid [M+H] and m/z = 196.0604, salicyluric acid [M+H]. For each treatment group, differences in raw intensities of aspirin catabolites between paired baseline and year three plasma samples were assessed using Wilcoxon Signed-Rank tests and visualized with box plots.
Statistical Analyses
Prior to statistical analyses, zero intensity values for metabolic features were replaced with the lowest value for that feature across all samples divided by 2 and then the data was log2-transformed. To identify metabolic features in plasma whose levels were associated with aspirin treatment we used multivariable linear regression. The year three log2-transformed ion intensity was the dependent variable and the predictor variable was the randomized aspirin treatment assignment. Models were adjusted for baseline log2-transformed ion intensity, age, sex, race (non-Hispanic white vs. other), and folate treatment assignment. Separately, two regression models were used to examine associations with 81 mg/day aspirin treatment compared to placebo or with 325 mg/day aspirin treatment compared to placebo in intention-to-treat analyses. Volcano plots were used to visualize results. Mummichog v2.0.6 (21) was used to identify metabolic pathways altered with aspirin treatment.
Metabolic features found to be associated with aspirin treatment were next examined for their associations with adenoma outcomes using multivariable Poisson regression models for binary data with common outcomes. Risk ratios (RR) and 95% confidence intervals (CIs) were estimated for associations with two-fold changes in ion intensity. In the regression models, adenoma outcome at year three was the dependent variable and the predictor variable was the year three log2-transformed ion intensity. Models were adjusted for age, sex, and race. Separately, three regression models were used to assess associations with risk of one or more adenoma, one or more advanced adenoma or high-risk findings.
In our analyses the number of independent estimates is uncertain because multiple metabolic features (adducts) can map to one metabolite, and metabolites in a pathway can be correlated. The Benjamini-Hochberg method was used to calculate false discovery rate (FDR) q-values with a 0.2 threshold for significance (22). However, in regression analyses of metabolic features associated with aspirin treatment, features were selected for subsequent analysis using a two-sided P-value threshold of <0.05 without adjustment for multiple testing to reduce type II error. This approach prioritizes discovery of novel metabolic features associated with aspirin treatment and enhances Mummichog analyses of metabolic pathways altered with the drug (21, 23). P-values calculated with Mummichog use permutation tests with sampling from Gamma distributions to account for clustered data; a threshold of P<0.05 was used to identify pathways altered with aspirin treatment to minimize type I error (21, 23). In regression analyses of metabolic features associated with adenoma outcomes only those features associated with aspirin treatment (two-sided P<0.05) were included, thereby limiting multiple testing, and FDR correction was performed. In analyses of changes in salicylic acid and salicyluric acid intensities between baseline and year three plasma samples, two-sided P<0.05 were considered statistically significant.
Spearman correlation analysis was used to assess metabolite-metabolite associations between natural log-transformed year 3 intensities among confirmed or tentatively identified metabolites that were increased with aspirin treatment and associated with adenoma outcomes. Analyses were conducted in R v3.5.1 and SAS v9.4.
Metabolite Annotation and Identification
For identification of metabolic features accurate mass and retention time were compared to authentic standards from an in-house library run under identical conditions using MS/MS. Additional tentative annotations were ascribed using Mummichog v2.0.6 (21) and xMSannotator v1.3.2 (24) with Human Metabolome Database v3.5. xMSannotator uses a multi-step approach based on m/z, retention time, isotopes, adducts, correlation across samples, and network and pathway associations to assign database matches into different categories: high, medium, or low confidence. Metabolite annotations and identities were classified per MSI criteria (25) level 1 (confirmed by MS/MS and co-elution with authentic standards) or level 2 (tentative annotation using xMSannotator with medium or high confidence).
Data Availability
The metabolomics data generated in this study are publicly available in the NIH Common Fund’s National Metabolomics Data Repository, the Metabolomics Workbench https://www.metabolomicsworkbench.org, as Project ID PR000730. The data can be accessed directly via its Project DOI: 10.21228/M89X1C.
RESULTS
Population Characteristics
Baseline characteristics of the 523 participants from the Aspirin/Folate Polyp Prevention Study whose blood plasma samples were included in this metabolomics analysis are shown in Table 1. The average age at enrollment was 58 years, 35% were female, and 86% were non-Hispanic whites. These demographic characteristics as well as risk factors for colorectal cancer, including body mass index, smoking status, alcohol use and family history of colorectal cancer, did not differ statistically significantly between the three aspirin treatment groups. Study and follow-up characteristics, including allocation to folate treatment (47% on average), length of follow-up (32 months on average) and occurrence of adenoma outcomes, also did not differ across aspirin treatments. Comparison of these characteristics across the two sub-sets of participants analyzed in runs 1 and 2 also didn’t show substantial differences (Supplementary Table S1).
Table 1.
