Abstract
Acute lung injury (ALI) is a potentially life-threatening, devastating disease with an extremely high rate of mortality. The underlying mechanism of ALI is currently unclear. In this study, we aimed to confirm the hub genes associated with ALI and explore their functions and molecular mechanisms using bioinformatics methods. Five microarray datasets available in GEO were used to perform Robust Rank Aggregation (RRA) to identify differentially expressed genes (DEGs) and the key genes were identified via the protein-protein interaction (PPI) network. Lipopolysaccharide intraperitoneal injection was administered to establish an ALI model. Overall, 40 robust DEGs, which are mainly involved in the inflammatory response, protein catabolic process, and NF-κB signaling pathway were identified. Among these DEGs, we identified two genes associated with ALI, of which the CAV-1/NF-κB axis was significantly upregulated in ALI, and was identified as one of the most effective targets for ALI prevention. Subsequently, the expression of CAV-1 was knocked down using AAV-shCAV-1 or CAV-1-siRNA to study its effect on the pathogenesis of ALI in vivo and in vitro. The results of this study indicated that CAV-1/NF-κB axis levels were elevated in vivo and in vitro, accompanied by an increase in lung inflammation and autophagy. The knockdown of CAV-1 may improve ALI. Mechanistically, inflammation was reduced mainly by decreasing the expression levels of CD3 and F4/80, and activating autophagy by inhibiting AKT/mTOR and promoting the AMPK signaling pathway. Taken together, this study provides crucial evidence that CAV-1 knockdown inhibits the occurrence of ALI, suggesting that the CAV-1/NF-κB axis may be a promising therapeutic target for ALI treatment.
Subject terms: Cell signalling, Respiratory tract diseases
Introduction
Acute Lung Injury (ALI) is a serious respiratory illness that is commonly secondary to the systemic inflammatory response [1, 2]. The causes of ALI include infection, trauma, aspiration, and transfusion [3]. ALI can lead to severe acute respiratory distress syndrome (ARDS). Abrupt onset, quick development, and poor prognosis have caused the ARDS death to be as high as 30–40% [4]. However, the pathology of ALI is complicated and has not been fully elucidated. There is no specific method of treatment for ALI [5]. Common therapies mainly include primary disease management, mechanical ventilation, administration of vasodilators, surfactants, antioxidants, glucocorticoids, and anti-inflammatory drugs [6]. However, due to the high death rate, these traditional therapies are not sufficient [7, 8]. Improving our understanding of the mechanisms involved in ALI and the identification of pathological modulators are crucial for the studies of ALI [9, 10]. Therefore, analysis via bioinformatics may help explore the potential mechanisms involved in ALI and help identify novel targets for use in future clinical research.
Caveolin-1 (CAV-1) is the primary constituent of caveolae and plays a crucial role in maintaining its shape, structure, and function [11, 12]. Studies have found that CAV-1 is involved in multiple processes, including transmembrane transport, endocytosis, lipid metabolism, and signal transduction [13–15]. Importantly, CAV-1 functions as a mediator of many signaling pathways that lead to the activation of nuclear factor kappa B (NF-κB) [16]. NF-κB is a multi-protein complex and NF-κBp65 (RELA) is one of its most important constituents. As a primary transcription factor, NF-κBp65 participates in the regulation of various inflammatory mediators [17–19], while inflammation plays an important role in the development of ALI [20, 21]. Garrean et al. [22] found that the knockdown of CAV-1 suppresses the activation of NF-κB and inhibits the infiltration of inflammatory cells, to reduce overall mortality. Meanwhile, it has also been reported that the promotion of CAV-1 expression suppresses MAPK and NF-κB activation, alleviating pulmonary inflammation [23]. However, the exact mechanism by which the CAV-1/NF-κB axis is involved in ALI is still not clear.
In this study, we used bioinformatics methods to analyze online clinical five datasets and found that the differential expression of CAV-1 and NF-κBp65 was important for ALI. Furthermore, we further validated the roles of CAV-1 and NF-κBp65 in vivo and in vitro, while we also explored the potential mechanisms by which CAV-1 could provide novel targets for ALI treatment (Fig. 1A).
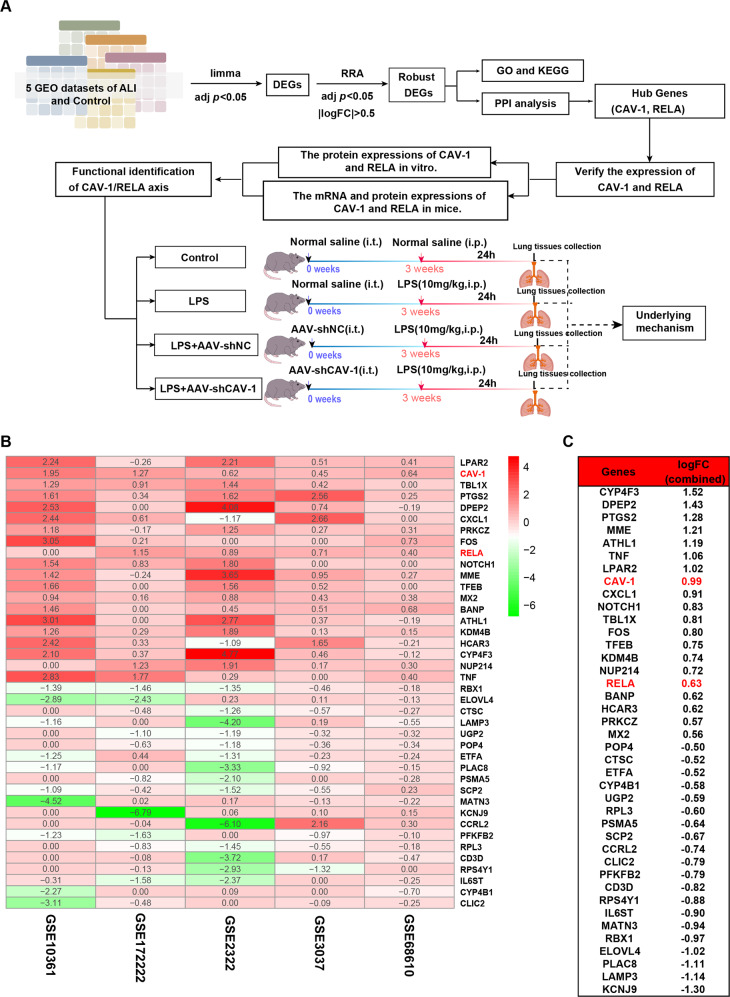
Fig. 1. The research scheme and RRA analysis of DEGs in ALI.
A Research scheme and a flow framework. We analyzed 5 ALI datasets using bioinformatics, screened out common differential genes, and performed GO, KEGG, and PPI analysis on these differential genes. The mRNA and protein expressions of the CAV-1 and RELA were verified in vitro and in vivo. The functions of CAV-1 and RELA were further verified to explore the potential mechanism in ALI. B Heatmap demonstrated the 40 robust DEGs between the ALI and control. Twenty genes were promoted and twenty genes were suppressed in ALI. Red shows upregulated gene expression and green shows downregulated gene expression in ALI. Numbers in the heatmap indicate log2 FC of genes in the five datasets compared with control groups. C The log2 FC of 40 DEGs was calculated from five datasets. The log2 FC sums of CAV-1 and RELA were 0.99 and 0.63, suggesting these two genes were significantly upregulated in ALI.
