Abstract
Introduction
Small cell lung cancer (SCLC) is a highly aggressive neuroendocrine tumor that is characterized by early acquired therapeutic resistance and modest benefit from immune checkpoint blockade (ICB). Repression of the major histocompatibility complex class I (MHC-I) represents a key mechanism driving resistance to T cell-based immunotherapies.
Methods
We evaluated the role of the lysine-specific demethylase 1 (LSD1) as a determinant of MHC-I expression, functional antigen presentation, and immune activation in SCLC in vitro and in vivo, through evaluation of both human SCLC cell lines and immunocompetent mouse models.
Results
We demonstrate that targeted inhibition of LSD1 in SCLC restores MHC-I cell surface expression and transcriptionally activates genes encoding the antigen presentation pathway. LSD1 inhibition further activates interferon signaling, induces tumor intrinsic immunogenicity, and sensitizes SCLC cells to MHC-I-restricted T cell cytolysis. Combination of LSD1 inhibitor with ICB augments the anti-tumor immune response in refractory SCLC models. Together, these data define a role for LSD1 as a potent regulator of MHC-I antigen presentation and provide rationale for combinatory use of LSD1 inhibitors with ICB to improve therapeutic response in SCLC.
Conclusions
Epigenetic silencing of MHC-I in SCLC contributes to its poor response to ICB. Our study identifies a previously uncharacterized role for LSD1 as a regulator of MHC-I antigen presentation in SCLC. LSD1 inhibition enables MHC-I-restricted T cell cytolysis, induces immune activation, and augments the anti-tumor immune response to ICB in SCLC.
Keywords: Small cell lung cancer, LSD1, epigenetics, antigen presentation
INTRODUCTION
Small cell lung cancer (SCLC) is a highly aggressive, poorly differentiated neuroendocrine tumor that represents 15% of all lung cancers1. Owing to a high proliferative rate, early widespread metastases and rapidly acquired therapeutic resistance, the prognosis for SCLC remains poor with an overall 5-year survival rate of <7%2. Development of new targeted therapies has been hindered by a paucity of driver mutations, and SCLC remains one of the two NCI-designated “recalcitrant” cancers3,4.
The strong association of SCLC with smoking history in ~98% of patients5 is reflected in its complex genetic landscape, which harbors a high prevalence of somatic mutations, both numeric and structural chromosomal abnormalities, and frequent alterations in microsatellite polymorphic repeats6-9. Consequently, SCLC exhibits a high tumor mutational burden10 that is associated with neoantigen frequency11-13 and, in other tumor types, a favorable response to immune checkpoint inhibitors14-17. However, clinical response to immune checkpoint blockade has been disappointing in patients with SCLC, with two anti-PD-L1 inhibitors extending median survival by only approximately 2 months in patients with newly diagnosed extensive stage disease, and no immunotherapy options granted FDA approval for treatment of recurrent disease18-27. This cumulative underperformance reflects the urgency to discover therapeutic targets that can enhance the efficacy and durability of immunotherapies in SCLC.
Suppression of the antigen presentation pathway represents a cell-intrinsic mechanism of resistance by which cancers can evade anti-tumor immunity. The majority of SCLC exhibit low or absent expression of MHC-I28-31. As a result, despite the presence of ample putative neoantigens, lack of antigen presentation might render these cells “invisible” to cytotoxic lymphocytes, leading to failure to engage T cell-mediated anti-tumor activities32,33. Cell surface expression of MHC-I in SCLC cell lines and subsequent recognition by cognate T cells can be induced by treatment with IFN-γ, indicating that the class I presentation machinery of SCLC is typically not mutated or functionally impaired but rather transcriptionally suppressed28. A recent study reported the observation of robust and durable responses to ICB in a minor subset of SCLC patients with high MHC-I expression, which was the only correlate identified that could predict survival after ICB treatment31. Together, these observations emphasize the potential translational impact of reactivating MHC-I antigen presentation as a therapeutic avenue for SCLC.
LSD1, encoded by KDM1A, removes methyl groups on mono- and di-methylated lysine 4 and 9 of histone H3 to regulate gene expression. LSD1 has been recently recognized as a potential therapeutic target in SCLC due to a tumor cell intrinsic function in growth and viability34. In a subset of SCLC, LSD1 inhibition was shown to activate NOTCH signaling and downregulate ASCL1, both of which control aspects of neuroendocrine development34-36. Recent studies have also kindled interests in LSD1 as an immunomodulatory target in some solid tumors37-40. However, its ability to alter the immunosuppressive microenvironment of SCLC has not been previously characterized. In this study, we identified LSD1 as a potent regulator of MHC-I-mediated antigen presentation and intrinsic immunogenicity in SCLC.
MATERIAL AND METHODS
Cell culture and cell lines
NCI-H69, DMS79, NCI-H146, SHP-77, NCI-H889, NCI-H1092, NCI-H209, NCI-H1963, NCI-H524, NCI-H82, NCI-H446, NCI-H526, NCI-H211, COR-L311, NCI-H196, and DMS114 were maintained in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. NCI-H841, NCI-H1048, and NCI-1694 were maintained in RPMI supplemented with 10% FBS, 1% penicillin/ streptomycin, 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30nM sodium selenite (Gemini Bio-Products Cat #400-145), 10nM hydrocortisone (Sigma-Aldrich Cat #H6909), and 10nM beta estradiol (Sigma-Aldrich Cat #E4389). NCI-H1341 were maintained in ACL-4 media without serum supplemented with 1% penicillin/streptomycin. A murine SCLC tumor cell line derived from a syngeneic genetically engineered mouse model (GEMM) is driven by biallelic loss of Trp53, Rb1 and cooperative deletion of the Rb family member p130 (RPP)41. Primary NY-ESO1 T cells were cultured in RPMI supplemented with 10% human serum (Gemini Bio Cat #100-512), 1% penicillin/streptomycin, 0.1% amphotericin, 1mM sodium pyruvate (Thermo Fisher Cat #11350070), non-essential amino acids (Millipore Sigma Cat #32160406), and Glutamax (Thermo Fisher Cat #35050061). All cell lines were cultured in a 5% CO2 incubator at 37°C and passaged every 2-3 days. Identities of cell lines were confirmed by STR profiling, and all lines were frequently tested for mycoplasma.
Cell viability assay
All cell lines were cultured in a 5% CO2 incubator at 37°C and passaged every 2-3 days. 1000 cells were seeded as triplicates in 96-well black opaque plates with lid (Corning Cat #3916) in 50μL normal growth media the day before drug treatment. Drug titrations were performed in normal growth media at 2X concentration. 50μL of drug-containing media was then added such that column 11 wells contained the highest drug concentration, column 3 contained the lowest, and column 2 contained vehicle instead of drug. Outside wells were filled with culture media (RPMI complete) to prevent precipitation. Drug concentrations are indicated in dose-response curves for ORY-1001 (Selleck Chemicals; Cat #S7795) or DMSO as vehicle. Cells were cultured in a 5% CO2 at 37°C for 7 days during which cells were passaged every 3 days and replenished with fresh media with the indicated concentration. After 7 days, Cell Titer Glo Luminescent Cell Viability Assay (Promega; Cat #G7570) was performed according to manufacturer’s instructions. Cell viability was calculated as percentage of average count of drug-treated cells and average count of vehicle-treated cells.
Immunoblots
Protein extraction and western blot were performed as previously described42. 30-5μg of protein was extracted and normalized using BCA Protein Assay Kit (Thermo Fisher Cat #23225) and diluted with NuPAGE LDS sample buffer 4X (Invitrogen Cat #NP0007), loaded on Bis-Tris Gel 4-12% (Invitrogen Cat. #NP0322) and electrophoresed before transferring with Trans-Blot Turbo RTA Mini LF PVDF Transfer Kit (Bio-Rad). Blots were incubated with blocking buffer at room temperature for 1 hour before overnight incubation with primary antibody. Blots were washed 4-6X with 1% PBST before secondary incubation with anti-rabbit, HRP-conjugated antibodies/ Chemiluminescent detection was performed on iBright Imaging Systems (Thermo Fisher). All antibodies used for this procedure are listed in Supplementary Table 1.
RT-qPCR
RNA extraction, reverse transcription and quantitative PCR were performed as followed. RNA extractions were performed using the RNeasy Plus Mini Kit (Qiagen, Cat #74034) according to manufacturer's instructions. Complementary DNA (cDNA) was synthesized from RNA by reverse transcription PCR using Superscript IV VILO Master Mix (Thermo Fisher Cat #11756050). Real time PCR was performed using TaqMan Fast Advanced Master Mix (Thermo Fisher Cat #4444556) following manufacturer's protocol. Triplicate PCR reactions were run on StepOnePlus Real-Time PCR System (Applied Biosystems) and 2−ΔΔ method was used to calculate comparative Ct and normalized to GAPDH. Fluorescent probes used in this study are included in Supplementary Table 2.
