Abstract
KRAS mutation in colorectal cancer (CRC) is associated with aggressive tumor behavior through increased invasiveness and higher rates of lung metastases, but the biological mechanisms behind these features are not fully understood. In this study, we show that KRAS mutant CRC upregulates integrin α6β4 through ERK/MEK signaling. Knocking-out integrin β4 specifically depleted the expression of integrin α6β4 and this resulted in a reduction in the invasion and migration ability of the cancer cells. We also observed a reduction in the number and area of lung metastatic foci in mice that were injected with integrin β4 knock-out KRAS mutant CRC cells compared to the mice injected with integrin β4 wild-type KRAS mutant CRC cells, while no difference was observed in liver metastases. Inhibiting integrin α6β4 in KRAS mutant CRC could be a potential therapeutic target to diminish the KRAS invasive phenotype and associated pulmonary metastasis rate.
Keywords: colorectal cancer, KRAS, ITGB4, ITGA6, metastasis
Introduction:
Integrins are a family of cell surface receptors that bind to extracellular matrix molecules to mediate cell adhesion and activate intracellular signaling pathways (1). They are composed of two transmembrane subunits, α and β, that combine to form distinct types of integrin that have varying specificity for ligands and play different biological roles (1,2). The β4 subunit, which heterodimerizes exclusively with the α6 subunit, functions as a receptor for the basement membrane protein laminin. In malignancy, it is thought to play alternative roles that promote carcinogenesis (1,3).
KRAS mutations are detected in up to 50% of all colorectal cancers (4–6) and are associated with worse prognosis and more aggressive tumor behavior (7,8). In the metastatic setting, treatment options are limited as KRAS mutant cancers are resistant to anti-EGFR therapies (9). In vitro studies using KRAS transformed kidney cancer cell lines have shown that KRAS upregulates integrin α6β4 to potentiate metastasis (10). Integrin α6β4 is involved in multiple signaling pathways such as PI3K, FAK, and c-Met to promote cell growth, migration, and invasion in carcinoma cells (11–13). Even though integrin α6β4 is overexpressed in several different cancers, its biological significance may be different by tumor type (14). In colorectal cancer (CRC), elevated levels of integrin α6β4 have been associated with high-risk features and poor survival, but its biologic role has not yet been fully characterized (15,16).
In this study, we demonstrate that KRAS mutant CRC cells overexpress integrin α6β4. Using a combination of in vitro and in vivo CRC models, we investigated how integrin α6β4 contributes to the aggressiveness of KRAS mutant tumors by examining its effects on cell proliferation, invasion, and migration.
Materials and methods:
TCGA PanCancer Atlas
ITGB4 mRNA expression profiles across 35 cancer types in TCGA PanCancer Atlas studies were visualized and downloaded using the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org, RRID:CVCL_2379).
CRC tissue staining and quantification
The study was performed under approval of the Institutional Review Board (no.16–1181) of Memorial Sloan Kettering Cancer Center and written informed consent was obtained from patients. All experiments were conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule). We used untreated stage I-III colon adenocarcinoma tissues from 38 patients that underwent resection at Memorial Sloan Kettering. Paraffin-embedded tissues were sequentially sectioned at 5-micron-thickness for H&E and immunofluorescence staining (IF). All stained slides were digitally scanned with Panoramic Flash Slide Scanner (3DHistech, Budapest, Hungary). Scanned data were reviewed by a pathologist who was blinded to the molecular data. Relevant regions of tumor core were annotated using CaseViewer (3DHistech, RRID:SCR_017654) with H&E as a guide. Tumor cells (E-cadherin-positive) expressing ITGB4 in annotated regions were quantified using custom macros written in Image J (RRID:SCR_003070, NIH, Bethesda, MD, USA). Percentages of positive cells in 6 tumor core regions were averaged per tissue section, and a Student’s t-test was performed for statistical analysis. For representative images, annotated regions were exported as TIFF in CaseViewer and processed in ImageJ.
Immunofluorescence staining
Five-micron paraffin-embedded tissue sections were deparaffinized and antigen retrieval was performed in pH 6.1 citrate buffer (Agilent Dako, S169984–2) by boiling for 20 minutes. Sections were blocked in 10% normal donkey serum (Millipore, S30-M) and 1% bovine serum albumin (Sigma, A2058) at room temperature for 1 h and immunostained overnight at 4 °C with primary antibodies followed by incubation for 2 h at room temperature with fluorophore-conjugated secondary antibodies (Invitrogen; 1:500). Cell nuclei were labeled with Hoechst dye (Sigma-Aldrich, B2261; 1 μg/ml). Following primary antibodies were used : E-cadherin (R&D, AF748-SP; 1:80, RRID:AB_355568) and ITGB4 (Abcam, ab110167, RRID:AB_10866385; 1:100) for CRC tissues; E-cadherin (BD, 610182, RRID:AB_397581; 1:125), ITGB4 (Abcam, ab110167, AB_10866385; 1:100), and Vimentin (Santa cruz, sc-7557, RRID:AB_793998; 1:50) for 3D matrix gels; GFP (Abcam, ab6673, RRID:AB_305643; 1:220) and ITGB4 (Abcam, ab182120; 1:222) for mouse tissues. Following secondary antibodies were used : donkey anti-mouse Alexa488 (Molecular Probes, A21202, RRID:AB_141607), donkey anti-rabbit Alexa488 (A21206, RRID:AB_2535792), donkey anti-rabbit Alexa546 (A10040, RRID:AB_2534016), donkey anti-rat Alexa594 (A21209, RRID:AB_2535795), donkey anti-goat Alexa546 (A11056, RRID:AB_142628), or donkey anti-goat Alexa647 (A21447, RRID:AB_141844) -conjugated secondary antibodies.
Cell line culture
Caco-2 cells, a microsatellite stable colorectal cancer cell line, with or without KRASG12V mutation (17) were cultured in MEM supplemented with 20% FCS, 1× nonessential amino acids, 2 mM L-glutamine, and penicillin–streptomycin. HCT-116 cells, a MSI colorectal cell line, with or without KRASG13D mutation (18) were kindly provided by Carlos Carmona-Fontaine (New York University) and cultured in McCoy’s 5A medium supplemented with 10% FCS and penicillin-streptomycin. Human normal colonic fibroblast cell line CCD-18Co (ATCC, CRL-1459, RRID:CVCL_2379) was cultured in MEM supplemented with 10% FCS, 1× nonessential amino acids, 2 mM L-glutamine, and penicillin–streptomycin. To generate ITGB4 overexpressing cells, Caco-2 cells were transduced with ITGB4 Lentiviral Activation Particle (h) (Santa cruz, sc-400573-LAC) according to the manufacturer’s instruction. Control Lentiviral Activation Particles (Santa cruz, sc-437282) were used as control. Transduced cells were selected in 5 μg/ml of puromycin (Sigma, P9620), 4 μg/ml of Blasticidin S HCl (Santa cruz, sc-495389), and 300 μg/ml of Hygromycin B (Santa cruz, sc-29067). Increased ITGB4 expression in Caco-2 cells was confirmed using immunofluorescence staining and western blotting. To generate ITGB4 knock out cells, HCT-116 cells were transfected with Integrin β4 (CRISPR/Cas9 KO plasmid (h2)) (Santa cruz, sc-400573-KO-2) and Integrin β4 HDR Plasmid (h2) (Santa cruz, sc-400573-HDR-2) using Lipofectamine 2000 (Invitrogen, 11668027) according to the manufacturer’s instruction. After 3 days, recombined cells were selected in 2 μg/ml of puromycin for 10 days and sorted with a FACS Aria instrument (BD Biosciences). RFP high populations were considered with double allelic recombination and sorted for further experiments. RFP low populations were considered with single allelic recombination and discarded. To determine the effect of KRAS signaling on ITGA6 and ITGB4 expression, HCT-116 cells were treated with 10 μM of MEK inhibitors PD0325901 (Sigma, PZ0162) or 10 μM of PI3K inhibitor LY294002 (Selleckchem, S1105) for 24 h and harvested for western blotting. To compare stability of ITGA6 expression, HCT-116 cells were treated with 50 μg/ml of cycloheximide (CHX; Santa Cruz, sc-3508) and harvested at 0, 6, 12, and 24 h after treatment for western blotting. For detection of phospho-FAK, HCT-116 cells were starved in 1% FBS/OPTI-MEM overnight and seeded into a laminin-coated plate (6 × 106 cells/ well) for 1 h. Laminin-coated plates were prepared by coating 6 well plates (Falcon, 353046) with 5 μg/ ml of human recombinant laminin 332 (BioLamina, LN332–0502) in 1× DPBS (Ca++/Mg++) for overnight at 4 °C. Before cell seeding, laminin solution was removed and plate was air dried. All cell lines were split twice or thrice weekly and were tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, LT27–217), and the latest test was performed in February, 2022 with negative results. Cells were passaged a maximum of 20 times.