Characteristics of Randomized Participants Selected for Plasma Metabolomics Study, by Aspirin Treatment Group (N=523)
| Placebo (N=180) |
81 mg Aspirin (N=177) |
325 mg Aspirin (N=166) |
Pa | |
|---|---|---|---|---|
| Baseline Characteristics: | ||||
| Age at Enrollment, Mean ± SD | 58.5 ± 9.5 | 57.5 ± 9.3 | 58.4 ± 9.2 | 0.55 |
| Female, N (%) | 62 (34.4) | 63 (35.6) | 59 (35.5) | 0.97 |
| Race/ethnicity, N (%) | 0.26 | |||
| White, Non-Hispanic | 152 (84.4) | 149 (84.2) | 151 (91.0) | |
| Black, Non-Hispanic | 12 (6.7) | 12 (6.8) | 3 (1.8) | |
| Hispanic | 10 (5.6) | 10 (5.7) | 5 (3.0) | |
| Other | 6 (3.3) | 6 (3.4) | 7 (4.2) | |
| Body-Mass Index, kg/m2, mean ± SD | 27.1 ± 4.1 | 27.2 ± 4.2 | 27.8 ± 4.5 | 0.28 |
| Current Smoker, N (%) | 25 (13.9) | 23 (13.1) | 22 (13.3) | 0.76 |
| Alcohol Use, N (%) | 119 (68.4) | 106 (62.0) | 101 (64.3) | 0.45 |
| Family History of Colorectal Cancer, N (%) | 51 (37.2) | 58 (39.5) | 60 (45.8) | 0.33 |
| Dietary Linoleic Acid Intake, gr/day, mean ± SD | 12.3 ± 6.8 | 10.9 ± 6.2 | 11.8 ± 7.1 | 0.17 |
|
| ||||
| Study and Follow-up Characteristics: | ||||
| Allocated to Folate Treatmentb, N (%) | 81 (49.1) | 85 (53.8) | 78 (53.1) | 0.66 |
| Study Follow-Up Time, Mean ± SDc | 32.7 ± 3.7 | 32.3 ± 2.5 | 32.5 ± 2.6 | 0.58 |
| ≥1 Adenoma at Year 3 Colonoscopy, N (%) | 75 (41.7) | 61 (34.5) | 64 (38.6) | 0.37 |
| ≥1 Advanced Adenoma at Year 3 Colonoscopyd, N (%) | 20 (11.1) | 12 (6.8) | 16 (9.6) | 0.36 |
| High-risk Findings at Year 3 Colonoscopye, N (%) | 29 (16.1) | 17 (9.6) | 20 (12.1) | 0.17 |
| Baseline plasma thaw history | 0.18 | |||
| 0 times | 71 (39.4) | 92 (52.0) | 72 (43.6) | |
| 1 time | 106 (58.9) | 83 (46.9) | 91 (55.2) | |
| 2 times | 3 (1.7) | 2 (1.1) | 2 (1.2) | |
| Year 3 plasma thaw history | 0.12 | |||
| 0 times | 33 (18.3) | 30 (17.0) | 33 (19.9) | |
| 1 time | 147 (81.7) | 146 (82.5) | 128 (77.1) | |
| 2 times | 0 | 1 (0.6) | 5 (3.0) | |
N’s for missing data for smoking status: 1 aspirin (81 mg), 1 aspirin (325 mg); alcohol use: 6 placebo, 6 aspirin (81 mg), 9 aspirin (325 mg); family history of colorectal cancer: 43 placebo, 30 aspirin (81 mg), 35 aspirin (325 mg); dietary linoleic acid intake: 5 placebo, 6 aspirin (81 mg), 8 aspirin (325 mg); baseline plasma thaw history: 1 aspirin (325 mg).
P-values for comparisons among the three treatment groups: Pearson chi-square tests for categorical variables and analysis of variance tests for continuous variables. Fisher’s exact test used for number of thaws.
Enrolled in study before randomization to folate began: 15 placebo, 19 aspirin (81 mg), 19 aspirin (325 mg).
Time in months from enrollment to year 3 end of treatment colonoscopy
Advanced adenomas are those with cancer, high-grade dysplasia, >25% villous component, or diameter ≥1 cm.
High-risk findings refers to ≥1 advanced adenomas or ≥3 adenomas of any type.
Levels of Aspirin Catabolites in Plasma: Salicylic Acid and Salicyluric Acid
Levels of salicylic acid and salicyluric acid, two major aspirin catabolites formed following aspirin ingestion (26), confirmed participant’s compliance with aspirin treatment allocations (Supplementary Figure S2). In year three compared to baseline plasma samples, levels of salicylic acid and salicyluric acid were increased among the participants allocated to 81 mg of aspirin (median change in raw intensities=4.68 X 106, P=2.8 X 10−10 and median change=1.33 X 106, P=7.6 X 10−21, respectively) and among those allocated to 325 mg of aspirin (median change=2.45 X 107, P=3.5 X 10−22 and median change=1.31 X 107, P=7.0 X 10−27 respectively), but not among participants allocated to placebo (median change=2.20 X 104, P=0.8 and median change=0, P=0.09, respectively).
Plasma Metabolic Features and Pathways Associated with Aspirin Treatment
Among 4,667 features analyzed in run 1 and 6,117 analyzed in run 2, there were 464 and 606 features, respectively, that were statistically significantly associated with aspirin treatment (either 81 mg or 325 mg) at a nominal P<0.05 (Supplementary Figure S1, step 3). Over both runs, roughly half of these features increased with aspirin treatment and half decreased, as visualized with volcano plots (Figure 2). There was limited overlap in features that increased or decreased with both doses of aspirin, as shown in Venn diagrams: 39 of 464 (8.4% in run 1) and 52 of 606 (8.6% in run 2)( Figure 2). Following correction for multiple testing, 4 features in run 1 and 10 in run 2 were associated with aspirin treatment with statistical significance (Supplementary Table S2), including salicyluric acid (m/z =196.0604, M+H) and salicylic acid (m/z =139.0389, M+H) as discussed above. The identities of the other features are unknown, but some could be unidentified forms of aspirin catabolites. The presence of more unidentified features in run 2 (see Figure 2 volcano plots, Supplementary Table S2) suggested the possibility of unknown technical issues in that run that could bias the findings. Thus, we focus on describing run 1 findings, and include run 2 findings as supplementary information.
Figure 2.