Materials and methods
GEO datasets retrieval and DEG identification
Microarray data were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Five datasets on the gene expression of ALI patients and healthy controls, including GSE68610 [24], GSE3037 [25], GSE172222 [26], GSE2322 [27], and GSE10361 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi] were obtained.
All data in the microarray datasets were processed using R language and the fold change between lung injury patients and controls were calculated. Identification of differentially expressed genes was performed using the R package, “limma”, and a p value < 0.05 was used to identify the differentially expressed genes (DEGs). Robust Rank Aggregation (RRA) was used to further identify robust DEGs, and a p value of <0.05 and log|FC| > 0.5 were considered to indicate significance.
Functional enrichment analysis of the DEGs
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of DEGs were performed using the R package, “Clusterprofiler”. A p value < 0.05 was considered to indicate statistical significance.
Construction of the PPI network
The tool for the retrieval of interacting genes (STRING, v11.5) is a database that can be used to search for and visualize networks of proteins and was used in this study to perform the protein-protein interaction (PPI) analysis. The networks were further visualized using the Cytoscape plug-in, “Cytohubba”, in which the degree of each node was illustrated using specific colors.
Construction of animal models and experimental procedures
Male 7–8 week old C57BL/6J mice were obtained from Hunan SJA Laboratory Animal CO., Ltd (Changsha, China). This study has been approved by the Ethics Committee for Animal Experiments of Hunan Normal University. 40 mice were randomly separated into four groups based on their weight. The four groups were the control, lipopolysaccharide (LPS), LPS + AAV-shNC, and LPS + AAV-shCAV-1 groups. The mice were treated with adeno-associated virus 6 through intratracheal instillation for 3 weeks, and the control group was treated with an equal amount of saline. Mice from each group received an intraperitoneal injection of LPS (10 mg/kg) and were killed 24 h later.
Lung edema
The level of lung edema in the mice was quantified by calculating the lung weight coefficient. After the mice were executed, whole lung tissues were immediately collected and weighed using an electronic balance. The lung weight coefficient was calculated as the wet weight of lung/the weight of the mouse × 100%.
Enzyme-linked immunosorbent assay (ELISA)
The expression levels of the cytokines, TNF-α, IL-1β, IL-6, and IL-18 were evaluated by ELISA following the manufacturer’s instructions (Beyotime, Shanghai, China). The evaluation was performed on 4 duplicates of each group to ensure constancy.
Haematoxylin-eosin (H&E) staining and histological assessment
Lung tissues of the mice were fixed using 4% formalin and embedded in paraffin followed by sectioning into 4-μm sections. The tissues were stained using H&E following standard procedures. The pathological characteristics of the tissues were evaluated by light microscopy. The degree of injury of lung tissue was divided into grades 0–4 [28]. Grade 0: no injury; grade 1: injury in <25% of the field of observation; grade 2: injury in 25–50% of the field of observation; grade 3: injury in 50–75% of the field of observation; grade 4: injury throughout the field of observation. At least five fields of observation were randomly selected and analyzed in each sample.
Immunohistochemistry analysis
Lung tissues were obtained from the mice and embedded in paraffin. After dewaxing and antigen thermal repair, 3% H2O2 solution was applied for 10 minutes to inactivate endogenous peroxidase, and 1% BSA was used for blocking. The primary antibodies of CAV-1, NF-κBp65, CD3, and F4/80 were incubated at 4 °C overnight, and corresponding secondary antibodies were added at room temperature and incubated for 2 h. Subsequently, the samples were incubated with DAB and hematoxylin staining solution. Finally, the samples were observed under a microscope and analyzed using ImageJ software (Bethesda, USA). The primary antibodies used are presented in Table S1.
Cell culture and small-interfering RNA transfection
Bone marrow-derived macrophages (BMDMs) obtained from the mice were cultured in DMEM medium containing 30% L929-cell conditioned medium, 10% fetal bovine serum, and 1% penicillin/streptomycin, and the cells were placed in a humidified atmosphere at 37 °C with 5% CO2 for 7 days [29]. On day 4, the cells were treated with fresh medium, and on day 7 cells were seeded into new plates and incubated overnight. According to the instructions provided by the manufacturer, the cells were transfected with CAV-1-siRNA and scrambled-siRNA using a Lipofectamine 2000 system. After 24 h, the cells were co-incubated with 1 μg/mL of LPS, and were harvested after 24 h of LPS administration.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from mouse lung tissues using TRIzol reagent following the manufacturer’s instructions. Reverse transcription into cDNA was performed using a HiScript III RT SuperMix Kit (Vazyme, China). Real-time PCR analysis was conducted to detect the expression levels of TNF-α, IL-1β, IL-6, IL-18, CAV-1, NF-κBp65, and GAPDH using the SYBR Green qPCR Kit (Vazyme, Nanjing, China). The 2−ΔΔCT method was used to quantify cDNA copy numbers. The mRNA levels were normalized to that of the reference gene, GAPDH. The primers used are presented in Table S2.
Western blotting analysis
Lung tissue and cells were collected, and protein was extracted using a RIPA lysis buffer, and protein concentrations were measured using BCA (Beyotime, China). The proteins were loaded onto 10–12% SDS-PAGE and transferred onto PVDF membranes, which were blocked using 5% non-fat milk at room temperature for 1 h, and incubated with primary antibodies overnight at 4 °C. After washing the membrane with TBST three times, the membrane was incubated with the secondary antibody at room temperature for 1 h. Finally, a chemiluminescence reagent imaging system was used to detect the bands, and all the bands were measured using ImageJ software (Bethesda, USA). The primary antibodies used are shown in Table S1.
Immunofluorescence analysis
The BMDMs were fixed using 4% paraformaldehyde, and permeated with 0.2% Triton X-100 for 10 minutes. Then, 5% bovine serum albumin was used for blocking for 1 h at room temperature. Subsequently, the cells were incubated with the primary antibodies, CAV-1 and NF-κBp65 (Table S1) overnight at 4°C, followed by incubation with the fluorescence secondary antibody for 1 h in the dark. The nuclei were stained using DAPI and the samples were sealed using the fluorescence quenching agent and observed under a confocal microscope (Leica, Germany).
Transmission electron microscopic analysis
The samples were prepared according to standard protocols. In brief, the BMDMs were fixed using 2.5% glutaraldehyde solution at 4 °C for 2 h and 2% osmium tetroxide solution for 2 h. Then the sample was dehydrated using an acetone gradient, the cells were embedded, cut into ultrathin sections, the autophagosomes were stained and observed under a transmission electron microscopic (HITA-CHI, Japan).
Statistical analyses
All the results are presented as mean ± SEM of at least four replicate experiments. Comparisons between groups were conducted using the Student’s t test or one-way ANOVA, and a p value of <0.05 was considered to indicate statistical significance.