Flow Cytometry
Human SCLC were blocked with TruFCX (BioLegend Cat#422302) and stained (30min, 4°C) with appropriate dilutions of anti-human HLA-A,B,C antibodies. Murine tumors were dissociated into single cells using GentleMACS according to standard protocol. Single cell suspensions were Fc-blocked with anti-CD16/32 (BioLegend Cat# 101320) and stained with appropriate anti-mouse antibodies. Antibody clone and dilutions are listed in Supplementary Table 3. Murine populations from Figure 5 were gated as follow: CD45-MHC-I+, CD45− PD-L1+, CD45+, CD45+CD3+ (total CD3), CD45+CD3+CD4+ (total CD4), CD45+CD3+CD8+ (total CD8), CD45+CD3+CD4+FoxP3+ (Tregs), CD45+CD3+CD8+CD69+ (early effector), CD45+CD3+CD8+CD44+CD25+ (late effector), CD45+CD3+CD8+GranzymeB+, CD45+CD3+CD8+PD1+TIM3+ (exhausted CD8), and CD45+CD3-NKp46+ (NK cells). Debris, doublets, and dead cells were excluded based on forward and side scatter, side scatter height vs side scatter width, forward scatter area vs forward scatter height, and Fixable Viability Dye Zombie NIR or Ghost Violet 510, unless otherwise indicated. Stained cells were acquired on a Cytek Aurora Flow Cytometer, and the data were analyzed using FlowJo software (version 10.6, BD Biosciences).
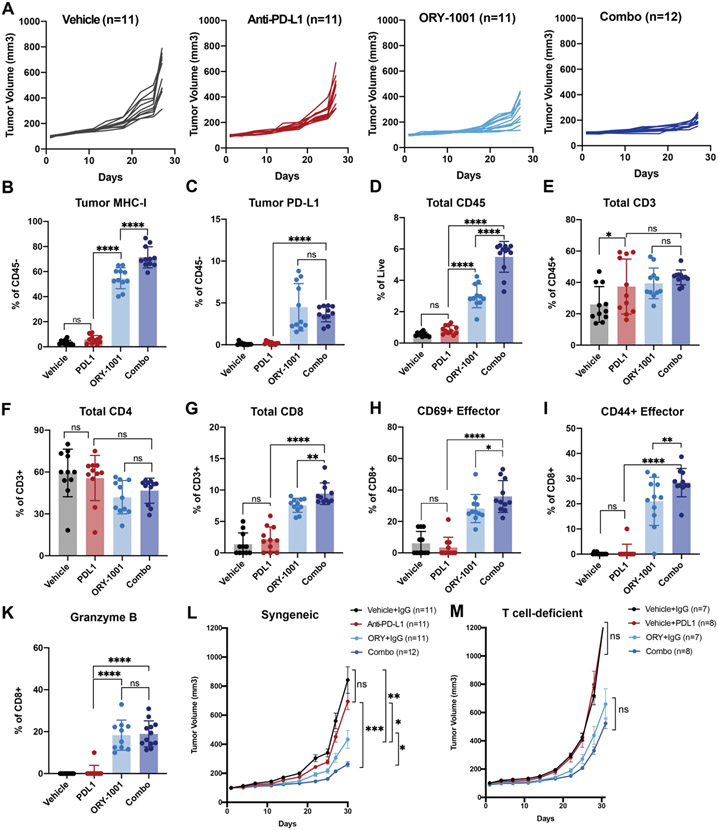
Figure 5: LSD1 inhibition rescues immune checkpoint blockade resistance and promotes anti-tumor immunity in a syngeneic SCLC model.
Rb1−/−, p53−/−, p130−/− RPP cells were subcutaneously grafted on B6129J female 6–8-week-old mice until tumors reached 80-120mm3 before treatment with control, ORY-1001 (50μg/animal/week), anti-PD-L1 antibody (300 μg/animal/week) or the combination. Tumors were collected and analyzed once control tumors reached 1200mm3. (A) Individual tumor growth curves in the four treatment groups: vehicle and isotype IgG (Vehicle) (n=11), vehicle and anti-PD-L1 antibody (anti-PD-L1) (n=11), ORY-1001 and isotype IgG (ORY-1001) (n=11), and ORY-1001 and anti-PD-L1 antibody (Combo) (n=12). (B-K) RPP tumors were dissociated into single cells. 2-5 million live cells were profiled by flow cytometry. Representative flow cytometry plots and summary data for the tumors are shown. Flow cytometry was performed to analyze levels of (B) MHC-I+CD45−, (C) PD-L1+CD45+, (D) total CD45+ infiltrating immune cells, (E) CD45+CD3+, (F) CD45+CD3+CD4+, (G) CD45+CD3+CD8+, (H) CD45+CD3+CD8+CD69+, (I) CD45+CD3+CD8+CD25+CD44+, and (K) CD45+CD3+CD8+GrzmB+. Percentages were normalized to live cells defined as Ghost Violet 510 negative staining. P values were calculated by one-way ANOVA analysis corrected for multiple comparisons with Tukey’s test; *P<0.032; ** P<0.0021 **; *** P< 0.0002; **** P<0.0001. (L) Tumor growth curves (mean ± SEM) of RPP tumors grafted on syngeneic B6129J background representing four treatment groups: vehicle and isotype IgG (Vehicle) (n=11), vehicle and anti-PD-L1 antibody (anti-PD-L1) (n=11), ORY-1001 and isotype IgG (ORY-1001) (n=11), and ORY-1001 and anti-PD-L1 antibody (Combo) (n=12). Data are representative of two independent experiments. P values were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (M) Tumor growth curves (mean ± SEM) of RPP tumors grafted on athymic nude background representing four treatment groups: vehicle and isotype IgG (Vehicle) (n=7), vehicle and anti-PD-L1 antibody (anti-PD-L1) (n=8), ORY-1001 and isotype IgG (ORY-1001) (n=7), and ORY-1001 and anti-PD-L1 antibody (Combo) (n=8). Data are representative of two independent experiments. P values were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
RNA sequencing
RNA extraction and sequencing was performed by Genewiz. Total RNA was extracted from fresh frozen cell pellet samples using QIAGEN RNeasy Plus Universal mini kit following manufacturer’s instructions (QIAGEN, Hilden, Germany). Extracted RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA).
RNA sequencing (RNAseq) libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina following manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were first enriched with Oligo(dT) beads. Enriched mRNAs were fragmented for 15 minutes at 94°C. First strand and second strand cDNAs were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3′ ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. The sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA).
The sequencing libraries were clustered on 2 lanes of a flow cell. After clustering, the flow cell was loaded on the Illumina HiSeq instrument (4000 or equivalent) according to manufacturer’s instructions. The samples were sequenced using a 2x150bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
RNA-sequencing analysis
Transcript counts and abundances were quantified from RNA-seq reads using Salmon v1.1.043. RNA-seq raw reads were mapped to 25-mer indexed hg38 genome by Salmon. With default settings, mapping validation (--validatemappings), bootstrapping with 30 re-samplings (--numBootstraps), sequence specific biases correction (--seqBias), coverage biases correction (--posBias) and GC biases correction (--gcBias) were also enabled. Transcript to gene mapping based on Ensembl 9244, normalized by size factor at gene level, and differential gene expression analyses were processed on Salmon output files using Sleuth v0.30.0 in gene mode45. Differentially expressed genes were identified using the Wald test. Genes were marked as significant if the False Discovery Rates, q, calculated using the Benjamini-Hochberg method, was less than 0.05, and beta (Sleuth-based estimation of log2 fold change) > 0.58, which approximately correlated to a log2 fold change of 1.5 in our data.
Gene set enrichment analysis (GSEA)46 was performed on full sets of differential gene expression results on the previously mentioned comparisons. Genes were ranked on p value scores computed as -log10(p value)*(sign of beta). Gene set annotations include all gene sets deposited in the Molecular Signatures Database (MSigDB v7.0.1)46,47. The significance level of enrichment was evaluated using permutation test and the p value was adjusted by Benjamini-Hochberg procedure. Any enriched gene sets with adjusted p value ≤ 0.05 were regarded as significant. This analyses were executed with the R package ClusterProfiler v3.18.148.