Organoids culture
shApc/Kraswt or shApc/KrasG12D mouse colonic organoids (19), in which the Apc gene can be conditionally suppressed using a doxycycline-regulated shRNA, were cultured in DMEM/F12 supplemented with 25% Wnt3a conditioned media, 50 ng/mL recombinant mouse EGF (Invitrogen, Carlsbad, CA, USA, PMG8043), 10% Noggin conditioned media, 10% R-spondin1 conditioned media, 1 mM N-acetyl cysteine (Sigma, A9165), 10 mM nicotinamide (Sigma, N0636), 10 mM HEPES pH 7.3 (Quality Biological, Gaithersburg, MD, USA, 118–089-721), 1× penicillin–streptomycin (GIBCO, 15140–122) 0.5 μg/mL doxycycline (Sigma,D9891) in Advanced DMEM/F12 (GIBCO, 12634–010). Apc−/−Trp53−/−Kraswt (AP) and Apc−/−Trp53−/−KrasG12D (APK) mouse colonic organoids were derived from Apcflox/flox; Trp53flox/flox and Apcflox/flox;LSL-KrasG12D/+; Trp53flox/flox mice with C57BL/6 background. Freshly isolated crypts were mixed with adenoviral Cre (Vector Biolabs, Malvern, PA, USA, 1045) and embedded in Matrigel (Corning, 356231) for loxP site recombination. Recombined organoids were selected and cultured in the above media with 0.1% BSA (Sigma, A2058) and without Wnt3a, R-spondin1 and doxycycline. ROCK inhibitor Y27632 (Sigma, Y0503) was added to the medium at 10 μM on the day of seeding. To generate Itgb4 knock out organoids, APK or APK-LM organoids were released from Matrigel using Cell Recovery Solution (Corning, 354253) and dissociated with TrypLE Express (Invitrogen, 12605028) for 3 min in 37 °C waterbath. Dissociated organoid pellets were resuspended with OPTI-MEM (GIBCO, 51985034) with Y27632 (10 μM) and seeded at low density into a 48 well plate (Falcon, 351178) coated with 25 μl/well of Matrigel. Organoids were transfected with Integrin β4 (CRISPR/Cas9 KO plasmid (m)) (Santa cruz, sc-431434) and Integrin β4 HDR Plasmid (m) (Santa cruz, sc-431434-HDR) using OMNIfect Transfection Reagent (TransOMIC, OTR1001) with OPTI-MEM media according to the manufacturer’s instruction. After 3 hours, wells were washed with OPTI-MEM and 50 μl of Matrigel was added to each well. After solidifying, 330 μl of APK culture media with Y27632 (10 μM) were added and organoids were cultured for 2 days before selection. Recombined cells were selected in 2 μg/ml of puromycin for 10 days and sorted with a FACS Aria instrument (BD Biosciences). RFP high populations were considered with double allelic recombination and sorted for further experiments. RFP low populations were considered with single allelic recombination and discarded. Sorted cells were cultured to grow as organoids and used for further experiments. For western blotting and quantitative real-time PCR, the mouse colonic organoids were released from Matrigel using Cell Recovery Solution and pellets harvested. All organoid lines were split twice or thrice weekly and were tested for mycoplasma contamination. Organoids were passaged a maximum of 20 times. Genotyping of shApc, Apc−/−, Trp53−/−, and KrasG12D alleles were performed by PCR using following primers : APC.3374-F 5’- GACTGTAATTTATTGGTGTTGC -3’, APC.3374-R 5’- CACCCTGAAAACTTTGCCCC -3’, Apc-Int13F2 5’- GAGAAACCCTGTCTCGAAAAAA -3’, Apc-Int14R4 5’- TTGGCAGACTGTGTATATAAGC -3’, KRAS G12D-F 5’- GTCTTTCCCCAGCACAGTGC -3’, Kras G12D-R 5’- CTCTTGCCTACGCCACCAGCTC -3’, p53-A 5’- CACAAAAACAGGTTAAACCCAG -3’, and p53-D 5’- GAAGACAGAAAAGGGGAGGG -3’.
Western blot analysis and co-immunoprecipitation
Cells were lysed in RIPA buffer (Thermo, 89900; Millipore, 20–188 for co-immunoprecipitation) supplemented with protease inhibitor (Sigma, P8340) and phosphatase inhibitor (Thermo, A32937) for 40 min on ice, and centrifuged at 4 °C for 10 min at 11,000×g. Supernatants were transferred to new tubes and protein concentrations were determined by Bio-Rad protein assay (Bio-Rad, 5000006). The proteins were denatured in NuPAGE LDS Sample Buffer (Invitrogen, NP0007) with reducing agent (Invitrogen, NP0004) at 70 °C for 10 min, separated on NuPAGE 4–12% Bis-Tris gel (Invitrogen, NP0322BOX) with NuPAGE MOPS SDS Running Buffer (Invitrogen, NP0001), and transferred to PVDF membrane (Bio-Rad, 162–0177) using NuPAGE Transfer Buffer (Invitrogen, NP0006–1) according to the manufacturer’s instruction. Membranes were blocked with 5% skim milk or 5% bovine serum albumin (Fisher Chemical, BP1600–000) in PBS with 0.1% Tween-20 (Sigma-Aldrich, P9416) for 1 hour at room temperature, incubated overnight with primary antibodies at 4 °C, followed by corresponding HRP-linked secondary antibodies for 1 hour at room temperature, and lastly incubated with Novex ECL Chemiluminescent Substrate Reagent Kit (Invitrogen, WP20005) according to the manufacturer’s instructions. Membranes were then exposed to X-ray film (Agfa, XC6A2) for different amounts of time. Films were scanned and band intensities were quantified using ImageJ (NIH, Bethesda, MD, USA). ITGA6 intensity was normalized to α-Tubulin intensity, and fold change was calculated by dividing normalized intensities by the respective normalized intensity of KRAS WT sample or zero-hour sample.
For co-immunoprecipitation, 1 mg of protein lysate were incubated with 4 μg of following primary antibodies : rat IgG2a kappa control IgG (Invitrogen, 14–4321-82) or rat IgG2a anti- ITGA6 (Santa Cruz, sc19622) for 1 h at 4 °C with gentle mixing on a rocker. Washed Protein G agarose beads (Millipore, 16–266) were added into the tube. After 1 h, beads were washed with RIPA buffer and then protein complexes were eluted using 100 μl of 1x SDS Sample Loading Buffer (bioWORLD, 21420018–1) by heating at 95 °C for 10 min. For input control, 20 μg of whole protein lysates were loaded into each lane.
The following primary antibodies were used for western blots : E-cadherin (BD, 610182, RRID:AB_397581; 1:10,000), ITGB4 (Santa Cruz, sc-374057, RRID:AB_10917002 and sc-9090, RRID:AB_2129021; 1:10,000 and 1:1,000), ITGA6 (abcam, ab181551; 1:10,000), ITGA6A (Millipore, MABT356; 1:1,000), ITGA6B (Millipore, MAB1358, RRID:AB_94180; 1:1,000), ITGB1 (Cell Signaling, 34971, RRID:AB_2799067; 1:1,000), phospho-ERK1/2 (Cell Signaling, 9106, RRID:AB_331768; 1:1,000), ERK1/2 (Cell Signaling, 9102, RRID:AB_330744; 1:1,000), phospho-AKT (Cell Signaling, 4051, RRID:AB_331158 and 4060, RRID:AB_2315049; 1:1,000), AKT (Cell Signaling, 9272, RRID:AB_329827; 1:1,000), K-Ras (Millipore, OP24, RRID:AB_2134115; 1:1,000), LAMC2 (Santa Cruz, sc-25341, RRID:AB_627873), Oct3B (Cell Signaling, 2788, RRID:AB_2167693; 1:1,000), CD24 (Santa Cruz, sc11406, RRID:AB_2072750; 1:1,000), CD44 (Cell Signaling, 3578, RRID:AB_2076463; 1:1,000), CD133 (MYBioSource, MBS462020, RRID:AB_2172873; 1:1,000), Gli (Santa Cruz, sc-20687, RRID:AB_2111764; 1:1,000), Zeb1 (NOVUS, NBP1–05987, RRID:AB_1556166; 1:1,000), Slug (Santa Cruz, sc-166476, RRID:AB_2191897; 1:1,000), Snail (Cell Signaling, 3895, RRID:AB_2191759; 1:1,000), phospho-FAK (Invitrogen, 700255, RRID:AB_2532307; 1:1,000), FAK (Millipore, 06–543, RRID:AB_11214316; 1:1,000), and α-Tubulin (Millipore, CP06, RRID:AB_2617116; 1:10,000).