Results
Identification of DEGs between ALI and normal controls using the GEO datasets
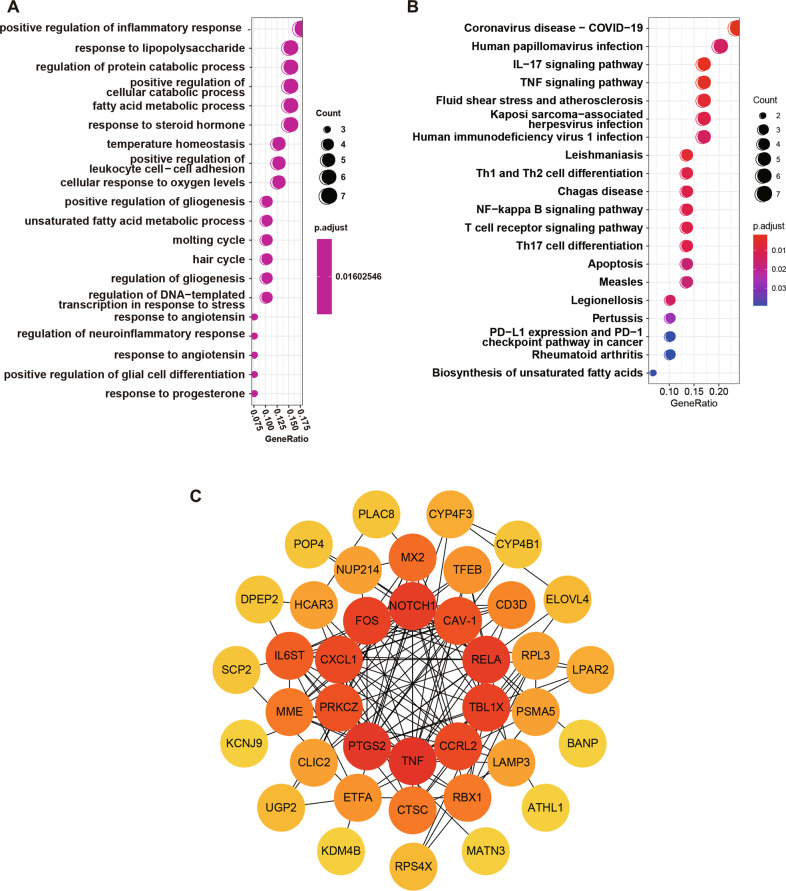
To identify the potential genes that are involved in ALI, five GEO datasets were analyzed and a total of 400 DEGs were identified. The subsequent RRA analysis found 40 robust DEGs, among which 20 genes were upregulated and the other 20 genes were downregulated (Table S3). In particular, CAV-1 and RELA expression levels were upregulated in the ALI groups (Fig. 1B, C). The functional enrichment analysis, included GO and KEGG pathways, and the most significantly enriched terms are shown in Fig. 2A, B. The 40 robust EDGs were primarily associated with the regulation of inflammatory response, response to lipopolysaccharide, protein catabolic process, and fatty acid metabolic process (Fig. 2A, Table S4). Meanwhile, the most related pathways included the IL-17 signaling pathway, TNF signaling pathway, and NF-κB signaling pathway (Fig. 2B, Table S5).
Fig. 2. Functional enrichment and PPI network construction of DEGs.
A, B Bubble plots show the results of GO functional enrichment and KEGG signaling pathway enrichment of DEGs. C The PPI network of the DEGs. Nodes represent proteins, edges represent interactions between proteins, and the intensity of the color indicates the degree of interaction of each protein. Proteins with more interaction were shown in the center of the network.
CAV-1 and RELA were recognized as hub genes related to ALI
We constructed the PPI network of the 40 robust DEGs using the STRING website. The network contained 40 nodes and 136 edges, and was visualized using Cytoscape software (Fig. 2C). The top 10 proteins with the most interactions were: TNF, PTGS2, RELA, CAV-1, NOTCH1, TBL1X, FOS, CCRL2, CXCL1, and PRKCZ. Ultimately, we selected CAV-1 and RELA for further analysis, but the functions and molecular mechanisms of involvement of these two genes in ALI is unclear.
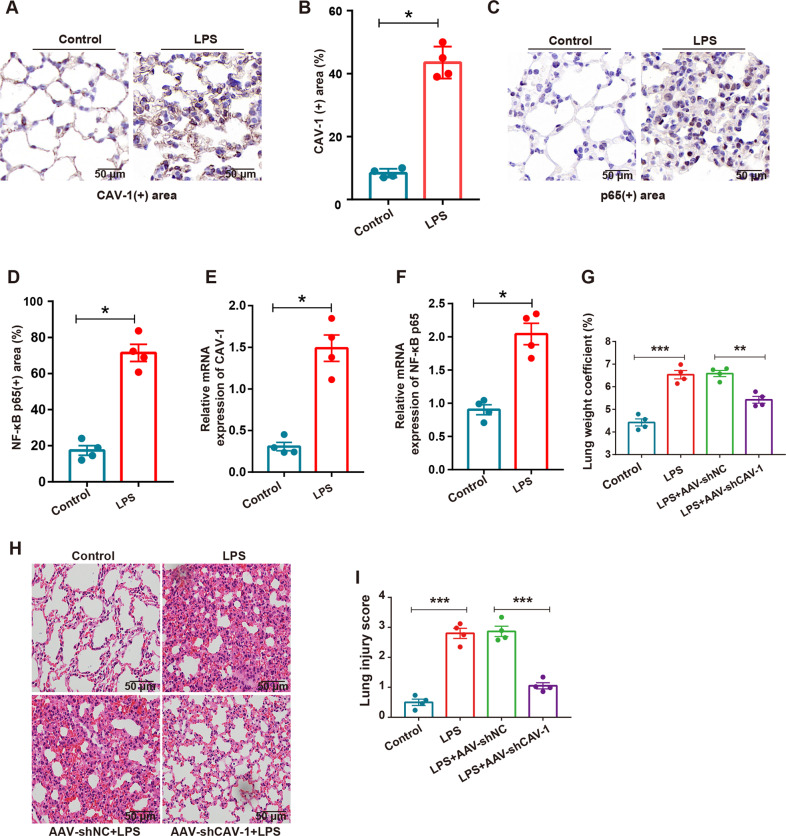
Verification of the expression levels of CAV-1 and NF-κBp65 in vivo
To confirm the expression levels of CAV-1 and NF-κBp65 in ALI, we detected the expression levels of CAV-1 and NF-κBp65 in lung tissue. Immunohistochemical staining and quantitative analysis showed that LPS significantly increased the expression of CAV-1 and NF-κBp65 in lung tissues of mice compared with the control group (Fig. 3A–D). The expression levels of CAV-1 and NF-κBp65 were detected using qRT-PCR analysis, and a significant increase in CAV-1 and NF-κBp65 expression levels were observed following LPS treatment compared with the control group (Fig. 3E, F). These results are consistent with the results of our previous bioinformatics research, indicating that CAV-1 and NF-κBp65 may play important roles in the development of ALI.
Fig. 3. Expression of CAV-1 and NF-κB in mouse model of ALI.