Pathways and gene sets of interest were procured from GSEA-msigdb database (https://www.gsea-msigdb.org/gsea/index.jsp) and Gene Ontology resources (http://geneontology.org/). Interested pathways were highlighted in H82 and H211 GSEA dot plots. The log of the normalized TPM values for significant genes within the interested pathways, were rescaled using a z-score transformation, and plotted in a heatmap using the ComplexHeatmap v2.7.1049 Library in R 4.0.450.
scRNA-seq analysis
To assess differential expression of KDM1A in SCLC tumors (Figure 1B-C), we profiled 54,313 SCLC transcriptomes from 21 SCLC tumors vs 3341 LUAD transcriptomes from 24 LUAD tumors51. We performed pseudo-bulk differential expression of KDM1A where we first aggregated UMI counts among cancer cells per sample to create the counts matrix U [samples x genes] and then performed differential expression between SCLC vs LUAD tumors using DESeq2 likelihood ratio test52. A recent comparative analysis53 showed that pseudo-bulk differential expression methods like DESeq2 perform well in scRNA-seq datasets by accounting for variation between biological replicates. Genes were considered significantly differentially expressed if the absolute log2 fold-change is greater than log2(1.5) and the Benjamini-Hochberg (BH) adjusted p-value is < 0.01. Based on this criteria, there were 6,886 differentially expressed genes (DEGs), including KDM1A (BH-adjusted p=3.578e-12). To visualize KDM1A expression between SCLC vs LUAD at the sample level, we normalized U [samples x genes] by the aggregated library size and scale by median library size, followed by log2 transform with a pseudo-count of 1.
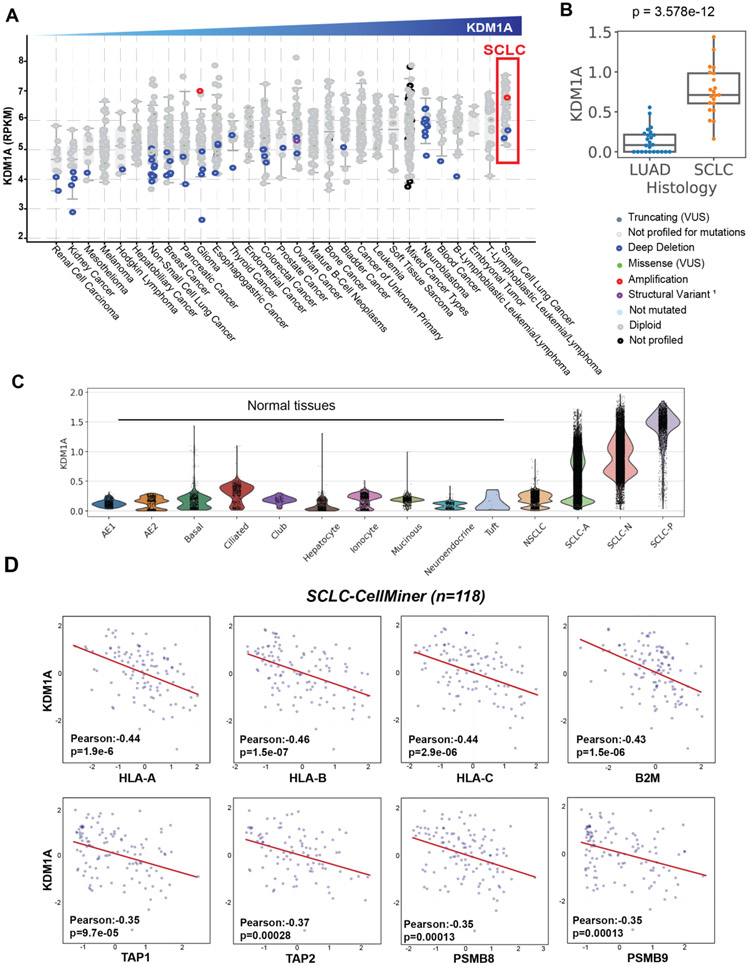
Figure 1: LSD1 expression is highly overexpressed in small cell lung cancer and inversely correlated with the expression of genes encoding the antigen presentation pathway.
(A) KDM1A mRNA expression was profiled in 29 unique cancer types; data from Cancer Cell Line Encyclopedia (CCLE) (Broad, 2019) (Downloaded from cBioPortal). (B) KDM1A expression was queried in primary tumors previously analyzed in the SCLC-Atlas51, consisting of LUAD (n=24) and SCLC (n=21) clinical samples. (C) KDM1A expression was profiled at the single-cell level from the SCLC-Atlas dataset51, consisting of 54,313 SCLC transcriptomes, 3341 LUAD transcriptomes and matched normal tissues. (D) Pearson’s correlations between LSD1 (KDM1A) expression and HLA-A, HLA-B, HLA-C, B2M, TAP1, TAP2, LMP-2 (PSMB8), LMP-7 (PSMB9) from SCLC-Global (downloaded from CellMiner). Values were calculated as RNAseq RPKM (log2(value+1)).
Lentiviral construction and synthesis
shRNA sense and antisense guide sequences were generated using splashRNA from coding sequence of KDM1A transcript, variant 1 (NM_001009999) following previously described protocols54,55. Four antisense constructs were generated from splashRNA: #1897 (TAAATAAAGGTTTGACTCGTGG), #392 (TTCTGAATAATACTCATCTTCT), #1227 (TAAGGTAAGATGTAGCTTCTAG) and #806 (TAAATAACTGTGAACTCGGTGG) and subsequently synthesized to include XhoI and EcoRI restriction sites. Cloning into LT3GEPIR vector backbone was performed using the following primers: miRE-Xho-short-fw (5′-AGAAGGCTCGAGAAGGTATATTGC-3′) and miRE-EcoPlasmid-rev (5′-GCTCGAATTCTAGCCCCTTGAAGTCCGAGG-3′). LT3GEPIR vector and primer sequences were provided as a gift from the Levine Lab.
NY-ESO1- and MART-1-GFP lentiviral plasmids were purchased from SinoBiological (Cat# HG15611-ACGLN and Cat #HG12253-ACGLN, respectively). Luciferase-GFP vectors were obtained from System Biosciences (Cat #LL310PA-1; pLL-CMV-rFLuc-T2A-GFP-mPGK-Puro). Lentiviral production and transduction were performed as previously described56. For transduced cell lines from Figure 4, H82, H211, H69 and T2 cells were first transduced with luciferase-neomycin vector and underwent neomycin selection for 7 days prior to transduction with NY-ESO1-GFP or MART-1-GFP.
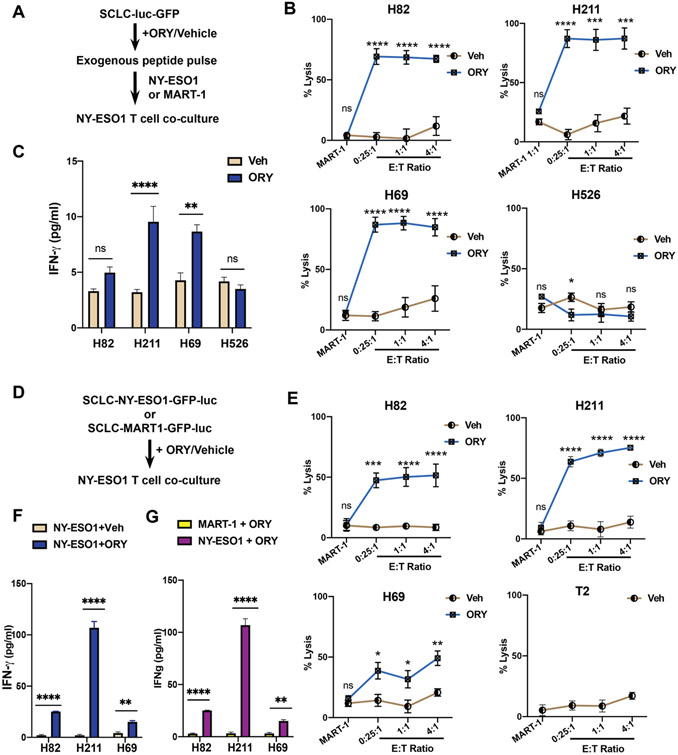
Figure 4: LSD1 inhibition sensitizes SCLC cells to T cell-mediated cytotoxicity.
(A) Workflow of T cell co-culture experiment with pulsed peptides. H82, H211, H69 and H526 expressing luciferase were treated with either 1μM ORY-1001 or vehicle for 10 days before short-term exposure to NY-ESO1 or MART-1 peptides (5μ/ml). NY-ESO1-reactive CD8+T cells were introduced at specific effector-to-target (E:T) ratio and co-incubated for 18 hours. (B) Luciferase assay was performed to determine the percentage of live cells remaining in each co-culture condition compared to control samples without effector cells. For cells pulsed with NY-ESO-1 antigens values were calculated at 0.25:1, 1:1 and 4:1 ratios. Cells pulsed with MART-1 antigen were co-cultured at a 1:1 ratio with NY-ESO-1+ CD8+ T cells. (C) Levels of human IFN-γ present in the culture media after co-incubation period were quantified using ELISA. (D) Workflow of T cell co-culture experiment with endogenous peptides. H82, H211, H69 and T2 cells expressing either NY-ESO1-luciferase or MART-1-luciferase were treated with either 1μM ORY-1001 or vehicle for 10 days before NY-ESO1 TCR+ CD8+ T cells were introduced at specific effector-to-target (E:T) ratio and co-incubated for 18 hours. (E) Percentage of live cells remaining in each co-culture condition compared to control samples without effector cells were assessed using a luciferase assay. For cells pulsed with NY-ESO1, cells were plated at 0.25:1, 1:1 and 4:1 E:T ratios. Cells pulsed with MART-1 antigen were co-cultured in 1:1 ratio with NY-ESO-1 TCR+ CD8+ T cells. T2 cells, which lack TAP transporters and express an empty HLA-A*02:01 on the cell surface, served as a negative control. (F-G) Levels of human IFN-γ present in the culture media after co-incubation period were quantified using ELISA. Comparison between treatment conditions was assessed in NY-ESO1-expressing cells lines in (F) or in MART-1- and NY-ESO1 expressing cells treated with 1μM ORY-1001 (G).