ITGA6 has two isoforms α6A (ITGA6A) and α6B (ITGA6B) which are C-terminal isoforms produced by alternative splicing of pre-messenger RNA (20). The proteins encoded by these transcripts are proteolytically cleaved into heavy and light chains and linked by a disulfide bond (21). Total ITGA6 was detected using ab181551 (116 kDa heavy chain band; observation around 120 kDa) and ITGA6A and ITGA6B were detected using MABT356 (30 kDa light chain band; observation around 25 kDa) and MAB1358 (30 kDa light chain band; observation around 25 kDa), respectively. Precision Plus Protein Dual Color Standards (Bio-Rad, 1610374) and Full Range Rainbow Recombinant Protein Weight Marker (Amersham, RPN800E) were used to estimate protein size.
The same blot was used for detection of ITGB4, ITGA6, phospho-AKT, AKT, phospho-ERK, ERK, K-Ras, and α-Tubulin after dividing into several sub-blots by cutting. Anti phospho-ERK and phospho-AKT antibodies were probed first and re-probed with total forms of ERK and AKT antibodies which derived from different species compared with phospho-form antibodies. For total ITGA6 and its two isoforms, the same amount of proteins were loaded into three gels and whole blots were used. If protein sizes are similar and antibodies are derived from the same species, different blots were used for probing.
RNA isolation and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Ambion, Austin, TX, USA, 15596018) and RNeasy Mini kit (Qiagen, 74104), and cDNA was synthesized using a TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA, N8080234). All targets were amplified using PerfeCTa FastMix II, Low ROX (Quanta BioSciences, 95120) and gene-specific Taqman primers and probe sets (Applied Biosystems) in ViiA 7 real-time PCR system (Applied Biosystems). Following primers are used : mouse Gapdh 4352339E, mouse Itgb4 Mm01266840_m1, human GAPDH Hs99999905, human ITGA6 Hs01041011_m1, human ITGB4 Hs00236216_m1. Fold change of gene expression was calculated after normalization to mouse Gapdh (mouse organoids) or human GAPDH (Caco-2 and HCT-116), using the 2−ΔΔCt method.
Proliferation assay
Caco-2 cells (3 × 103 cells/ well) or HCT-116 cells (4 × 103 cells/ well) were seeded in quadruplicate into 96-well plates on the day before the experiments. At day 0 and 3, cells were incubated with 0.5 mg/ml of MTT solution (Sigma-Aldrich, M2128) at 37°C for 2 h. The medium was then removed and 100 μl of DMSO was added to dissolve the formazan crystal. The absorbance of formazan was detected at 540-nm wavelength with a reference wavelength at 650 nm for background using a SpectraMax M4 plate reader (Molecular Devices). MTT values at day 3 were normalized to day 0. All experiments were repeated three times.
Mouse organoids were dissociated with TrypLE Express and filtered through 30-μm nylon mesh (Miltenyi, 130–041-407). Dissociated mouse organoids (1.5 × 103 cells/ 25 μl of Matrigel/ well) were seeded in triplicate in 24-well suspension cell culture plate (Greiner Bio-One, 662102) on 3 days before the experiments. At day 0 and 3, organoids were incubated with WST-1 (Roche, 05015944001) at 37°C for 4 h. The absorbance of formazan in culture media was detected at 440-nm wavelength with a reference wavelength at 650-nm for background using a SpectraMax M4 plate reader (Molecular Devices). WST-1 values at day 3 were normalized to day 0. All experiments were repeated three times.
Migration and invasion assay using transwell
For our migration assay, HCT-116 cells (2 × 105 cells) in 100 μl of OPTI-MEM (Gibco, 51985034) were plated into a 0.8-μm pore sized 24-well Transwell inserts (Corning, 3422) and 600 μl of OPTI-MEM with 10% FBS was added to a lower chamber. For invasion assay, Transwell inserts were coated with 10% Matrigel (Corning, 356231) before cell seeding. After 24 hours, cells were stained with Hemacolor Stain Set (Sigma-Aldrich, 65044) according to manufacturer’s instruction. Non-migrating or -invading cells were removed using a wet cotton swab from the top surface of membrane. Images of migrating or invading cells on the bottom surface of membrane were captured on 100× fields (Olympus, IX71). Cell numbers were counted in at least six fields per insert from four independent experiments.
Organotypic invasion assay
The invasion assay using organotypic culture system was modified from previous studies (22–24). To make acellular layer, each well of 24 well plate (Falcon, 353047) was coated with 250-μl of collagen gel (2 mg/ml; Corning, 354236) and incubated at 37 °C for 10 min. To make cellular layer, CCD-18Co fibroblasts at 5 × 104 cells/0.5 ml diluted in collagen and Matrigel (Corning, 356231) mixture (each 2 mg/ml) with 10% FBS were added onto the precoated collagen layer and incubated at 37°C in CO2 incubator for 30 min. After polymerization, 500 μl of CCD-18Co culture media was added and incubated at 37°C in CO2 incubator with the medium being replaced every 2–3 days. After 6 days, the culture media was removed from the well and then 4 × 105 HCT-116 cells in 15 μl of HCT-116 culture media were added to the top layer and incubated at 37°C in CO2 incubator for 2 h. After incubation 250 μl of HCT-116 culture media was added and the culture was incubated for an additional 2 days at 37°C in CO2 incubator. To make air-exposed tissue model, sterile grid (Sigma, S0770) was placed in a 60-mm culture dish (Falcon, 353004) and covered with a sterile filter paper (Whatman, WHA10311804). The dish was filled with CCD-18Co growth media and the 3D matrix gels were mounted onto the grids. The medium was gently removed so the bottom of the matrix is in contact with media to generate an air/liquid interface. Air-exposed 3D matrix gels were incubated at 37°C in CO2 incubator and the culture media was replaced every 2–3 days. After 12 days of invasion, the 3D matrix gels were fixed in 4% paraformaldehyde (Thermo Fisher Scientific, J19943-K2) overnight and processed using standard histological methods. Immunofluorescence stained sections were scanned with Panoramic Flash Slide Scanner (3DHistech, Budapest, Hungary) and analyzed using Image J software (NIH, Bethesda, MD, USA). For quantification, the distance of the deepest invading cells from the lower boundary of the noninvasive cells (maximum invasion depth), the number of invading cells, and E-cadherin-positive invasion area was measured in 100× field. Four fields per duplicate gels from five independent experiments were used for quantification.
Flank tumor model
All animal experiments were performed in accordance with the Memorial Sloan Kettering Institutional Animal Care and Use Committee (IACUC) under protocol number 06–07-012. HCT-116 cells were resuspended in ice-cold PBS with 50% Matrigel and injected subcutaneously into the right and left flanks of female athymic nude mice (The Jackson Laboratory, Bar Harbor, ME, USA) (5 × 106/100 μl/site). Mouse organoids were released from Matrigel using cell recovery solution, dissociated with TrypLE Express, and filtered through 100-μm nylon mesh (Falcon, 352360). Dissociated mouse organoid were resuspended in ice-cold PBS with 50% Matrigel (Corning, 356237) and injected subcutaneously into the right and left flanks of female C57BL/6 mice (The Jackson Laboratory) (2 × 105/100 μl/site). Tumor sizes were measured using a handheld imaging device (Peira TM900) according to the manufacturer’s protocol. Tumor tissues were harvested and fixed in 4% paraformaldehyde overnight and processed using standard histological methods. Tumor images of H&E staining were captured using Olympus BX46 microscope with an Olympus DP21 camera.
Endoluminal transplantation of mouse organoids
Mouse organoids were released from Matrigel using cell recovery solution, dissociated with TrypLE Express, and filtered through 100-μm nylon mesh (Falcon, 352360). Dissociated mouse organoids were resuspended in ice-cold PBS with 5% Matrigel (Corning, 354253) to a concentration of 6×106 cells/ml. Under the anesthesia with 2% isoflurane, small caliber brush (Karl Storz, 27650C) was inserted through the anus of an anesthetized female C57BL/6 mouse (The Jackson Laboratory, Bar Harbor, ME, USA) and the rectal mucosa were mechanically disrupted by gently pulling the brush out and in 2–5 times. After mechanical injury, 50 μl of mouse organoid suspension was injected slowly via anus into the mice using a p200 pipette. The anuses were then sealed by 5 μl of Vetbond tissue adhesive (3M) for 6 h and then the Vetbond was removed. The progression of endoluminal tumor was checked at 2 and 4 weeks after transplantation using small animal endoscopy (Karl Storz Endoscope, El Segundo, CA, USA). Rectum tissues were harvested at 4 weeks and fixed in 4% paraformaldehyde overnight and processed using standard histological methods. Existence of tumors was confirmed in H&E stained tissue sections. Tumor size was analyzed using Image J software and expressed as a percentage of Tumor area within a total lumen area. Rounded endoscopic area in captured images was considered as a total lumen area.