A–D The expression levels of CAV-1 and NF-κBp65 in lung tissues were analyzed by IHC. Scale bar = 50 μm. E, F qRT-PCR was performed to detect the levels of CAV-1 and NF-κBp65 in lung tissues. G The degree of pulmonary edema in each group was evaluated by lung weight coefficient. H, I H&E staining was used to detect the pathological changes in lung tissue and the score of morphological injury of lung tissue in each group. Scale bar = 50 μm. Results were represented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
CAV-1 silencing ameliorated LPS-induced lung injury in mice
To explore the relationship between CAV-1 and the development of ALI, we treated ALI model mice with AAV carrying shCAV-1 to silence the expression of CAV-1. Compared with the control group, qRT-PCR and western blotting analysis confirmed that the expression of CAV-1 in the AAV-shCAV-1 group decreased by ~80% (Fig. S1A, B). The severity of lung injury was evaluated using the lung weight coefficient and H&E. The lung weight coefficient of LPS-induced ALI was significantly elevated, compared with the control. AAV-shCAV-1 injection could significantly decrease the lung weight coefficient after treatment with LPS (Fig. 3G). Accordingly, in the CAV-1 silenced mice treated with LPS, H&E staining showed that the infiltration of inflammatory cells decreased significantly, and the alveolar walls were significantly thinner (Fig. 3H, I). Overall, these results indicate that blocking CAV-1 could reduce ALI severity in the mice.
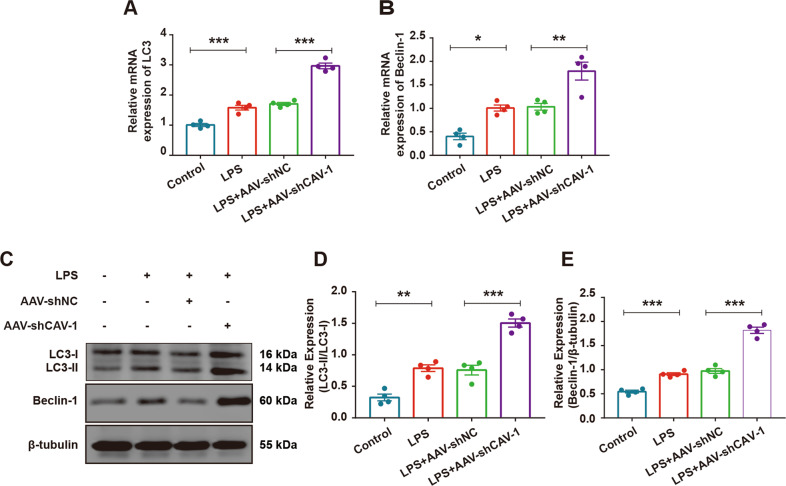
CAV-1 silencing could promote autophagy in the LPS- induced lung injury model mice
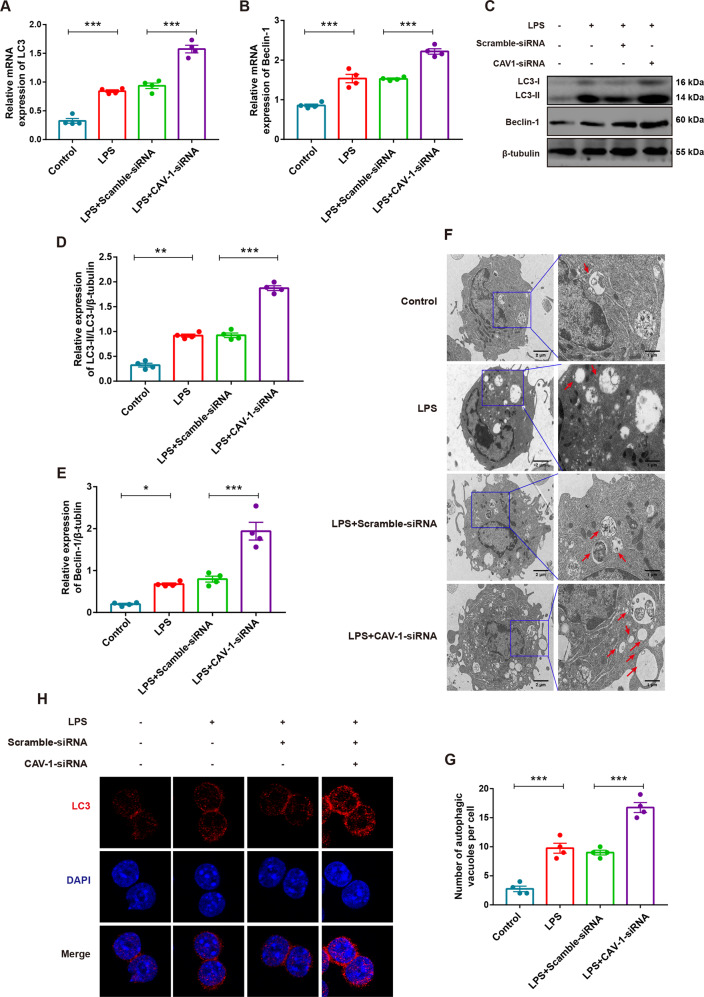
Autophagy plays an important role in the occurrence and development of ALI. To investigate the effect of CAV-1 on autophagy in the mice, we silenced CAV-1 in the mice using AAV-shCAV-1 to observe its influence on autophagy-related proteins. The results showed that both the mRNA and the protein levels of LC3II/I, Beclin-1, and Atg5 increased and p62 decreased after LPS stimulation. Meanwhile, the expression levels of LC3II/I, Beclin-1, and Atg5 were further upregulated and p62 was significantly downregulated after the administration of AAV-shCAV-1 (Fig. 4A–E, Fig. S2A–F), suggesting that CAV-1 silencing could promote autophagy and ameliorate LPS-induced lung injure in mice.
Fig. 4. Knocking down of CAV-1 improved autophagy and ameliorated ALI.
A, B qRT-PCR demonstrated the mRNA levels of LC3 and Beclin-1 in the lung tissues. C The protein levels of LC3II/I and Beclin-1 were examined by Western blotting in lung tissues. D, E Quantification of LC3 and Beclin-1 protein bands. Results were represented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
CAV-1 silencing inhibited the pulmonary inflammatory response
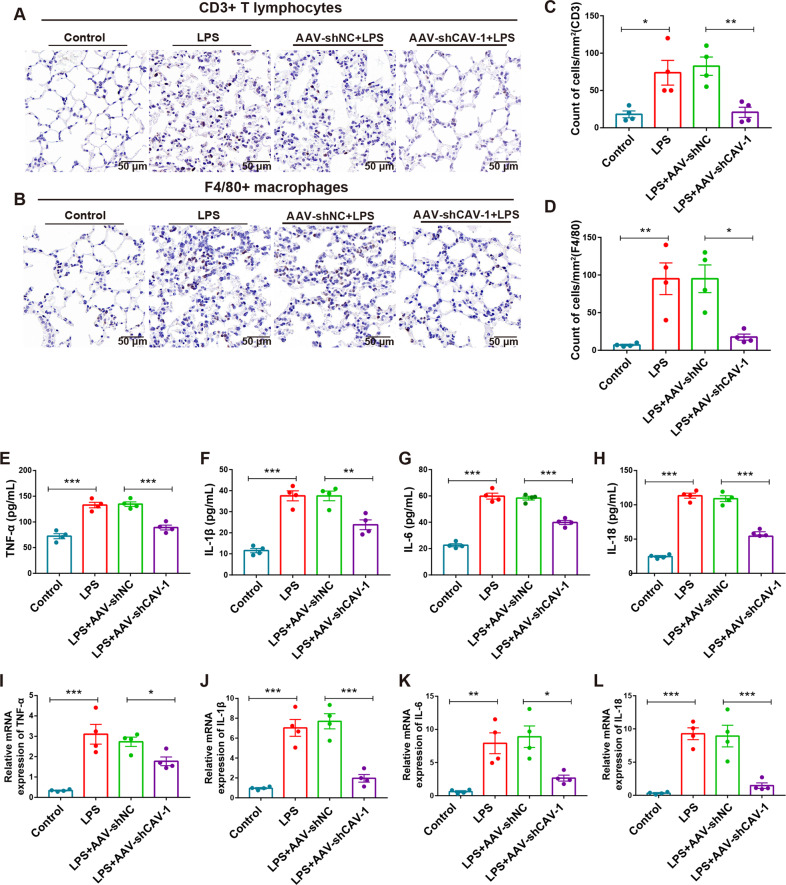
Subsequently, to explore the immune mechanism of CAV-1 in LPS-induced ALI, we evaluated whether CAV-1 silencing could regulate pulmonary inflammation. According to our immunohistochemistry results of the mice lung tissues obtained from each group, LPS treatment increased the infiltration of CD3+ T lymphocytes compared with the control, while AAV-shCAV-1 administration significantly reversed the accumulation of CD3+ T cells (Fig. 5A, C). Similar results were observed with the amount of F4/80+ macrophages (Fig. 5B, D), indicating that CAV-1 knockdown prevented immune cell infiltration. Meanwhile, we evaluated the secretion of proinflammatory cytokines in ALI. The levels of IL-1β, IL-6, IL-18, and TNF-α were upregulated in the LPS-treated mice compared with control mice, while AAV-shCAV-1 injection could significantly decrease these expression levels (Fig. 5E–H). Similar results were observed with the mRNA expression levels of the proinflammatory cytokines in lung tissues (Fig. 5I–L). These results indicate that the silencing CAV-1 using AAV-shCAV-1 could suppress the excessive immune response in LPS-induced ALI.