Cytotoxicity Assay
Luciferase-GFP SCLC cell lines were plated into 6 well plates and treated with 1μM ORY-1001 for 10 days in complete RMPI media. Media was replenished every 3 days. Commercially available human NY-ESO1-TCR CD8+ T cells were isolated from a healthy CMV seropositive patient, pre-activated with autologous monocytes pulsed with NY-ESO1 peptides (Cellero Cat##1093-4908OC20) and subsequently cultured in RPMI media supplemented with human serum and IL-2 (100IU/ml). After treatment, SCLC cells were plated onto 96-well white opaque plates with lid (Thermo Fisher Cat #136101). For exogenous peptide assay (Figure 4A), cells were pulsed with 5μg/ml of NY-ESO-1 peptide (SLLMWITQC) or MART-1/MLANA peptide (ELAGIGILTV) for 4 hours prior to T cell co-culture. For endogenous peptide assay, NY-ESO-1-luc-GFP or MART-1-luc-GFP were pretreated with ORY-1001 or vehicle before co-incubation. In both assays, co-incubation with NY-ESO-1-TCR CD8+ T cells were performed at appropriate Effector:Target ratio as indicated in the Figure Legend. After 18 hours, culture media was collected for ELISA. Promega Luciferin was added to each well of the plate, incubated for 20 minutes and luminescence signal was recorded on the Synergy HT1 (Biotek).
ELISA
90μl of co-culture media was collected from each well, centrifuged to remove debris and subjected to quantitative human IFN-gamma ELISA performed using the Quantikine ELISA Immunoassay (R&D Systems, Cat #DIF50C) according to manufacturer’s instruction. ELISA plates were read using a microplate reader set at 450nm with wavelength correction set to 540nm.
Mouse models
All animal experiments were approved by and used in accordance with animal care guidelines from the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee. Female B6129J/S mice (6-8 weeks old) were purchased from Taconic Biosciences. Athymic nude mice (inbred; Cat# 002019) were purchased from the Jackson Laboratory. All animals were housed in accredited facilities under pathogen-free conditions. Prior to all experiments, purchased mice were allowed one week to acclimate to housing conditions at Memorial Sloan Kettering Cancer Center Animal Facility.
Animal model and treatment
4×106 RPP single cells were injected subcutaneously in a 1:1 mixture of PBS and Matrigel (Thermo Fisher #CB40234) into the left flanks of 6-week-old female B6129F1 mice (Taconic). Treatment commenced either at seven days after injection or once the mean tumor volume reached approximately 100 mm3, unless otherwise indicated in the figure legend. Mice were randomized and treated once weekly with either vehicle (0.5% Methylcellulose; oral gavage) and rat IgG2b isotype (300μg/animal/week, intraperitoneal) (BioXCell Cat #BE009), vehicle and anti-PD-L1 antibody (300μg/animal/week, intraperitoneal) (BioXCell Cat #BE0101), ORY-1001 (50μg/animal/week, oral gavage) and isotyp(, or a combination of anti-PD-L1 and ORY-1001. Tumors were measured twice a week using calipers and their volumes were calculated as width2 x length x 0.5. Statistical significance was determined by Student t-test.
Resource availability
Further information and requests for resources, reagents, and data should be directed to and will be fulfilled by the Lead Contact, Charles M. Rudin (rudinc@mskcc.org).
RESULTS
LSD1 expression is inversely correlated with expression of antigen presentation genes in SCLC
To investigate LSD1 expression in SCLC relative to other cancer types, we queried expression of the KDM1A gene in 1,153 cell lines of the Broad Cancer Cell Line Encyclopedia (CCLE) Portal57,58. KDM1A expression is more highly expressed in SCLC than in any of the other 28 cancer types (Figure 1A). To corroborate this cell line observation in primary tumors, we leveraged our recently published single-cell atlas of SCLC tumors51 comprising 21 SCLC, 24 lung adenocarcinoma (LUAD) and 4 normal lung samples, which confirmed higher differential expression of KDM1A specifically in SCLC (Figure 1B-C). Mutational analyses of the CCLE dataset supported overexpression as the major pathological feature as mutations of KDM1A are rare (<2%) (Figure 1A).
Recent studies have demonstrated that transcriptional silencing of MHC-I genes can be epigenetically mediated59. To explore the relationship between LSD1 overexpression and antigen presentation deficiency in SCLC, we analyzed correlative expressions of KDM1A and genes encoding the antigen presentation pathway from three SCLC cell line datasets: CCLE (n=50)58,60, University of Texas Southwestern (UTSW) 2017 dataset (SCLC Consortium cBioPortal) (n=70)61 and the Sanger/Massachusetts General Hospital (Genomics of Drug Sensitivity in Cancer [GDSC]) (n=74) 62 (Supplementary Figure 1A-C). In all three databases, we discovered a significant inverse correlation between transcript levels of KDM1A and HLA-A, HLA-B, HLA-C, and B2M, as well as the peptide transporters (TAP1, TAP2, TAPBP) and subunits of the immunoproteasomes (PSMB8 or LMP-2, PSMB9 or LMP-7). To collectively interpret correlation from these datasets, we utilized the publicly available CellMiner-SCLC toolkit (https://discover.nci.nih.gov/SclcCellMinerCDB/)63, which integrates and normalizes microarray and RNA-seq from five independent sources into a dataset labelled “SCLC-Global” (Figure 1D). Integrating different methodologies, these datasets uniformly demonstrated a strong negative correlation between KDM1A and multiple key genes required for antigen presentation, leading us to posit that LSD1 overexpression might play a role in silencing MHC-I-mediated antigen presentation in SCLC.
LSD1 inhibition rescues MHC-I expression in SCLC cell lines
We validated transcriptomic data from CCLE with cell surface expression of MHC-I in 10 different SCLC models and confirmed that the majority of SCLC express low MHC-I (Supplementary Figure 2A-B). Two models with the lowest B2M mRNA transcripts, H211 and H82, also displayed the lowest MHC-I cell surface expression (Supplementary Figure 2A-B). In contrast, H196 had strongest cell surface level of MHC-I, in agreement with its transcriptional assessment (Supplementary Figure 2A-B).
To perturb LSD1 demethylase function, we employed the irreversible LSD1 inhibitor, ORY-1001. This structural derivative of tranylcypromine targets the H3-binding pocket of LSD1 through irreversible covalent modification of FAD cofactor, leading to inactivation of catalytic functions64. Prior studies have indicated effective inhibitory concentrations of ORY-1001 at nanomolar range34,64. Importantly, we did not observe substantial in vitro growth inhibition of 20 human SCLC cell lines after treatment with increasing concentrations of ORY-1001 up to 10 μM (Supplementary Figure 2C). We compared the efficacy of ORY-1001 to another FAD-dependent LSD1 inhibitor, GSK-LSD1, and found that the inability of ORY-1001 to induce cell death after 7 days was similarly observed with GSK-LSD1 (Supplementary Figure 2D). Interestingly, H211 and H82 ranked among the top 5 cell lines with highest KDM1A expression yet retained 90-100% cell viability following treatment with 1μM ORY-1001 (Supplementary Figure 2A-D). Another SCLC cell line, H69, similarly exhibited strong anti-correlation of KDM1A and B2M expression (Supplementary Figure 2B-D). We employed these three models to further assess the immunomodulatory effects of LSD1 inhibition.
To determine whether LSD1 inhibition elicits concentration- or time-dependent effects, we initially evaluated changes in total cell surface MHC-I expression of H211 cells following treatment with varying doses of ORY-1001 over time (Supplementary Figure 2E). We detected upregulation of MHC-I expression starting at 7 days, which further intensified after 10 days with relatively minimal dose-dependency (Supplementary Figure 2E). Changes in MHC-I level were not observed earlier than 7 days at any concentration (Supplementary Figure 2E). These data suggest that latency, rather than concentration, might primarily contribute to LSD1-specific regulation of downstream targets in SCLC.