Metastases models
Apc−/−Trp53−/−KrasG12D mouse colonic organoids were infected with retrovirus MSCV-LUC-IRES EGFP, encoding luciferase and EGFP, using standard infection protocols and FACS-sorted for EGFP-positive cells to establish APK LucEGFP cells. 2.5×105 cells of APK LucEGFP were injected intra-splenically to seed the liver through portal circulation. Tumor growth was followed by in vivo bioluminescence (IVIS Spectrum) and mice with established metastatic disease were euthanized. Metastatic lesions were dissected, minced and incubated in a 1 mg/ml collagenase V (Sigma, C9263) solution in HBSS for 1 hour at 37 °C. Cell suspension was then filtered through a 100 μm mesh, centrifuged and cells were resuspended in Matrigel and cultured in APK organoids culture media. Organoids cultures were passaged and liver-metastatic APK organoids (APK-LM) were established and used for generating APK-LM/ITGB4-KO organoids. To develop lung metastatic tumors, 3×105 cells/200 μl of APK-LM or APK-LM/ITGB4-KO organoids were injected into the tail vein of female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; The Jackson Laboratory, Bar Harbor, ME, USA). Tumor growth was followed by in vivo bioluminescence (IVIS Spectrum) and tissues were harvested at 5 weeks after injection. Lung and liver tissues were fixed in 4% paraformaldehyde overnight and processed using standard histological methods. Serial tissue sections were stained with H&E and with antibodies. Anti-GFP antibody stained sections were used to assess the existence of organoid cells in the tissue. Tumor regions were identified in H&E stained sections using CaseViewer with GFP as a guide. The number of metastatic tumor nodules (metastatic foci) were counted on H&E-stained section and the total area (metastatic area) of tumor nodules in each section were measured using CaseViewer.
Statistical analysis
Statistical analyses were performed using one-way ANOVA for Fig.3B, 3C, 3E–H, multiple t-test for Fig. 3I and unpaired two-tailed Student’s t-test for the other comparisons. All the analyses were performed using GraphPad Prism version 6.0 (RRID:SCR_002798). P values of <0.05 were considered significant.
Figure 3.
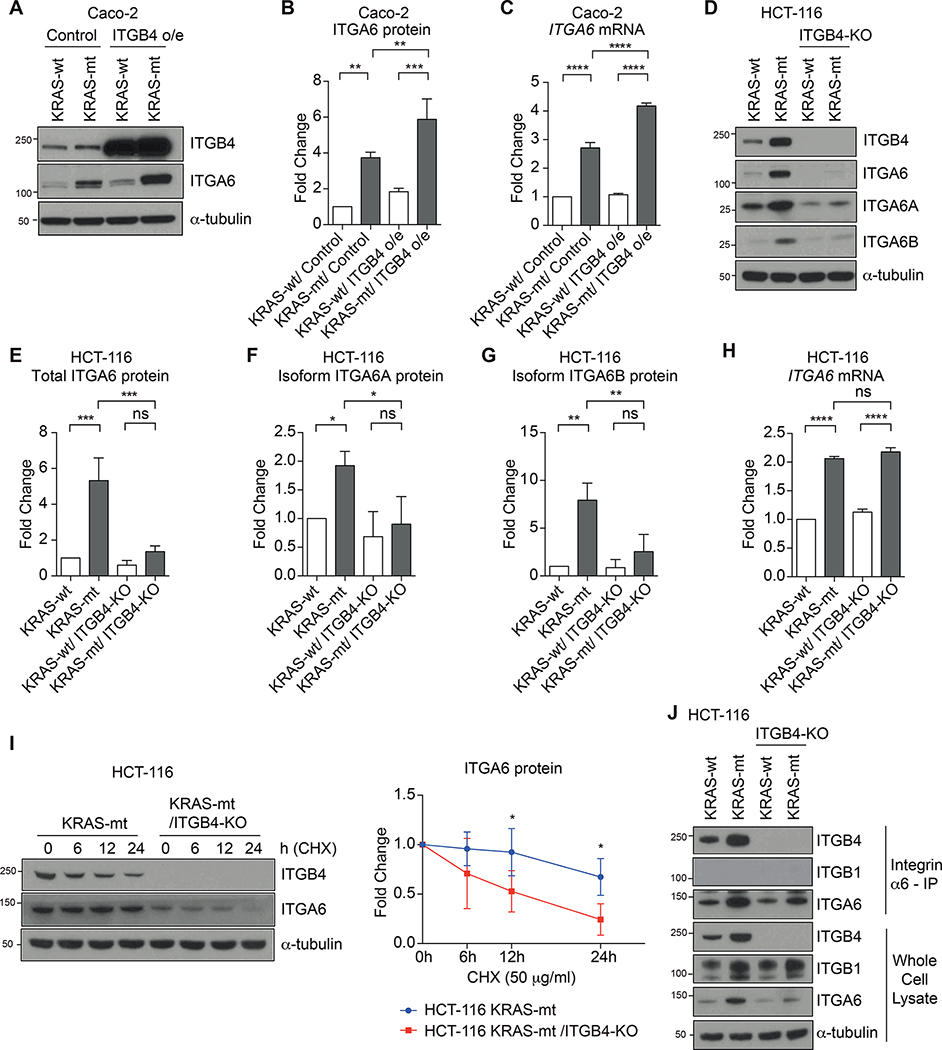
ITGB4 is necessary for the stability of ITGA6 protein in HCT-116 cells. A and B, Western blot analysis (A) and quantification (B) of total ITGA6 protein in KRAS-wt and -mt Caco-2 cells with or without ITGB4 overexpression (ITGB4 o/e). B, Bar graphs show the mean ± SD of three independent experiments. C, Quantitative RT-PCR analysis of ITGA6 mRNA expression in Caco-2 cells. Bar graph shows the mean ± SD of three independent experiments. (A-C) Caco-2 cells (5 × 105 cells/ 100 mm dish) were cultured for 3 days. D, Western blot analysis of total ITGA6 protein and two splice isoforms (ITGA6A and ITGA6B) in KRAS-wt and -mt HCT-116 cells with or without ITGB4-knockout (KO). E-G, Quantification of total ITGA6 protein (E), splice isoform ITGA6A protein (F), and splice isoform ITGA6B protein (G) in HCT-116 cells using western blot in (D). Bar graphs show the mean ± SD of three independent experiments. H, Quantitative RT-PCR analysis of ITGA6 mRNA expression in HCT-116 cells. Bar graph shows the mean ± SD of three independent experiments. D-H, HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 3 days. I, Western blot analysis of ITGA6 protein stability in KRAS-mt HCT-116 cells with or without ITGB4-KO. HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 2 days and treated with protein synthesis inhibitor, cycloheximide (CHX, 50 μg/ml), and harvested at indicated time points. Graph shows the mean ± SD of four independent experiments. Each ITGA6 protein level was normalized to respective untreated control (0 h). Multiple t-test. *, P < 0.05. J, Co-immunoprecipitation of ITGB4 and ITGB1 with anti-ITGA6 antibody from HCT-116 cells. HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 3 days. Representative of three independent experiments. Whole cell lysates were used as input control. B, C and E-H, One-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; o/e, overexpression.
Data Availability
The data analyzed in this study were obtained from the Memorial Sloan Kettering Cancer Center cBioPortal : http://www.cbioportal.org
Results:
Integrin β4 is overexpressed in KRAS mutated CRC
Colorectal adenocarcinomas exhibit the seventh highest level of integrin β4 (ITGB4) expression among 33 cancer types sequenced by The Cancer Genome Atlas (Fig. 1A) and ITGB4 is overexpressed in KRAS mutant vs. KRAS wild type CRC (P= 0.001) (Fig. 1B).
Figure 1.