Fig. 5. Knockdown of CAV-1 could reduce the infiltration of CD3+ T cells and F4/80+ macrophages and inhibit proinflammatory cytokines.
A, C The expression of CD3 protein in T cells was detected by immunohistochemistry and the quantification of CD3+ T cells. B, D The protein levels of F4/80 was determined by IHC and quantification of F4/80+ cells. Scale bar = 50 μm. E–H The secretion of TNF-α, IL-1β, IL-6, and IL-18 in serum were determined by ELISA. I–L qRT-PCR analysis of TNF-α, IL-1β, IL-6, and IL-18 in lung tissues. Results were represented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
CAV-1 knockdown upregulated the level of autophagy in the BMDMs
To investigate the role of CAV-1 in BMDMs, two pairs of CAV-1-siRNA were transfected into BMDMs to interfere with the expression of CAV-1. As shown in Fig. S3A, B, qRT-PCR analysis and western blotting showed that the expression of CAV-1 decreased by ~85% in the CAV-1-siRNA group, compared with the control groups. CAV-1 and autophagy play vital regulatory roles in ALI. To more closely elucidate the function of CAV-1 on autophagy, CAV-1 was knocked down in the LPS-treated cells using CAV-1-siRNA. The mRNA and protein levels of LC3II/I and Beclin-1 were upregulated, compared with the control groups. The knockdown of CAV-1 could further increase these mRNA and protein expression levels after LPS treatment (Fig. 6A–E). Furthermore, cells in the LPS groups exhibited a larger number of autophagosomes in BMDMs, compared with the control groups. Autophagosome formation increased significantly in the CAV-1 knockdown BMDMs after LPS treatment (Fig. 6F, G). Meanwhile, immunofluorescence assays showed that autophagy was activated by increasing the formation of LC3 puncta after stimulation by LPS, compared with the controls. CAV-1 knockdown could markedly increase the level of LC3 after treatment with LPS (Fig. 6H). These results suggest that the knockdown of CAV-1 promotes the activation of autophagy in BMDMs.
Fig. 6. Knockdown of CAV-1 activated autophagy in BMDMs.
A, B The mRNA levels of LC3 and Beclin-1 by qRT-PCR in BMDMs. C The protein expression of LC3II/I and Beclin-1 were assessed by western blotting in BMDMs. D, E Quantification of LC3 and beclin-1 expression. F The formation of autophagosomes in different groups of BMDMs was observed by TEM. Red arrows indicate autophagosomes. G Quantitative analysis of BMDMs autophagosomes number in each group. H Immunofluorescence analysis of LC3 (red) expression in BMDMs (magnification ×63). Results were represented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
CAV-1 knockdown inhibited the AKT/mTOR and promoted AMPK signaling pathways
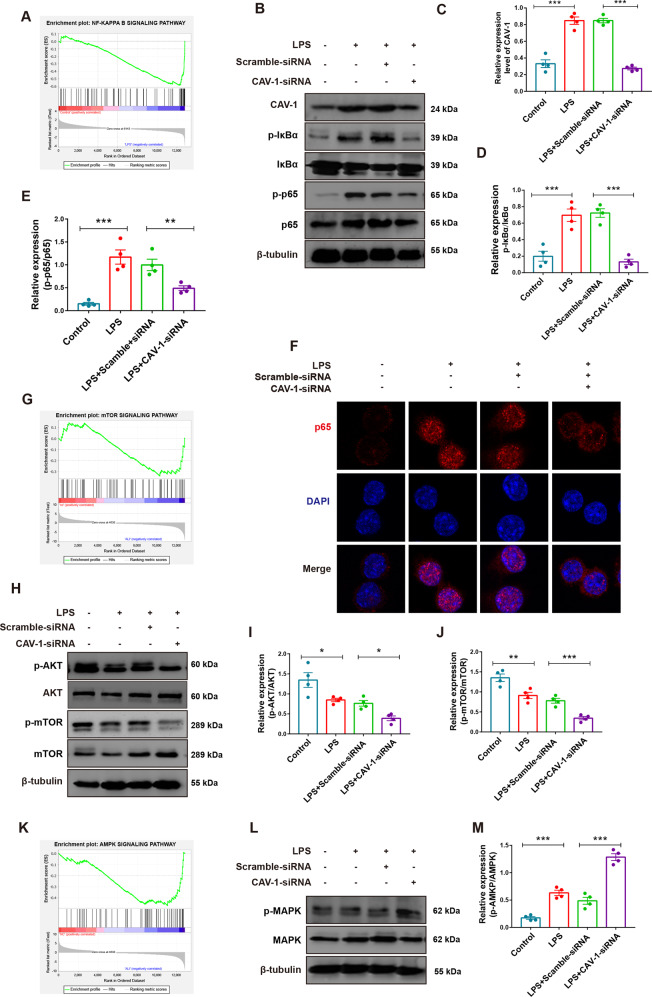
Autophagy-related pathways AKT/mTOR and AMPK play important roles in many diseases. To elucidate the precise mechanism by which CAV-1 promotes ALI, we determined whether CAV-1 expression was associated with the NF-κB, AKT/mTOR, and AMPK signal pathways, which are crucial regulators of autophagy. Gene set enrichment analysis showed that NF-κB, mTOR, and AMPK signaling pathway was significantly enriched in ALI group (Fig. 7A, G, K). Western blotting revealed that CAV-1, IκBα, and NF-κBp65 were significantly activated after LPS administration, compared with the control, and this effect was significantly attenuated by CAV-1 knockdown (Fig. 7B–E). Meanwhile, we analyzed the nuclear translocation of NF-κBp65 using immunofluorescence staining, and CAV-1 knockdown significantly reduced LPS-induced nuclear translocation of NF-κBp65 (Fig. 7F). Furthermore, AKT and mTOR phosphorylation levels in the LPS exposed DMBMs decreased, and these phosphorylation levels were more significantly attenuated after CAV-1 knockdown (Fig. 7H–J). Subsequently, the phosphorylation level of AMPK increased significantly after LPS administration, while transfection with CAV-1-siRNA effectively reduced the level of AMPK phosphorylation (Fig. 7L, M). Taken together, we demonstrated that CAV-1 knockdown could increase autophagy in part by inhibiting the NF-κB and AKT/mTOR pathway and promoting the AMPK pathway.