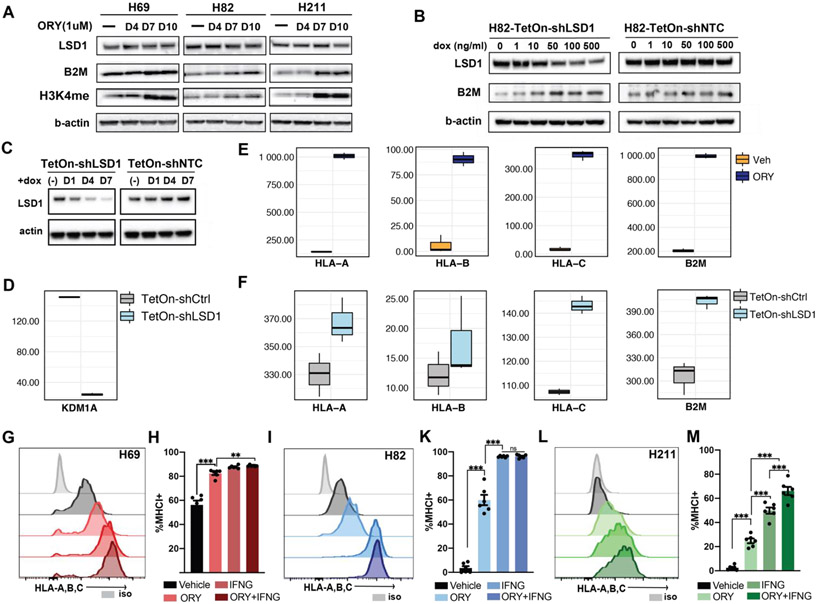
To validate that the observed MHC-I enhancement is LSD1-specific and not a cell line-specific event, we evaluated expression of mono-methylated H3K4 (H3K4me) in three biological replicates of H211, H69, and H82 after treatment with ORY-1001 (Figure 2A). As expected, ORY-1001 does not affect LSD1 protein level but specifically inhibited its enzymatic activity, resulting in increased H3K4me at days 7 and 10, but not at day 4 relative to vehicle treatment (Figure 2A). The observed accumulation of H3K4me correlates with a significant increase in B2M expression after 7 days, but not earlier, in agreement with what we had observed by flow cytometry (Figure 2A; Supplementary Figure 2E). To substantiate that MHC-I upregulation is indeed an LSD1-specific phenomenon, we performed a Tet-On doxycycline-inducible shRNA knockdown of LSD1 expression. Consistent with ORY-1001 treatment, LSD1 knockdown, but not non-targeting control, increased B2M protein level in H82 cell line in a dose-dependent manner (Figure 2B). Similarly, inactivation of LSD1 by decreasing transcript and protein level (Figure 2C-D) or by catalytic inhibition (Figure 2E) both markedly upregulated HLA-A, -B, -C and B2M transcripts (Figure 2F).
Figure 2: LSD1 abrogation induces MHC-I expression in SCLC.
(A) Immunoblots of H69, H211 and H82 treated with 1μM ORY-1001 for 4, 7 and 10 days before cell lysates were collected and blotted for LSD1, B2M, di-methylated H3K4 and beta-actin protein expression. (B) Doxycycline-inducible shRNA knockdown of LSD1 (sh#1227) or non-targeting control (NTC) was performed on H82 to assess LSD1, B2M and beta-actin expression after LSD1 suppression. (C) Immunoblots of LSD1 protein level were analyzed following doxycycline-inducible shRNA knockdown in H211. (D) mRNA transcript level of KDM1A indicates shRNA knockdown of LSD1 in H211. Transcriptional activation of HLA-A, -B, -C and B2M genes were observed following (E) catalytic inhibition by ORY-1001 or (F) inducible knockdown of LSD1. Flow cytometry analysis was performed on (G) H69, (I) H211 and (L) H82 cell lines treated with either 1μM ORY-1001 for 10 days, 10ng/ml IFN-γfor 24 hours, or its combination to assess cell-surface expression of MHC-I by HLA-A,B,C antibodies. (H; K; M) Levels of increased MHC-I expression were quantitated against baseline expression from two independent experiments. Statistical significance was performed with two-way ANOVA using Geisser-Greenhouse correction; ns p>0.12; * p<0.033; ** p<0.002; and *** p<0.001.
Next, we evaluated the functional effect of LSD1 inhibition in inducing MHC-I expression. Exposure to low-dose IFN-γ has been shown to induce MHC-I expression in SCLC cell lines28,31,59, and an impairment in IFN-γ signaling is associated with ICB resistance65,66. We then compared the intensity of MHC-I re-activation by LSD1 inhibition with that of IFN-γ (10ng/ml) treatment alone or a combination of both. Treatment with ORY-1001 was able to restore MHC-I expression to a level near to that of IFN-γ effect in all three models (Figure 2G-M). In H211, which is less sensitive to IFN-γ, combination treatment with ORY-1001 and IFN-γ induced higher MHC-I expression than IFN-γ alone (Figure 2L-M). Together, these data demonstrate that perturbation of LSD1 activities via pharmacological inhibition or RNA interference can significantly de-repress MHC-I cell surface expression in SCLC.
LSD1 inhibition transcriptionally activates antigen presentation and induces intrinsic immunogenicity
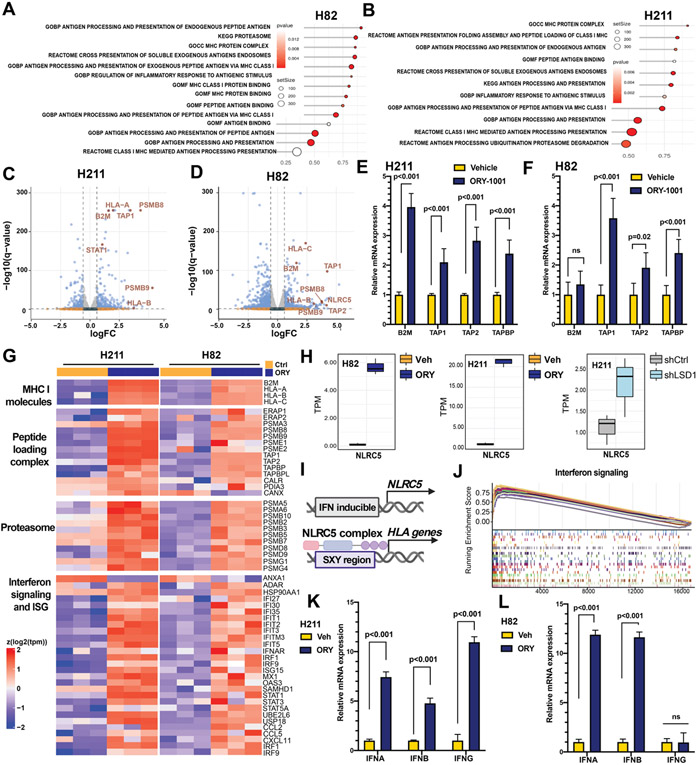
We next conducted transcriptomic analysis to determine the extent to which antigen processing and presentation (APP) machinery was re-activated upon LSD1 inhibition. Gene set enrichment analysis (GSEA) revealed significant enrichment of gene sets defining the APP machinery in both H82 and H211 following LSD1 inhibition by ORY-1001 (Figure 3A-B). Analyses of differentially expressed genes (DEGs) highlighted those of the endogenous peptide antigen presentation pathway, including HLA-A, HLA-B, HLA-C, B2M, TAP1/2, PSMB8/9, to be among the most significantly upregulated genes following LSD1 inhibition (Figure 3C-D). We further validated the role of LSD1 as a transcriptional regulator of key regulators of antigen presentation in H211 and H82 by RT-qPCR (Figure 3E-F).
Figure 3: Transcriptomic characterization of SCLC cells after LSD1 inhibition.
Cells treated with 1 μM ORY-1001 or vehicle for 10 days were isolated and subjected to RNA-seq. Pathway enrichment analyses for antigen presentation pathways (APP) are shown as dot plots for (A) H82 and (B) H211. Each dot represents a gene set significantly enriched after LSD1 inhibition. Red indicates significant p values. Dot size reflects number of genes in each gene set. (C-D) Volcano plots showing differential gene expression as beta value (Sleuth-based estimation of log2 fold change) in (C) H211 and (D) H82. APP-related genes are highlighted in red. (E-F) Increased relative mRNA expression of APP-related genes were confirmed by RT-qPCR for (E) H211 and (F) H82. (G) Heatmap reporting mRNA expression of MHC-I molecules, MHC-I peptide complex, proteasomal components and combined interferon signaling and interferon-stimulated genes in H82 and H211 cell lines treated with 1μM ORY ORY-1001 or vehicle (Ctrl). Each column represents an independent replicate. (H) Transcript levels of NLRC5 following LSD1 inactivation in H82 (left; ORY-1001 vs vehicle treatment), H211 (middle; ORY-1001 vs vehicle treatment) and H211 (right; shLSD1 vs shNTC). (I) Schematics showing transactivation of HLA genes by NLRC5 (J) Gene set enrichment analyses for interferon signaling were represented as enrichment scores sorted by rank list. (J-K) Increased relative mRNA expression of interferon genes were confirmed by RT-qPCR for (J) H211 and (K) H82.