Increased expression of ITGB4 in KRAS mutant CRC. A, The expression profile of ITGB4 mRNA across cancer types in TCGA PanCancer Atlas studies. B, ITGB4 mRNA expression in CRC by KRAS status in TCGA PanCancer Atlas studies. C, Western blot analysis of ITGB4 protein in human CRC cell lines. Caco-2 cells (5 × 105 cells/100 mm dish) and HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 3 days. Data are representative of three independent experiments. D, Quantitative RT-PCR analysis of Itgb4 mRNA expression in mouse colonic organoids. Bar graph shows the mean ± SD of four independent experiments. E, Western blot analysis of ITGB4 protein in mouse colonic organoids. Data are representative of three independent experiments. D and E, Mouse colonic organoids (at density of 3 × 103 cells/ 50 μl of Matrigel/ 24 well) were cultured for 5 days. F, IF staining and quantification of ITGB4 and E-cadherin positive cells in human CRC tissues. Each dot represents one CRC patient. Bar graphs show the mean ± SD for each group (KRAS-wt, n=24; KRAS-mt, n=14). Scale bars, 100 μm. D and F, Student’s t test. *, P < 0.05; ns, not significant.
We utilized two common CRC cell lines (Caco-2 and HCT-116) and created both KRAS wild-type (KRAS-wt) and mutant (KRAS-mt) lines. As Caco-2 is a KRAS-wt cell line, we introduced KRAS G12V mutation via retrovirus to generate a KRAS-mt line. Conversely, as HCT-116 is KRAS-mt (KRAS G13D), KRAS G13D mutation was removed by adeno-associated virus to create a KRAS-wt line. In both cell lines, overactivation of KRAS led to the overexpression of ITGB4 at the protein level (Fig. 1C). We validated these findings in vitro by growing KRAS-wt and KRAS-mt murine colonic organoids in 3-dimensional culture. We derived KRAS-wt (shApc) and KRAS-mt (shApc/KrasG12D) colonic organoids from a previously published genetically engineered mouse model (GEMM) in which the Apc gene can be conditionally suppressed using a doxycycline-regulated shRNA (19,25). We also used an additional set of KRAS-wt and KRAS-mt mouse organoids with inactivated TP53 (Apc−/−Trp53−/−Kraswt and Apc−/−Trp53−/−KrasG12D) because these combined alterations are predominant in colorectal cancer (26–28). In both sets of organoids, we observed that ITGB4 expression increased when KRAS was mutated (Figs. 1, D and E). We then performed immunofluorescence staining to measure the expression levels of ITGB4 in CRC patient tissue samples (24 KRAS-wt and 14 KRAS-mt, Table S1). ITGB4 expression was higher in KRAS-mt vs. KRAS-wt tumors (p=0.01) (Fig. 1F). By contrast, no significant differences were observed based on sex or tumor location.
To explore the mechanisms by which KRAS upregulates ITGB4 in CRC cell lines, we examined the levels of ITGB4 and its dimerization partner ITGA6 after inhibiting the MEK/ERK and PI3K/AKT pathway using 10 μM of MEK inhibitor PD0325901 or 10 μM of PI3K inhibitor LY294002 in KRAS-wt and KRAS-mt HCT-116 cells (Fig. 2). These high concentrations were used to ensure full inhibition of the two signaling pathways. Consistent with the results shown in Fig. 1, the levels of ITGB4 were increased in KRAS-mt HCT-116 cells compared to KRAS-wt, and ITGA6 and p-ERK were also increased (Fig. 2A). Inhibition of the MEK/ERK pathway decreased both mRNA and protein expression of both ITGB4 and ITGA6 (Figs. 2, A–C). In contrast to p-ERK, the phosphorylation of AKT was not increased in KRAS-mt HCT-116 cells compared to KRAS-wt HCT-116 cells (Fig. 2A). Inhibition of the PI3K pathway resulted in an increase of ITGB4 mRNA expression (Fig. 2D), whereas the opposite is shown with decreasing ITGA6 mRNA level after the addition of PI3K inhibitor (Fig. 2E). However, only minimal differences were observed in protein levels of ITGB4 and ITGA6 by PI3K inhibition compared to untreated control (Fig. 2A). Overall, these results indicate that KRAS upregulates ITGB4 and ITGA6 expression at transcriptional level primarily via the MEK/ERK pathway and not the PI3K-AKT pathway.
Figure 2.

Mutant KRAS increases ITGB4 and ITGA6 expression in HCT-116 cells through the MEK/ERK signaling pathway. A-E, Western blot analysis (A) and quantitative RT-PCR analysis (B-E) of ITGB4 and ITGA6 expression in HCT-116 cells treated with MEK kinase inhibitor (PD0325901, 10 μM) or PI3K inhibitor (LY294002, 10 μM). HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 2 days and treated with the inhibitors for 24 h. A, Data are representative of three independent experiments. B-E, Bar graphs show the mean ± SD of three independent experiments. Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Integrin β4 regulates the protein stability of integrin α6
As functional integrin α6β4 is a dimer formed of ITGA6 and ITGB4, we investigated what consequences experimentally manipulating the levels of ITGB4 would have on the expression of ITGA6. Consistent with our finding in KRAS-mt HCT-116 cells (Fig. 2), we found that KRAS-mt Caco-2 also expressed higher levels of ITGB4 and ITGA6 compared to KRAS-wt Caco-2 (Figs. 3, A–C). After overexpression of ITGB4 (ITGB4 o/e) using CRISPR/Cas9-mediated target gene activation system, mRNA and protein levels of ITGA6 were increased in KRAS-mt Caco-2 but not in KRAS-wt Caco-2 (Figs. 3, A–C). Protein analysis further showed a slight increase in ERK phosphorylation for KRAS-mt Caco-2 cells but not in KRAS-wt Caco-2 cells after overexpression of ITGB4 (Fig. S1A). A previous study has shown that ITGB4 induces phosphorylation of ERK after ligation of laminin-332 (29). To investigate if laminin-332 is the reason for the increase of ERK phosphorylation, we examined the expression level of LAMC2 (one of Laminin-332 chains) in Caco-2 cell lysates and supernatants, and observed that KRAS-mt Caco-2 cells secrete higher levels of LAMC2 compared to KRAS-wt (Fig S1C).
KRAS-mt HCT-116 also displayed overexpression of both ITGB4 and ITGA6 compared to KRAS-wt HCT-116, as previously shown (Figs. 3, D and E). As the two cytoplasmic isoforms of ITGA6 (ITGA6A or ITGA6B) have been implicated to play opposing roles in epithelial-to-mesenchymal transition in breast cancer (30), we first explored whether KRAS-mt CRC cells preferentially upregulate one of the isoforms. We found that both ITGA6A and ITGA6B isoforms were overexpressed in KRAS-mt HCT-116, indicating that KRAS signaling does not preferentially select for a specific ITGA6 isoform (Figs. 3, D, F and G). We then knocked out (KO) ITGB4 by CRISPR in both KRAS-wt HCT-116 and KRAS-mt HCT-116 cells (Figs. 3, D–H) to study how this would alter the expression levels of ITGA6. The mRNA level of ITGA6 was found to be elevated in KRAS-mt HCT-116 despite knocking out ITGB4 (Fig. 3H). ERK phosphorylation was also not altered in KRAS-mt HCT-116 cells after knocking out ITGB4, and HCT-116 itself does not secrete LAMC2 (Fig. S1, B and C). Compared to the ITGA6 mRNA, the ITGA6 protein levels were diminished in both the KRAS-wt and KRAS-mt/ITGB4-KO HCT-116 cell lines (Figs. 3, D and E). To investigate whether this discrepancy in the mRNA and protein levels of ITGA6 in KRAS-mt/ITGB4-KO cells was related to changes in the post-translational stability of ITGA6 protein, we inhibited protein synthesis in both KRAS-mt and KRAS-mt/ITGB4-KO HCT-116 cells using cycloheximide and probed for ITGA6 protein levels over a time course of 0–24 hours. Without the presence of ITGB4, we found that ITGA6 levels diminished faster, indicating that ITGB4 plays a role in maintaining the protein stability of ITGA6 (Fig. 3I). As ITGA6 can also dimerize with integrin β1 (ITGB1) to form integrin α6β1, another distinct laminin receptor, we explored whether knocking out ITGB4 would lead to changes in the expression of integrin α6β1. In the whole cell lysate, we found increased expression of ITGB1 in both the KRAS-mt and KRAS-mt/ITGB4-KO HCT-116 cells relative to the KRAS-wt lines (Fig. 3J). Using an antibody against ITGA6, we also immunoprecipitated proteins that were bound to ITGA6 from the whole cell lysate. As expected, in both KRAS-wt and KRAS-mt cells, ITGB4 co-precipitated with ITGA6, whereas this disappeared in the cell lines with ITGB4-KO (Fig. 3J). Deficiency of ITGB4 did not lead to an abundance of integrin α6β1, indicating that our manipulation of ITGB4 caused changes specific to integrin α6β4 (Fig. 3J).