Fig. 7. Knockdown of CAV-1 inhibited autophagy-related signaling pathways in BMDMs.
A GSEA analysis of NF-κB signaling pathway in ALI samples. B Western blotting analysis of CAV-1, p-IκBα, IκBα, p-NF-κBp65, and NF-κBp65 protein levels in BMDMs. C–E Quantification of these protein bands. F Confocal laser immunofluorescence showed NF-κBp65 nuclear translocation in BMDMs from different groups (magnification ×63). G GSEA analysis of mTOR signaling pathway in ALI sample. H, L The expression of p-AKT, AKT, p-mTOR, mTOR, p-AMPK, and AMPK were detected by western blotting in BMDMs. I, J, M Quantification of these protein bands. K GSEA analysis of AMPK signaling pathway in ALI samples. Results were represented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
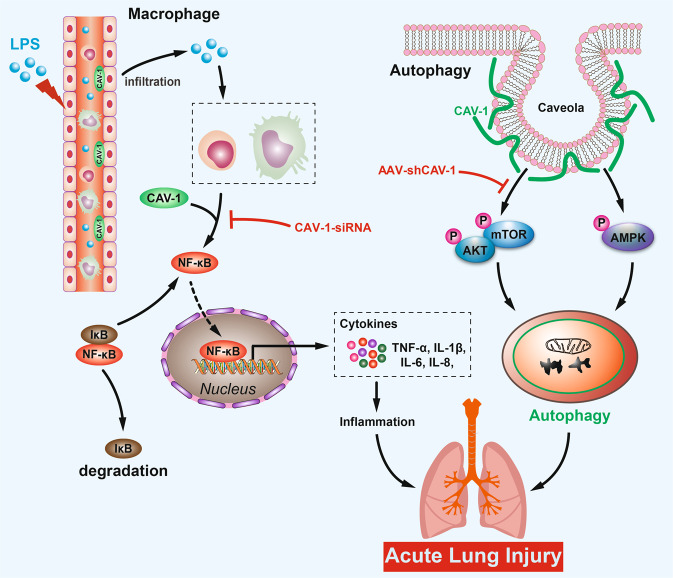
In this study, we found that activation of the CAV-1/NF-κB axis was associated with LPS-induced inflammatory response and autophagy in ALI. Inhibition of the CAV-1/NF-κB axis through CAV-1 knockdown could markedly reduce the severity of LPS-induced ALI by promoting the autophagy-related pathway, AMPK, and by inhibiting the activation of the AKT/mTOR pathway and the inflammatory response caused by the infiltration of CD4/80+ macrophages and CD3+ T lymphocytes. Taken together, these results indicate that the CAV-1/NF-κB axis could be a potential therapeutic target for ALI (Fig. 8).
Fig. 8. Molecular mechanism of CAV-1-mediated LPS-induced ALI development.
CAV-1 expression is upregulated in LPS-induced ALI, which recruits CD3+ T lymphocytes and F4/80+ macrophages to lung tissue, thereby triggering inflammatory response. Meanwhile, upregulated CAV-1 could promote AKT/mTOR and inhibit AMPK to downregulate autophagy, resulting in ALI. Conversely, knockdown of CAV-1 ameliorates ALI.
We first used bioinformatics analysis to identify CAV-1 and NF-κB as key genes among the DEGs that were significantly upregulated in ALI. CAV-1 is a protein located on cell membranes that can regulate cell signaling pathways through certain intracellular signal transduction molecules [30]. Each signaling pathway converges in NF-κB, which is a key pathway of the inflammatory response that contributes to the occurrence of ALI [31, 32]. He et al. [33] found that the CAV-1/NF-κB pathway could increase macrophage infiltration into lung tissue and promote the occurrence of ALI, while CAV-1 deficiency aggravated ALI. The CAV-1/NF-κB axis is also involved in various pathological processes, such as tumorigenesis [34], inflammatory response [32], aerobic glycolysis [35], autophagy [36], and apoptosis, and may be a promising therapeutic target for ALI treatment. At present, few studies have reported that CAV-1 is associated with ALI, and its precise mechanism is not fully understood [37]. Our in vivo and in vitro experiments identified that the hyperactivation of the CAV-1/NF-κB axis was associated with ALI, which was consistent with the results of the bioinformatics analysis. Subsequently, CAV-1 knockdown was used to explore its molecular mechanism in ALI through autophagy and inflammatory response.
Immune cell infiltration and inflammatory responses play an important role in ALI [38, 39]. In sepsis-induced ALI, immune cells migrate through the vessels into the alveoli, causing pulmonary endothelial cell damage that leads to ALI [20]. Leukocytes and macrophages play a crucial part in ALI development [40]. Meanwhile, the infiltration of innate immune cells contributes to the early-stage secretion of cytokines, including TNF-α, IL-1β, IL-6, IL-10, and IL-18, which further induced an excessive immune response in the alveoli to aggravate ALI [41–43]. CD3+ T cells are also involved in ALI through both direct tissue damage and indirect injury by producing inflammatory factors [44]. CAV-1 and NF-κB have been reported to participate in the development of ALI [22]. Garrean et al. [22] reported that CAV-1 could control the activation of NF-κB, which promotes the inflammation response in ALI. Xu L et al. [23] showed that Dex administration promoted the expression of CAV-1 and suppressed the activation of NF-κB, thus reducing LPS-induced pulmonary injury. However, the precise mechanism by which the CAV-1/NF-κB axis is involved in ALI remains unclear. In our study, we showed that the infiltration of CD3+ T cells and F4/80+ macrophages increased after LPS administration, which is consistent with the results of previous studies. Importantly, CAV-1 knockdown by AAV-shCAV-1 in mice suppressed elevated recruitment of T cells and macrophages. Meanwhile, LPS administration promoted the secretion of TNF-α, IL-1β, IL-6, and IL-18, but AAV-shCAV-1 decreased the expression levels of these cytokines as well as their mRNA level expression. Interestingly, we verified that the knockdown of CAV-1 reduced the activation and nuclear translocation of NF-κBp65. Considering that NF-κB is a crucial mediator of inflammatory factor transcription, we suggest that the CAV-1/NF-κB axis may regulate immune infiltration and cytokine secretion.