We next assessed additional biological processes associated with antigen presentation by MHC-I molecules. Preceding peptide:MHC-I assembly, multi-subunit immunoproteasomes must degrade large cytosolic polypeptides into short peptides suitable for transport to the endoplasmic reticulum by TAP transporters and for binding to MHC-I molecules. We profiled gene expression of MHC-I molecules, comprised of the alpha chain and B2M, as well as the MHC-I peptide loading complex (PLC). The latter includes the ER-bound chaperones calnexin (CANX), ERp47 (PDIA3), calreticulin (CALR), tapasin (TAPBP), and the TAP transporters (TAP1, TAP2), together required for antigenic peptide:MHC-I binding and stabilization. We found that LSD1 inhibition transcriptionally activates most genes encoding the MHC-I PLC (Figure 3G). Among the most differentially expressed genes, the NOD-like receptor CARD domain containing 5 (NLRC5) gene, a member of the NLR family of regulators of inflammatory responses, was significantly activated following both ORY-1001 treatment and shRNA knockdown (Figure 3H). NLRC5 has been recognized as a key MHC-I trans-activator, capable of forming an enhanceosome bound to the conserved SXY motif in MHC class I promoters to regulate transcription of HLA and B2M genes67-69 (Figure 3I). Induction of NLCR5 suggests that re-expression of MHC-I-genes resulted from transcriptional activation.
Interestingly, the promoter of NLRC5 has been previously reported to contain an IFN-γ-activating sequence70, suggesting that LSD1 inactivation might also activate interferon signaling. Consistent with this hypothesis, gene set enrichment and DEGs of H82 and H211 following LSD1 inhibition revealed consistent induction of innate immune-related processes, including response to type I and type II interferons and interferon-stimulated genes reflecting pan-inflammatory processes (Figure 3G, 3J-L). We also observed induced expression of interferon-α receptor IFNAR, a common receptor recognizing both IFN-α and IFN-β interferons (Figure 3G). IFNAR signals through STAT1 to interact with IRF9, which together binds to the promoters of interferon stimulated genes (ISGs). Indeed, expression of STAT1, IRF9 and ISGs are all induced following LSD1 inhibition (Figure 3G). Significant enrichment of interferon response signaling in LSD1-inactivated cells was further confirmed by GSEA (Figure 3J), and by RT-qPCR, demonstrating consistent induction of IFNA and IFNB, and cell line-selective induction in IFNG in H82 (Figure 3K-L). Collectively, these data demonstrate that LSD1 perturbation transcriptionally upregulates MHC-I antigen presentation machinery and primes SCLC into an immunogenic phenotype.
LSD1 inhibition leads to loss of neuroendocrine signatures in neuroendocrine SCLC
Recent studies have established the consensus that the intrinsic capability of SCLC to become immunogenic is tightly linked to its neuroendocrine (NE) signatures31,71-73. >70% of SCLC express a high-NE phenotype73. We postulated that LSD1 inhibition not only induces autonomous immunogenicity but can also alter NE lineage-specific signatures. To interrogate whether LSD1 can play a role in regulating NE-specific gene expression, we queried a previously defined NE signature gene set73 (Supplementary Table 4) in both non-NE and high-NE SCLC cell lines. Following LSD1 inhibition, the high-NE cell line H82 exhibited coordinate suppression of most NE genes, consistent with differentiation into a low-NE phenotype (Supplementary Figure 3A). In contrast, the non-NE cell line H211 showed no difference in NE signatures despite an evident shift in antigen presentation, suggesting that a low-NE phenotype might be a prerequisite for induction of immunogenicity (Supplementary Figure 3A; Figure 3).
SCLC tumors have also been recently recognized as biologically heterogenous subtypes defined by the differential expression of transcription factors ASCL1, NEUROD1, POU2F3, and YAP174. Intra-tumoral heterogeneity with co-expression of more than one of these factors and tumor evolution from one predominant subtype to another have been observed in clinical and experimental models. To further characterize the phenotypic changes of H82, we leveraged our recently published subtype-specific signatures51 (Supplementary Table 5) to our transcriptomic analysis. To our surprise, we noticed that following inhibition, the cells appear to lose all of their NEUROD1-specific, lineage-defined signatures, further supporting a phenotypic switch induced by loss of LSD1 activities (Supplementary Figure 3B). Additional studies demonstrated that LSD1 inhibition can activate NOTCH signaling34 and downregulate ASCL135 in SCLC models, both of which have been associated with non-NE de-differentiation. In agreement with these data, we also observed induction of NOTCH signaling in both low-NE and high-NE models, suggesting that NOTCH activation might occur concurrently with changes in immunogenicity (Supplementary Figure 4). Collectively, these data demonstrate that the phenotypic switch between high-NE and low-NE SCLC can be epigenetically regulated and coupled with induction of tumor-intrinsic immunogenicity.
LSD1 inhibition rescues antigen-specific T cell-mediated recognition and cytolysis
To explore the functional consequences of cell-surface MHC-I re-expression, we next assessed the effects of LSD1 inhibition on the antigen-specific cytotoxic T cell response. H82, H211 and H69 all express the HLA-A*02:01 alleletype (Supplementary Figure 5A). We generated luciferase-expressing derivatives of these lines and treated them with ORY-1001 or vehicle. These cells were subsequently pulsed with HLA-A*02:01-restricted NY-ESO1 peptide antigen (SLLMWITQC), which preferentially binds to cell surface HLA-A2 with high affinity. Following antigen pulse, we co-cultured these SCLC cells with commercially available cognate CD8 T lymphocytes (CTLs) cells recognizing the HLA-A2-NY-ESO1 peptide complex (Figure 4A). These primary CTLs have been pre-activated and expanded with autologous monocytes pulsed with NYESO1 peptides. After 18 hours, we measured luciferase activity to assess for antigen-specific T cell killing. At baseline, H69 was more effectively lysed than H211 and H82, consistent with their relative expression of MHC-I (Supplementary Figure 2A-B; Supplementary Figure 5C). In contrast to the vehicle-treated cells, markedly enhanced cytotoxicity was observed in H82, H211, and H69 pretreated with ORY-1001 (Figure 4B). At higher T cell (effector) to tumor (target) ratios (E:T), we noticed low cytolysis activity in the vehicle treated group, which is consistent with the low-level presence of MHC-I in these cell lines (Figure 4B; Supplementary Figure 2A). As a negative control, we employed the SCLC cell line H526, in which baseline MHC-I expression could be augmented by LSD1 inhibition almost up to IFN-γ-treated level, but which lacks HLA-A*02:01 (Supplementary Figure 5B). We did not observe any T cell-dependent lysis of H526 even after LSD1 inhibition, substantiating the notion that NY-ESO1 presentation was HLA-A2-restricted (Figure 4B).
To confirm that the observed T cell recognition was specific to NY-ESO1 peptide, we pulsed SCLC cells with another HLA-A*02:01-restricted antigen, MART-1/MLANA peptide (ELAGIGILTV) prior to co-incubation with NY-ESO-1 CTLs. MART-1-pulsed SCLC failed to be cleared by NY-ESO1 CTLs in either treatment group, confirming that the observed T cell-mediated killing activity was indeed antigen-specific (Figure 4B). Because CD8 T cells release IFN-γ after recognition of the MHC-I-peptide complex and during lysis of target cells, we performed ELISA on the co-culture media to quantify IFN-γ levels after target clearance. IFN-γ was significantly released in the media of H211 and H69 cells pretreated with ORY-1001 (Figure 4C). A similarly elevated level of IFN-γ was detected in H82 co-culture after LSD1 inhibition though the difference was not statistically significant. As expected, ORY-1001 pretreatment did not increase IFN-γ levels in the co-culture media of H526 cells (Figure 4C). Together, these results further support that the MHC-I induction by LSD1 inhibition was sufficient to enable MHC-I restricted, antigen-specific T cell lysis.
Transcriptional activation of TAP-dependent antigen presentation machinery (Figure 3A-F) led us to hypothesize that LSD1 inhibition could restore endogenous peptide processing in SCLC. These SCLC cell lines were previously shown to be antigen presentation deficient but TAP-competent28. To test our hypothesis, we engineered the SCLC cells to stably express either NY-ESO1-luciferase or MART1-luciferase endogenous peptides. We also employed the T2 cell line, which lacks a functional TAP complex and expresses an empty HLA-A*02:01 on the cell surface, as an additional control75. To test the inducibility of endogenous peptide processing by LSD1 inhibition, we subjected these transduced cell lines to LSD1 inhibition or vehicle treatment before co-incubation with NY-ESO1-specific T cells (Figure 4D). After the co-culture period, we were able to detect lysis of target cells in ORY-1001-treated cells but not in vehicle-treated controls (Figure 4E). Absence of T cell killing was observed in both TAP-deficient T2 cells and MART1-expressing cells, indicating that T cell recognition was dependent on functional endogenous peptide processing (Figure 4E). Consistent with these results, IFN-γ induction was observed in the co-culture media of ORY-1001-treated NY-ESO1-expressing SCLC relative to vehicle-treated NY-ESO1-specific populations (Figure 4F), or to ORY-1001-treated MART-1-expressing cells (Figure 4G). Together, our data demonstrate that inhibition of LSD1 is necessary and sufficient to activate the endogenous antigen processing and presentation machinery of SCLC, which allows for recognition and subsequent clearance by cognate CD8 T cells in an antigen-specific manner.