Integrin β4 does not play a significant role in CRC cell proliferation
We confirmed an overactive KRAS signaling pathway in the KRAS-mt/ITGB4-KO HCT-116 along with a reduction in the levels of ITGB4 and ITGA6 expression compared to the levels in the parental KRAS-mt HCT-116 cells (Fig. 4A). While KRAS-mt HCT-116 had a higher cell proliferation rate compared to KRAS-wt HCT-116, knocking out ITGB4 did not alter the cell proliferation rates of HCT-116 cells (Fig. 4B). We also generated ITGB4 knockout organoids from Apc−/−Trp53−/−KrasG12D (APK) organoids, but did not observe any changes in cell proliferation (Fig. 4C). Inducing overexpression of ITGB4 in Caco-2 cells did not affect cell proliferation (Fig. 4D). These data suggest that integrin α6β4 is not crucial for tumor intrinsic cell growth.
Figure 4.

Knock out of ITGB4 does not affect the proliferation rate and the tumor growth of KRAS mutant HCT-116 cells and mouse colonic organoids. A, Western blot analysis of ERK1/2 and AKT phosphorylation in ITGB4 knockout HCT-116 cells. HCT-116 cells (1 × 106 cells/ 100 mm dish) were cultured for 3 days. Data are representative of three independent experiments. B-D, Proliferation rates of HCT-116 cells (B), mouse organoids (C), and Caco-2 (D) were determined at day3 by MTT, WST-1, and MTT, respectively. Bar graphs show the mean ± SD of three independent experiments. MTT or WST-1 values at day 3 were normalized to day 0 of each cell line. Student’s t test. *, P < 0.05; ns, not significant. E and F, HCT-116 cells were injected into the both flanks of nude mice subcutaneously. Tumor growths were measured on indicated days. E, Growth of HCT-116 KRAS-mt tumors with or without ITGB4-KO. Graph shows the mean ± SD for each group (HCT-116 KRAS-mt, n=10; HCT-116 KRAS-mt/ITGB4-KO, n=10 tumors from each 5 mice). F, Representative H&E staining images of tumor tissues. Scale bars, 50 μm. G and H, Mouse organoids were injected into both flanks of C57BL/6 mice subcutaneously. Tumor growths were measured on indicated days. G, Growth of APK and APK/ITGB4-KO in C57BL/6 mice. Graph shows the mean ± SD for each group (APK, n=8 tumors from 4 mice; APK/ITGB4-KO, n=9 tumors from 5 mice). H, Representative H&E staining images of tumor tissues. Scale bars, 50 μm.
To study the role of integrin α6β4 in cell extrinsic mechanisms related to tumor proliferation in vivo, we implanted KRAS-mt HCT-116 cells (n=10 mice) and KRAS-mt/ITGB4-KO HCT-116 cells (n=10 mice) subcutaneously in the flank of athymic nude mice and monitored tumor volume (Figs. 4, E and F). We also performed a similar experiment by implanting Apc−/−Trp53−/−KrasG12D (APK) organoids (n=8 mice) and Apc−/−Trp53−/−KrasG12DItgb4−/− (APK/ITGB4-KO) organoids (n=9 mice) (Figs. 4, G and H). In both models, knocking out ITGB4 did not lead to changes in the tumor growth rate (Figs. 4, E and G).
The aggressive migratory and invasive properties of KRAS mutant CRC requires integrin β4
We also investigated the potential role of ITGB4 in modulating cell migration and invasion of KRAS mutant tumors. In transwell migration assay experiments, we found that KRAS-mt HCT-116 cells have an increased ability to migrate compared to KRAS-wt HCT-116 cells (161 vs 55 migrating cells) (Fig. 5A). However, knocking out ITGB4 in KRAS-mt HCT-116 significantly reduced the cell’s migratory phenotype to a level seen in KRAS-wt HCT 116. (Fig. 5A). Next, we performed transwell invasion assays with matrigel and observed that the number of cells invading matrigel were higher in KRAS-mt HCT-116 compared to KRAS-wt HCT-116 (56 vs 8 cells) (Fig. 5B). KRAS-mt/ITGB4-KO HCT-116, on the other hand, displayed similar invasive potential as KRAS-wt HCT-116 (Fig. 5B). To further investigate the role of ITGB4 on cell invasion, we cultured a monolayer of HCT-116 cancer cells on top of a 3D matrix composed of collagen, matrigel, and fibroblasts, and then embedded this culture system in paraffin blocks after 12 days. We then performed immunofluorescence staining on sections of this matrix to compare the invasive abilities of KRAS-wt HCT116, KRAS-mt HCT-116 as well as KRAS-mt/ITGB4-KO HCT-116 (Figs. 5, C–F). Using E-cadherin as a marker for the epithelial cancer cells, we quantified the maximum invasion depth, invasion area, and the number of invading cancer cells in the matrix. We also stained for ITGB4 and verified overexpression of ITGB4 in KRAS-mt vs KRAS-wt HCT-116 and the lack of ITGB4 expression in KRAS-mt/ITGB4-KO HCT-116 cells (Fig. 5C). KRAS-mt HCT-116 cells demonstrated a significantly greater maximum depth of invasion compared to KRAS-wt HCT-116 cells (243 vs 139 μm; P=0.0002). The maximum invasion depth observed in KRAS-mt/ITGB4-KO HCT-116 was similar to KRAS-mt HCT-116 (Fig. 5D). The tumor invasion area was 1.5 fold greater in KRAS-mt HCT-116 vs KRAS-wt HCT-116 cells (P=0.021) (Fig. 5E). KRAS-mt/ITGB4-KO HCT-116 had a significantly smaller tumor invasion area compared to KRAS-mt HCT-116 (Fig. 5E). Finally, we quantified the number of cancer cells invading the matrix. KRAS-mt cells had a higher number of invading cells versus KRAS-wt cells (Fig. 5F), although this was not statistically significant. Knocking out ITGB4 in KRAS-mt HCT-116 diminished the number of invading cells compared to KRAS-mt HCT-116 (Fig. 5F).
Figure 5.

Knock out of ITGB4 decreases migration and invasion of KRAS mutant HCT-116 cells in vitro. A-B, Transwell migration assay (A) and transwell invasion assay (B) of HCT116 cells. HCT-116 cells were seeded on the transwell membrane without (A) or with (B) matrigel coating. After 24 h, cells on the opposite surface of the transwell membrane were fixed, stained, and quantified. Bar graphs show the mean ± SD of four independent experiments. Representative images are shown below. Scale bars, 100 μm. C, 3D invasion assay of HCT-116 cells in organotypic culture system. HCT-116 cells were seeded onto 3D gels composed of collagen, matrigel, and fibroblasts. After 10–12 days, gels were fixed and paraffin-embedded, and cells invading into the gel were stained and quantified. Representative images of IF staining of E-cadherin, ITGB4, and Vimentin are shown. Scale bars, 100 μm. D-F, Quantification of maximum invasion depth (D), total invasion area (E), and total invaded cell numbers (F) of 3D invasion assay in (C). Bar graphs show the mean ± SD of five independent experiments. A, B and D-F, Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
As it has been shown that ITGB4 enriched breast cancer cells display stem-cell like markers and are more mesenchymal-like (31), we investigated whether these factors were contributing to the invasive phenotype seen in our experiments. However, we did not observe changes in the cancer-stemness markers or markers suggestive of an epithelial-mesenchymal transition (EMT) in both KRAS-wt and KRAS-mt HCT-116 cells with or without ITGB4 (Fig. S2A). Taken together, these results indicate that the overexpression of ITGB4 in KRAS-mt CRC plays a crucial role in promoting cancer migration and invasion and that this is not directly related to cancer-stemness or EMT.
Knocking out integrin β4 reduces cancer cell invasiveness and metastatic potential in vivo
To validate our findings from in vitro experiments, we used murine models to study whether knocking out ITGB4 in KRAS-mt cancer cells will impair its invasive and metastatic properties. We generated Apc−/−Trp53−/−KrasG12DItgb4−/− organoids (APK/ITGB4-KO) from Apc−/−Trp53−/−KrasG12D (APK) murine colonic organoids by knocking out ITGB4 using CRISPR. We hypothesized that the Apc−/−Trp53−/−KrasG12D would engraft more efficiently and form viable tumors whereas the Apc−/−Trp53−/−KrasG12DItgb4−/− organoids would not. We then transplanted these colonic organoids to the rectum of C57Bl/6J mice after mechanical disruption of the endoluminal mucosa (Fig. 6A). 5/15 (33%) of mice transplanted with APK organoids engrafted tumor, whereas 0/14 (0%) of mice transplanted with APK/ITGB4-KO organoids engrafted tumor (Fig. 6, B–D). As the proliferation rate of APK compared to APK/ITGB4-KO organoids were found to be no different (Fig. 4C), this result can be explained by the reduction in the invasive potential that we observed when knocking out ITGB4.