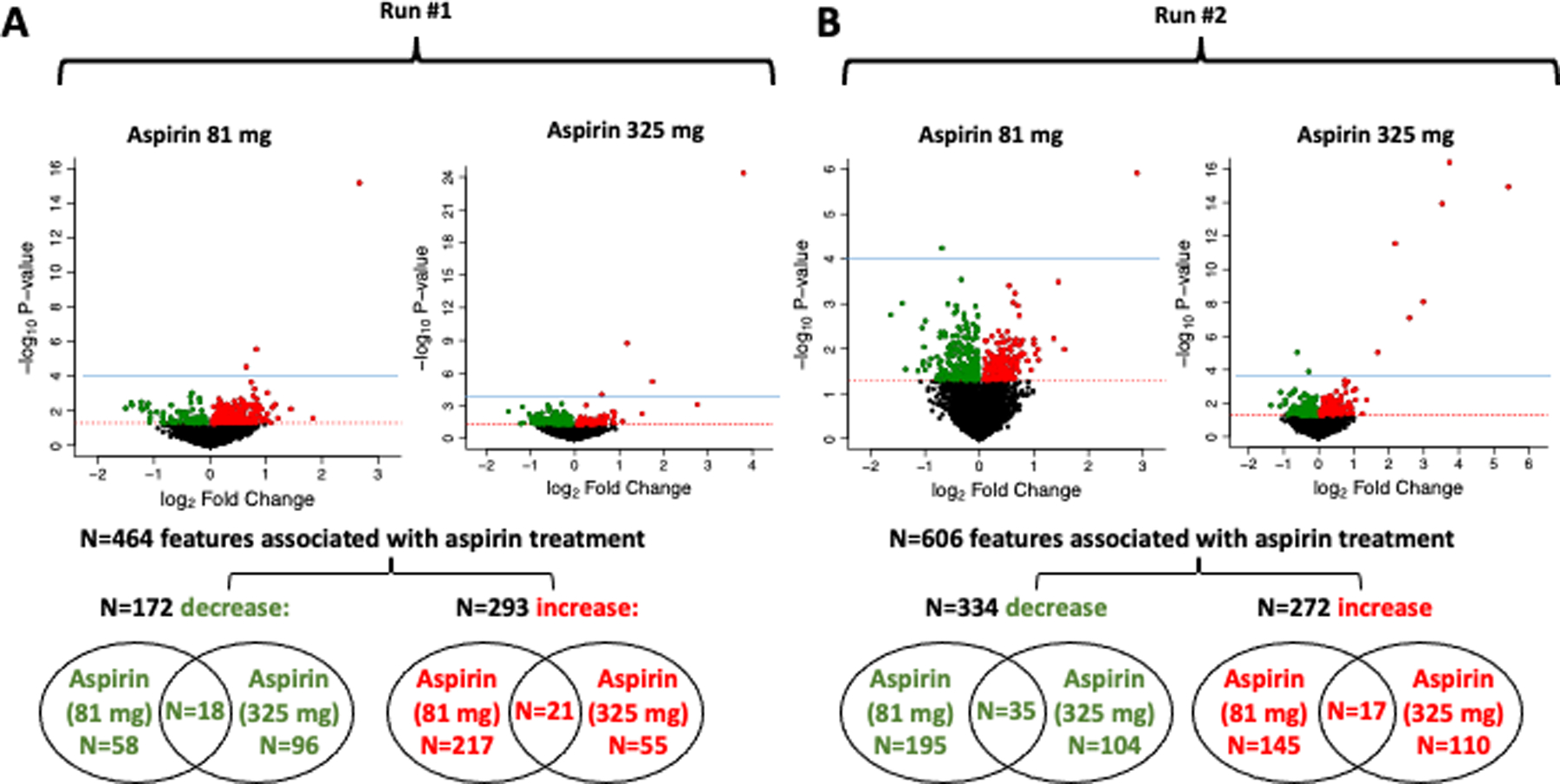
Blood metabolites associated with aspirin treatment. A, Run 1 dataset. B, Run 2 dataset. Linear regression was used to estimate the change in year 3 metabolite intensities in the aspirin treated group compared to the placebo group, adjusting for age, sex, race, and folate treatment. Volcano plots depict for each feature the magnitude of the change on the x-axis (log2fold change with aspirin treatment vs. placebo) and the statistical significance on the y-axis (-log10p-value for the association with aspirin treatment). Color-coded features statistically significantly increased (red) or decreased (green) with aspirin treatment (P<0.05). Features significant at FDR (q<0.2) are above the blue dotted line. Venn diagrams indicate overlap of significant features across the two aspirin treatment groups (81 or 325 mg) for those that either decrease or increase with treatment. In Run #1, one feature was both increased with 81 mg aspirin and decreased with 325 mg aspirin, so total number of features associated with aspirin treatment is N=464 instead of N=465.