LSD1 inhibition sensitizes SCLC tumors to anti-PD-L1 blockade response
The restoration of antigen presentation and activation of intrinsic immunogenicity by LSD1 inhibition highlights the possibility that targeting LSD1 might enhance sensitivity of SCLC tumors to T cell-based immunotherapies. To explore this idea, we utilized SCLC tumors derived from a syngeneic genetically engineered mouse model (GEMM) driven by biallelic loss of Trp53, Rb1 and cooperative deletion of the Rb family member p130 (RPP)41. This SCLC RPP model exhibits primary resistance to anti-PD-L1 therapy (Figure 5A). To characterize the effects of LSD1 inhibition on anti-tumor response, we initiated treatment once the average tumors volumes reached 80-120 mm3 (Figure 5A). Cohorts of tumor-bearing mice were randomized into treatment with control vehicle, anti-PD-L1 (300μg/week), ORY-1001 (50μg/week), or combination of dual inhibition. Compared to vehicle and anti-PD-L1 therapy, ORY-1001 significantly slowed tumor growth (Figure 5A). The addition of ORY-1001 to anti-PD-L1 regimen further enhanced tumor reduction, suggestive of a combinatorial effect (Figure 5A). These results indicate that targeting LSD1 can sensitize SCLC tumors to PD-L1 blockade.
Tumors derived from combination treatment demonstrated significantly higher MHC-I expression than other groups, with tumors from the ORY-1001-treated cohort also exhibiting higher MHC-I compared to either control or anti-PD-L1-treated tumors (Figure 5B). These data agree with our in vitro observation that treatment with LSD1 inhibitor can upregulate H2-Kb expression in RPP cells (Supplementary Figure 6A-B). Expression of tumor PD-L1 was also elevated in the ORY-1001-treated groups while the addition of anti-PD-L1 elicited no significant difference (Figure 5C). To further understand the basis of the observed combinatorial antitumor effect, we profiled different subsets of tumor-infiltrating immune cells. We observed significantly more CD45+ cells in ORY-1001-treated, and even more in combination-treated tumors, suggesting cooperative recruitment of infiltrating immune cells (Figure 5D). While we did not observe any overall changes in CD3+ or CD4+ T cell populations (Figure 5E-F), we recorded significantly greater infiltration of CD8+ CTLs (Figure 5G). Subsets of CD69+ early activated and CD25+CD44+ effector cells were significantly more enriched in cohorts treated with ORY-1001 (Figure 5H-I). Similarly, we also observed higher infiltration of granzyme-B+ effector CD8+ (Figure 5K), suggesting that the enhanced anti-tumor response might be largely dependent on cytotoxic T cell activities. No evident toxicity was recorded at this dose (Supplementary Figure 6C). Even though there were no significant differences in CD4+ T cells across treatment levels (Figure 5E), more infiltrating CD4+Tregs and exhausted CD8+ were present in cohorts given LSD1 inhibitor though these levels varied widely, reflecting more robust immune cell activities (Supplementary Figure 6D-E). To examine whether the anti-tumor effect of combination treatment was T cell-dependent, we performed the same experiments in athymic nude mice that have deficient mature T cell activities and observed a more rapid tumor growth, attributable to the absence of immunosurveillance (Figure 5L-M). We observed a complete loss of the combinatorial effect previously seen with dual inhibition of LSD1 and PD-L1 (Figure 5L), suggesting that the anti-tumor immune activity was largely mediated by effector CTLs. The slight reduction of tumor volume observed in the ORY-1001-treated nude mouse cohort can be accounted by residual T cell activities in this model76 or by MHC-I-independent NK cell activities, as NK cells are also present at higher level in the dual treatment group (Supplementary Figure 5F). Taken together, these data strongly support that LSD1 inhibition can overcome immune evasion in SCLC, reactivate a robust antitumor immune response and sensitize resistant tumors to ICB therapies.
DISCUSSION
Immune checkpoint blockade therapies have revolutionized cancer treatment and resulted in major shifts in standard of care treatments for many solid tumors. Antigen presentation by MHC-I is an essential mechanism by which these T cell-based immunotherapies can achieve efficacy and durability. Despite an abundance of mutation-derived neoantigens, SCLC tumors exhibit low or absent of MHC-I expression which may contribute to an immunosuppressive phenotype and resultant failure to achieve benefit from ICB therapy in the majority of SCLC patients31. Therapeutic reactivation of antigen presentation represents a salient strategy to enhance sensitivity to ICB in SCLC. Previous work has reported that inhibition of EZH2, an epigenetic modifier, can induce MHC-I expression in neuroendocrine tumors including SCLC31,59. In this study, we have identified LSD1 as another potent epigenetic regulator of MHC-I-mediated antigen presentation. We demonstrate a strong inverse correlation of KDM1A and genes involved in MHC-I presentation and further show that selective inhibition of LSD1 can significantly restore MHC-I cell surface expression, transcriptionally activate the antigen presentation pathway, trigger intrinsic interferon signaling and amplify tumoral immunogenicity. Furthermore, we provide evidence that pharmacological inhibition of LSD1 is sufficient to rescue MHC-I-restricted cytotoxic T cell recognition and lysis in several SCLC models. Combination treatment of LSD1 inhibitor with anti-PD-L1 resulted in enhanced tumor growth inhibition in a primary resistant model of SCLC. Importantly, we also demonstrate markedly higher MHC-I expression, enhanced recruitment of immune cell infiltrates and significantly more cytotoxic and activated CD8+ T cells in tumors treated with LSD1 inhibitor. Together, our findings nominate LSD1 inhibition as a viable approach to restore MHC-I expression, attenuate immune suppression, and enhance SCLC sensitivity to the standard-of-care immunotherapies.
Most SCLC tumors have low or absent MHC-I antigen presentation and have strong neuroendocrine features. We demonstrated that LSD1 inhibition can induce the conversion of high-NE SCLC into a low-NE phenotype, with notable signatures of high immunogenicity and activation of NOTCH signaling. Previous work has associated exogenous NOTCH activation in vitro with increased MHC-I expression in SCLC.71 Inactivating mutations of NOTCH in 25% of SCLC represent a hurdle to immunosensitize this tumor exclusively through this pathway9. We detected loss of NEUROD1-specific signatures in a NEUROD1-driven SCLC model, which might have therapeutic implication as NEUROD1 tumors often exhibit a particularly immune-cold subtype51,77. We further provide evidence of enhanced immunogenicity in low-NE SCLC model, consistent with the idea that the immune-phenotypic switch of SCLC might coincide with a subtype shift from high- into low-NE status.
LSD1 inhibition together with PD-L1 blockade in immunocompetent GEMM-derived SCLC inhibited but did not abrogate tumor growth, which may be in part due to the low mutational burden of GEMM tumors relative to that of SCLC in patients, which is generally attributable to long-term exposure to tobacco carcinogens. A greater response might be anticipated in immunocompetent models with a neoantigen burden that more closely approximate to that of the human disease. Another limitation of this study relates to the potential effects of LSD1 inhibition on the endogenous functions of immune cells, which was not the focus of this study. It is possible that LSD1 inhibition can elicit T cell-specific effects, i.e. reversing exhausted-related functional impairment78,79 which might contribute to the observed anti-tumor response.
Our findings contribute to the emerging data nominating epigenetic targets as immune modulators. LSD1 inhibition has been reported to activate transcription of endogenous double-stranded RNA (dsRNA) and interferon signaling in breast cancer cell lines and to enhance response to ICB in melanoma mouse models80. This induction of tumor-intrinsic immunity by LSD1 inhibition resembles that reported in other epigenetic regulators, including DNA-methyltransferase 1 (DNMT1) and EZH2, in other cancer types81-84. We supplement this theme by demonstrating that LSD1 inhibition can rescue antigen presentation in MHC-I-deficient SCLC tumors and elicit cooperative benefits when combined with ICB. This study provides further support for the hypothesis that deficiency in antigen presentation can be therapeutically targeted and epigenetically reversed through pharmacological means. Of direct translational relevance, several LSD1 small molecule inhibitors are being actively investigated in patients with SCLC. Our study provides pre-clinical support for clinical trials combining LSD1 inhibition with immune checkpoint blockade as a promising approach to overcome poor response in SCLC, with potential implications for other MHC-I-deficient tumors.