Figure 6.

Knock out of ITGB4 decreases orthotopic implantation rate of APK organoids. A-D, APK and APK/ITGB4-KO organoids were transplanted into the lumen of C57BL/6 mouse rectum. Tumor growth was assessed by endoscopy at 2 and 4 weeks after transplantation, and rectal tissues were collected after second endoscopy (APK, n=15; APK/ITGB4-KO, n=14). A, Schematic illustration of endoluminal tumor model. B, Representative endoscopic images and H&E staining of rectums with adenocarcinoma. Scale bar : 200 and 50 μm. C, Graph shows the percentage of tumor burden at 4 weeks after transplantation. Fisher’s exact test. P = 0.0421. D, Graph shows the percentage of tumor size within lumen. Mean ± SEM for each group (APK, n=5; APK/ITGB4-KO, n=14).
We investigated the metastatic potential of Itgb4−/− versus Itgb4wt KRAS-mt cells after injecting them into the tail veins of mice (Fig. 7A). We hypothesized that the Apc−/−Trp53−/−KrasG12D organoids would metastasize to lung and liver parenchyma more readily compared to the Apc−/−Trp53−/−KrasG12DItgb4−/− organoids. We used APK-LM organoids, that were generated from secondary liver metastases of APK organoids, and NSG mice as these are known to exhibit higher metastatic potential as measured by quantitative features such as the metastatic tumor area (32) and the metastatic occurrence rate (33). We also created an ITGB4 deficient line (APK-LM/ITGB4-KO). We injected 11 mice with APK-LM and 11 mice with APK-LM/ITGB4-KO. No significant differences were observed in the gross number of mice that developed lung metastasis (10/11 in APK-LM vs. 10/11 in APK-LM-ITGB4-KO) or liver metastasis (10/11 in APK-LM vs. 8/11 in APK-LM-ITGB4-KO). However, the number of pulmonary metastatic foci was significantly lower in mice injected with APK-LM/ITGB4-KO versus mice injected with APK-LM (3.6 vs 20.4 foci; P=0.002) (Figs. 7, B and C). Additionally, the area of the pulmonary metastatic lesions was also lower in mice injected with APK-LM/ITGB4-KO versus mice injected with APK-LM (0.9 vs 3.3 mm2) (Figs. 7, B and C). The number of hepatic metastases and the associated area, on the other hand, were no different between APK-LM and APK-LM/ITGB4-KO injected mice (Figs. 7, D and E).
Figure 7.

Knock out of ITGB4 decreases number of pulmonary metastatic foci of APK organoids. A-E, APK-LM and APK-LM/ITGB4-KO organoids derived from liver metastasis were transplanted into NSG mice via tail vein. Lung and liver tissues were collected at 5 weeks after transplantation. A, Schematic illustration of lung metastatic tumor model. B, Representative images of H&E staining and IF staining of lung tissues. GFP is a marker of transplanted organoids. Scale bar : 2000, 200, and 50 μm. C, Quantification of metastatic foci and metastatic area in (B) are shown. D, Representative images of H&E staining of liver. Scale bar : 2000, 500, and 50 μm. (E) Quantification of metastatic foci and metastatic area in (D) are shown. C and E, Each dot in the individual graphs represents one mouse. Bar graphs show the mean ± SD for each group (APK-LM, n=11; APK-LM/ITGB4-KO, n=11). Student’s t test. **, P < 0.01; ns, not significant.
As ITGB4 has been shown to induce the phosphorylation of focal adhesion kinase (FAK) to promote anchorage independent growth in vitro and pulmonary metastasis in vivo of hepatocellular carcinoma cells (34), we explored whether this was also part of the underlying mechanism in our CRC model. In Figure S1C, we measured the expression level of LAMC2, one of laminin-332 chains, in HCT-116 cell lysates and supernatants. Since these do not express or secrete LAMC2, they need exogenous laminin-332 to activate integrin α6β4. To examine the level of p-FAK, KRAS-wt and -mt HCT-116 cells with or without ITGB4-KO were cultured in the regular culture plates or Laminin-332 coated culture plates (Fig. S2B). The level of p-FAK was very weak in KRAS-wt HCT-116 cells grown without laminin-332, and strongly increased when cultured with laminin-332 (Fig. S2B). However, the expression of p-FAK was not altered by KRAS mutation or ITGB4-KO in HCT-116 cells in both culture conditions (Fig. S2B), suggesting that p-FAK is not associated with the effects of ITGB4.
Discussion:
Colorectal adenocarcinomas with a KRAS mutation are more invasive and likely to metastasize to the lung and the brain (35–37) but the molecular mechanisms behind this observation are not completely understood. Using the transcriptomic data of CRC patients from TCGA as well as immunofluorescence staining of patient tissues, we have shown that KRAS-mt CRC is associated with upregulation of ITGB4. We have also demonstrated, using multiple in vitro and in vivo models, that KRAS-mt CRC upregulates integrin α6β4 through ERK/MEK signaling and that this contributes to its invasive and metastatic potential.
Targeted deletion of Itga6 in intestinal epithelial cells (IECs) from mice has been shown to result in both ITGA6 and ITGB4 proteins being removed from IECs (38), implying stabilizing function of ITGA6 for ITGB4 protein. Our data also shows that ITGB4 regulates the stability of ITGA6 in KRAS-mt HCT-116 cells (Fig. 3, D–I). Without ITGB4, the mRNA level of ITGA6 was not changed (Fig. 3, H) but protein level was decreased in KRAS-mt HCT-116 cells (Fig. 3, D–E). However, the stabilizing effect of ITGB4 on ITGA6 was not observed in KRAS-mt Caco-2 cells. After overexpression of ITGB4, mRNA and protein levels of ITGA6 were increased in KRAS-mt Caco-2 cells (Fig. 3, A–C). We speculate that the mechanism for this discrepancy between Caco-2 and HCT-116 cells is through laminin-332 mediation of the ERK signaling pathway. In Figure S1C, we have examined the expression level of LAMC2 (one of Laminin-332 chains) in HCT-116 cell lysates and supernatants, and observed that they do not express or secrete LAMC2. In addition, p-ERK level of KRAS-mt HCT-116 was not increased after ITGB4-KO. It is therefore unlikely that self-generated laminin-332 affects the signaling pathways in HCT-116 cells by ligation to integrin α6β4. In Caco-2 cells, however, tiny amounts of LAMC2 were expressed and the secretion of LAMC2 was slightly increased by KRAS-mt (Fig. S1C). Moreover, p-ERK signaling is slightly increased in KRAS-mt/ITGB4 overexpressing Caco-2 cells (Fig. S1A). Thus, it is possible that overexpressed ITGB4 stabilizes ITGA6 protein and the increased ITGA6/ITGB4 dimeric receptor binds with secreted laminin-332 and increases p-ERK pathway, resulting in higher ITGA6 expression at the transcriptional level (Fig. 2). We confirmed that knocking out ITGB4 results in the depletion of integrin α6β4 without altering the levels of integrin α6β1. We did not observe changes in EMT or cancer stemness markers when knocking out ITGB4. The tumor proliferation rate was also unchanged. Depleting ITGB4 in KRAS-mt CRC cells reduced the locally invasive properties and the extent of lung metastases in vivo.
Integrin α6β4 has been shown to promote local invasion in breast cancer by activating PI3K (11). Furthermore, ITGB4 has been shown to amplify ErbB2 signaling to promote tumorigenesis (39). While integrin α6β4 can activate these pathways, in the context of an oncogenic KRAS mutation, it seems that KRAS is what regulates the expression of integrin α6β4 through the ERK/MEK pathway in CRC. Our results showing integrin α6β4-mediated increase of invasion and pulmonary metastatic foci in KRAS-mt CRC are consistent with previous reports in other cancer types (29,40). In our experiment with the orthotopic murine model, knocking out ITGB4 in KRAS-mt CRC inhibited endoluminal tumor engraftment, which we interpreted as a result of disabling the locally invasive properties. In another in vivo experiment, we knocked out ITGB4 and observed a reduction in the extent of pulmonary metastatic foci, but did not observe the same phenomenon relative to liver metastases. While a detailed characterization of the mechanisms underlying this observation is beyond the scope of our study, work by other groups using breast and pancreatic cancer cells has shown that pre-treating mice with tumor-derived microvesicles enriched in integrin α6β4 before inoculating mice with cancer cells led to preferential metastasis to the lung (41). Akin to our results, they reported that knocking out ITGB4 in these cells significantly reduced the lung metastasis rate. As KRAS-mt CRC cells have been reported to shed microvesicles enriched in ITGB4 (42), the upregulation of integrin α6β4 in these microvesicles could lead to the higher incidence of lung metastasis compared to KRAS-wt tumors. Additional experiments need to be performed to test this hypothesis.