Metabolic pathways that were altered in response to aspirin treatment in run 1 are shown in Figure 3. Carnitine shuttle metabolism was altered with both doses of aspirin. Linoleate, glycerophospholipid, saturated fatty acids beta-oxidation and arachidonic acid metabolism were only altered with the 81 mg aspirin dose. Aspartate and asparagine metabolism was only altered with the 325 mg aspirin dose. Different pathways (tryptophan, tyrosine and purine metabolism) were identified in run 2 (Supplementary Figure S3).
Figure 3.
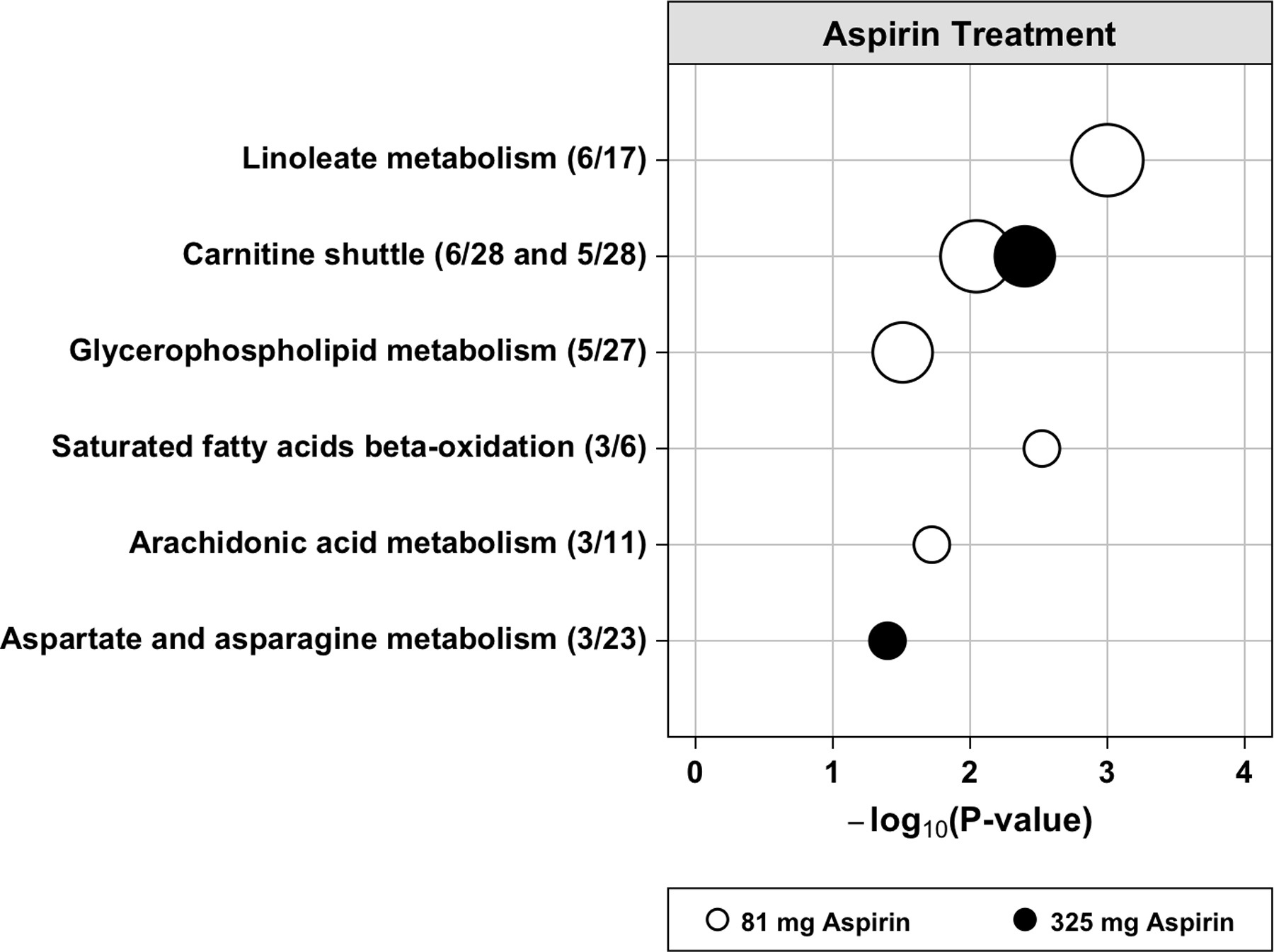
Dysregulated metabolic pathways associated with aspirin treatment in blood plasma in run 1. The vertical axis represents the pathways (circles) with the radius representing the number of hits (significant metabolic features). The horizontal axis represents the negative log10 of the gamma adjusted P-values for each pathway with at least 3 hits. The open circles are for 81 mg aspirin and the solid circles are for 325 mg aspirin treatment. In parentheses next to each pathway name is the number of hits divided by the pathway size (total number of features detected in the pathway).