Supplementary Material
ACKNOWLEDGEMENTS
This research was funded by the Geoffrey Beene Foundation (EMN), the Yasuda Medical Foundation (HT), NIH R01 CA258784, Congressionally Directed Medical Research Programs (DOD-IITRA) LC190161, Parker Institute for Cancer Immunotherapy grant, LCFA-BMS/ILC Foundation Young Investigator Research Awards in Translational Immuno-oncology (TS), NIH K08 CA248723 (AC), NIH R01 CA197936, R35 CA263816, U24 CA213274 the Druckenmiller Center for Lung Cancer Research, and the Van Andel Institute – Stand Up to Cancer Epigenetics Dream Team grant (CMR). Stand Up to Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. We thank Dr. Scott Lowe at Memorial Sloan Kettering Cancer Center for providing critical reagents. We also acknowledge the use of the MSKCC Flow Cytometry Core Facility, the MSKCC Antitumor Assessment Core, and the Integrated Genomics Core, which are funded by MSKCC Support Grant/Core Grant (P30 CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
SFC is a consultant for and holds equity interest in Imago Biosciences. TS receives research funding from Jazz Pharmaceuticals. CMR has consulted regarding oncology drug development with AbbVie, Amgen, Ascentage, Astra Zeneca, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Jazz, Lilly, Pfizer, PharmaMar, Syros, and Vavotek. CMR serves on the scientific advisory boards of Bridge Medicines, Earli, and Harpoon Therapeutics.
REFERENCES
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA. Cancer J. Clin 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Wang S et al. Survival changes in patients with small cell lung cancer and disparities between different sexes, socioeconomic statuses and ages. Sci. Rep 7, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarado-Luna G & Morales-Espinosa D Treatment for small cell lung cancer, where are we now?-A review. Transl. Lung Cancer Res. 5, 26–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt EB & Jalal SI Small Cell Lung Cancer. in Lung Cancer: Treatment and Research (ed. Reckamp KL) 301–322 (Springer International Publishing, 2016). doi: 10.1007/978-3-319-40389-2_14. [DOI] [PubMed] [Google Scholar]
- 5.Varghese AM et al. Small-cell lung cancers in patients who never smoked cigarettes. J. Thorac. Oncol. 9, 892–896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet 44, 1104–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudin CM et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet 44, 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesch B et al. Cigarette smoking and lung cancer - Relative risk estimates for the major histological types from a pooled analysis of case - Control studies. Int. J. Cancer 131, 1210–1219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George J et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (80-. ). 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le DT et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topalian SL, Drake CG & Pardoll DM Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL, Taube JM, Anders RA & Pardoll DM Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pardoll DM The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonia SJ et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 17, 883–895 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Spigel DR et al. Second-Line Nivolumab in Relapsed Small-Cell Lung Cancer: CheckMate 331. Ann. Oncol, xxx, (2021). [DOI] [PubMed] [Google Scholar]
- 20.Ott PA et al. Pembrolizumab in patients with extensive-stage small-cell lung cancer: Results from the phase Ib KEYNOTE-028 study. J. Clin. Oncol 35, 3823–3829 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Chung HC et al. Pembrolizumab After Two or More Lines of Previous Therapy in Patients With Recurrent or Metastatic SCLC: Results From the KEYNOTE-028 and KEYNOTE-158 Studies. J. Thorac. Oncol 15, 618–627 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Rudin CM et al. Pembrolizumab or Placebo Plus Etoposide and Platinum as First-Line Therapy for Extensive-Stage Small-Cell Lung Cancer: Randomized, Double-Blind, Phase III KEYNOTE-604 Study. J. Clin. Oncol 38, 2369–2379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reck M et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line therapy in extensivedisease-small-cell lungcancer: Results from a randomized, double-blind, multicenter phase 2 trial. Ann. Oncol 24, 75–83 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Reck M et al. Phase III randomized trial of ipilimumab plus etoposide and platinum versus placebo plus etoposide and platinum in extensive-stage small-cell lung cancer. J. Clin. Oncol 34, 3740–3748 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Horn L et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med 379, 2220–2229 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Paz-Ares L et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 394, 1929–1939 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Arriola E et al. Outcome and biomarker analysis from a multicenter phase 2 study of ipilimumab in combination with carboplatin and etoposide as first-line therapy for extensive-stage SCLC. J. Thorac. Oncol 11, 1511–1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Restifo NP et al. Identification of human cancers deficient in antigen processing. J. Exp. Med 177, 265–272 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle Austin, et al. Markedly Decreased Expression of Class I Histocompatibility antigens, protein, and mRNA in human small-cell lung cancer. J. Exp. Med 161, 1135–1151 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funa K, Gazdar AF, Minna JD & Linnoila RI Paucity of beta 2-microglobulin expression on small cell lung cancer, bronchial carcinoids and certain other neuroendocrine tumors. Lab. Invest 55, 186–93 (1986). [PubMed] [Google Scholar]
- 31.Mahadevan NR et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov. XX, candisc.0913.2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science (80-. ). 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Zaretsky JM et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augert A et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal 12, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sriuranpong V et al. Notch Signaling Induces Rapid Degradation of Achaete-Scute Homolog 1. Mol. Cell. Biol 22, 3129–3139 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito T et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127, 3913–3921 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Sheng W et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 174, 549–563.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin Y et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 38, 390–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan AHY et al. Lysine-specific histone demethylase 1A regulates macrophage polarization and checkpoint molecules in the tumor microenvironment of triple-negative breast cancer. Front. Immunol 10, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng W, Liu Y, Chakraborty D, Debo B & Shi Y Simultaneous Inhibition of LSD1 and TGFβ Enables Eradication of Poorly Immunogenic Tumors with Anti-PD-1 Treatment. Cancer Discov. 11, 1970 LP–1981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaffer BE et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 70, 3877–3883 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wohlhieter CA et al. Concurrent Mutations in STK11 and KEAP1 Promote Ferroptosis Protection and SCD1 Dependence in Lung Cancer. Cell Rep. 33, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zerbino DR et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pimentel H, Bray NL, Puente S, Melsted P & Pachter L Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu G, Wang LG, Han Y & He QY ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Team, R. R: A Language and Environment for Statistical Computing.
- 51.Chan JM et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39, 1479–1496.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love Michael, Huber Wolfgang, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squair JW et al. Confronting false discoveries in single-cell differential expression. Nat. Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fellmann C et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Zuber J et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol 29, 79–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wohlhieter CA et al. Concurrent Mutations in STK11 and KEAP1 Promote Ferroptosis Protection and SCD1 Dependence in Lung Cancer. Cell Rep. 33, 108444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nusinow DP et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burr ML et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 36, 385–401.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gazdar AF, Girard L, Lockwood WW, Lam WL & Minna JD Lung cancer cell lines as tools for biomedical discovery and research. J. Natl. Cancer Inst 102, 1310–1321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garnett MJ et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tlemsani C et al. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep. 33, 108296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maes T et al. ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell 33, 495–511.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Benci JL et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeda K et al. IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat. Commun 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner TB et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. U. S. A 107, 13794–13799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi KS & Van Den Elsen PJ NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol 12, 813–820 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Neerincx A, Castro W, Guarda G & Kufer TA NLRC5, at the heart of antigen presentation. Front. Immunol 4, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuenzel S et al. The Nucleotide-Binding Oligomerization Domain-Like Receptor NLRC5 Is Involved in IFN-Dependent Antiviral Immune Responses. J. Immunol 184, 1990–2000 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Roper N et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat. Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dora D et al. Neuroendocrine subtypes of small cell lung cancer differ in terms of immune microenvironment and checkpoint molecule distribution. Mol. Oncol 14, 1947–1965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai L et al. Cell-autonomous immune gene expression is repressed in pulmonary neuroendocrine cells and small cell lung cancer. Commun. Biol 314 (2021) doi: 10.1101/2020.07.28.225763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudin CM et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ninkovic T et al. Identification of O-glycosylated decapeptides within the MUC1 repeat domain as potential MHC class I (A2) binding epitopes. Mol. Immunol 47, 131–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stutman O Tumor Development after 3-Methylcholanthrene in Immunologically Deficient Athymic-Nude Mice. Science (80-. ). 183, 534–536 (1974). [DOI] [PubMed] [Google Scholar]
- 77.Best SA et al. Harnessing Natural Killer Immunity in Metastatic SCLC. J. Thorac. Oncol 15, 1507–1521 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Tu WJ et al. Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch. Front. Immunol 11, 1–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y et al. LSD1 inhibition sustains T cell invigoration with a durable response to PD-1 blockade. Nat. Commun 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheng W et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 174, 549–563.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiappinelli KB et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cañadas I et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med 24, 1143–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roulois D et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zingg D et al. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep. 20, 854–867 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Boegel S, Löwer M, Bukur T, Sahin U & Castle JC A catalog of HLA type, HLA expression, and neoepitope candidates in human cancer cell lines. Oncoimmunology 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.