In conclusion, KRAS-mt CRC upregulates integrin α6β4, which promotes cell invasion and migration. Since depleting integrin α6β4 blunts the invasive properties and the extent of pulmonary metastatic foci, integrin α6β4 may be a potential target for therapy in KRAS-mt CRC.
Supplementary Material
Implications:
Knocking-out Integrin β4 (ITGB4), which is overexpressed in KRAS mutant colorectal cancer and promotes tumor aggressiveness, diminishes local invasiveness and rates of pulmonary metastasis.
Acknowledgments:
This study was supported by the National Institutes of Health, National Cancer Institute R01 Grant CA090559. F.M.B. was supported by a GMTEC Postdoctoral Fellowship, an Memorial Sloan Kettering’s Translational Research Oncology Training Fellowship (5T32CA160001-08), and a Young Investigator Award by the Edward P. Evans Foundation. We acknowledge the use of services provided by the Molecular Cytology Core Facility, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA008748-53), and the Antitumor Assessment Core Facility, funded by the Institutional Core Grant (CCSG P30 CA008748-53). We thank Carlos Carmona-Fontaine (New York University) for providing us with KRAS-wt and KRAS-mt HCT-116 cell lines and Alan Hall for providing us with KRAS-wt and KRAS-mt Caco-2 cell lines. We also thank Arthur Gelmis for editorial assistance. The authors have no conflicting financial interests.
Footnotes
Disclosure of Potential Conflicts of Interest:
JGA has received honorarium for being a consultant with the following: Medtronics, Ethicon J&J, Da Vinci Intuitive Surgical. JJS has received travel support from Intuitive Surgical Inc. for fellow education (2015) and has served as a clinical advisor for Guardant Health, Inc (2019). The other co-authors have no conflicts of interest to disclose.
References:
- 1.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. [DOI] [PubMed] [Google Scholar]
- 2.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–32. [DOI] [PubMed] [Google Scholar]
- 3.Lipscomb EA, Mercurio AM. Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev. 2005;24:413–23. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serebriiskii IG, Connelly C, Frampton G, Newberg J, Cooke M, Miller V, et al. Comprehensive characterization of RAS mutations in colon and rectal cancers in old and young patients. Nat Commun. 2019;10:3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125–136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. [DOI] [PubMed] [Google Scholar]
- 8.Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, et al. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13:12153–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Myllymäki SM, Gao P, Devarajan R, Kytölä V, Nykter M, et al. Oncogenic K-Ras upregulates ITGA6 expression via FOSL1 to induce anoikis resistance and synergizes with αV-Class integrins to promote EMT. Oncogene. 2017;36:5681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw LM, Rabinovitz I, Wang HH, Toker A, Mercurio AM. Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell. 1997;91:949–60. [DOI] [PubMed] [Google Scholar]
- 12.Trusolino L, Bertotti A, Comoglio PM. A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107:643–54. [DOI] [PubMed] [Google Scholar]
- 13.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–26. [DOI] [PubMed] [Google Scholar]
- 14.Mercurio AM, Rabinovitz I. Towards a mechanistic understanding of tumor invasion--lessons from the alpha6beta 4 integrin. Semin Cancer Biol. 2001;11:129–41. [DOI] [PubMed] [Google Scholar]
- 15.Falcioni R, Turchi V, Vitullo P, Navarra G, Ficari F, Cavaliere F, et al. Integrin Beta-4 expression in colorectal-cancer. Int J Oncol. 1994;5:573–8. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Jiang X, Wang G, Zhai C, Liu Y, Li H, et al. ITGB4 is a novel prognostic factor in colon cancer. J Cancer. 2019;10:5223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magudia K, Lahoz A, Hall A. K-Ras and B-Raf oncogenes inhibit colon epithelial polarity establishment through up-regulation of c-myc. J Cell Biol. 2012;198:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161:1539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogervorst F, Kuikman I, van Kessel AG, Sonnenberg A. Molecular cloning of the human alpha 6 integrin subunit. Alternative splicing of alpha 6 mRNA and chromosomal localization of the alpha 6 and beta 4 genes. Eur J Biochem. 1991;199:425–33. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenberg A, Hogervorst F, Osterop A, Veltman FE. Identification and characterization of a novel antigen complex on mouse mammary tumor cells using a monoclonal antibody against platelet glycoprotein Ic. J Biol Chem. 1988;263:14030–8. [PubMed] [Google Scholar]
- 22.Young AIJ, Law AMK, Castillo L, Chong S, Cullen HD, Koehler M, et al. MCL-1 inhibition provides a new way to suppress breast cancer metastasis and increase sensitivity to dasatinib. Breast Cancer Res. 2016;18:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timpson P, McGhee EJ, Erami Z, Nobis M, Quinn JA, Edward M, et al. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. J Vis Exp. 2011;e3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King H, Thillai K, Whale A, Arumugam P, Eldaly H, Kocher HM, et al. PAK4 interacts with p85 alpha: implications for pancreatic cancer cell migration. Sci Rep. 2017;7:42575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dow LE, Nasr Z, Saborowski M, Ebbesen SH, Manchado E, Tasdemir N, et al. Conditional reverse tet-transactivator mouse strains for the efficient induction of TRE-regulated transgenes in mice. PLoS ONE. 2014;9:e95236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duldulao MP, Lee W, Nelson RA, Li W, Chen Z, Kim J, et al. Mutations in specific codons of the KRAS oncogene are associated with variable resistance to neoadjuvant chemoradiation therapy in patients with rectal adenocarcinoma. Ann Surg Oncol. 2013;20:2166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow OS, Kuk D, Keskin M, Smith JJ, Camacho N, Pelossof R, et al. KRAS and Combined KRAS/TP53 Mutations in Locally Advanced Rectal Cancer are Independently Associated with Decreased Response to Neoadjuvant Therapy. Ann Surg Oncol. 2016;23:2548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datta J, Smith JJ, Chatila WK, McAuliffe JC, Kandoth C, Vakiani E, et al. Coaltered Ras/B-raf and TP53 Is Associated with Extremes of Survivorship and Distinct Patterns of Metastasis in Patients with Metastatic Colorectal Cancer. Clin Cancer Res. 2020;26:1077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Giancotti FG. Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell. 2004;6:471–83. [DOI] [PubMed] [Google Scholar]
- 30.Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, et al. Regulated splicing of the α6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep. 2014;7:747–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bierie B, Pierce SE, Kroeger C, Stover DG, Pattabiraman DR, Thiru P, et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci USA. 2017;114:E2337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roe J-S, Hwang C-I, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170:875–888.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai E, Nakayama M, Oshima H, Kouyama Y, Niida A, Fujii S, et al. Combined mutation of apc, kras, and tgfbr2 effectively drives metastasis of intestinal cancer. Cancer Res. 2018;78:1334–46. [DOI] [PubMed] [Google Scholar]
- 34.Leng C, Zhang Z-G, Chen W-X, Luo H-P, Song J, Dong W, et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett. 2016;376:188–96. [DOI] [PubMed] [Google Scholar]
- 35.Yaeger R, Cowell E, Chou JF, Gewirtz AN, Borsu L, Vakiani E, et al. RAS mutations affect pattern of metastatic spread and increase propensity for brain metastasis in colorectal cancer. Cancer. 2015;121:1195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira AAL, Rego JFM, Morris V, Overman MJ, Eng C, Garrett CR, et al. Association between KRAS mutation and lung metastasis in advanced colorectal cancer. Br J Cancer. 2015;112:424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Unable to find information for 12395477.
- 38.De Arcangelis A, Hamade H, Alpy F, Normand S, Bruyère E, Lefebvre O, et al. Hemidesmosome integrity protects the colon against colitis and colorectal cancer. Gut. 2017;66:1748–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J, Cai B, Zeng M, Hao Y, Giancotti FG, Fu BM. Integrin β4 signaling promotes mammary tumor cell adhesion to brain microvascular endothelium by inducing ErbB2-mediated secretion of VEGF. Ann Biomed Eng. 2011;39:2223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pouliot N, Nice EC, Burgess AW. Laminin-10 mediates basal and EGF-stimulated motility of human colon carcinoma cells via alpha(3)beta(1) and alpha(6)beta(4) integrins. Exp Cell Res. 2001;266:1–10. [DOI] [PubMed] [Google Scholar]
- 41.Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demory Beckler M, Higginbotham JN, Franklin JL, Ham A-J, Halvey PJ, Imasuen IE, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study were obtained from the Memorial Sloan Kettering Cancer Center cBioPortal : http://www.cbioportal.org