Plasma Metabolic Features and Pathways Associated with Adenoma Risk
The primary aim of this work was to identify metabolites or metabolic pathways that may help to explain the chemopreventive effects of aspirin in the colorectum. Therefore, we next assessed whether the changes in levels of metabolic features due to aspirin treatment at either dose were also associated with risk of colorectal adenoma outcomes in a direction that could explain the chemopreventive effects of aspirin. We found that changes in levels of 143 metabolic features were statistically significantly (nominal P<0.05) associated with one or more of the adenoma outcomes (any adenoma, advanced adenoma, or high-risk outcomes) in a direction consistent with a potential chemopreventive effect for aspirin (77 features in run 1 and 66 in run 2, Supplementary Table S3).
Although identities of many of these metabolic features are unknown, findings in run 1 for eighteen features corresponding to thirteen metabolites that could be confirmed or tentatively identified (MSI level 1 or 2) are shown in Table 2, (See Supplementary Table S4 for run 2 findings.) These features were increased due to 81 mg aspirin treatment and were also associated with a reduced risk of adenoma outcomes. They are all linked to glycerophospholipid, linoleate, and arachidonic acid metabolism pathways and located upstream of aspirin inhibition of COX (Supplementary Figure S4), including: linoleic acid, α- or γ-linolenic acid (these two compounds are indistinguishable with the methods used), trihydroxyoctadecenoic acid (TriHOME), dihydroxyoxooctadecenoic acid, arachidonoylcarnitine, phosphatidylcholine PC(38:5), and several forms of lysophosphatidylcholine (LysoPC) and lysophosphatidylethanolamine (LysoPE). After correcting for multiple testing, four of these metabolites were statistically significantly associated with risk of advanced adenomas: LysoPC(P-16:0), LysoPE(20:1), LysoPC(P-18:0), and lysoPE(22:1); and one with high-risk outcomes: TriHOME (Table 2). Correlation analysis (Figure 4) among these identified metabolites indicates that levels of all the lysophospholipids were positively correlated (Spearman correlation coefficient range 0.33–0.86). In addition, levels of the two linoleate metabolites, TriHOME and dihydroxyoxooctadecenoic acid, were highly correlated. These metabolites were all identified in run 1, but for the sub-set that was also able to be identified in run 2, we did not find consistent trends suggesting an increase from aspirin treatment and an association with decreased risk of adenoma outcomes (Supplementary Figure S5). Overall, six of the metabolites identified in Table 2 are components of pathways significantly altered with aspirin treatment (Table 2 footnote f, Supplementary Table S5). Sensitivity analyses adjusting for participants’ dietary linoleic acid intake and for the thaw status of blood samples used in the metabolomics measurement did not meaningfully alter our findings.
Table 2.
Plasma Metabolites that may Contribute to the Chemopreventive Effects of Aspirin in the Colorectum (Run 1)
| Metabolite Namea | MSI level | Adduct | m/z | RT | FC (95% CI)b 81 mg Aspirin | FC (95% CI)b 325 mg Aspirin | RR (95% CI)c Any Adenomad | RR (95% CI)c Advancedd | RR (95% CI)c High-Riskd |
|---|---|---|---|---|---|---|---|---|---|
| α- or γ-Linolenic acid | 1 | M+H | 279.2318f | 448 | 1.19 (1.05–1.36) | 1.09 (0.97–1.24) | 0.88 (0.76–1.03) | 0.77 (0.56–1.07) | 0.74 (0.56–0.97) |
| Linoleic acid | 1 | M+H | 281.2474f | 441 | 1.14 (1.01–1.28) | 1.08 (0.95–1.23) | 0.94 (0.81–1.08) | 0.72 (0.58–0.91) | 0.77 (0.63–0.95) |
| TriHOMEe | 2 | 13C M+H | 332.2511 | 407 | 1.35 (1.07–1.70) | 1.04 (0.80–1.35) | 0.93 (0.87–0.99) | 0.93 (0.78–1.11) | 0.91 (0.79–1.04) |
| M+Na | 353.2297f | 406 | 1.28 (1.03–1.59) | 0.96 (0.74–1.26) | 0.95 (0.89–1.01) | 0.87 (0.76–1.00) | 0.86 (0.79–0.93) | ||
| Dihydroxy-oxo-octadecenoic acid | 2 | M+Na | 351.2146f | 405 | 1.25 (1.03–1.53) | 0.99 (0.80–1.23) | 0.96 (0.88–1.05) | 0.92 (0.76–1.11) | 0.87 (0.76–0.99) |
| Arachidonoyl-carnitine | 2 | M+H | 448.3422f | 460 | 1.22 (1.01–1.47) | 1.16 (0.96–1.40) | 0.91 (0.84–0.98) | 0.92 (0.73–1.16) | 0.87 (0.73–1.03) |
| LysoPC(P-16:0)e | 2 | 13C M+H | 481.3484 | 480 | 1.09 (1.01–1.18) | 1.00 (0.91–1.11) | 0.85 (0.72–1.00) | 0.60 (0.48–0.74) | 0.70 (0.53–0.91) |
| LysoPC(P-18:1) | 2 | M+H | 506.3605 | 519 | 1.07 (1.00–1.14) | 1.00 (0.95–1.06) | 0.73 (0.53–0.99) | 0.49 (0.27–0.91) | 0.69 (0.38–1.26) |
| 13C M+H | 507.3641 | 522 | 1.07 (1.01–1.15) | 0.99 (0.93–1.05) | 0.73 (0.55–0.96) | 0.49 (0.27–0.92) | 0.77 (0.43–1.39) | ||
| LysoPE(20:1)e | 2 | M+H | 508.3398 | 460 | 1.09 (1.01–1.18) | 1.06 (0.97–1.15) | 0.98 (0.78–1.22) | 0.52 (0.33–0.81) | 0.65 (0.43–0.99) |
| LysoPC(P-18:0)e | 2 | M+H | 508.3763f | 504 | 1.16 (1.01–1.33) | 1.02 (0.90–1.17) | 0.97 (0.84–1.12) | 0.70 (0.60–0.82) | 0.82 (0.67–1.00) |
| 13C M+H | 509.3796f | 507 | 1.18 (1.04–1.34) | 1.05 (0.94–1.18) | 0.91 (0.79–1.05) | 0.65 (0.54–0.77) | 0.80 (0.62–1.01) | ||
| LysoPE(20:0) | 2 | M+H | 511.3591 | 488 | 1.11 (1.01–1.21) | 1.03 (0.94–1.13) | 0.96 (0.79–1.15) | 0.67 (0.47–0.96) | 0.74 (0.54–1.00) |
| 13C M+H | 512.3621 | 488 | 1.21 (1.02–1.23) | 1.04 (0.95–1.15) | 0.98 (0.82–1.17) | 0.68 (0.49–0.95) | 0.78 (0.57–1.07) | ||
| M+Na | 532.3379 | 488 | 1.10 (1.00–1.21) | 1.04 (0.95–1.15) | 0.94 (0.79–1.12) | 0.64 (0.47–0.88) | 0.75 (0.55–1.01) | ||
| LysoPE(22:1)e | 2 | M+H | 536.3716 | 496 | 1.12 (1.03–1.22) | 1.08 (0.99–1.18) | 0.84 (0.69–1.04) | 0.47 (0.33–0.67) | 0.61 (0.41–0.91) |
| LysoPE(22:0) | 2 | M+H | 538.3871 | 542 | 1.13 (1.02–1.24) | 1.05 (0.94–1.18) | 0.88 (0.76–1.03) | 0.66 (0.49–0.89) | 0.75 (0.56–1.00) |
| PC(38:5) | 2 | M+H | 830.5669 | 560 | 1.13 (1.03–1.23) | 1.06 (0.98–1.16) | 0.83 (0.71–0.96) | 1.19 (0.81–1.75) | 1.00 (0.70–1.44) |
Features whose levels increased with at least one dose of aspirin treatment (P<0.05, bold type) and are also inversely associated with risk of at least one of the adenoma outcomes (P<0.05, bold type) are presented; only MSI level 1 and 2 features are included. Abbreviations: MSI, Metabolomics Standards Initiative; m/z, mass-to-charge ratio; RT, retention time in seconds; FC; fold change; RR, risk ratio; CI, confidence interval; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine; TriHOME, trihydroxy-octadecenoic acid.
Some isobaric metabolites are not distinguishable with the mass spectroscopy methods used and multiple identities are given. For each metabolite, MSI identification confidence level and adduct forms are indicated in the columns to the right.
Linear regression was used to estimate the fold change in year 3 ion intensity in the aspirin treatment group compared to the placebo group, adjusting for baseline ion intensity, age, sex, race, and folate treatment.
Poisson regression was used to assess the association of risk of adenoma outcomes at year 3 with a two-fold change in year 3 ion intensity, adjusting for age, sex, and race.
Any adenoma refers to ≥1 adenoma of any type. Advanced refers to adenomas with cancer, high-grade dysplasia, >25% villous component, or diameter ≥1 cm. High-risk refers to ≥1 advanced adenoma or ≥3 adenomas of any type.
Indicates metabolites with statistically significant associations with adenoma outcomes (RR shown in italics and bold) after correction for multiple testing.
Metabolic features (m/z’s) that are components of pathways significantly altered with aspirin treatment.
Figure 4.