Autophagy, a process by which proteins and damaged organelles are degraded, plays a critical role in maintaining the homeostasis of the intracellular environment and cell survival [45, 46]. Autophagy is reportedly relevant in multiple processes, including cell renovation, inflammation, and pathogen defense [47], and plays a crucial role in various diseases, such as neurodegenerative disorders and cancers [48]. In the process of autophagy formation, the cytoplasmic LC3-I was transformed into LC3II, and the expression of autophagy markers Atg5 and Beclin-1 increased [49]. Meanwhile, p62 could be degraded by proteolytic enzymes in autophagolysosomes, and the expression of p62 is negatively correlated with autophagy activity [50, 51]. In recent years, autophagy has been shown to play a crucial role in lung diseases [52]. Selective autophagy is associated with NF-κB activation and induces inflammation, which plays a part in ALI [53]. In addition, autophagy could directly participate in ALI by regulating NALP3-dependent inflammation [54]. Chen et al. [55] demonstrated that CAV-1 could bind to ATG12-ATG5, and that the knockdown of CAV-1 led to the dissociation of the complex and induced autophagy in lung epithelial cells. However, the mechanism by which CAV-1 and autophagy are involved in ALI remains to be explored. In our study, in vivo, and in vitro experiments showed that CAV-1 knockdown promoted autophagy-related proteins LC3II/I, Beclin-1, Atg5 and downregulation of p62 expression in ALI. To further investigate the mechanism involved, we examined the activation of autophagy-related pathways. Our results indicated that CAV-1 downregulation suppressed the AKT/mTOR pathway and activated autophagy, while the AMPK pathway was promoted.
In conclusion, this study confirmed the role of CAV-1/NF-κB in ALI, and demonstrated the mechanism by which CAV-1 knockdown upregulated autophagy to attenuate LPS-induced ALI. Nevertheless, the relationship between CAV-1 and autophagy in ALI is still not clear. Therefore, future research should focus on clarifying the detailed mechanism to support its clinical application.
Conclusions
Taken together, this study showed that activation of the CAV-1/NF-κB axis in LPS-induced ALI was associated with a decrease in autophagy and an increase in inflammation in the lung tissue. CAV-1 knockdown could block the CAV-1/NF-κB axis to significantly suppress ALI by inhibiting the autophagy-related signaling pathway, AKT/mTOR, and promoting AMPK activation, as well as inhibiting the inflammatory response caused by the infiltration of CD3+ and F4/80+ cells. However, further functional experiments need to be performed to gain a deeper understanding of the underlying mechanisms and support the blockade of the CAV-1/NF-κB axis, which may provide broad prospects for the future clinical treatment of ALI.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China [no. 81100054].
Author contributions
LHQ, YL, and LS contributed to the concept of the manuscript, and experiments and edited it. TY performed the preparation of the paper. QF, CC, YJZ, WTL, ZQL, and YYC participated in data analysis and experiments. All authors reviewed and approved the final version of the manuscript.
Data availability
All data required for the conclusion of the manuscript can be found in the article. Additional raw data related to this study are available on request from corresponding authors.
Competing interests
The authors declare no competing interests.
Ethics statement
All animal experiments were approved by the Medical Ethics Committee of Hunan Normal University.
Footnotes
Edited by Dr Yufang Shi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lihua Qu, Yi Li, Chao Chen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-05134-8.
References
- 1.Li W, Long L, Yang X, Tong Z, Southwood M, King R, et al. Circulating BMP9 protects the pulmonary endothelium during inflammation-induced lung injury in mice. Am J Respir Crit Care Medi. 2021;203:1419–30. doi: 10.1164/rccm.202005-1761OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, Wei Y, Zhu G, Zhao H, Zhang X. Design, synthesis and structure-activity relationship studies of 4-indole-2-arylaminopyrimidine derivatives as anti-inflammatory agents for acute lung injury. Eur J Med Chem. 2021;225:113766. doi: 10.1016/j.ejmech.2021.113766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler AP, Bernard GR. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet (London, England) 2007;369:1553–64. doi: 10.1016/S0140-6736(07)60604-7. [DOI] [PubMed] [Google Scholar]
- 4.Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. Lancet (London, England) 2021;398:622–37. doi: 10.1016/S0140-6736(21)00439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su R, Zhang Y, Zhang J, Wang H, Luo Y, Chan HF, et al. Nanomedicine to advance the treatment of bacteria-induced acute lung injury. J Mater Chem B. 2021;9:9100–15. doi: 10.1039/D1TB01770E. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita CM, Lewis JF. Emerging therapies for treatment of acute lung injury and acute respiratory distress syndrome. Expert Opin Emerg Drugs. 2012;17:1–4. doi: 10.1517/14728214.2012.667800. [DOI] [PubMed] [Google Scholar]
- 7.Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, et al. Acute respiratory distress syndrome. Nat Rev Dis Prim. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J, Huang K, Xu S, Garcia JGN, Wang C, Cai H. Targeting NOX4 alleviates sepsis-induced acute lung injury via attenuation of redox-sensitive activation of CaMKII/ERK1/2/MLCK and endothelial cell barrier dysfunction. Redox Biol. 2020;36:101638. doi: 10.1016/j.redox.2020.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He YQ, Zhou CC, Yu LY, Wang L, Deng JL, Tao YL, et al. Natural product derived phytochemicals in managing acute lung injury by multiple mechanisms. Pharmacol Res. 2021;163:105224. doi: 10.1016/j.phrs.2020.105224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao K, He W, Guan W, Hou F, Yan P, Xu J, et al. Mesenchymal stem cells reverse EMT process through blocking the activation of NF-κB and Hedgehog pathways in LPS-induced acute lung injury. Cell Death Dis. 2020;11:863. doi: 10.1038/s41419-020-03034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Ariotti N, Rae J, Liang H, Tillu V, Tee S, et al. Caveolin-1 and cavin1 act synergistically to generate a unique lipid environment in caveolae. J Cell Biol. 2021;220:e202005138. [DOI] [PMC free article] [PubMed]
- 12.Tang W, Li Y, Li Y, Wang Q. Caveolin-1, a novel player in cognitive decline. Neurosci Biobehav Reviews. 2021;129:95–106. doi: 10.1016/j.neubiorev.2021.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 14.Parton RG, Tillu VA, Collins BM. Caveolae. Curr Biol. 2018;28:R402–r405. doi: 10.1016/j.cub.2017.11.075. [DOI] [PubMed] [Google Scholar]
- 15.Nwosu ZC, Ebert MP, Dooley S, Meyer C. Caveolin-1 in the regulation of cell metabolism: a cancer perspective. Mol Cancer. 2016;15:71. doi: 10.1186/s12943-016-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parton RG. Caveolae: structure, function, and relationship to disease. Annu Rev Cell Dev Biol. 2018;34:111–36. doi: 10.1146/annurev-cellbio-100617-062737. [DOI] [PubMed] [Google Scholar]
- 17.Gross CM, Kellner M, Wang T, Lu Q, Sun X, Zemskov EA, et al. LPS-induced acute lung injury involves NF-κB-mediated downregulation of SOX18. Am J Respir Cell Mol Biol. 2018;58:614–24. doi: 10.1165/rcmb.2016-0390OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Wang X, Wei X, Zheng SW, Lenhart BJ, Xu P, et al. Multiplex quantitative detection of SARS-CoV-2 specific IgG and IgM antibodies based on DNA-assisted nanopore sensing. Biosens Bioelectron. 2021;181:113134. doi: 10.1016/j.bios.2021.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209. doi: 10.1038/s41392-020-00312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar V. Pulmonary innate immune response determines the outcome of inflammation during pneumonia and sepsis-associated acute lung injury. Front Immunol. 2020;11:1722. doi: 10.3389/fimmu.2020.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang JS, Kim KH, Park J, Kim SM, Cho H, Lee Y, et al. Glucosamine improves survival in a mouse model of sepsis and attenuates sepsis-induced lung injury and inflammation. J Biol Chem. 2019;294:608–22. doi: 10.1074/jbc.RA118.004638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrean S, Gao XP, Brovkovych V, Shimizu J, Zhao YY, Vogel SM, et al. Caveolin-1 regulates NF-kappaB activation and lung inflammatory response to sepsis induced by lipopolysaccharide. J Immunol. 2006;177:4853–60. doi: 10.4049/jimmunol.177.7.4853. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Li T, Chen Q, Liu Z, Chen Y, Hu K, et al. The α2AR/Caveolin-1/p38MAPK/NF-κB axis explains dexmedetomidine protection against lung injury following intestinal ischaemia-reperfusion. J Cell Mol Med. 2021;25:6361–72. doi: 10.1111/jcmm.16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang X, Abbott J, Cheng L, Colby JK, Lee JW, Levy BD, et al. Human mesenchymal stem (stromal) cells promote the resolution of acute lung injury in part through lipoxin A4. J Immunol. 2015;195:875–81. doi: 10.4049/jimmunol.1500244. [DOI] [PubMed] [Google Scholar]
- 25.Silva E, Arcaroli J, He Q, Svetkauskaite D, Coldren C, Nick JA, et al. HMGB1 and LPS induce distinct patterns of gene expression and activation in neutrophils from patients with sepsis-induced acute lung injury. Intens Care Med. 2007;33:1829–39. doi: 10.1007/s00134-007-0748-2. [DOI] [PubMed] [Google Scholar]
- 26.Saren G, Wong A, Lu YB, Baciu C, Zhou W, Zamel R, et al. Ischemia-reperfusion injury in a simulated lung transplant setting differentially regulates transcriptomic profiles between human lung endothelial and epithelial cells. Cells 2021;2713:10. [DOI] [PMC free article] [PubMed]
- 27.Coldren CD, Nick JA, Poch KR, Woolum MD, Fouty BW, O’Brien JM, et al. Functional and genomic changes induced by alveolar transmigration in human neutrophils. Am J Physiol Lung Cell Mol Physiol. 2006;291:L1267–1276. doi: 10.1152/ajplung.00097.2006. [DOI] [PubMed] [Google Scholar]
- 28.Smith KM, Mrozek JD, Simonton SC, Bing DR, Meyers PA, Connett JE, et al. Prolonged partial liquid ventilation using conventional and high-frequency ventilatory techniques: gas exchange and lung pathology in an animal model of respiratory distress syndrome. Crit Care Med. 1997;25:1888–97. doi: 10.1097/00003246-199711000-00030. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen HCB, Adlanmerini M, Hauck AK, Lazar MA. Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature. 2020;584:286–90. doi: 10.1038/s41586-020-2576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin F, Pei L, Zhang Q, Han W, Jiang S, Lin Y, et al. Ox-LDL induces endothelial cell apoptosis and macrophage migration by regulating caveolin-1 phosphorylation. J Cell Physiol. 2018;233:6683–92. doi: 10.1002/jcp.26468. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Qu L, Li Y, Chen C, He W, Shen L, et al. Glycyrrhizic acid alleviates lipopolysaccharide (LPS)-induced acute lung injury by regulating angiotensin-converting enzyme-2 (ACE2) and caveolin-1 signaling pathway. Inflammation. 2022;45:253–66. doi: 10.1007/s10753-021-01542-8. [DOI] [PubMed] [Google Scholar]
- 32.Tsai TH, Tam K, Chen SF, Liou JY, Tsai YC, Lee YM, et al. Deletion of caveolin-1 attenuates LPS/GalN-induced acute liver injury in mice. J Cell Mol Med. 2018;22:5573–82. doi: 10.1111/jcmm.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He R, Yuan X, Lv X, Liu Q, Tao L, Meng J. Caveolin-1 negatively regulates inflammation and fibrosis in silicosis. J Cell Mol Med. 2022;26:99–107. doi: 10.1111/jcmm.17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Wang C, Dong W, Su Y, Ma Z. WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell Biol Int. 2021;45:1269–77. doi: 10.1002/cbin.11570. [DOI] [PubMed] [Google Scholar]
- 35.Jiao L, Wang S, Zheng Y, Wang N, Yang B, Wang D, et al. Betulinic acid suppresses breast cancer aerobic glycolysis via caveolin-1/NF-κB/c-Myc pathway. Biochem Pharmacol. 2019;161:149–62. doi: 10.1016/j.bcp.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Xue W, Wang J, Jiang W, Shi C, Wang X, Huang Y, et al. Caveolin-1 alleviates lipid accumulation in NAFLD associated with promoting autophagy by inhibiting the Akt/mTOR pathway. Eur J Pharmacol. 2020;871:172910. doi: 10.1016/j.ejphar.2020.172910. [DOI] [PubMed] [Google Scholar]
- 37.Cai L, Yi F, Dai Z, Huang X, Zhao YD, Mirza MK, et al. Loss of caveolin-1 and adiponectin induces severe inflammatory lung injury following LPS challenge through excessive oxidative/nitrative stress. Am J Physiol Lung Cell Mol Physiol. 2014;306:L566–573. doi: 10.1152/ajplung.00182.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K, Leist SR, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 2018;9:e01753-18. [DOI] [PMC free article] [PubMed]
- 39.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–39. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye C, Li H, Bao M, Zhuo R, Jiang G, Wang W. Alveolar macrophage - derived exosomes modulate severity and outcome of acute lung injury. Aging. 2020;12:6120–8. doi: 10.18632/aging.103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukhopadhyay S, Hoidal JR, Mukherjee TK. Role of TNFalpha in pulmonary pathophysiology. Respir Res. 2006;7:125. doi: 10.1186/1465-9921-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X, Xiu H, Zhang S, Zhang G. The role of macrophages in the pathogenesis of ALI/ARDS. Mediators inflamm. 2018;2018:1264913. doi: 10.1155/2018/1264913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskin DL, Malaviya R, Laskin JD. Role of macrophages in acute lung injury and chronic fibrosis induced by pulmonary toxicants. Toxicol Sci. 2019;168:287–301. doi: 10.1093/toxsci/kfy309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin S, Wu H, Wang C, Xiao Z, Xu F. Regulatory T cells and acute lung injury: cytokines, uncontrolled inflammation, and therapeutic implications. Front Immunol. 2018;9:1545. doi: 10.3389/fimmu.2018.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, He P, Huang Y, Li YF, Lu J, Li M, et al. Selective autophagy of intracellular organelles: recent research advances. Theranostics. 2021;11:222–56. doi: 10.7150/thno.49860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaaf MB, Houbaert D, Meçe O, Agostinis P. Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ. 2019;26:665–79. doi: 10.1038/s41418-019-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 48.Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–24. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 51.Yang R, Song Z, Wu S, Wei Z, Xu Y, Shen X. Toll-like receptor 4 contributes to a myofibroblast phenotype in cardiac fibroblasts and is associated with autophagy after myocardial infarction in a mouse model. Atherosclerosis. 2018;279:23–31. doi: 10.1016/j.atherosclerosis.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–71. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, et al. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2014;306:L1016–1025. doi: 10.1152/ajplung.00268.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data required for the conclusion of the manuscript can be found in the article. Additional raw data related to this study are available on request from corresponding authors.