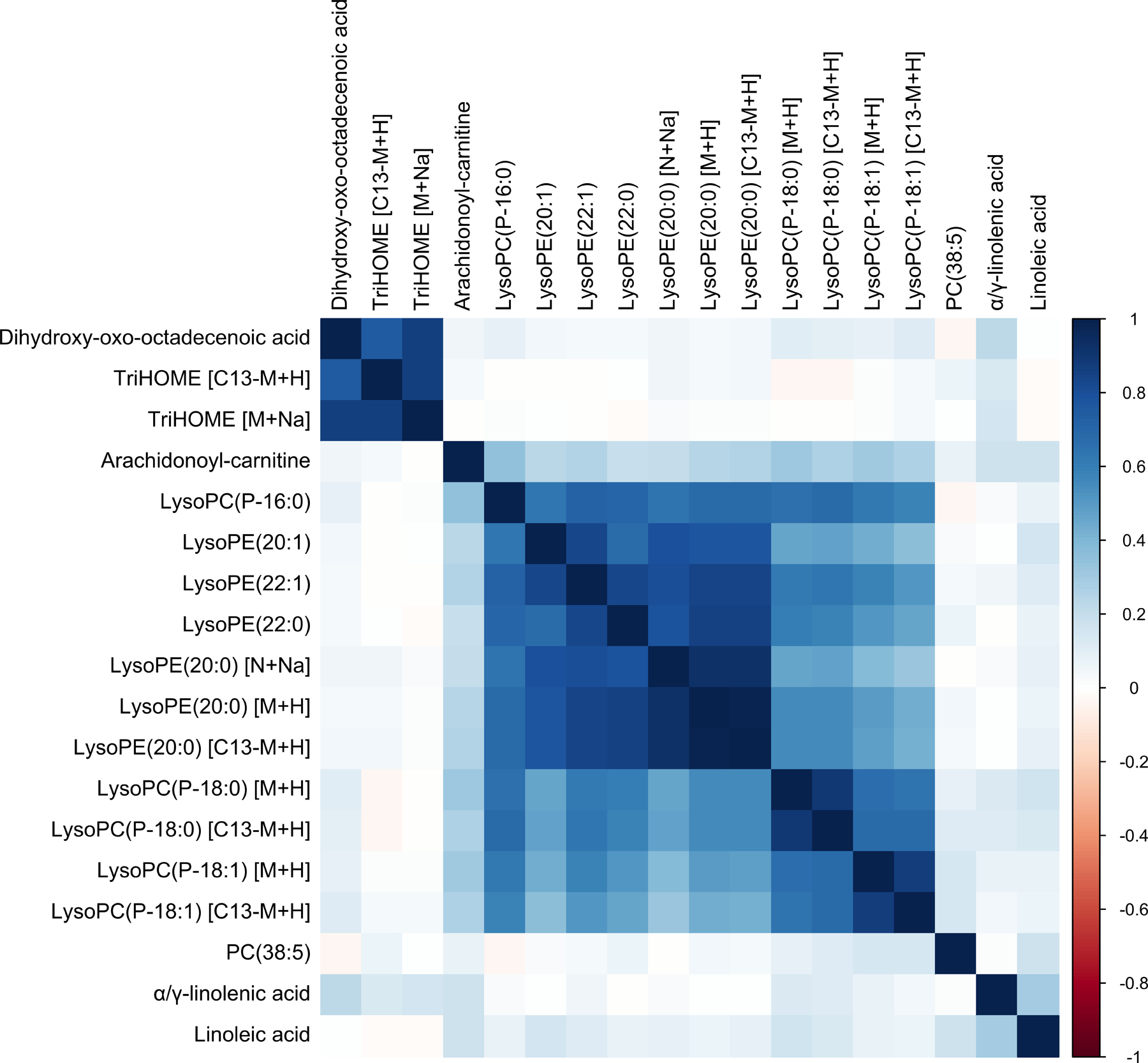
Metabolite-metabolite correlation analysis. Spearman correlations between natural log-transformed year 3 intensities for metabolites that increased with aspirin treatment and were also associated with reduced adenoma risk (Table 2). Positive correlations are in blue and negative correlations are in red. Metabolites are ordered by hierarchical clustering.
DISCUSSION
We performed an untargeted metabolomics investigation of pre- and post-aspirin treatment blood plasma samples from participants in a randomized clinical trial to identify metabolites and metabolic pathways that may be informative of the chemopreventive effects of aspirin for colorectal adenomas. We found metabolic pathways altered with aspirin treatment included linoleate and glycerophospholipid metabolism for the 81 mg/day dose and carnitine shuttle metabolism for both doses. Furthermore, we observed that levels of numerous metabolites with confirmed or tentative identities in the glycerophospholipid and linoleate metabolism pathways increased with 81 mg aspirin treatment and were associated with reduced risk of adenoma outcomes. These included linoleic acid, which was also identified in our metabolomics analysis using colon tissue specimens from participants in this trial (11), as well as several other metabolites with statistically significant associations after correction for multiple testing: lysoPC(P-16:0), lysoPC(P-18:0), LysoPE(20:1), lysoPE(22:1) and TriHOME. Our novel findings link lysophospholipids and linoleate metabolites upstream of cyclooxygenase inhibition to the chemoprevention of colorectal adenomas by aspirin.
There have been few prior investigations of the metabolomic effects of aspirin in humans. The mechanism of variability in the anti-platelet response to aspirin was examined in a subset of up to 205 participants from the Hereditary and Phenotype Intervention Heart Study treated for two weeks with 81 mg/day aspirin (27–29). These analyses measured up to 180 serum metabolites and implicated the purine pathway (27), serotonin (28), and oxylipid profiles (29) in the variability to aspirin response. A more recent analysis of 11 platelet metabolites was unable to distinguish between aspirin response groups in a small case-control study (30). A broader metabolomics analysis of aspirin’s effects measured 2007 urine metabolites from seven healthy volunteers treated for one week with 100 mg/day aspirin and found reduced levels of glutamine, acyl-carnitines, and histidine and purine pathway metabolites (31). In a small, randomized placebo-controlled cross-over trial of 40 healthy participants treated for 60 days with 325 mg/day aspirin, plasma concentrations of the oncometabolite 2-hydroxyglutarate were reduced among 363 plasma metabolites analyzed suggesting a potential chemopreventive mechanism (32). Compared to this prior research, our previous analysis in colon tissue (11) and our present analysis in blood plasma employed larger samples sizes, broader metabolomics analyses and uniquely linked aspirin’s metabolic effects to colorectal adenoma chemoprevention.
Linoleic acid levels in stool and blood have been associated with colorectal cancer risk in previous studies. Linoleic acid levels were lower in stool from patients with colorectal cancer compared to controls (33–37) and higher genetically-predicated levels of circulating linoleic acid was associated with reduced colorectal cancer risk (38, 39). These observations are consistent with our findings that aspirin treatment altered the linoleate metabolism pathway and increased linoleic acid levels in blood (present analysis) and in colon tissue (11), and that these increases were associated with reduced adenoma risk. However, in contrast to our findings, a previous study found aspirin treatment (81 mg/day) reduced linoleic acid (27) and slightly reduced 9,12,13-TriHOME (29) levels in serum. These differences will require further investigation but could be related to the much shorter duration of aspirin treatment (14 days) compared with our clinical trial (three years) or the lack of a placebo control group in their study.
Recently, a rigorous metabolomics analysis of plasma from colorectal cancer cases and controls found inverse associations with colorectal cancer for several lysoPCs including lysoPC(P-16:0) (40), which is one of the lysophospholipids that we found to be increased in plasma with aspirin treatment and inversely associated with adenoma risk. In a subsequent analysis of that metabolomics data, levels of PC(38:5) were associated with colorectal cancer stage: lower in stage 3–4 versus stage 1–2 (41). In our analysis, PC(38:5) was also increased with aspirin treatment and associated with reduced adenoma risk. Although it is unknown how levels of PC(38:5) in adenomas compare to different stages of colorectal cancer, further research may shed light on potential effects of aspirin in cancer progression (42). Finally, a lysophospholipid [LysoPE(18:1)] was also implicated in our colon tissue analysis (11), although in the opposite direction, supporting the potential importance of aspirin’s effects on these compounds.
Aspirin may increase levels of linoleic acid and its metabolites through mechanisms involving cyclooxygenase inhibition, which could increase arachidonic acid levels and thereby reduce its synthesis from linoleic acid, increasing linoleic acid levels and the formation of alternative linoleate metabolites (Supplementary Figure S4). Our observation that levels of arachidonoyl carnitine increased with aspirin treatment in blood, and in colon tissue in our prior analysis (11), provides support for this hypothesis. The mechanism for an increase in plasma levels of the various lysophospholipids that we identified is uncertain. However, changes in linoleic acid levels and related metabolites may have complex effects upstream on the modulation of glycerophospholipid metabolism. In contrast to previous research highlighting effects of aspirin downstream of cyclooxygenase inhibition via reductions in levels of anti-inflammatory prostaglandins (4, 8), our novel findings provide evidence that metabolites upstream of cyclooxygenase inhibition may be important to the chemopreventive effects of aspirin in the colorectum.
To our knowledge our study is unique in directly comparing the metabolomics effects of two different doses of aspirin in one study population. The variation that we observed by dose is plausible because aspirin uniquely incorporates two bioactive components in one molecule, a reactive acetyl moiety and a salicylate group, with different cellular targets and dose-response relationships (26). In our clinical trial, the 81 mg dose of aspirin appeared to be more efficacious than the 325 mg dose in preventing colorectal adenomas (10), although there is insufficient evidence to support our finding from a meta-analysis of randomized trials (43) and from observational studies that lack information on tablet dose (44, 45). In the present analysis, we observed that metabolites in linoleate and glycerophospholipid metabolism pathways associated with lower adenoma risk were increased with the 81 mg/day dose of aspirin but not statistically significantly associated with the 325 mg/day dose. Thus, these findings provide mechanistic evidence for a more beneficial profile for the lower 81 mg dose in adenoma prevention. An inverse dose response for aspirin has also been observed for ovarian cancer risk (46) and will be an important issue to address to optimize aspirin use for chemoprevention.
Major strengths of our work include the rigorous design of the randomized clinical trial with two doses of aspirin assessed and long-term follow-up of adenoma outcomes. In addition, measurement variation was reduced by performing a paired analysis of pre- and post-treatment blood plasma samples allowing individuals to serve as their own control. Finally, careful selection of participants for inclusion in the metabolomics analysis used detailed information on pre-study aspirin use, treatment compliance and non-protocol aspirin use during the study. This enabled the selection of all pre-study plasma samples and post-treatment samples from the placebo group with minimal aspirin exposure, as well optimizing exposure to the appropriate dose of aspirin in the post-treatment plasma samples from the two treatment groups.
There are also some limitations. Blood samples were only collected at baseline and year three, so we do not have data confirming self-reported treatment adherence from measurements of aspirin catabolites at earlier time points. The timing of year three blood collection after aspirin intake was not consistent between study participants, which likely added some variability in our metabolomics measurements. Another potential source of variability is that blood samples used for metabolomics measurements had undergone different numbers of freeze-thaw cycles, although analyses that allowed for this gave very similar results. Also, consistent with a current challenge in untargeted metabolomics, many metabolic features potentially linked to aspirin’s chemopreventive effects in our analysis are unidentified at this time.
An important limitation of this research is that due to the large number of pre- and post-treatment plasma samples that met our selection criteria, our analysis was divided into two independent runs but the run 2 findings did not replicate those from run 1. Although the two runs used separate sub-sets of participants, there were no noticeable differences in demographics or factors related to colorectal cancer risk raising concern about the potential for artifactual findings. We can offer no compelling explanation for the distinct findings, and can’t rule out the impact of chance and measurement variability. Nonetheless, our previous colon tissue analysis (11) provides support for the pathways and a few of the metabolites identified in run 1, leading us to speculate that the lack of replication may be due to unknown technical problems in run 2. This interpretation is further supported by the biological plausibility of our findings (as illustrated in Supplementary Figure S4) as well as consistency with prior research implicating some of the same metabolites in colorectal cancer risk.
Given these considerations our findings should be considered hypothesis-generating. With technical improvements in the field of metabolomics (47) some of these limitations may be addressed with better bioinformatics tools and analytical instrumentation to enable identification of unknowns as well as replication and validation of the present findings with increased sample sizes. Future investigation should include rigorous biospecimen collection and metabolomics analyses targeting linoleate and glycerophospholipid metabolism pathways in a population at increased risk with randomized aspirin exposure and long-term follow-up of precancerous or cancerous outcomes. To facilitate future research using the extensive metabolomics resource that we generated here, our data has been deposited into the metabolomics workbench repository (https://www.metabolomicsworkbench.org) where it is available to other scientists.
In conclusion, using a global untargeted metabolomics approach to analyze blood plasma samples from participants in a randomized clinical trial of aspirin for adenoma chemoprevention, we found that increases in lysophospholipids and linoleate metabolites in metabolic pathways upstream of cyclooxygenase inhibition appeared to be linked to reduced risk of colorectal adenomas due to aspirin treatment. Our novel findings expand on prior targeted research, which has focused on prostaglandins located downstream of cyclooxygenase inhibition as potential mediators of aspirin’s anticancer effects.
Supplementary Material
PREVENTION RELEVANCE.
This research used metabolomics, an innovative discovery-based approach, to identify molecular changes in human blood that may help to explain how aspirin use reduces the risk of colorectal neoplasia in some individuals. Ultimately, this work could have important implications for optimizing aspirin use in the prevention of colorectal cancer.
ACKNOWLEDGMENTS
The authors would like to thank the participants, investigators, and staff of the Aspirin/Folate Polyp Prevention Study for their valuable contributions.
Financial Support:
This research was funded by the National Cancer Institute at the National Institutes of Health (R01CA188038 to E. L. Barry and R01CA059005 to J. A. Baron). The study biorepository was supported by the National Institute of General Medical Sciences at the National Institutes of Health (P20GM104416 to M. R. Karagas). Aspirin and placebo tablets were provided by the Bayer Corporation.
Footnotes
Conflicts of Interest: Together with the Trustees of Dartmouth College, John A. Baron holds a use patent, not currently licensed, for the chemopreventive use of aspirin for colorectal cancer. All the other authors declare no potential conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713–32. [DOI] [PubMed] [Google Scholar]
- 3.Ricciotti E, FitzGerald GA. Aspirin in the Prevention of Cardiovascular Disease and Cancer. Annu Rev Med 2021;72:473–95. [DOI] [PubMed] [Google Scholar]
- 4.Drew DA, Cao Y, Chan AT. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat Rev Cancer 2016;16:173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosetti C, Santucci C, Gallus S, Martinetti M, La Vecchia C. Aspirin and the risk of colorectal and other digestive tract cancers: an updated meta-analysis through 2019. Ann Oncol 2020;31:558–68. [DOI] [PubMed] [Google Scholar]
- 6.Bibbins-Domingo K, Force USPST. Aspirin Use for the Primary Prevention of Cardiovascular Disease and Colorectal Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med 2016;164:836–45. [DOI] [PubMed] [Google Scholar]
- 7.Task Force Issues Draft Recommendation Statement on Aspirin Use to Prevent Cardiovascular Disease U.S. Preventive Services Task Force Bulletin; 2021. [Google Scholar]
- 8.Drew DA, Chan AT. Aspirin in the Prevention of Colorectal Neoplasia. Annu Rev Med 2021;72:415–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med 2003;348:891–9. [DOI] [PubMed] [Google Scholar]
- 11.Barry EL, Fedirko V, Uppal K, Ma C, Liu K, Mott LA, et al. Metabolomics Analysis of Aspirin’s Effects in Human Colon Tissue and Associations with Adenoma Risk. Cancer Prev Res (Phila) 2020;13:863–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. J Am Med Assoc 2007;297:2351–9. [DOI] [PubMed] [Google Scholar]
- 13.Block G, Thompson FE, Hartman AM, Larkin FA, Guire KE. Comparison of two dietary questionnaires validated against multiple dietary records collected during a 1-year period. J Am Diet Assoc 1992;92:686–93. [PubMed] [Google Scholar]
- 14.Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012;295:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013;9:S132–S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci 2015;148:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25:1930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 20.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 23.Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 2016;29:1956–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem 2017;89:1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, et al. The metabolomics standards initiative (MSI). Metabolomics 2007;3:175–8. [Google Scholar]
- 26.Schror K. Acetylsalicylic Acid: Wiley-Blackwell; 2016. p. 66–74. [Google Scholar]
- 27.Yerges-Armstrong LM, Ellero-Simatos S, Georgiades A, Zhu H, Lewis JP, Horenstein RB, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther 2013;94:525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellero-Simatos S, Lewis JP, Georgiades A, Yerges-Armstrong LM, Beitelshees AL, Horenstein RB, et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacometrics Syst Pharmacol 2014;3:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellero-Simatos S, Beitelshees AL, Lewis JP, Yerges-Armstrong LM, Georgiades A, Dane A, et al. Oxylipid Profile of Low-Dose Aspirin Exposure: A Pharmacometabolomics Study. J Am Heart Assoc 2015;4:e002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang JY, Lee SH, Chen YC, Wu CK, Chuang JY, Lo SC, et al. Metabolomic Analysis of Platelets of Patients With Aspirin Non-Response. Front Pharmacol 2019;10:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Minno A, Porro B, Turnu L, Manega CM, Eligini S, Barbieri S, et al. Untargeted Metabolomics to Go beyond the Canonical Effect of Acetylsalicylic Acid. J Clin Med 2019;9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liesenfeld DB, Botma A, Habermann N, Toth R, Weigel C, Popanda O, et al. Aspirin Reduces Plasma Concentrations of the Oncometabolite 2-Hydroxyglutarate: Results of a Randomized, Double-Blind, Crossover Trial. Cancer Epidemiol Biomarkers Prev 2016;25:180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, Chan EC. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther 2014;15:389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis 2014;35:2089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One 2016;11:e0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Wang J, Rao B, Deng L. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med 2017;13:2848–54. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.May-Wilson S, Sud A, Law PJ, Palin K, Tuupanen S, Gylfe A, et al. Pro-inflammatory fatty acid profile and colorectal cancer risk: A Mendelian randomisation analysis. Eur J Cancer 2017;84:228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khankari NK, Banbury BL, Borges MC, Haycock P, Albanes D, Arndt V, et al. Mendelian Randomization of Circulating Polyunsaturated Fatty Acids and Colorectal Cancer Risk. Cancer Epidemiol Biomarkers Prev 2020;29:860–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geijsen A, Brezina S, Keski-Rahkonen P, Baierl A, Bachleitner-Hofmann T, Bergmann MM, et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int J Cancer 2019;145:1221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geijsen A, van Roekel EH, van Duijnhoven FJB, Achaintre D, Bachleitner-Hofmann T, Baierl A, et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int J Cancer 2020;146:3256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNeil JJ, Gibbs P, Orchard SG, Lockery JE, Bernstein WB, Cao Y, et al. Effect of Aspirin on Cancer Incidence and Mortality in Older Adults. J Natl Cancer Inst 2021;113:258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole BF, Logan RF, Halabi S, Benamouzig R, Sandler RS, Grainge MJ, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst 2009;101:256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan AT, Giovannucci EL, Schernhammer ES, Colditz GA, Hunter DJ, Willett WC, et al. A prospective study of aspirin use and the risk for colorectal adenoma. Ann Intern Med 2004;140:157–66. [DOI] [PubMed] [Google Scholar]
- 45.Chudy-Onwugaje K, Huang WY, Su LJ, Purdue MP, Johnson CC, Wang L, et al. Aspirin, ibuprofen, and reduced risk of advanced colorectal adenoma incidence and recurrence and colorectal cancer in the PLCO Cancer Screening Trial. Cancer 2021;127:3145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnard ME, Poole EM, Curhan GC, Eliassen AH, Rosner BA, Terry KL, et al. Association of Analgesic Use With Risk of Ovarian Cancer in the Nurses’ Health Studies. JAMA Oncol 2018;4:1675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinu FR, Goldansaz SA, Jaine J. Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites 2019;9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics data generated in this study are publicly available in the NIH Common Fund’s National Metabolomics Data Repository, the Metabolomics Workbench https://www.metabolomicsworkbench.org, as Project ID PR000730. The data can be accessed directly via its Project DOI: 10.21228/M89X1C.


